Abstract
Background
The larval nervous system of the solitary tunicate Ciona is a simple model for the study of chordate neurodevelopment. The development and connectivity of the Ciona motor ganglion have been studied in fine detail, but how this important structure develops in other tunicates is not well known.
Methods and Results
By comparing gene expression patterns in the developing MG of the distantly related tunicate Molgula occidentalis, we found that its patterning is highly conserved compared to the Ciona MG. MG neuronal subtypes in Molgula were specified in the exact same positions as in Ciona, though the timing of subtype-specific gene expression onset was slightly shifted to begin earlier, relative to mitotic exit and differentiation. In transgenic Molgula embryos electroporated with Dmbx reporter plasmids, we were also able to characterize the morphology of the lone pair of descending decussating neurons (ddNs) in Molgula, revealing the same unique contralateral projection seen in Ciona ddNs and their putative vertebrate homologs the Mauthner cells. Although Dmbx expression labels the ddNs in both species, cross-species transgenic assays revealed significant changes to the regulatory logic underlying Dmbx transcription. We found that Dmbx cis-regulatory DNAs from Ciona can drive highly specific reporter gene expression in Molgula ddNs, but Molgula sequences are not active in Ciona ddNs.
Conclusions
This acute divergence in the molecular mechanisms that underlie otherwise functionally conserved cis-regulatory DNAs supports the recently proposed idea that the extreme genetic plasticity observed in tunicates may be attributed to the extreme rigidity of the spatial organization of their embryonic cell lineages.
Electronic supplementary material
The online version of this article (10.1186/s13227-018-0107-0) contains supplementary material, which is available to authorized users.
Background
Tunicates have a long history as tractable laboratory organisms for the study of embryonic development [1]. Most tunicate larvae develop rapidly but invariantly, according to highly stereotyped cell lineages. Furthermore, many also possess highly compact genomes and are quite amenable to a wide variety of molecular assays and perturbations. More recently, tunicates have also begun emerging as model organisms for developmental neurobiology [2]. The complete connectome of the larva of the tunicate Ciona intestinalis, the second connectome ever mapped after that of the nematode C. elegans [3], revealed the synaptic connections of all 177 neurons of the central nervous system (CNS) and all 54 neurons of the peripheral nervous system (PNS) [4, 5]. With 231 total neurons (CNS and PNS combined), the Ciona larval nervous system is one of the smallest ever described, smaller than even the nervous system of the C. elegans hermaphrodite (302 neurons).
The Ciona connectome further revealed specific neural circuits that are conserved between tunicates and their sister group, the vertebrates, including a putative homolog of the Mauthner cell/C-start escape response circuit of fish [6]. In Ciona, the putative Mauthner cell (M-cell) homologs are a single pair of descending decussating neurons (ddNs), which correspond to the A12.239 pair of cells of the motor ganglion (MG), a cluster of 30 neurons that comprise a central pattern generator for the swimming behavior of the larva [7], and proposed to be homologous to a combination of the vertebrate hind brain and spinal cord [8]. The development of the ddNs and the rest of the MG have been studied in some detail, revealing gene expression patterns and transcriptional regulatory networks that are shared with hindbrain and spinal cord development in vertebrates [9–11]. These close parallels are especially striking considering the obvious, drastic reduction in size and complexity of the Ciona MG relative to the corresponding regions in vertebrates. However, it was found that cell fate specification and transcriptional patterning in the Ciona MG depends largely on cell–cell contact-dependent signaling within the neural tube by the Delta/Notch and Ephrin/Eph pathways, not on gradients of secreted, long-range morphogens as in the vertebrate spinal cord [11, 12].
Tunicates have also garnered recent attention for the fact that their extremely reduced, stereotyped cell lineages are highly conserved even between distantly related species. In spite of highly elevated mutation rates genome sequence divergence, and deep evolutionary timescales [13–16], the embryos of distantly related solitary tunicates like Ciona and Molgula (estimated divergence 390 million years apart) [15] are nearly indistinguishable [17]. We previously compared the development of the cardiopharyngeal mesoderm between Ciona robusta and Molgula occidentalis and found that there were virtually no differences in cell lineage and gene expression. However, we did find that underneath this seemingly conserved developmental program, there were considerable cryptic functional differences, resulting in cis-regulatory “unintelligibility” between Ciona and Molgula. In other words, homologous cis-regulatory elements driving identical gene expression patterns were partially or completely non-functional in cross-species transgenic assays. This phenomenon was ascribed to a particularly acute form of developmental system drift (DSD) [18]. It was recently proposed that the acute DSD observed in solitary tunicate evolution may be directly related to their mode of development, which depends primarily on invariant, contact-dependent intercellular signaling for cell fate induction. This geometric constraint would have relaxed constraints on genome evolution imposed by the relatively intricate and immutable transcriptional networks and cis-regulatory logics required for inductive events in species with larger, more variable embryos (i.e., the “Geometry vs. Genes” paradigm) [19].
Here we report that the phenomenon of acute DSD also extends to neurodevelopment in relatively late phases of tunicate embryogenesis. By surveying the development of the M. occidentalis MG, we show that transcription factor expression patterns, or “codes,” in the developing MG are identical between Molgula and Ciona, indicating near-perfect conservation of arrangement of neuronal subtypes. However, cross-species reporter assays revealed acute DSD in the regulation of Dmbx transcription in homologous ddN precursor cells. Our results support a “Geometry vs. Genes” model to explain the emergence of DSD in the tunicate MG, given its patterning by invariant cell–cell contacts and the parsimony of its final configuration as inferred by the Ciona connectome.
Methods
Molgula occidentalis genome assembly improvement
Libraries prepared for the version of the Molgula occidentalis genome (ELv1.2) currently available on ANISEED (https://www.aniseed.cnrs.fr/) [17, 20, 21] were reused to reassemble the genome. The original 3 DNA libraries of paired-end reads had insert sizes of 300–400, 650–750, and 875–975 base pairs (NCBI SRA ID# PRJNA253689) [17]. Reads were first filtered to a kmer coverage of 100X using the Khmer suite [22]. Assembly was done using Oases (v 0.2.08) [23] at various word lengths (k), with k = 67 being selected as the assembly to continue the downstream analyses. After reassembly (version “k67”), additional scaffolding was done using Redundans [24] producing the version “k67_R” (available at https://osf.io/3crup/). The ANISEED genome (ELv1.2) was scaffolded with Redundans as well, for comparison (version “ANISEED_R”). First redundant contigs were detected and selectively removed, and next genome fragments were joined using the previously mentioned libraries from the initial assembly. Finally, gapped regions of the scaffold were filled using these 3 paired-end libraries. After reassembly and additional scaffolding, the scaffolding quality was examined using REAPR (v1.0.17) [25] mapping all three libraries to back to the assembly.
Molgula occidentalis transcriptome sequencing
Gravid Molgula occidentalis (Traustedt, 1883) adults were collected and shipped by Gulf Specimen Marine Lab (Panacea, FL). Eggs were fertilized as previously described [17]. M. occidentalis embryos and larvae were grown at ~ 24 °C and collected into three separate stage pools: 0.33 hours post-fertilization (hpf), 7, and 16 hpf. Total RNA was extracted from each pooled sample using RNAqueous Total RNA Isolation kit (ThermoFisher). PolyA+ mRNAs were selected using oligo d(T)25 magnetic beads (New England Biolabs) following two rounds of the manufacturer’s protocol. Directional RNAseq libraries were prepared according to a modified version of the protocol contained at: https://wikis.nyu.edu/pages/viewpage.action?pageId=24445095 [26]. First-strand synthesis was performed using Super Script III (ThermoFisher), primed with oligo d(T) and random hexamer primers. Second-strand synthesis was performed using dUTP instead of dTTP for directional (strand-specific) sequencing [27]. Samples were processed and ligated to NETFLEX DNA barcode adapters (BioO Scientific) for multiplex sequencing. Adapter dimers were removed using AMPure beads (Agencourt). Samples were then treated with uracil-DNA glycosylase, amplified using 12 cycles of PCR, and purified once with a minElute kit (Qiagen) and once with AMPure beads. Samples were sequenced by Illumina 2000 2 × 50 bp runs on two lanes (all three samples were multiplex-sequenced in each lane). Resulting sequences will be deposited in the NCBI SRA database (accession number pending).
Transcriptome assembly and gene model improvement
Paired-end sequences generated above were used to generate gene model. Reads were quality filtered/trimmed using trimmomatic (v0.33) [28] and the following parameters “MINLEN:25, and sliding window of 4 with a minimum score of 5 (ILLUMINACLIP:TruSeq3-PE.fa:2:30:10 SLIDINGWINDOW:4:5 MINLEN:25).” Each of three embryonic stage libraries (0.33, 7, and 16 hpf) were individually mapped to the various genome versions using hisat2 (v 2.1.0) [29], with the default parameters. The sam files generated from hisat2 were then sorted and converted to bam using samtools (v 1.5) [30] and merged using PicardTools with java version 1.8 (http://broadinstitute.github.io/picard/). The merged bam file was then processed using StringTie (v 1.3.3b) [31] to create gene models. Gene models were then extracted using the gffread utility from cufflinks [32] and evaluated using BUSCO for completeness [33]. Transcripts/gene models have been deposited at OSF (https://osf.io/3crup/).
Fertilization, dechorionation, and electroporation of embryos
M. occidentalis eggs were fertilized, dechorionated, and/or electroporated as previously described [17]. Ciona robusta (intestinalis type A) adults were collected and shipped by M-REP (San Diego, CA) and eggs dechorionated, fertilized, and electroporated as previously described [34]. Dmbx cis-regulatory DNAs from two Molgula species (M. occidentalis and M. oculata) were cloned intro reporter plasmids as illustrated in supplemental sequences files (Additional file 1). Cirobu.Dmbx>Unc-76::Venus and Cirobu.Fgf8/17/18>H2B::mCherry reporter plasmids were published previously [10, 11].
mRNA probe synthesis and in situ hybridization
Templates for antisense riboprobes for in situ hybridization were amplified by PCR or SMARTer 3’/5’-RACE (Clontech) from cDNA libraries or from genomic DNA (see Additional file 1 for details on each sequence). Template sequences were cloned either using TOPO-TA cloning (ThermoFisher) into pCRII dual promoter vector, or using restriction enzyme cloning into pCiProbe (see Additional file 1) linearized NotI-EcoRI. In vitro transcription of labeled riboprobes and two-color fluorescent in situ hybridization were performed as previously described [35].
Results and discussion
M. occidentalis transcriptome assembly and gene prediction
We recently sequenced the genomes of 3 species in the genus Molgula: M. occidentalis, M. oculata, and M. occulta [17], which can be browsed freely on the Tunicate molecular biology database ANISEED (https://www.aniseed.cnrs.fr/) [21]. Of these, M. occidentalis emerged as a valuable species for comparative studies of tunicate development, mainly because their zygotes can be transfected with plasmid DNAs via electroporation, much like the major tunicate laboratory model species in the Ciona genus (C. intestinalis, C. robusta, and C. savignyi). To help establish additional molecular tools for developmental studies in M. occidentalis, we assembled a transcriptome based on RNAseq of 3 stages of embryonic development. These were then used to predict a new set of gene models found in the M. occidentalis genome sequence.
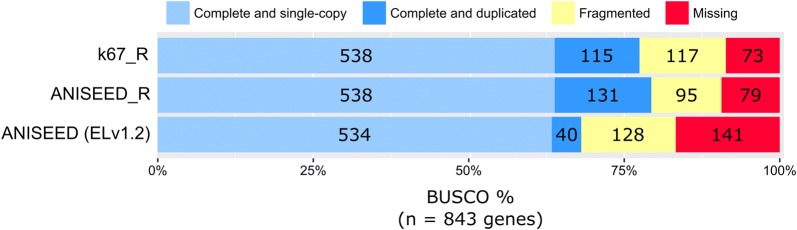
To do this, we also reassessed our previous M. occidentalis genome assemblies. In previous assemblies, some of the low-coverage regions were removed using khmer to reduce assembly fragmentation [22]. To preserve the information contained in these low-coverage regions, we re-assembled the genome from the original raw sequencing reads (see “Methods” section for details). While this new assembly initially had an N50 of only 519 bp and over 680,000 scaffolds, additional scaffolding decreased the number of scaffolds to 30,188, with 20,616 of those (68%) being over 1 kb in length (Table 1). As a result of this better scaffolding, the N50 also increased from 519 to 18,312. The gap filling process decreased the number of missing bases (Ns) from 5,510,404 to 189,152. The end result is the “k67_R” genome assembly version, which we have deposited at OSF (https://osf.io/3crup/). While these procedures improved the genome assembly statistics, we wanted to ensure that the additional gene content was still preserved. To check this, we mapped mRNA reads and produced gene models to be tested using BUSCO [33]. This allowed us to compare our new gene models against a defined collection of highly conserved sequences expected to be present in all metazoans, as a measure of assembly completeness. Our BUSCO results (Fig. 1) indicated that fewer reference genes were missing (73 missing out of 843 total) in the new “k67_R” assembly than in the ANISEED ELv1.2 version (141/843 missing) or even a version of the ANSEED assembly with additional scaffolding performed (“ANISEED_R,” 79/843 missing). This indicates that these new versions of the genome assembly and their corresponding gene models should enhance the identification and cloning of sequences to use as molecular tools for M. occidentalis.
Table 1.
M. occidentalis genome reassembly statistics
| Assembly version | k67_R | ANISEED_R | ANISEED (ELv1.2) |
|---|---|---|---|
| Total length (bp) | 207,168,883 | 242,783,314 | 262,547,660 |
| N’s | 189,152 | 885,908 | 33,473,590 |
| Scaffolds | 30,188 | 16,148 | 51,761 |
| Scaffolds ≥ 1 kb | 20,616 | 13,751 | 21,251 |
| Longest scaffold (bp) | 250,303 | 235,023 | 229,829 |
| N50 (bp) | 18,312 | 35,542 | 26,298 |
Fig. 1.
Benchmarking Universal Single-Copy Orthologs (BUSCO) in M. occidentalis. BUSCO was used to asses gene model predictions based on the previously published and released M. occidentalis genome assembly (ANISEED ELv1.2) and on the re-assemblies from this study (k67_R and ANISEED_R). BUSCO is used as a measure of completeness of gene model sets predicted from genome assemblies, searching for 843 highly conserved genes that should be present in single copies in > 90% of metazoan genomes. There are five categories of gene recovery: complete and single-copy, complete and duplicated, fragmented, and missing. Complete genes are those that preserve ~ 95% of the gene length. Single-copy genes are those with only one copy in the gene model set and duplicated genes are those with multiple copies identified either by gene duplication or assembly errors. Fragmented genes are those recovered with less than 95% of the gene length, and missing genes are those that are not found to be present at all
Gene expression patterns in the M. occidentalis motor ganglion
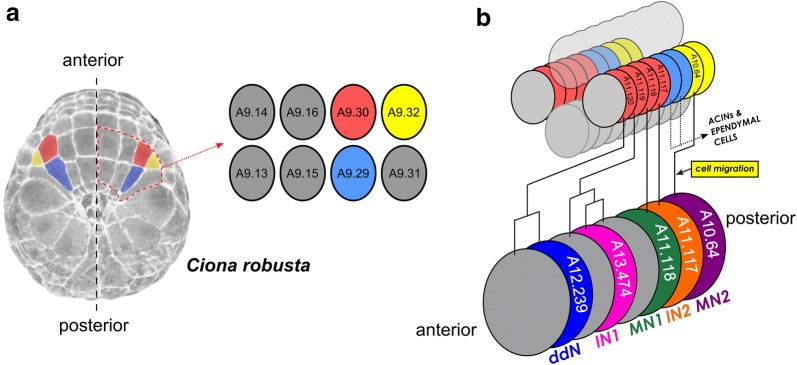
The Ciona MG is comprised of 22 neurons and includes a “core” MG of 5 morphologically and molecularly distinct left/right pairs of neurons [4]. In the remainder of this study, we will only refer to the neurons on one side, for simplicity. The core MG is derived from the A9.30 and A9.32 blastomeres of the neural plate (Fig. 2a) that will give rise to cells along the lateral rows of the neural tube after neurulation and neural tube closure [36–39]. At the level of the MG, the neural tube of the Ciona embryo is formed by only four single-file rows of cells oriented along the anterior–posterior (A–P) axis: a dorsal row (roof plate), a ventral row (floor plate), and two neurogenic lateral rows from which most of the core MG neurons are specified (Fig. 2b). Ascending contralateral inhibitory neurons (ACINs) derived from the A9.29 blastomeres [40] have been traditionally excluded from the MG based on their more posterior location at the base of the tail. However, these are likely indispensable cogs in the MG central pattern generator, driving left/right alternation of tail contractions during swimming by glycinergic neurotransmission [7]. Remaining MG neurons are poorly studied, and their development is largely unknown. In this study, we focused on the “core” MG neurons, those derived from the A9.30 and A9.32 lineages, because these lineages have been the most thoroughly studied MG lineages [9, 10, 39]. Expression patterns of developmental regulators and provisional gene regulatory networks have been documented in these lineages [9], and the induction events responsible for specifying the 5 distinct types of neurons in this core MG have also been elucidated [10, 11]. Thus, the MG is a perfect starting point for a comparative study on the evolution of neurodevelopment in tunicates.
Fig. 2.
Motor ganglion lineages and neuron subtype configuration in Ciona. a Dorsal view of a late gastrula-stage Ciona robusta embryo highlighting in false color the bilaterally symmetric right/left pairs of blastomeres in the vegetal neural plate that give rise to the “core” Motor Ganglion and Anterior Caudal Inhibitory Neurons (ACINs) of the nerve cord: A9.30 (red), A9.32 (yellow), and A9.29 (blue). Black dashed line indicated embryo midline. Red dashed outline denotes the right vegetal neural plate annotated in the inset diagram, which indicates the exact cell identities according to the established cell lineage nomenclature [64]. Embryo image adapted from the Four-dimensional Ascidian Body Atlas website (FABA, http://chordate.bpni.bio.keio.ac.jp/faba/1.4/top.html) [65]. b Diagram representing a section of the dorsal hollow neural tube derived from the neural plate after the process of neurulation. Cells of the neural tube are color-coded according to their lineages using the same color scheme as the neural plate diagram in (a). The neural tube is comprised exactly of four single-file rows of cells: 1 dorsal row, 2 lateral rows, and 1 ventral row. Cells of the lateral rows contributing to the bilaterally symmetric motor ganglion are: A11.117-A11.120, derived from A9.30, and A10.64, derived from A9.32. The A9.29 lineage, which gives rise to ACINs and ependymal cells posterior to the MG, is not illustrated in great detail, for the sake of simplicity. At bottom is a diagram of the 5 core MG neuron types. Black lines indicate their descent patterns from those cells at top. ddN = descending decussating neuron, IN1 = interneuron 1, IN2 = interneuron 2, MN1 = motor neuron 1, MN2 = motor neuron 2. Non-neuronal cells are gray. Cells are identical on both left and right sides
We previously showed that the overall development of the M. occidentalis embryo is very similar to the development of the Ciona embryo [17]. An in-depth study of the cardiopharyngeal mesoderm (B7.5 cell lineage) revealed perfect conservation of precise cell divisions and fate specification events, with only minimal differences in morphogenesis and timing of gene expression. However, other tissues beyond the cardiopharyngeal mesoderm were not surveyed. In this study, we sought to extend our understanding of tunicate evolution by comparing the developing nervous systems of M. occidentalis and C. robusta (intestinalis type A). In situ mRNA hybridization (ISH) for neural marker Celf3/4/5 (formerly Etr-1) in M. occidentalis neurula embryos revealed the CNS developing from the lateral rows of the dorsal hollow neural tube, as in Ciona [41] (Fig. 3a). Later, at tailbud stages, ISH for the neuronal transcription factors Neurogenin, Onecut, and Ebf revealed ongoing neuronal specification in the brain, MG, and bipolar tail neurons of the tail (Fig. 3b–d), also identical to the expression of these regulators in Ciona embryos [41, 42]. Two-color ISH of Ebf and the cholinergic marker Slc18a3 (also known as vesicular acetylcholine transporter, or VAChT) [43] revealed the earliest differentiating neurons of the larval CNS, the motor neurons of the MG (Fig. 3d). Given that no significant differences were revealed between Molgula and Ciona using these broad markers of neural fate, we focused our attention specifically to the developing MG, where we would be able to analyze more subtype-specific gene expression and cell fates.
Fig. 3.
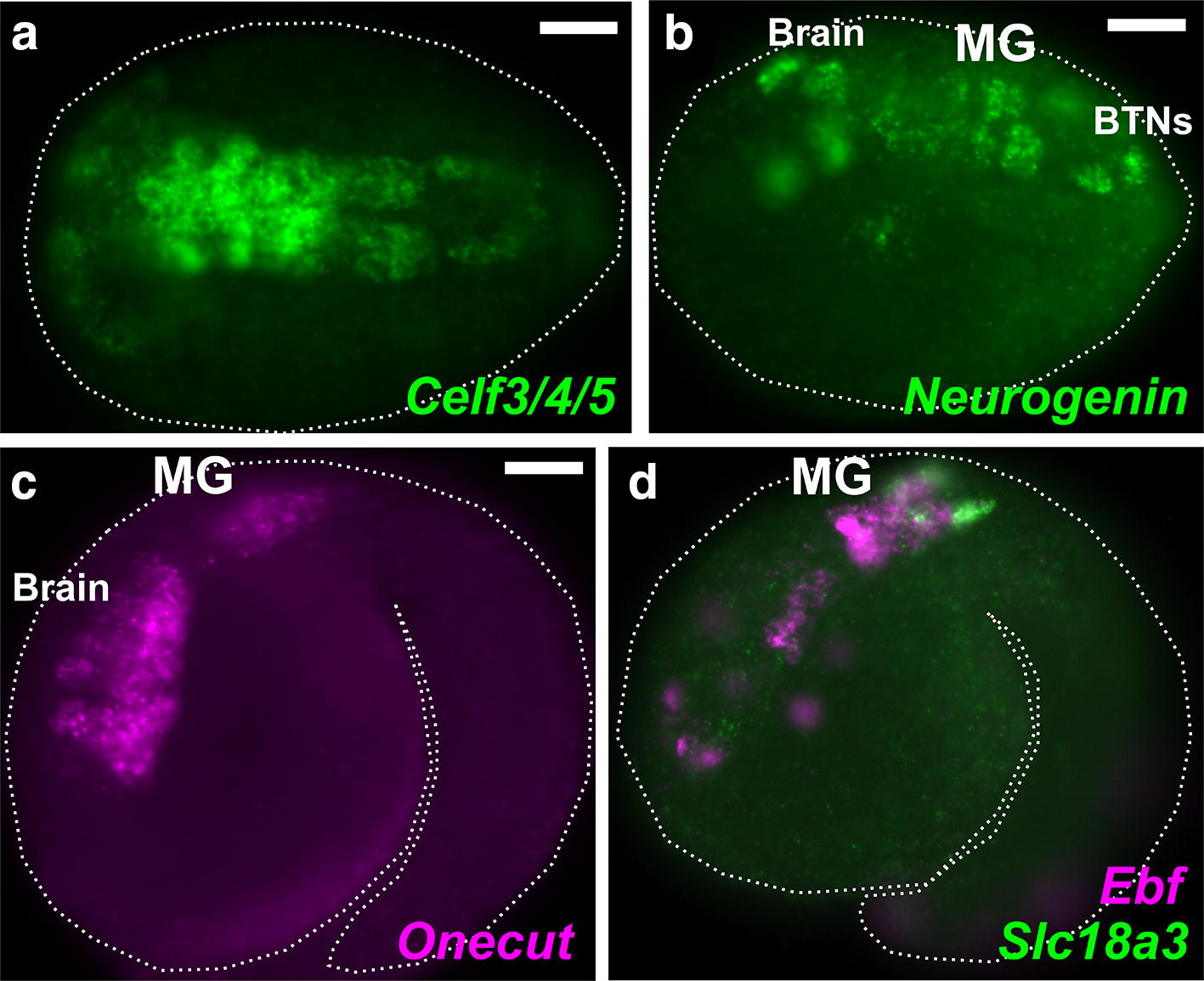
The developing central nervous system of Molgula occidentalis. a Dorsal view of a neurula stage Molgula occidentalis embryo. Green shows fluorescent in situ hybridization for Celf3/4/5 (also known as Etr-1) transcription. b In situ hybridization for Neurogenin in initial tailbud embryo, showing transcription in the embryonic brain, motor ganglion (MG), and bipolar tail neurons (BTNs). c In situ hybridization for Onecut, expressed in brain and MG. d Two-color fluorescent in situ hybridization showing co-expression of Ebf (magenta) and Slc18a3 (also known as VAChT, green) in the MG. Embryo outlined by dotted lines. Anterior is to the left in all images. All scale bars = 25 µm
In C. robusta, the expression patterns of mRNAs encoding transcription factors are dynamically regulated in the developing MG [9, 44]. Some of these patterns comprise a conserved “code” of homeodomain-containing genes that are differentially expressed along the dorsal–ventral (D-V) developing spinal cord of vertebrate embryos [11]. However, in the developing Ciona MG, the neurogenic domain of neural tube is restricted to the single-file lateral rows of cells. Therefore, the D-V code of the vertebrate spinal cord is transposed along the A-P axis in Ciona. From these neural progenitors expressing different combinations, or codes, of these homeodomain proteins, the 5 distinct neuronal subtypes of the Ciona MG are born. We sought to characterize the expression patterns of homologous genes in the developing MG of M. occidentalis. We were able to identify these readily by performing BLAST against our transcriptome assembly, which revealed clear orthologs for the following genes: Mnx, Vsx, Islet, Nk6, Pax3/7, and Dmbx. An additional gene Lhx3/4.a had already been identified and characterized in our previous study [17].
Motor neuron and interneuron identities
In Ciona, 4 of the 5 core MG neuron subtypes are marked by either Mnx or Vsx [9, 10]. These are orthologs of conserved transcription factors that specify motor neurons (HB9, MNR2, etc.) or interneurons (Chox10, CHX10, Ceh-10, etc.), respectively. Two-color ISH revealed a neatly alternating Vsx-Mnx-Vsx-Mnx pattern on one side of the developing MG of M. occidentalis, around 8.5 hours post-fertilization (hpf) (Fig. 4a). This alternating pattern closely mirrors the alternation of motor neuron (MN) and interneuron (IN) subtypes in the Ciona MG: IN1-MN1-IN2-MN2. In the Ciona MG, the latter three are the first MG neurons to differentiate, corresponding to cells A11.118 (MN1), A11.117 (IN2), and A10.64 (MN2), which are consecutively arrayed with no intervening cells between them. This appears to be the case in M. occidentalis also. However, in Ciona there are additional non-neuronal cells intercalated between IN1 and MN1, as a result of continued proliferation in the anterior MG that ultimately gives rise to IN1 (see Fig. 2b). In Ciona, Vsx expression is restricted to post-mitotic interneurons and is not observed in A11.119, the grandmother cell of IN1 [10]. In M. occidentalis, the lack of any discernible gap between the anterior-most Vsx+ and Mnx+ cells at first suggests that Vsx transcription starts in the A11.119 progenitor cell itself, a clear example of transcriptional “priming” of cell fate [45]. Indeed, we observed Vsx expression in two anterior cells when observed at slightly later stages, suggesting that A11.119 divided after the onset of Vsx activation (Fig. 4b). Together with evidence for earlier Dmbx expression in the A11.120 progenitor cell (see below), these data suggest that the expression of certain MG neuron subtype-specific transcription factors is already primed in M. occidentalis MG progenitors. This heterochronic shift may be related to the ~ 10% faster development of Molgula relative to Ciona [17].
Fig. 4.
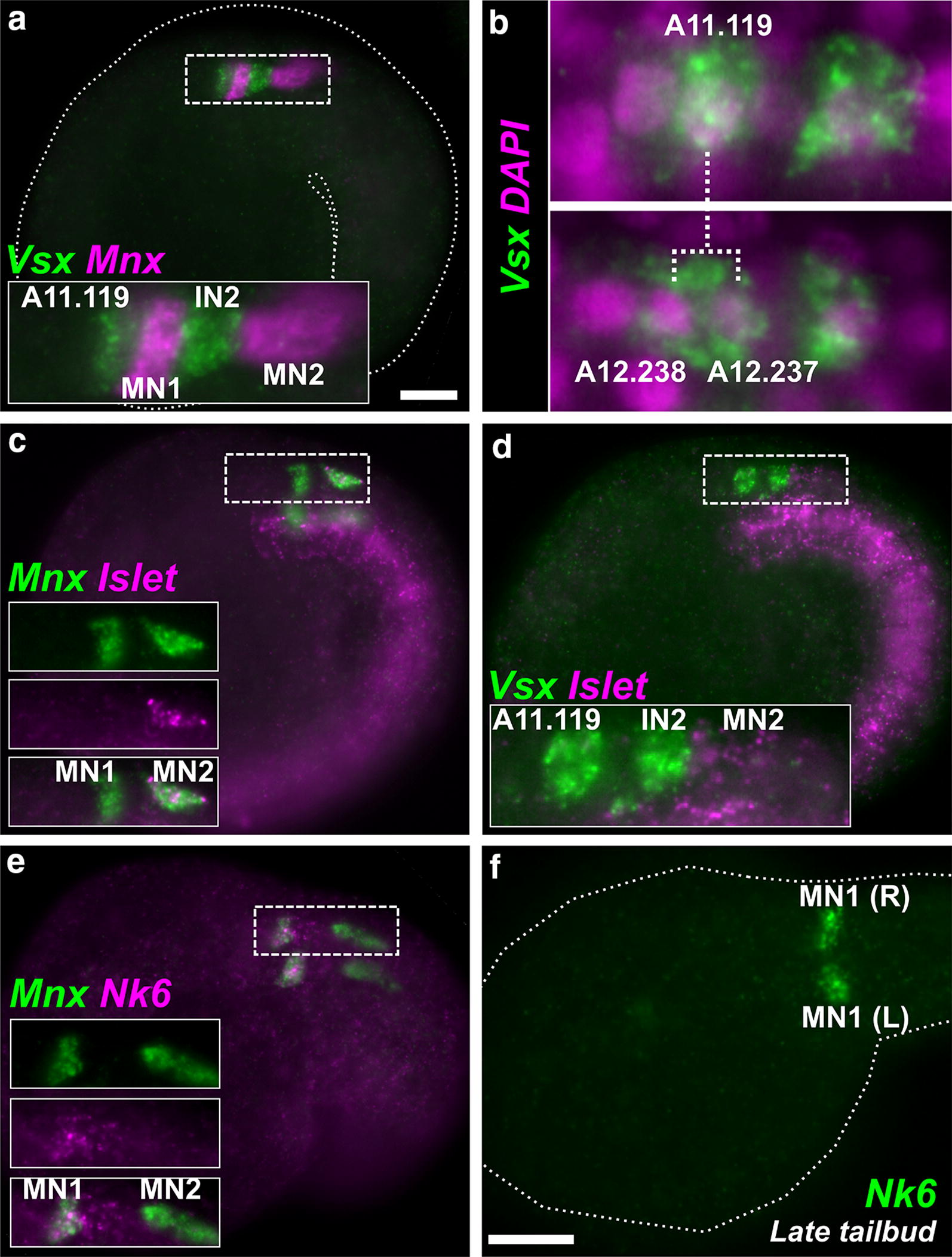
Motor neurons and interneurons in the MG. a Lateral view of mid-tailbud stage Molgula occidentalis embryo (dotted outline). Two-color fluorescent in situ hybridization reveals alternating expression of interneuron marker Vsx (green) in undifferentiated A11.119 interneuron progenitor cell and interneuon 2 (IN2) and motor neuron marker Mnx (magenta) in motor neurons 1 and 2 (MN1, MN2). Only one side of embryo is shown. Inset is a magnified view of dashed box. b In situ hybridization for Vsx (green) in embryos at two successive stages. Vsx is initially activated in A11.119 (top panel), which divides and gives rise to daughter cells A12.238 and A12.237, both of which continue to express Vsx at this stage. These data confirm that Vsx is activated in the A11.119 cell in M. occidentalis, which is not the case in Ciona (see text for details). Nuclei are counterstained with DAPI (magenta). c Two-color fluorescent in situ hybridization for Mnx (green) and MN2 marker Islet (magenta), revealing identities of MN1 (Mnx+/Islet-) and MN2 (Mnx+/Islet+) cells. Boxed area magnified in insets. d Two-color fluorescent in situ hybridization for Vsx (green) and Islet (magenta). Only one side of embryo is in focus. e Two-color fluorescent in situ hybridization for Mnx (green) and MN1 marker Nk6 (magenta), confirming identities of MN1 (Mnx+/Nk6+) and MN2 (Mnx+/Nk6-). f Nk6 in situ hybridization (green) in late tailbud embryo, showing sharpened expression in MN1 cells on both sides of the MG (R = right side and L = left side). All scale bars = 25 µm
Islet expression reveals MN2 (A10.64 cell)
From the Mnx ISH, MN2 appeared to be the posterior-most cell of the core MG. The identity of this cell was confirmed by two-color ISH with Islet, a marker of MN2 fate in Ciona (Fig. 4c, d). In Ciona, MN2 is the only core MG neuron that is not derived from the A9.30 lineage. Long identified as the A10.57 cell derived from the A9.29 lineage, MN2 was recently revealed in fact to be the A10.64 cell of the A9.32 lineage instead and ultimately derived from the A8.16 neuromesodermal lineage that also gives rise to the secondary tail muscles of the larva [39]. Despite its origin from further posterior in the embryo, MN2 becomes associated with the MG thanks to a dramatic, anterior migration along the outside of the neural tube, leapfrogging over the entire A9.29 lineage [39]. The migrating MN2 is elongated along the A-P axis due to the extension of a leading edge that ultimately contacts the A9.30 lineage. Upon contacting the A9.30 lineage, MN2 extends its axon posteriorly, retaining an elongated cell body and forming en passant synapses with the dorsal band of muscle cells down the length of the tail [46]. In M. occidentalis, the elongated morphology of MN2 was readily apparent by ISH, in which fluorescent signal fills most of the cell bodies to reveal their shapes. Therefore, we conclude that, in M. occidentalis, the specification and morphogenesis of MN2 are highly conserved.
Nk6 expression is refined and restricted to MN1
While MN2 controls tail contractions in a graded manner [47], the other major MN in Ciona is MN1, the A11.118 cell. This neuron was shown to form large, leaf-like (“frondose”) motor endplates at the base of the tail [10]. While MN2 s are proposed to exert graded motor control during maneuvering, the all-or-none flexions of the tail that drive swimming are thought to be triggered instead by MN1 s. This cell is characterized by Mnx expression without co-expression of Islet. In Ciona, MN1 is also marked by late, sustained expression of Nk6 close to hatching, even though this gene is expressed broadly throughout the posterior A9.30 lineage earlier in development [11]. In M. occidentalis, we found this also to be true. Nk6 expression was seen in A11.119, A11.118 (MN1), and A11.117 (IN2) at the mid-tailbud stage (Fig. 4d), but later was seen only in a single pair of cells in the embryo, presumably MN1s (Fig. 4e). This further confirms the highly conserved nature of the posterior MG and the specification of MN1 by sustained Nk6 expression.
Subfunctionalization of Lhx3/4 paralogs
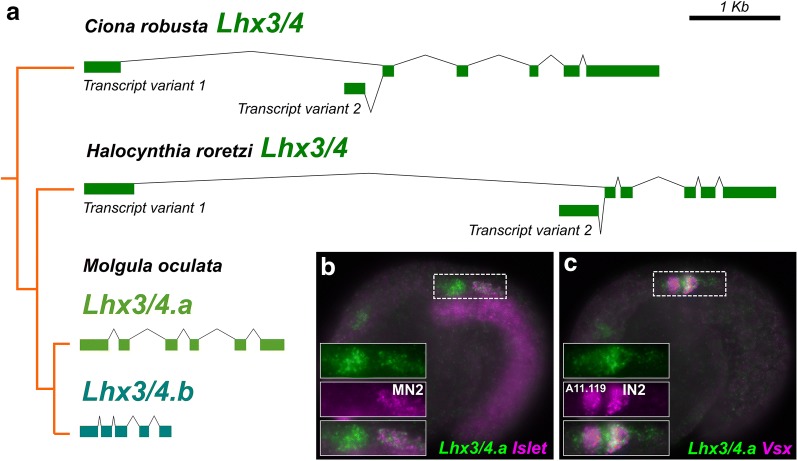
In Ciona and the stolidobranch tunicate Halocynthia roretzi, a single Lhx3/4 is transcribed as two alternative isoforms, transcript variants 1 and 2, originating from alternate promoters (Fig. 5a) [48, 49]. In both Ciona and Halocynthia, transcript variant 1 is expressed in MG precursors, while transcript variant 2 is expressed in early vegetal pole cells and is required for endoderm and cardiopharyngeal mesoderm specification [49, 50]. We previously found that in all three Molgula genomes sequenced to date, the ancestral tunicate Lhx3/4 gene was duplicated, resulting in two paralogs, termed Lhx3/4.a and Lhx3/4.b (Fig. 5a) [17]. In M. occidentalis, their expression patterns and putative functions appear to mirror those of the two different transcript variants identified in Ciona/Halocynthia: Lhx3/4.a is expressed in the MG, while Lhx3/4.b is only expressed in the early vegetal pole cells [17]. Lhx3/4.a is the more conservative paralog, retaining an ancestral C-terminal peptide motif, which has been lost in Lhx3/4.b [17]. Given that Halocynthia is more closely related to Molgula than to Ciona [51], this suggests a Molgula-specific duplication of Lhx3/4, followed by subfunctionalization through partitioning of the expression domains of the ancestral transcript variants in addition to protein-coding changes.
Fig. 5.
Molgula-specific duplication and subfunctionalization of Lhx3/4. a Diagram depicting Lhx3/4 genes in the genomes of Ciona robusta, Halocynthia roretzi, and Molgula oculata, with their phylogenetic relationships indicated by the orange tree. All diagrams are at the same scale (scale bar is at top right). Exons indicated as colored bars, introns are depicted by crooked lines. Both Ciona and Halocynthia have a single Lhx3/4 gene that is transcribed from two distinct promoters to give rise to two transcript variants. In both species, variant 2 is involved in vegetal pole patterning in the early embryo, while variant 1 is involved in motor ganglion patterning. In all three Molgula species sequenced, Lhx3/4 has been duplicated, giving rise to paralogs Lhx3/4.a and Lhx3/4.b. Loci from M. oculata are shown here (instead of M. occidentalis) because its genome is the least fragmented Molgula genome. M. occidentalis Lhx3/4 genes are similar in structure to their M. oculata orthologs, but assembly errors near the 3’ end of M. occidentalis Lhx3/4.b (not shown) prevented us from using sequences from this species in our diagram. b Two-color fluorescent in situ hybridization for Lhx3/4.a (green) and Islet (magenta) showing co-expression in MN2. c Two-color fluorescent in situ hybridization for Lhx3/4.a (green) and Vsx (magenta) showing strong co-expression in IN2, and diffuse Lhx3/4.a expression throughout the MG
Here we looked more closely at the expression of Lhx3/4.a in the developing MG of M. occidentalis. In Ciona, Lhx3/4 is initially expressed in the posterior MG (MN1, IN2, and more weakly in MN2) and later in IN1 [9, 44]. In Halocynthia, Lhx3/4 was also detected earlier in development, in the A9.30 blastomere [52]. In M. occidentalis, we found Lhx3/4.a expressed throughout the entire MG at the mid-tailbud stage, though more strongly in MN1 and IN2 (Fig. 5b, c). Higher expression in these cells is shared with Ciona and Halocynthia, and the weaker expression in the anterior MG may reflect fading expression from earlier stages as in Halocynthia. Based on these results, we propose that Lhx3/4.a has, for the most part, conserved the MG-specific expression pattern and putative function of the ancestral Lhx3/4 gene, more specifically those of its transcript variant 1.
Pax3/7 and Dmbx delineate the descending decussating neurons (ddNs)
The anterior-most neuron in the core MG of Ciona is the descending decussating neuron (ddN), corresponding to the A12.239 cell. The recently mapped Ciona intestinalis connectome revealed that these neurons are likely homologous to giant reticulospinal Mauthner cells (M-cells) of vertebrates and situated in a neural circuit that is topologically very similar to M-cell escape circuits [6]. In Ciona, the ddN is specified by a gene regulatory network involving the transcription factors Pax3/7 and Dmbx [11]. To summarize, Pax3/7 is sufficient and necessary for the specification of the A11.120 progenitor, the mother cell of the ddN, and directly activates Dmbx expression in the posterior daughter cell of A11.120, which becomes the ddN (A12.239). Dmbx in turn encodes a repressor that mediates an FGF-regulated switch for differentiation [11]. Upon differentiating, ddNs project their nascent axons across the midline, perpendicular to the A-P axis. Thus, as their name implies, they are the only core MG neurons that decussate.
We therefore sought to characterize the expression patterns of Pax3/7 and Dmbx in M. occidentalis. ISH revealed Pax3/7 and Dmbx expression in a single cell in the MG, just anterior to the Vsx-expressing cell identified as A11.119 (Fig. 6a, b). These data indicate that Pax3/7 and Dmbx are co-expressed in A11.120, the mother cell of the ddN. While Pax3/7 expression in A11.120 is shared between Molgula and Ciona, Dmbx expression in M. occidentalis appears to be shifted earlier relative to mitotic exit of the neuron. In Ciona, transcription of Dmbx is not detected in the A11.120 cell but is activated later, specifically in the ddN, by combinatorial action of Pax3/7 (inherited from the mother cell), and Neurogenin [11].
Fig. 6.
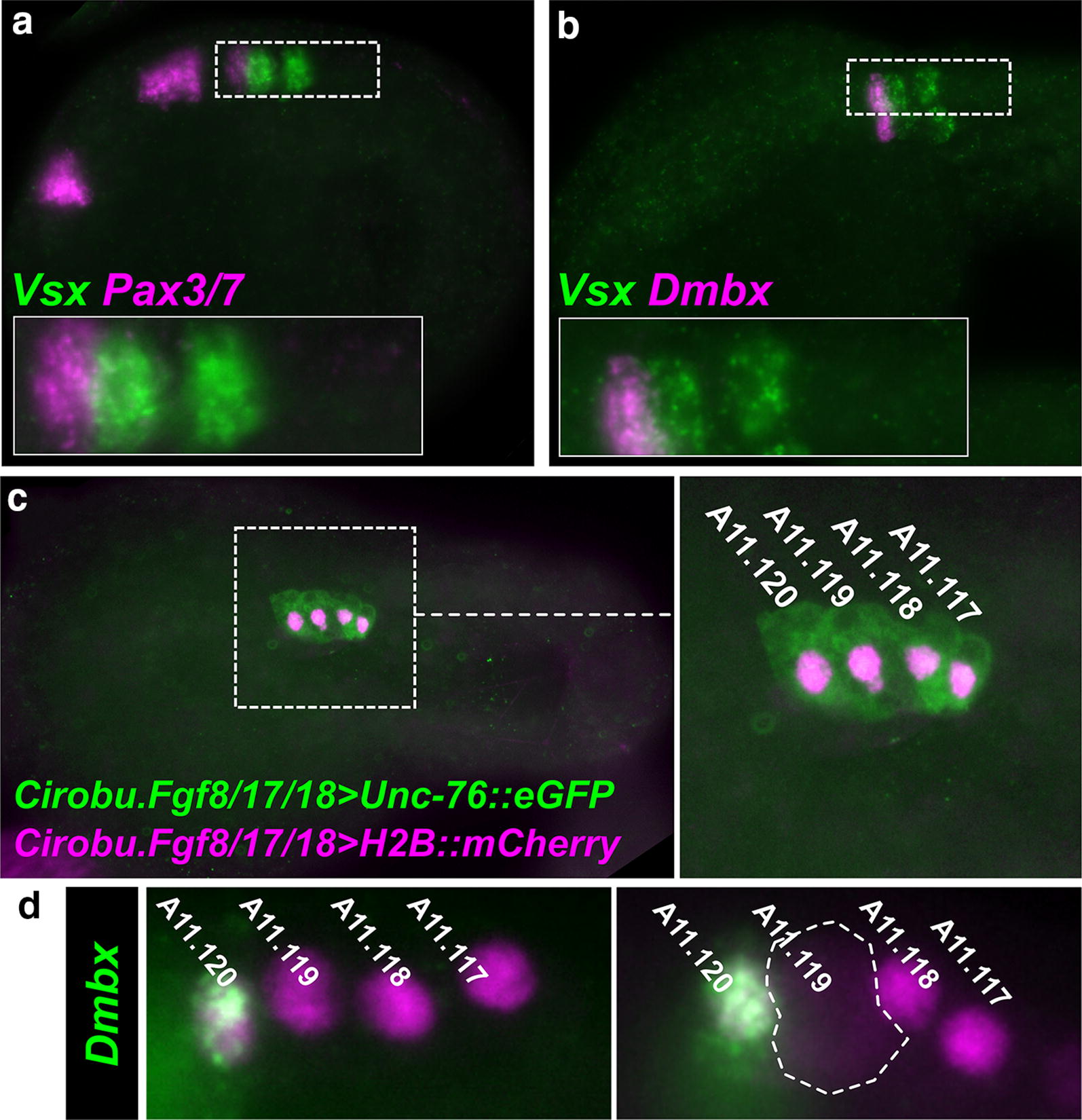
Pax3/7 and Dmbx activation in A11.120. a Two-color fluorescent in situ hybridization for Vsx (green) and Pax3/7 (magenta) showing expression of Pax3/7 anterior and adjacent to Vsx expression, shown in Fig. 4 to correspond to the A11.119 cell. Pax3/7 is thus deduced to be expressed in the sister cell of A11.119, A11.120. This is identical to its expression pattern in Ciona. b Two-color fluorescent in situ hybridization for Vsx (green) and Dmbx (magenta) also showing expression of Dmbx in A11.120 like Pax3/7. This is different from the situation in Ciona, in which Dmbx is detected only later, in the descending decussating neuron (ddN, equivalent to A12.239, the posterior daughter cell of A11.120). c M. occidentalis early tailbud embryo electroporated with Ciona robusta (Cirobu) Fgf8/17/18 reporter plasmids, labeling the A9.30 lineage on one side, with H2B::mCherry-labeled nuclei in magenta and Unc-76::eGFP-labeled cell bodies in green. Inset at right showing higher magnification of boxed area, showing all four descendants of A9.30, arrayed from anterior to posterior. d In situ hybridization of Dmbx coupled to immunostaining of beta-galactosidase (magenta nuclei) driven by Cirobu.Fgf8/17/18>nls::lacZ reporter plasmid, revealing that Dmbx is strongly activated in the A11.120 cell, the mother cell of the ddN. Right panel shows A11.119 entering mitosis indicated by nuclear envelope breakdown and diffusion on nuclearly localized beta-galactosidase (dashed outline). A11.119 appears to divide before the other cells in the MG at this stage, which is also the case in Ciona. This further confirms the precocious activation of Vsx in this cell relative to Ciona (see Fig. 4)
To confirm that Dmbx is expressed in the A11.120 cells of M. occidentalis, we sought to determine its expression pattern at cellular resolution. We turned to electroporation to transfect embryos with a reporter construct that could selectively label the A9.30 lineage and allow us to combine ISH with immunofluorescence. To this end, we used the Fgf8/17/18 promoter from C. robusta [9] to drive expression of histone 2B::mCherry (Cirobu.Fgf8/17/18>H2B::mCherry) in the A9.30 lineage of electroporated M. occidentalis embryos. Activity of this reporter plasmid was relatively weak and infrequent in M. occidentalis embryos, but managed to label the 4 cells of the developing MG of M. occidentalis, equivalent to the cells A11.120, A11.119, A11.118, and A11.117 (from anterior to posterior) (Fig. 6c), and ISH + immunofluorescence revealed Dmbx expression in A11.120 at this stage (Fig. 6d). Taken together, these data indicate a M. occidentalis-specific priming of Dmbx transcription in the progenitor of the ddN, similar to the priming of Vsx in the progenitor of IN1 in this species as well (see above).
Conserved morphogenesis and axon projection of ddNs in M. occidentalis
Given the unique morphology and axon trajectory of the ddNs in Ciona and their likely homology to M-cells in a conserved, pan-chordate escape network [6], we asked whether their morphogenesis is conserved in M. occidentalis. To label differentiated ddNs, we electroporated M. occidentalis embryos with a Dmbx reporter plasmid containing ~ 1.4 Kb of genomic DNA sequence 5’ to the ATG corresponding to the predicted start codon of the Dmbx mRNA from the related species M. oculata (Additional file 1), fused to the Unc-76::eGFP fluorescent reporter gene to label axons uniformly [53]. This construct was sufficient to label the ddNs of M. occidentalis (Fig. 7a, b). Unc-76::eGFP expression in the ddNs revealed a polarization along the medial–lateral axis, perpendicular to the A-P, resulting in their unique axon trajectory initially straight across the midline, then abruptly turning 90° to descend along the outside of the neural tube toward the tail. This closely mirrors the processes of axonogenesis and axon guidance that result in the conserved axon trajectory of the ddNs in Ciona [10]. This suggests that ddN morphogenesis is an ancient and highly conserved process in tunicates, which also shares features with M-cell morphogenesis in vertebrates [54].
Fig. 7.
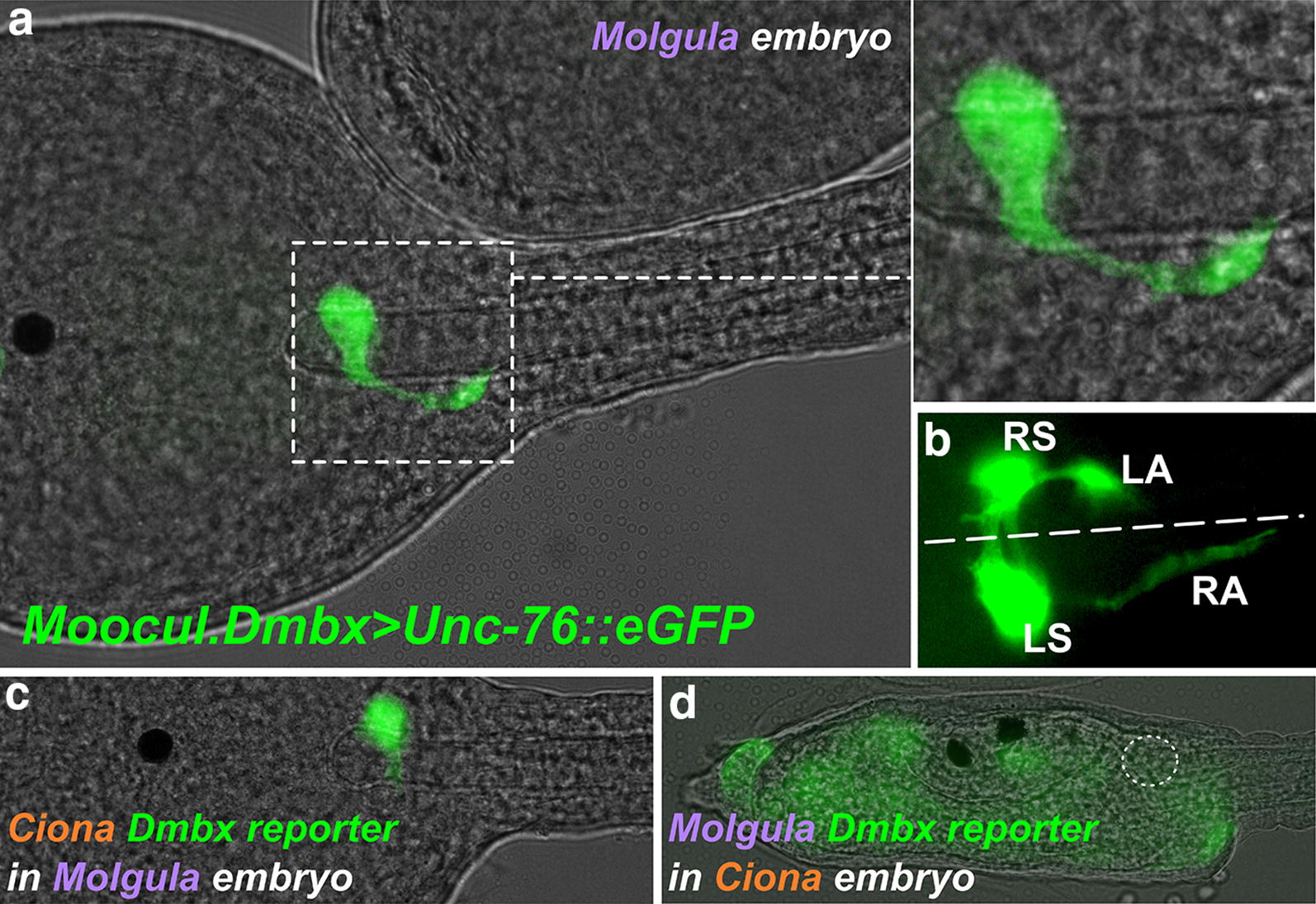
Dmbx reporter assays reveal conserved morphogenesis of the ddN and asymmetric unintelligibility of cis-regulatory sequences. a Dorsal view of a Molgula occidentalis embryo electroporated with a Molgula oculata (Moocul) Dmbx>Unc-76::eGFP reporter plasmid, specifically labeling (in green) a developing descending decussating neuron (ddN) on one side of the embryo. Inset at top right shows magnified view of boxed area, showing the conserved axon trajectory of the ddN, which is initially perpendicular to the anterior–posterior axis and then turns 90 degrees to continue posteriorly toward the tail. b Another embryo electroporated with Moocul.Dmbx>Unc-76::eGFP, this time labeling both right and left ddNs. Both axons cross the midline (dashed line) to continue down toward the tail on the other side. RS: right soma, RA: right axon, LS: left soma, LA: left axon. c M. occidentalis embryo electroporated with Ciona robusta Dmbx>Unc-76::Venus reporter, which is activated precisely and robustly in the ddN (green). d Ciona robusta embryo electroporated with Molgula oculata Dmbx>Unc-76::eGFP reporter, which is not active in this species’ ddN (approximate location indicated by dashed circle). Note that non-specific expression (green) can be seen in other unrelated tissues and cell types (palps, mesenchyme, etc.), suggesting that transcription/translation of GFP was not globally affected (see text for details)
Cross-species Dmbx reporter assays reveal asymmetric developmental system drift (DSD)
In our comparison of cardiopharyngeal development between Ciona and Molgula, we discovered pervasive, acute developmental system drift (DSD) [18] that has resulted in cross-species incompatibility of orthologous cis-regulatory DNAs that regulate identical gene expression patterns in the tunicate cardiopharyngeal mesoderm [17]. Here we tested whether DSD might also underlie the highly conserved gene expression patterns seen in the developing MG. We found that the previously identified C. robusta Dmbx driver [10] was sufficient to activate reporter gene expression in M. occidentalis ddNs (Fig. 7c). However, the M. oculata Dmbx reporter plasmid, used to label M. occidentalis ddNs, was non-functional in Ciona embryos (Fig. 7d). Thus, there is an asymmetry in the intelligibility of Dmbx cis-regulatory sequences: the Ciona cis-regulatory sequence works in Molgula, but not vice versa. This incompatibility is probably not due to basal promoter incompatibility since widespread non-specific M. oculata Dmbx>GFP expression was clearly visible in a variety of cell types in Ciona (Fig. 7d). Furthermore, the incompatibility is also not likely due to differences in trans-splicing (resulting in a failure to translate the fluorescent reporter protein), since we found that Dmbx is also trans-spliced in M. occidentalis (Additional file 1: Fig. S1).
One possible explanation for this asymmetry is that, in Molgula, Dmbx is initially activated in the A11.120 cell, the mother cell of the ddN (see Fig. 6d). In Ciona, Dmbx is only detected later, in the post-mitotic ddN [10], and is activated by a combination of Neurogenin and Pax3/7 and depends critically on the presence of a Pax3/7 binding site located in a cis-regulatory element that controls Dmbx transcription in the ddN [11]. Therefore, it is possible that an alternate trans-regulatory logic drives the earlier activation of Dmbx that we observe in Molgula, considering that Pax3/7 and Dmbx are transcribed simultaneously in M. occidentalis MG, not sequentially as in Ciona. The required factors for this earlier Pax3/7-independent activation of Dmbx in M. occidentalis might therefore be absent from the Ciona MG, resulting in lack of activation of the Molgula reporters in Ciona. In contrast, we show that both Pax3/7 (see Fig. 6a) and Neurogenin (see Fig. 3b) expression patterns are conserved in M. occidentalis, which would allow for proper activation of the Ciona Dmbx reporter plasmid in this species.
Alternatively, the incompatibility we observed may not be due to divergent trans-regulatory logic. In fact, we previously showed that the trans-regulatory logic of cross-species incompatibility of Mesp reporter plasmids between Ciona and Molgula is in part due to coevolution of the Tbx6-related transcription factor and its target Mesp promoter [17]. Thus, it is possible that activation of Dmbx by Pax3/7 and Neurogenin is conserved in Molgula and that the cross-species incompatibility of our Dmbx reporter plasmids is instead due to significant cis/trans coevolution between these trans-activating factors and the Dmbx cis-regulatory sequence. Regardless of the exact underlying molecular mechanism, the asymmetric intelligibility of Dmbx cis-regulatory sequences we have uncovered is yet another example of acute DSD between Ciona and Molgula, which stands in stark contrast to the overall conservation of MG development that we have documented in this study.
Conclusions
Here we have documented the deep conservation of regulatory gene expression patterns that establish the precise configuration of neuronal subtypes in the MG of the tunicate larva (summarized in Fig. 8). Our data suggest that the circuitry of the MG, as revealed by the recently completed C. intestinalis connectome [4], may be as rigidly conserved as the embryonic development of the larva itself. We did not observe any indication that certain MG neuron subtypes identified in Ciona are missing in M. occidentalis, or that additional subtypes (not observed in Ciona) are specified in M. occidentalis. Not only does there appear to be a 1-to-1 correspondence of core MG neurons between Ciona and M. occidentalis, their position in the MG is not altered. The only differences observed are precocious transcription of certain markers in M. occidentalis (e.g., Vsx, Dmbx), which may be related to the faster developmental rate of this species relative to Ciona spp. These observations suggest that the MG of the solitary tunicate larva has changed very little in nearly 400 million years, the estimated time of divergence between Ciona and Molgula [15]. As such, the tunicate larval MG likely represents a minimal but ancient and exquisitely adapted central pattern generator for swimming behavior.
Fig. 8.
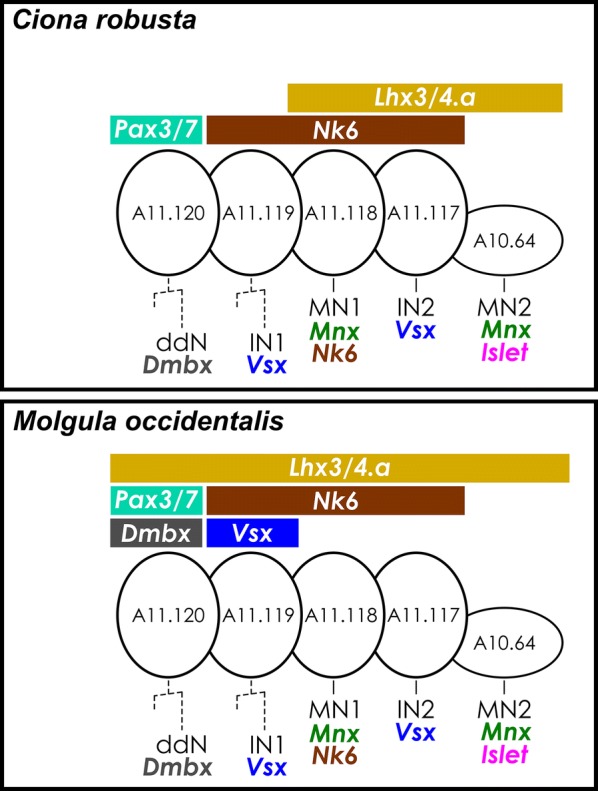
Summary of transcription factor expression patterns in Ciona and Molgula. Comparison of transcription factor expression patterns in the developing MG of Ciona robusta and Molgula occidentalis, summarizing the findings of the current study. C. robusta patterns are from references [10, 11, 35]. Gene expression domains during the early patterning of MG progenitors at the mid-tailbud stage are indicated by colored bars. MG neuron subtype-specific gene expression (ddN, IN1, MN1, IN2, MN2) indicated below. Dashed lines represent additional cell divisions before final specification and differentiation of ddN and IN1, which were resolved in C. robusta but not in M. occidentalis. The major differences observed in M. occidentalis are: early expression of Dmbx in A11.120 and Vsx in A11.119, and broader expression of Lhx3/4.a relative to Lhx3/4 in C. robusta
Our finding of acute, asymmetric DSD of Dmbx regulation hints at a possible cause for divergence in the regulation of seemingly identical gene expression patterns. In this case, both C. robusta and M. occidentalis specify a single pair of Dmbx+ ddNs. However, precocious transcriptional priming of Dmbx in the ddN progenitor (A11.120) in M. occidentalis may be the result of an alternate regulatory mechanism that does not operate in homologous cells of Ciona. Thus, while the spatial pattern of ddN specification is highly conserved, likely constrained by the invariance of the MG and its developmental lineages, there have been changes to the temporal dynamics of this process. Although the developmental timing of the ddN is identical between Molgula and Ciona in terms of number of mitotic generations, Molgula development is accelerated on an absolute timescale (e.g., hours post-fertilization). This accelerated developmental rate on an absolute timescale may have required very different regulatory strategies simply to maintain the same output (i.e., specification of the A12.239 pair of cells as the ddNs). Although this hypothesis remains untested, it will be interesting to investigate in the future whether the root of the many cis/trans incompatibilities observed between these geometrically identical embryos lies mostly in their different absolute developmental rates. This would be an extension of the models for how DSD arises, posited by True and Haag [18] and later expanded upon [55–57]. Stabilizing selection would have maintained the same exact MG neuron arrangement, while compensatory changes would be needed to account for the different absolute developmental rates. Unfortunately, the extreme divergence of tunicate noncoding sequences (Additional file 1: Fig. S2) means phylogenetic footprinting is often uninformative when searching for functional transcription factor binding sites in orthologous cis-regulatory elements. This makes comparative cis-regulatory functional studies very difficult to pursue in distantly related tunicates, such as Ciona and Molgula.
The remarkable conservation of embryonic cell lineages and geometries between distantly related tunicates has been recently proposed to also underlie the extreme divergence of tunicate genomes [19]. Because gene expression and cell fates are induced by very precise and invariant cell–cell contact events in tunicate embryos, the gene regulatory networks involved in their regulation have been allowed to drift, freed from the constraints imposed by the mechanisms required by larger, more complex and variable embryos. In other words, precision and robustness of gene expression in tunicates arise from the precision and robustness of the embryo itself and not its gene networks. In larger animals (e.g., vertebrates), this precision and robustness can only come from more precise and robust gene regulatory networks instead. Additionally, while the invariant geometry of the tunicate embryo may have served to release the genome from these constraints, it may have imposed a different set of evolutionary constraints. Because of their invariance, tunicate embryos are unable to compensate for errors in cell fate specification and likely experienced strong selective pressure to maintain precise embryo geometries even as some evolved to develop more rapidly on an absolute timescale, like M. occidentalis. It is perhaps due to this evolutionary ratchet that Oikopleura dioica, the tunicate with the fastest rate of development and most geometrically constrained embryo, also has the most fragmented, highly derived genome as well [58–63]. Future comparative studies among the tunicates promise to shed further light on the fascinating interplay between embryonic form and genetic architecture in evolution.
Additional file
Additional file 1. Supplemental sequences, information, and figures.
Authors’ contributions
EKL and AS planned the study, did the experiments, interpreted the results, and wrote the paper. The work was supported by funds awarded to AS. Both authors read and approved the final manuscript.
Acknowledgements
We thank Rodoniki Athanasiadou and David Gresham for helping us with the RNAseq. We are also grateful for the constant support from the MoEvoDevo collaborative network: Billie Swalla, C. Titus Brown, Lionel Christiaen, Filomena Ristoratore, and Claudia Racioppi. Work in the A.S. laboratory is funded by NIH award R00 HD084814. The authors have no competing interests to declare.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datasets supporting the conclusions of this article are available in the Open Science Framework (OSF) repository, https://osf.io/3crup/, and the Sequence Read Archive (SRA), accession number pending.
Consent for publication
All authors consent to publication.
Ethics approval and consent to participate
Not applicable.
Funding
Work in the A.S. laboratory is funded by NIH award R00 HD084814.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s13227-018-0107-0) contains supplementary material, which is available to authorized users.
Contributor Information
Elijah K. Lowe, Email: elijahlowe@gmail.com
Alberto Stolfi, Email: alberto.stolfi@biosci.gatech.edu.
References
- 1.Satoh N. Developmental genomics of ascidians. Hoboken: Wiley; 2013. [Google Scholar]
- 2.Nishino A. Morphology and physiology of the ascidian nervous systems and the effectors. In: Sasakura Y, editor. Transgenic ascidians. Singapore: Springer; 2018. pp. 179–196. [DOI] [PubMed] [Google Scholar]
- 3.White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1986;314(1165):1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- 4.Ryan K, Lu Z, Meinertzhagen IA. The CNS connectome of a tadpole larva of Ciona intestinalis (L.) highlights sidedness in the brain of a chordate sibling. Elife. 2016;5:1–34. doi: 10.7554/eLife.16962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ryan K, Lu Z, Meinertzhagen IA. The peripheral nervous system of the ascidian tadpole larva: types of neurons and their synaptic networks. J Comp Neurol. 2018;526(4):583–608. doi: 10.1002/cne.24353. [DOI] [PubMed] [Google Scholar]
- 6.Ryan K, Lu Z, Meinertzhagen IA. Circuit homology between decussating pathways in the Ciona larval CNS and the vertebrate startle-response pathway. Curr Biol. 2017;27(5):721–728. doi: 10.1016/j.cub.2017.01.026. [DOI] [PubMed] [Google Scholar]
- 7.Nishino A, Okamura Y, Piscopo S, Brown ER. A glycine receptor is involved in the organization of swimming movements in an invertebrate chordate. BMC Neurosci. 2010;11(1):6. doi: 10.1186/1471-2202-11-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wada H, Saiga H, Satoh N, Holland PW. Tripartite organization of the ancestral chordate brain and the antiquity of placodes: insights from ascidian Pax-2/5/8, Hox and Otx genes. Development. 1998;125(6):1113–1122. doi: 10.1242/dev.125.6.1113. [DOI] [PubMed] [Google Scholar]
- 9.Imai KS, Stolfi A, Levine M, Satou Y. Gene regulatory networks underlying the compartmentalization of the Ciona central nervous system. Development. 2009;136(2):285–293. doi: 10.1242/dev.026419. [DOI] [PubMed] [Google Scholar]
- 10.Stolfi A, Levine M. Neuronal subtype specification in the spinal cord of a protovertebrate. Development. 2011;138(5):995–1004. doi: 10.1242/dev.061507. [DOI] [PubMed] [Google Scholar]
- 11.Stolfi A, Wagner E, Taliaferro JM, Chou S, Levine M. Neural tube patterning by Ephrin, FGF and Notch signaling relays. Development. 2011;138(24):5429–5439. doi: 10.1242/dev.072108. [DOI] [PubMed] [Google Scholar]
- 12.Hudson C, Ba M, Rouvière C, Yasuo H. Divergent mechanisms specify chordate motoneurons: evidence from ascidians. Development. 2011;138(8):1643–1652. doi: 10.1242/dev.055426. [DOI] [PubMed] [Google Scholar]
- 13.Tsagkogeorga G, Cahais V, Galtier N. The population genomics of a fast evolver: high levels of diversity, functional constraint, and molecular adaptation in the tunicate Ciona intestinalis. Genome Biol Evol. 2012;4(8):852–861. doi: 10.1093/gbe/evs054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsagkogeorga G, Turon X, Galtier N, Douzery EJ, Delsuc F. Accelerated evolutionary rate of housekeeping genes in tunicates. J Mol Evol. 2010;71(2):153–167. doi: 10.1007/s00239-010-9372-9. [DOI] [PubMed] [Google Scholar]
- 15.Delsuc F, Philippe H, Tsagkogeorga G, Simion P, Tilak M-K, Turon X, López-Legentil S, Piette J, Lemaire P, Douzery EJ. A phylogenomic framework and timescale for comparative studies of tunicates. BMC Biol. 2018;16(1):39. doi: 10.1186/s12915-018-0499-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kocot KM, Tassia MG, Halanych KM, Swalla BJ. Phylogenomics offers resolution of major tunicate relationships. Mol Phylogenet Evol. 2018;121:166–173. doi: 10.1016/j.ympev.2018.01.005. [DOI] [PubMed] [Google Scholar]
- 17.Stolfi A, Lowe EK, Racioppi C, Ristoratore F, Brown CT, Swalla BJ, Christiaen L. Divergent mechanisms regulate conserved cardiopharyngeal development and gene expression in distantly related ascidians. eLife. 2014;3:e03728. doi: 10.7554/eLife.03728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.True JR, Haag ES. Developmental system drift and flexibility in evolutionary trajectories. Evol Dev. 2001;3(2):109–119. doi: 10.1046/j.1525-142x.2001.003002109.x. [DOI] [PubMed] [Google Scholar]
- 19.Guignard L, Fiuza U-M, Leggio B, Faure E, Laussu J, Hufnagel L, Malandain G, Godin C, Lemaire P. Contact-dependent cell communications drive morphological invariance during ascidian embryogenesis. bioRxiv. 2018;238741. [DOI] [PubMed]
- 20.Brozovic M, Martin C, Dantec C, Dauga D, Mendez M, Simion P, Percher M, Laporte B, Scornavacca C, Di Gregorio A. ANISEED 2015: a digital framework for the comparative developmental biology of ascidians. Nucl Acids Res. 2015;44(D1):D808–D818. doi: 10.1093/nar/gkv966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brozovic M, Dantec C, Dardaillon J, Dauga D, Faure E, Gineste M, Louis A, Naville M, Nitta KR, Piette J. ANISEED 2017: extending the integrated ascidian database to the exploration and evolutionary comparison of genome-scale datasets. Nucl Acids Res. 2017;46(D1):D718–D725. doi: 10.1093/nar/gkx1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crusoe MR, Alameldin HF, Awad S, Boucher E, Caldwell A, Cartwright R, Charbonneau A, Constantinides B, Edvenson G, Fay S, et al. The khmer software package: enabling efficient nucleotide sequence analysis. F1000Research. 2015;4:900. doi: 10.12688/f1000research.6924.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schulz MH, Zerbino DR, Vingron M, Birney E. Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics. 2012;28(8):1086–1092. doi: 10.1093/bioinformatics/bts094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pryszcz LP, Gabaldón T. Redundans: an assembly pipeline for highly heterozygous genomes. Nucl Acids Res. 2016;44(12):e113. doi: 10.1093/nar/gkw294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hunt M, Kikuchi T, Sanders M, Newbold C, Berriman M, Otto TD. REAPR: a universal tool for genome assembly evaluation. Genome Biol. 2013;14(5):R47. doi: 10.1186/gb-2013-14-5-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neymotin B, Athanasiadou R, Gresham D. Determination of in vivo RNA kinetics using RATE-seq. RNA. 2014;20(10):1645–1652. doi: 10.1261/rna.045104.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parkhomchuk D, Borodina T, Amstislavskiy V, Banaru M, Hallen L, Krobitsch S, Lehrach H, Soldatov A. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucl Acids Res. 2009;37(18):e123. doi: 10.1093/nar/gkp596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pertea M, Pertea GM, Antonescu CM, Chang T-C, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33(3):290. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31(19):3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 34.Christiaen L, Wagner E, Shi W, Levine M. Isolation of sea squirt (Ciona) gametes, fertilization, dechorionation, and development. Cold Spring Harb Protoc. 2009;2009(12):pdb-prot5344. doi: 10.1101/pdb.prot5344. [DOI] [PubMed] [Google Scholar]
- 35.Ikuta T, Saiga H. Dynamic change in the expression of developmental genes in the ascidian central nervous system: revisit to the tripartite model and the origin of the midbrain-hindbrain boundary region. Dev Biol. 2007;312:631–643. doi: 10.1016/j.ydbio.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 36.Cole AG, Meinertzhagen IA. The central nervous system of the ascidian larva: mitotic history of cells forming the neural tube in late embryonic Ciona intestinalis. Dev Biol. 2004;271(2):239–262. doi: 10.1016/j.ydbio.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 37.Nicol D, Meinertzhagen I. Development of the central nervous system of the larva of the ascidian, Ciona intestinalis L: I. The early lineages of the neural plate. Dev Biol. 1988;130(2):721–736. doi: 10.1016/0012-1606(88)90363-6. [DOI] [PubMed] [Google Scholar]
- 38.Nicol D, Meinertzhagen I. Development of the central nervous system of the larva of the ascidian, Ciona intestinalis L: II. Neural plate morphogenesis and cell lineages during neurulation. Dev Biol. 1988;130(2):737–766. doi: 10.1016/0012-1606(88)90364-8. [DOI] [PubMed] [Google Scholar]
- 39.Navarrete IA, Levine M. Nodal and FGF coordinate ascidian neural tube morphogenesis. Development. 2016;143(24):4665–4675. doi: 10.1242/dev.144733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishitsuji K, Horie T, Ichinose A, Sasakura Y, Yasuo H, Kusakabe TG. Cell lineage and cis-regulation for a unique GABAergic/glycinergic neuron type in the larval nerve cord of the ascidian Ciona intestinalis. Dev Growth Differ. 2012;54(2):177–186. doi: 10.1111/j.1440-169X.2011.01319.x. [DOI] [PubMed] [Google Scholar]
- 41.Satou Y, Takatori N, Yamada L, Mochizuki Y, Hamaguchi M, Ishikawa H, Chiba S, Imai K, Kano S, Murakami SD, et al. Gene expression profiles in Ciona intestinalis tailbud embryos. Development. 2001;128(15):2893–2904. doi: 10.1242/dev.128.15.2893. [DOI] [PubMed] [Google Scholar]
- 42.Mazet F, Hutt JA, Milloz J, Millard J, Graham A, Shimeld SM. Molecular evidence from Ciona intestinalis for the evolutionary origin of vertebrate sensory placodes. Dev Biol. 2005;282(2):494–508. doi: 10.1016/j.ydbio.2005.02.021. [DOI] [PubMed] [Google Scholar]
- 43.Takamura K, Egawa T, Ohnishi S, Okada T, Fukuoka T. Developmental expression of ascidian neurotransmitter synthesis genes. Dev Genes Evol. 2002;212(1):50–53. doi: 10.1007/s00427-001-0205-0. [DOI] [PubMed] [Google Scholar]
- 44.Ikuta T, Saiga H. Dynamic change in the expression of developmental genes in the ascidian central nervous system: revisit to the tripartite model and the origin of the midbrain-hindbrain boundary region. Dev Biol. 2007;312:631–643. doi: 10.1016/j.ydbio.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 45.Razy-Krajka F, Lam K, Wang W, Stolfi A, Joly M, Bonneau R, Christiaen L. Collier/OLF/EBF-dependent transcriptional dynamics control pharyngeal muscle specification from primed cardiopharyngeal progenitors. Dev Cell. 2014;29(3):263–276. doi: 10.1016/j.devcel.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Imai JH, Meinertzhagen IA. Neurons of the ascidian larval nervous system in Ciona intestinalis: I. Central nervous system. J Comp Neurol. 2007;501(3):316–334. doi: 10.1002/cne.21246. [DOI] [PubMed] [Google Scholar]
- 47.Nishino A, Baba SA, Okamura Y. A mechanism for graded motor control encoded in the channel properties of the muscle ACh receptor. Proc Natl Acad Sci. 2011;108(6):2599–2604. doi: 10.1073/pnas.1013547108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kobayashi M, Takatori N, Nakajima Y, Kumano G, Nishida H, Saiga H. Spatial and temporal expression of two transcriptional isoforms of Lhx3, a LIM class homeobox gene, during embryogenesis of two phylogenetically remote ascidians, Halocynthia roretzi and Ciona intestinalis. Gene Expr Patterns. 2010;10(2):98–104. doi: 10.1016/j.gep.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 49.Christiaen L, Stolfi A, Davidson B, Levine M. Spatio-temporal intersection of Lhx3 and Tbx6 defines the cardiac field through synergistic activation of Mesp. Dev Biol. 2009;328(2):552–560. doi: 10.1016/j.ydbio.2009.01.033. [DOI] [PubMed] [Google Scholar]
- 50.Takatori N, Kumano G, Saiga H, Nishida H. Segregation of germ layer fates by nuclear migration-dependent localization of not mRNA. Dev Cell. 2010;19(4):589–598. doi: 10.1016/j.devcel.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 51.Tsagkogeorga G, Turon X, Hopcroft RR, Tilak M-K, Feldstein T, Shenkar N, Loya Y, Huchon D, Douzery EJ, Delsuc F. An updated 18S rRNA phylogeny of tunicates based on mixture and secondary structure models. BMC Evol Biol. 2009;9(1):187. doi: 10.1186/1471-2148-9-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Katsuyama Y, Okada T, Matsumoto J, Ohtsuka Y, Terashima T, Okamura Y. Early specification of ascidian larval motor neurons. Dev Biol. 2005;278(2):310–322. doi: 10.1016/j.ydbio.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 53.Dynes JL, Ngai J. Pathfinding of olfactory neuron axons to stereotyped glomerular targets revealed by dynamic imaging in living zebrafish embryos. Neuron. 1998;20(6):1081–1091. doi: 10.1016/S0896-6273(00)80490-0. [DOI] [PubMed] [Google Scholar]
- 54.Kimmel CB, Model PG. Developmental studies of the Mauthner cell. In: Faber DS, Korn H, editors. Neurobiology of the Mauthner cell. New York: Raven Press; 1978. pp. 183–220. [Google Scholar]
- 55.Pavlicev M, Wagner GP. A model of developmental evolution: selection, pleiotropy and compensation. Trends Ecol Evol. 2012;27(6):316–322. doi: 10.1016/j.tree.2012.01.016. [DOI] [PubMed] [Google Scholar]
- 56.Johnson NA, Porter AH. Evolution of branched regulatory genetic pathways: directional selection on pleiotropic loci accelerates developmental system drift. Genetica. 2007;129(1):57–70. doi: 10.1007/s10709-006-0033-2. [DOI] [PubMed] [Google Scholar]
- 57.Landry CR, Wittkopp PJ, Taubes CH, Ranz JM, Clark AG, Hartl DL. Compensatory cis-trans evolution and the dysregulation of gene expression in interspecific hybrids of Drosophila. Genetics. 2005;171:1813–1822. doi: 10.1534/genetics.105.047449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Denoeud F, Henriet S, Mungpakdee S, Aury J-M, Da Silva C, Brinkmann H, Mikhaleva J, Olsen LC, Jubin C, Cañestro C. Plasticity of animal genome architecture unmasked by rapid evolution of a pelagic tunicate. Science. 2010;330(6009):1381–1385. doi: 10.1126/science.1194167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fujii S, Nishio T, Nishida H. Cleavage pattern, gastrulation, and neurulation in the appendicularian, Oikopleura dioica. Dev Genes Evol. 2008;218(2):69–79. doi: 10.1007/s00427-008-0205-4. [DOI] [PubMed] [Google Scholar]
- 60.Seo H-C, Kube M, Edvardsen RB, Jensen MF, Beck A, Spriet E, Gorsky G, Thompson EM, Lehrach H, Reinhardt R. Miniature genome in the marine chordate Oikopleura dioica. Science. 2001;294(5551):2506. doi: 10.1126/science.294.5551.2506. [DOI] [PubMed] [Google Scholar]
- 61.Seo H-C, Edvardsen RB, Maeland AD, Bjordal M, Jensen MF, Hansen A, Flaat M, Weissenbach J, Lehrach H, Wincker P. Hox cluster disintegration with persistent anteroposterior order of expression in Oikopleura dioica. Nature. 2004;431(7004):67. doi: 10.1038/nature02709. [DOI] [PubMed] [Google Scholar]
- 62.Edvardsen RB, Lerat E, Maeland AD, Flåt M, Tewari R, Jensen MF, Lehrach H, Reinhardt R, Seo H-C, Chourrout D. Hypervariable and highly divergent intron–exon organizations in the chordate Oikopleura dioica. J Mol Evol. 2004;59(4):448–457. doi: 10.1007/s00239-004-2636-5. [DOI] [PubMed] [Google Scholar]
- 63.Nishida H. Development of the appendicularian Oikopleura dioica: culture, genome, and cell lineages. Dev Growth Diff. 2008;50(s1):S239–S256. doi: 10.1111/j.1440-169X.2008.01035.x. [DOI] [PubMed] [Google Scholar]
- 64.Conklin EG. The organization and cell-lineage of the ascidian egg. Science. 1906;13:340–344. [Google Scholar]
- 65.Hotta K, Mitsuhara K, Takahashi H, Inaba K, Oka K, Gojobori T, Ikeo K. A web-based interactive developmental table for the ascidian Ciona intestinalis, including 3D real-image embryo reconstructions: I. From fertilized egg to hatching larva. Dev Dyn. 2007;236(7):1790–1805. doi: 10.1002/dvdy.21188. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Supplemental sequences, information, and figures.
Data Availability Statement
The datasets supporting the conclusions of this article are available in the Open Science Framework (OSF) repository, https://osf.io/3crup/, and the Sequence Read Archive (SRA), accession number pending.