Graphical abstract

Keywords: HCV, Genotypes, NS5B, 5′ UTR, Phylogenetic tree, Sofosbuvir
Abstract
Chronic hepatitis C virus (HCV) infection is a main health problem in Egypt causing high rates of mortalities. Egypt has the highest HCV prevalence in the world, with specific HCV subtypes epidemic and circulating extensively in the country. Different antiviral therapy protocols have been implemented for treating Egyptian HCV patients. Due to the limited data about HCV in Egypt, this study aimed to genotype HCV strains circulating in the Nile Delta Damietta governorate and to investigate the variation in the nonstructural 5B (NS5B) region targeted by the newly approved antiviral drugs. Thirty HCV samples from treatment-naïve patients were genotyped by restriction fragment length polymorphism. Some samples were genotyped by direct sequencing of their 5′ untranslated region (UTR) and NS5B regions. Phylogenetic analysis was also performed on the sequences of their NS5B regions. Fourteen new sequences have been deposited in the GenBank database. Results showed that subtype 4a was prevalent in addition to subtype 1g. None of the previously reported NS5B substitutions were detected in the sequenced isolates from treatment-naïve patients, which may be a good predictor for efficient treatment of HCV Egyptian patients with Sofosbuvir. Further studies on Sofosbuvir treated-HCV Egyptian patients are required to investigate whether any NS5B substitutions can confer resistance to treatment.
Introduction
Hepatitis C virus (HCV) is the main cause of chronic liver disease and it is an epidemic pathogen worldwide. It is estimated that around 71 million people are chronically infected with HCV and approximately 400 thousand people die each year mostly because of liver cirrhosis and hepatocellular carcinoma [1]. HCV is a blood-borne, positive sense, single-stranded RNA virus. Its RNA strand is approximately 9.6 kb with a long open reading frame encoding 3 structural (C, E1, E2) and 7 non-structural (P7, NS2, NS3, 4A, NS4B, NS5A, NS5B) proteins [2]. HCV strains are classified into 7 genotypes (1–7) and several subtypes based on the phylogenetic and sequence analyses of the complete viral genomes [3]. At the nucleotide level, HCV genotypes differ from each other by 31–33% and the subtypes within the same genotype differ from each other by 20–25% [4]. HCV genotypes are geographically distributed worldwide, with HCV genotypes 1, 2 and 3 widely distributed and subtypes 1a, 1b and 2a specifically called epidemic subtypes [5], [6]. Genotype 3 circulates mainly in south Asia, genotype 4 in central Africa and Middle East, genotype 5 in Southern Africa, genotype 6 in South East Asia and genotype 7 in Congo [7], [8], [9].
Egypt records the highest HCV prevalence worldwide [10]. According to the Demographic Health Survey (DHS) of 2015, seroprevalence of HCV was 10% compared to 14.7% in 2008 [11]. It has been reported that around 93% of infections are due to genotype 4 [12]. HCV subtype 4a became epidemic and extensively distributed in Egypt due to the unsafe injections during the anti-schistosomal public health campaigns in the past [13]. Subtype 4a has been reported as the predominant subtype in Egypt in some studies [14], [15], [16], while infection with genotype 1 was thought to never exceed 10% [15]. In Egypt, an increased risk of hepatocellular carcinoma was significantly associated with the infection with HCV subtype 4o [17]. HCV genotype 4 responsiveness to treatment with PEG-INF/RBV is better than genotype 1 but worse than genotype 2 and 3 [18]. However, genotype 4 became the most difficult to treat genotype after the effective response of genotype 1 to protease inhibitors [19].
PEG-INF/RBV treatments have been implemented in Egypt for long time and they achieved sustained virological response (SVR) of approximately 40–69% [20]. One of the recently approved new treatments for HCV in Egypt is Sofosbuvir [21]. Sofosbuvir plus Ribavirin (interferon-free regime) for 12 or 24 week became the effective treatment protocol for both naïve and previously treated Egyptian patients [22]. Combination of Elbasvir (NS5A inhibitor) and Grazoprevir (NS3/4A protease inhibitor) for treating genotypes 1 and 4 produced high SVR rates [23]. HCV genotyping is clinically important in predicting the efficiency of antiviral therapy, in determining the duration of treatment [24] and, also, in detecting HCV molecular epidemiology [25]. This study aims to genotype HCV strains circulating in Damietta Governorate and to investigate the variation in HCV NS5B region that is targeted by the newly approved antiviral drugs.
Patients and methods
Blood sampling
Blood samples were collected in EDTA-containing vacuotainer tubes from 30 HCV-infected but treatment-naïve patients from Damietta governorate. The study has been approved by our institution board (5/2/2/1) and all patients have provided an informed consent.
Viral RNA extraction
Viral RNA was extracted from all blood samples by GeneJET Viral DNA/RNA Purification Kit (#K0821, Thermo Scientific, Waltham MA, USA) according to the manufacturer’s instructions. The purified RNA was immediately used or stored at −20 °C.
Polymerase chain reaction (PCR)
All viral RNA samples were converted into cDNA using RevertAid™ H Minus First Strand cDNA Synthesis Kit (#K1631, Thermo Scientific, Waltham MA, USA). PCRs were performed to amplify 302 nt-fragment of the 5′ UTR (47–348) using the forward primer 5′-GTGAGGAACTACTGTCTTCACGCAG-3′ and the reverse primer 5′-TGCTCATGGTGCACGGTCTACGAGA-3′ [15], and to amplify 381 nt-fragment of the NS5B region (8256–8636) using the forward primer 5′-TATGAYACCCGCTGYTTTGAC-3′ and the reverse primer 5′-CCTGGTCATAGCCTCCGTGAA-3′ [17]. Each PCR included 25 µL of Maxima Hot Start PCR master mix kit (#K1051, Thermo Scientific, Waltham MA, USA), 2 µL of the forward primer, 2 µL of the reverse primer and 4 µL of the cDNA sample. Each reaction was brought to a final volume of 50 µL with nuclease-free water. Reactions were loaded to a thermal cycler (Multigene, Labnet, Edison NJ, USA) and subjected to 40 cycles of 30 s at 95 °C, 30 s at the optimized annealing temperature and 40 s at 72 °C. An initial activation step (4 min at 95 °C) and a final extension step (10 min at 72 °C) were also included in the program.
Restriction digestion
PCR products of the 5′ UTR region were digested using two combinations of restriction endonucleases; MvaI/HinfI and RsaI/HaeIII. Restriction reactions were run in 2.5% agarose gels for 40 min in TBE (#B52, Thermo Scientific, Waltham MA, USA) buffer, stained for 25 min with Ethidium Bromide and the gels were documented in Photo Doc-IT Imaging system (UVP, Upland CA, USA).
DNA sequencing
PCR products of the 5′ UTR and NS5B regions were purified using GeneJET Gel extraction kit (#K0691, Thermo Scientific, Waltham, MA USA) and sequenced using the standard Sanger method on ABI 3730XL DNA Sequencer at Macrogen sequencing services (Macrogen, Seol, South Korea). Chromatograms were checked and corrected using a sequence viewer. The genotype of each sample was determined by comparing its sequence with those of HCV prototypes deposited in the GenBank database, followed by further genetic analysis.
Phylogenetic analysis
The phylogenetic tree has been constructed using Neighbor-Joining method and the evolutionary distances were computed using the Kimura 2-parameter method with discrete gamma distribution using MEGA7 software. Bootstrap values were determined using 1000 replicates. 281 HCV sequences of other HCV isolates retrieved from HCV database and the GenBank database were used for constructing the phylogenetic tree.
Results
Genotyping by restriction fragment length polymorphism (RFLP)
Viral RNA was extracted from 30 HCV-infected patients, cDNA was synthesized and a 302 nt-fragment of HCV 5′ UTR was amplified by PCR. PCR products were subjected to double restriction digestion reactions using 2 pairs of enzymes; MvaI/HinfI and RsaI/HaeIII. Digestion of the 30 patient samples with MvaI/HinfI resulted in a clear ∼ 177 bp band and 2 close bands (∼56 and ∼69 bp) in 28 samples (Fig. 1a samples 2–8), while in the other 2 samples one separate 129 bp band, 2 close bands (∼63 bp and ∼69 bp) and ∼41 bp band have been obtained (Fig. 1a, sample 1). On the other hand, double digestion with RsaI/HaeIII resulted in 2 separate bands (114–118 bp and 58–61 bp) in the 30 patient samples (Fig. 1b). Based on this and compared to the digestion pattern of known genotypes 1 and 4 (Fig. 1c), collectively, these results show the presence of at least 2 different HCV genotypes; most probably genotype 1 (represented by sample 1 in Fig. 1) and genotype 4 (represented by samples 2–8 in Fig. 1).
Fig. 1.
RFLP of 5′ UTR for 8 representative HCV samples and some control samples. A: double digestion with MvaI/HinfI. B: double digestion with RsaI/HaeIII. Wells designated “P” show undigested PCR products and wells designated “R” show digested products. “M” designates the well containing a 50 bp DNA ladder. Sample 1 represents type 1 cleavage pattern (Isolate EGDAM112) and samples 2–8 represent type 4 cleavage pattern (Isolates EGDAM113, EGDAM114, EGDAM115, EGDAM116, EGDAM117, EGDAM118 and EGDAM119 respectively). C: Restriction digestion pattern of known genotypes 1 and 4 with the same restriction enzymes, in addition to “no template” control reactions. From the ladder side, digestion with MvaI/HinfI is followed by RsaI/HaeIII.
Variability of 5′ UTR and NS5B sequences
Out of the 30 HCV samples under study, 10 samples (2 samples thought to be genotype 1 and 8 samples thought to be genotype 4 based on RFLP results) were selected for sequencing their 5′ UTR and NS5B regions to confirm the RFLP results and to subtype them. The obtained 5′ UTR and NS5B sequences were compared to the sequences in the NCBI database using basic local alignment search tool (BLAST). BLAST results (Table 1) show that 6 samples (Isolates EGDAM006, EGDAM072, EGDAM082, EGDAM089, EGDAM091 and EGDAM136) had the highest identities to the 5′ UTR and NS5B regions of subtype 4a. One isolate (EGDAM025) had the highest identity to the 5′ UTR region of subtype 4a but the NS5B of subtype 1b/1g. Two isolates (EGDAM032 and EGDAM099) had the highest identity to the 5′ UTR region of subtype 4t/4b but the NS5B of subtype 4a. One isolate (EGDAM112) had the highest identity to the 5′ UTR region of subtype 1g and the NS5B of subtype 1g/1b. It is worthy to note that, according to RFLP results, isolates EGDAM025 and EGDAM112 were thought to be type 1, while the other 8 isolates were thought to be type 4.
Table 1.
BLAST results of 5′ UTR and NS5B regions of 10 HCV isolates.
| Isolate | N5 region |
NS5B region |
||||
|---|---|---|---|---|---|---|
| Highest identical sequences | Genotype | Identity% | Highest identical sequences | Genotype | Identity% | |
| EGDAM006 | KY283130.1 | 4a | 203/204(99%) | FN668607.1 | 4a | 281/294(96%) |
| KT735185.1 | 4a | 203/204(99%) | AB470051.1 | 4a | 280/293(96%) | |
| EGDAM025 | KY283130.1 | 4a | 204/204(100%) | FJ807054.1 | 1b | 257/266(97%) |
| KT735185.1 | 4a | 204/204(100%) | AY548727.1 | 1g | 290/302(96%) | |
| EGDAM032 | FJ839869.1 | 4t | 203/204(99%) | AB470053.1 | 4a | 273/293(93%) |
| FJ462435.1 | 4b | 203/204(99%) | LC109166.1 | 4a | 281/302(93%) | |
| EGDAM072 | KY283130.1 | 4a | 204/204(100%) | AB470053.1 | 4a | 272/293(93%) |
| KT735185.1 | 4a | 204/204(100%) | LC109166.1 | 4a | 280/302(93%) | |
| EGDAM082 | KY283130.1 | 4a | 204/204(100%) | EF694502.1 | 4a | 281/294(96%) |
| KT735185.1 | 4a | 204/204(100%) | LC109141.1 | 4a | 287/302(95%) | |
| EGDAM089 | KM587624.1 | 4a | 202/204(99%) | EF694393.1 | 4a | 277/292(95%) |
| KY283130.1 | 4a | 201/204(99%) | DQ911222.1 | 4a | 287/305(94%) | |
| EGDAM091 | KY283130.1 | 4a | 204/204(100%) | JN203160.1 | 4a | 292/305(96%) |
| KT735185.1 | 4a | 204/204(100%) | AB470006.1 | 4a | 277/293(95%) | |
| EGDAM099 | FJ839869.1 | 4t | 204/204(100%) | FN668602.1 | 4a | 271/293(92%) |
| FJ462435.1 | 4b | 204/204(100%) | EF694502.1 | 4a | 271/293(92%) | |
| EGDAM112 | KJ009288.1 | 1g | 204/204(100%) | AY548727.1 | 1g | 290/302(96%) |
| AM910652.2 | 1g | 204/204(100%) | FJ807054.1 | 1b | 254/266(95%) | |
| EGDAM0136 | KY283130.1 | 4a | 199/204(98%) | EF694476.1 | 4a | 275/293(94%) |
| KT735185.1 | 4a | 199/204(98%) | LC109250.1 | 4a | 280/299(94%) | |
The nucleotide variance rate in the 5′ UTR region is less than the nucleotide variance rate in the NS5B region (Table 1). Analysis of nucleotide variance in the 5′ UTR region, compared to the reference subtype 4a (Y11604) isolated previously from Egypt, revealed that G dominates the position 150 instead of T, and T dominates the position 284 instead of C in the 10 sequenced isolates (Table 2). The isolates EGDAM112 and EGDAM136 had the highest nucleotide variation rate among the 10 sequenced isolates. The position 183 contained C instead of T in 3 isolates. It is worthy to mention that the 2 isolates EGDAM025 and EGDAM112, which are thought to be of the same genotype by RFLP, differ in their 5′ UTR sequence. The isolate EGDAM136 which is thought to be type 4 by RFLP, like the other 7 isolates, has many polymorphic sites in its 5′ UTR region.
Table 2.
Nucleotide variance in the 5′ UTR region of 10 HCV isolates. Sequences are compared to the reference sequence of type 4a (Y11604) isolated from Egypt.
 |
*Nucleotide numbers are based on the prototype H77 (NC_004102). R (A, g); Y (C, T); K (g, T).
Analysis of the amino acid variance in the NS5B region (Table 3), compared to the reference genotype Y11604, reveals the dominance of E258 instead of D, S282 instead of T and G307 instead of A in the 10 sequenced isolates. In 6 isolates, A/T replaced V235. The isolates EGDAM025 and EGDAM112 had a very similar NS5B amino acid sequence which is very different from that of the other 8 isolates (Table 3).
Table 3.
Amino acid variance in the NS5B region of 10 HCV isolates. Amino acids 226–327 are compared to amino acids of the reference sequence of type 4a (Y11604) isolated from Egypt.
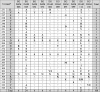 |
*Amino acid numbers are based on the prototype H77 (NC_004102).
NS5B sequences of the 10 isolates were submitted to the GenBank the under the accession numbers MF371339-MF371348. Sequences of the 5′ UTR region of the isolates EGDAM006, EGDAM032, EGDAM089 and EGDAM136 were submitted to the GenBank under the accession numbers MF497264-MF497267.
Phylogenetic analysis
Because of the very close results of the NS5B sequencing (but not the 5′ UTR sequencing) and RFLP results and the discrepant subtyping of genotype 1 based on BLAST results, NS5B sequences of the 10 sequenced isolates were subjected to phylogenetic analysis. A 203 nt-segment covering the middle of NS5B region of the 10 sequenced isolates was aligned with 281 sequences including several reference genome sequences in addition to sequences from Egyptian HCV isolates, and a Neighbor-Joining phylogenetic tree was constructed (Fig. 2). The tree revealed that 8 of the 10 studied isolates clustered with different isolates of HCV subtype 4a from Egypt. The isolates EGDAM025 and EGDAM112 clustered with each other and with different isolates of subtype 1 g from Egypt and with subtype 1 g from Spain.
Fig. 2.
Neighbor-Joining phylogenetic tree of NS5B sequences (positions 8256–8636) of seven representative HCV genotypes. Bootstrap values based on 1000 replicates are shown next to the branches; bootstrap values more than 50% only are shown. Sequences are labeled to the right of each branch in the order: GenBank accession number, isolate name, genotype/subtype and country. Sequences from reference genomes are referred to with “Ref” after the subtype name. Sequences of the current study are underlined and italic.
Discussion
In this study, 30 HCV samples from Damietta, the most northern governorate in the Nile Delta of Egypt, have been genotyped by RFLP using 2 combinations of restriction enzymes; MvaI/HinfI and RsaI/HaeIII. These combinations have been used previously for RFLP genotyping of HCV [17], [26]. Several cleavage patterns were reported due to the absence of some enzyme cleavage sites or the creation of extra cut sites. For HCV type 4a isolated previously from Egypt (Y11604), the typical MvaI/HinfI cleavage pattern is 56, 69 and 177 bp fragments, while the typical RsaI/HaeIII cleavage pattern is 7, 9, 53, 58, 61 and 114 bp fragments. For HCV type 1 (NC_004102), the typical MvaI/HinfI cleavage pattern is 41, 63, 69 and 129 bp fragments, while the typical RsaI/HaeIII cleavage pattern is 9, 61, 114 and 118 bp fragments. Compared to these cleavage patterns, 2 out of 30 HCV samples studied here (6.7%) were thus determined to be type 1 while the other 28 samples (93.3%) were determined to be type 4.
RFLP has been considered as a reliable, inexpensive and useful HCV genotyping method in the regions endemic for HCV genotype 4 [15]. Previous RFLP genotyping studies of HCV in Egypt reported the presence of genotype 4 (91%), genotype 1a (1%), genotype 1b (1%) and other strains that couldn’t be typed (7%) [15]. Abdel-Hamid et al. [17] presented slightly different RFLP results; genotype 4 (73%), genotype 1 (2%), genotype 3 (1%) and other strains that couldn’t be typed (12%).
In the Mediterranean countries, subtype 1b is the most prevalent HCV subtype [27]. In Lebanon and Tunisia, genotype 1 is the predominant genotype followed by genotype 4, while in Syria genotype 4 is the prevalent genotype followed by genotype 1 and genotype 5 [28]. In Turkey, subtype 1b is the most prevalent subtype followed by 1a and 3a [29]. In Spain, subtype 1b is the predominant subtype followed by genotypes 1a, 3 and 4 [30].
Because RFLP is a general genotyping method and is not reliable for HCV subtyping, 10 HCV samples including the 2 different genotypes determined as types 1 and 4 by RFLP were further genotyped by direct sequencing of their 5′ UTR and NS5B regions.
BLAST results of the 5′ UTR region typed 7 isolates as subtype 4a, 2 isolates as subtype 4 t/4b and 1 isolate as subtype 1 g. This 5′ UTR subtyping is obviously discrepant with RFLP-based genotyping. BLAST results of NS5B typed 8 isolates as subtype 4a and 2 isolates as subtype 1 g/1b; results that clearly support RFLP genotyping. The 5′ UTR region is the most conserved region in the HCV genome [31] and it lacks considerable variability and thus leads to subtyping errors [32]. Therefore, direct sequencing of NS5B, core and envelope regions became increasingly reliable for genotyping [32].
The sequenced part of the 5′ UTR region in the current study, nucleotides 103–306, covers a part of internal ribosome entry site (IRES) which consists of 4 domains (II-IV) and is responsible for the regulation of Cap-independent translation of HCV [33]. Nucleotide variance data in the current work showed that most of the mutations (35.7%) were localized in the stem-loop IIIb (Nucleotides 172–227). El Awady et al. [34] detected 19 mutations in the patients with SVR and in breakthrough patients. Mutations in IIIa/b were associated with reduced RNA stability, while IIId mutations were suggested to increase RNA stability. The thermodynamic stability of RNA secondary structure is considered as a significant but not sufficient parameter for predicting viral stabilization, or response to IFNα [34].
Compared to the reference type 4a (Y11604) isolated previously from Egypt, the nucleotide G dominated the position 150 instead of T, and T dominated the position 284 instead of C in the 5′ UTR region of the 10 sequenced isolates. Hemeida et al. [35] sequenced 3 HCV samples from 3 non-responders from Sohag governorate in Egypt and reported the presence of 7 nucleotide variations at the positions 74, 92, 112, 113, 133, 172 and 180 in the 5′ UTR region. None of these variations has been recorded in our sequenced isolates. Nucleotide substitution in HCV 5′ UTR may affect viral translation and viral sensitivity to interferon [36]. Zekri et al. [37] reported the mutation G160A in the 5′ UTR as a unique mutation among non-responders. This position corresponds to position 243 in the current study, which was found to be polymorphic (A/G) in the 2 isolates EGDAM006 and EGDAM089. Unfortunately, interferon has been extensively used as the main treatment method for Egyptian HCV patients, recorded 40–69% SVR [20] and thus left many patients non cured and struggling with HCV side effects.
The amino acid sequences of NS5B of the isolates EGDAM025 and EGDAM112, which were subtyped 1g/1b by NS5B BLAST, were very similar to each other but very different from the other 8 isolates subtyped 4a. Because BLAST results of the NS5B typed 2 isolates as subtype 1g/1b, we further analyzed the 10 NS5B sequences of this study by phylogenetic analysis. Neighbor-Joining phylogenetic tree further confirmed that 8 of the 10 sequenced isolates belong to subtype 4a while 2 isolates clustered with subtype 1g. Current results are consistent with most of the previous studies on Egyptian patients. In south Egypt, the predominant subtype was HCV-4a followed by subtype 1g, subtype 4l, then 4n and 4o [38]. In Ismailia governorate, subtype 4a was the predominant subtype, followed by subtype 1g, and subtype 4o [16]. In Sharkia governorate, the prevalent subtype was 4a, followed by 4o, 1g and 4n [14]. The predominant subtype was 4a, followed by subtype 4m, 4o, 4n and 4p in Alexandria governorate [39]. In the current study, subtypes other than 4a and 1g couldn’t be detected most likely because of the small number of patients.
Although recombination is important for RNA viruses to generate a genetic variation; recombination is rare in HCV [40] suggesting that it is rare in vivo [41]. Some natural inter-genotypic, intra-genotypic and intra-subtype recombinations have been identified [42]. However, it is unlikely that the isolate EGDAM025 could be a recombinant strain from types 1 and 4 because the homology of the 5′ UTR between EGDAM025 and type 1b is 95% while the homology with type 1g is 97%. For NS5B, the homology between EGDAM025 and type 1b is 87.5% while the homology with type 1g is 92%. Based on reviewing the literature, genotype 4/1 recombination has never been detected previously in Egypt or in any other country.
Compared to the reference genotype 4a (Y11604), this study revealed the dominance of the amino acids E258 instead of D, S282 instead of T and G307 instead of A in the NS5B region of the 10 sequenced isolates. In 6 isolates, A/T replaced V235. NS5B is the RNA-dependent RNA polymerase (RdRp) [43] that is responsible for viral genome replication. The sequenced part of NS5B region in the current study (amino acids 226–327) covers a part of NS5B palm region of RdRp. RdRp lacks the proofreading activity and thus leads to high RNA mutation rates of around 10−4–10−5 per base pair [44].
Like all polymerases, HCV NS5B resembles a cupped right hand and is configured into three domains; palm, fingers and thumb [45]. The palm domain contains 5 motifs (A to E) and is considered to be the most conserved region [46] compared with the two other NS5B domains (fingers and thumb domains). The sequenced NS5B region in this study covers two motifs of the palm domain; motif B and motif C, and it covers two amphipathic α-helices responsible for connecting the palm and fingers domains. Motif B contains conserved residues responsible for sugar selection [47] and if these amino acids mutated, NS5B polymerase activity would be abolished [48]. In motif C, the highly conserved GDD motif among all HCV genotypes [47] plays a role in nucleotidyl transfer reaction [46]. No amino acid substitutions were detected in these conserved amino acid residues indicating a non-defective activity of the HCV polymerase among Egyptian patients.
The mutation S282T has been reported to confer resistance against the antiviral drugs of the family 2′-C-methyl modified ribonucleosides [49]. This mutation alters the conformation of the enzyme’s catalytic site [50] and severely compromises viral fitness among different HCV genotypes [51]. S282T mutation was not detected in the 10 sequenced isolates in the current study. The absence of S282T mutation can, therefore, be considered a good predictor for the responsiveness of HCV Egyptian patients to the Sofosbuvir-based therapy.
It has been reported that the amino acids C316 and V321 are in close proximity to the catalytic triad of the HCV NS5B polymerase (D220, D318, and D319) and, therefore, the changes at these positions may alter the conformation of the active site [52] and interfere with the ability of Sofosbuvir to enter the active site [53]. The amino acid substitutions C316N, L320F, and V321I have been reported to confer resistance to Sofosbuvir in several clinical studies [52]. None of these substitutions were detected in our sequenced isolates and this, again, may be a good predictor for Sofosbuvir efficiency in treating HCV Egyptian patients.
The position 307 is close from the catalytic site residues of NS5B polymerase (317–319) and it is not known whether the dominance of this position with G instead of A in the sequenced isolates in this study affects the patients response to Sofosbuvir. It is worthy to report that Sofosbuvir and Ribavirin treatment for either 12 or 24 weeks was successful in treating Egyptian patients infected with HCV genotype 4 where SVR was 90% and 77% for patients receiving 24 and 12 weeks of therapy respectively [22].
Further studies on a large cohort of HCV Egyptian patients are, therefore, required to investigate the substitutions that regulate the response/resistance to the massively used Sofosbuvir in Egypt.
Conclusions
Genotyping of HCV infecting patients in Damietta Governorate revealed the predominance of HCV subtype 4a followed by subtype 1g. This study recorded 10 new HCV isolates and their partial sequences were deposited in the GenBank database. NS5B amino acid substitutions in the positions affecting polymerase activity and in the positions affecting resistance to Sofosbuvir were absent in the sequenced isolates in this study. Provided further studies on Sofosbuvir-treated HCV patients, absence of such substitutions could be a good predictor for efficient treatment of Egyptian patients with Sofosbuvir.
Study limitations
Due to limitation of funding, sample size in this study was small. Additional studies using larger sample size are inevitable to confirm the current findings.
Conflict of interest
The authors have declared no conflict of interest.
Acknowledgments
Authors would like to thank Zoology Department and Faculty of Science, Damietta University for partial funding of this work through facilitating the use of Biotechnology and Physiology laboratory during this study.
Footnotes
Peer review under responsibility of Cairo University.
References
- 1.Hepatitis C, fact sheet N 164. [database on the Internet]; 2017. Available from: <http://www.who.int/mediacentre/factsheets/fs164/en/>.
- 2.Bartenschlager R., Vogt P.K. Springer; 2013. Hepatitis C virus: from molecular virology to antiviral therapy. [Google Scholar]
- 3.Smith D.B., Bukh J., Kuiken C., Muerhoff A.S., Rice C.M., Stapleton J.T. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment web resource. Hepatology. 2014;59(1):318–327. doi: 10.1002/hep.26744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simmonds P., Bukh J., Combet C., Deléage G., Enomoto N., Feinstone S. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology. 2005;42(4):962–973. doi: 10.1002/hep.20819. [DOI] [PubMed] [Google Scholar]
- 5.Magiorkinis G., Magiorkinis E., Paraskevis D., Ho S.Y., Shapiro B., Pybus O.G. The global spread of hepatitis C virus 1a and 1b: a phylodynamic and phylogeographic analysis. PLoS Med. 2009;6(12):e1000198. doi: 10.1371/journal.pmed.1000198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pybus O.G., Cochrane A., Holmes E.C., Simmonds P. The hepatitis C virus epidemic among injecting drug users. Infect Genet Evol. 2005;5(2):131–139. doi: 10.1016/j.meegid.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 7.Pybus O.G., Barnes E., Taggart R., Lemey P., Markov P.V., Rasachak B. Genetic history of hepatitis C virus in East Asia. J Virol. 2009;83(2):1071–1082. doi: 10.1128/JVI.01501-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wantuck J., Ahmed A., Nguyen M. The epidemiology and therapy of chronic hepatitis C genotypes 4, 5 and 6. Aliment Pharmacol Ther. 2014;39(2):137–147. doi: 10.1111/apt.12551. [DOI] [PubMed] [Google Scholar]
- 9.Murphy D.G., Sablon E., Chamberland J., Fournier E., Dandavino R., Tremblay C.L. Hepatitis C virus genotype 7, a new genotype originating from central Africa. J Clin Microbiol. 2015;53(3):967–972. doi: 10.1128/JCM.02831-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.The Polaris Observatory HCV Collaborators Global prevalence and genotype distribution of hepatitis C virus infection in 2015: a modelling study. Lancet Gastroenterol Hepatol. 2017;2(3):161–176. doi: 10.1016/S2468-1253(16)30181-9. [DOI] [PubMed] [Google Scholar]
- 11.Ministry of Health and Population [Egypt], El-Zanaty and Associates [Egypt] and ICF International. Egypt Health Issues Survey 2015. Cairo, Rockville, MD: Ministry of Health and Population, ICF International; 2015.
- 12.Kamal S.M., Nasser I.A. Hepatitis C genotype 4: What we know and what we don't yet know. Hepatology. 2008;47(4):1371–1383. doi: 10.1002/hep.22127. [DOI] [PubMed] [Google Scholar]
- 13.Frank C., Mohamed M.K., Strickland G.T., Lavanchy D., Arthur R.R., Magder L.S. The role of parenteral antischistosomal therapy in the spread of hepatitis C virus in Egypt. The Lancet. 2000;355(9207):887–891. doi: 10.1016/s0140-6736(99)06527-7. [DOI] [PubMed] [Google Scholar]
- 14.Fakhr A.E., Pourkarim M.R., Maes P., Atta A.H., Marei A., Azab M. Hepatitis C Virus NS5B Sequence-Based Genotyping Analysis of Patients From the Sharkia Governorate, Egypt. Hepat Mon. 2013;13(12):e12706. doi: 10.5812/hepatmon.12706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ray S.C., Arthur R.R., Carella A., Bukh J., Thomas D.L. Genetic epidemiology of hepatitis C virus throughout Egypt. J Infect Dis. 2000;182(3):698–707. doi: 10.1086/315786. [DOI] [PubMed] [Google Scholar]
- 16.Youssef A., Yano Y., Utsumi T., Serwah A.-H., Hayashi Y., abd El-alah E.M. Molecular epidemiological study of hepatitis viruses in Ismailia, Egypt. Intervirology. 2009;52(3):123–131. doi: 10.1159/000219385. [DOI] [PubMed] [Google Scholar]
- 17.Abdel-Hamid M., El-Daly M., Molnegren V., El-Kafrawy S., Abdel-Latif S., Esmat G. Genetic diversity in hepatitis C virus in Egypt and possible association with hepatocellular carcinoma. J Gen Virol. 2007;88(5):1526–1531. doi: 10.1099/vir.0.82626-0. [DOI] [PubMed] [Google Scholar]
- 18.Varghese R., Al-Khaldi J., Asker H., Fadili A., Al Ali J., Hassan F. Treatment of chronic hepatitis C genotype 4 with peginterferon alpha-2a plus ribavirin. Hepatogastroenterology. 2008;56(89):218–222. [PubMed] [Google Scholar]
- 19.Benhamou Y., Moussalli J., Ratziu V., Lebray P., De Backer K., De Meyer S. Telaprevir activity in treatment-naive patients infected hepatitis C virus genotype 4: a randomized trial. J Infect Dis. 2013;208(6):1000–1007. doi: 10.1093/infdis/jit274. [DOI] [PubMed] [Google Scholar]
- 20.Abdel-Ghaffar T.Y., Sira M.M., El Naghi S. Hepatitis C genotype 4: the past, present, and future. World J Hepatol. 2015;7(28):2792–2810. doi: 10.4254/wjh.v7.i28.2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilead Sciences. SOVALDI® (sofosbuvir) tablets, for oral use Initial U.S. 2015. <http://www.accessdata.fda.gov/drugsatfda_docs/label/2015/204671s002lbl.pdf>.
- 22.Doss W., Shiha G., Hassany M., Soliman R., Fouad R., Khairy M. Sofosbuvir plus ribavirin for treating Egyptian patients with hepatitis C genotype 4. J Hepatol. 2015;63(3):581–585. doi: 10.1016/j.jhep.2015.04.023. [DOI] [PubMed] [Google Scholar]
- 23.El Kassas Mohamed, Elbaz Tamer AELY, Esmat Gamal. Elbasvir and grazoprevir for chronic hepatitis C genotypes 1 and 4. Expert Rev Clin Pharmacol 2016;9(11):1413–21. [DOI] [PubMed]
- 24.Cavalcante L.N., Lyra A.C. Predictive factors associated with hepatitis C antiviral therapy response. World J Hepatol. 2015;7(12):1617–1631. doi: 10.4254/wjh.v7.i12.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rajhi M., Ghedira K., Chouikha A., Djebbi A., Cheikh I., Yahia A.B. Phylogenetic analysis and epidemic history of hepatitis C Virus genotype 2 in Tunisia, North Africa. PLoS ONE. 2016;11(4):e0153761. doi: 10.1371/journal.pone.0153761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McOmish F., Yap P., Dow B., Follett E., Seed C., Keller A. Geographical distribution of hepatitis C virus genotypes in blood donors: an international collaborative survey. J Clin Microbiol. 1994;32(4):884–892. doi: 10.1128/jcm.32.4.884-892.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeytinli Ü.O., Yücel F.M., Dinçer Ş.D., Yanilmaz Ö., Aksaray S., Özdil K. Distribution of hepatitis C virus genotypes in the region of İstanbul Northern Anatolian Association of Public Hospitals. Viral Hepatitis J. 2017;23(1):10–13. [Google Scholar]
- 28.Sadeghi F., Salehi-Vaziri M., Almasi-Hashiani A., Gholami-Fesharaki M., Pakzad R., Alavian S.M. Prevalence of hepatitis C virus genotypes among patients in countries of the eastern mediterranean regional office of WHO (EMRO): a systematic review and meta-analysis. Hepat Mon. 2016;16(4):e35558. doi: 10.5812/hepatmon.35558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sağlik İ., Mutlu D., Öngut G., Inan D., Öğünç D., Can S.R. Distribution of hepatitis C virus genotypes among patients with chronic hepatitis C infection in Akdeniz University Hospital, Antalya, Turkey: a five-year evaluation. Mikrobiyol Bul. 2014;48(3):429–437. doi: 10.5578/mb.7685. [DOI] [PubMed] [Google Scholar]
- 30.Aguilera A., Navarro D., Rodríguez-Frias F., Viciana I., Martínez-Sapiña A., Rodríguez M. Prevalence and distribution of hepatitis C virus genotypes in Spain during the 2000–2015 period (the GEHEP 005 study) J Viral Hepat. 2017;24:725–732. doi: 10.1111/jvh.12700. [DOI] [PubMed] [Google Scholar]
- 31.Shi ST, Lai MM. HCV 5′ and 3′ UTR: when translation meets replication. In: Tan S-L, editor. Hepatitis C Viruses: Genomes and molecular biology: horizon bioscience; 2006. p. 49–87. [PubMed]
- 32.Murphy D.G., Willems B., Deschênes M., Hilzenrat N., Mousseau R., Sabbah S. Use of sequence analysis of the NS5B region for routine genotyping of hepatitis C virus with reference to C/E1 and 5′ untranslated region sequences. J Clin Microbiol. 2007;45(4):1102–1112. doi: 10.1128/JCM.02366-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang C., Sarnow P., Siddiqui A. A conserved helical element is essential for internal initiation of translation of hepatitis C virus RNA. J Virol. 1994;68(11):7301–7307. doi: 10.1128/jvi.68.11.7301-7307.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.El Awady M.K., Azzazy H.M., Fahmy A.M., Shawky S.M., Badreldin N.G., Yossef S.S. Positional effect of mutations in 5'UTR of hepatitis C virus 4a on patients' response to therapy. World J Gastroenterol. 2009;15(12):1480–1486. doi: 10.3748/wjg.15.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hemeida A.A.O.M., El-Shahat M., Hashem M.H., Mahmoud A., Dahi H. Genetic variations in a conserved 5'-untranslated region of hepatitis C Virus isolated from Egypt. Int J Virol. 2011;7(3):91–99. [Google Scholar]
- 36.Yasmeen A., Siddiqui A.A., Hamid S., Sultana T., Jafri W., Persson M.A. Genetic variations in a well conserved 5′-untranslated region of hepatitis C virus genome isolated in Pakistan. J Virol Methods. 2009;160(1):38–47. doi: 10.1016/j.jviromet.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 37.Zekri A.R.N., El-Din H.M.A., Bahnassy A.A., Khaled M.M., Omar A., Fouad I. Genetic distance and heterogenecity between quasispecies is a critical predictor to IFN response in Egyptian patients with HCV genotype-4. Virol J. 2007;4(1):16. doi: 10.1186/1743-422X-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elkady A., Tanaka Y., Kurbanov F., Sugauchi F., Sugiyama M., Khan A. Genetic variability of hepatitis C virus in South Egypt and its possible clinical implication. J Med Virol. 2009;81(6):1015–1023. doi: 10.1002/jmv.21492. [DOI] [PubMed] [Google Scholar]
- 39.Genovese D., Dettori S., Argentini C., Villano U., Chionne P., Angelico M. Molecular epidemiology of hepatitis C virus genotype 4 isolates in Egypt and analysis of the variability of envelope proteins E1 and E2 in patients with chronic hepatitis. J Clin Microbiol. 2005;43(4):1902–1909. doi: 10.1128/JCM.43.4.1902-1909.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yun Z., Lara C., Johansson B., Lorenzana de Rivera I., Sönnerborg A. Discrepancy of hepatitis C virus genotypes as determined by phylogenetic analysis of partial NS5 and core sequences. J Med Virol. 1996;49(3):155–160. doi: 10.1002/(SICI)1096-9071(199607)49:3<155::AID-JMV1>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 41.Viazov S., Widell A., Nordenfelt E. Mixed infection with two types of hepatitis C virus is probably a rare event. Infection. 2000;28(1):21–25. doi: 10.1007/s150100050005. [DOI] [PubMed] [Google Scholar]
- 42.Morel V., Fournier C., Francois C., Brochot E., Helle F., Duverlie G. Genetic recombination of the hepatitis C virus: clinical implications. J Viral Hepat. 2011;18(2):77–83. doi: 10.1111/j.1365-2893.2010.01367.x. [DOI] [PubMed] [Google Scholar]
- 43.Behrens S.-E., Tomei L., De Francesco R. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 1996;15(1):12–22. [PMC free article] [PubMed] [Google Scholar]
- 44.Holland JJd, De La Torre JC, Steinhauer DA. RNA virus populations as quasispecies. In: Holland JJ, editor. Genetic diversity of RNA viruses. Springer; 1992. p. 1–20. [DOI] [PubMed]
- 45.Choi KH. Viral polymerases. In Viral molecular machines. In: Rossmann MG, Rao VB, editors. Viral molecular machines. Springer Science: NY, USA; 2012. p. 267–304.
- 46.Lesburg C.A., Cable M.B., Ferrari E., Hong Z., Mannarino A.F., Weber P.C. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat Struct Mol Biol. 1999;6(10):937–943. doi: 10.1038/13305. [DOI] [PubMed] [Google Scholar]
- 47.O'Reilly E.K., Kao C.C. Analysis of RNA-dependent RNA polymerase structure and function as guided by known polymerase structures and computer predictions of secondary structure. Virology. 1998;252(2):287–303. doi: 10.1006/viro.1998.9463. [DOI] [PubMed] [Google Scholar]
- 48.Lohmann V., Körner F., Herian U., Bartenschlager R. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J Virol. 1997;71(11):8416–8428. doi: 10.1128/jvi.71.11.8416-8428.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Migliaccio G., Tomassini J.E., Carroll S.S., Tomei L., Altamura S., Bhat B. Characterization of resistance to non-obligate chain-terminating ribonucleoside analogs that inhibit hepatitis C virus replication in vitro. J Biol Chem. 2003;278(49):49164–49170. doi: 10.1074/jbc.M305041200. [DOI] [PubMed] [Google Scholar]
- 50.Dutartre H., Bussetta C., Boretto J., Canard B. General catalytic deficiency of hepatitis C virus RNA polymerase with an S282T mutation and mutually exclusive resistance towards 2′-modified nucleotide analogues. Antimicrob Agents Chemother. 2006;50(12):4161–4169. doi: 10.1128/AAC.00433-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lam A.M., Espiritu C., Bansal S., Steuer H.M.M., Niu C., Zennou V. Genotype and subtype profiling of PSI-7977 as a nucleotide inhibitor of hepatitis C virus. Antimicrob Agents Chemother. 2012;56(6):3359–3368. doi: 10.1128/AAC.00054-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Donaldson E.F., Harrington P.R., O'Rear J.J., Naeger L.K. Clinical evidence and bioinformatics characterization of potential hepatitis C virus resistance pathways for sofosbuvir. Hepatology. 2015;61(1):56–65. doi: 10.1002/hep.27375. [DOI] [PubMed] [Google Scholar]
- 53.Ito J., Suda G., Yamamoto Y., Nagasaka A., Furuya K., Kumagai K. Prevalence and characteristics of naturally occurring sofosbuvir resistance-associated variants in patients with hepatitis C virus genotype 1b infection. Hepatol Res. 2016;46(13):1294–1303. doi: 10.1111/hepr.12685. [DOI] [PubMed] [Google Scholar]




