Salmonella enterica serovars are associated with diarrhea and gastroenteritis and are a helpful model for understanding host-pathogen mechanisms. Salmonella enterica serovar Typhimurium regulates the distribution of O antigen (OAg) and presents a trimodal distribution based on Wzy polymerase and the WzzST (long-chain-length OAg [L-OAg]) and WzzfepE (very-long-chain-length OAg [VL-OAg]) copolymerases; however, several mechanisms regulating this process remain unclear.
KEYWORDS: Salmonella, LPS, O antigen, VisP, lipopolysaccharide
ABSTRACT
Salmonella enterica serovars are associated with diarrhea and gastroenteritis and are a helpful model for understanding host-pathogen mechanisms. Salmonella enterica serovar Typhimurium regulates the distribution of O antigen (OAg) and presents a trimodal distribution based on Wzy polymerase and the WzzST (long-chain-length OAg [L-OAg]) and WzzfepE (very-long-chain-length OAg [VL-OAg]) copolymerases; however, several mechanisms regulating this process remain unclear. Here, we report that LPS modifications modulate the infectious process and that OAg chain length determination plays an essential role during infection. An increase in VL-OAg is dependent on Wzy polymerase, which is promoted by a growth condition resembling the environment of Salmonella-containing vacuoles (SCVs). The virulence- and stress-related periplasmic protein (VisP) participates in OAg synthesis, as a ΔvisP mutant presents a semirough OAg phenotype. The ΔvisP mutant has greatly decreased motility and J774 macrophage survival in a colitis model of infection. Interestingly, the phenotype is restored after mutation of the wzzST or wzzfepE gene in a ΔvisP background. Loss of both the visP and wzzST genes promotes an imbalance in flagellin secretion. L-OAg may function as a shield against host immune systems in the beginning of an infectious process, and VL-OAg protects bacteria during SCV maturation and facilitates intramacrophage replication. Taken together, these data highlight the roles of OAg length in generating phenotypes during S. Typhimurium pathogenesis and show the periplasmic protein VisP as a novel protein in the OAg biosynthesis pathway.
INTRODUCTION
Bacterium-host interaction is a natural synergistic relationship. However, this interaction may diverge, leading to distinct consequences that range from essential and beneficial cooperation to deadly outcomes. Salmonella is a human pathogen that is responsible for intestinal infections and typhoid fever. Salmonellosis is one of the most common and broadly found foodborne illnesses in the world. Yearly estimates include approximately 10 million human cases worldwide, resulting in more than 100,000 deaths (1). Salmonella enterica serovar Typhimurium is a major foodborne pathogen responsible for gastroenteritis and complications such as serious invasive nontyphoidal Salmonella (iNTS) disease and is frequently reported in sub-Saharan regions (2, 3). S. Typhimurium is acquired through contaminated food ingestion and has mechanisms to survive the low-pH milieu and bile salts of the stomach, allowing it to reach the intestine (4). Once in the intestinal lumen, S. Typhimurium employs its flagella, which constitute a complex multiprotomeric structure, to move toward a nutrient-rich environment, such as the epithelial mucosa, and thus establish a colonization process (5, 6). The goblet cells in the normal intestine accumulate multiple mucous storage vesicles. S. Typhimurium can benefit from this propitious environment by inducing inflammation to increase the secretion of mucin and glycoconjugate, high-energy nutrient sources, by the goblet cells (5, 7). The release of glycoconjugates from goblet cells creates an attractant gradient for S. Typhimurium, causing S. Typhimurium to move in its direction and hence initially attach to epithelial cells (5).
The inflammatory process can be initiated by different S. Typhimurium membrane structures, such as flagellins, which are flagellum subunits (8–10), type 3 secretion system (TTSS) structural proteins (11–16), lipopolysaccharide (LPS) (17, 18), and other effector proteins encoded in Salmonella pathogenicity islands (SPI) (19–22). Fimbriae and other adhesins help S. Typhimurium initiate contact with epithelial cells by binding to membrane mannosidic glycoproteins, which anchor the bacterium (23, 24).
Bacterium-host cell interactions enable the SPI-1 TTSS to inject effector proteins into the epithelial cell, which modify the cell's structure by ruffling its membrane, culminating in bacterial engulfment (25). S. Typhimurium can live and replicate within the intracellular environment by manipulating cellular vesicle trafficking to obtain nutrients (4). Intracellular settling and colonization are classically mediated by SPI-2 TTSS protein effectors (4, 26, 27) and by SPI-3 proteins associated with metabolic adaptations, such as ion transport (28). Cytokines released by epithelial cells into the bloodstream in response to an inflammatory process are responsible for recruiting polymorphonuclear cells (PMN) such as neutrophils to eliminate pathogens (21, 29). Therefore, the epithelial cell invasion route helps S. Typhimurium evade an immune response. However, S. Typhimurium can overcome macrophage phagocytosis by employing the SPI-2 TTSS machinery, which enables bacterial survival and intracellular replication (4).
The bacterial membrane plays an essential role in all of these pathogenic processes. Its structure provides protection against host defense mechanisms (30, 31), physical stability for multiprotomeric complexes such as flagella (32, 33), fimbriae (34), and the TTSS (35), and bacterial homeostasis by controlling ion transport (36–38) and sensing environment stimuli (39–41). Individual bacterial membranes activate cascade responses to better adapt themselves to different situations (42–51). LPS is the major constituent of the outer leaflet of the outer membrane (OM) of S. Typhimurium. LPS is composed of three chemically distinct parts: the membrane-embedded lipid A molecule, which is the hydrophobic and immunogenic portion of LPS that anchors LPS to the OM; the oligosaccharide core; and the highly immunogenic O antigen (OAg). The OAg molecule is a polysaccharide chain composed of four-sugar repeat units (RU) that externally protrude from the bacterial membrane (52). Gram-negative bacteria synthesize the OAg RU linked to undecaprenyl-phosphate (UndP), an essential bacterial lipid that “carries” various hydrophilic precursors across the membrane, on the cytoplasmic side of the inner membrane (IM). The flippase Wzx then transports UndP-linked OAg RU to the periplasmic face of the IM. The OAg RU are then used as building blocks for chain elongation, a process mediated by the Wzy polysaccharide polymerase (53–55). Initially, Wzy binds to the UndP-linked RU and then adds another RU or a growing chain of an OAg to the educing end of the OAg RU in a catalytic distributive mechanism (53, 54, 56). In S. Typhimurium, the Wzz polysaccharide copolymerases (PCPs) then serve as a molecular ruler that determines the final length of the OAg chains. S. Typhimurium OAg chains follow a trimodal distribution (35) of short (S-OAg) (less than 16 RU), long (L-OAg) (16 to 35 RU), and very long (VL-OAg) (more than 100 RU) forms. The WzzST and WzzfepE proteins regulate L-OAg (57–59) and VL-OAg (60) synthesis, respectively.
Recently, our group reported that a novel membrane protein, virulence- and stress-related periplasmic protein (VisP), enables S. Typhimurium survival upon encountering stress factors during pathogenesis (61). VisP has an important role in intramacrophage survival and interacts with the lipid A modification enzyme LpxO (61), an Fe3+/α-ketoglutarate-dependent dioxygenase (62, 63) related to macrophage evasion (17, 52). VisP was initially identified as a member of the bacterial oligonucleotide/oligosaccharide-binding fold (BOF) family (61) and was predicted to bind oligosaccharides, such as N-acetylglucosamine and N-acetylmuramic acid (64).
In this study, we investigated the role of VisP in the Wzy-dependent pathway of OAg biosynthesis and determined the importance and consequences of the longer forms of OAg chains for S. Typhimurium pathogenesis. Furthermore, we also examined how different OAg forms directly affect protection against host defenses during in vivo infections and other pathogenic mechanisms, such as flagellum-mediated motility.
RESULTS
OAg changes are driven by intracellular conditions.
S. Typhimurium is a facultative intracellular pathogen. LPS is one of a number of known pathogen-associated molecular patterns (PAMPs), and bacteria are recognized by the innate immune system via interactions with these PAMPs. We performed OAg extraction from cells cultured under different growth conditions to observe different luminal or intracellular conditions that trigger OAg chain length changes in S. Typhimurium. LB medium was employed as a rich nutrient condition to mimic the intestinal lumen (44, 65). We also harvested bacteria from N-minimal medium under nutrient-depleted conditions, such as low Mg2+ (10 μM) and phosphate (1 mM KH2PO4) concentrations, at a mildly acidic pH of 5.0; these conditions are similar to those of the Salmonella-containing vacuole (SCV) environment (39, 46, 66–71). The OAg profile of the wild-type (WT) strain presented significant increases in VL- and S-OAg after growth in N-minimal medium compared to the levels after growth in LB (Fig. 1A, lanes 2 and 3; see Fig. S1A and B in the supplemental material). Under both growth conditions, the three distinct modal forms (VL-, L-, and S-OAg) were well distributed; however, the VL and S bands were the most intense and were easily distinguished under N-minimal growth conditions (Fig. 1A, lanes 2 and 3, and S1A and B). Apparently, low-nutrient conditions, such as the SCV environment, are favorable to more prominent bands of VL-OAg than the lumen conditions of S. Typhimurium. Similar VL-OAg chain augmentation was previously described for S. Typhimurium C5 growing in inactivated guinea pig serum and under iron-limited conditions (31). We measured the expression levels of OAg chain biosynthesis pathway genes to better understand the observed changes in OAg final length. The expression of the wzy gene, which encodes the OAg polysaccharide polymerase (53–55, 57, 72), was assessed under both growth conditions for the WT strain, the ΔvisP, ΔwzzfepE, and ΔwzzST single mutants, and the ΔvisP wzzfepE and ΔvisP wzzST double mutants (Fig. 1B). The OAg profile of the WT strain (Fig. 1A, lanes 2 and 3, and S1A and B) matched wzy gene expression levels because wzy was upregulated under the N-minimal growth condition (Fig. 1B). Therefore, S. Typhimurium may remodel its OM landscape within SCVs in response to its environment by increasing the VL- and S-OAg forms, according to the observations under growth conditions resembling SCVs (N-minimal medium). Furthermore, this remodeling may be mediated by Wzy polymerase overexpression.
FIG 1.
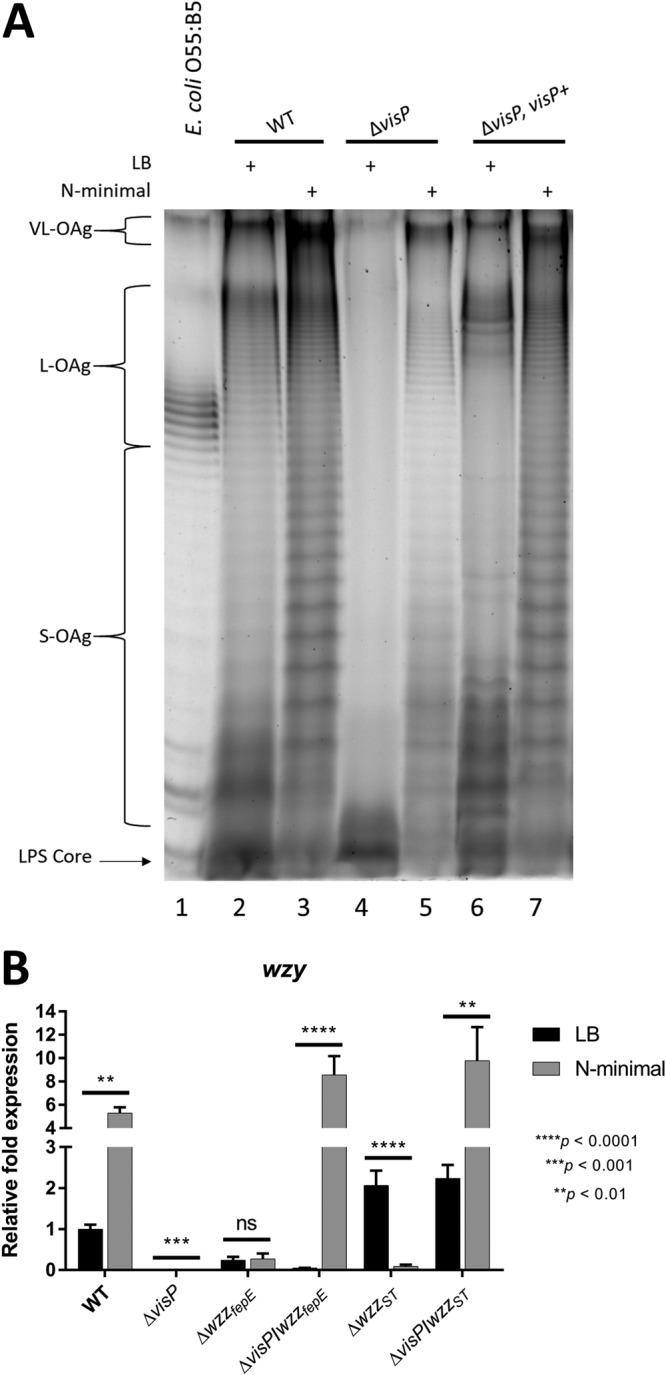
OAg changes are driven by intracellular conditions. (A) Electrophoresis profile of LPS in LB and N-minimal media. E. coli strain O55:B5 (lane 1) and the S. Typhimurium WT (lanes 2 and 3), ΔvisP (lanes 4 and 5), and complemented visP+ (lanes 6 and 7) strains were used. (B) qRT-PCR analysis of wzy polymerase in LB and N-minimal media. Relative fold expression in the WT, ΔvisP, ΔwzzfepE, ΔwzzST, ΔvisP wzzfepE, and ΔvisP wzzST strains is shown. **, P < 0.01; ****, P < 0.0001.
Periplasmic VisP changes the OAg final structure.
VisP is a periplasmic protein that is important for virulence and stress responses (61) and was initially described as a BOF family member (61, 64). OAg chain biosynthesis by the Wzy-dependent pathway occurs mainly in the periplasmic environment (52); based on this location, we assessed whether VisP plays a role in OAg chain formation. The ΔvisP mutant strain exhibited a noticeable decrease in OAg chains in both growth media (Fig. 1A, lanes 4 and 5, and S1C and D). In LB medium, this mutant appeared to have a single RU in LPS (Fig. 1A, lane 4, and S1C), a rough-like phenotype. The complemented visP+ strain had a restored OAg profile that was similar to the WT profile and presented an increase in both the VL and S forms in N-minimal medium; however, these levels were lower than those in the WT strain (Fig. 1A, lanes 6 and 7, and S1E and F). Strains with double mutations of the visP and PCP genes (wzzST and wzzfepE) were designed to address the role of VisP as a protein important for the assembly of OAg at the final length observed here (Fig. 1A) and to investigate the extremely low wzy expression levels in the ΔvisP single mutant (Fig. 1B). Again, wzy expression levels were highly upregulated in N-minimal growth medium compared with those in LB medium for both double mutants (ΔvisP wzzfepE and ΔvisP wzzST) and the WT strain (Fig. 1B). However, in LB medium, wzy expression levels diverged between the double mutants; compared to that in the WT, wzy expression was downregulated in the ΔvisP ΔwzzfepE double mutant (similar to the case for the ΔvisP single mutant) and upregulated in the ΔvisP ΔwzzST double mutant (Fig. 1B). Hence, the extreme downregulation of wzy in the absence of VisP was reversed by deleting one of the PCP genes, with a major response to wzzST cdeletion under nutrient-rich growth conditions. Next, we further evaluated macrophage-bacterium interactions under different conditions to explore the aspects of OAg assembly during intracellular environment survival.
Changes in intracellular stress conditions and role of PCP in pathogenesis.
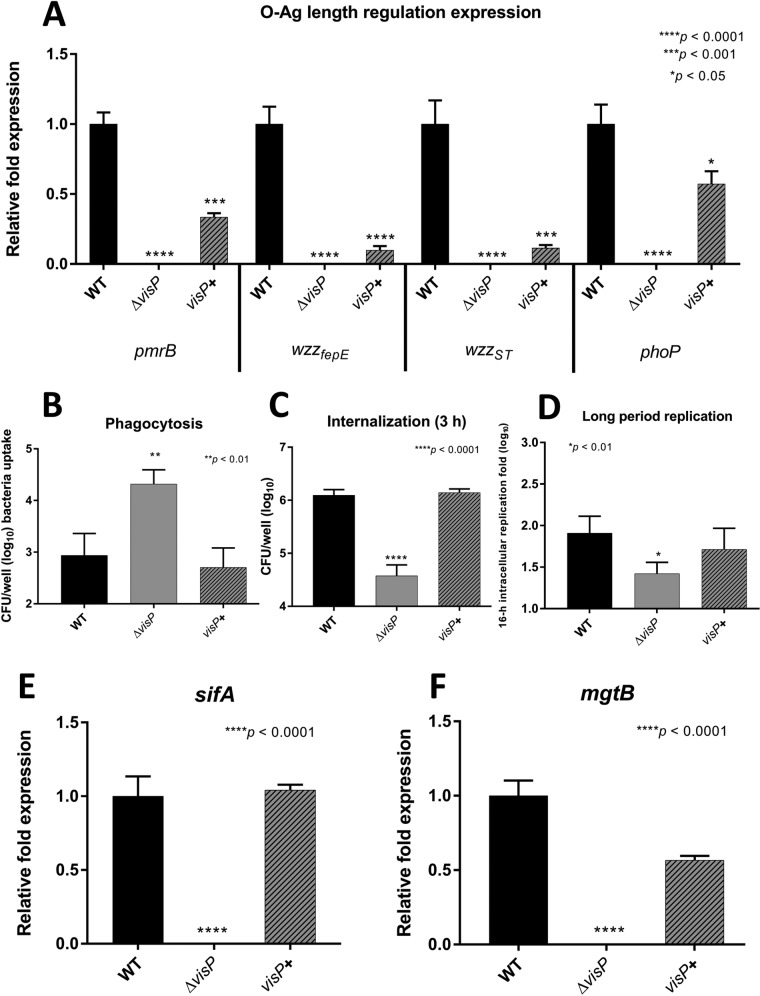
The pmrB gene, also known as basS, encodes a two-component system (TCS) sensor kinase that transcriptionally regulates chemical modifications of LPS upon sensing different environmental levels of Fe3+, Al3+, and acidic pH (73–75). Thus, pmrB plays a role in the control of the expression of the PCP genes wzzST and wzzfepE (76). In S. Typhimurium, the transcriptional regulation of the wzy gene has not been elucidated. The phoP gene encodes a global TCS sensor kinase that regulates bacterial LPS remodeling through interplay with PmrAB (77). This TCS also helps regulate metal uptake (40, 78, 79), SPI-2 virulence effector expression (46), and resistance to cationic antimicrobial peptides (80, 81). All of the genes cited above exhibited undetectable expression levels in the ΔvisP mutant background, in contrast to WT (Fig. 2A). Complementation with visP partially restored the expression of pmrB, the two PCP genes, and phoP (Fig. 2A). Therefore, VisP has a transcriptional effect on the genes of the Wzy-dependent OAg chain biosynthesis pathway, which is reflected in the OAg profile (Fig. 1A, lane 4). As the absence of VisP results in modifications of LPS, the importance of VisP was then evaluated during intracellular survival within J774 macrophages. We performed distinct macrophage cell survival assays (67, 82, 83) to analyze the different stages of bacterium-macrophage interactions during the infection process. First, we performed a phagocytosis assay to quantify the bacterial load directly engulfed by J774 macrophages without extra time after bacterium-macrophage interactions and associated the load with the OAg pattern observed in the ΔvisP mutant (Fig. 1A, lanes 4 and 5). The absence of longer OAg forms in the ΔvisP mutant resulted in an increase in macrophage bacterial uptake of more than one order of magnitude compared to WT levels (Fig. 2B). A similar result was observed for the ΔwaaL mutant lacking OAg, which had been previously reported (35) as evidence of the role of OAg chains in macrophage evasion. Complementation with the visP gene restored the phenotype to WT levels (Fig. 2B). We then evaluated macrophage uptake and survival during a 3-h intracellular replication assay. In contrast to the results of the previous assay, the ΔvisP single mutant showed a statistically significant decrease in J774 macrophage internalization of 1.5 orders of magnitude compared to that in the WT (Fig. 2C), as previously reported (61). visP gene complementation restored the WT phenotype (Fig. 2C). An extended 16-h intramacrophage replication assay was subsequently performed (Fig. 2D) to evaluate OM landscape remodeling in the SCV environment, which is similar to N-minimal growth conditions (Fig. 1A). The semirough OAg ΔvisP mutant strain exhibited a decrease of half an order of magnitude compared to the WT strain (Fig. 2D), a much smaller difference than that in the previous assay (Fig. 2C). ΔvisP mutant complementation restored the WT intramacrophage replication phenotype (Fig. 2D). After observing these different phenotypes, gene expression levels from specific targets associated with intramacrophage survival and replication were evaluated to investigate whether the observed differences were direct effects of OAg and other SCV environmental features. Initially, we assessed the expression levels of sifA, an important SPI-2 TTSS effector related to SCV maintenance and the formation of cellular Salmonella-inducing filaments (84), under N-minimal bacterial growth conditions. The ΔvisP mutant presented extremely low levels of sifA expression compared with the WT levels (Fig. 2E). Expression levels similar to WT were restored by visP complementation (Fig. 2E). The SPI-3 gene mgtB encodes an Mg2+ transporter ATPase related to intracellular adaptation and survival (85, 86). We observed very low mgtB expression levels in the ΔvisP mutant under N-minimal growth conditions compared to the WT levels (Fig. 2F). The complemented visP+ strain showed partially restored mgtB expression to WT levels (Fig. 2F). The transcriptional downregulation of the genes sifA and mgtB was in accordance with the reduced 3-h intracellular replication phenotype of the ΔvisP mutant, indicating a clear decrease in S. Typhimurium virulence. Furthermore, the absence of the OAg chain reduced the protection for this strain against macrophage-mediated phagocytosis. However, the ΔvisP mutant was able to remodel its OM under conditions resembling the SCV environment, producing longer OAg forms, but this production did not reach WT levels (Fig. 1A, lane 5, and S1D). The phenotypic difference between the ΔvisP and WT strains decreased after 16 h of intracellular replication when they were compared in a 3-h assay, even though the ΔvisP mutant downregulated intramacrophage survival-related genes. This late adaptation of the ΔvisP strain may be related to longer-chain OAg recovery under growth conditions resembling the SCV environment, which may confer greater protection against macrophage-mediated phagocytosis.
FIG 2.
Roles of OAg and VisP in S. Typhimurium pathogenesis. (A) qRT-PCR analysis of genes involved in the OAg biosynthesis pathway. relative fold expression of the pmrB, wzzfepE, wzzST, and phoP genes in the WT, ΔvisP, and complemented visP+ strains grown in LB is shown. (B to D) J774 phagocytosis (B), internalization (C), and long-period replication (D) of the WT, ΔvisP, and complemented visP+ strains. (E and F) qRT-PCR analysis of the sifA (N-minimal growth condition) (E) and mgtB (LB growth condition) (F) genes. Relative fold expression in the WT, ΔvisP, and complemented visP+ strains is shown. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Role of VisP-dependent PCP in the final OAg structure.
The PCPs, together with Wzy polymerase, are responsible for finalizing OAg chain assembly. Initially, we addressed the two PCP single mutants, ΔwzzST and ΔwzzfepE, during wzy gene expression analysis to better understand the role of VisP and the PCPs in this process under nutrient-rich growth conditions (LB medium). Compared to the results in the WT strain, the change in wzy expression in the ΔvisP wzzST double mutant was similar to those in the respective PCP single mutants, in contrast to the ΔvisP wzzfepE double mutant, which exhibited lower wzy expression than the ΔwzzfepE single mutant (Fig. 1B). VisP and the PCPs participate in wzy transcriptional regulation; however, in the case of WzzST, the effect of PCP appeared to supersede that of VisP under LB growth conditions. Environmental conditions influenced the effects of VisP and PCP on wzy transcription, since under N-minimal growth conditions, wzy overexpression occurred only in the absence of both VisP and any OAg PCP, resembling WT levels (Fig. 1B). Moreover, these three proteins appear to generate a balance in which VisP, WzzST, and WzzfepE are important during wzy expression. PCP gene expression levels in both double mutants compared to the WT under different growth conditions were evaluated to explore the transcriptional regulation of the remaining PCP. The ΔvisP wzzfepE mutant exhibited a decrease in wzzST levels in LB medium, whereas in N-minimal medium, its expression increased compared to that in the WT strain (Fig. 3A). In contrast, the ΔvisP ΔwzzST mutant displayed wzzfepE expression levels similar to those observed in the WT strain in LB medium, whereas the wzzfepE expression level greatly decreased in N-minimal medium (Fig. 3A). Differentiated intestinal conditions also play an essential role in the absence of VisP, providing partial complementation of the VL and S forms in the ΔvisP ΔwzzST mutant under N-minimal growth conditions (Fig. 3B). Transcriptionally, N-minimal medium appeared to favor the WzzST PCP, whereas wzzfepE expression was decreased in this medium. LB medium favored WzzfepE, whereas wzzST was more highly expressed under N-minimal growth conditions, but this expression always occurred in the absence of VisP. However, the VL and S forms were clearly phenotypically distinguishable under N-minimal conditions, which may be correlated with wzy overexpression (Fig. 1B). Together, these results indicate complementary functions of the two PCPs based on distinct surroundings and nutrient conditions.
FIG 3.
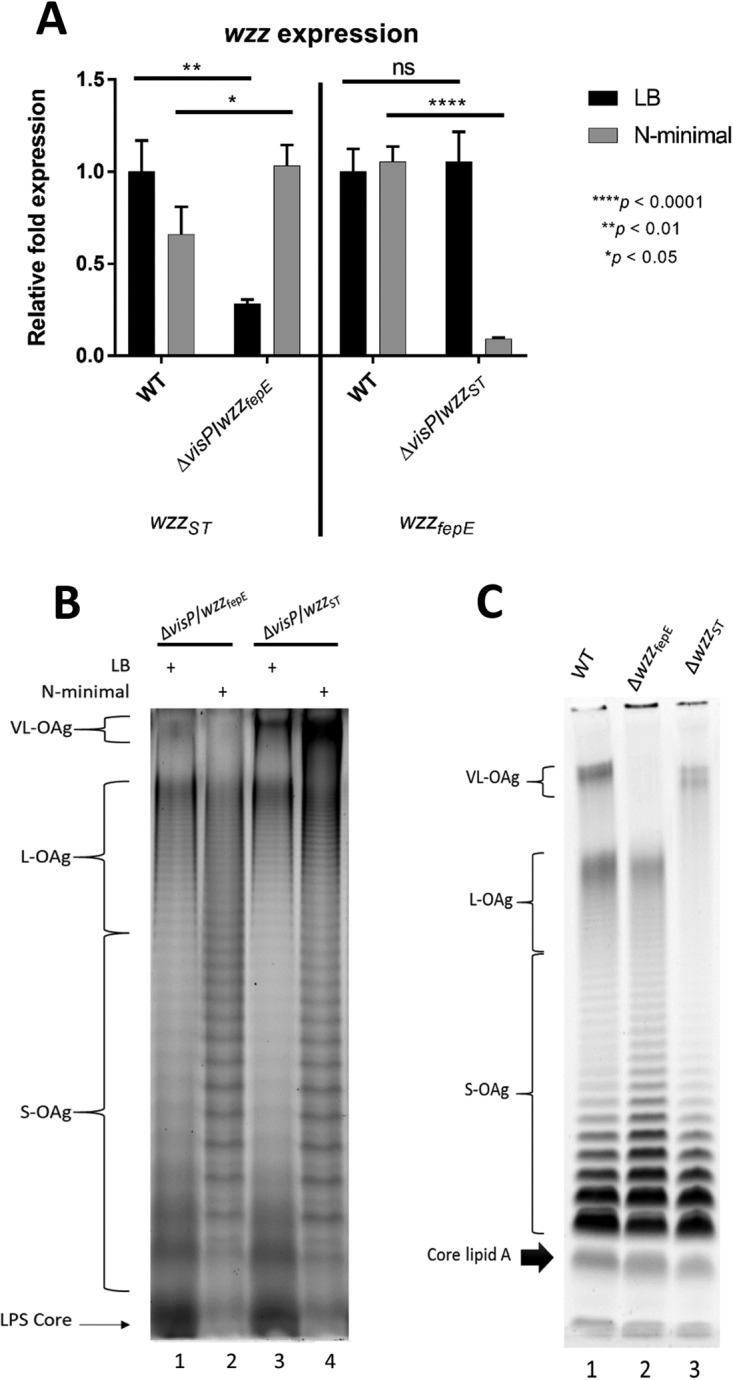
The interplay of VisP and PCP determines the final length of OAg. (A) qRT-PCR analysis of wzz expression in LB and N-minimal media. Relative fold expression in the WT, ΔvisP wzzfepE, and ΔvisP wzzST strains is shown. (B) High-resolution electrophoresis (15% SDS-PAGE) and staining with a ProQ Emerald 300 LPS gel staining kit (Thermo Fisher, Waltham, MA, USA) of the LPS profiles of the ΔvisP wzzfepE (lanes 1 and 2) and ΔvisP wzzST (lanes 3 and 4) strains grown in LB and N-minimal media. (C) Gradient SDS-PAGE (4 to 12%) of the LPS profile stained with a ProQ Emerald 300 LPS gel staining kit (Thermo Fisher, Waltham, MA, USA), showing the core lipid A and different OAg chain lengths in the WT, ΔwzzfepE, and ΔwzzST strains, all grown in LB. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Novel OAg length control.
The ΔvisP mutant was previously reported to have differentiated lipid A modification mediated by the LpxO enzyme (61). The ΔvisP single mutant also showed a clearly distinct OAg phenotype with different patterns depending on the growth condition (Fig. 1A). The two double mutants with mutations of visP and one of the genes encoding a PCP protein showed differentiated OAg profiles compared with those of their respective single PCP gene mutants (Fig. 3B and C). The ΔvisP wzzfepE strain did not show the VL-OAg banding pattern (Fig. 3B, lanes 1 and 2), consistent with the ΔwzzfepE phenotype (Fig. 3C, lane 2) and the absence of WzzfepE. Surprisingly, the ΔvisP wzzST mutant showed an unprecedented phenotype in both media (Fig. 3B, lanes 3 and 4), with restored L forms compared to that in the ΔwzzST single mutant (Fig. 3C, lane 3). This finding contradicts the established consensus on WzzST-dependent L-OAg chain formation (57–59); WzzfepE is the remaining PCP in this background and may exert this function by cross-complementation. Moreover, the S forms were very noticeable between different growth conditions; specifically, the S forms showed a well-defined profile in N-minimal medium (Fig. 3B). Together, these results suggest that the novel role of WzzfepE seems to be observed only in the absence of periplasmic VisP.
Longer OAg facilitates intestinal colonization.
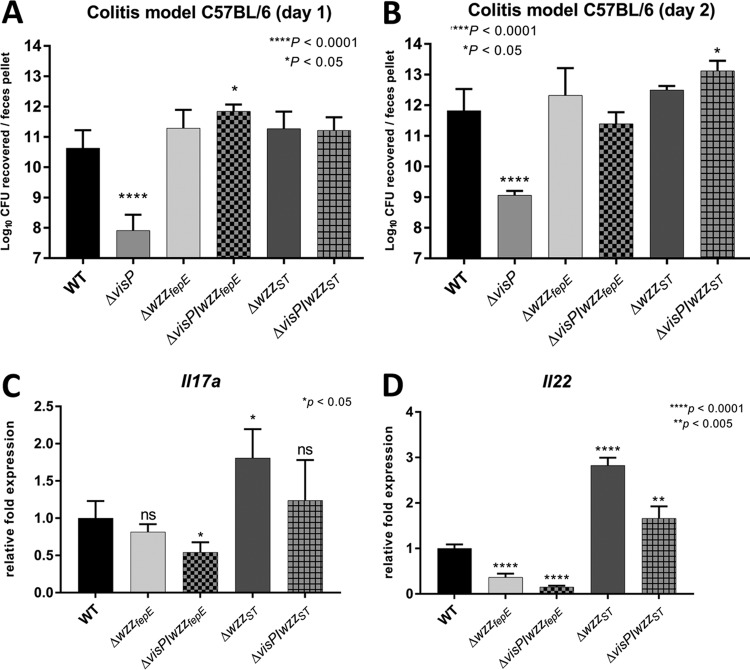
S. Typhimurium causes gastroenteritis and promotes colitis in murine models (87, 88). Therefore, we evaluated the effects of OAg modal length patterns in an S. Typhimurium colitis model via the oral route in streptomycin-pretreated C57BL/6 mice (89). First, the semirough OAg phenotype (90, 91), which was observed in the ΔvisP mutant, was attenuated by more than two orders of magnitude during in vivo colonization compared with the WT levels, as noted at both 1 and 2 days postinfection (p.i.) (Fig. 4A and B). These results were similar to those previously described in a BALB/c mouse colitis infection model (61). In the in vivo assays, the ΔvisP wzzfepE double mutant showed an increase in colonization of more than 1 order of magnitude in comparison with WT levels at day 1 p.i. (Fig. 4A). These data demonstrate that the ΔvisP wzzfepE mutant strain lacking VL-OAg was better able than the WT to adapt to host defenses and intestinal colonization in vivo at day 1 p.i. (Fig. 4A). However, this mutant showed a decrease in colonization of half of an order of magnitude at day 2 compared to the level at day 1 p.i. (Fig. 4A and B). The ΔvisP wzzST mutant presented a more prominent two-order-of-magnitude increase between days 1 and 2 p.i. during murine intestinal colonization (Fig. 4A and B). This limited colonization may be due to the absence of VL-OAg in the ΔvisP wzzfepE mutant strain. The only exception was the ΔvisP wzzST double mutant strain, which showed higher colonization at day 2 p.i. than at day 1 p.i. (Fig. 4A and B). In addition to intestinal colonization fitness, we measured the transcriptional levels of two distinct interleukins, IL-17A and IL-22, to better assess the induction of inflammation by the WT, the ΔwzzfepE and ΔwzzST single mutants, and the ΔvisP wzzfepE and ΔvisP wzzST double mutants at 2 days p.i. in the colon (Fig. 4C and D). IL-17A mediates neutrophil recruitment (92, 93) and suppresses S. Typhimurium colonization in enteric mucosa by inducing the production of antimicrobial peptides at the epithelial surface (94). The transcriptional levels of Il17a in the ΔwzzfepE and ΔvisP wzzST strains were similar to those in the WT, whereas ΔwzzST mutant infection resulted in a 2-fold increase (Fig. 4C). The ΔvisP wzzfepE double mutant exhibited a 2-fold reduction in Il17a expression in the colon compared to that in the WT strain (Fig. 4C), which may be related to the success of the initial intestinal colonization at day 1 p.i. (Fig. 4A). IL-22 is responsible for inducing the production of antimicrobial proteins in the mucosa, which sequester metal ions required for the mechanism of evasion by S. Typhimurium, allowing this pathogen to outcompete the microbiota and enhancing its colonization (95). Interestingly, both strains lacking VL-OAg, the ΔwzzfepE and ΔvisP wzzfepE mutants, induced less Il22 expression than the WT (Fig. 4D), whereas the ΔwzzST and ΔvisP wzzST strains exhibited higher induction of Il22 expression than the WT (Fig. 4D). During Salmonella-induced colitis, VL-OAg also plays a role in bile salt resistance (96). Accordingly, we performed a bile resistance assay (97) comparing previous LB growth with N-minimal growth. The WT and ΔvisP backgrounds showed growth recovery after 40 min at a lethal bile salt concentration (see Fig. S2A and B in the supplemental material) only after previous N-minimal growth, which enhances VL-OAg chains (Fig. 1A). After 1 h at a lethal bile salt concentration, the longer-OAg-chain-deficient ΔvisP strain exhibited a two-order-of-magnitude decrease in CFU compared with that in the WT (Fig. S2C). The low bile salt resistance may diminish the bacterial load of the ΔvisP strain reaching the intestines and affect the day 1 p.i. intestinal colonization levels (Fig. 4A). The higher levels of IL-22 production and the trimodal OAg pattern, similar to that of the WT (Fig. 1A and 3C [lane 1]), may explain the ΔvisP wzzST strain colonization fitness at 2 days p.i. (Fig. 4B), and the performance of this strain here confirms the importance of the L and VL OAg forms during the in vivo infectious process.
FIG 4.
Longer OAg facilitates intestinal colonization. A colitis model of infection was used in C57BL/6 mice. (A) Day 1 p.i. The log10 CFU of the WT, ΔvisP, ΔwzzfepE, ΔwzzST, ΔvisP wzzfepE, and ΔvisP wzzST strains recovered from each feces pellet are shown. (B) Day 2 p.i. The log10 CFU of the WT, ΔvisP, ΔwzzfepE, ΔwzzST, ΔvisP wzzfepE, and ΔvisP wzzST strains recovered from each feces pellet are shown. (C and D) qRT-PCR analysis of Il17a (C) and Il22 (D) performed with RNA extracted from the mouse colon. Relative fold expression in the WT, ΔwzzfepE, ΔwzzST, ΔvisP wzzfepE, and ΔvisP wzzST strains causing colitis is shown. *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
Flagellar changes with different OAgs.
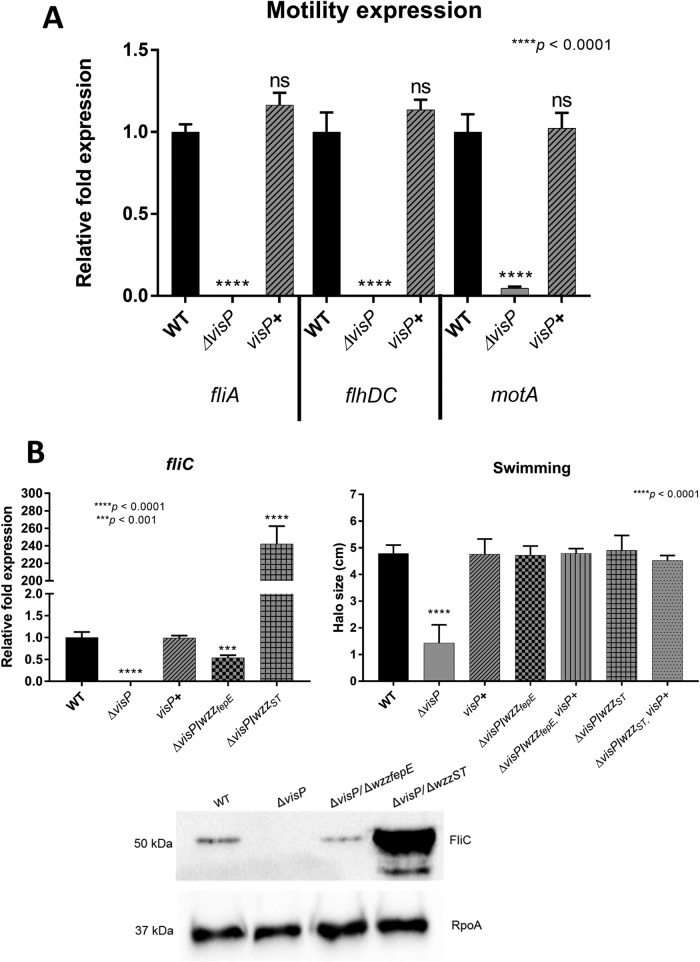
Flagella play an important role in S. Typhimurium pathogenesis by helping the bacteria move toward nutrient-rich environments and eliciting inflammation processes in the gut (5, 6, 8–10). The flagellum regulon is formed by 25 operons (over 67 genes) divided into 3 regulatory classes and is highly controlled by external factors. The class I operon flhDC produces FlhD and FlhC, components of the heteromultimeric complex FlhD4C2, which activates class II transcription through σ70 and autorepresses flhDC transcription. Hook basal body assembly is dependent on class II genes. After its completion, late substrates, such as FlgM, are exported from the cell, and σ28 starts the transcription of class III promoters. Class III genes include structural filament genes, such as fliC and fljB, and genes from the chemosensory pathway (98). Flagellar gene expression was employed to better evaluate the full role of flagellar structure and regulation via quantitative real-time reverse transcription-PCR (qRT-PCR) of essential flagellar genes from this complex regulon, such as the structural flagellin gene fliC (98), the master regulator flhDC (99, 100), genes such as fliA (101), which are responsible for class 3 transcription, and the gene encoding flagellar proton motive force rotation, motA (102, 103). The ΔvisP mutant showed very low levels of all tested flagellar genes (Fig. 5A and B), and the complemented visP+ strain restored WT expression levels. We performed a swimming assay in semisolid agar to assess the motility of the strains and evaluate bacterial movement in vitro; after 8-h assays, five strains presented phenotypes similar to that of the WT (Fig. 5B). The ΔvisP single mutant showed decreased motility compared to the WT levels. The complemented visP+ strain exhibited restored motility (Fig. 5B). The Salmonella flagellin FliC, which constitutes the flagellar filament subunit, was evaluated in terms of FliC protein levels via an immunoblotting assay with an anti-FliC monoclonal antibody and fliC gene expression levels via qRT-PCR. The FliC protein was not detected in the ΔvisP strain. The ΔvisP wzzfepE strain presented a slightly reduced amount of flagellin relative to WT levels, and the ΔvisP wzzST strain overproduced flagellin FliC at abundant levels (Fig. 5B). The protein levels matched fliC gene expression levels in all strains (Fig. 5B). Considering the OAg forms, the semirough OAg ΔvisP strain appeared to present less-functional flagella, whereas the strains with more prominent VL-OAg, such as the ΔvisP wzzST strain, which have excess flagellin, did not seem to employ this abundant flagellin in a more efficient way than the WT strain (Fig. 5B).
FIG 5.
Changes in flagellar gene expression with different OAgs. (A) qRT-PCR analysis of genes involved in flagellar assembly and motility. relative fold expression of the fliA, flhDC, and motA genes in the WT, ΔvisP, and complemented visP+ strains grown in LB is shown. (B) qRT-PCR of the flagellin gene fliC, swimming motility assay, and immunoblotting performed with anti-FliC and anti-RpoA (control) monoclonal antibodies. ***, P < 0.001; ****, P < 0.0001.
DISCUSSION
S. Typhimurium pathogenesis within the host depends on a complex assortment of factors mediated by stimulus recognition, such as bacterial movement across the lumen and mechanisms of evasion of host immune responses, to ultimately establish intestinal colonization. S. Typhimurium can elicit inflammation and benefits from this process. The host response through the innate immune system, among other defense machineries, recruits macrophage cells to combat the pathogen. Thus, S. Typhimurium has vast resources for colonization, as do other very successful bacterial pathogens, which employ several different virulence traits as an arsenal to strike back. The bacterial membrane plays an essential role in this process, providing protection against host defense mechanisms and physical stability to multiprotomeric complexes to support bacterial homeostasis and sensing of surrounding stimuli, which all culminate in metabolic and structural changes.
Here, we have demonstrated the outstanding mechanism by which S. Typhimurium differentially shapes its OM landscape under conditions that resemble the intracellular environment. Clearly, this mechanism involves modifications of distinct OAg chain patterns, with increases in the VL-OAg (>100 RU) and S-OAg (less than 16 RU) forms (Fig. 1A) of LPS. These environmental stimulus-mediated LPS changes were observed by Lateenmaki and colleagues (104), who also measured OAg profiles extracted from macrophage-bacterium cocultures with more evident VL-OAg augmentation and, conversely, L-OAg reduction. Other growth conditions, such as with inactivated guinea pig sera and iron-limited media, also augment VL-OAg chains in S. Typhimurium, although the specific stimuli were not identified (31). The length of longer OAg forms is controlled by the PCP proteins WzzST and WzzfepE in a possible interaction with Wzy polymerase, as previously described in Shigella flexneri (105). However, the underlying mechanism remains unclear. The distribution of OAg polymerization catalysis occurs via Wzy in a “catch-and-release” process in which Wzy adds each OAg RU or oligomer to a single RU chain end without remaining bound to the growing chain (53, 54, 56). This process results in a random OAg chain length distribution controlled by the OAg RU availability surrounding Wzy; therefore, short OAg forms should be most prevalent. The PCP WzzST is the molecular ruler responsible for L-OAg forms, shifting the random distribution of OAg chains by approximately 30 RU (58). WzzfepE is a second PCP molecular ruler that shifts the OAg distribution to chain lengths larger than 100 RU (see Fig. S1 in the supplemental material), as previously described (60). N-minimal medium is commonly employed for the expression of intracellular virulence traits, such as SPI-2 TTSS effectors (65), because it resembles the SCV environment. Here, we observed that wzy overexpression is mediated by N-minimal condition stimuli (Fig. 1B). Taken together, longer OAg chains result from the increase in Wzy that arises from the OAg polymer elongation rate, which is due to its distributive mechanism of action (54) as predicted by Michaelis-Menten enzyme kinetics. Therefore, WT S. Typhimurium increases VL-OAg by upregulating wzy, as observed previously in S. flexneri (106), which increases OAg chain elongation; however, it maintains WzzfepE levels and reduces WzzST levels, as PCP proteins may act as competitors in Wzy interactions.
Based on the OAg profile, we identified a potential novel role of periplasmic VisP as a missing piece in the Wzy-dependent OAg biosynthesis pathway puzzle. These findings provide novel insights into the complex mechanism of defined L-OAg and VL-OAg formation in S. Typhimurium. The absence of VisP induced a semirough OAg phenotype in LB nutrient-rich growth medium (Fig. 1A, lane 4), which was caused by Wzy and PCP deficiency (Fig. 1B). However, the Wzx flippase and WaaL OAg ligase do not appear to be affected, nor is the OM LPS export apparatus, as evidenced by the presence of LPS with at least a single OAg RU, such as in the S-OAg form (Fig. 1A, lane 4). The absence of VisP appears to broadly affect S. Typhimurium transcription, based on the observed downregulation of OAg biosynthesis pathway genes, the SPI-2 TTSS effector gene sifA, SPI-3 mgtB, and flagellum regulation (Fig. 1B, 2A, E, and F, and 5A and B). This transcriptional modulation could be associated with a possible signal transduction pathway mediated indirectly by VisP, as this periplasmic protein is regulated by the QseC sensor kinase and has a cryptic AraC-like regulator encoded on the same operon as YgiV (61). Two other very important TCSs of S. Typhimurium are implicated, namely, PhoPQ and PmrAB, which are represented here by the expression of phoP and pmrB; these gene levels were equally downregulated in the ΔvisP mutant (Fig. 2A). Previously, these two TCSs were not considered important for sensor kinase QseC regulation (44). Our results indicate that TCSs may exhibit cross talk under specific conditions that are dependent on the membrane functions examined here, such as OAg chain length determination. Moreover, the probable VisP signal transduction effect is associated with PCP proteins and is exemplified by the transcriptional regulation of wzy in the ΔvisP single mutant compared with that in the ΔvisP wzzfepE and ΔvisP wzzST double mutants (Fig. 1B). A recent structural study of the Escherichia coli PCP proteins FepE and WzzE and Salmonella enterica WzzST characterized a conserved G-rich motif on the cytoplasmic part of the second transmembrane domain (TM2) as a putative tyrosine kinase (pfam13807) (107). The N-terminal cytoplasmic region immediately before the transmembrane helix is highly conserved among PCPs and could be a potential domain for interactions with cytoplasmic proteins and signal transduction pathways (107). Current evidence suggests that VisP and both PCPs, WzzST and WzzfepE, work together during Wzy expression, which is the major control factor for OAg length. Double mutation of visP with wzzST or wzzfepE resulted in increased wzy expression compared with that in the OAg semirough ΔvisP mutant. Unexpectedly, L-OAg formation was WzzST independent in the ΔvisP wzzST double mutant (Fig. 3B and S1I and J), in contrast to expectations for OAg length determinants based on the previously described WzzST-dependent L-OAg biosynthetic pathway (57–59). Based on this finding, we presume that WzzfepE has a novel role during L-OAg biosynthesis in the absence of VisP via cross-complementation. Recently, enterobacterial common antigen (ECA) was first characterized in Shigella sonnei as linked to LPS (more specifically, to lipid A) anchored to the OM (108). Therefore, the L-OAg modality observed here in the absence of WzzST in the ΔvisP wzzST strain may be ECALPS chains with similar lengths of L-OAg. The hypothesis presented here is supported by the similar molecular weights of ECA and OAg RU, approximately 607 and 606 Da, respectively. Similar to the OAg system of S. Typhimurium, the system for ECA binding to LPS has its own polymerase (WzyE) and PCP protein (WzzE), which have been characterized only by genomic studies (109). Together, these findings indicate bacterial membrane heterogeneity and the simultaneous formation of glycolipid structures in the periplasm, a transient and rich environment with many distinct proteins that may interact with one another and participate in similar synthesis processes.
The ΔvisP single mutant displayed a clear deficiency during mouse gut colonization in our colitis infectious model (Fig. 4). The absence of VisP causes strong acidic pH susceptibility, as previous in vitro tests have shown that its absence in a ΔvisP mutant affects survival under acidic conditions (61). In E. coli, longer OAg forms maintain membrane stability and low pH tolerance (110). Furthermore, the ΔvisP strain is defective in bile salt resistance (see Fig. S2C in the supplemental material), in which OAg longer forms, primarily VL-OAg, have an important role (96). Thus, the semirough OAg phenotype of the ΔvisP strain may increase its susceptibility to the low pH of the stomach, as well as bile salts, reducing the bacterial load that reaches the intestine. Moreover, a phenotype with reduced levels of highly immunogenic proteins, such as FliC flagellin, may eliminate the ability of the ΔvisP strain to elicit inflammation processes, thus affecting its initial colonization ability (5, 6, 8–16, 19–22, 29, 111).
As described by Barthel and colleagues, the induction of the inflammation process by S. Typhimurium in a streptomycin-pretreated mouse colitis model was observed starting at 8 h p.i. based on the presence of PMN in the mouse intestinal lumen (89). Thereafter, they observed a large increase in PMN cell transmigration to the intestinal lumen from 20 h to 48 h p.i. (89) as a clear consequence of the inflammation process (5, 6, 8–16, 19–22, 29, 111). Therefore, at a few hours p.i., the host started to attack the pathogen through cells of the innate immune system. OAg chains function indispensably to trick macrophage recognition and consequent phagocytosis (Fig. 2B). The OAg rough phenotype is caused by complete OAg absence in the OM as a result of impairment of Wzx flippase or WaaL ligase function (35, 112–117). The semirough OAg phenotype is characterized by LPS with only one OAg RU, which occurs due to Wzy malfunction (90). Both phenotypes result in a defective Gram-negative bacterial membrane, which enables more efficient antibody opsonization and activation of the complement cascade, culminating in increased macrophage-mediated phagocytosis (29, 30, 35, 55, 90, 118), as observed here in the ΔvisP strain (Fig. 2B).
S. Typhimurium must adapt itself to sense phagosome differences in acidification and nutrient starvation after macrophage-mediated phagocytosis, such as low Mg2+ concentrations, through TCSs such as PmrAB and PhoPQ. Together, these TCSs activate cascade responses to defend against host enzymes and counterattack by coating the SCVs (28, 40, 77–79, 81, 82, 119, 120). The modulation of these responses triggers SPI-2 T3SS, and its gene expression is essential because it encodes effector proteins that are injected into host cells to modulate and manipulate the cell machinery for the bacterium's benefit (69–71, 84).
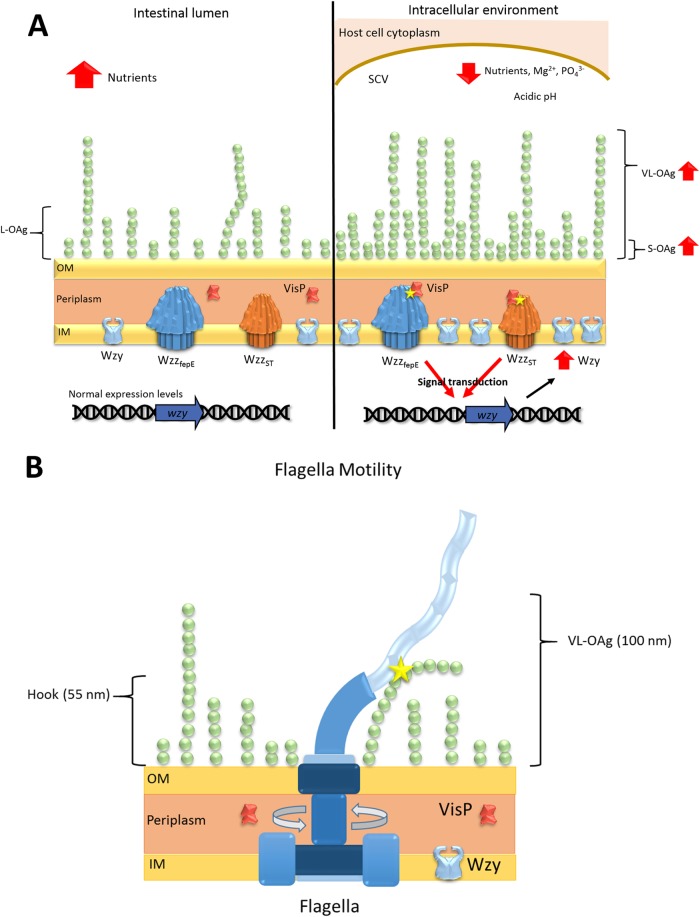
We showed that the long forms of OAg in S. Typhimurium play important roles within macrophages (Fig. 2). Initially, we demonstrated that S. Typhimurium OM remodeling is driven by SCV environments resembling stimulus conditions, resulting in increased OAg chains, primarily the VL-OAg pattern (Fig. 1A). The ΔvisP strain has a semirough OAg phenotype and is less protected within macrophages, as reflected in its intracellular survival levels (Fig. 2C). However, the ΔvisP mutant is able to produce longer OAg forms under conditions resembling SCVs, but this production occurs to a lesser extent than it does in the WT strain (Fig. 1A). Although the ΔvisP strain presents low expression of important intramacrophage survival genes, such as sifA and mgtB, its OM is remodeled with OAg long forms in the SCV, conferring sufficient protection to reduce its phenotypic differences from the WT strain in a 16-h assay (Fig. 2D). Moreover, the SPI-2 TTSS is assembled over longer periods, and some of its structures are detected only after 5 h upon SPI-2 gene expression induction (121). S. Typhimurium in intracellular macrophages may overproduce VL-OAg chains, which may help the SPI-2 TTSS build and promote the stability of the SseBC appendage structure, a gigantic structure that is larger than 160 nm (121). Furthermore, the SPI-2 TTSS is sheathed by protein effectors during and after its assembly, approaching SCV membranes and reaching the macrophage cytoplasm to exert its function in SCV maintenance (121). Consequently, VL-OAg chains may also help these effector proteins move across the SPI-2 TTSS. An analogous function for VL-OAg was proposed in giant adhesin SiiE secretion, which pulls its structure through ionic forces mediated by Ca2+ cations present at the OAg chains (34). L-OAg chains employ an initial protective role and may provide more support for VL-OAg, maintaining LPS stability as a whole. More importantly, longer OAg forms confer strong protection against macrophage phagocytosis to S. Typhimurium, allowing the bacterium to survive in the phagosome and thus creating a propitious environment for its replication. Taking all these observations into consideration, we propose a model in which S. Typhimurium strengthens its protection by increasing OAg longer forms inside the phagosome through the overproduction of Wzy (Fig. 6A). This process occurs via transcriptional control of wzy, which may be mediated by VisP and PCP protein interactions in the periplasm, creating a signal transduction cascade ignited by SCV environmental stimuli (Fig. 6A).
FIG 6.
Proposed models of OAg outer-leaf remodeling and flagellum-OAg interaction. (A) Model of OAg final chain length in the intestinal lumen (left) and intracellular environment (right). (B) S. Typhimurium flagellum interaction with VL-OAg chains.
All PCP mutant strains caused Il17a expression levels in a mouse colitis model that were similar to those in the WT, with a slight reduction for wzzfepE mutants (Fig. 4C). In this manner, we presume that the levels of intestinal inflammation and, more importantly, levels of neutrophil transmigration to the intestinal lumen are comparable for all strains. Nonetheless, distinct Il22 transcription patterns were observed, with a significant decrease in mice infected by strains lacking VL-OAg (Fig. 4D). IL-22 induces the production of antimicrobial proteins responsible for metal ion starvation, such as lipocalin-2 (93). S. Typhimurium can overcome this host defense mechanism and thus outcompete the affected microbiota (95). Since infection with strains presenting VL-OAg resulted in greater Il22 expression, this OAg modal length may induce the expression of Il22, culminating in more successful intestinal colonization. The higher expression rate of Il22 combined with a more protected OM presenting a WT-like OAg profile (Fig. 3B) could explain the increased level of intestinal colonization by the ΔvisP wzzST strain at 48 h p.i. (Fig. 4B).
Bacterial motility may be considered one of the most complex assembly mechanisms in bacterial structures because it includes a regulon network of more than 50 distinct genes (98) that are involved and directly influenced by environmental differences in the expression of the flagellins FliC and FljB of S. Typhimurium. Here, we also observed a role of VisP during bacterial motility, as the absence of periplasmic VisP caused a direct defect during swimming. After gene complementation, the swimming ring size was restored to WT levels (Fig. 5B). The ΔvisP single mutant had very low expression levels of the master regulator flhDC, the class 2 sigma factor fliA, the class 3 gene motA, and the flagellin gene fliC. The FlhD4C2 heteromultimeric complex for flhDC expression starts the flagellum assembly cascade, which is followed by class 2 and 3 flagellum operons. The fliA gene encodes a sigma factor that activates the transcription of class 3 genes. After hook completion, FlgM is exported from the cell and allows σ28 to start class 3 transcription. The flagellin FliC is the main filament component and is also exported via the flagellar T3SS (122). The MotAB system of flagella is responsible for the crucial motility of the flagella and is assembled via a Sec-dependent pathway (98). These two proteins form the ion-conducting stator complexes necessary for flagellar rotation (102, 103).
Mutation of either wzz gene in the ΔvisP mutant background restored flagellum gene expression to WT levels and the presence of the FliC protein in bacterial membrane extracts (Fig. 5B). Previously, PCP proteins have been shown to have a dual regulatory role in motility networks in bacterial LPS modifications. The WzzST protein is required to maintain the balance of 4-aminoarabinose and phosphoethanolamine in Salmonella lipid A modifications, which are mediated by ArnT and EptA, respectively (76). In Campylobacter jejuni, a gene encoding a phosphoethanolamine transferase was directly correlated with lipid A modifications and the flagellar rod protein FlgG (123).
The ΔvisP single mutant lacked all flagellum parameters evaluated here, such as the expression of various flagellum genes and reduced production of FliC flagellin. These deficiencies may be directly linked to its attenuation in our in vivo and in vitro tests, such as in phagocytosis and short- and long-period macrophage survival. The ΔvisP wzzST double mutant exhibited prominent overproduction of the flagellin FliC compared to WT levels; however, this overproduction apparently did not influence its motile phenotype (Fig. 5B). The presence of FliC protein indicates mature flagella, and overproduction of the filament suggests that the ΔvisP wzzST double mutant strain presents more flagella or longer filaments than the WT, although this change may not be reflected as a more efficient swimming mechanism (Fig. 5B). Thus, we propose that VL-OAgs with sizes over 100 nm (60) physically interact with the rotating flagellum, as the hook structure is smaller than approximately 55 nm (124), to reach the maximum plateau of motility efficiency even in the presence of more flagella or longer filaments (Fig. 6B). Previous studies have demonstrated the importance of LPS for surface or swarming motility (125), mainly due to the wettability of LPS. Moreover, the increase in expression levels of OAg-related genes occurs concomitantly with the expression of class 3 motility genes and, consequently, flagellum maturation (126). Wang et al. have also shown that the overexpression of the iron uptake system (126), encoded adjacent to the genome location of wzzfepE (60), is directly affected by different iron concentrations (31). Important LPS structural assembly genes, such as rfaC, impact the secretion of proteins in S. Typhimurium, such as SipA, FlgK, FliC, FliD, SipC, InvJ, and FlgL, thereby hampering protein translocation, which may be enhanced by the absence of OAg (127). In addition, proteins related to LPS modification also perform posttranslational modifications of flagellar structural proteins, such as phosphoethanolamine addition to FlgG catalyzed by Cj0256 in Campylobacter jejuni (123) and flagellin O-glycosylation by the protein encoded by rmlB in Burkholderia pseudomallei (128). Similarly, we hypothesize that the ΔvisP wzzST mutant strain may overproduce FliC flagellin in a correlated mechanism with PCP WzzfepE in the absence of both VisP and the other PCP WzzST, supporting the importance of OAg chain structures during the secretion of other membrane proteins.
The periplasmic protein VisP appears to be essential for cell membrane maintenance, virulence, and proper flagellum function. In addition, the ΔvisP wzzfepE double mutant was also able to partially restore flagellin FliC and fliC gene expression (Fig. 5B). Together, these data show that VisP has a novel and important role during flagellum expression that warrants further study and that both Wzz PCPs presented here may function as cofactors to restore flagellar function in S. Typhimurium via FliC. The VL forms (ΔvisP wzzST) apparently increase the overproduction of FliC flagellin, which may be correlated with the size of the LPS chains. Moreover, the L forms (ΔvisP wzzfepE) induce greater production of FliC than in the ΔvisP single mutant; therefore, there is a link between the longer forms of OAg and flagellum production that is not fully understood and requires clarification.
The bacterial membrane is the first line of interaction with the host. Determining how it senses and responds to host cell signals by changing its structure and composition is the initial step toward fully understanding this complex relationship. A better understanding of these processes will shed light on how hosts and pathogens have coevolved and how we can develop new therapies to dismantle pathogen defenses. In this work, we identified an S. Typhimurium response mechanism against macrophage phagocytosis. The next step will be to elucidate how each piece of this complex process exerts its function, which will enable the design of target-specific drugs to block their activity.
MATERIALS AND METHODS
Ethics statement.
All procedures were performed under supervision of a veterinarian with the approval of the Animal Ethics Committee of the School of Pharmaceutical Sciences (CEUA/FCF/CAr no. 23/2016). The study was conducted in accordance with the Brazilian Law for Animal Research no. 11.794, 8 October 2008, decree no. 6.899, 15 July 2009.
Strains and plasmids.
All strains and plasmids used in this study are listed in Table S1 in the supplemental material. Recombinant DNA and molecular biology techniques were performed as previously described (129). The oligonucleotides used in this study are listed in Table S2 in the supplemental material.
Construction of isogenic mutants.
Construction of isogenic nonpolar S. Typhimurium SL1344 wzzfepE and wzzST single mutants and visP wzzfepE and visP wzzST double knockout mutants was achieved by using λ Red mutagenesis (130). The visP wzzfepE and visP wzzST double mutants were complemented with the visP gene cloned into the vector pBADMycHisA (KpnI and EcoRI) (61) to generate strains 184 and 185.
Macrophage infection assays.
Intramacrophage phagocytosis assays were performed as previously described (35). In the survival assay, the cells were washed and incubated at 37°C with 5% CO2 for 3 h without antibiotics (67, 82, 83). After extracellular bacteria killing in the intramacrophage replication assay, the cells were washed and incubated at 37°C with 5% CO2 for 16 h with 15 μg/ml gentamicin (35).
Motility assays.
The swimming motility behaviors of S. Typhimurium were assessed in semisolid plates at 37°C for 8 h, as previously described (44, 131).
qRT-PCR.
Quantitative real-time reverse transcription-PCR (qRT-PCR) was performed as previously described (132).
Colitis model infection with S. Typhimurium.
Mice (C57BL/6, 7 to 9 weeks old, female) were infected orally for the colitis model as previously described (89).
Western blot assay.
Culture supernatants were obtained from strains grown statically in LB medium at 37°C overnight. The medium was removed by centrifugation, and the cells were resuspended in 10 ml of phosphate-buffered saline (PBS) plus 100 μl of phenylmethylsulfonyl fluoride (100 mM). Bacteria were lysed by sonication, pelleted by centrifugation (4,200 × g, 10 min, 4°C), and analyzed by SDS-PAGE. Proteins were separated by SDS-PAGE and electrophoretically transferred to nitrocellulose membranes via a semidry transfer cell (Bio-Rad, Hercules, CA). Immunoblots were probed with monoclonal anti-flagellin FliC (InvivoGen, San Diego, CA) (1:1,000) and anti-RpoA (Santa Cruz Biotechnology, Dallas, TX) (1:5,000). Bound antibody was detected with the horseradish peroxidase-conjugated secondary antibody anti-mouse IgG (1:2,500) (Promega, Fitchburg, WI), followed by enhanced chemiluminescence substrate (ECL; Promega, Fitchburg, WI) and detection in ChemiDoc MP (Bio-Rad, Hercules, CA).
Analysis of LPS profile.
Samples were grown overnight aerobically in LB medium at 37°C. Cell cultures were diluted 1:100 and grown on LB or N-minimal medium at 37°C and 200 rpm until an optical density at 600 nm (OD600) of 1.0 was reached, and then 1 ml of cells was harvested and centrifuged at 15,000 × g for 5 min. The pellets were resuspended in lysis buffer (2% β-mercaptoethanol, 2% SDS, and 10% glycerol in 0.1 M Tris-HCl, pH 6.8). The suspension was boiled for 10 min and then treated with 1.25 μl of proteinase K (20 mg/ml) overnight at 55°C. LPS was separated on 15% SDS-polyacrylamide gels using a Laemmli buffer system at 25 mA and visualized by gel staining using the ProQ Emerald 300 LPS gel staining kit (Thermo Fisher Scientific, Waltham, MA).
Bile salt resistance assay.
Bacteria were grown in LB or N-minimal medium until they reached exponential phase (OD600 of 0.6) and then were centrifuged at 4,000 × g for 10 min. The pellets were resuspended in LB broth with 30% bile salts. OD600 readings were acquired every 20 min for 1 h, with the first measurement occurring after resuspension (97). To compare the two different strains, samples initially grown in LB broth were washed twice with PBS immediately after exposure to bile salts for 1 h. The samples were serially diluted and plated for CFU enumeration.
Supplementary Material
ACKNOWLEDGMENTS
We thank all faculty members and technicians of the Biological Sciences Department at São Paulo State University for assistance in setting up the Moreira laboratory and especially for sharing reagents and equipment for our initial work. In particular, we thank Sandro Valentini and Fernando Pavan for their assistance.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00319-18.
REFERENCES
- 1.World Health Organization. 2013. Salmonella (non-typhoidal), fact sheet no. 139. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Feasey NA, Cain AK, Msefula CL, Pickard D, Alaerts M, Aslett M, Everett DB, Allain TJ, Dougan G, Gordon MA, Heyderman RS, Kingsley RA. 2014. Drug resistance in Salmonella enterica ser. Typhimurium bloodstream infection, Malawi. Emerg Infect Dis 20:1957–1959. doi: 10.3201/eid2011.141175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ley B, Le Hello S, Lunguya O, Lejon V, Muyembe JJ, Weill FX, Jacobs J. 2014. Invasive Salmonella enterica serotype Typhimurium infections, Democratic Republic of the Congo, 2007-2011. Emerg Infect Dis 20:701–704. doi: 10.3201/eid2004.131488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haraga A, Ohlson MB, Miller SI. 2008. Salmonellae interplay with host cells. Nat Rev Microbiol 6:53–66. doi: 10.1038/nrmicro1788. [DOI] [PubMed] [Google Scholar]
- 5.Stecher B, Barthel M, Schlumberger MC, Haberli L, Rabsch W, Kremer M, Hardt WD. 2008. Motility allows S. Typhimurium to benefit from the mucosal defence. Cell Microbiol 10:1166–1180. doi: 10.1111/j.1462-5822.2008.01118.x. [DOI] [PubMed] [Google Scholar]
- 6.Stecher B, Hapfelmeier S, Muller C, Kremer M, Stallmach T, Hardt WD. 2004. Flagella and chemotaxis are required for efficient induction of Salmonella enterica serovar Typhimurium colitis in streptomycin-pretreated mice. Infect Immun 72:4138–4150. doi: 10.1128/IAI.72.7.4138-4150.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wadhams GH, Armitage JP. 2004. Making sense of it all: bacterial chemotaxis. Nat Rev Mol Cell Biol 5:1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- 8.Salazar-Gonzalez RM, McSorley SJ. 2005. Salmonella flagellin, a microbial target of the innate and adaptive immune system. Immunol Lett 101:117–122. doi: 10.1016/j.imlet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 9.Lai MA, Quarles EK, Lopez-Yglesias AH, Zhao X, Hajjar AM, Smith KD. 2013. Innate immune detection of flagellin positively and negatively regulates salmonella infection. PLoS One 8:e72047. doi: 10.1371/journal.pone.0072047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayashi F, Smith KD, Ozinsky A, Hawn TR, Yi EC, Goodlett DR, Eng JK, Akira S, Underhill DM, Aderem A. 2001. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410:1099–1103. doi: 10.1038/35074106. [DOI] [PubMed] [Google Scholar]
- 11.Franchi L, Amer A, Body-Malapel M, Kanneganti TD, Ozoren N, Jagirdar R, Inohara N, Vandenabeele P, Bertin J, Coyle A, Grant EP, Nunez G. 2006. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in salmonella-infected macrophages. Nat Immunol 7:576–582. doi: 10.1038/ni1346. [DOI] [PubMed] [Google Scholar]
- 12.Rayamajhi M, Zak DE, Chavarria-Smith J, Vance RE, Miao EA. 2013. Mouse NAIP1 detects the type III secretion system needle protein. J Immunol 191:3986–3989. doi: 10.4049/jimmunol.1301549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miao EA, Mao DP, Yudkovsky N, Bonneau R, Lorang CG, Warren SE, Leaf IA, Aderem A. 2010. Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc Natl Acad Sci U S A 107:3076–3080. doi: 10.1073/pnas.0913087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miao EA, Alpuche-Aranda CM, Dors M, Clark AE, Bader MW, Miller SI, Aderem A. 2006. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nat Immunol 7:569–575. doi: 10.1038/ni1344. [DOI] [PubMed] [Google Scholar]
- 15.Sun YH, Rolan HG, Tsolis RM. 2007. Injection of flagellin into the host cell cytosol by Salmonella enterica serotype Typhimurium. J Biol Chem 282:33897–33901. doi: 10.1074/jbc.C700181200. [DOI] [PubMed] [Google Scholar]
- 16.Yang J, Zhao Y, Shi J, Shao F. 2013. Human NAIP and mouse NAIP1 recognize bacterial type III secretion needle protein for inflammasome activation. Proc Natl Acad Sci U S A 110:14408–14413. doi: 10.1073/pnas.1306376110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khan SA, Everest P, Servos S, Foxwell N, Zahringer U, Brade H, Rietschel ET, Dougan G, Charles IG, Maskell DJ. 1998. A lethal role for lipid A in Salmonella infections. Mol Microbiol 29:571–579. doi: 10.1046/j.1365-2958.1998.00952.x. [DOI] [PubMed] [Google Scholar]
- 18.Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, Freudenberg M, Ricciardi-Castagnoli P, Layton B, Beutler B. 1998. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science 282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 19.Zhang S, Santos RL, Tsolis RM, Mirold S, Hardt WD, Adams LG, Bäumler AJ. 2002. Phage mediated horizontal transfer of the sopE1 gene increases enteropathogenicity of Salmonella enterica serotype Typhimurium for calves. FEMS Microbiol Lett 217:243–247. doi: 10.1111/j.1574-6968.2002.tb11482.x. [DOI] [PubMed] [Google Scholar]
- 20.Raffatellu M, Wilson RP, Chessa D, Andrews-Polymenis H, Tran QT, Lawhon S, Khare S, Adams LG, Bäumler AJ. 2005. SipA, SopA, SopB, SopD, and SopE2 contribute to Salmonella enterica serotype Typhimurium invasion of epithelial cells. Infect Immun 73:146–154. doi: 10.1128/IAI.73.1.146-154.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keestra AM, Godinez I, Xavier MN, Winter MG, Winter SE, Tsolis RM, Bäumler AJ. 2011. Early MyD88-dependent induction of interleukin-17A expression during Salmonella colitis. Infect Immun 79:3131–3140. doi: 10.1128/IAI.00018-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hapfelmeier S, Ehrbar K, Stecher B, Barthel M, Kremer M, Hardt WD. 2004. Role of the Salmonella pathogenicity island 1 effector proteins SipA, SopB, SopE, and SopE2 in Salmonella enterica subspecies 1 serovar Typhimurium colitis in streptomycin-pretreated mice. Infect Immun 72:795–809. doi: 10.1128/IAI.72.2.795-809.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kingsley RA, Santos RL, Keestra AM, Adams LG, Baumler AJ. 2002. Salmonella enterica serotype Typhimurium ShdA is an outer membrane fibronectin-binding protein that is expressed in the intestine. Mol Microbiol 43:895–905. doi: 10.1046/j.1365-2958.2002.02805.x. [DOI] [PubMed] [Google Scholar]
- 24.Dorsey CW, Laarakker MC, Humphries AD, Weening EH, Baumler AJ. 2005. Salmonella enterica serotype Typhimurium MisL is an intestinal colonization factor that binds fibronectin. Mol Microbiol 57:196–211. doi: 10.1111/j.1365-2958.2005.04666.x. [DOI] [PubMed] [Google Scholar]
- 25.Gerlach RG, Cláudio N, Rohde M, Jäckel D, Wagner C, Hensel M. 2008. Cooperation of Salmonella pathogenicity islands 1 and 4 is required to breach epithelial barriers. Cell Microbiol 10:2364–2376. doi: 10.1111/j.1462-5822.2008.01218.x. [DOI] [PubMed] [Google Scholar]
- 26.Poirier V, Av-Gay Y. 2015. Intracellular growth of bacterial pathogens: the role of secreted effector proteins in the control of phagocytosed microorganisms. Microbiol Spectr doi: 10.1128/microbiolspec.VMBF-0003-2014. [DOI] [PubMed] [Google Scholar]
- 27.Liss V, Hensel M. 2015. Take the tube: remodelling of the endosomal system by intracellular Salmonella enterica. Cell Microbiol 17:639–647. doi: 10.1111/cmi.12441. [DOI] [PubMed] [Google Scholar]
- 28.Blanc-Potard AB, Groisman EA. 1997. The Salmonella selC locus contains a pathogenicity island mediating intramacrophage survival. EMBO J 16:5376–5385. doi: 10.1093/emboj/16.17.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keestra-Gounder AM, Tsolis RM, Baumler AJ. 2015. Now you see me, now you don't: the interaction of Salmonella with innate immune receptors. Nat Rev Microbiol 13:206–216. doi: 10.1038/nrmicro3428. [DOI] [PubMed] [Google Scholar]
- 30.Murray GL, Attridge SR, Morona R. 2006. Altering the length of the lipopolysaccharide O antigen has an impact on the interaction of Salmonella enterica serovar Typhimurium with macrophages and complement. J Bacteriol 188:2735–2739. doi: 10.1128/JB.188.7.2735-2739.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murray GL, Attridge SR, Morona R. 2005. Inducible serum resistance in Salmonella typhimurium is dependent on wzz(fepE)-regulated very long O antigen chains. Microbes Infect 7:1296–1304. doi: 10.1016/j.micinf.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 32.Parker CT, Kloser AW, Schnaitman CA, Stein MA, Gottesman S, Gibson BW. 1992. Role of the rfaG and rfaP genes in determining the lipopolysaccharide core structure and cell surface properties of Escherichia coli K-12. J Bacteriol 174:2525–2538. doi: 10.1128/jb.174.8.2525-2538.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morgenstein RM, Clemmer KM, Rather PN. 2010. Loss of the waaL O-antigen ligase prevents surface activation of the flagellar gene cascade in Proteus mirabilis. J Bacteriol 192:3213–3221. doi: 10.1128/JB.00196-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barlag B, Hensel M. 2015. The giant adhesin SiiE of Salmonella enterica. Molecules 20:1134–1150. doi: 10.3390/molecules20011134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holzer SU, Schlumberger MC, Jackel D, Hensel M. 2009. Effect of the O-antigen length of lipopolysaccharide on the functions of type III secretion systems in Salmonella enterica. Infect Immun 77:5458–5470. doi: 10.1128/IAI.00871-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nikaido H, Vaara M. 1985. Molecular basis of bacterial outer membrane permeability. Microbiol Rev 49:1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nikaido H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev 67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Galdiero S, Falanga A, Cantisani M, Tarallo R, Della Pepa ME, D'Oriano V, Galdiero M. 2012. Microbe-host interactions: structure and role of Gram-negative bacterial porins. Curr Protein Pept Sci 13:843–854. doi: 10.2174/138920312804871120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deiwick J, Nikolaus T, Erdogan S, Hensel M. 1999. Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol Microbiol 31:1759–1773. doi: 10.1046/j.1365-2958.1999.01312.x. [DOI] [PubMed] [Google Scholar]
- 40.Soncini FC, Garcia Vescovi E, Solomon F, Groisman EA. 1996. Molecular basis of the magnesium deprivation response in Salmonella typhimurium: identification of PhoP-regulated genes. J Bacteriol 178:5092–5099. doi: 10.1128/jb.178.17.5092-5099.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sperandio V, Torres AG, Kaper JB. 2002. Quorum sensing Escherichia coli regulators B and C (QseBC): a novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in E. coli. Mol Microbiol 43:809–821. doi: 10.1046/j.1365-2958.2002.02803.x. [DOI] [PubMed] [Google Scholar]
- 42.Anderson JK, Smith TG, Hoover TR. 2010. Sense and sensibility: flagellum-mediated gene regulation. Trends Microbiol 18:30. doi: 10.1016/j.tim.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hughes DT, Terekhova DA, Liou L, Hovde CJ, Sahl JW, Patankar AV, Gonzalez JE, Edrington TS, Rasko DA, Sperandio V. 2010. Chemical sensing in mammalian host-bacterial commensal associations. Proc Natl Acad Sci U S A 107:9831–9836. doi: 10.1073/pnas.1002551107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moreira CG, Weinshenker D, Sperandio V. 2010. QseC mediates Salmonella enterica serovar Typhimurium virulence in vitro and in vivo. Infect Immun 78:914–926. doi: 10.1128/IAI.01038-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pullinger GD, van Diemen PM, Dziva F, Stevens MP. 2010. Role of two-component sensory systems of Salmonella enterica serovar Dublin in the pathogenesis of systemic salmonellosis in cattle. Microbiology 156:3108–3122. doi: 10.1099/mic.0.041830-0. [DOI] [PubMed] [Google Scholar]
- 46.Fass E, Groisman EA. 2009. Control of Salmonella pathogenicity island-2 gene expression. Curr Opin Microbiol 12:199–204. doi: 10.1016/j.mib.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gunn JS. 2008. The Salmonella PmrAB regulon: lipopolysaccharide modifications, antimicrobial peptide resistance and more. Trends Microbiol 16:284–290. doi: 10.1016/j.tim.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 48.Juarez-Rodriguez MD, Torres-Escobar A, Demuth DR. 2013. ygiW and qseBC are co-expressed in Aggregatibacter actinomycetemcomitans and regulate biofilm growth. Microbiology 159:989–1001. doi: 10.1099/mic.0.066183-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khajanchi BK, Kozlova EV, Sha J, Popov VL, Chopra AK. 2012. The two-component QseBC signalling system regulates in vitro and in vivo virulence of Aeromonas hydrophila. Microbiology 158:259–271. doi: 10.1099/mic.0.051805-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Unal CM, Singh B, Fleury C, Singh K, Chávez de Paz L, Svensäter G, Riesbeck K. 2012. QseC controls biofilm formation of non-typeable Haemophilus influenzae in addition to an AI-2-dependent mechanism. Int J Med Microbiol 302:261–269. doi: 10.1016/j.ijmm.2012.07.013. [DOI] [PubMed] [Google Scholar]
- 51.Wang X, Wang Q, Yang M, Xiao J, Liu Q, Wu H, Zhang Y. 2011. QseBC controls flagellar motility, fimbrial hemagglutination and intracellular virulence in fish pathogen Edwardsiella tarda. Fish Shellfish Immunol 30:944–953. doi: 10.1016/j.fsi.2011.01.019. [DOI] [PubMed] [Google Scholar]
- 52.Raetz CR, Whitfield C. 2002. Lipopolysaccharide endotoxins. Annu Rev Biochem 71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Woodward R, Yi W, Li L, Zhao G, Eguchi H, Sridhar PR, Guo H, Song JK, Motari E, Cai L, Kelleher P, Liu X, Han W, Zhang W, Ding Y, Li M, Wang PG. 2010. In vitro bacterial polysaccharide biosynthesis: defining the functions of Wzy and Wzz. Nat Chem Biol 6:418–423. doi: 10.1038/nchembio.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao G, Wu B, Li L, Wang PG. 2014. O-antigen polymerase adopts a distributive mechanism for lipopolysaccharide biosynthesis. Appl Microbiol Biotechnol 98:4075–4081. doi: 10.1007/s00253-014-5552-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Daniels C, Vindurampulle C, Morona R. 1998. Overexpression and topology of the Shigella flexneri O-antigen polymerase (Rfc/Wzy). Mol Microbiol 28:1211–1222. doi: 10.1046/j.1365-2958.1998.00884.x. [DOI] [PubMed] [Google Scholar]
- 56.Robbins PW, Bray D, Dankert BM, Wright A. 1967. Direction of chain growth in polysaccharide synthesis. Science 158:1536–1542. doi: 10.1126/science.158.3808.1536. [DOI] [PubMed] [Google Scholar]
- 57.Batchelor RA, Haraguchi GE, Hull RA, Hull SI. 1991. Regulation by a novel protein of the bimodal distribution of lipopolysaccharide in the outer membrane of Escherichia coli. J Bacteriol 173:5699–5704. doi: 10.1128/jb.173.18.5699-5704.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Batchelor RA, Alifano P, Biffali E, Hull SI, Hull RA. 1992. Nucleotide sequences of the genes regulating O-polysaccharide antigen chain length (rol) from Escherichia coli and Salmonella typhimurium: protein homology and functional complementation. J Bacteriol 174:5228–5236. doi: 10.1128/jb.174.16.5228-5236.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morona R, van den Bosch L, Manning PA. 1995. Molecular, genetic, and topological characterization of O-antigen chain length regulation in Shigella flexneri. J Bacteriol 177:1059–1068. doi: 10.1128/jb.177.4.1059-1068.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Murray GL, Attridge SR, Morona R. 2003. Regulation of Salmonella typhimurium lipopolysaccharide O antigen chain length is required for virulence; identification of FepE as a second Wzz. Mol Microbiol 47:1395–1406. doi: 10.1046/j.1365-2958.2003.03383.x. [DOI] [PubMed] [Google Scholar]
- 61.Moreira CG, Herrera CM, Needham BD, Parker CT, Libby SJ, Fang FC, Trent MS, Sperandio V. 2013. Virulence and stress-related periplasmic protein (VisP) in bacterial/host associations. Proc Natl Acad Sci U S A 110:1470–1475. doi: 10.1073/pnas.1215416110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gibbons HS, Lin S, Cotter RJ, Raetz CR. 2000. Oxygen requirement for the biosynthesis of the S-2-hydroxymyristate moiety in Salmonella typhimurium lipid A. Function of LpxO, A new Fe2+/alpha-ketoglutarate-dependent dioxygenase homologue. J Biol Chem 275:32940–32949. [DOI] [PubMed] [Google Scholar]
- 63.Gibbons HS, Reynolds CM, Guan Z, Raetz CR. 2008. An inner membrane dioxygenase that generates the 2-hydroxymyristate moiety of Salmonella lipid A. Biochemistry 47:2814–2825. doi: 10.1021/bi702457c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ginalski K, Kinch L, Rychlewski L, Grishin NV. 2004. BOF: a novel family of bacterial OB-fold proteins. FEBS Lett 567:297–301. doi: 10.1016/j.febslet.2004.04.086. [DOI] [PubMed] [Google Scholar]
- 65.Bustamante VH, Martinez LC, Santana FJ, Knodler LA, Steele-Mortimer O, Puente JL. 2008. HilD-mediated transcriptional cross-talk between SPI-1 and SPI-2. Proc Natl Acad Sci U S A 105:14591–14596. doi: 10.1073/pnas.0801205105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vazquez-Torres A, Xu Y, Jones-Carson J, Holden DW, Lucia SM, Dinauer MC, Mastroeni P, Fang FC. 2000. Salmonella pathogenicity island 2-dependent evasion of the phagocyte NADPH oxidase. Science 287:1655–1658. doi: 10.1126/science.287.5458.1655. [DOI] [PubMed] [Google Scholar]
- 67.Pfeifer CG, Marcus SL, Steele-Mortimer O, Knodler LA, Finlay BB. 1999. Salmonella typhimurium virulence genes are induced upon bacterial invasion into phagocytic and nonphagocytic cells. Infect Immun 67:5690–5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lober S, Jackel D, Kaiser N, Hensel M. 2006. Regulation of Salmonella pathogenicity island 2 genes by independent environmental signals. Int J Med Microbiol 296:435–447. doi: 10.1016/j.ijmm.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 69.Hensel M, Shea JE, Waterman SR, Mundy R, Nikolaus T, Banks G, Vazquez-Torres A, Gleeson C, Fang FC, Holden DW. 1998. Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol Microbiol 30:163–174. doi: 10.1046/j.1365-2958.1998.01047.x. [DOI] [PubMed] [Google Scholar]
- 70.Figueira R, Holden DW. 2012. Functions of the Salmonella pathogenicity island 2 (SPI-2) type III secretion system effectors. Microbiology 158:1147–1161. doi: 10.1099/mic.0.058115-0. [DOI] [PubMed] [Google Scholar]
- 71.Cirillo DM, Valdivia RH, Monack DM, Falkow S. 1998. Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol Microbiol 30:175–188. doi: 10.1046/j.1365-2958.1998.01048.x. [DOI] [PubMed] [Google Scholar]
- 72.Wong DK, Morris C, Lam TL, Wong WK, Hackett J. 1999. Identification of O-antigen polymerase transcription and translation start signals and visualization of the protein in Salmonella enterica serovar Typhimurium. Microbiology 145:2443–2451. doi: 10.1099/00221287-145-9-2443. [DOI] [PubMed] [Google Scholar]
- 73.Wosten MM, Kox LF, Chamnongpol S, Soncini FC, Groisman EA. 2000. A signal transduction system that responds to extracellular iron. Cell 103:113–125. doi: 10.1016/S0092-8674(00)00092-1. [DOI] [PubMed] [Google Scholar]
- 74.Delgado MA, Mouslim C, Groisman EA. 2006. The PmrA/PmrB and RcsC/YojN/RcsB systems control expression of the Salmonella O-antigen chain length determinant. Mol Microbiol 60:39–50. doi: 10.1111/j.1365-2958.2006.05069.x. [DOI] [PubMed] [Google Scholar]
- 75.Chen HD, Groisman EA. 2013. The biology of the PmrA/PmrB two-component system: the major regulator of lipopolysaccharide modifications. Annu Rev Microbiol 67:83–112. doi: 10.1146/annurev-micro-092412-155751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Farizano JV, Pescaretti Mde lM, López FE, Hsu FF, Delgado MA. 2012. The PmrAB system-inducing conditions control both lipid A remodeling and O-antigen length distribution, influencing the Salmonella Typhimurium-host interactions. J Biol Chem, 287:38778–38789. doi: 10.1074/jbc.M112.397414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kato A, Groisman EA. 2004. Connecting two-component regulatory systems by a protein that protects a response regulator from dephosphorylation by its cognate sensor. Genes Dev 18:2302–2313. doi: 10.1101/gad.1230804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Choi E, Groisman EA, Shin D. 2009. Activated by different signals, the PhoP/PhoQ two-component system differentially regulates metal uptake. J Bacteriol 191:7174–7181. doi: 10.1128/JB.00958-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Garcia Vescovi E, Soncini FC, Groisman EA. 1996. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell 84:165–174. doi: 10.1016/S0092-8674(00)81003-X. [DOI] [PubMed] [Google Scholar]
- 80.Pilonieta MC, Erickson KD, Ernst RK, Detweiler CS. 2009. A protein important for antimicrobial peptide resistance, YdeI/OmdA, is in the periplasm and interacts with OmpD/NmpC. J Bacteriol 191:7243–7252. doi: 10.1128/JB.00688-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bader MW, Sanowar S, Daley ME, Schneider AR, Cho U, Xu W, Klevit RE, Le Moual H, Miller SI. 2005. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell 122:461–472. doi: 10.1016/j.cell.2005.05.030. [DOI] [PubMed] [Google Scholar]
- 82.Detweiler CS, Monack DM, Brodsky IE, Mathew H, Falkow S. 2003. virK, somA and rcsC are important for systemic Salmonella enterica serovar Typhimurium infection and cationic peptide resistance. Mol Microbiol 48:385–400. doi: 10.1046/j.1365-2958.2003.03455.x. [DOI] [PubMed] [Google Scholar]
- 83.Fierer J, Eckmann L, Fang F, Pfeifer C, Finlay BB, Guiney D. 1993. Expression of the Salmonella virulence plasmid gene spvB in cultured macrophages and nonphagocytic cells. Infect Immun 61:5231–5236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Beuzon CR, Meresse S, Unsworth KE, Ruiz-Albert J, Garvis S, Waterman SR, Ryder TA, Boucrot E, Holden DW. 2000. Salmonella maintains the integrity of its intracellular vacuole through the action of SifA. EMBO j 19:3235–3249. doi: 10.1093/emboj/19.13.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Snavely MD, Miller CG, Maguire ME. 1991. The mgtB Mg2+ transport locus of Salmonella typhimurium encodes a P-type ATPase. J Biol Chem 266:815–823. [PubMed] [Google Scholar]
- 86.Snavely MD, Gravina SA, Cheung TT, Miller CG, Maguire ME. 1991. Magnesium transport in Salmonella typhimurium. Regulation of mgtA and mgtB expression. J Biol Chem 266:824–829. [PubMed] [Google Scholar]
- 87.Wallis TS, Galyov EE. 2000. Molecular basis of Salmonella-induced enteritis. Mol Microbiol 36:997–1005. doi: 10.1046/j.1365-2958.2000.01892.x. [DOI] [PubMed] [Google Scholar]
- 88.Santos RL, Zhang S, Tsolis RM, Kingsley RA, Adams LG, Baumler AJ. 2001. Animal models of Salmonella infections: enteritis versus typhoid fever. Microbes Infect 3:1335–1344. doi: 10.1016/S1286-4579(01)01495-2. [DOI] [PubMed] [Google Scholar]
- 89.Barthel M, Hapfelmeier S, Quintanilla-Martínez L, Kremer M, Rohde M, Hogardt M, Pfeffer K, Rüssmann H, Hardt WD. 2003. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect Immun 71:2839–2858. doi: 10.1128/IAI.71.5.2839-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Collins LV, Attridge S, Hackett J. 1991. Mutations at rfc or pmi attenuate Salmonella typhimurium virulence for mice. Infect Immun 59:1079–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nevola JJ, Stocker BA, Laux DC, Cohen PS. 1985. Colonization of the mouse intestine by an avirulent Salmonella typhimurium strain and its lipopolysaccharide-defective mutants. Infect Immun 50:152–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kolls JK, Linden A. 2004. Interleukin-17 family members and inflammation. Immunity 21:467–476. doi: 10.1016/j.immuni.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 93.Blaschitz C, Raffatellu M. 2010. Th17 cytokines and the gut mucosal barrier. J Clin Immunol 30:196–203. doi: 10.1007/s10875-010-9368-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mayuzumi H, Inagaki-Ohara K, Uyttenhove C, Okamoto Y, Matsuzaki G. 2010. Interleukin-17A is required to suppress invasion of Salmonella enterica serovar Typhimurium to enteric mucosa. Immunology 131:377–385. doi: 10.1111/j.1365-2567.2010.03310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Behnsen J, Jellbauer S, Wong CP, Edwards RA, George MD, Ouyang W, Raffatellu M. 2014. The cytokine IL-22 promotes pathogen colonization by suppressing related commensal bacteria. Immunity 40:262–273. doi: 10.1016/j.immuni.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Crawford RW, Keestra AM, Winter SE, Xavier MN, Tsolis RM, Tolstikov V, Baumler AJ. 2012. Very long O-antigen chains enhance fitness during Salmonella-induced colitis by increasing bile resistance. PLoS Pathog 8:e1002918. doi: 10.1371/journal.ppat.1002918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Prouty AM, Brodsky IE, Falkow S, Gunn JS. 2004. Bile-salt-mediated induction of antimicrobial and bile resistance in Salmonella typhimurium. Microbiology 150:775–783. doi: 10.1099/mic.0.26769-0. [DOI] [PubMed] [Google Scholar]
- 98.Chevance FF, Hughes KT. 2008. Coordinating assembly of a bacterial macromolecular machine. Nat Rev Microbiol 6:455–465. doi: 10.1038/nrmicro1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yanagihara S, Iyoda S, Ohnishi K, Iino T, Kutsukake K. 1999. Structure and transcriptional control of the flagellar master operon of Salmonella typhimurium. Genes Genet Syst 74:105–111. doi: 10.1266/ggs.74.105. [DOI] [PubMed] [Google Scholar]
- 100.Wang S, Fleming RT, Westbrook EM, Matsumura P, McKay DB. 2006. Structure of the Escherichia coli FlhDC complex, a prokaryotic heteromeric regulator of transcription. J Mol Biol 355:798–808. doi: 10.1016/j.jmb.2005.11.020. [DOI] [PubMed] [Google Scholar]
- 101.Ohnishi K, Kutsukake K, Suzuki H, Iino T. 1990. Gene fliA encodes an alternative sigma factor specific for flagellar operons in Salmonella typhimurium. Mol Gen Genet 221:139–147. doi: 10.1007/BF00261713. [DOI] [PubMed] [Google Scholar]
- 102.Blair DF, Berg HC. 1990. The MotA protein of E. coli is a proton-conducting component of the flagellar motor. Cell 60:439–449. doi: 10.1016/0092-8674(90)90595-6. [DOI] [PubMed] [Google Scholar]
- 103.Berg HC. 2003. The rotary motor of bacterial flagella. Annu Rev Biochem 72:19–54. doi: 10.1146/annurev.biochem.72.121801.161737. [DOI] [PubMed] [Google Scholar]
- 104.Lahteenmaki K, Kyllonen P, Partanen L, Korhonen TK. 2005. Antiprotease inactivation by Salmonella enterica released from infected macrophages. Cell Microbiol 7:529–538. doi: 10.1111/j.1462-5822.2004.00483.x. [DOI] [PubMed] [Google Scholar]
- 105.Nath P, Morona R. 2015. Detection of Wzy/Wzz interaction in Shigella flexneri. 161:1797–1805. doi: 10.1099/mic.0.000132. [DOI] [PubMed] [Google Scholar]
- 106.Carter JA, Jimenez JC, Zaldivar M, Alvarez SA, Marolda CL, Valvano MA, Contreras I. 2009. The cellular level of O-antigen polymerase Wzy determines chain length regulation by WzzB and WzzpHS-2 in Shigella flexneri 2a. Microbiology 155:3260–3269. doi: 10.1099/mic.0.028944-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tocilj A, Munger C, Proteau A, Morona R, Purins L, Ajamian E, Wagner J, Papadopoulos M, Van Den Bosch L, Rubinstein JL, Fethiere J, Matte A, Cygler M. 2008. Bacterial polysaccharide co-polymerases share a common framework for control of polymer length. Nat Struct Mol Biol 15:130–138. doi: 10.1038/nsmb.1374. [DOI] [PubMed] [Google Scholar]
- 108.Gozdziewicz TK, Lugowski C, Lukasiewicz J. 2014. First evidence for a covalent linkage between enterobacterial common antigen and lipopolysaccharide in Shigella sonnei phase II ECALPS. J Biol Chem 289:2745–2754. doi: 10.1074/jbc.M113.512749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kroger C, Dillon SC, Cameron AD, Papenfort K, Sivasankaran SK, Hokamp K, Chao Y, Sittka A, Hebrard M, Handler K, Colgan A, Leekitcharoenphon P, Langridge GC, Lohan AJ, Loftus B, Lucchini S, Ussery DW, Dorman CJ, Thomson NR, Vogel J, Hinton JC. 2012. The transcriptional landscape and small RNAs of Salmonella enterica serovar Typhimurium. Proc Natl Acad Sci U S A 109:E1277–E1286. doi: 10.1073/pnas.1201061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Novoa-Garrido M, Steinum TM, Marolda CL, Valvano MA, Sorum H. 2009. Reduced lipopolysaccharide O antigen expression, increased acid susceptibility and multicellular behaviour in an Escherichia coli isolate after long-term in vitro exposure to formic acid. Microb Ecol Health Dis 21:87–94. [Google Scholar]
- 111.Darwin KH, Miller VL. 1999. InvF is required for expression of genes encoding proteins secreted by the SPI1 type III secretion apparatus in Salmonella typhimurium. J Bacteriol 181:4949–4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Liu D, Cole RA, Reeves PR. 1996. An O-antigen processing function for Wzx (RfbX): a promising candidate for O-unit flippase. J Bacteriol 178:2102–2107. doi: 10.1128/jb.178.7.2102-2107.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Feldman MF, Marolda CL, Monteiro MA, Perry MB, Parodi AJ, Valvano MA. 1999. The activity of a putative polyisoprenol-linked sugar translocase (Wzx) involved in Escherichia coli O antigen assembly is independent of the chemical structure of the O repeat. J Biol Chem 274:35129–35138. doi: 10.1074/jbc.274.49.35129. [DOI] [PubMed] [Google Scholar]
- 114.Marolda CL, Tatar LD, Alaimo C, Aebi M, Valvano MA. 2006. Interplay of the Wzx translocase and the corresponding polymerase and chain length regulator proteins in the translocation and periplasmic assembly of lipopolysaccharide o antigen. J Bacteriol 188:5124–5135. doi: 10.1128/JB.00461-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kaniuk NA, Vinogradov E, Whitfield C. 2004. Investigation of the structural requirements in the lipopolysaccharide core acceptor for ligation of O antigens in the genus Salmonella: WaaL “ligase” is not the sole determinant of acceptor specificity. J Biol Chem 279:36470–36480. doi: 10.1074/jbc.M401366200. [DOI] [PubMed] [Google Scholar]
- 116.Abeyrathne PD, Daniels C, Poon KK, Matewish MJ, Lam JS. 2005. Functional characterization of WaaL, a ligase associated with linking O-antigen polysaccharide to the core of Pseudomonas aeruginosa lipopolysaccharide. J Bacteriol 187:3002–3012. doi: 10.1128/JB.187.9.3002-3012.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hong Y, Reeves PR. 2016. Model for the controlled synthesis of O-antigen repeat units involving the WaaL ligase. mSphere 1:00074–15. doi: 10.1128/mSphere.00074-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Houppert AS, Bohman L, Merritt PM, Cole CB, Caulfield AJ, Lathem WW, Marketon MM. 2013. RfaL is required for Yersinia pestis type III secretion and virulence. Infect Immun 81:1186–1197. doi: 10.1128/IAI.01417-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zwir I, Latifi T, Perez JC, Huang H, Groisman EA. 2012. The promoter architectural landscape of the Salmonella PhoP regulon. Mol Microbiol 84:463–485. doi: 10.1111/j.1365-2958.2012.08036.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Richards SM, Strandberg KL, Conroy M, Gunn JS. 2012. Cationic antimicrobial peptides serve as activation signals for the Salmonella Typhimurium PhoPQ and PmrAB regulons in vitro and in vivo. Front Cell Infect Microbiol 2:102. doi: 10.3389/fcimb.2012.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chakravortty D, Rohde M, Jager L, Deiwick J, Hensel M. 2005. Formation of a novel surface structure encoded by Salmonella pathogenicity island 2. EMBO j 24:2043–2052. doi: 10.1038/sj.emboj.7600676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Osterman IA, Dikhtyar YY, Bogdanov AA, Dontsova OA, Sergiev PV. 2015. Regulation of flagellar gene expression in bacteria. Biochemistry (Mosc) 80:1447–1456. doi: 10.1134/S000629791511005X. [DOI] [PubMed] [Google Scholar]
- 123.Cullen TW, Trent MS. 2010. A link between the assembly of flagella and lipooligosaccharide of the Gram-negative bacterium Campylobacter jejuni. Proc Natl Acad Sci U S A 107:5160–5165. doi: 10.1073/pnas.0913451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hirano T, Yamaguchi S, Oosawa K, Aizawa S. 1994. Roles of FliK and FlhB in determination of flagellar hook length in Salmonella typhimurium. J Bacteriol 176:5439–5449. doi: 10.1128/jb.176.17.5439-5449.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Toguchi A, Siano M, Burkart M, Harshey RM. 2000. Genetics of swarming motility in Salmonella enterica serovar typhimurium: critical role for lipopolysaccharide. J Bacteriol 182:6308–6321. doi: 10.1128/JB.182.22.6308-6321.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wang Q, Frye JG, McClelland M, Harshey RM. 2004. Gene expression patterns during swarming in Salmonella typhimurium: genes specific to surface growth and putative new motility and pathogenicity genes. Mol Microbiol 52:169–187. doi: 10.1111/j.1365-2958.2003.03977.x. [DOI] [PubMed] [Google Scholar]
- 127.Crhanova M, Malcova M, Mazgajova M, Karasova D, Sebkova A, Fucikova A, Bortlicek Z, Pilousova L, Kyrova K, Dekanova M, Rychlik I. 2011. LPS structure influences protein secretion in Salmonella enterica. Vet Microbiol 152:131–137. doi: 10.1016/j.vetmic.2011.04.018. [DOI] [PubMed] [Google Scholar]
- 128.Scott AE, Twine SM, Fulton KM, Titball RW, Essex-Lopresti AE, Atkins TP, Prior JL. 2011. Flagellar glycosylation in Burkholderia pseudomallei and Burkholderia thailandensis. J Bacteriol 193:3577–3587. doi: 10.1128/JB.01385-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 130.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sperandio V, Torres AG, Jarvis B, Nataro JP, Kaper JB. 2003. Bacteria-host communication: the language of hormones. Proc Natl Acad Sci U S A 100:8951–8956. doi: 10.1073/pnas.1537100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Walters M, Sperandio V. 2006. Autoinducer 3 and epinephrine signaling in the kinetics of locus of enterocyte effacement gene expression in enterohemorrhagic Escherichia coli. Infect Immun 74:5445–5455. doi: 10.1128/IAI.00099-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.