Abstract
Neuronal ELAV (nELAV) proteins are RNA-binding proteins which play a physiological role in controlling gene expression in memory formation, and their alteration may contribute to cognitive impairment associated with neurodegenerative pathologies such as Alzheimer’s disease (AD). Indeed, we found that the content of nELAV proteins is significantly decreased along with clinical dementia progression in the hippocampi of AD brains, where it inversely correlates with the amount of amyloid-β (Aβ). To check the direct influence of Aβ on nELAV, we performed in vitro experiments using human SH-SY5Y cells, finding that Aβ1–42 specifically determines nELAV proteins reduction. Since ADAM10 mRNA has the predicted sequences targeted by nELAV, we investigated whether Aβ, through nELAV proteins, could originate a vicious circle affecting amyloid-β protein precursor (AβPP) processing. Immunoprecipitation experiments showed that indeed nELAV proteins bind to ADAM10 mRNA and that this binding is disrupted by Aβ1–42 exposure, resulting in a decreased ADAM10 protein expression. ADAM10 protein diminution was also found in AD hippocampi. These data show for the first time the involvement of nELAV in AD pathology and suggest that their alteration may affect genes implicated in AβPP processing.
Keywords: ADAM10, Alzheimer’s disease, amyloid-β, amyloid-β protein precursor processing, nELAV
INTRODUCTION
The RNA-binding proteins ELAV (Embryonic Lethal Abnormal Vision) appear to be implicated in the regulation of all the key steps of mRNA metabolism, from pre-mRNA splicing to mRNA transport, stability and translation [1–3]. In vertebrates, HuB (a.k.a. Hel-N1), HuC and HuD represent the neuron-specific members of the family (nELAV proteins), while HuR (a.k.a. HuA) is ubiquitously expressed. ELAV proteins act post-transcriptionally by preferentially binding to adenine and uridine-rich elements (AREs) found in the 3’ untranslated region (3’ UTR) of some mRNAs, and enhance gene expression by increasing the cytoplasmic stability and rate of translation of ARE-containing mRNAs (for a review, see [2]). We previously demonstrated a physiological role for nELAV proteins in controlling gene expression in spatial memory, a form of memory in which the hippocampus plays a predominant role [4,5]. Moreover, hippocampal HuD upregulation was documented in another non-spatial learning paradigm, fear conditioning [6]. Since ELAV proteins play a pleiotropic role in several cellular events by stabilizing ARE-containing mRNAs, their derangement may have implications in neurodegenerative pathologies characterized by a loss of memory and multiple dysfunctions such as Alzheimer’s disease (AD). Within this context, it was of interest to investigate whether AD is associated with a loss of ELAV proteins in regions involved in the memory circuitry and whether amyloid-β (Aβ) itself may have direct actions on the ELAV network.
Moreover, among the various genes involved in the pathogenesis of AD, we found that ADAM10 bears, in the 3’UTR of its mRNA, clear ARE signatures, thus representing a putative target of ELAV proteins. ADAM10 is a member of the ADAM (A Disintegrin and Metalloproteinase) family of integral membrane proteins that act as α-secretases (see [7]). Literature data indicate significantly reduced ADAM10 protein levels in platelets of sporadic AD patients coupled with reduced soluble AβPPα (sAβPPα) levels in both platelets and cerebrospinal fluid [8]. These observations underscored the interest to investigate whether changes in ELAV proteins may be linked to a defective ADAM10 expression. Accordingly, in the present paper, we explored the possibility that nELAV proteins are altered in the hippocampus of AD patients, which indeed was the case. We also explored whether the changes were associated with Aβ burden and ADAM10 expression. Moreover, in a cellular model, the human neuroblastoma SH-SY5Y cells, we investigated the effect of Aβ on nELAV proteins, the binding of nELAV proteins to ADAM10 mRNA and the effect of Aβ exposure on this binding and on ADAM10 expression.
MATERIALS AND METHODS
Patients selection criteria
Human postmortem brain samples from AD and age-matched non-AD cases were obtained from the Alzheimer’s Disease Brain Bank of the Mount Sinai School of Medicine. The cases selected had either no significant neuropathological features or only neuropathological features associated with AD [9,10]. A multistep approach based on cognitive and functional status during the last 6 months of life was applied to the assignment of CDRs [11] as previously reported [9, 10]. Samples were divided into groups on the basis of their CDRs as follows: CDR 0, non-demented (n = 20); CDR 0.5, at high risk of developing AD dementia, also defined as MCI (Mild Cognitive Impaired) (n = 13); CDR 1, mild dementia (n =15); CDR 2, moderate dementia (n = 14); CDR 5, severe dementia (n = 24). Patients information included in this study is summarized in Table 1.
Table 1.
Summary of the post-mortem brain hippocampi
| CDR | N | Age at death (years) Mean ± S.D |
Female/Male | PMI (minutes) Mean ± S.D. |
|---|---|---|---|---|
| 0 | 20 | 77± 15 | 13/7 | 709 ± 585 |
| 0.5 | 13 | 87 ± 8 | 7/6 | 388 ± 323 |
| 1 | 15 | 82± 11 | 8/7 | 493 ± 702 |
| 2 | 14 | 87± 7 | 12/2 | 298 ± 181 |
| 5 | 24 | 83± 10 | 17/7 | 341 ± 255 |
PMI: postmortem interval time.
All studies involving human samples were approved by the Institutional Review Board of Mount Sinai School of Medicine, New York, USA.
Quantification of Aβ peptide content
Hippocampal samples were treated according to a previous published method [12]. Briefly, samples were first extracted with TBS (20 mM Tris and 137 mM Na-Cl pH 7.6 with protease inhibitor cocktail), followed by ultracentrifugation at 4°C for one hour (100,000 × g). The supernatant was analyzed using a commercially available ELISA kit (BioSource, Camarillo, CA) according to the manufacturer’s instruction. The pellet was extracted with 70% formic acid, neutralized with 2 M Tris-HCl (pH 11) in the presence of protease inhibitor cocktail (Roche Biochemicals, Indianapolis, IN) and centrifuged at 4°C and 100,000 × g for 1 hour. The content of formic acid soluble amyloid peptide was quantified by ELISA assay.
Cell cultures and in vitro treatments
SH-SY5Y human neuroblastoma cells were grown in Eagle’s minimum essential medium (MEM) supplemented with 10% fetal calf serum (FCS), penicillin/streptomycin, non-essential amino acids and sodium pyruvate (1 mM) at 37°C in an atmosphere of 5% CO2 and 95% humidity. SH-SY5Y human neuroblastoma cells were exposed to the solvent (DMSO) or to 1 µM Aβ1–42 or the reverse Aβ40–1 peptides for 24 hours.
MTT assay
SH-SY5Y cells were plated 50,000 cells/well in a 96-wells plate. Cell viability was determined by 3-(4,5-dimethylthiazol-2-y1)-2,5-diphenyltetrazolium bromide (MTT, Sigma) assay, an indicator of mitochondrial activity. Following cell treatments, MTT solution (1 mg/ml) was added to the medium and incubated at 37°C for 4 hours. Finally, the lysis buffer (20% SDS in 50% dimethylformamide) was added to the 96-wells plate and the cells incubated at 37°C overnight. Absorbance was measured at 495nm in a UV spectrophotometer and the results were expressed as a percentage of the absorbance of the treated samples in comparison to the control.
Immunocytochemistry
Cells plated on coverslips were treated as described above. Then cells were processed as reported in [13]. Cells were then incubated for 1 hour at room temperature with the monoclonal antibody 6E10 recognizing the Aβ protein, diluted 1:50 in PBS/1% BSA solution. Cells were then washed with PBS solution and incubated for 1 hour with the rabbit anti-IgG antibody RPE (R-phycoerythrin-conjugated; DakoCytomation, Denmark) diluted 1:200 in PBS/1% BSA solution. After the labeling procedures, cells were mounted up-side-down on glass slides, in a drop of mounting medium with DAPI (Vectashield, USA).
Preparation of the cytoskeletal fractions
The cytoskeletal fractions from brain samples and SH-SY5Y human neuroblastoma cells were separated accordingly to a published method [14] with slight modifications. 20 µl of total homogenate were stored while the majority was centrifuged at 2,500 × g for 10 minutes in a bench-centrifuge. The supernatant was centrifuged at 100,000 × g for 1 hour. The pellet was resuspended with the homogenization buffer containing 1% Triton X-100 sonicated, incubated for 45 minutes, and centrifuged at 100,000 × g for 1 hour. The resulting pellet constitutes the cytoskeleton. Protein contents of all the samples were determined by the Bradford’s method.
Western blotting
Total lysates and the cytoskeletal fractions were processed as previously described [14]. The following antibodies were diluted in 6% milk in TBS-T Buffer (10 mM Tris-HCl, 100 mM NaCl, 0.1% Tween, pH 7.5): anti-nELAV (1:1000; Santa Cruz, USA), anti-ADAM10 (1:500; Abcam, England), anti-α-tubulin (1:1000; Chemicon, Italy) and 6E10 (1:500; Chemicon, Italy). The horseradish peroxidase-conjugated secondary antibodies (Molecular Probes, OR, USA) were diluted in 6% milk in TBS-T Buffer. The nitrocellulose membranes signals were detected by chemiluminescence. The specificities of all the antibodies were evaluated performing preliminary immunoblottings in the presence of the secondary antibodies alone. Experiments were performed at least three times for each different cell preparation. Statistical analysis of western blotting data was performed on the densitometric values obtained with the NIH image software 1.61 (downloadable at http://rsb.info.nih.gov/nih-image).
Immunoprecipitation followed by RNA extraction
SH-SY5Y human neuroblastoma cytoskeletal fraction was obtained as described above and immunoprecipitation was performed according to a previously published protocol with minor modifications [13]. Immunoprecipitation was performed using 1 µg of anti-Hu antibody (Santa Cruz, USA) per 50 µg of proteins diluted with an equal volume of 2× Immunoprecipitation Buffer [2% Triton X-100, 300 mM NaCl, 20 mM Tris-HCl (pH 7.4), 2 mM EDTA, 2 mM EGTA, 0.4 mM sodium vanadate, protease inhibitor cocktail and a RNAase inhibitor] in presence of 50 µl of protein A/G plus agarose (Santa Cruz Biotech, CA, USA). The samples were finally subjected to RNA extraction. The negative control was obtained in the same conditions, but in the presence of an irrelevant antibody with the same isotype of the specific immunoprecipitating antibody. 100 µl of the immunoprecipitation mix were immediately collected from each sample and used as “input signals” to normalize the RT-PCR data.
Real-time quantitative RT-PCR
Total RNA was extracted from SH-SY5Y cells and from immunoprecipitated pellets and relative “input signals” by the Trizol reagent (Invitrogen, CA, USA), treated with DNase and subjected to reverse transcription following standard procedures. PCR amplifications were carried out using the Lightcycler instrument (Roche, Germany), as previously described [13], with primers designed by using the PRIMER3 software (http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi). Primer sequences were as follows: ADAM10, 5’-GGGACACATGAGACGCTAACT-3’ (upstream), 5’-AATTCCACCTGG-TCTGAGGA-3’ (downstream); GAPDH, 5’-CAGCAAGAGCACAAGAGGAAG-3’ (upstream), 5’-CAACTGTGAGGAGGGGAGATT-3’ (downstream); RPL6, 5’-AGATTA-CGGAGCAGCGCAAGATTG-3’ (upstream), 5’ - GCAAACACAGATCGCAGGTAGCCC-3’ (downstream). The GAPDH and RPL6 mRNAs were chosen to check the specificity of the nELAV binding to ADAM10 (not shown). Treatments were repeated at least 3 times, and for each treatment RT-PCR experiments were performed on samples run in duplicate.
Data analysis
For statistical analysis, the GraphPad Instat statistical package (version 3.05 GraphPad software, San Diego, CA, USA) was used. The data were analyzed by analysis of variance (ANOVA) followed, when significant, by an appropriate post hoc comparison test. When Bartlett’s test indicated non-homogeneity of variance, a non-parametric test, i.e., the Kruskall-Wallis ANOVA followed by Dunn’s test was applied. Differences were considered statistically significant when p values ≤ 0.05.
RESULTS
The nELAV are decreased in postmortem AD patients
Considering the key involvement of nELAV proteins in memory processes, we wanted to assess whether their content was altered in AD, a pathology associated with cognitive deficits. Keeping in mind that the cytoskeleton is the subcellular compartment where nELAV are mostly activated [13], we performed western blotting analyses on cytoskeletal fractions from hippocampal samples of patients characterized by increasing dementia rating (CDR 0.5, CDR 1, CDR 2, CDR 5) relative to neurological control cases (CDR 0). Case characteristics are summarized in Table 1. Analysis of variance indicated that there were no significant differences among the various CDR groups with respect to postmortem interval and age at death. Cause of death was reviewed for all the patients to rule out the possibility that intercurrent infection or other events would affect study measures; major causes of death of patients enrolled in the study included acute cardiac failure, ventricular fibrillation, cardiopulmonary arrest, myocardial infarct, congestive heart failure, cancer, and pneumonia.
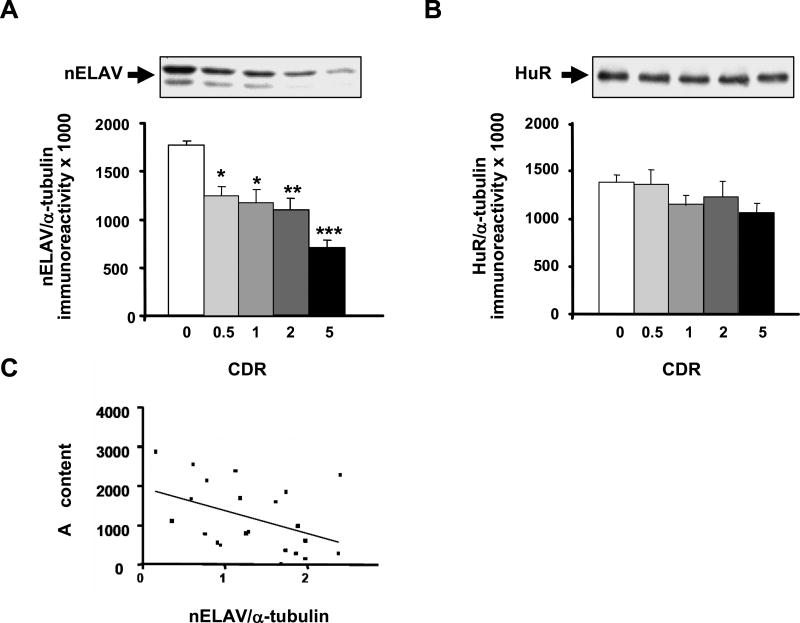
As shown in Fig. 1, nELAV protein levels decreased along with clinical dementia progression. Interestingly, significant lower levels were already present in CDR 0.5, showing the highest reduction in CDR 5 (see Fig. 1A). The diminution of nELAV content becomes so dramatic in severe AD that a significant difference is also observed between CRD 0.5 and CDR 5 (p < 0.05, Dunn’s test). These results are further supported by the evidence that nELAV levels inversely correlated with CDR scores (r = −0.6468; p < 0.0001). In contrast, the ubiquitously expressed ELAV HuR protein was not modified (Fig. 1B). Most interestingly, we found that the content of hippocampal nELAV proteins inversely correlate with the hippocampal content of TBS-soluble Aβ (r = −0.432; p < 0.05; Fig. 1C) and the formic acid soluble Aβ (r = −0.326; p < 0.05) as a function of progression of clinical dementia. These data suggest that Aβ, among other effects, may have a causal role in decreasing nELAV proteins content.
Fig. 1.
nELAV but not HuR protein levels are altered in the hippocampus from AD patients. (Upper) Representative Western blots of nELAV (A) and HuR (B) in the hippocampal cytoskeleton from control (CDR 0) and AD patients with a different progression of the disease (CDR 0.5, CDR 1, CDR 2, CDR 5). (Lower) Mean grey levels ratios (mean ± S.E.M.) of nELAVs/α-tubulin (A) and HuR/α-tubulin (B) immunoreactivities measured by Western blotting in the same samples. *p < 0.05, **p < 0.01, ***p < 0.001, Dunn’s test, n = 13–24. (C) nELAV proteins inversely correlate with Aβ content. Total Aβ content in the cytoskeleton of AD hippocampi was evaluated by ELISA assay. High levels of Aβ correspond to lower nELAVs/α-tubulin ratios. The solid line represents the best-fit correlation between nELAVs/α-tubulin ratios and Aβ content.
Aβ intracellular accumulation induces a reduction of nELAV in SH-SY5Y cells
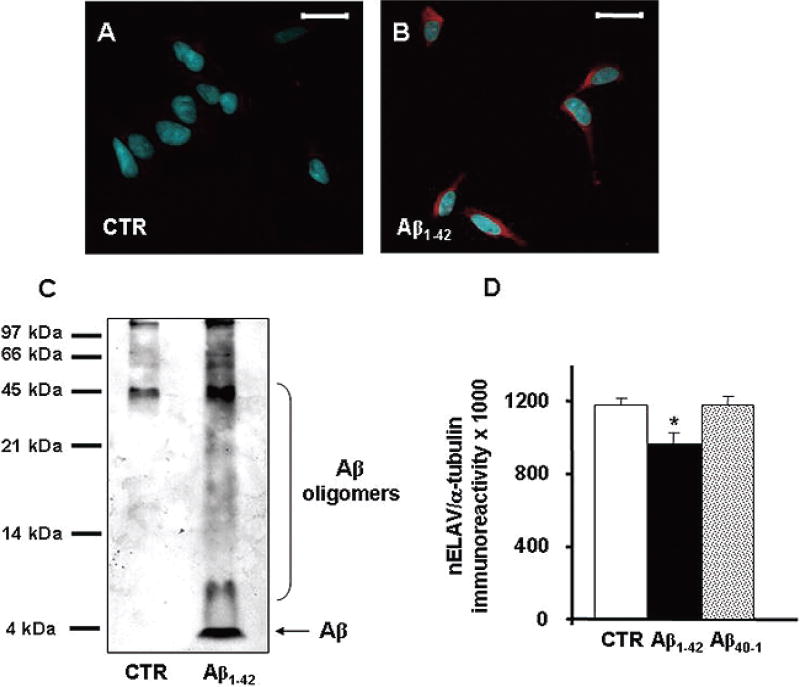
To better investigate the direct influence of Aβ on nELAV proteins, we performed in vitro experiments using a well characterized cellular model such as SHSY5Y human neuroblastoma cells. The major products of the amyloidogenic pathway are Aβ1–40, the most abundant fragment released in physiological conditions, and Aβ1–42, the most hydrophobic and toxic peptide predominant in AD plaques [15]; this latter evidence led us to focus our experiments on the Aβ1–42 peptide. Treatment of SH-SY5Y human neuroblastoma cells with 1 µM Aβ1–42 for 24 hours induced a cytoplasmic accumulation of this amyloid peptide, as shown in Fig. 2B, without any significant induction of mitochondrial damage as measured by the MTT assay (not shown). Besides the incorporation of exogenous Aβ, we cannot exclude that the intracellular accumulation of Aβ may be also due to an increase in the cleavage of endogenous AβPP. Following Aβ1–42 exposure, immunoblots performed on total cell homogenates confirmed the presence of intracellular monomeric Aβ (around at 4kDa) and self-assembled Aβ oligomers, visible as high-molecular weight bands (Fig. 2C). Aβ1–42 determined a clear decrease of nELAV levels in total cell lysates while no effect was observed with the reverse, inactive, Aβ40–1 peptide (Aβ1–42 vs control: −18%; p < 0.05, Dunnett’s Multiple Comparisons test, n = 5–6). Similar results to those obtained for Aβ1–42 on nELAV protein levels were achieved with Aβ1–40 at the same concentration of 1 µM (not shown). Further examination of cell fractions showed that a reduction in nELAV protein levels was evident following Aβ1–42 challenge especially in the cytoskeleton, as shown in Fig. 2D.
Fig. 2.
(A–B) Aβ is taken up by SH-SY5Y neuroblastoma cells. Confocal fluorescence representative images of SH-SY5Y control (A) and treated with 1 µMAβ1–42 (B) for 24 hours. Aβ staining (red) is almost undetectable in control cells, while in treated cells is spread all around the cytoplasm. Nuclei are stained with DAPI (blue). Scale bar: 20 µm. (C) Aβ aggregates in multiple assembly states. Representative Western blotting on the total homogenate from control and Aβ1–42 treated SH-SY5Y neuroblastoma cells. The 6E10 antibody recognizes the amino acid residues 1–17 of the Aβ sequence and is able to detect Aβ monomer and various Aβ aggregates with different molecular weights. (D) Aβ challenge reduces nELAV protein levels. (Upper) Representative Western blot of nELAV in the cytoskeleton from control and Aβ1–42 treated SH-SY5Y neuroblastoma cells. (Lower) Mean grey levels ratios (mean ± S.E.M.) of nELAV/α-tubulin immunoreactivities measured by Western blotting in the cytoskeletal fractions from the same samples. **p < 0.01, Student t-test, n = 6. (Colours are visible in the online version of the article www.iospress.nl.)
nELAV proteins specifically bind the ADAM10 mRNA and this binding is disrupted following Aβ exposure
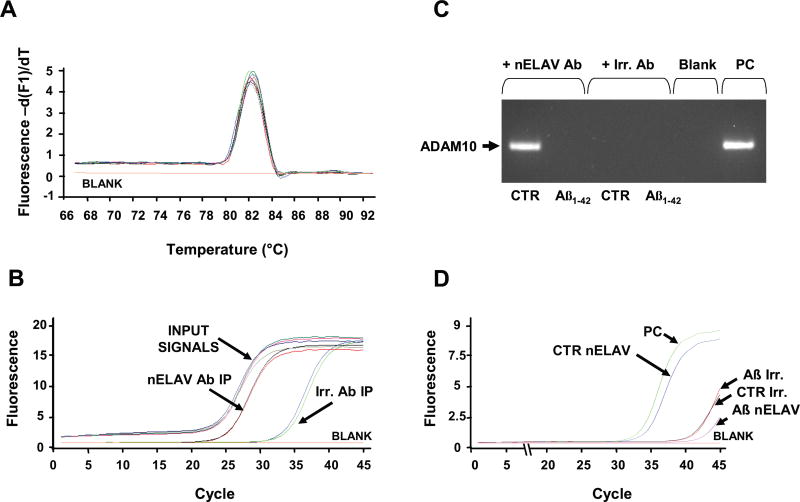
ELAV proteins can form ribonucleoproteic complexes that associate to the translational machinery through the cytoskeleton [16] and have been demonstrated to enhance translation of target mRNAs [17–19]. A reduction of nELAV content may then lead to a decreased protein expression of the target mRNA. The mRNA of ADAM10, the best characterized α-secretase, contains in its 3’UTR computationally predicted ARE sequences, thus leading us to hypothesize that this mRNA could be one of the nELAV targets; we also hypothesized that Aβ might interfere with the binding between nELAV proteins and ADAM10 mRNA. To first test the hypothesis that nELAV proteins can interact with ADAM10 mRNA, we extracted the bound mRNA from the immunoprecipitated nELAV proteins and performed real-time quantitative RT-PCR experiments for the ADAM10 mRNA. Immunoprecipitation with an irrelevant antibody of the same isotype of the nELAV antibody was used as a negative control. The content of ADAM10 mRNA in the immunoprecipitated nELAV pellet was significantly higher (about 300-fold, see Fig. 3B) in comparison to the negative control, suggesting the existence of a specific binding of nELAV proteins to the ADAM10 mRNA. To evaluate the specificity of the nELAV binding to ADAM10 mRNA, all the immunoprecipitates were also subjected to real-time RT-PCR with primers for GAPDH and Ribosomal Protein L6 (RPL6), since they do not contain ARE sequences. As expected, the GAPDH and RPL6 mRNAs were almost undetectable in the immunoprecipitated nELAV proteins (not shown).
Fig. 3.
nELAV proteins specifically bind the ADAM10 mRNA. (A) Melting curve analysis of the Input Signals and of the immunoprecipitated pellets (IP) from nELAV and from the irrelevant antibody (Irr.). All the samples yield only one peak resulting from the specific amplification product corresponding to ADAM10. For the blank sample, the template was replaced with PCR-grade water. (B) Amplification plots showing the increase in fluorescence from nELAV IP versus Irr. IP; nELAV IP contains a much higher amount of ADAM10 template in comparison to Irr. IP [287.5 fold change calculated on the basis of the respective cycle threshold (Ct) means; nELAV IP Ct: 24.8; Irr. IP Ct: 33.2]. All the Input Signals (IS) have the same amplification plot [nELAV IS Ct: 23.6; Irr. IS Ct: 23.8], indicating that the amount of the starting template is the same for both the immunoprecipitating nELAV and Irr. antibodies. (C) Aβ challenge disrupts nELAV proteins binding to ADAM10 mRNA. Representative agarose gel following real time quantitative RT-PCR coupled with immunoprecipitation experiment using anti-nELAV antibody (+nELAV Ab) or an irrelevant antibody (+Irr. Ab), with the same isotype of nELAVs, as a negative control. The band corresponding to ADAM10 is detectable only in control SH-SY5Y cells immunoprecipitated with anti-nELAV. A cDNA obtained from a total mRNA extract was utilized as a positive control (PC). In the blank sample the template was replaced with PCR-grade water. (D) Amplification plots relative to ADAM10 contained in the samples run in Figure 3C. The plots show that nELAV Aβ1–42 treated cells IP contains a much less amount of ADAM10 template in comparison to nELAV control cells IP [respective cycle threshold (Ct): nELAV IP control, Ct: 31.7; nELAV IP Aβ1–42 treated Ct: >41; Irr. IP control Ct: >41; Irr. IP Aβ1–42 treated Ct: >41; PC Ct: 30.6].
We then tested functional changes in the binding of nELAV proteins to ADAM10 mRNA by performing the immunoprecipitation experiment after Aβ1–42 treatment. Indeed, while the ADAM10 mRNA from control cells was detectable by real-time quantitative RT-PCR, no signal was observed following exposure to the peptide (see the amplified products run in an agarose gel in Fig. 3C and the relative curves in Fig. 3D).
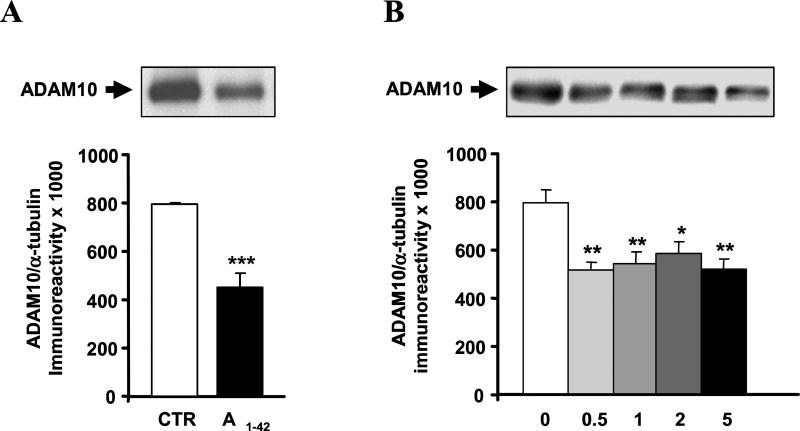
We therefore measured ADAM10 protein levels following the exposure to Aβ1–42 and the reverse Aβ40–1 peptide as control. The data show that Aβ1–42 induces a diminution in the levels ofADAM10 protein, while no effect was observed with the reverse peptide (Aβ1–42 vs control: −28%; p<0.05, Dunnett’s Multiple Comparisons test, n = 5). Similar results to those obtained for Aβ1–42 on ADAM10 protein levels were achieved with Aβ1–40 at the same concentration of 1 µM (not shown). Further examination of cell fractions showed a statistically significant reduction of ADAM10 protein levels in the cytoskeletal compartment following the exposure to Aβ1–42 for 24 hours (see Fig. 4A). We additionally tested another α-secretase, ADAM17, finding that its levels were instead unchanged (not shown).
Fig. 4.
(A) Aβ treatment affects ADAM10 protein levels. (Upper) Representative Western blotting of ADAM10 in the cytoskeleton from control and Aβ1–42 treated SH-SY5Yneuroblastoma cells. (Lower) Mean grey levels ratios (mean ± S.E.M.) of ADAM10/α-tubulin immunoreactivities measured by Western blotting in the cytoskeleton from the same samples. ***p < 0.001, Student t-test, n = 6. (B) ADAM10 protein levels are altered in the hippocampus from AD patients. (Upper) Representative Western blot of ADAM10 in the hippocampal cytoskeleton from control (CDR 0) and AD patients with a different progression of the disease (CDR 0.5, CDR 1, CDR 2, CDR 5). (Lower) Mean grey levels ratios (mean ± S.E.M.) of ADAM10/α-tubulin immunoreactivities measured by Western blotting in the hippocampal cytoskeleton from the same samples. *p < 0.05, **p < 0.01, Dunnett’s Multiple Comparison test, n = 13–24.
ADAM10 protein levels are reduced in AD patients
The in vitro data indicate that Aβ-mediated decreases in nELAV proteins possibly result in an impaired translation of the target ADAM10 mRNA, as suggested by the finding that ADAM10 protein levels are reduced. Considering that in AD patients we observed a decrease in nELAV protein content, we also explored whether ADAM10 protein was altered in the hippocampal tissues. As shown in Fig. 4B, ADAM10 protein levels are significantly diminished in CDR 0.5 patients (−35%) and remained low in all subjects independently from CDR.
DISCUSSION
In the present work, we found that nELAV proteins are decreased in postmortem patients along with AD progression. Indeed, we observed a reduction, already present in CDR 0.5 subjects, which became more dramatic with the worsening of cognitive decline. Conversely, we did not observe changes in HuR levels, the ELAV protein ubiquitously expressed, thus indicating that only nELAV proteins are affected in AD pathology. These findings are in agreement with the role, evidenced by us and by others, of nELAV proteins in memory processes. In fact, we previously reported that following spatial learning there is a specific increase of nELAV proteins in the hippocampus (the brain area mainly involved in spatial memory) of rodents trained in different behavioral tests [4,5]. The key role of nELAV proteins in memory was additionally confirmed by experiments showing, on one side, a significant impairment in spatial memory performance of mice treated with a HuC antisense oligonucleotide [5] and, on the other side, an improvement in cognitive function following the treatment of mice with a compound [20] that we showed to increase HuD expression in vitro and in vivo [13].
From postmortem measurements in the hippocampi we also found an inverse correlation between nELAV proteins and Aβ content thus suggesting that the amyloid peptide could directly affect nELAV protein levels. As known, Aβ peptide is produced through the amyloidogenic pathway involving the β and γ-secretase enzymes. In physiological conditions, the majority of the produced Aβ fragment is characterized by a length of 40 residues (Aβ1–40), with a minor fraction represented by the longer form Aβ1–42. However, the Aβ1–42 peptide is more hydrophobic and more prone to self-aggregate with respect to Aβ1–40 [21]; moreover, Aβ1–42 is the predominant product in the cerebral plaques of AD patients [15,22]. A large body of evidence recently demonstrated that Aβ also accumulates intracellularly and again that the prevalent intracellular form in AD is Aβ1–42 (see for a review [23]) thus stressing the major role played by Aβ1–42 in AD pathogenesis. Within this context, intraneuronal Aβ immunoreactivity has been found in MCI patients in the hippocampus and in the entorhinal cortex [24], the two brain areas that are more prone to develop pathological alterations typical of an early AD. Additionally, it has been reported in Down syndrome subjects that plaque formation follows temporally intracellular Aβ accumulation [25]. These findings are in agreement with data obtained from transgenic mice and are in favor of the idea that intracellular accumulation of Aβ may represent an early stage in AD pathology, preceding the extracellular deposition of Aβ in plaques (see [23]). Intracellular Aβ can be generated within the cell or taken up extracellularly; it is likely that a dynamic equilibrium exists between the intracellular and the extracellular pools of Aβ. Inside the cells Aβ accumulation may have various pathological effects, such as disruption of synaptic activity, proteasome dysfunction, mitochondrial deficit, calcium dyshomeostasis, and may also facilitate tau hyperphosphorylation (see [23]). All these interferences with neuronal cell physiology may contribute to alteration of cognition, as suggested by the cognitive impairment observed in 3×Tg-AD mice that already manifest intraneuronal Aβ accumulation when they are 4 months old [26].
It was then of great interest to investigate the direct effect of Aβ1–42 on nELAV proteins in a well characterized cellular model, the human neuroblastoma SHSY5Y cells. First we found that following Aβ challenge SH-SY5Y cells were able to uptake the extracellular peptide. Besides the monomeric form, we observed that intracellular Aβ is present in multiple assembly states with different molecular weights, including oligomers. The Aβ oligomers, which have been reported to be the most pathological aggregation form of Aβ, are able to impair cognitive function [27], to inhibit long-term potentiation [28], and to disrupt memory in transgenic mouse models [29]. In our conditions, the direct exposure of the cells to Aβ1–42 induced a decrease of nELAV protein levels, especially in the cytoskeleton, an active site of protein synthesis [30]. The effect was specific for the amyloidogenic peptide, since the control reverse peptide did not cause any alteration. These results suggest that Aβ1–42 may contribute to the nELAV impairment observed in the hippocampus of postmortem AD patients, although in vivo additional mechanisms may take part to the nELAV alteration. Considering that nELAV are RNA-binding proteins, their deficit may affect the protein expression of specific target mRNAs. Interestingly, we found that the mRNA of ADAM10, the best characterized α-secretase implicated in the non-amyloidogenic processing of AβPP, bears in its 3’UTR multiple putative AREs of various types. This feature classifies the ADAM10 mRNA as a potential target of nELAV proteins. We tested this hypothesis in our cellular model through the detection of this mRNA in the nELAV-targeted enriched transcripts by immunoprecipitation experiments coupled with real-time quantitative RT-PCR. The experiments show for the first time that ADAM10mRNA represents a nELAV target and that these RNA-binding proteins can play a role in the post-transcriptional regulation of ADAM10 expression. In addition and very interestingly, we found that the binding between nELAV proteins and the ADAM10 mRNA is disrupted as a function of Aβ1–42 treatment. Considering that ELAV proteins can form ribonucleoproteic complexes that associate to the translational machinery through the cytoskeleton [16] and have been demonstrated to enhance translation of target mRNAs [17–19], a disruption of the binding of nELAV proteins to ADAM10 mRNA can lead to a reduction in its translation. This event indeed seemed to be the case, and, in fact, following Aβ1–42 challenge we observed a deficit of ADAM10 protein expression, again especially in the cytoskeleton, that possibly relies in alteration of the nELAV mediated translational control, although the involvement of post-translational events could not be excluded. Among the α-secretases, the effects of Aβ are specific for ADAM10, as the protein levels of another α-secretase, ADAM17, did not show changes. Further investigation is needed to understand why these α-secretases are differently affected by Aβ. In light of the in vitro data, we also explored the amount of ADAM10 protein in postmortem brain samples and found that it was indeed reduced. The observation that this deficit is already very evident in CDR 0.5 subjects, without further changes with the progression of the disease, suggests that the reduction of nELAV proteins in CDR 0.5 is a threshold event sufficient to determine a marked decrease in ADAM10 protein content. Impairment in ADAM10 protein levels may in turn imply a deficit in the non-amyloidogenic cleavage of AβPP, resulting in a reduced release of the neuroprotective and neurotrophic sAβPPα [31,32]. Considering that the amyloidogenic and the non-amyloidogenic pathways are mutually excluding [33], the deficit in the non-amyloidogenic processing of AβPP might determine a further increase in Aβ production by the cell, thus possibly leading to a vicious circle where the newly generated Aβ additionally contributes to down-regulate nELAV and ADAM10 proteins. However, further investigations would be requested to confirm this hypothesis.
Taken together, these results raise the possibility that molecules able to trigger nELAV-mediated post-transcriptional control may restore the deficit in ADAM10 expression associated with AD. This work for the first time describes nELAV as a new molecular target that may be, among others, directly affected by Aβ and associated with the cognitive impairment accompanying AD pathogenesis.
Acknowledgments
The authors wish to thank Prof. Schinelli and Dr. Paolillo for their help and kindness for the performance of real-time RT-PCR experiments. M.A. has been partially supported by a fellowship from the Fondo Sociale Europeo – Regione Lombardia.
References
- 1.Keene JD. Why is Hu where? Shuttling of early-response-gene messenger RNA subsets. Proc Natl Acad Sci USA. 1999;96:5–7. doi: 10.1073/pnas.96.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pascale A, Amadio M, Quattrone A. Defining a neuron: neuronal ELAV proteins. Cell Mol Life Sci. 2008;65:128–40. doi: 10.1007/s00018-007-7017-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perrone-Bizzozero N, Bolognani F. Role of HuD and other RNA-binding proteins in neural development and plasticity. J Neurosci Res. 2002;68:121–126. doi: 10.1002/jnr.10175. [DOI] [PubMed] [Google Scholar]
- 4.Pascale A, Gusev PA, Amadio M, Dottorini T, Govoni S, Alkon DL, Quattrone A. Neuronal ELAV proteins enhance mRNA stability by a PKC alpha-dependent pathway. Proc Natl Acad Sci USA. 2004;101:1217–1222. [Google Scholar]
- 5.Quattrone A, Pascale A, Noguès X, Zhao W, Gusev P, Pacini A, Alkon DL. Posttranscriptional regulation of gene expression in learning by the neuronal ELAV-like mRNA-stabilizing proteins. Proc Natl Acad Sci USA. 2001;98:11668–11673. doi: 10.1073/pnas.191388398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolognani F, Merhege MA, Twiss J, Perrone-Bizzozero NI. Dendritic localization of the RNA-binding protein HuD in hippocampal neurons: association with polysomes and upregulation during contextual learning. Neurosci Lett. 2004;371:152–157. doi: 10.1016/j.neulet.2004.08.074. [DOI] [PubMed] [Google Scholar]
- 7.Allinson TM, Parkin ET, Turner AJ, Hooper NM. ADAMs family members as amyloid precursor protein alpha-secretases. J Neurosci Res. 2003;74:342–352. doi: 10.1002/jnr.10737. [DOI] [PubMed] [Google Scholar]
- 8.Colciaghi F, Borroni B, Pastorino L, Marcello E, Zimmermann M, Cattabeni F, Padovani A, Di Luca M. [alpha]-Secretase ADAM10 as well as [alpha]APPs is reduced in platelets and CSF of Alzheimer disease patients. Mol Med. 2002;8:67–74. [PMC free article] [PubMed] [Google Scholar]
- 9.Haroutunian V, Perl DP, Purohit DP, Marin D, Khan K, Lantz M, Davis KL, Mohs RC. Regional distribution of neuritic plaques in the nondemented elderly and subjects with mild Alzheimer disease. Arch Neurol. 1998;55:1185–1191. doi: 10.1001/archneur.55.9.1185. [DOI] [PubMed] [Google Scholar]
- 10.Haroutunian V, Purohit DP, Perl DP, Marin D, Khan K, Lantz M, Davis KL, Mohs RC. Neurofibrillary tangles in nondemented elderly subjects and mild Alzheimer disease. Arch Neurol. 1999;56:713–718. doi: 10.1001/archneur.56.6.713. [DOI] [PubMed] [Google Scholar]
- 11.Berg L. Clinical Dementia Rating (CDR) Psychopharmacol Bull. 1988;24:637–639. [PubMed] [Google Scholar]
- 12.Kawarabayashi T, Younkin LH, Saido TC, Shoji M, Ashe KH, Younkin SG. Age-dependent changes in brain, CSF, and plasma amyloid (beta) protein in the Tg2576 transgenic mouse model of Alzheimer’s disease. J Neurosci. 2001;21:372–381. doi: 10.1523/JNEUROSCI.21-02-00372.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pascale A, Amadio M, Scapagnini G, Lanni C, Racchi M, Provenzani A, Govoni S, Alkon DL, Quattrone A. Neuronal ELAV proteins enhance mRNA stability by a PKCα-dependent pathway. Proc Natl Acad Sci USA. 2005;102:12065–12070. doi: 10.1073/pnas.0504702102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pascale A, Fortino I, Govoni S, Trabucchi M, Wetsel WC, Battaini F. Functional impairment in protein kinase C by RACK1 (receptor for activated C kinase 1) deficiency in aged rat brain cortex. J Neurochem. 1996;67:2471–2477. doi: 10.1046/j.1471-4159.1996.67062471.x. [DOI] [PubMed] [Google Scholar]
- 15.Younkin SG. The role of A beta 42 in Alzheimer’s disease. J Physiol Paris. 1998;92:289–292. doi: 10.1016/s0928-4257(98)80035-1. [DOI] [PubMed] [Google Scholar]
- 16.Antic D, Keene JD. Messenger ribonucleoprotein complexes containing human ELAV proteins: interactions with cytoskeleton and translational apparatus. J Cell Sci. 1998;111:183–197. doi: 10.1242/jcs.111.2.183. [DOI] [PubMed] [Google Scholar]
- 17.Antic D, Lu N, Keene JD. ELAV tumor antigen, Hel-N1, increases translation of neurofilament M mRNA and induces formation of neurites in human teratocarcinoma cells. Genes Dev. 1999;13:449–461. doi: 10.1101/gad.13.4.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jain RG, Andrews LG, McGowan KM, Pekala PH, Keene JD. Ectopic expression of Hel-N1, an RNA-binding protein, increases glucose transporter (GLUT1) expression in 3T3-L1 adipocytes. Mol Cell Biol. 1997;17:954–962. doi: 10.1128/mcb.17.2.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mazan-Mamczarz K, Galban S, Lopez de Silanes I, Martindale JL, Atasoy U, Keene JD, Gorospe M. RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc Natl Acad Sci USA. 2003;100:8354–8359. doi: 10.1073/pnas.1432104100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Etcheberrigaray R, Tan M, Dewachter I, Kuiperi C, Van der Auwera I, Wera S, Qiao L, Bank B, Nelson TJ, Kozikowski AP, Van Leuven F, Alkon DL. Therapeutic effects of PKC activators in Alzheimer’s disease transgenic mice. Proc Natl Acad Sci USA. 2004;101:11141–11146. doi: 10.1073/pnas.0403921101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jarrett JT, Berger EP, Lansbury PT., Jr The carboxy terminus of the beta amyloid protein is critical for the seeding of amyloid formation: implications for the pathogenesis of Alzheimer’s disease. Biochemistry. 1993;32:4693–4697. doi: 10.1021/bi00069a001. [DOI] [PubMed] [Google Scholar]
- 22.Gandy S. The role of cerebral amyloid beta accumulation in common forms of Alzheimer disease. J Clin Invest. 2005;115:1121–1129. doi: 10.1172/JCI25100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laferla FM, Green KN, Oddo S. Intracellular amyloid-beta in Alzheimer’s disease. Nat Rev Neurosci. 2007;8:499–509. doi: 10.1038/nrn2168. [DOI] [PubMed] [Google Scholar]
- 24.Gouras GK, Tsai J, Naslund J, Vincent B, Edgar M, Checler F, Greenfield JP, Haroutunian V, Buxbaum JD, Xu H, Greengard P, Relkin NR. Intraneuronal Abeta42 accumulation in human brain. Am J Pathol. 2000;156:15–20. doi: 10.1016/s0002-9440(10)64700-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gyure KA, Durham R, Stewart WF, Smialek JE, Troncoso JC. Intraneuronal Abeta precedes development of amyloid plaques in Down syndrome. Arch Pathos Lab Med. 2001;125:489–492. doi: 10.5858/2001-125-0489-IAAPDO. [DOI] [PubMed] [Google Scholar]
- 26.Billings LM, Oddo S, Green KN, McGaugh JL, LaFerla FM. Intraneuronal Abeta causes the onset of early Alzheimer’s disease-related cognitive deficits in transgenic mice. Neuron. 2005;45:675–688. doi: 10.1016/j.neuron.2005.01.040. [DOI] [PubMed] [Google Scholar]
- 27.Cleary JP, Walsh DM, Hofmeister JJ, Shankar GM, Kuskowski MA, Selkoe DJ, Ashe KH. Natural oligomers of the amyloid-beta protein specifically disrupt cognitive function. Nat Neurosci. 2005;8:79–84. doi: 10.1038/nn1372. [DOI] [PubMed] [Google Scholar]
- 28.Walsh DM, Klyubin I, Fadeeva JV, Cullen WK, Anwyl R, Wolfe MS, Rowan MJ, Selkoe DJ. Naturally secreted oligomers of amyloid beta protein potently inhibit hippocampal long-term potentiation in vivo. Nature. 2002;416:535–539. doi: 10.1038/416535a. [DOI] [PubMed] [Google Scholar]
- 29.Lesne S, Koh MT, Kotilinek L, Kayed R, Glabe CG, Yang A, Gallagher M, Ashe KH. A specific amyloid-beta protein assembly in the brain impairs memory. Nature. 2006;440:352–357. doi: 10.1038/nature04533. [DOI] [PubMed] [Google Scholar]
- 30.Jansen RP. RNA-cytoskeletal associations. FASEB J. 1999;13:455–466. [PubMed] [Google Scholar]
- 31.Mattson MP, Cheng B, Culwell AR, Esch FS, Lieberburg I, Rydel RE. Evidence for excitoprotective and intraneuronal calcium-regulating roles for secreted forms of the beta-amyloid precursor protein. Neuron. 1993;10:243–254. doi: 10.1016/0896-6273(93)90315-i. [DOI] [PubMed] [Google Scholar]
- 32.Mucke L, Abraham CR, Masliah E. Neurotrophic and neuroprotective effects of hAPP in transgenic mice. Ann N Y Acad Sci. 1996;777:82–88. doi: 10.1111/j.1749-6632.1996.tb34405.x. [DOI] [PubMed] [Google Scholar]
- 33.Racchi M, Govoni S. The pharmacology of amyloid precursor protein processing. Exp Gerontol. 2003;38:145–157. doi: 10.1016/s0531-5565(02)00158-4. [DOI] [PubMed] [Google Scholar]