Abstract
SEFIR domain-containing proteins are crucial for mammalian adaptive immunity. As a unique intracellular signaling domain, the SEFIR-SEFIR interactions mediate physical protein-protein interactions in the immune signaling network, especially the IL-17- and IL-25-mediated pathways. However, due to the lack of structural information, the detailed molecular mechanism for SEFIR-SEFIR assembly remains unclear. In the present study, we solved the crystal structures of a prokaryotic SEFIR domain from Bacillus cereus F65185 (BcSEFIR), where the SEFIR domain is located at the N terminus. The structure of BcSEFIR revealed two radically distinct SEFIR-SEFIR interaction modes. In the asymmetric form, the C-terminal tail of one SEFIR binds to the helix αA and βΒ-αΒ’ segment of the other one, while in the symmetric form, the helices ηC and αE and the DE-segment compose the interprotomer interface. The C-terminal tail of BcSEFIR, critical for asymmetric interaction, is highly conserved among the SEFIR domains of Act1 orthologs from different species, in particular three absolutely conserved residues that constitute an EXXXXPP motif. In the symmetric interaction mode, the most significant contacts made by residues on helix αE are highly conserved in Act1 SEFIR domains, constituted an RLI/LXE motif. The two novel SEFIR-SEFIR interaction modes might explain the structural basis for SEFIR domain-mediated complex assembly in signaling pathways.
Keywords: SEFIR, dimer, self-association, bacterial, STIR
Introduction
SEFIR [SEF (similar expression to fibroblast growth factor genes) and IL-17R] domain-containing proteins, including interleukin-17 receptor (IL-17R) family (IL-17RA to IL-17RE) and their adaptor protein NF-kB activator 1 [Act1; also known as connection to I-kB kinase and stress-activated protein kinase (CIKS)], are crucial for mammalian adaptive immunity (1–6). These SEFIR-domain-containing proteins have been shown to be associated with many autoimmune diseases in both humans and mice (2,3,7–11). Upon stimulation, SEFIR-domain-containing receptors, such as IL-17R (IL-17RA and IL-17RC) and IL-25R (IL-17RA and IL-17RB), recruit the cytoplasmic adaptor protein Act1 via heterotypic SEFIR-SEFIR interaction (7,12–14). Act1 can also form oligomers via homotypic SEFIR-SEFIR interaction (15–17). Therefore, SEFIR domain is well known as a functional signaling domain to mediate protein-protein interaction.
SEFIR domain shares limited sequence homology with Toll/interleukin-1 receptor/resistance (TIR) domain, which has also been proved to mediate protein-protein interactions in various signal transduction pathways (14,18–22). SEFIR-domain- and TIR-domain-containing proteins together constitute a large superfamily named STIR (SEFIR/TIR) (1). Of all the STIRs, many TIR domains structures, including mammalian, plant and bacterial TIR domains have been determined (23–29), but only two monomer structures of SEFIR domain (human IL-17RA-SEFIR and mouse IL-17RB-SEFIR) were reported (17,30). These two SEFIR structures share similar α/β fold with TIR domains, and display canonical STIR folding as a compact globular architecture comprised of a central parallel β-sheet wrapped by several α-helices (17,30). However, the monomer structures could not provide structural information for SEFIR-SEFIR assembly mode, the major function of this domain. Previous studies have suggested that SEFIR domain has specific structural elements for IL-17R signaling compared with TIR domain (17,28,31). Moreover, mutagenesis experiments demonstrated the distinct interfaces for Act1-SEFIR to interact with IL-17RA-SEFIR or form oligomers (31,32). Therefore, the detailed structural information for SEFIR-SEFIR interaction has been of great interest and importance.
The SEFIR-domain-containing proteins have been found throughout the animal kingdom and even in prokaryotes (1,16,33,34). Comparative and phylogenetic sequence analyses suggested that bacterial SEFIR homologues may be acquired through lateral gene transfer from eukaryotes (33). Prokaryotic SEFIR domain-containing proteins with various domain architectures occur in a wide range of bacteria, almost certainly play diverse roles in bacterial physiology. SEFIR domains may function as general protein-protein interaction domains for diverse physiological actions. To elucidate the detailed SEFIR-SEFIR interaction modes, we have determined crystal structures of a prokaryotic SEFIR domain, unraveling two novel SEFIR-SEFIR interaction forms (asymmetrical and symmetrical) mediated by distinct binding sites. This study provides the structural basis for SEFIR-mediated protein-protein interactions.
Results
Crystal Structures of BcSEFIR
Multiple sequence alignment indicates that SEFIR domains from different proteins share an extended C-terminal tail region (up to 18 residues in human Act1) (Fig. 1A), which has not been observed in previously reported SEFIR structures (17,30). While the SEFIR domain has been implicated in protein-protein interactions, the function of this C-terminal tail remains unclear. We aimed to investigate the structure and function of this tail region in the context of SEFIR domain by using a prokaryotic SEFIR domain from Bacillus cereus F65185 (BcSEFIR) as a model, which is located at the N- terminus of the protein.
Figure 1. Crystal structures of BcSEFIR.
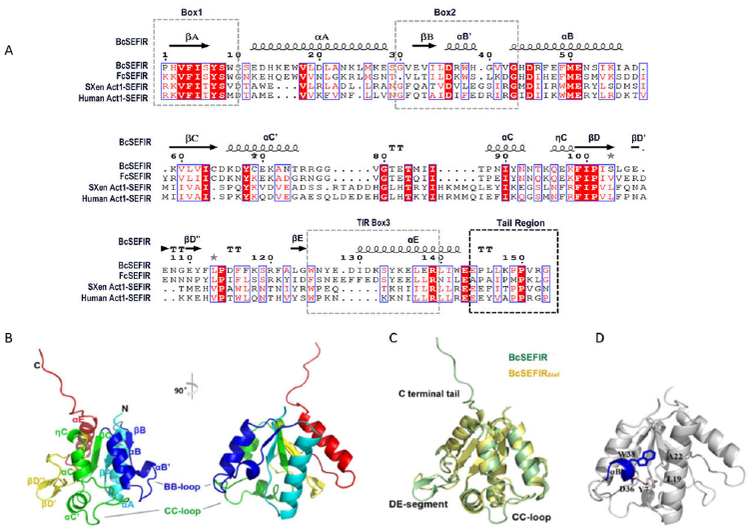
(A) Alignment of the SEFIRs in different species. Secondary structures shown above is BcSEFIR(β-sheets, black arrows; a-helices, cylinders). SEFIR domain form Bacillus cereus, Flavobacterium cauens, Xenopus (Silurana) tropicalis, human are used in the alignment. (B) Overall structure of BcSEFIR depicted in cartoon representation with secondary elements colored differently and labeled appropriately. (C) Superposition of BcSEFIR and BcSEFIRΔtail structures. Regions with conformational difference are labeled. (D) The short helix αB’ (blue) is tethered by hydrophobic interactions and hydrogen-bond. Residue Asp-36 in helix αB’ forms a hydrogen-bond with Tyr-7 from AA-loop.
According to the multiple sequence alignment (Fig. 1A), two constructs containing BcSEFIRΔtail (amino acid residues 1–143) and BcSEFIR (residues 1–153) were designed. Their structures were determined in high resolution of 2.0 Å and 1.8 Å, respectively. All the residues in the constructs could be readily traced, including those located at the loop regions. As expected, the overall structure of BcSEFIR exhibits a folding of typical α/β domains like other STIR superfamily members, which was characterized by a central parallel β-sheet flanked by several α-helices on both sides (Fig. 1B). Superposition of BcSEFIR and BcSEFIRΔtail showed that truncation of C-terminal tail had limited influence on the conformation of the globular core of BcSEFIR (r.m.s. deviation of 0.82 Å for all atoms), except for minor conformational differences at the regions around CC-loop or DE-segment (Fig. 1C), probably due to different molecular packing patterns in two crystal forms. The C-terminal 10-residue tail of BcSEFIR does not attach to the globular α/β core or adopt a disordered state with high flexibility. Instead, the tail extended outward to form extensive interactions with an adjacent BcSEFIR molecule in the crystal.
The central β-sheet of BcSEFIR consists of five β-strands (βA–βE) and is slightly twisted. In the β-sheet of a typical TIR domain, such as TLR1-TIR and TLR2-TIR (35), each β-strand is followed by only one α-helix (αA–αE). However, BcSEFIR adopts a more complex structural configuration, where the region between two adjacent β-strands contains more secondary structural elements than a single α-helix (Fig. S1). The BC-segment located between βB and βC strands possesses two α-helices (αB’ and αB), while CD-segment is composed by two α-helices (αC’ and αC) and one 310-helix (ηC). The DE-segment is even more unusual, comprising no helices, but a β-hairpin formed by two short β-strands (βD’ and βD”) (Fig. S1). The BB-loop and CC-loop (important for STIR-STIR interactions (31,35–39)) is equivalent to the loops linking αB’ and αB, and αC’ and αC, respectively (Fig. 1B). These two regions are fully exposed to solvent in BcSEFIR domain. The short αB’ helix in BcSEFIR adopts a stable and rigid conformation through interactions with helix αA and αB (Fig. 1D). Residue Trp-38 on helix αB’ is associated with a hydrophobic platform containing residues Leu-19 and Ala-22 on helix αA. In addition to the intensive hydrophobic interactions, residue Asp-36 on helix αB’ is hydrogen bonded to Tyr-7 on AA-loop (2.68 Å). Interestingly, the Tyr-7 and Asp-36 are highly conserved in Act1-SEFIR (Fig. S2), indicating that this hydrogen bond could be formed in Act1-SEFIR, tethering aB’ helix to adopt a stable conformation.
Structural Comparison of BcSEFIR with IL-17Rs-SEFIR
Although BcSEFIR has low amino acid sequence similarity with the SEFIR domains of IL-17RB (14.6% identity) and IL-17RA (18.3% identity), they indeed share similar overall topology. Superposition of BcSEFIR and IL-17RB-SEFIR gave an r.m.s. deviation of 2.27 Å for 107 aligned Cα atoms (Fig. 2A), indicating that bacterial SEFIR domains share similar fold with mammalian SEFIR. Between these two structures, major differences lie in the BC-, CD- and DE-segments. The CC-loop of IL-17RB-SEFIR cannot be observed in its crystal structure, due to its high flexibility. Accordingly, a model was proposed with crossed CC- and DD-loops, exhibiting an unusual knot topology (Fig. 2C) (17). However, in the structures of BcSEFIR, all loop regions are well ordered without any knot topology formed.
Figure 2. Superposition of BcSEFIR with other SEFIR domain structures.
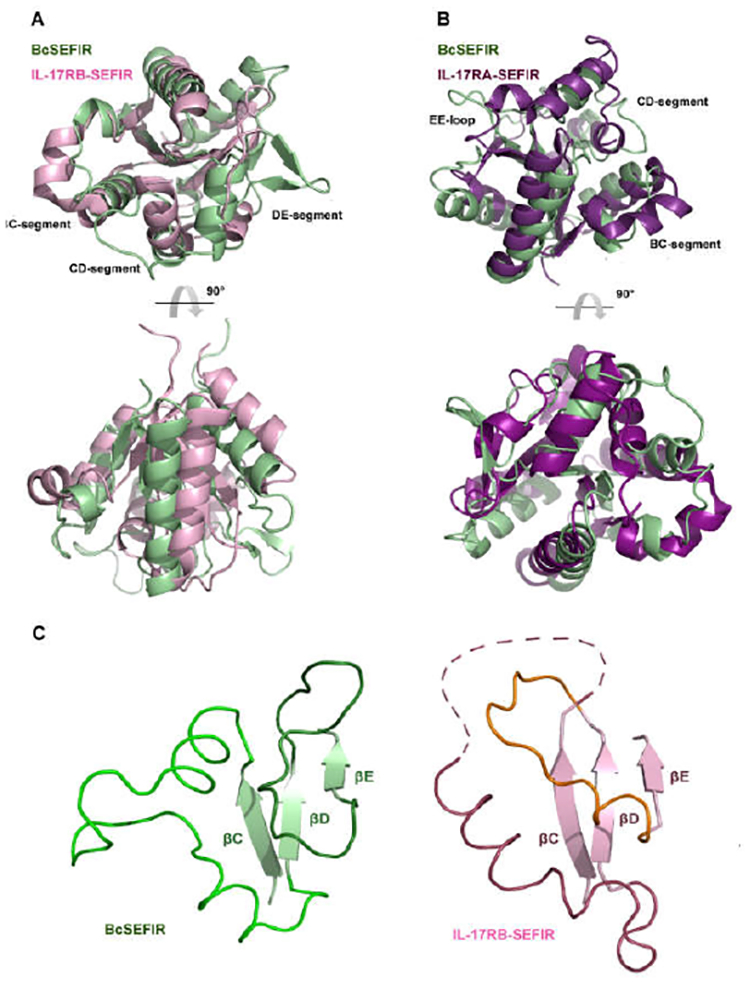
(A) Superposition of BcSEFIR and IL-17RB SEFIR domain (PDB code: 3VBC). (B) Superposition of BcSEFIR and IL-17RA SEFIR domain (PDB code: 4NUX). (C) The segment between βC and βD is entirely ordered in BcSEFIR structure and does not form the unusual knot topology as suggested by the structure of IL-17RB-SEFIR.
The structure of BcSEFIR partially resembles that of IL-17RA-SEFIR, with an r.m.s. deviation of 1.92 Å over 119 aligned Cα atoms. Although the core structural elements of these two structures share canonical STIR folding, the helices, β-sheets and loops connecting them in BcSEFIR differ from IL-17RA-SEFIR in their lengths and orientations. The significant differences between the two structures he in CD-segment (αCC’ins, αC’, CC loop) and DE-segment (αD, DD loop) (Fig. 2B).
Asymmetric interaction mode observed in BcSEFIR structure
The crystal structure of BcSEFIR contains only one protein molecule per asymmetric unit, but we noticed an interesting interaction form between two adjacent BcSEFIR domains. The C-terminal tail (CT) of one BcSEFIR (referred to as the tail-presenting) attaches to the cleft between the αA helix and βΒ-αΒ’ segment of the adjacent BcSEFIR molecule (referred to as the tail-docking) via extensive interactions, where most of the BcSEFIR CT residues are involved (Fig. 3). Multiple sequence alignment of SEFIR domains from different species (Fig. 1A) indicates that the extended tails at the C-terminus are ubiquitous in eukaryotic, implicating potential biological importance of the extended C-terminal tails.
Figure 3.
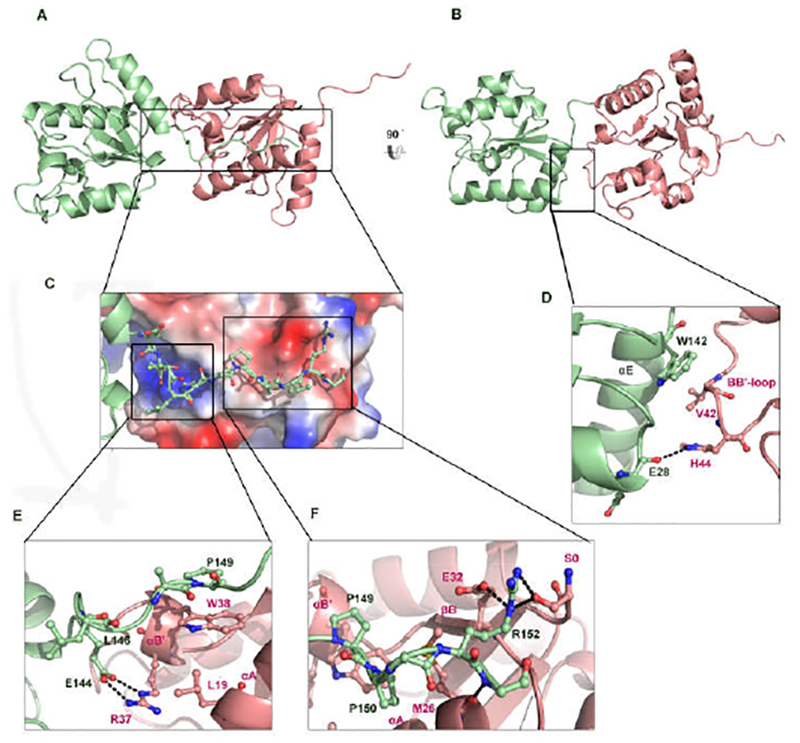
The interface formed by the protruding tail of BcSEFIR. (A) In this asymmetric dimer of BcSEFIR, the tail-presenting molecule is colored in light green, as wellas the tail-docking one in light pink. Helices αA, αB, αB′ are involved in the interaction. Among them, αB, αB′ play the most important role. Interface formed by thetail portion is highlighted with boxes. (B) The asymmetric dimer is shown in different orientation, interface is highlighted with a box. (C) The zoomed-in view of the protruding tail attaching to the adjacent BcSEFIR molecule. (D) Detailed interactions of the αE and αA helices in the tail-presenting BcSEFIR with the αB in the taildocking BcSEFIR. (E) Detailed interactions of the N-terminal tail and intermediate portion in the tail-presenting BcSEFIR with the αA, αB′ helices in the tail-docking BcSEFIR. Interacting residues are labeled, hydrogen-bonds are shown as dashed lines. (F) Detailed interactions of the tail C-terminal portion in the tail-presenting BcSEFIR with the αA, αB′ helices and βB in the tail-docking BcSEFIR, where interacting residues are labeled, and hydrogen-bonds are shown as dashed lines.
At the C-terminus of BcSEFIR, Arg-152 is the key residue mediating the anchoring of this tail portion (Fig. 3F.) An ionic bond is formed between this Arg-152 and the βΒ Glu-32 of the tail-docking subunit. There are other two hydrogen-bonds between this Arg-152 and the Ser-0 at the very N-terminus of the tail-docking BcSEFIR. It is important to note that the Ser-0 is an artificially introduced amino acid residue, while the natural residue valine at this position might not form hydrogen-bonds via its side chain. At the N-terminus of BcSEFIR CT, the key residue is Glu-144 that forms two hydrogen-bonds with the αB’ Arg-37 of the tail-docking subunit (Fig. 3E). BcSEFIR CT also exhibits hydrophobic interactions between its intermediate portion (Leu-146-Pro-150) and the small hydrophobic patch in the cleft (including Leu-19, Met-26 on αA and Trp-38 on αB’) (Fig. 3E & F). Because of those extensive interactions, the C-terminal tail of the tail-presenting BcSEFIR is tightly fixed at the surface of the taildocking BcSEFIR. The BcSEFIR CT is conserved among Act1 SEFIR domains, the three absolutely conserved residues including Glu-144, Pro-149, Pro-150 constitute an EXXXXPP motif (Fig. S3). Additionally, the αE and αA helices of the tail-presenting BcSEFIR interact with the αB of the tail-docking BcSEFIR, further stabilizing the asymmetric SEFIR-SEFIR association (Fig. 3B). Hydrophobic interactions play an important role at this interface, where the αE Trp-142 of the tail-presenting BcSEFIR and the αB Val-42 of the tail-docking BcSEFIR are the central residues. Notably, there is also a hydrogen-bond between the side chain nitrogen atom of αB His-44 from the tail-docking subunit and the main chain oxygen atom of αA Glu-28 from the tail-presenting subunit (Fig. 3D).
Symmetric interaction mode observed in BcSEFIRΔtail structure
Without the C-terminus ‘tail’, two BcSEFIR,Δtail molecules form a dimer in a symmetric manner distinguished from BcSEFIR, exhibiting a distinct dimerization interface (Fig. 4A). The area of the BcSEFIRΔtail dimerization interface is ~958 Å2, approximately 1.5-fold of the asymmetric interface of BcSEFIR (~662 Å2). The symmetric BcSEFIRΔtail dimer exhibits a canonical pattern of protein-protein interactions: a hydrophobic core patch surrounded by hydrogen-bond connections. The hydrophobic core is mainly comprised by the Phe-121 at the C-terminal of βΕ strand and the Leu-140 of αE helix. The benzene ring of Phe-121 is inserted into the hydrophobic patch of the opposite BcSEFIRΔtail surface, forming a solid association (Fig. 4A). Several hydrogen-bonds are built between the αE helix of one BcSEFIRΔtail (residues Lys-135, Arg-139 and Glu-143) and the DE-segment of the opposite one (residues Asn-108 and Arg-120), as well as between the two ηC helices from each side (residues Gln-96 and Glu-97). The interacting residues Arg-139, Leu-140, Glu-143 are highly conserved in Act1 SEFIR domains, constituted an RLI/LXE motif (Fig. 1A). Taken together, the characterization of two distinct SEFIR-SEFIR interaction forms (asymmetrical and symmetrical) illustrates two plausible interaction modes via distinct binding sites on SEFIR domains (Fig. 5A & B). In addition, the symmetric interface found in BcSEFIRΔtail cannot be observed in BcSEFIR crystal lattice. And there is no conserved interface in the two structures. There are several other SEFIR-SEFIR domain interfaces observed in the both BcSEFIR and BcSEFIRΔtail crystal lattice. However, most are crystallization contacts with much smaller interfaces as well as less interacting residues (Table S1), which may not represent physiological interaction.
Figure 4. Symmetric dimer of BcSEFIRΔtail.
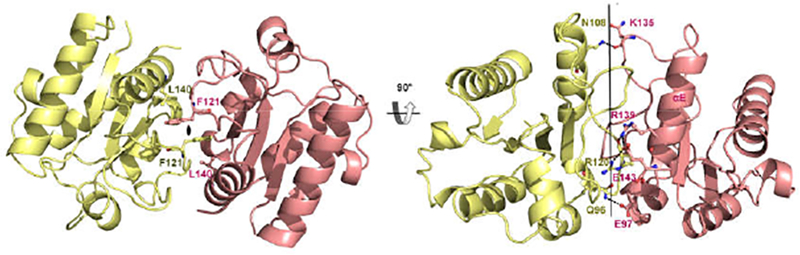
(A) Two SEFIR domains interacted with each other symmetrically in the structure of BcSEFIRΔtail, residues which form a central hydrophobic core are labeled. (B) The symmetric dimer is shown in different orientation, interacting residues are labeled, hydrogen-bonds are shown as dashed lines.
Figure 5. Two distinct SEFIR-SEFIR interfaces.
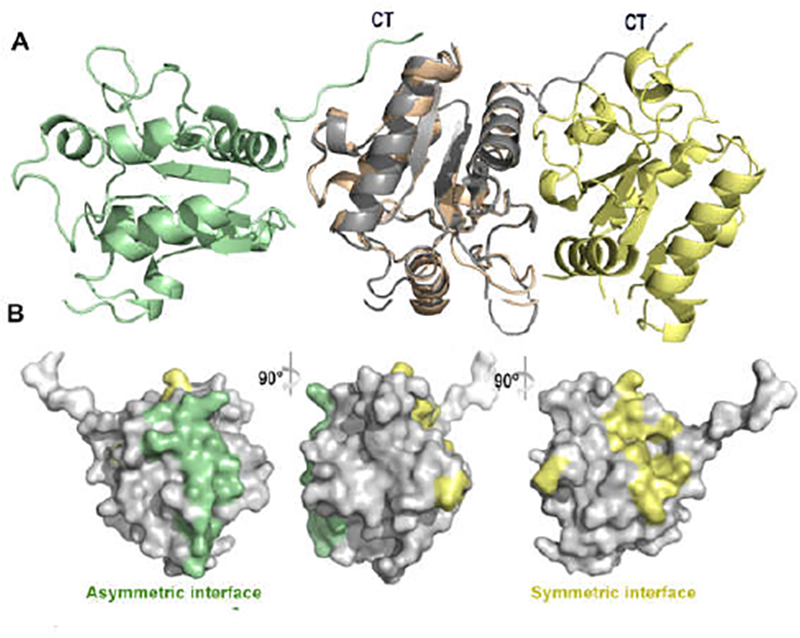
(A) Two distinct SEFIR-SEFIR interaction modes are aligned together, asymmetric dimer are coloured in green and grey, symmetric dimer are coloured in pink and yellow. C- terminal tail region are labled as CT. (B) The asymmetric (green colour showed) and symmetric interfaces (yellow colour showed) are located at different regions on the surface of SEFIR domain.
To quantitatively investigate the BcSEFIR-SEFIR interaction in solution, we performed Surface Plasmon Resonance (SPR) experiment. The binding affinities between BcSEFIR-BcSEFIR, BcSEFIRΔtail-BcSEFIRΔtail, BcSEFIR-BcSEFIRΔtail were all assessed (Fig. S5). These SPR results were consistent with the SEFIR-SEFIR interacion modes we observed in crystals, that BcSEFIR domain has different binding sites, and the tail region doesn’t disturb the binding of the other interface. It has been reported that most isolated recombinant mammalian STIR domains formed monomers in solution, such as IL-17RA-, IL-17RB SEFIR and TIR domain-containing adaptor proteins (17,30,40–43). In our binding assay, BcSEFIR domain also showed weak affinity for self-association in solution. Moreover, in the gel filtration chromatography, BcSEFIR was eluted as monomer in solution (Fig. S6). These results suggested that weak interaction and transient oligomers of STIR domains may be a general feature of this superfamily.
Validation of key residues for BcSEFIR-SEFIR interactions
To validate the significance of the two SEFIR-SEFIR interaction modes observed in the crystals, mutagenesis experiments were conducted to identify the structure-function relationship. Key residues on asymmetric binding site (Met-26, Arg-37, Trp-38, His-44, Trp-142, Glu-144, Arg-152) or symmetric binding site (Gln-96, Glu-97, Asn-108, Arg-120, Phe-121, Lys-135, Arg-139, Glu-143) were mutated to alanine. By co-immunoprecipitation experiments, we found that mutation of asymmetric binding site significantly compromised the self-association of BcSEFIR while it had minimum impact on the BcSEFIRΔtail symmetric interaction. On the other hand, mutation of symmetric binding site greatly reduced the dimerization of BcSEFIRΔtail. but with little impact on self-association of BcSEFIR (Fig. 6A &B). These results provide supporting evidence for the two interaction interfaces revealed by our crystal structures, which is consistent with the proposed two modes of interaction (asymmetrical and symmetrical). Additionally, the mutagenesis experiment also suggesting that with the tail region, SEFIR domain will prefer to form dimer through the asymmetric interface, as without the tail region, symmetric interaction is the dominant interaction mode.
Figure 6. Co-immuniprecipitation assays.
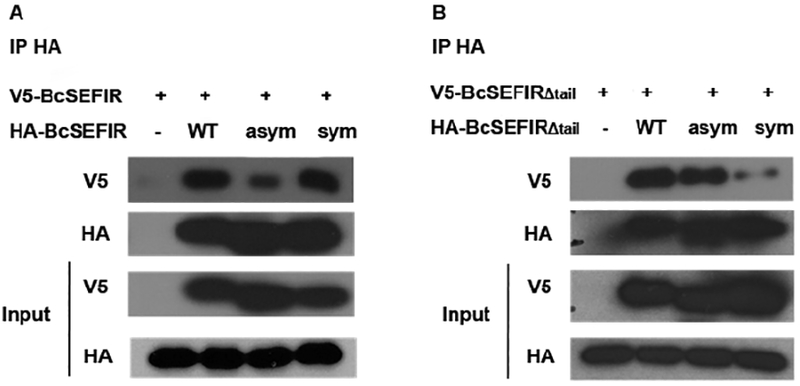
(A) The ability of V5-BcSEFIR to co-immunoprecipitate with the HA-mutations or wild type HA-BcSEFIR was shown. Mutagenesis on asymmetric interface markedly reduced BcSEFIR self-association. (B) The ability of V5-BcSEFIRΔtail to co-immunoprecipitate with the HA-mutations or wild type HA-BcSEFIRΔtail was shown. Mutagenesis on symmetric interface markedly reduced BcSEFIRΔtail self-association. WT, wild type recombinant protein. Asym, mutant with key interacting amino acids on asymmetric binding site substituted by Alanine. Sym, mutant with key interacting amino acids on symmetric binding site substituted by Alanine.
Discussion
Both SEFIR family and TIR family belong to the STIR superfamily, of which the members share similar α/β fold and are known to mediate physical protein-protein interactions in cytosolic signal transductions. Several TIR-TIR complex structures have been reported, including human, plant and microbial TIR domains (e.g. TLR2, TLR6, TLR10, MAL/TIRAP, IL-1RAPL, L6, RPS4, RRS1TcpB, PdTIR etc.)(38,40–48). Structural analysis combined with biochemical data revealed that TIR domain forms various different kinds of dimers. For example, the homodimer of TLR10-TIR adopts a symmetric association mode, where the interactions at the homodimeric interface are mainly contributed by BB-loop and aC helix from both sides (Fig. S7 D)(42). The C713S mutant of TLR2-TIR was used for crystallization and unraveled an asymmetric homodimerization interface, where an inter-molecular disulfide bond is formed between the Cys-750 of one subunit and the Cys-640 of the other one (Fig. S7 A)(38). In bacterial TcpB-TIR, the structural elements involved in these interfaces include the DD and EE loops and the aD and aE helices (Fig. S7 F)(47). In this study, we determined the crystal structures of BcSEFIR from a prokaryotic species, Bacillus cereus F65185, and revealed two novel SEFIR-SEFIR interaction modes (asymmetrical and symmetrical) mediated by two distinct interaction interfaces on SEFIR domains, which are totally different from any previously reported TIR-TIR complex structures. We further validate these two binding sites through site-directed mutagenesis. Our data showed that mutation of asymmetric binding site significantly compromised the self-association of BcSEFIR without substantial impact on BcSEFIRΔtail, symmetric interaction. Meanwhile, mutation of symmetric binding site impaired the self-association of BcSEFIRΔtail, but not the dimerization of BcSEFIR (Fig. 6A & B), suggesting the two interaction interfaces on SEFIR domain are physiologically relevant to BcSEFIR domain self-association. We performed SPR experiments to quantitatively investigate the BcSEFIR-SEFIR interaction in solution, found that BcSEFIR domain showed weak affinity for self-association in solution (Fig. S5). The SPR results were consistent with the SEFIR-SEFIR interacion modes we observed in crystals, that BcSEFIR domain has different binding sites, and the tail region doesn’t disturb the binding of the other interface. Jointly, our structural and biochemical data demonstrate two novel SEFIR-SEFIR interaction modes.
The asymmetric interaction mode implicates that SEFIR domains can create a fiber-like oligomeric structure as observed in the crystals (Fig. S4), We cannot rule oue the possibility that this configuration might be the result of crystal packing in the high protein concentration during crystallization. Notably, SEFIR domains appear to have the ability to form oligomers larger than dimer in vivo, as well as in vitro yeast two-hybrid and co-immunoprecipitation experiments of Act1 SEFIR domain, which is critical for Act1-mediated downstream signaling (15). Recently, Bostjan Kobe et al reported that TIR domains of the TLR adaptors MAL and MyD88 can self-assembles into filaments, consisting of two parallel strands of TIR-domain subunits in a head-to-tail arrangement with two major types of asymmetric interaction(49). Although the assemblies mediated by Act1-SEFIR are remain structurally elusive, previous studies have shown that the helix aB’ and aB in Act1 SEFIR domain are essential for homotypic SEFIR-SEFIR interactions, since deletion part of the helix αB’ and αB (residues 425–432, dashed underline shows in Fig. 1A) in Act1 SEFIR domain greatly reduced the self-association without significant impact on the heterotypic interaction of Act1 with IL-17RA-SEFIR (17,31). In this study, our crystal structures indicate that helix αB’ and αB are part of the asymmetric interaction interface for creation of fiber-like oligomeric structure. One thing to be noted is that the helix αB’ in all three SEFIR structures is locked into a rigid conformation by hydrophobic interactions and hydrogen bond.(17,30). The helix αB’ in IL-17RA-, IL-17RB forms hydrogen bond with helix αB, the related residues are conserved among IL-17Rs-SEFIR and Act1-SEFIR (Fig. S2, green boxes). While the helix αB’ in BcSEFIR forms hydrogen bond with AA-loop, the related residues are also strictly conserved among IL-17Rs-SEFIR and Act1-SEFIR (Fig. S2C, red boxes). The residues involved in hydrophobic interactions are also conserved, indicating that αB’ in Act1-SEFIR is tightly tethered. The stable conformation of helix αB’ suggested that αB’ may play important roles during SEFIR domain assembly.
According to the multiple sequence alignment, the BcSEFIR tail region is highly conserved among Act1 SEFIR domains, including three absolutely conserved residues that constitute an EXXXXPP motif (Fig. S3). The highly conserved two adjacent proline residues form the core of the hydrophobic interactions on the tail. As to the binding surface of the tail-docking molecule, Asn-23 and Trp-38 are the most critical hydrophobic residues interact with the proline residues on the tail region, which are conserved in Act1-SEFIRs (Fig. S3, showed in green boxes). Conserved hydrophobic residues Leu-19, Gly-30, Val-31 are also involved in the interaction with tail region. The highly conserved glutamate residue in EXXXXPP motif is the N-terminal anchoring point of the tail in asymmetric interaction mode, it forms two hydrogen-bonds and one ionic bond with Arg-37 in BcSEFIR, but in Act1-SEFIR this arginine residue is substituted by isoleucine or valine, which cannot form hydrogen-bonds (Fig. S3, showed in blue boxes). In the symmetrical interaction mode, interacting residues on helix αE are conserved and constituted an RLI/LXE motif near the C-terminus of Act1 SEFIR domains. The absolutely conserved residues Arg-139, Leu-140, Glul43 interact with the Arg-120 from the opposite molecule, form three hydrogen bonds and one salt bridge (Fig. 3B). In Act1-SEFIR, Arg-120 is substituted by threonine (Fig. 1A), with adjacent residues of asparagine and histidine, which hydrogen might also be formed.
In summary, our study reveals that there are two distinct SEFIR-SEFIR interface (asymmetrical and symmetrical) exist on BcSEFIR domain, illustrates two plausible modes for SEFIR-mediated protein-protein interactions. However, future studies are required to determine whether these interaction modes are shared by Act1-SEFIR in IL-17 signaling.
Materials and Methods
Cloning, expression, and purification
The two coding sequences of BcSEFIRΔtail (residues 1–143) and BcSEFIR (residues 1–153) were cloned into a modified pET28a vector which have a fused SUMO protein with 6×His tag at N terminal. The cells were grown at 37 °C to an optical density of 0.6 and then induced with 0.2 mM isopropyl-D-thiogalactopyranoside at 16 °C overnight. Bacterial pellets were then resuspended in lysis buffer (20 mM Tris- HCl pH 8.0, 150 mM NaCl) and lysed by sonication at 4 °C. The fusion proteins were purified by Ni-affmity chromatography, followed by digestion of protease Ulp1 to remove N terminal 6×His tag and fused sumo, then proteins were purified by anion exchange chromatograph for further investigations. Selenomethionine (SeMet)- substituted BcSEFIR was expressed in M9 minimal medium supplemented with amino acids (50) and purified similarly to the native protein using the procedures described above. Mutations DNA sequences were synthesized commercially. For the immunoprecipitation experiments, the BcSEFIR or mutations DNA sequences with a N-terminal HA-tag or V5-tag were generated by PCR methods. The new DNA inserts were subsequently subcloned into the plasmid described before. The HA-tagged or V5-tagged proteins were expressed and purified as native proteins.
Crystallization
The BcSEFIRΔtail and BcSEFIR were both crystallized at 16 °C using the hanging-drop vapor-diffusion method by equilibrating a mixture containing 1 μl of protein solution (8 mg/ml in a buffer of 20 mM Tris-HCl pH 8.0, 150 mM NaCl) and 1 μl of reservoir solution (0.2 M NaCl, 0.1 M imidozale pH 8.0, 30% PEG 8000) against 1 ml of reservoir solution. After one week, single crystals formed and were flash-frozen in liquid nitrogen for future data collection. The crystal of selenomethionine-labeled BcSEFIR grew in a different reservoir solution (0.2 M Lithium sulfate monohydrate, 0.1 M Bis-Tris pH 5.5, 25% PEG 3350).
Data collection, phasing and model refinement
Diffraction data for BcSEFIR crystals were collected at beamline BL19U1 of Shanghai Synchrotron Radiation Facility. Diffraction data for BcSEFIRΔtail crystals were collected using Rigaku FR-E/VariMax HR/Raxis IV++ (Rigaku, Japan) at the Structural Biology Core Facility (Institute of Biophysics, Chinese Academy of Sciences). Data were indexed and scaled with HKL2000 (51). Phasing problems of BcSEFIR was solved by using multiwavelength anomalous diffraction method (52), and there are three selenium sites in each asymmetric unit in the structure. Then the structure of BcSEFIRΔtail was solved by using molecular replacement method performed on PHENIX.phascr (53), with the structure of BcSEFIR as a search model. Refinement was carried out with PHENIX.refine (53) and COOT (54). All the statistics of data collection and refinement are listed in Table 1. The completeness in the high resolution shell for BcSEFIRΔtail is relatively low. Coordinates and structure factors have been deposited in the Protein Data Bank with accession numbers 5Y8F and 5Y8E for structures of BcSEFIRΔtail and BcSEFIR, respectively.
Immunoprecipitation
For co-immunoprecipitation assays to validate the structure-function relationship, 0.5 μg purified V5-tagged BcSEFIR or BcSEFIRΔtail were mixed 1:1 with HA-tagged mutations or wild type BcSEFIRs for 30 min in 4 °C in immunoprecipitation (IP) buffer (20 mM Tris-HCl pH 8.0, 150 mM NaCl). Protein G dynabeads were added to each sample, then incubated with anti-HA antibody (Cell Signalling Technology) at 4 °C overnight, beads were washed four times in IP buffer, boiled in Faemmli sample buffer, and analyzed by SDS-PAGE/immunoblotting. Blots were labeled using anti-V5 and anti-HA antibodies (Invitrogen), while detection was carried out using ECL reagents (GE Healthcare).
Supplementary Material
Table 1.
Data collection and refinement statistics
| BcSEFIR | BcSEFIR∆tail | |
|---|---|---|
| Crystallographic data | ||
| dmin (Å) | 1.80 (1.80–1.83) | 2.00 (2.00–2.05) |
| Wavelength (Å) | 0.9793 | 1.5418 |
| Measured reflections | 108121 (3104) | 271348 (1654) |
| Average redundancy | 4.1 (4.1) | 2.8 (2.0) |
| Mean I/ σ (I) | 11.2 (8.6) | 12.7 (2.2) |
| Completeness (%) | 99.4 (99.2) | 94.9 (67.3) |
| Rmergea | 0.097 (0.218) | 0.079 (0.391) |
| Refinement statistics | ||
| Bragg spacing (Å) | 35.33–1.80 | 29.18–2.00 |
| Space group | P21 | P21 |
| Cell parameters | ||
| a (Å) | 38.74 | 37.86 |
| b (Å) | 61.67 | 68.02 |
| c (Å) | 39.27 | 54.94 |
| Reflections in working set | 14504 | 16937 |
| Reflections in test set | 765 | 917 |
| R cryst b | 0.1668 | 0.1857 |
| R free c | 0.1963 | 0.2219 |
| r.m.s.d. bonds (Å) | 0.006 | 0.004 |
| r.m.s.d. angles (°) | 0.963 | 0.982 |
| Average B-factor (Å2) | 22.48 | 25.38 |
| No. of residues | 154 | 288 |
| No. of waters | 223 | 346 |
Values in parentheses indicate the corresponding statistics in the highest resolution shell.
Rmerge = (Ii − ‹Ii›|)/Ii|, where Ii is the integrated intensity of a given reflection.
Rcryst = (‖Fo| − |Fc‖)/|Fo|, where Fo and Fc denote observed and calculated structure factors, respectively.
Rfree is equivalent to Rcryst, but calculated using randomly chosen 10% reflections as the test set, which were excluded from the refinement process.
Acknowledgements
We are very grateful to the staff of the Structural Biology Core Facility (Institute of Biophysics, Chinese Academy of Sciences) for their technical assistance, especially to Ms. Ya Wang, Mr. Yi Han and Mr. Jianhui Li. This work was supported by the National Natural Science Foundation of China (Project No. 31400638, 31271491 and 31470792), and Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDB08030102).
References
- 1.Novatchkova M, Leibbrandt A, Werzowa J, Neubuser A, and Eisenhaber F (2003) The STIR-domain superfamily in signal transduction, development and immunity. Trends Biochem Sci 28, 226–229 [DOI] [PubMed] [Google Scholar]
- 2.Sonder SU, Saret S, Tang W, Sturdevant DE, Porcella SF, and Siebenlist U (2011) IL-17-induced NF-kappaB activation via CIKS/Act1: physiologic significance and signaling mechanisms. J Biol Chem 286, 12881–12890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pisitkun P, Claudio E, Ren N, Wang H, and Siebenlist U (2010) The adaptor protein CIKS/ACT1 is necessary for collagen-induced arthritis, and it contributes to the production of collagen-specific antibody. Arthritis Rheum 62, 3334–3344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sonder SU, Paun A, Ha HL, Johnson PF, and Siebenlist U (2012) CIKS/Act1-mediated signaling by IL-17 cytokines in context: implications for how a CIKS gene variant may predispose to psoriasis. J Immunol 188, 5906–5914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li X, Commane M, Nie H, Hua X, Chatterjee-Kishore M, Wald D, Haag M, and Stark GR (2000) Act1, an NF-kappa B-activating protein. Proc Natl Acad Sci U S A 97, 10489–10493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moynagh PN (2005) The NF-kappaB pathway. J Cell Sci 118, 4589–4592 [DOI] [PubMed] [Google Scholar]
- 7.Chang SH, Park H, and Dong C (2006) Act1 adaptor protein is an immediate and essential signaling component of interleukin-17 receptor. J Biol Chem 281, 35603–35607 [DOI] [PubMed] [Google Scholar]
- 8.Kang Z, Wang C, Zepp J, Wu L, Sun K, Zhao J, Chandrasekharan U, DiCorleto PE, Trapp BD, Ransohoff RM, and Li X (2013) Act1 mediates IL-17-induced EAE pathogenesis selectively in NG2+ glial cells. Nat Neurosci 16, 1401–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giltiay NV, Lu Y, Cullen JL, Jorgensen TN, Shlomchik MJ, and Li X (2013) Spontaneous loss of tolerance of autoreactive B cells in Act1-deficient rheumatoid factor transgenic mice. J Immunol 191, 2155–2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ryzhakov G, Blazek K, Lai CC, and Udalova IA (2012) IL-17 receptor adaptor protein Act1/CIKS plays an evolutionarily conserved role in antiviral signaling. J Immunol 189, 4852–4858 [DOI] [PubMed] [Google Scholar]
- 11.DeSelm CJ, Takahata Y, Warren J, Chappel JC, Khan T, Li X, Liu C, Choi Y, Kim YF, Zou W, and Teitelbaum SL (2012) IL-17 mediates estrogen-deficient osteoporosis in an Act1-dependent manner. J Cell Biochem 113, 2895–2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Claudio Estefania*, S. U. S, Saret Sun*, Carvalho Gabrielle*,2, Ramalingam Thirumalai R+, W. TA, Chariot Alain*,3, Garcia-Perganeda Antonio*,4, Leonardi Antonio*, Paun Andrea*, Chen Amy#, Ren Nina Y.*, Wang Hongshan*, and Siebenlist Ulrich*. (2009) The adaptor protein CIKS/Act1 is enssential for IL-25-mediated allergic airway inflammation. J Immunol [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaffen SL (2009) Structure and signalling in the IL-17 receptor family. Nat Rev Immunol 9, 556–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fitzgerald KA, Palsson-McDermott EM, Bowie AG, Jefferies CA, Mansell AS, Brady G, Brint E, Dunne A, Gray P, Harte ΜT, McMurray D, Smith DE, Sims JE, Bird TA, and O’Neill LA (2001) Mal (MyD88-adapter-like) is required for Toll-like receptor-4 signal transduction. Nature 413, 78–83 [DOI] [PubMed] [Google Scholar]
- 15.Mauro C, Vito P, Mellone S, Pacifico F, Chariot A, Formisano S, and Leonardi A (2003) Role of the adaptor protein CIKS in the activation of the IKK complex. Biochemical and Biophysical Research Communications 309, 84–90 [DOI] [PubMed] [Google Scholar]
- 16.Ryzhakov G, Blazek K, and Udalova IA (2011) Evolution of vertebrate immunity: sequence and functional analysis of the SEFIR domain family member Act1. J Mol Evol 72, 521–530 [DOI] [PubMed] [Google Scholar]
- 17.Zhang B, Liu C, Qian W, Han Y, Li X, and Deng J (2013) Crystal structure of IL-17 receptor B SEFIR domain. J Immunol 190, 2320–2326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iwasaki A, and Medzhitov R (2004) Toll-like receptor control of the adaptive immune responses. Nat Immunol 5, 987–995 [DOI] [PubMed] [Google Scholar]
- 19.Watters TM, Kenny EF, and O’Neill LA (2007) Structure, function and regulation of the Toll/IL-1 receptor adaptor proteins. Immunol Cell Biol 85, 411–419 [DOI] [PubMed] [Google Scholar]
- 20.von Bernuth H, Picard C, Jin Z, Pankla R, Xiao H, Ku CL, Chrabieh M, Mustapha IB, Ghandil P, Camcioglu Y, Vasconcelos J, Sirvent N, Guedes M, Vitor AB, Herrero-Mata MJ, Arostegui JI, Rodrigo C, Alsina L, Ruiz-Ortiz E, Juan M, Fortuny C, Yague J, Anton J, Pascal M, Chang ΗH, Janniere L, Rose Y, Gariy BZ, Chapel H, Issekutz A, Marodi L, Rodriguez-Gallego C, Banchereau J, Abel L, Li X, Chaussabel D, Puel A, and Casanova JL (2008) Pyogenic bacterial infections in humans with MyD88 deficiency. Science 321, 691–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dangl JL, Horvath DM, and Staskawicz BJ (2013) Pivoting the plant immune system from dissection to deployment. Science 341, 746–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Medzhitov R, Preston-Hurlburt P, and Janeway CA Jr. (1997) A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature 388, 394–397 [DOI] [PubMed] [Google Scholar]
- 23.Ve T, Williams SJ, and Kobe B (2015) Structure and function of Toll/interleukin-1 receptor/resistance protein (TIR) domains. Apoptosis 20, 250–261 [DOI] [PubMed] [Google Scholar]
- 24.Halabi S, Sekine E, Verstak B, Gay NJ, and Moncrieffe MC (2017) Structure of the Toll/Interleukin-1 Receptor (TIR) Domain of the B-cell Adaptor That Links Phosphoinositide Metabolism with the Negative Regulation of the Toll-like Receptor (TLR) Signalosome. J Biol Chem 292, 652–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang X, Bernoux M, Bentham AR, Newman TE, Ve T, Casey LW, Raaymakers TM, Hu J, Croll TI, Schreiber KJ, Staskawicz BJ, Anderson PA, Sohn KH, Williams SJ, Dodds PN, and Kobe B (2017) Multiple functional self-association interfaces in plant TIR domains. Proc Natl Acad Sci U S A 114, E2046–E2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams SJ, Yin L, Foley G, Casey LW, Outram MA, Ericsson DJ, Lu J, Boden M, Dry IB, and Kobe B (2016) Structure and Function of the TIR Domain from the Grape NLR Protein RPV1. Front Plant Sci 7, 1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Enokizono Y, Kumeta H, Funami K, Horiuchi M, Sarmiento J, Yamashita K, Standley DM, Matsumoto M, Seya T, and Inagaki F (2013) Structures and interface mapping of the TIR domain-containing adaptor molecules involved in interferon signaling. Proc Natl Acad Sci U S A 110, 19908–19913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hyun KG, Lee Y, Yoon J, Yi H, and Song JJ (2016) Crystal structure of Arabidopsis thaliana SNC1 TIR domain. Biochem Biophys Res Commun 481, 146–152 [DOI] [PubMed] [Google Scholar]
- 29.Nishimura ΜT, Anderson RG, Cherkis KA, Law TF, Liu QL, Machius M, Nimchuk ZL, Yang L, Chung EH, El Kasmi F, Hyunh M, Osborne Nishimura E, Sondek JE, and Dangl JL (2017) TIR-only protein RBA1 recognizes a pathogen effector to regulate cell death in Arabidopsis. Proc Natl Acad Sci U S A 114, E2053–E2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang B, Liu C, Qian W, Han Y, Li X, and Deng J (2014) Structure of the unique SEFIR domain from human interleukin 17 receptor A reveals a composite ligand-binding site containing a conserved alpha-helix for Act1 binding and IL-17 signaling. Acta Crystallogr D Biol Crystallogr 70, 1476–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu C, Swaidani S, Qian W, Kang Z, Sun P, Han Y, Wang C, Gulen MF, Yin W, Zhang C, Fox PL, Aronica M, Hamilton TA, Misra S, Deng J, and Li X (2011) A CC’ loop decoy peptide blocks the interaction between Act1 and IL-17RA to attenuate IL-17-and IL-25-induced inflammation. Sci Signal 4, ra72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boisson B, Wang C, Pedergnana V, Wu L, Cypowyj S, Rybojad M, Belkadi A, Picard C, Abel L, Fieschi C, Puel A, Li X, and Casanova JL (2013) An ACT1 mutation selectively abolishes interleukin-17 responses in humans with chronic mucocutaneous candidiasis. Immunity 39, 676–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu B, Gong J, Liu L, Li T, Wei T, and Bai Z (2012) Evolution of prokaryotic homologues of the eukaryotic SEFIR protein domain. Gene 492, 160–166 [DOI] [PubMed] [Google Scholar]
- 34.Wu B, Jin M, Gong J, Du X, and Bai Z (2011) Dynamic evolution of CIKS (TRAF3IP2/Act1) in metazoans. Dev Comp Immunol 35, 1186–1192 [DOI] [PubMed] [Google Scholar]
- 35.Xu Y, Tao X, Shen B, Horng T, Medzhitov R, Manley JL, and Tong L (2000) Structural basis for signal transduction by the Toll/interleukin-1 receptor domains. Nature 408, 111–115 [DOI] [PubMed] [Google Scholar]
- 36.Toshchakov VY, Szmacinski H, Couture LA, Lakowicz JR, and Vogel SN (2011) Targeting TLR4 signaling by TLR4 Toll/IL-1 receptor domain-derived decoy peptides: identification of the TLR4 Toll/IL-1 receptor domain dimerization interface. J Immunol 186, 4819–4827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toshchakov VU, Basu S, Fenton MJ, and Vogel SN (2005) Differential involvement of BB loops of toll-IL-1 resistance (TIR) domain-containing adapter proteins in TLR4-versus TLR2-mediated signal transduction. J Immunol 175, 494–500 [DOI] [PubMed] [Google Scholar]
- 38.Tao X, Xu Y, Zheng Y, Beg AA, and Tong L (2002) An extensively associated dimer in the structure of the C713S mutant of the TIR domain of human TLR2. Biochem Biophys Res Commun 299, 216–221 [DOI] [PubMed] [Google Scholar]
- 39.Williams SJ, Sohn KH, Wan L, Bernoux M, Sarris PF, Segonzac C, Ve T, Ma Y, Saucet SB, Ericsson DJ, Casey LW, Lonhienne T, Winzor DJ, Zhang X, Coerdt A, Parker JE, Dodds PN, Kobe B, and Jones JD (2014) Structural basis for assembly and function of a heterodimeric plant immune receptor. Science 344, 299–303 [DOI] [PubMed] [Google Scholar]
- 40.Valkov E, Stamp A, Dimaio F, Baker D, Verstak B, Roversi P, Kellie S, Sweet MJ, Mansell A, Gay NJ,Martin JL, and Kobe B (2011) Crystal structure of Toll-like receptor adaptor MAL/TIRAP reveals the molecular basis for signal transduction and disease protection. Proc Natl Acad Sci U S A 108, 14879–14884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin Z, Lu J, Zhou W, and Shen Y (2012) Structural insights into TIR domain specificity of the bridging adaptor Mal in TLR4 signaling. PloS one 7, e34202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nyman T, Stenmark P, Flodin S, Johansson I, Hammarstrom M, and Nordlund P (2008) The crystal structure of the human toll-like receptor 10 cytoplasmic domain reveals a putative signaling dimer. TheJournal of biological chemistry 283, 11861–11865 [DOI] [PubMed] [Google Scholar]
- 43.Alaidarous M, Ve T, Casey LW, Valkov E, Ericsson DJ, Ullah MO, Schembri MA, Mansell A, Sweet MJ, and Kobe B (2014) Mechanism of bacterial interference with TLR4 signaling by Brucella Toll/interleukin-1 receptor domain-containing protein TcpB. The Journal of biological chemistry 289, 654–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jang TH, and Park ΗH (2014) Crystal structure of TIR domain of TLR6 reveals novel dimeric interface of TIR-TIR interaction for toll-like receptor signaling pathway. J Mol Biol 426, 3305–3313 [DOI] [PubMed] [Google Scholar]
- 45.Chan SL, Mukasa T, Santelli E, Low LY, and Pascual J (2010) The crystal structure of a TIR domain from Arabidopsis thaliana reveals a conserved helical region unique to plants. Protein Sci 19, 155–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bernoux M, Ve T, Williams S, Warren C, Hatters D, Valkov E, Zhang X, Ellis JG, Kobe B, and Dodds PN (2011) Structural and functional analysis of a plant resistance protein TIR domain reveals interfaces for self-association, signaling, and autoregulation. Cell Host Microbe 9, 200–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Snyder GA, Deredge D, Waldhuber A, Fresquez T, Wilkins DZ, Smith PT, Durr S, Cirl C, Jiang J, Jennings W, Luchetti T, Snyder N, Sundberg EJ, Wintrode P, Miethke T, and Xiao TS (2014) Crystal structures of the Toll/Interleukin-1 receptor (TIR) domains from the Brucella protein TcpB and host adaptor TIRAP reveal mechanisms of molecular mimicry. The Journal of biological chemistry 289, 669–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Khan JA, Brint EK, O’Neill LA, and Tong L (2004) Crystal structure of the Toll/interleukin-1 receptor domain of human IL-1RAPL. The Journal of biological chemistry 279, 31664–31670 [DOI] [PubMed] [Google Scholar]
- 49.Ve T, Vajjhala PR, Hedger A, Croll T, DiMaio F, Horsefield S, Yu X, Lavrencic P, Hassan Z, Morgan GP, Mansell A, Mobli M, O’Carroll A, Chauvin B, Gambin Y, Sierecki E, Landsberg MJ, Stacey KJ, Egelman EH, and Kobe B (2017) Structural basis of TIR-domain-assembly formation in MAL- and MyD88-dependent TLR4 signaling. Nat Struct Mol Biol [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, and Clardy J (1993) Atomic stmctures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J Mol Biol 229, 105–124 [DOI] [PubMed] [Google Scholar]
- 51.Otwinowski Z, and Minor W (1997) [20] Processing of X-ray diffraction data collected in oscillation mode, in Methods in Enzymology, Academic Press, pp 307–326 [DOI] [PubMed] [Google Scholar]
- 52.Kahn R, Fourme R, Bosshard R, Chiadmi M, Risler JL, Dideberg O, and Wery JP (1985) Crystal structure study of Opsanus tau parvalbumin by multiwavelength anomalous diffraction. FEBS Lett 179, 133–137 [DOI] [PubMed] [Google Scholar]
- 53.Zwart PH, Afonine PV, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, McKee E, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Storoni LC, Terwilliger TC, and Adams PD (2008) Automated structure solution with the PHENIX suite. Methods Mol Biol 426, 419–435 [DOI] [PubMed] [Google Scholar]
- 54.Emsley P, and Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


