Abstract
Purpose
Early prediction of treatment outcomes represents an essential step towards increased treatment efficacy and survival in patients with hepatocellular carcinoma (HCC). In this study, we performed two-dimensional electrophoresis (2-DE) followed by protein profiling to identify biomarkers predictive of therapeutic outcomes in patients with HCC who received liver-directed therapy (LDTx) involving local radiotherapy (RT), and studied the underlying mechanisms of the identified proteins.
Materials and Methods
2-DE analysis was conducted by pooling sera from patients with a good or poor prognosis; serum proteomic profiles of the two groups were compared and analyzed using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Identified proteins were confirmed via enzyme-linked immunosorbent assay. An invasion assay was performed after overexpression and knockdown of target protein in Huh7 cells.
Results
Levels of inter-alpha inhibitor H4 (ITIH4), fibrinogen gamma chain, keratin 9/1 complex, carbonic anhydrase I, and carbonmonoxyhemoglobin S were changed by more than 4-fold in response to LDTx. In particular, pre-LDTx ITIH4 expression was more than 5-fold higher in patients with a good prognosis, compared to patients with a poor prognosis. The migration ability of Huh7 cells was significantly suppressed and enhanced by ITIH4 overexpression and knockdown, respectively. The tumors of patients with HCC and a good prognosis expressed high levels of ITIH4, compared to those of patients with a poor prognosis.
Conclusion
Taken together, ITIH4 may be a potential therapeutic target that could inhibit cancer metastasis, as well as a prognostic marker for patients with HCC who are receiving LDTx.
Keywords: Prognosis, Hepatocellular carcinoma, Liver-directed therapy, ITIH4, Biomarkers
Introduction
Hepatocellular carcinoma (HCC), which has a high rate of recurrence and unsatisfactory treatment outcomes, is a critical global health issue [1,2]. To achieve better oncologic outcomes, a personalized approach involving the prediction of therapeutic results, followed by application, has been proposed [3,4] and has led to suggestions of the significance of biomarkers. The concept of biomarkers is not new in the field of oncology. These markers, which are defined as biological molecules found in the blood, other body fluids, or tissues that indicate a normal or abnormal process, have been extensively studied in various cancers [5,6]. However, investigative studies of biomarkers for HCC are complex because of the molecular diversity of HCC itself, as well as associated conditions related to inflammation and cirrhosis that distinguish HCC from other solid malignancies.
Alpha-fetoprotein (AFP) is a well-known prognostic marker of HCC, and elevated levels of this protein correlate with an advanced disease stage and aggressiveness in approximately 70% of cases [7]. Nevertheless, AFP does not yield satisfactory predictions of treatment outcomes, particularly in cases of AFP-negative HCC.
Active searches for biomarkers have been performed recently with the aims of developing and applying these concepts to clinical trials of biomarker-based systemic agents [8]. Although this approach is valuable, biomarkers predictive of clinical outcomes also seem important for patients with HCC, particularly those with localized disease but a heterogeneous prognosis. The development of high-throughput serum proteomics techniques has made it possible to identify serum biomarkers of HCC [9].
In this study, we investigated possible biomarkers that could predict the clinical outcomes of patients with HCC, particularly those with locally advanced disease. Analysis of sera from patients with HCC was performed to two-dimensional electrophoresis (2-DE) followed by the identification of possible biomarkers via protein profiling; in addition, the underlying mechanisms of these biomarkers were elucidated.
Materials and Methods
1. Preparation of serum samples from patients with HCC
This prospectively conducted study conformed to the ethical guidelines of the Declaration of Helsinki; it was approved by the Institutional Review Board of Yonsei Severance Hospital (4-2008-0212). Between September 2008 and December 2009, a total of 51 patients treated with liver-directed therapy (LDTx) involving local radiotherapy (RT; median dose, 45 Gy in 5 weeks) concurrently with hepatic arterial infusion chemotherapy with 5-fluorouracil, as described in a previous report [10], were prospectively enrolled in this study. Blood samples were collected before the start and at the end of the RT schedule. We selected patients for inclusion in good (n=10) and poor prognosis groups (n=10). The 10 patients in the good prognosis group did not exhibit treatment failure during the follow-up period, whereas those in the poor prognosis group exhibited early treatment failure after the completion of RT. Treatment failures were defined as follows: local failure, recurrence at the tumor site; regional failure, recurrence in the liver that was not treated with RT; and distant metastasis, recurrence outside the liver.
2. Depletion of majorly abundant proteins using an immunoaffinity column
The six most abundant proteins (albumin, transferrin, IgG, IgA, haptoglobin, and antitrypsin) in serum or plasma were depleted using a Multiple Affinity Removal Column (MARC; Agilent, Wilmington, DE). A 4.6-mm×50-mm MARC with a binding capacity for 20 μL of human plasma was used. Chromatographic separation of the abundant proteins by MARC was performed using a mobile phase reagent kit according to a standard LC protocol provided by the manufacturer. Briefly, each crude human serum and plasma sample was diluted five times with Buffer A containing protease inhibitors (COMPLETE, Roche, Basel, Switzerland) and filtered through a 0.22-μm spin filter via centrifugation at 16,000 ×g and room temperature for 1-2 minutes. The sample was injected into the MARC, and flow-through fractions were collected and stored at ‒20°C until use. To resolve depleted plasma proteins on 2-DE gels, flow-through fractions from MARC were pooled and precipitated with a precooled solution of 10% trichloroacetic acid for 1 hour at ‒20°C. After washing with ice-cold acetone, pellets were resolublized in 2-DE sample buffers.
3. 2-DE and image analysis
2-DE was performed as described previously. First, aliquots in sample buffer (7 M urea, 2 M thiourea, 4.5% CHAPS, 100 mM DTE, 40 mM Tris, pH 8.8) were applied to immobilized pH 3-10 nonlinear gradient strips (Amersham Biosciences, Uppsala, Sweden). Isoelectric focusing was performed at 80,000 Vh. The second dimension was analyzed on 9%-16% linear gradient polyacrylamide gels (18 cm×20 cm× 1.5 mm) at a constant 40 mA per gel for approximately 5 hours. After protein fixation in 40% methanol and 5% phosphoric acid for 1 hour, the gels were stained with CBB G-250 for 12 hours. The gels were subsequently destained with H2O, scanned using a Bio-Rad GS710 densitometer (Richmond, CA), and converted into electronic files that were analyzed using the Image Master Platinum 5.0 image analysis program (Amersham Biosciences).
4. Matrix-assisted laser desorption/ionization time-of-flight
Protein spots were excised from gels with a sterile scalpel and placed into Eppendorf tubes. Proteins were digested with trypsin (Promega, Madison, WI) as previously described [11]. For matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (MS) analysis, peptides were concentrated using a POROS R2, Oligo R3 column (Applied Biosystems, Foster City, CA). After washing the column with 70% acetonitrile, 100% acetonitrile, and 50 mM ammonium bicarbonate, samples were applied to the R2, R3 column and eluted with cyano-4-hydroxycinamic acid (Sigma, St. Louis, MO) in 70% acetonitrile and 2% formic acid onto the MALDI plate (Opti-TOF 384-well Insert, Applied Biosystems) [12]. MALDI-TOF MS analysis was performed on a 4800 MALDI-TOF/TOF Analyzer (Applied Biosystems) equipped with a 355-nm Nd:YAG laser. The pressure in the TOF analyzer was approximately 0.00010132 Pa. Mass spectra were obtained in reflectron mode with an accelerating voltage of 20 kV and sum of 500 laser pulses, and calibrated using the 4700 calibration mixture (Applied Biosystems). Data Explorer 4.4 (PerSeptive Biosystems, Framingham, MA) was used for monoisotopic mass data acquisition and extraction.
5. Mascot database search
The mascot algorithm (Matrix Science, Inc., Boston, MA) was used to identify peptide sequences present in a protein sequence database. The following database search criteria were used: taxonomy, Homo sapiens(NCBInr database downloaded on Nov 1, 2012); fixed modification, carbamidomethylated (+57) at cysteine residues, variable modification, and oxidized (+16) at methionine residues; maximum allowed missed cleavage, 1; and MS tolerance, 100 ppm. Only peptides yielded by trypsin digestion were considered.
6. Serum levels of inter-alpha inhibitor 4 in patients with HCC
Serum inter-alpha inhibitor 4 (ITIH4) levels were measured using an enzyme-linked immunosorbent assay (ELISA) according to the manufacturer's instructions (Cloud-Clone Corp., Houston, TX). Serum samples were collected < 1 week before and after the completion of LDTx.
7. Cell culture
The human HCC cell line Huh7 was maintained in Dulbecco’s minimal essential medium (GIBCO, Carlsbad, CA) supplemented with 10% (v/v) fetal bovine serum (GIBCO), 100 U/mL penicillin, and 100 μg/mL streptomycin (GIBCO) at 37°C in a humidified atmosphere of 5% CO2 and 95% air.
8. Expression constructs
For ITIH4 knockdown, pLKO-shITIH4 control and MISSION shRNA clones were purchased from Sigma-Aldrich (SHCLNG-NM_002218).
For ITIH4 overexpression, a pCNS-D2 human ITIH4 mammalian expression vector and control mock vector were purchased from the Korea human gene bank (http://genbank.kribb.re.kr/).
9. In vitro gene transfection
For lentivirus production and transduction, 60%-90% confluent 293T cells in antibiotic-free medium were transfected with pLKO-shITIH4 control or gene-specific shRNA clones along with packaging plasmids. After 24 hours, medium containing transfection reagent was replaced with fresh medium containing antibiotics. At 48-hour post-transfection, cultured medium harvest and transduction in Huh7 pLKO-shITIH4 control and MISSION shRNA clones lentiviral virion. Transduced cells were selected in medium containing 10 μg/mL of puromycin (GIBCO) for 6 days, and individual puromycin-resistant clones were isolated. These clones were propagated and screened for ITIH4 expression via reverse transcription‒polymerase chain reaction (RT-PCR). Stable knockdown Huh7 clones were maintained in medium containing 5 μg/mL of puromycin.
To generate ITIH4 overexpressing cells, Huh7 cells were cultured to 70%-80% confluence in 60-mm dishes. Cells were transfected with the empty or ITIH4 expression plasmid using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) in accordance with the manufacturer’s recommendations. Briefly, in each dish, plasmid DNA (4 μg) and 12 μL of Lipofectamine were separately diluted in 100 μL Opti-MEM media, mixed, and incubated at room temperature for 20 minutes. The cells were refreshed with Opti-MEM media after removal of the regular supplemented media. The mixed DNA-Lipofectamine complexes were added to each dish containing cells. The cells were incubated at 37°C for 8 hours to facilitate transfection, after which growth media containing 10% serum was added. After transfection, cells were selected in medium containing 20 μg/mL of blasticidin (GIBCO) for 6 days, and individual blasticidin-resistant clones were isolated. These clones were propagated and screened for ITIH4 expression via RT-PCR. Stable Huh7 overexpressing clones were maintained in medium containing 5 μg/mL of puromycin.
10. RT-PCR
Total RNA was extracted from Huh7 cells using TRIzol reagent (Life Technologies, Carlsbad, CA). One microgram of total RNA was reverse transcribed to cDNA using an Omniscript RT Kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. cDNA (1 μL) was subsequently amplified via polymerase chain reaction (PCR) using a TaKaRa PCR Thermal Cycler Dice Gradient (Takara Bio Inc., Shiga, Japan). The following PCR protocol was used: a 4-minute denaturation step at 94°C and 29 cycles comprising 30 seconds of denaturation at 94°C, 15 seconds of annealing at 57°C, and 30 seconds of elongation at 72°C. The following primers were used: ITIH4, 5′-GGAGCAAACTGTCTCCGCAT-3′ and 5′-AGAAAGTGG AACCTGAGTGGAC-3′; GAPDH, 5′-GGCTGCTTTTAACTCTGGTA-3′ and 5′-ACTTGATTT TGGAGGGATCT-3′.
11. Cell viability assay
Cell viability was evaluated using the using EZ-Cytox Cell Viability Assay WST-1 Kit (Daeillab Service, Seoul, Korea) according to the manufacturer’s instructions. Briefly, 100 μL of cell suspension (1×104 cells per well) were added to each well of a 96-well plate and cultured overnight. Next, 10 μL of EZ-Cytox solution was added to each well, followed by a 2-hour incubation at 37°C. Absorbance was measured by spectrophotometry (VersaMax ELISA Microplate Reader, Molecular Devices, Eugene, OR) at 450 nm. Cell viability was determined as a percentage by comparing the absorbance of transfected and normal Huh7 cells.
12. In vitro migration assay
The invasiveness of stable shITIH4 knockdown or ITIH4 overexpressing cells was evaluated in 24-well chambers. The invasion assay was performed using transwell cell culture chambers (BD Biosciences, Bedford, MA) according to the manufacturer's instructions. Briefly, a transwell with an 8-μm-diameter pore membrane was coated with 20 μL of collagen (500 μg/mL) and incubated overnight. Cells (2×104) in 200 μL of serum-free medium were seeded into the upper chamber of each transwell, and the lower chamber was filled with 1 mL of RPMI-1640 containing 10% fetal bovine serum to induce chemotaxis. After a 24-hour incubation period at 37°C in a humidified 5% CO2 atmosphere, the cells were fixed in methanol and stained with hematoxylin and eosin, and cells that had invaded through the pores to the lower surfaces of the filters were counted under a microscope. Values were calculated by averaging the total numbers of cells from three different, randomly selected fields.
13. ITIH4 expression in tumors of patients with HCC who received LDTx
Tumor tissues were obtained from patients with HCC who underwent surgical resection at Severance Hospital. To investigate ITIH4 expression, specimens of liver tumors and normal liver tissues were biopsied from liver cancer patients. To generate frozen sections, tumor samples fixed in 4% formalin were snap-frozen over liquid nitrogen in Tissue-Tek OCT compound (Sakura Finetek, Tokyo, Japan) and cryosectioned (15 μm). Microwave antigen retrieval was performed in citrate buffer (pH 6.0). Specimens were blocked in 3% hydrogen peroxide for 15 minutes to inactivate peroxidases, followed by washing in phosphate-buffered saline and blocking for 1 hour with 3% goat serum at room temperature. Samples were incubated with a polyclonal mouse antibody against ITIH4 (1:100, CL1858, Novus Biologicals, Littleton, CO) and a biotinylated secondary antibody (Dako, Carpinteria, CA). For visualization, samples were incubated with peroxidase-labeled streptavidin (Dako) and 3,3′-diaminobenzidine (Dako, Glostrup, Denmark). Slides were subsequently counterstained with hematoxylin.
14. Statistical analysis
The statistical significances of differences in the mean ITIH4 according to patient and tumor characteristics were determined using chi-square test and Mann-Whitney U test. Survival rates were evaluated using the Kaplan-Meier method, and correlations of between failure-free survival and clinical factors as continuous variables were analyzed using a Cox hazard proportional model. A Cox stepwise regression model was used for a multivariate analysis.
Results
1. Study population and characteristics
Among the 51 treated patients, 10 representative patients each with a good prognosis or poor prognosis were selected according to clinical outcomes after LDTx (Fig. 1). The characteristics of the two study groups are presented in Table 1. The median ages of the good and poor prognosis groups were 63 years (range, 40 to 75 years) and 51 years (range, 39 to 72 years), respectively. Most patients had International Union Against Cancer stage II, III, and IVA HCC at the time of diagnosis. Seven patients in the good prognosis group (70%) and six patients in the poor prognosis group (60%) were treatment-naïve before LDTx. Treatment-non-naïve patients underwent transarterial chemoembolization or transarterial chemoinfusion before LDTx. The median serum AFP levels of patients in the good and poor prognosis groups were 2,639 ng/mL (range, 1.85 to 83,000 ng/mL) and 13,902 ng/mL (range, 5.08 to 83,000 ng/mL), respectively. The pre-LDTx levels of protein induced by vitamin K antagonist-II in the same groups were 336 mAU/mL (range, 19 to 2,000 mAU/mL) and 1,121 mAU/mL (range, 37 to 2,000 mAU/mL), respectively. The corresponding median total prescribed RT doses were 50 Gy (range, 45 to 54 Gy) and 49.3 Gy (range, 45 to 54 Gy), respectively. The treatment failure patterns of each study group are described in Table 2. No treatment failures were reported in the good prognosis group during follow-up. In the poor prognosis group, although local failures occurred in three patients, most treatment failures were regional failures (9/10) or distant metastasis (10/10), and poor prognosis group showed worse overall survival at 1 year and 2 years than good prognosis group (20% vs. 40%, 10% vs. 40%).
Fig. 1.
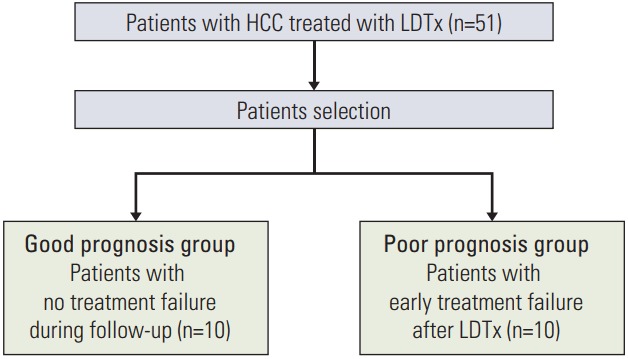
Flow chart of patient selection. HCC, hepatocellular carcinoma; LDTx, liver-directed therapy.
Table 1.
Characteristics of patients
| Characteristic | Good prognosis (n=10) | Poor prognosis (n=10) | p-value |
|---|---|---|---|
| Age, median (range, yr) | 63 (40-75) | 51 (39-72) | 0.492 |
| Sex | |||
| Male | 9 | 8 | > 0.99 |
| Female | 1 | 2 | |
| Viral type | |||
| B | 5 | 10 | 0.033 |
| C | 2 | 0 | |
| NBNC | 3 | 0 | |
| Child-Pugh class | |||
| A | 10 | 9 | > 0.99 |
| B | 0 | 1 | |
| Previous treatment | |||
| No | 7 | 6 | > 0.99 |
| Yes | 3 | 4 | |
| Stage | |||
| II | 3 | 4 | > 0.99 |
| III | 4 | 4 | |
| IVA | 3 | 2 | |
| AFP, median (range, ng/mL) | 2,639 (1.85 to > 10,043) | 13,902 (5.08 to > 10,043) | 0.314 |
| PIVKA-II, median (range, mAU/mL) | 336 (19 to > 2,000) | 1,121 (37 to > 2,000) | 0.661 |
NBNC, non-B non-C; AFP, α-fetoprotein; PIVKA-II, protein induced by vitamin K absence-II.
Table 2.
Treatment failure patterns of patients
| Patients (n=10) | Local failure | PFS | Intrahepatic failure | PFS | Extra-hepatic failure | PFS |
|---|---|---|---|---|---|---|
| Good prognosis | ||||||
| P1 | No | 26.4 | No | 26.4 | No | 26.4 |
| P2 | No | 60.4 | No | 60.4 | No | 60.4 |
| P3 | No | 60.9 | No | 60.9 | No | 60.9 |
| P4 | No | 3.5 | No | 3.5 | No | 3.5 |
| P5 | No | 12 | No | 12 | No | 12 |
| P6 | No | 4.4 | No | 4.4 | No | 4.4 |
| P7 | No | 7.3 | No | 7.3 | No | 7.3 |
| P8 | No | 6.5 | No | 6.5 | No | 6.5 |
| P9 | No | 5.8 | No | 5.8 | No | 5.8 |
| P10 | No | 51 | No | 51 | No | 51 |
| Poor prognosis | ||||||
| P1 | No | 2.7 | Yes | 1.1 | Yes | 0.1 |
| P2 | Yes | 7.7 | Yes | 3.4 | Yes | 0 |
| P3 | No | 8.6 | No | 8.6 | Yes | 0 |
| P4 | No | 9 | Yes | 1 | Yes | 1 |
| P5 | No | 24.5 | Yes | 0 | Yes | 15.4 |
| P6 | Yes | 0.9 | Yes | 0.9 | Yes | 0.9 |
| P7 | Yes | 6.1 | Yes | 0 | Yes | 4.9 |
| P8 | No | 5.7 | Yes | 3.2 | Yes | 0 |
| P9 | No | 7.4 | Yes | 3.1 | Yes | 0 |
| P10 | No | 17.8 | Yes | 0 | Yes | 8.5 |
Local failure, failure in the tumor site; PFS, progression-free survival; P, patient.
2. 2-DE assessment of differentially expressed proteins in sera of patients with HCC according to prognosis
To identify a useful post-LDTx prognostic serum biomarker, pre- and post-LDTx serum samples were collected from each patient and pooled in both study groups. The six most abundant serum proteins were removed using a MARS column as described above to facilitate the detection of low-abundance proteins. Following 2-DE, 481 and 317 spots were detected in pre- and post-LDTx samples, respectively, from the good prognosis group, of which 41 exhibited significant (more than 2-fold) differences in expression between pre- and post-LDTx levels. In the poor prognosis group, 362 and 357 spots were detected in pre- and post-LDTx samples, respectively, and 33 spots were found to be significantly differentially expressed (more than 2-fold) between pre- and post-LDTx samples (Fig. 2).
Fig. 2.
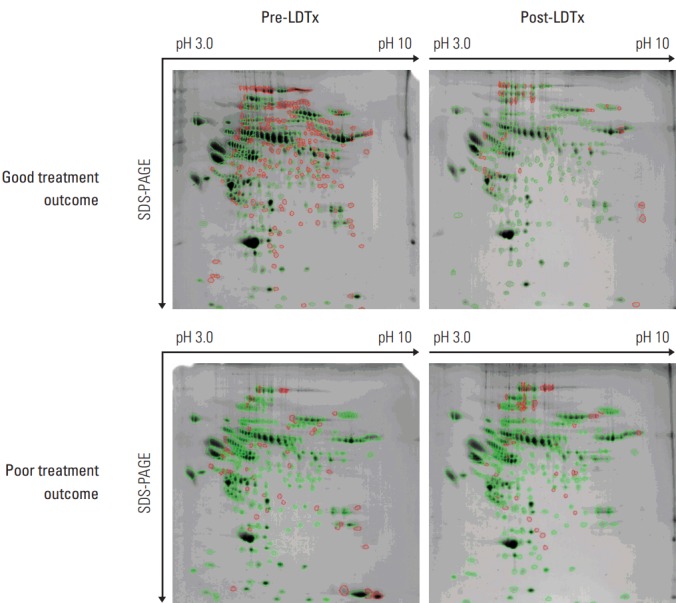
Differentially expressed proteins identified in pre- and post-LDTx samples according to treatment outcomes in patients with hepatocellular carcinoma. Protein spots were identified using two-dimensional electrophoresis. Green and red circles indicate paired and non-paired spots, respectively. LDTx, liver-directed therapy; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis.
3. MALDI-TOF analysis of differentially expressed protein spots
All 74 differentially expressed proteins identified by comparing the pre- and post-LDTx samples of the good and poor prognosis groups were selected via imaging analysis. Subsequently, 30 proteins were identified by MALDI-TOF/TOF MS from among these differentially expressed protein spots, and a NCBI Reference Sequence human protein databases search was conducted (Table 3). As shown in Table 3, 4 of these proteins—ITIH4, inter-alpha-trypsin inhibitor heavy chain H4 isoform 2 precursor, keratin 9/1 complex, and fibrinogen gamma chain—were expressed in post-LDTx samples at levels 4-fold higher or more than those in pre-LDTx samples from the good prognosis group. In contrast, 3 proteins—carbonic anhydrase I (CA1), unnamed protein, and human carbonmonoxyhemoglobin S—were downregulated by more than 4-fold in post-LDTx samples from the poor prognosis group, compared to pre-LDTx samples. In particular, the pre-LDTx level of ITIH4 in the good prognosis group was more than 5-fold higher than that in the poor prognosis group (data not shown). These results suggest that ITIH4 may be an important biomarker that could predict better treatment outcomes of patients receiving LDTx for HCC.
Table 3.
Serum biomarkers identified via comparison of pre- and post-LDTx in patients with HCC who had a good or poor prognosis
| Patient group | Protein No. | Protein name | Fold change | Category | p-value |
|---|---|---|---|---|---|
| Good prognosis | 258 | Inter-alpha (globulin) inhibitor H4 (plasma kallikrein-sensitive glycoprotein) | 6.1 | Cancer | < 0.05 |
| 278 | Inter-alpha-trypsin inhibitor heavy chain H4 isoform 2 precursor | 5.7 | Cancer | ||
| 400 | Gi|119581148 (keratin 9)+gi|186772 (keratin 1) | 4.6 | Cancer | < 0.05 | |
| 427 | Fibrinogen gamma chain, isoform CRA_j | 5.4 | Cancer | ||
| Poor prognosis | 2,353 | Chain B, X ray structure of the complex between carbonic anhydrase I and the phosphonate antiviral (CA1) | ‒4.7 | Metabolism | < 0.05 |
| 2,412 | Unnamed protein product | ‒5.5 | Cancer | ||
| 2,477 | Chain D, crystalline human carbonmonoxy hemoglobin S | ‒19.9 | Metabolism | < 0.05 |
LDTx, liver-directed therapy; HCC, hepatocellular carcinoma.
Table 4.
ELISA confirmation using serum samples from each patient in the good prognosis group
| Patient | Good prognosis (ITIH4) |
|||
|---|---|---|---|---|
| Aa) (ng/mL) | Bb) (ng/mL) | B-A (ng/mL) | Fold change | |
| P1 | 47.9 | 253.3 | 205.4 | 5.3 |
| P2 | 84.6 | 82.5 | ‒2.1 | 1 |
| P3 | 12.8 | 244 | 231.1 | 19 |
| P4 | 34 | 201.1 | 167.1 | 5.9 |
| P5 | 51.9 | 182.1 | 130.2 | 3.5 |
| P6 | 53.1 | 214.6 | 161.5 | 4 |
| P7 | 56.4 | 206.4 | 150 | 3.7 |
| P8 | 13.4 | 176.1 | 162.7 | 13.1 |
| P9 | 35.2 | 40.3 | 5.2 | 1.1 |
| P10 | 36.8 | 217.4 | 180.6 | 5.9 |
| Average | 42.6 | 181.8 | 139.2 | 6.3 |
ELISA, enzyme-linked immunosorbent assay; ITIH4, inter-alpha inhibitor 4; P, patient.
Pre-radiotherapy,
Post-radiotherapy.
4. Up-regulation of serum ITIH4 in post-LDTx level compare to pre-LDTx of patients with good prognosis
To confirm the 2-DE results and further validate ITIH4 as a possible prognostic biomarker of HCC, serum samples from all patients with a good prognosis were subjected to ELISA. The mean serum ITIH4 concentration was significantly higher in post-LDTx samples when compared with pre-LDTx samples (181.8±20.39 pg/mL vs. 42.6±5.78 pg/mL) (Table 4). Most patients in the good prognosis group exhibited a post-LDTx increase in ITIH4 levels relative to the pre-LDTx levels, consistent with the 2-DE analysis results.
It may cause a question on whether ITIH4 could be simply hepatitis B virus (HBV)‒associated protein. However, in a separate experiment on patients with diverse viral background (B, C, and non-B non-C) all patients showed increased postTx ITIH4 comparing to preTx regardless of viral type, suggesting ITIH4 could be an independent moiety from B-virus (S1 Fig.).
5. ITIH4 as a suppressor of migration and metastasis in HCC
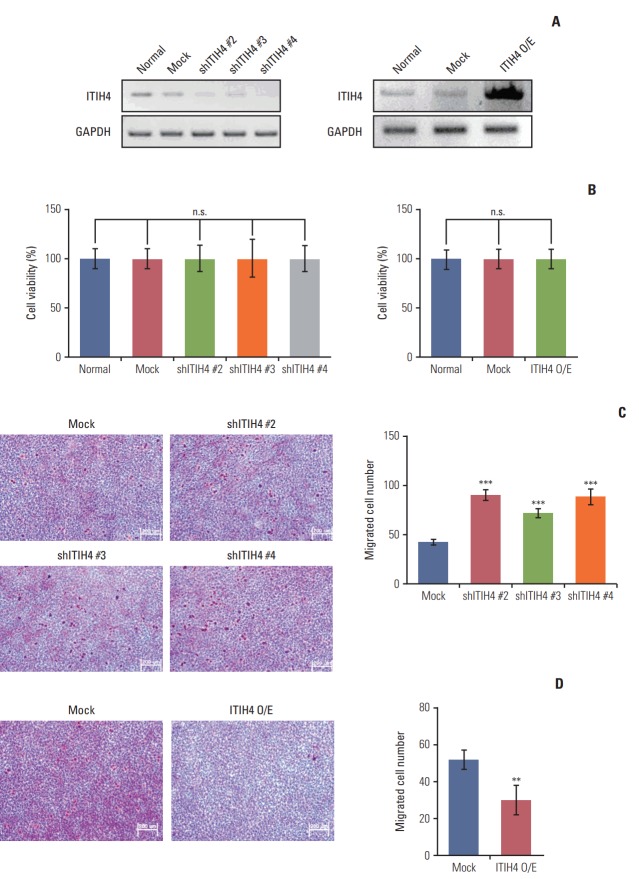
To investigate the role of ITIH4 in HCC, we generated ITIH4 knockdown Huh7 cells (shITIH4) or ITIH4 overexpressing Huh7 cells (ITIH4 O/E) (Fig. 3A) and subjected these cells to a transwell invasion assay. ITIH4 knockdown or overexpression did not affect cell viability (Fig. 3B). As shown in Fig. 3C, the invasiveness of Huh7 cells was increased by ITIH4 knockdown. In consistent with this result, ITIH4 overexpression reduced the invasiveness of Huh7 cells relative to controls (Fig. 3D). These results indicate that ITIH4 can suppress the invasion and migration of HCC.
Fig. 3.
Inter-alpha inhibitor 4 (ITIH4) inhibited the migration of Huh7 cells. (A) ITIH4 mRNA expression in Huh7 cells. ITIH4 was downregulated in Huh7 cells infected with ITIH4 shRNA lentiviral vectors. For ITIH4 overexpression, Huh7 cells were transfected with the ITIH4 pCNS-D2 vector. (B) Cell viability of ITIH4 knockdown or overexpressing Huh7 cells. (C) Migration ability increased in response to ITIH4 downregulation in Huh7 cells (×200). (D) Migration ability decreased in ITIH4 overexpressing Huh7 cells (×200). Data are representative of the means and standard deviations of three independent experiments. n.s., not significant. **p < 0.01, ***p < 0.001 compared to control.
6. ITIH4 expression in patients’ tumor tissue
Finally, we examined ITIH4 expression in tumor tissues from patients with HCC. As shown in Fig. 4A, ITIH4 expression in tumor tissues was significantly higher than that in normal liver tissues from the same patient. As demonstrated by the examples in Fig. 4B, higher ITIH4 expression was observed in a patient with a good prognosis (patient 1), compared to the patient with a poor prognosis (patient 2). These results supported other results in this study on the role of ITIH4 as a prognostic marker for HCC.
Fig. 4.
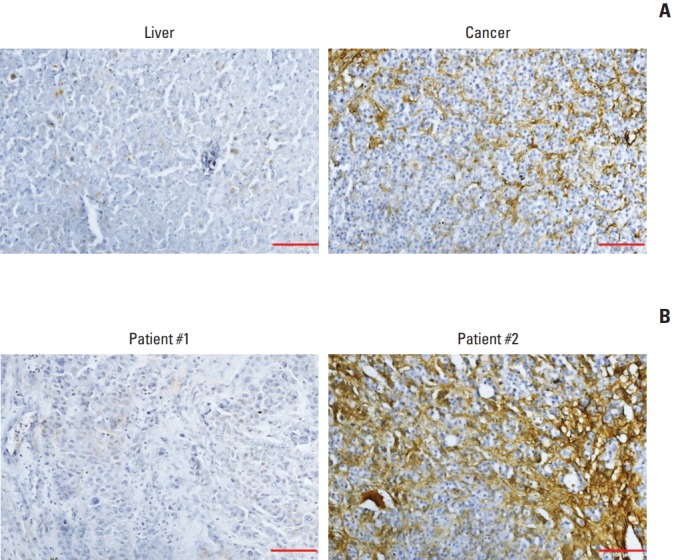
Immunohistochemical analysis of inter-alpha inhibitor H4 (ITIH4) in surgical specimens of human hepatocellular carcinoma. (A) ITIH4 expression was significantly higher in cancer tissues than in non-tumor liver tissues (×200). (B) Higher levels of ITIH4 were detected in the tumors of patients with a good prognosis (representative: patient 1), compared with tumors of patients with a poor prognosis (representative: patient 2) (×200).
Discussion
In the present study, we performed a 2-DE analysis to search for potential serum-based biomarkers that could predict the treatment outcomes of patients with HCC after LDTx. We identified four proteins—ITIH4, inter-alpha-trypsin inhibitor heavy chain H4 isoform 2 precursor, keratin 9/1 complex, and fibrinogen gamma chain—for which the post-LDTx expression levels were more than 4-fold higher than the pre-LDTx levels in the good prognosis group. In the poor prognosis group, we identified three proteins—CA1, unnamed protein, and human carbonmonoxy hemoglobin S—for which the post-LDTx levels decreased by more than 4-fold relative to the pre-LDTx levels. Moreover, pre-LDTx levels of ITIH4 were more than 5-fold higher in patients with a good prognosis, compared to those with a poor prognosis. We selected three proteins—ITIH4, keratin 9/1, and CA1—that are related to cancer progression as well as present high variation among the identified total proteins for validation as useful biomarkers. Serum levels of the selected-target proteins were evaluated in each patient via ELISA. As results, ITHI4 exhibited the most consistency as a potential prognostic factor for HCC treatment than other candidate proteins when all results were combined. The keratin 9/1 ELISA results were not consistent with the 2-DE analysis results. In patients with a poor prognosis, post-LDTx serum CA1 levels decreased by an average of two fold relative to pre-LDTx levels (data not shown). Furthermore, patients with HCC and a good prognosis exhibited higher levels of ITIH4 in tumors, compared the levels in tumors from patients with a poor prognosis.
ITIH4 is a member of a liver-restricted serine protease inhibitor. This protein belongs to the ITIH family, the members of which are found at relatively high concentrations in human plasma [13]. ITIH family members have been reported to contribute to extracellular matrix stability and have been shown to be involved in processes such as tumor cell migration and metastasis [14]. Hamm et al. [15] reported that multiple human solid cancers frequently exhibited ITIH4 downregulation, which might be associated with the initiation and progression of these malignancies.
Indeed, we found that ITIH4 overexpression could inhibit the invasiveness of Huh7 cells, whereas knockdown of ITIH4 increased the migration of these cells. These results indicate that ITIH4 plays an important role in the invasiveness and migration of HCC.
Recently, ITIH4 was identified as a valuable diagnostic indicator in patients with HBV-associated HCC. The expression of this protein is significantly lower in patients with HBV-associated HCC than in those with liver cirrhosis and hepatitis [16]. ITIH4 is mainly expressed in the liver and is secreted into the blood [17]. In this study, ITIH4 was expressed at higher levels in tumors than in normal liver tissues, suggesting that serum ITIH4 levels are mainly tumor-derived. ITIH4 was also expressed at significantly higher levels in the tumors of patients with HCC who had a good prognosis, compared to those with a poor prognosis. In addition, most patients with a poor prognosis having low levels of ITIH4 exhibited worse outcomes in terms of either intrahepatic or extrahepatic failure-free survival in this study. This finding suggests that ITIH4 identified via serum protein profiling could serve as a potential prognostic factor in LDTx-treated patients with HCC.
However, this study has some limitations, including the small number of patients who were treated with LDTx and the lack of validation in patients with HCC at different disease stages. Also, it is unclear whether ITIH4 is the marker predicting prognosis just in patients who receive radiation therapy or whether the marker can generally predict the prognosis in HCC patients. Since ITIH4 was identified as the marker predicting prognosis through comparative analysis between pre-RT and post-RT in patients with liver cancer, ITIH4 seems to be a biomarker to predict prognosis in patients receiving RT. Nevertheless, as mentioned above, ITIH4 was a diagnostic and prognostic indicator in HBV-associated HCC. Accordingly, ITIH4 might be a biomarker in HCC patients who don’t receive RT as well as HCC patients who receive RT. In our study, preTx level of ITIH4 was higher in the good prognosis group compared to poor prognosis, suggesting that ITIH4 per se would be a biomarker regardless of the treatment. Therefore, additional large-scale studies are needed to confirm regarding this issue in various HCC stages.
In conclusion, we identified ITIH4 as a novel biomarker that could predict clinical outcomes in patients with locally advanced HCC who were receiving LDTx. Given its involvement in tumor progression and invasion, as well as metastasis, serum ITIH4 appears to be an important mediator related to the clinical outcomes of locally advanced HCC.
Acknowledgments
This work was supported by the Mid-career Researcher Program (NRF-2014R1A2A1A11054463) through the National Research Foundation of Korea funded by the Ministry of Science.
Footnotes
Conflict of interest relevant to this article was not reported.
Electronic Supplementary Material
Supplementary materials are available at Cancer Research and Treatment website (http://www.e-crt.org).
References
- 1.Park JW, Koh YH, Kim HB, Kim HY, An S, Choi JI, et al. Phase II study of concurrent transarterial chemoembolization and sorafenib in patients with unresectable hepatocellular carcinoma. J Hepatol. 2012;56:1336–42. doi: 10.1016/j.jhep.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 2.Llovet JM, Fuster J, Bruix J, Barcelona-Clinic Liver Cancer Group The Barcelona approach: diagnosis, staging, and treatment of hepatocellular carcinoma. Liver Transpl. 2004;10(2 Suppl 1):S115–20. doi: 10.1002/lt.20034. [DOI] [PubMed] [Google Scholar]
- 3.Duffy MJ, Crown J. A personalized approach to cancer treatment: how biomarkers can help. Clin Chem. 2008;54:1770–9. doi: 10.1373/clinchem.2008.110056. [DOI] [PubMed] [Google Scholar]
- 4.Baumann M, Krause M, Overgaard J, Debus J, Bentzen SM, Daartz J, et al. Radiation oncology in the era of precision medicine. Nat Rev Cancer. 2016;16:234–49. doi: 10.1038/nrc.2016.18. [DOI] [PubMed] [Google Scholar]
- 5.National Cancer Institute . Bethesda, MD: National Cacner Institute; 2015. NCI dictionary of cancer terms [Internet] [cited 2015 Apr 6]. Available from: https://www.cancer.gov/publications/dictionaries/cancer-terms?cdrid=45618. [Google Scholar]
- 6.Sargent DJ, Conley BA, Allegra C, Collette L. Clinical trial designs for predictive marker validation in cancer treatment trials. J Clin Oncol. 2005;23:2020–7. doi: 10.1200/JCO.2005.01.112. [DOI] [PubMed] [Google Scholar]
- 7.Bialecki ES, Di Bisceglie AM. Diagnosis of hepatocellular carcinoma. HPB (Oxford) 2005;7:26–34. doi: 10.1080/13651820410024049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henry NL, Hayes DF. Cancer biomarkers. Mol Oncol. 2012;6:140–6. doi: 10.1016/j.molonc.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao HJ, Chen YJ, Zuo D, Xiao MM, Li Y, Guo H, et al. Quantitative proteomic analysis for high-throughput screening of differential glycoproteins in hepatocellular carcinoma serum. Cancer Biol Med. 2015;12:246–54. doi: 10.7497/j.issn.2095-3941.2015.0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim KH, Kim MS, Chang JS, Han KH, Kim DY, Seong J. Therapeutic benefit of radiotherapy in huge (≥10 cm) unresectable hepatocellular carcinoma. Liver Int. 2014;34:784–94. doi: 10.1111/liv.12436. [DOI] [PubMed] [Google Scholar]
- 11.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–8. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 12.Choi BK, Cho YM, Bae SH, Zoubaulis CC, Paik YK. Single-step perfusion chromatography with a throughput potential for enhanced peptide detection by matrix-assisted laser desorption/ionization-mass spectrometry. Proteomics. 2003;3:1955–61. doi: 10.1002/pmic.200300558. [DOI] [PubMed] [Google Scholar]
- 13.Chandler KB, Brnakova Z, Sanda M, Wang S, Stalnaker SH, Bridger R, et al. Site-specific glycan microheterogeneity of inter-alpha-trypsin inhibitor heavy chain H4. J Proteome Res. 2014;13:3314–29. doi: 10.1021/pr500394z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kobayashi H, Gotoh J, Hirashima Y, Fujie M, Sugino D, Terao T. Inhibitory effect of a conjugate between human urokinase and urinary trypsin inhibitor on tumor cell invasion in vitro. J Biol Chem. 1995;270:8361–6. doi: 10.1074/jbc.270.14.8361. [DOI] [PubMed] [Google Scholar]
- 15.Hamm A, Veeck J, Bektas N, Wild PJ, Hartmann A, Heindrichs U, et al. Frequent expression loss of inter-alpha-trypsin inhibitor heavy chain (ITIH) genes in multiple human solid tumors: a systematic expression analysis. BMC Cancer. 2008;8:25. doi: 10.1186/1471-2407-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noh CK, Kim SS, Kim DK, Lee HY, Cho HJ, Yoon SY, et al. Inter-alpha-trypsin inhibitor heavy chain H4 as a diagnostic and prognostic indicator in patients with hepatitis B virus-associated hepatocellular carcinoma. Clin Biochem. 2014;47:1257–61. doi: 10.1016/j.clinbiochem.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 17.Huang L, Yoneda M, Kimata K. A serum-derived hyaluronan-associated protein (SHAP) is the heavy chain of the inter alpha-trypsin inhibitor. J Biol Chem. 1993;268:26725–30. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.