Summary
Many community- and hospital-acquired bacterial infections are caused by antibiotic-resistant pathogens. Methicillin-resistant Staphylococcus aureus (MRSA) predisposes humans to invasive infections that are difficult to eradicate. We designed a closed-loop gene network programming mammalian cells to autonomously detect and eliminate bacterial infections. The genetic circuit contains human Toll-like receptors as the bacterial sensor and a synthetic promoter driving reversible and adjustable expression of lysostaphin, a bacteriolytic enzyme highly lethal to S. aureus. Immunomimetic designer cells harboring this genetic circuit exhibited fast and robust sense-and-destroy kinetics against live staphylococci. When tested in a foreign-body infection model in mice, microencapsulated cell implants prevented planktonic MRSA infection and reduced MRSA biofilm formation by 91%. Notably, this system achieved a 100% cure rate of acute MRSA infections, whereas conventional vancomycin treatment failed. These results suggest that immunomimetic designer cells could offer a therapeutic approach for early detection, prevention, and cure of pathogenic infections in the post-antibiotic era.
Video Abstract
Keywords: synthetic gene circuit, Toll-like receptor, lysostaphin, cell therapy, encapsulation, methicillin-resistant Staphylococcus aureus, implant-associated infection, biofilm, antibiotic resistance, murine tissue cage infection model
Graphical Abstract
Highlights
-
•
A closed-loop gene network with bacterial sense-and-destroy actuation
-
•
Direct diagnosis of implant-associated infections through blood biomarkers
-
•
Early prevention of MRSA infection, as well as biofilm formation, in vivo
-
•
Curing acute MRSA infections as an alternative to antibiotic therapy
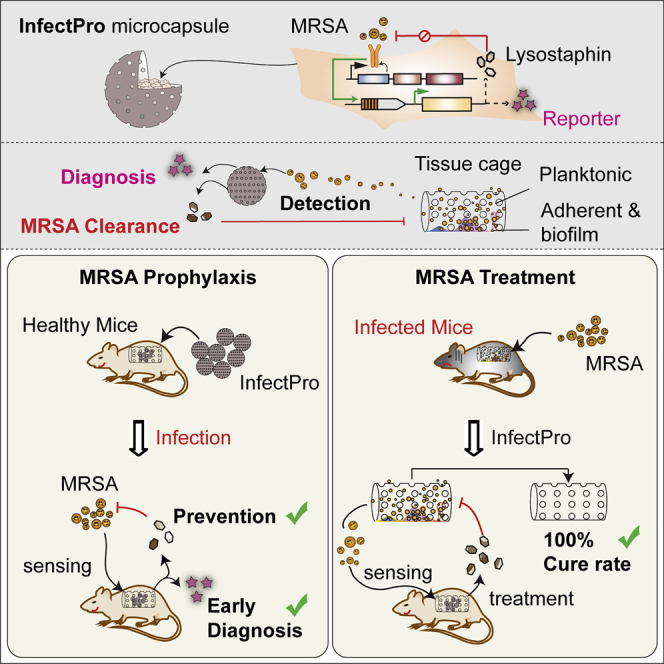
Encapsulated mammalian cells with a synthetic gene circuit to sense and respond to MRSA infection could provide potential prophylactic, diagnostic, or therapeutic options for medical implant-associated infections.
Introduction
Infectious disease is the leading cause of mortality and morbidity worldwide, claiming about 15 million lives each year (Fauci and Morens, 2012). Staphylococcus aureus is one of the most frequent pathogens, accounting for 11.6 million outpatient and emergency room visits and nearly 500,000 hospitalizations per year in the United States alone (Klein et al., 2007, McCaig et al., 2006). This organism causes a variety of human diseases, ranging from skin and soft tissue infections to life-threatening infections, such as bacteremia, pneumonia, osteomyelitis, endocarditis, meningitis, and sepsis (Tong et al., 2015). The high morbidity associated with S. aureus has further increased with the rising prevalence of strains that exhibit broad antibiotic resistance, such as methicillin-resistant S. aureus (MRSA) (Dantes et al., 2013), which causes more deaths annually (∼19,000) than any other single infectious agent in the United States; indeed, the number of deaths caused by MRSA exceeds that associated with HIV/AIDS, hepatitis, and influenza combined (Boucher and Corey, 2008). As these staphylococcal strains are highly virulent and are increasingly becoming resistant to every clinically available antibiotic (Stryjewski and Corey, 2014), alternative therapies are urgently needed.
One particularly important unmet medical need for anti-S. aureus therapies is to treat implant-associated infections (IAIs) (Darouiche, 2004). IAIs account for half of the 2 million cases of nosocomial infections that occur each year in the United States (Darouiche, 2004) and are one of the most feared and difficult-to-treat medical complications, causing high morbidity and mortality, and leading to substantial healthcare costs (Kapadia et al., 2016). S. aureus is the leading cause of IAI and is particularly adept at infecting foreign bodies within the human host (Del Pozo and Patel, 2009). This organism is able to persist on implant surfaces, forming biofilms, which are sessile communities of microcolonies encased in an extracellular matrix that adheres to biomedical implants (Bjarnsholt et al., 2013). Infections associated with biofilms are difficult to treat due to the presence of biomaterials that can reduce the inoculum of S. aureus required to establish an infection by a factor of more than 100,000 (Puhto et al., 2014), and it is estimated that sessile bacteria in biofilms are over 1,000-fold less sensitive to antibiotics than their planktonic counterparts (Sutherland, 2001). Therefore, most implants that are infected by S. aureus have to be surgically removed to achieve a definite cure, leading to a poor patient outcome and considerable economic burden (Darouiche, 2004).
Human innate immune response is the first line of defense against infectious microbes (Akira et al., 2006). Early recognition of S. aureus is initiated by pattern recognition receptors (PRRs) on epithelial cells and innate phagocytic cells (Fournier and Philpott, 2005). Toll-like receptor 2 (TLR2) has emerged as the most important of these PRRs in detecting extracellular S. aureus (Fournier and Philpott, 2005). TLR2 recognizes lipoproteins, lipoteichoic acid, and peptidoglycan embedded in the staphylococcal cell envelope by forming heterodimers with TLR1 (Jin et al., 2007) or TLR6 (Kang et al., 2009), and the pathogen recognition is facilitated by a CD14 co-receptor (Nilsen et al., 2008). Upon stimulation, TLR2 and TLR1 or TLR6 initiate downstream signaling events that lead to the translocation of nuclear factor κB (NF-κB) and the production of proinflammatory cytokines and chemokines that recruit phagocytes to the site of infection for the disposal of pathogens (Akira et al., 2006)). However, S. aureus is a well-adapted pathogen that has evolved many mechanisms for thwarting the human immune response, ranging from blocking neutrophil chemotaxis, lysing leukocytes, and avoiding phagocytosis to resisting phagocytic killing and surviving within host cells (Foster et al., 2014). In this study, instead of using the detect-deflect-destroy policy employed by the innate immunity, we apply a direct sense-and-destroy strategy based on engineering of a synthetic genetic circuit that expresses lysostaphin under the regulation of human TLR2, TLR1, TLR6, and CD14. Lysostaphin is a bacteriocin that kills most known staphylococcal species (von Eiff et al., 2003). It is an endopeptidase that enzymatically cleaves the specific cross-linking polyglycine bridges in the cell walls of staphylococci (Schindler and Schuhardt, 1964). The bactericidal efficacy of lysostaphin was reported to be higher than those of human native antimicrobials and broad-spectrum antibiotics, including penicillin (Schaffner et al., 1967), oxacillin (Kiri et al., 2002), and vancomycin (Placencia et al., 2009). It is also effective against biofilms (Kokai-Kun et al., 2009, Hertlein et al., 2014) and has been widely tested in various animal models (Dajcs et al., 2000, Hertlein et al., 2014, Kokai-Kun et al., 2003, Kokai-Kun et al., 2007, Patron et al., 1999) and in humans (Davies et al., 2010, Harris et al., 1967, Stark et al., 1974).
Here, we show that this synthetic gene network can be implemented into a variety of rodent and human cells, providing an autonomous, self-regulated staphylococcal sensing-and-eliminating function. We also show that in a foreign-body infection mouse model, for which therapeutic options are extremely limited, this closed-loop synthetic gene network operating inside microencapsulated designer-cell implants could effectively protect mice from biofilm-forming MRSA infection and cure acute MRSA infection.
Results
Design of a Synthetic Gene Circuit for Infectious Microbial Sensing
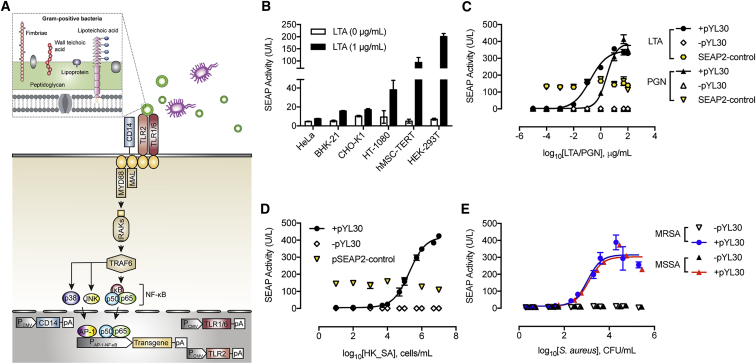
Infectious Microbial Sensing Gene Network (Infectsen) consists of a synthetic bacteria-responsive signaling cascade in which the constitutive co-expression of human TLR2 (hTLR2; pYL30, PhCMV-hTLR2-pA), human TLR1 (hTLR1; pYL126, PhCMV-hTLR1-pA), human TLR6 (hTLR6; pYL125, PhCMV-hTLR6-pA), and human CD14 (hCD14; pYL25, PhCMV-hCD14-pA) senses extracellular cell-wall components of gram-positive bacteria and triggers a MYD88-dependent signaling pathway that results in activation and translocation of AP-1 and NF-κB into the nucleus (Akira and Takeda, 2004). By connecting the signaling activation of hTLR2, hTLR1, hTLR6, and hCD14 to a carefully tuned chimeric promoter (PAP-1 × 5-NF-κB × 5-IFNβmin) containing AP-1 and NF-κB response elements, the presence of extracellular microbial components could be directly linked to the expression of a specific target gene (Figure 1A). Since human embryonic kidney 293 cells (HEK293) cells already express hTLR1 and hTLR6 endogenously (Godefroy et al., 2014), we characterized the microbial sensor device in HEK293 cells without ectopic expression of hTLR1 and hTLR6. To optimize the gene network, we first screened for synthetic NF-κB-responsive promoters with different NF-κB-binding sites and minimal promoters (Figure S1A). We then validated the signaling activation through AP-1 by incorporating AP-1 binding elements (Figure S1B). Subsequently, to obtain the best overall transgene induction, we compared a series of promoters containing different combinations of AP-1/NF-κB-binding sequences upstream of several minimal promoter variants (Figure S1C). We also showed that system activation is greatly influenced by the expression strength of hTLR2 driven by different constitutive promoters (Figure S1D). In summary, the combinatorial assembly of 5 repeats of AP-1 and NF-κB followed by an IFNB minimal promoter (pYL3, PAP-1 × 5-NF-κB × 5-IFNβmin-SEAP-pA) yielded the greatest signal transduction in response to stimulation of pYL30 (PhCMV-hTLR2-pA), endogenous hTLR1 and 6, and pYL25 (PhCMV-hCD14-pA) (Infectsen) (Figure S1).
Figure 1.
Design of a Transcription-Control Device Activated by Infectious Microbes (Infectsen)
(A) Gram-positive bacterial cell walls comprise multiple peptidoglycan layers, wall teichoic acids linked to peptidoglycan (PGN), and lipoteichoic acid (LTA) linked to the cytoplasmic membrane. Constitutively expressed human TLR2, TLR1, TLR6, and the CD14 co-receptor recognize PGN, LTA, and lipoproteins from the bacterial cell walls. Upon stimulation, TLR2 undergoes conformational changes required for its association with Mal and MYD88, and the subsequent formation of a complex of IRAKs and TRAF6 is induced. TRAF6 then undergoes phosphorylation and ubiquitylation, which induces the phosphorylation of both mitogen-activated protein kinases p38 and JNK, and the IKK complex, allowing AP-1 and NF-κB to translocate into the nucleus, bind to the synthetic promoters containing AP-1- and NF-κB-responsive elements (PAP-1-NF-κB), and initiate the expression of target genes. Mal, MYD88 adaptor-like, also known as TIR domain containing adaptor protein; MYD88, myeloid differentiation primary response 88; IRAKs, interleukin 1 receptor-associated kinase 1; TRAF6, TNF receptor-associated factor 6; JNK, JUN N-terminal kinase; IKK complex, inhibitor of nuclear factor kappa B-kinase complex; AP-1, activator protein 1; and NF-κB, nuclear factor kappa B.
(B) Validation of Infectsen in different mammalian cells. Cells were transfected with pYL25 (PhCMV-hCD14-pA)/pYL30 (PhCMV-hTLR2-pA)/pYL126 (PhCMV-hTLR1-pA)/pYL125 (PhCMV-hTLR6-pA)/pYL3 (PAP-1 × 5-NF-κB × 5-IFNβmin-SEAP-pA) and incubated with 1 μg/mL LTA for 24 hr before measuring SEAP.
(C–E) Sensitivity and adjustability of Infectsen to staphylococcal cell-wall components LTA and PGN (C), heat-killed Staphylococcus aureus (HK_SA) (D), and live Staphylococcus aureus MRSA and MSSA (E). HEK293 cells were transfected with pYL25/pYL30/pYL3 (+pYL30), pYL25/pcDNA3.1(+)/pYL3 (-pYL30), or pcDNA3.1/pSEAP2-control (SEAP2-control) and incubated with various ligands at increasing concentrations. SEAP expression was assayed after 24 hr.
All data are means ± SD (n = 3).
See also Figure S1.
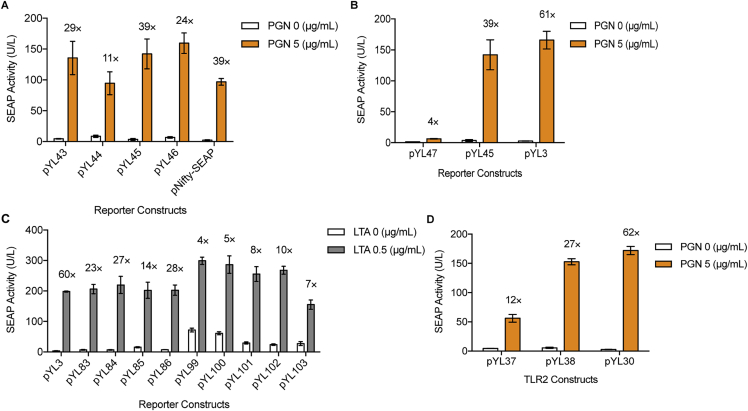
Figure S1.
Optimization of Bacteria-Regulated Transgene Expression with Different Promoter Configurations, Related to Figure 1
(A) Selection of NF-κB binding sites and minimal promoters. SEAP-expressing vectors driven by different promoters containing 5 repeats of NF-κB binding site (GGGGACTTTCC) (pYL43, PNF-κB × 5min-SEAP-pA; pYL45, PNF-κB × 5-IFNβmin-SEAP-pA; pNifty-SEAP, PNF-κB × 5-ELAM-SEAP-pA) or 5 repeats of NF-κB’ binding sequence (GGGAATTTCC) (pYL44, PNF-κB’ × 5min-SEAP-pA; pYL46, PNF-κB’ × 5-IFNβmin-SEAP-pA) were co-transfected with pYL30/pYL25 (PhCMV-hTLR2-pA/PhCMV-hCD14-pA) in HEK293 cells. SEAP activity was assayed in the supernatant of cells after 24 hr in the presence of 5 μg/mL PGN.
(B) Validation of signal transactivation through AP-1-responsive promoters. pYL30/pYL25-transgenic HEK293 cells were co-transfected with NF-κB-responsive reporter plasmids with or without AP-1 binding site (pYL3, PAP-1 × 5-NF-κB × 5-IFNβmin-SEAP-pA; pYL45, PNF-κB × 5-IFNβmin-SEAP-pA; pYL47, PAP-1 × 5-IFNβmin-SEAP-pA). SEAP activity was profiled at 24 hr after exposing cells with or without 5 μg/mL PGN.
(C) Screening of AP-1/NF-κB-responsive promoters for best transgene induction. HEK293 cells were co-transfected with pYL30/pYL25 and SEAP-encoding plasmids controlled by composite promoters varying in binding sequences, number of repeats and minimal promoters (pYL3, PAP-1 × 5-NF-κB × 5-IFNβmin-SEAP-pA; pYL83, P[NF-κB’-NF-κB]1-IFNβmin-SEAP-pA; pYL84, P[NF-κB’-NF-κB]2-IFNβmin-SEAP-pA; pYL85, P[NF-κB’-NF-κB]1min-SEAP-pA; pYL86, P[NF-κB’-NF-κB]2min-SEAP-pA; pYL99, P[NF-κB’- NF-κB]1-hCMVmin1-SEAP-pA; pYL100, P[NF-κB’- NF-κB]1-hCMVmin2-SEAP-pA; pYL101, PAP-1 × 5-NF-κB × 5-hCMVmin1-SEAP-pA; pYL102, PAP-1 × 5-NF-κB × 5-hCMVmin2-SEAP-pA; pYL103, PAP-1 × 5-NF-κB × 5-hCMVmin3-SEAP-pA). SEAP activity was profiled at 24 hr after exposing cells with or without 0.5 μg/mL LTA.
(D) Screening for constitutive promoters driving hTLR2 expression. pYL25/pYL3-transgenic HEK293 cells were cotransfected with a plasmid encoding hTLR2 expression unit driven by different promoters (pYL30, PhCMV-hTLR2-pA; pYL37, Pmin-hTLR2-pA; pYL38, PSV40-hTLR2-pA).
Cell were induced with 5 μg/mL PGN and assayed for SEAP activity after 24 hr. Fold induction of SEAP expression levels is shown above each figure bar. All data are means ± SD (n = 3).
The versatility of the optimized gene circuit was assessed by co-transfection of pYL30, pYL126, pYL125, pYL25 and pYL3 into a set of mammalian and human cell lines. Consistent secreted alkaline phosphatase (SEAP) induction by lipoteichoic acid (LTA) indicated that the system was functional in all tested cell lines, including stem-cell-derived hMSC-hTERT, suggesting broad applicability of this gene control device (Figure 1B). Different expression profiles among cell lines may be due to differences in the transfection efficiency, availability of receptor and adaptor proteins, cellular composition of downstream signaling effectors and regulators, and protein expression and secretion capacities. Considering the basal expression level, maximum expression level and induction fold, we selected HEK293 for further characterization (Figure 1B). Infectsen cells were highly sensitive to either cell-wall components of S. aureus: LTA, PGN (Figure 1C), or heat-killed whole S. aureus (HK_SA) (Figure 1D). The transcriptional activation of SEAP expression was regulated dose dependently and initiated at 103 cells/mL of HK_SA (Figure 1D). A lower inoculum of 102 colony-forming unit (CFU)/mL of live S. aureus was already sufficient to activate gene expression (Figure 1E), owing to the replication of bacteria in the cell culture medium. All transcriptional activations, however, were greatly diminished without hTLR2 expression (Figures 1C–1E), indicating that the system is hTLR2-dependent. Furthermore, the metabolism or viability of the cells as scored by constitutive SEAP expression (pSEAP2-control, PSV40-SEAP-pA) was not impacted over the entire concentration range of bacterial components or whole bacteria examined (Figures 1C and 1D).
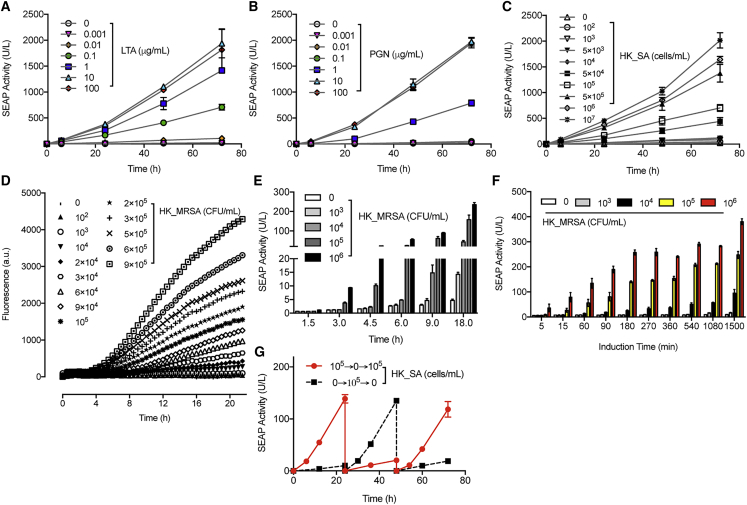
Characterization of the gene expression kinetics of Infectsen cells, cultivated for extended periods of time in medium containing specific concentrations of staphylococcal cell-wall components (LTA, PGN) or whole bacteria (HK_SA), revealed consistently low leakiness and a robust, dose-dependent induction of SEAP over 3 days (Figures 2A–2C), indicating a direct correlation between transgene expression and the microbial concentration and duration of infection. In-depth real-time monitoring of the Staphylococcus-responsive production of intracellular fluorescent reporter gene turboGFP further confirmed the high sensitivity to different concentrations of heat-killed, methicillin-resistant S. aureus (HK_MRSA) and rapid transgene switching of Infectsen (Figure 2D). Importantly, the switching time of the system is strongly dependent on the bacterial dosage; the higher the inoculum, the faster the transgene is switched on (Figure 2E). Furthermore, when Infectsen was exposed to HK_MRSA at varying doses for different pulse durations, high bacteria levels rapidly activated transgene expression, whereas lower levels required a longer induction pulse period to reach significant SEAP production (Figure 2F). Finally, gene expression could be switched on and off in response to the presence or absence of heat-killed S. aureus (Figure 2G).
Figure 2.
Expression Kinetics, Responsiveness, and Reversibility of Infectsen
(A–C) Induction kinetics of SEAP expression with increasing concentrations of LTA (A), PGN (B), and HK_SA (C) in HEK293 cells transfected with pYL25/pYL30/pYL3 during cultivation for 72 hr.
(D) Real-time monitoring with a micro-plate reader of intracellular fluorescent GFP production in HEK293 cells transfected with pYL25/pYL30/pYL58 (PAP-1 × 5-NF-κB × 5-IFNβmin-tGFP-dest1-pA) and induced by heat-killed MRSA (HK_MRSA). Plot are means ± SD (n = 4).
(E) Responsiveness of Infectsen to different bacterial doses and induction time in terms of SEAP expression. pYL25/pYL30/pYL3-transgenic HEK293 cells were cultivated with different concentrations of HK_MRSA and assayed for SEAP expression at indicated time points.
(F) Impact of bacterial concentrations and induction pulses on SEAP expression. pYL25/pYL30/pYL3-transgenic HEK293 cells were incubated with various amounts of HK_MRSA for the indicated periods. Cells were washed once before medium change and sampled at 25 hr for SEAP production.
(G) Reproducibility and reversibility of Infectsen. pYL25/pYL30/pYL3-transfected HEK293 cells were cultivated in the presence (6 hr) or absence of 105 cells/mL HK_SA for 72 hr. Every 24 hr, the culture medium was exchanged, and the cell density was readjusted.
All data are means ± SD (n ≥ 3).
Design and Killing Dynamics of an Infectious Microbial Protection Device
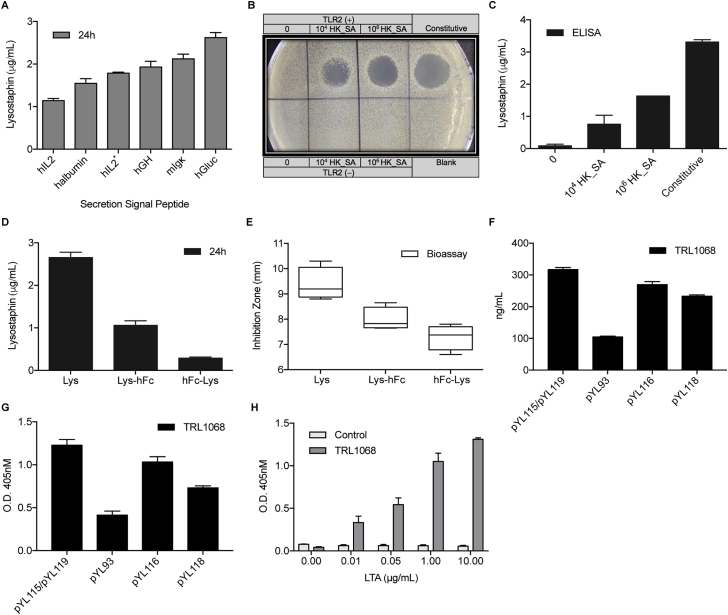
To obtain staphylococcal killing effects, we replaced SEAP with lysostaphin as a therapeutic output under the control of human TLR2, TLR1, TLR6, and CD14 (Infectpro). Functional expression of Lysostaphin in eukaryotic cells was achieved by introducing two base-pair substitutions converting Asn125 and Asn232 to glutamine residues from the mature lysostaphin sequence (Kerr et al., 2001). Since signal peptide influences protein production in mammalian cells (Knappskog et al., 2007), we screened six signal peptides of different origin and selected that derived from Gaussia luciferase in a humanized form for maximal lysostaphin production (Figure S2A). The enzymatic activity of HEK293-expressed lysostaphin was determined by bactericidal plate assay (Figure S2B) and levels of lysostaphin expression were quantified in cell culture supernatant, showing dose-dependent responsiveness upon HK_SA induction (Figure S2C). Further modifications of the therapeutic output can be achieved by fusing the human immunoglobulin G (IgG)-derived Fc (fragment crystallizable) region to the C terminus of lysostaphin to increase its serum half-life (Czajkowsky et al., 2012) (Figures S2D and S2E) or by co-expressing lysostaphin and TRL1068, a human monoclonal antibody that binds and disrupts biofilm (Estellés et al., 2016), to specifically target biofilm-associated infections (Figures S2F–S2H).
Figure S2.
Lysostaphin Secretion, Expression, and Further Modifications, Related to Figure 3
(A) Screening for optimal signal peptide that facilitates lysostaphin secretion. HEK293 cells were transfected with plasmids encoding constitutive lysostaphin expression directed by various signal peptides for secretion. hGLuc (pYL48, PhCMV-hGLuc-Lys-pA); hGH (pYL33, PhCMV-hGH-Lys-pA); hIL2 (pYL49, PhCMV-hIL2-Lys-pA); hIL2∗ (pYL51, PhCMV-hIL2∗-Lys-pA); halbumin (pYL50, PhCMV-halbumin-Lys-pA); mIgκ (pYL52, PhCMV-mIgκ-Lys-pA). Lysostaphin levels were quantified by ELISA after 24 hr.
(B) Spot-on-lawn assay for staphylolytic activity. Lytic zones developed on a lawn of S. carnosus at 24 hr after sample application. Samples (30 μL) were derived from HEK293 cells transfected with pYL25/pYL30/pYL75 (TLR2 (+)), pcDNA3.1(+)/pYL48 (Constitutive), pYL25/pcDNA3.1(+)/pYL75 (TLR2 (−)) and pYL25/pYL30/pYL3 (Blank), and induced with 104 and 106 cells/mL of HK_SA for 48 hr.
(C) Lysostaphin expression in HEK293 cells transfected with pYL25/pYL30/pYL75 (PAP-1 × 5-NF-κB × 5-IFNβmin-hGLuc-Lys-pA) or pcDNA3.1(+)/pYL48 (Constitutive), and induced with 104 and 106 cells/mL of HK_SA for 48 hr.
(D) Expression of lysostaphin fusion proteins with human IgG-Fc fragment (hFc). HEK293 cells were transfected with lysostaphin (Lys; pYL48, PhCMV-hGLuc-Lys-pA), Lysostaphin C-terminally fused with hFc (Lys-hFc; pYL72, PhCMV-hGLuc-Lys-hFc-pA) or Lysostaphin rN-terminally fused with hFc (hFc-Lys; pYL69, PhCMV-hGLuc-hFc-Lys-pA), and cultivated for 24 hr before sampling for lysostaphin expression.
(E) Antimicrobial activity of lysostaphin-fusion proteins. Bioassay toward MRSA was performed with cell supernatants from Lys (pYL48)-, Lys-hFc (pYL72)-, or hFc-Lys (pYL69)-transgenic HEK293 cells after 24 hr.
(F) The influence of linker sequence on the expression of human TRL1068. HEK293 cells were transfected with pYL115/pYL119 (PhCMV-hTRL1068-HC-pA/PhCMV-hTRL1068-LC-pA), pYL93 (PhCMV-hTRL1068-HC-RKRRP2A-LC-pA), pYL116 (PhCMV-hTRL1068-HC-RKRRF2A-LC-pA) or pYL118 (PhCMV-hTRL1068-HC-RAKRF2A-LC-pA), and sampled after 24 hr for the determination of hTRL1068 expression.
(G) Validation of hTRL1068 activity. Titration ELISA assay was used for the verification of the binding of TRL1068 to the epitope (AARKGRNPQTGKEID) contained in Staphylococcus aureus histone-like DNA-binding protein. Samples were derived from HEK293 cells transfected with pYL115/pYL119, pYL93, pYL116, or pYL118 after 24 hr.
(H) Dose-dependent regulation on hTLR1068 activity. Titration ELISA assay of TRL1068 binding to the epitope contained in Staphylococcus aureus histone-like DNA-binding protein.
Cell supernatants were taken from non-transfected HEK293 cells (Control) or cells transfected with pYL25/pYL30/pYL117 (P[NF-κB’-NF-κB]1-IFNβmin-hTRL1068-HC-RKRRF2A-LC-pA) (TRL1068) and induced with LTA at increasing concentrations for 72 hr. All data are means ± SD (n ≥ 3).
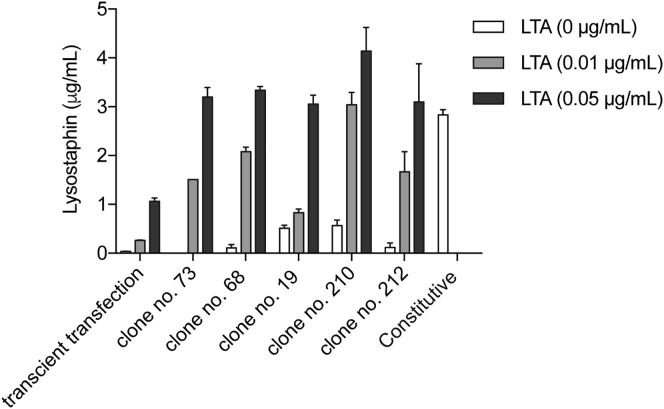
To ensure consistent performance over time, we constructed a stable chromosomal integration of pYL25/pYL30/pYL75 (PAP-1 × 5-NF-κB × 5-IFNβmin-hGLuc-Lys-pA) in HEK293 cells. Stable Infectpro monoclonal cell populations were selected and profiled for lysostaphin expression after induction with LTA (Figure S3). The monoclonal no. 73 cell population of Infectpro was the best-performing transgenic cell line, combining high induction fold of lysostaphin expression, superior absolute lysostaphin levels, and improved sensitivity compared to transient transfection of the same genetic components (Figure S3).
Figure S3.
Stably Transgenic Cell Lines of Infectpro, Related to Figure 3
Transgene expression of monoclonal HEK293 cells stably expressing pYL97/pYL98/pYL112 (ITR-PhEF1α-hCD14-pA:PRPBSA-dTomato-P2A-BlastR-pA/ITR-PhEF1α-hTLR2-pA:PRPBSA-BFP-P2A-PuroR-pA-ITR/ITR-PAP-1 × 5-NF-κB × 5-IFNβmin-hGLuc-Lys-pA:PRPBSA-EGFP-P2A-ZeoR-pA-ITR) (Infectpro). Stable cells were cultivated with LTA at the indicated concentrations for 24 hr, and then lysostaphin levels in the cell culture supernatant were quantified. All data are means ± SD (n = 3).
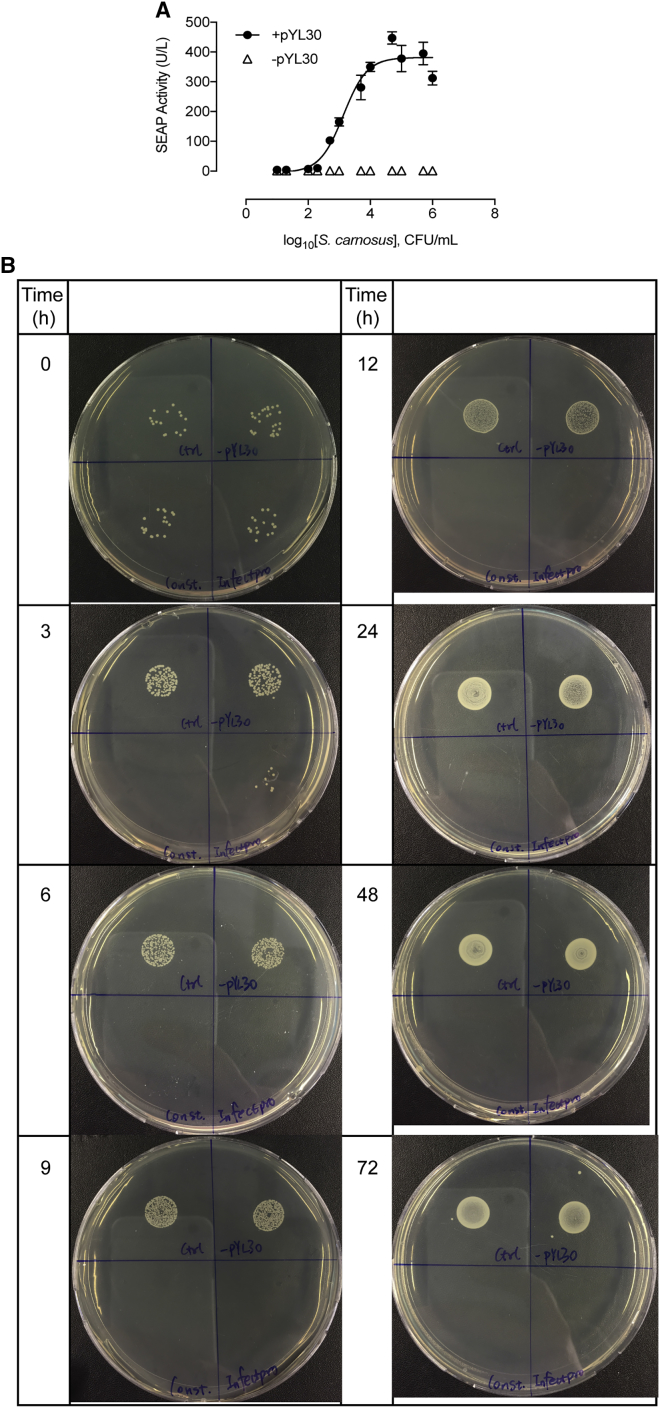
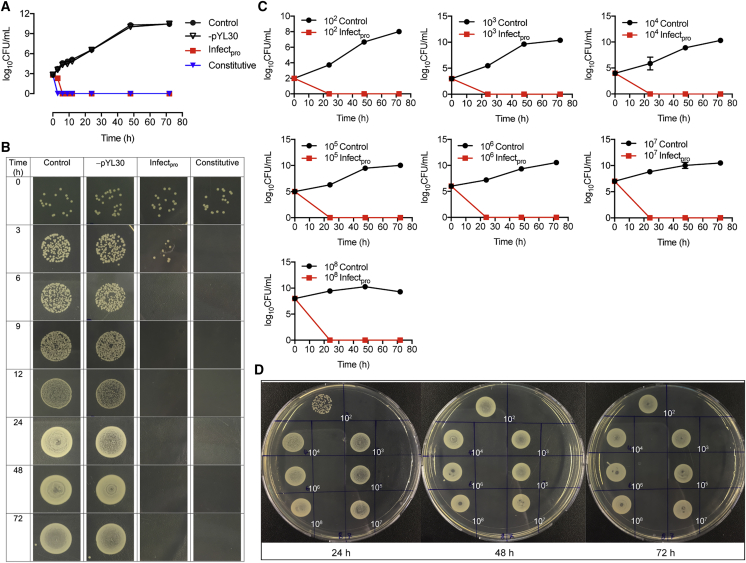
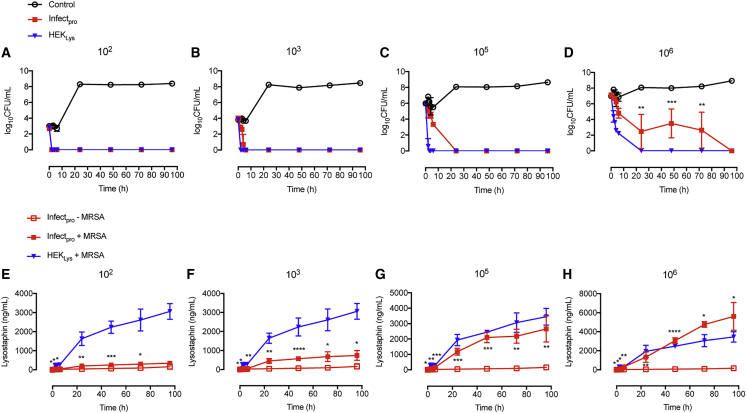
The killing dynamics of Infectpro toward live staphylococci were first evaluated using Staphylococcus carnosus, an apathogenic strain of the Staphylococcus genus. S. carnosus has a similar peptidoglycan composition to S. aureus (Schleifer and Fischer, 1982), rendering this organism an excellent candidate for the verification of lysostaphin activity (Strauss et al., 1998). Stimulation of Infectsen cells with live S. carnosus at increasing concentrations exhibited a similar SEAP induction profile to that obtained with live S. aureus (Figure S4A). A time-kill study of Infectpro against S. carnosus showed a complete elimination of staphylococcal growth at 6 hr after an initial inoculum of 103 CFU/mL (Figure 3A). Constitutive cells transfected with pcDNA3.1(+)/pYL48 (PhCMV-hGLuc-Lys-pA) for constant lysostaphin expression were able to inhibit all bacterial growth at 3 hr after inoculation while control cells and cells without hTLR2 expression showed increasing bacterial growth with time (Figure 3A). The time-dependent killing effects were also shown on agar plates (Figure 3B). To evaluate the bactericidal effects of Infectpro toward S. carnosus at different inocula, Infectpro cells were incubated with increasing concentrations of S. carnosus from 102 to 108 CFU/mL, and assayed for bacterial growth in the cell culture medium over 3 days (Figure 3C). Complete clearance of S. carnosus was found at all tested concentrations with Infectpro (Figure 3C). Again, bacterial load was captured by plate imaging (Figure 3D); the results were well correlated with the quantitative findings. Furthermore, the bactericidal effects of Infectpro cells against MRSA at 102, 103, 105, and 106 CFU/mL were examined for 4 days (Figures 4A–4D). Infectpro cleared bacterial growth at all inocula levels, albeit at a later time point compared to cells stably transgenic for constitutive lysostaphin production (HEKLys) (Figure S6A) (Figures 4A–4D); this difference is due to the detection-based delay in ramping up lysostaphin production (Figures 4E–4H).
Figure S4.
S. carnosus Sense-and-Killing Dynamics, Related to Figure 3
(A) S. carnosus-controlled SEAP expression. HEK293 cells transfected with pYL25/pYL30/pYL3 (+pYL30) or pYL25/pcDNA3.1(+)/pYL3 (−pYL30) were incubated overnight with S. carnosus culture at increasing concentrations. SEAP expression was assayed after 24 hr. All data are means ± SD (n = 3).
(B) Bactericidal effect of Infectpro against Staphylococcus carnosus over time. HEK293 cells transfected with pYL25/pYL30/pYL3 (Control: upper left), pYL25/pcDNA3.1(+)/pYL75 (-pYL30: upper right), pcDNA3.1(+)/pYL48 (Constitutive: lower left) or Infectpro (Infectpro: lower right) were incubated with live S. carnosus culture at 103 CFU/mL and sampled at the indicated time points for plate assay.
Figure 3.
Staphylococcal Sense-and-Destroy Function In Vitro
(A and B) Time course of bactericidal effect of Infectpro against Staphylococcus carnosus. HEK293 cells transfected with pYL25/pYL30/pYL3 (control), pYL25/pcDNA3.1(+)/pYL75 (-pYL30), pcDNA3.1(+)/pYL48 (constitutive), or Infectpro were incubated with live S. carnosus culture at 103 CFU/mL and sampled at the indicated time points for bacterial quantification (A) and agar plate imaging (B). Original pictures are shown in Figure S4B.
(C and D) Dose-dependent killing of S. carnosus. HEK293 cells transfected with pYL25/pYL30/pYL3 (control) or Infectpro were incubated with live S. carnosus at various concentrations and sampled at every 24 hr for bacterial counting (C) and plate assay (control, left; Infectpro, right) (D).
All data are means ± SD (n = 3).
See also Figures S2, S3, and S4.
Figure 4.
MRSA Sense-and-Destroy Function In Vitro
(A–D) Bactericidal kinetics of Infectpro against MRSA. HEK293 cells transfected with pYL25/pYL30/pYL3 (control), HEKLys (constitutive lysostaphin production), or Infectpro were incubated with MRSA culture at 102 CFU/mL (A), 103 CFU/mL (B), 105 CFU/mL (C), or 106 CFU/mL (D). Bacterial growth was determined over time.
(E–H) Lysostaphin expression levels in culture samples of (A)–(D). HEK293 cells transfected with pYL25/pYL30/pYL3 (control), HEKLys (constitutive lysostaphin production), or Infectpro were incubated with MRSA at 102 CFU/mL (E), 103 CFU/mL (F), 105 CFU/mL (G), or 106 CFU/mL (H). Cell culture supernatant was sampled at indicated time points for lysostaphin quantification.
Significant differences between Infectpro and control groups were analyzed using the Student’s t test (A–H). All data are means ± SD (n = 3).
Figure S6.
Control Experiments, Related to Figure 6
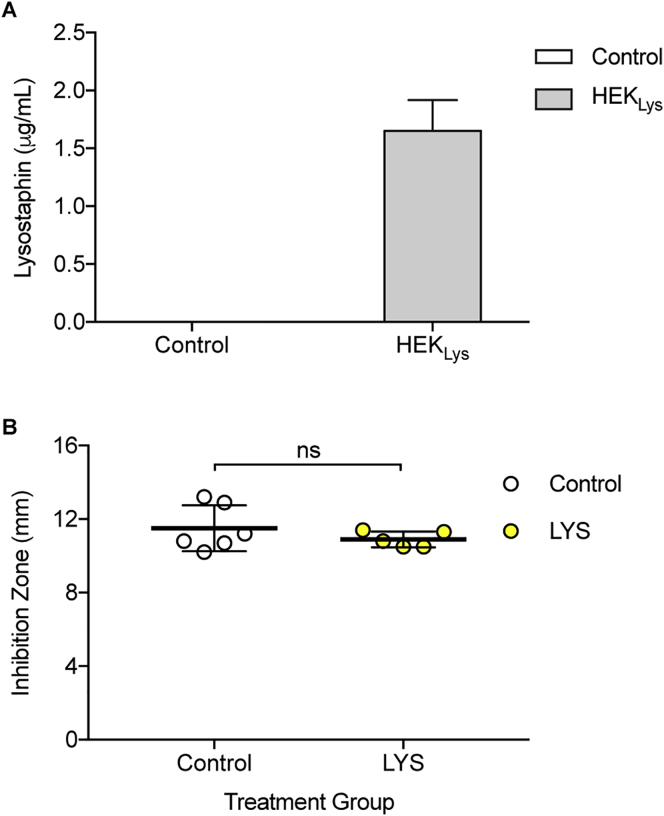
(A) Lysostaphin production in HEK293 cells stably expressing pYL128 (ITR-PhCMV-hGLuc-Lys-pA:PRPBSA-ZeoR-P2A-mRuby-pA-ITR) (HEKLys) versus blank HEK293 cells (Control) after 24 hr. Data are means ± SD.
(B) Inhibition zones of MRSA strains surviving from the treatment groups (Control, n = 6; LYS, n = 5) using a disk diffusion assay. Data are means ± SD; significant difference by two-tailed t test (ns: not significant).
Bactericidal Effects In Vivo
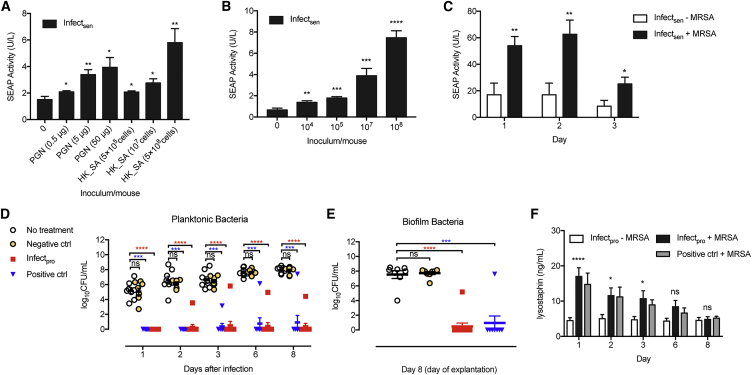
After validation of both alginate-microencapsulated Infectsen and Infectpro cells in culture (Figures S5A and S5B), we tested the performance of cell capsules in vivo. Mice receiving microencapsulated Infectsen cells showed dose-dependent SEAP expression in the blood in response to PGN and HK_SA (Figure 5A) or live S. carnosus (Figure 5B) after 24 hr. Control cells without hTLR2 expression were devoid of SEAP induction in mice at both low and high inocula of live S. carnosus (Figure S5C). Next, to determine the efficacy of Infectpro toward S. aureus, we employed a murine model of foreign-body infection that closely mimics human implant-associated infection (Nowakowska et al., 2014). In the prophylactic study, each mouse was injected with cell capsules prior to infection with MRSA. Tissue cage fluid from mice receiving Infectsen cells showed significant SEAP induction (Figure 5C), confirming the diagnostic value of the system in vivo. Tissue cage fluid from the Infectpro group showed cleared planktonic bacteria in 10 out of 11 mice 1 day after infection; the other mouse started showing infection on day 2 with a consistently lower bacterial number than the average of the control group (Figure 5D). As with the positive control group, planktonic bacterial growth in the tissue cage fluid was completely eliminated in 7 out of 8 mice; the other mouse showed infection on day 3 (Figure 5D). In contrast, cells deficient in lysostaphin expression showed neither bactericidal effects nor clearance (Figure 5D). Consistent results were also obtained for the growth of adherent bacteria on the tissue cage after explantation (Figure 5E). Except for the one infected mouse from each group (Infectpro and positive control) that displayed adherent bacterial growth, all other mice in both groups showed no biofilm formation (Figure 5E). The infection prevention rate reached 91% for the Infectpro group and 88% for the positive control group because of regrowth of the pathogen in 1 cage of each group after cage explantation. Furthermore, levels of lysostaphin expression in tissue cage fluid were gradually downregulated with the elimination of infection, and were reduced to the background level at the day of explantation (Figure 5F), confirming the closed-loop control characteristic of the Infectpro device.
Figure S5.
Control Experiments, Related to Figure 5
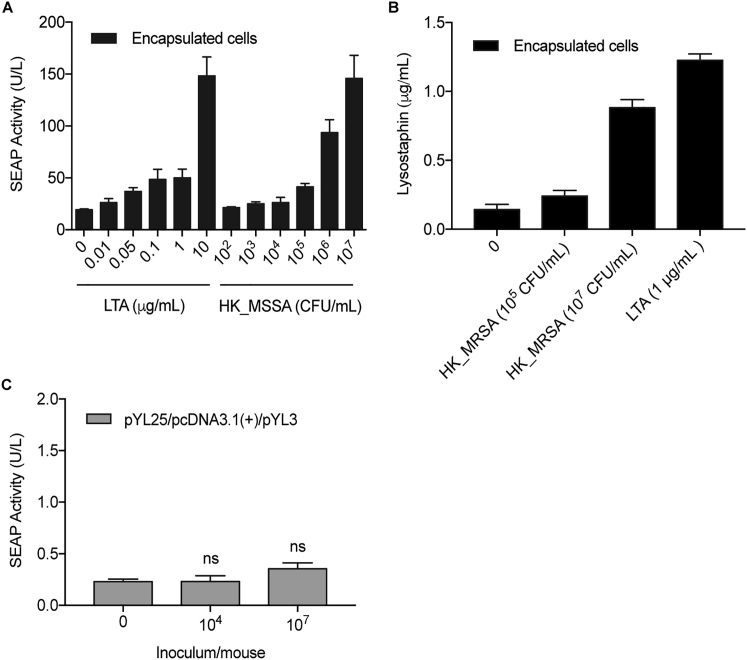
(A) In vitro SEAP induction profiles of cells after encapsulation. HEK293 cells transfected with pYL25/pYL30/pYL3 were encapsulated and induced with increasing doses of LTA or heat-killed MSSA (HK_MSSA). SEAP activity was assayed after 24 hr. Data are means ± SD (n = 3).
(B) In vitro lysostaphin induction profiles of cells after encapsulation. HEK293 cells transfected with pYL25/pYL30/pYL75 were cultivated with heat-killed MRSA (HK_MRSA) or LTA at the indicated concentrations. Lysostaphin expression levels in the culture supernatant were determined after 24 hr. Data are means ± SD (n = 3).
(C) Transgene expression in mice induced by live S. carnosus (n = 7). Mice implanted intraperitoneally with microencapsulated pYL25/pcDNA3.1(+)/pYL3-transgenic HEK293 cells received intraperitoneal injection of S. carnosus culture at the indicated concentrations. Blood SEAP activity was assayed after 24 hr. Data are means ± SEM; significant difference by two-tailed t test (ns: not significant).
Figure 5.
Prevention of MRSA Infection by Infectpro’s Sense-and-Destroy Function in Mice
(A and B) Dose-dependent transgene expression in mice induced by PGN (n = 6), HK_SA (n = 6) (A), or live S. carnosus (n = 7) (B). Mice implanted intraperitoneally with microencapsulated pYL25/pYL30/pYL3-transgenic HEK293 cells received an intraperitoneal injection of inducers at various concentrations. Blood SEAP activity was assayed after 24 hr.
(C) SEAP expression in tissue cage fluid from a murine foreign-body infection model. Mice were treated with microencapsulated pYL25/pYL30/pYL3-transgenic HEK293 cells into each cage and infected with live MRSA culture at 1.3 × 103 CFU/cage. Infectsen + MRSA (n = 8); Infectsen − MRSA (n = 7).
(D–F) Quantification of bacterial load and lysostaphin in mice receiving no treatment (no treatment, n = 8), implanted with microencapsulated pYL25/pYL30/pYL3-transfected HEK293 cells (negative ctrl, n = 8), microencapsulated pcDNA3.1(+)/pYL48-transfected HEK293 cells (positive ctrl, n8) or Infectpro (n8) prior to MRSA infection. (D) Planktonic MRSA present in tissue cage fluid of mice. (E) Adherent MRSA recovered from tissue cage biofilms in mice. (F) Lysostaphin expression in tissue cage fluid during 8 days.
Student’s t test (A–C and F) and Mann-Whitney U test (D and E). All data are means ± SEM.
See also Figure S5.
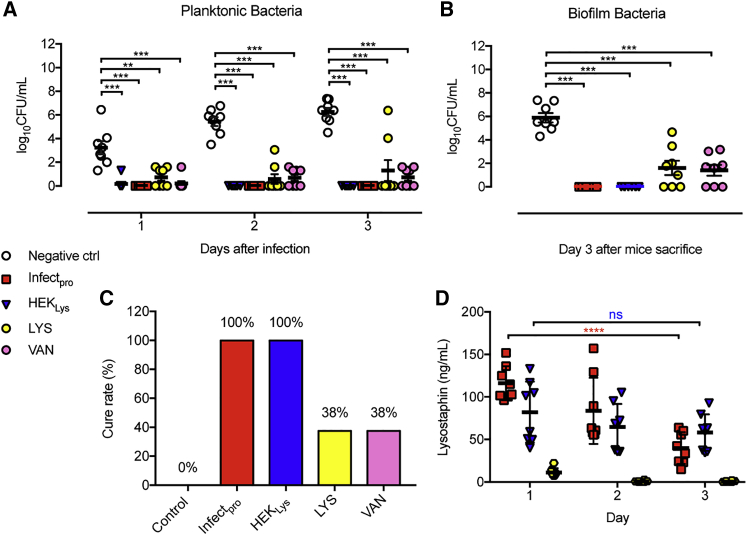
To further evaluate Infectpro’s potential for the treatment of acute MRSA foreign-body infections, MRSA-infected mice were implanted with microencapsulated Infectpro designer cells. Analysis of the tissue cage fluid of MRSA-infected mice showed that Infectpro and constitutive lyosostaphin-producing cells (HEKLys) (Figure S6A) were equally effective in clearing planktonic bacteria in all mice (Figure 6A). In contrast, treatment with vancomycin or recombinant lysostaphin only cleared planktonic bacteria in 4 out of 8 and 6 out of 8 infected mice after three days, respectively (Figure 6A). Similar results were also obtained for the treatment of bacterial biofilms in the tissue cages. While all animals treated with Infectpro and HEKLys were cleared of MRSA biofilm, adherent MRSA was still detected in 5 out of 8 mice treated with vancomycin or lysostaphin (Figure 6B). These treatment failures were not associated with the emergence of resistance to vancomycin (there was no increase in minimum inhibitory concentration [MIC] ≤ 2 μg/mL; guidelines by the Clinical and Laboratory Standards Institute, 2017) or lysostaphin (Figure S6B). Remarkably, the cell-based treatment strategies (Infectpro and HEKLys) achieved 100% cure rates of MRSA foreign-body infections, whereas vancomycin and lysostaphin treatment failed to eliminate infections, achieving only a 38% cure rate in each case (Figure 6C). Again, lysostaphin expression levels in the tissue cage fluid were gradually downregulated in the Infectpro group, while they remained constant in the HEKLys treatment group (Figure 6D). In summary, performance validation in mice suggests that this anti-MRSA cell-based therapy was highly effective for diagnosis, prevention, and cure of difficult-to-treat staphylococcal infections.
Figure 6.
Curing Acute MRSA Infections in Mice
(A and B) Bacterial load of MRSA-infected mice treated with Infectpro. Infected mice were treated with microencapsulated pYL25/pYL30/pYL3-transgenic HEK293 cells (negative ctrl), HEKLys, Infectpro, recombinant lysostaphin (LYS, 5 mg/kg, once), or vancomycin (VAN, 200 mg/kg, every 12 hr). Planktonic MRSA in tissue cage fluid of treated mice (A). Adherent MRSA in tissue cage biofilms of treated mice (B).
(C) Treatment efficacies among different groups expressed as cure rate.
(D) Lysostaphin levels in tissue cage fluid during 3 days.
Mann-Whitney U test (A and B) and Student’s t test (D). All data are means ± SEM (n = 8).
See also Figure S6.
Discussion
Bacterial resistance is overwhelming our ability to cope with infection, and development of new antimicrobials is failing to meet the need (Marston et al., 2016). In fact, addressing the threat of antimicrobial resistance is a perpetual challenge, because antimicrobial resistance is a predictable outcome of all antimicrobial use (Fauci and Marston, 2014). It is not just “inappropriate” use that selects for resistance. Rather, the speed with which resistance spreads is driven by microbial exposure to all antimicrobials (Spellberg et al., 2013). Thus, interventions that aim to prevent infections from occurring in the first place will reduce the need for antimicrobials and slow the appearance of resistance, thereby prolonging the useful lives of all available antimicrobials (Spellberg et al., 2013). Although vaccines hold great promise for restricting the need for antibiotics and preventing antibiotic-resistant infections, no antistaphylococcal vaccine has yet successfully passed clinical trials (Mohamed et al., 2017). Much of the difficulty stems from the lack of biomarkers for protective human immunity, the problems of translating findings from animal models into clinical trials, and safety concerns associated with unbalanced immune stimulations (Proctor, 2012). Moreover, it appears to be impossible to create a single universal vaccine against the broad range of virulence factors and diseases produced by a plethora of bacterial strains (Proctor, 2012).
The results of this study show that our cell-based approach (Infectpro) can prevent (by 91%) and cure (by 100%) multi-drug resistant bacterial infections (MRSA) in a murine model of implant-associated infections. This finding suggests potential clinical usefulness of the system, because once infection is established, especially in the case of biofilm-associated implant infections, standard antibiotic therapies (e.g., vancomycin, the first-line antibiotic for treatment of MRSA infections) are often no longer effective, and surgical procedures are required to extract and replace the infected device (Del Pozo and Patel, 2009). The mortality associated with MRSA infections is particularly high among patients with cardiovascular implants, such as prosthetic heart valves (Wang et al., 2007) and aortic grafts (Smeds et al., 2016). Although less severe, infections associated with orthopedic devices and ventricular shunts often result in serious disabilities, and infections of mammary and penile implants can cause major disfigurement and psychological trauma (Darouiche, 2004). On the other hand, prevention of antibiotic-resistant infections before symptoms appear would be particularly beneficial to patients with compromised immune functions (e.g., due to HIV, organ transplantation, cancer, and/or treatment with immunosuppressive medication), in whom infections are often life threatening. Furthermore, Infectpro effectively addresses the challenge of diagnosing implant-associated infection. Standard microbiological diagnosis of implant-associated infections requires large sample size, long culture time, and isolation of the organisms from periprosthetic tissue and/or explanted implants (Trampuz et al., 2007). We show here that application of our cellular device enables tissue fluid or blood to be used for measurement of reporter protein levels to enable detection and diagnosis of infection at the site of infection.
The availability of a closed-loop gene network to regulate therapeutic agent release in mammalian cells should also facilitate implementation of novel antiinfectives. Protein therapeutics such as lysins, antibodies, and antimicrobial peptides represent good alternatives to conventional antibiotics (Czaplewski et al., 2016): (1) they can have multiple and alternative mechanisms of action that are less prone to antimicrobial resistance (Fitzgerald-Hughes et al., 2012), (2) they can have narrow antibacterial spectra, thereby sparing non-pathogenic members of the human microbiome, which is essential to human health and infection control (Khosravi et al., 2014), and (3) they can have immunomodulatory and/or endotoxin-neutralizing effects (Finlay and Hancock, 2004) in addition to microbial killing. Furthermore, protein peptides are amenable to modifications that increase their potency (Peng et al., 2017) and half-life (Dennis et al., 2002) or can be used in combination therapy to achieve synergistic effects (Desbois et al., 2013) and multifunctional properties (Becker et al., 2016), hence reducing the chances of resistance development (Holmes et al., 2016). However, key unresolved issues regarding toxicity, stability, and method of delivery of the intact agent to sites of action are major reasons for the lack of success in systemic applications (Fitzgerald-Hughes et al., 2012). Indeed, we found that in contrast to cell-based treatment with Infectpro or HEKLys, injection of recombinant lysostaphin was not sufficient to cure bacterial infections in mice, probably because of the poor pharmacokinetics of lysostaphin (Walsh et al., 2003), a general disadvantage of protein therapeutics. Our cellular device provides a means to achieve dosing control and adjustment on a self-regulated basis, as well as infection-targeted direct delivery, affording high local tissue levels of active agent without systemic effects, together with access to difficult-to-reach regions such as avascular joint or bone prostheses.
Although alginate-poly-(L-lysine)-alginate microcapsules can remain functional for at least several years (Elliott et al., 2007) and have been licensed for human clinical trials (ClinicalTrials.gov NCT01379729, NCT00790257, NCT01739829), key challenges facing the translation of designer cell-based therapies into first-in-human applications include treatment economics, the use of autologous cells, non-invasive monitoring of cell stability and performance over extended periods of time, and strategies to replace or remove cells when required (Orive et al., 2015). However, these challenges do not seem insurmountable, and already several strategies, such as safety switches (Di Stasi et al., 2011, Santos et al., 2013), encapsulation material with improved biocompatibility (Vegas et al., 2016a, Vegas et al., 2016b), and real-time implant monitoring (Barnett et al., 2011, Catena et al., 2010), are being introduced to improve the safety and efficacy of cellular grafts and to allow the maintenance of cellular function for extended periods of time in humans.
We believe that the gene circuit described here could serve as a pioneering model that would be readily adaptable into different clinical settings and applicable to populations at high risk of infections, including immunologically compromised individuals, as well as patients undergoing surgery or with indwelling medical devices. We envision that this cell-based anti-MRSA approach could serve as a valuable alternative to current antibiotic therapies and an example to stimulate development of next-generation antiinfectives in the post-antibiotic era.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-lysostaphin antibody | Linscott’s Directory, AICBiotech | cat# PAb102; lot no. J101 |
| Goat anti-human IgG Fcγ fragment | Jackson ImmunoResearch | cat# 109-005-008; lot no. 122635; AB_2337534 |
| Goat anti-human IgG-alkaline phosphatase antibody | Sigma | cat# A9544-25ML; lot no. 096M4793V; AB_258459 |
| Bacterial strains | ||
| S. carnosus | Spanish Type Culture Collection (CECT), Valencia, Spain | CECT no. 4491 |
| Methicillin-resistant S. aureus (MRSA) | ATCC | ATCC 43300 |
| Methicillin-susceptible S. aureus (MSSA) | ATCC | ATCC 35556 |
| Chemicals, Materials, Peptides, and Recombinant Proteins | ||
| Purified LTA from Staphylococcus aureus (LTA) | InvivoGen, LabForce AG, Switzerland | cat# tlrl-pslta |
| PGN from Staphylococcus aureus (PGN) | InvivoGen, LabForce AG, Switzerland | cat# tlrl-pgns2 |
| Heat-killed Staphylococcus aureus (HKSA) | InvivoGen, LabForce AG, Switzerland | cat# tlrl-hksa |
| Polyethyleneimine max (PEI) | Polysciences, Warrington, USA | cat# 24765-2 |
| 4-Nitrophenyl phosphate, disodium salt, hexahydrate (pNPP) | Acros Organics, Geel, Belgium | cat# 128860100 |
| Recombinant lysostaphin | AMBI PRODUCTS LLC, Lawrence, NY | cat# LSPN-50 |
| Vancomycin | Teva Pharma | lot no. 166977 |
| Alginate | BÜCHI Labortechnik AG | cat# 11061528 |
| TMB | Thermo Scientific | cat# 34028 |
| Antibiotic assay disc | Whatman | cat# 2017-006 |
| Critical Commercial Assays | ||
| SEAP reporter gene assay | Roche Diagnostics GmbH, Mannheim, Germany | cat# 11779842001 |
| Lightening-link HRP | Innova Biosciences | cat# 701-0030 |
| Experimental Models: Cell Lines | ||
| Human: HEK293T | ATCC | CRL-11268 |
| Hamster: BHK-21 | ATCC | CCL-10 |
| Human: HeLa | ATCC | CCL-2 |
| Human: HT-1080 | ATCC | CCL-121 |
| Human: hMSC-TERT | Simonsen et al., 2002 | RRID: CVCL_Z015 |
| Hamster: CHO-K1 | ATCC | CCL-61 |
| Experimental Models: Organisms/Strains | ||
| Mouse: WT: OF1 mice | Charles River Laboratories | Crl:OF1 |
| Mouse: WT: C57BL/6 mice | Janvier Labs, France | stock no. 000664 |
| Oligonucleotides | ||
| Primers and oligonucleotides, see Table S1 | This paper | N/A |
| Recombinant DNA | ||
| Plasmids construction and design, see Table S1 | This paper | N/A |
| Software and Algorithms | ||
| Prism V7.0a | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| Benchling | Benchling platform | https://benchling.com |
Contact for Reagent and Resource Sharing
Requests for further information or resources and reagents should be directed to and will be fulfilled by the Lead Contact, Martin Fussenegger (fussenegger@bsse.ethz.ch). Plasmids containing the human TRL1068 cDNA used and generated in this study are subject to restrictions under a simple material transfer agreement (MTA) with Trellis Bioscience.
Experimental Model and Subject Details
Mice
Dose-dependent transgene expression in mice induced by PGN, HK_SA or live S. carnosus were performed according to the directives of the European Community Council (2010/63/EU), approved by the French Republic and performed in 2012 by Ghislaine Charpin-El Hamri (license no. 69266309) at the Universitéde Lyon, F-69622 Lyon, France. Six to eight week-old native healthy female wild-type OF1 mice (oncins France souche 1) were kept in a pathogen-free environment in a 12 hr light/dark cycle (light from 6 am to 6 pm) in a temperature-controlled room (24°C). Six to seven mice were randomly assigned to experimental groups, which were not involved in previous procedures and housed in standardized cages with unrestricted access to regular mouse chow and water.
Mouse model of foreign body infection (John et al., 2011, Nowakowska et al., 2014) was established with the approval of the Kantonale Veterinaeramt Basel-Stadt (permit no. 1710). Experiments were conducted according to the regulations of Swiss veterinary law. Healthy wild-type female C57BL/6 mice at 13 weeks age (JanvierLabs) kept under specific-pathogen-free conditions (Biosafety level 2) in the animal house of the Department of Biomedicine, University Hospital Basel, were used for the MRSA infection experiments. They were housed in a 12 hr light/dark cycle (light from 7 am to 7 pm) in a temperature-controlled room (24°C) with free access to regular mouse chow and water. Mice were randomly assigned to experimental groups, which were not involved in previous procedures.
Cell types and culture conditions
Human embryonic kidney cells (HEK293, female), baby hamster kidney cells (BHK-21, male), human cervical adenocarcinoma cells (HeLa, female), human fibrosarcoma cells (HT-1080, male) and human bone marrow stromal cells transgenic for the catalytic subunit of human telomerase (hMSC-TERT, male) were cultivated in Dulbecco’s modified Eagle’s medium (DMEM, Life Technologies, cat# 31966047) supplemented with 10% (v/v) fetal calf serum (FCS, Sigma-Aldrich, St. Louis, MO, USA, cat# F7524) and 1% (v/v) penicillin/streptomycin solution (Biowest, Nuaillé, France; cat. no. L0022-100). Chinese hamster ovary cells (CHO-K1, female) were grown in ChoMaster HTS (Cell Culture Technologies GmbH, Gravesano, Switzerland, art. no. CHTS-8) supplemented with 5% (v/v) FCS and 1% (v/v) penicillin/streptomycin solution. All cell lines were cultivated at 37°C in a humidified atmosphere of 5% CO2 in air. Cell line authentication information was supplied by ATCC. Information on the sex of the cell line was obtained from ATCC, https://web.expasy.org/cellosaurus/CVCL_0317, and Shah et al. (2014).
Staphylococcal strains and growth conditions
S. carnosus were cultivated aerobically in Luria Broth (LB) medium with shaking at 37°C. Methicillin-susceptible S. aureus (MSSA) and methicillin-resistant S. aureus (MRSA) were aerobically grown in tryptic soy broth (TSB) (Becton Dickinson and Company, Allschwil, Switzerland) overnight at 37°C without shaking.
Method Details
Cell transfection
For transfection of HEK293 and hMSC-TERT cells, a 1:3 DNA:PEI mixture of 0.525 μg DNA (pYL25:pYL30:pYL126:pYL125:pYL3 = 1:2:1:1:2) and 1.575 μL PEI (1 mg/mL in water, pH 7.0) was incubated for 15 min at room temperature before being added dropwise to 2 105 cells/mL seeded in cell culture plates 18 hr before transfection. After 7 hr, the medium was changed and cells were induced with different ligands. BHK-21, CHO-K1, HeLa and HT-1080 cells were seeded at 1 105 cells/mL and transfected with a 1:4 DNA:PEI ratio. Transgene expression was profiled at indicated time points after adding inducer reagents.
Bacterial inoculum and quantification
For in vitro sense-and-destroy study, MRSA overnight culture was washed twice with PBS (Bichsel, Interlaken, Switzerland) before being inoculated into cell culture medium. For the in vivo prevention study, MRSA overnight culture was washed twice with PBS before infecting mice at desired concentrations. For the in vivo acute infection, MRSA overnight culture was washed twice with 0.9% saline (Bichsel, Interlaken, Switzerland) for the inoculation of mice with adjusted concentrations as required. To prepare bacteria in the exponential-growth phase, the overnight culture was diluted 1:100 and further incubated for 6 hr at 37°C, followed by two washes with 0.9% saline, and the concentration was adjusted as required.
The CFU (colony forming unit) per milliliter (mL) was determined by plating aliquots from appropriate dilutions on MHA agar plates, followed by colony counting after 24 hr of incubation at 37°C.
Generation of stable cell lines
The polyclonal HEKhTLR2 population, transgenic for stable expression of human TLR2, was constructed by co-transfecting 2 × 105 HEK293 cells/mL with 1 μg/mL pYL98 (ITR-PhEF1α-hTLR2-pA:PRPBSA-BFP-P2A-PuroR-pA-ITR) and 50 ng/mL of the Sleeping Beauty transposase expression vector pCMV-T7-SB100 (PhCMV-SB100X-pA). After selection with 5 μg/mL puromycin for two passages, the surviving population was FACS-sorted into three different subpopulations based on blue-fluorescence intensities. The subpopulation with low 15% BFP intensity showed the best induction profile and was used for follow-up experiments.
The polyclonal HEKhCD14/hTLR2 population, transgenic for stable expression of human CD14 and human TLR2, was generated by co-transfecting 2 × 105 HEKhTLR2 cells/mL with 1 μg/mL pYL97 (ITR-PhEF1α-hCD14-pA:PRPBSA-dTomato-P2A-BlastR-pA) and 50 ng/mL of pCMV-T7-SB100. After selection with 15 μg/mL blasticidin, the surviving population was FACS-sorted into three different subpopulations based on red-fluorescence intensities. The subpopulation with the top 15% dTomato intensity showed the best performance and was used for follow-up experiments.
The monoclonal Infectpro cell line, transgenic for bacteria-stimulated lysostaphin expression, was constructed by co-transfecting HEKhCD14/hTLR2 cells with 1 μg/mL pYL112 (ITR-PAP-1 × 5-NF-κB × 5-IFNβmin-hGLuc-Lys-pA:PRPBSA-EGFP-P2A-ZeoR-pA-ITR) and 50 ng/mL of pCMV-T7-SB100, followed by selection in culture medium containing 1 mg/mL zeocin. The surviving population with the highest 4% EGFP expression levels was subjected to FACS-mediated single-cell sorting. Clone no. 73 showing optimal LTA-inducible lysostaphin expression was used for all follow-up studies.
HEKLYS, stably transgenic for constitutive lysostaphin expression, was constructed by co-transfecting 2 × 105 HEK293 cells/mL with 1 μg/mL pYL128 (ITR-PhCMV-hGLuc-Lys-pA:PRPBSA-ZeoR-P2A-mRuby-pA-ITR) and 50 ng/mL of the Sleeping Beauty transposase expression vector pCMV-T7-SB100 followed by stable selection using 1 mg/mL zeocin.
FACS-mediated single-cell sorting
HEK293 cells expressing EGFP (488nm laser, 530/30 emission filter) BFP (405 nm laser, 450/50 bandpass filter) or dTomato (561nm laser, 586/15 emission filter) were sorted using a Becton Dickinson LSRII Fortessa flow cytometer (Becton Dickinson, Allschwil, Switzerland) while excluding dead cells and cell doublets. Untreated HEK293 cells or parental polyclonal populations were used as negative controls.
Quantification of target gene expression
SEAP
Production of the human placental secreted alkaline phosphatase (SEAP) in culture was quantified by means of light absorbance measurement using a p-nitrophenyl phosphate-based assay (Schlatter et al., 2002). In brief, 100 μL of culture medium was heat-inactivated for 30 min at 65°C and centrifuged for 30 s at 15000 g. Subsequently, 80 μL of the heat-inactivated supernatant was transferred to a well of a 96-well plate containing 100 μL 2 × SEAP assay buffer (2 M diethanolamine, 2 mM L-homoarginine hydrochloride, 1 mM MgCl2, pH 9.8). After addition of 20 μL 120 mM 4-nitrophenyl phosphate (pNPP) to each well, the increase in light absorbance was measured at 405 nm every 30 s up to 20 min. Serum levels of SEAP were quantified using a chemiluminescence-based assay (SEAP reporter gene assay).
Fluorescence
The cellular fluorescence of cell populations cultured in an atmosphere of 5% CO2 in air at 37°C in 96-well plates was profiled every 30 min using a monochromatic Tecan Infinite M200 PRO plate reader (Tecan Group Ltd, Männedorf, Switzerland) at excitation and emission wavelengths of 482/502 nm, respectively. The autofluorescence of mock-transfected cells was subtracted from the fluorescence of cells containing Infectsen components.
Lysostaphin
Levels of lysostaphin were determined using enzyme-linked immunosorbent assay (ELISA). 96-well EIA/RIA plates were coated overnight at 4°C with 100 μL of 4 μg/mL rabbit polyclonal anti-lysostaphin antibody in coating buffer (0.1 M sodium carbonate, pH 9.5). The plates were then washed with PBS-0.05% Tween 20 (PBST) 3 times, blocked for 4 hr at 37°C with 200 μL 3% BSA in PBS, and again washed 3 times. Serial dilutions of standards (recombinant lysostaphin) and samples (1:5 dilution) in dilution buffer (PBS-0.01% Tween 80 and 0.1% BSA) were added to the wells and the plates were incubated at 37°C for 1 hr, then washed with PBST 5 times. Polyclonal anti-lysostaphin antibody conjugated to horseradish peroxidase (HRP) (lightening-link HRP) at 1 μg/mL in dilution buffer (100 μL) was added and incubation was continued for 1 hr at 37°C, followed by washing 10 times with PBST and incubation with TMB for colorimetric detection. Lysostaphin was quantified by reading the absorbance at 370 nm using an Envision 2104 multilabel (Perkin Elmer).
Human IgG ELISA
TRL1068 expression levels were quantified using sandwich ELISA. 96-well EIA/RIA plates were coated with 5 μg/mL goat anti-human IgG Fcγ fragment in coating buffer (0.1 M sodium carbonate, pH 9.5) at 4°C overnight. Plates were blocked with 200 mL per well PBS/3% BSA, pH 7.4, for 1 hr at 22°C. Human IgG standard (0.976-125 ng/mL) and samples (100 μL per well) were diluted (PBS-0.01% Tween 80 and 0.1% BSA) and incubated for 1.5 hr at 22°C. Plates were washed three times with 200 mL per well PBS containing 0.05% Tween-20 and then incubated with 100 mL per well of a goat anti-human IgG-alkaline phosphatase antibody (dilution 1:5000) for 1 hr at 22°C. After three washing steps (200 mL PBS/0.05% Tween-20), the plates were developed by incubation with 100 mL per well of pNPP substrate diluted in assay buffer (0.1 M ethanolamine-HCL, 5 mM MgCl2, pH 9.8; 1 mg/mL) at 22°C for 6 min. TRL1068 was quantified at 405 nm using an Envision 2104 multilabel reader (Perkin Elmer).
HU binding assay of TRL1068
Streptavidin-coated plates (Thermo Scientific Pierce, cat no. 15500) were washed (PBS-0.05% Tween 20) four times prior to use. The plates were then coated with biotinylated epitope from S. aureus histone-like DNA-bindig protein (Trellis Bioscience) at 10 μM in dilution buffer (PBS-0.01% Tween 80 and 0.1% BSA) for 2 hr with shaking at room temperature (RT). Samples from cell supernatant and epitope peptide standard were then added to each well and incubated on an orbital shaker for 30 min at RT. The plates were washed 4 times and incubated with goat anti-human IgG-alkaline phosphatase antibody (dilution 1:1000) on an orbital shaker for 30 min at RT. After washing for 4 times, plates were developed by incubation with pNPP substrate diluted in buffer (0.1 M ethanolamine-HCL, 5 mM MgCl2, pH 9.8; 1 mg/mL) at RT for 3 min and measured at 405 nm using an Envision 2104 multilabel reader (Perkin Elmer).
Antimicrobial assays
Time-kill study
0.5 105 Infectpro and HEK293 cells transfected with pYL25/pYL30/pYL3 (Control), pYL25/pcDNA3.1(+)/pYL75 (-pYL30) or pcDNA3.1(+)/pYL48 (Constitutive) were seeded per well in a 48-well plate one day before inoculation. After transfection for overnight, cell culture medium was changed to that without antibiotics, and appropriate amounts of S. carnosus overnight culture were added to each well. At different time points after induction, cell mixtures were collected for bacterial counting by serial dilution plating to determine bactericidal activity. Samples (30 μL) were also dropped on agar plates and incubated for 24 hr at 37°C, and then the plates were imaged.
Bactericidal effects against MRSA
3 independent MRSA overnight cultures were washed twice in PBS and diluted as required for inocula triplicates. Infectpro, HEKLys or control cells were seeded one day before. After transfection for overnight, cell culture medium was changed to that without antibiotics, and different concentrations of MRSA culture was added to each well for up to 96 hr at 37°C. Bacterial growth was quantified through serial plating, and lysostaphin was quantified by ELISA.
Spot-on-lawn assay
After 24 hr incubation, spots were imaged. Cleared spots indicate lytic activity. An exponentially growing culture of S. carnosus was spread on the plate, and 30 μL of lysostaphin-containing sample was spotted on the air-dried plate. Overnight incubation results in growth of a bacterial lawn, whereas antimicrobial activity of a protein is indicated by a clear lysis zone within the lawn, where growth has been prevented.
Bioassay
1.5 104 CFU of a MRSA culture was added to MHA medium and used to fill bioassay dishes. Samples of 100 μL cell supernatant from transfected HEK293 cells were applied in punched holes in the agar plate. The plate was incubated for 20 hr at 37. Following incubation, the diameter of inhibition zones around the various holes was measured. Results are the means of four individual experiments.
Emergence of antimicrobial resistance in vivo
Surviving MRSA were sampled from explanted tissue cages and screened for the emergence of resistance to vancomycin or lysostaphin during in vivo treatment. Colonies collected from bacterial subcultures were suspended to the turbidity of a McFarland 0.5 standard, and spread on Mueller-Hinton agar plates containing 2 μg/mL and 3 μg/mL of vancomycin. Plates were incubated at 37°C and profiled for bacterial growth after 20 hr. Resistance to lysostaphin was determined using a disk diffusion assay (50 μg/disc) (Kusuma and Kokai-Kun, 2005). The plates were incubated at 37°C and profiled for bacterial growth after 20 hr. Inhibition zones were compared between the treatment and control groups.
Cell encapsulation
Cells were encapsulated in 400 μm alginate-PLL (poly-L-lysine)-alginate beads using the Inotech Encapsulator Research IE-50R (Buechi Labortechnik AG, Flawil, Switzerland) according to the manufacturer’s protocol. The parameters were as follows: 25 mL syringe operated at a flow rate of 500 units, 200 μm nozzle with a vibration frequency of 1,024 Hz and bead dispersion voltage of 1.2 kV, stirrer speed set at 4.5 units.
Animal testing
Transgene induction
To test the bacteria-controlled transgene expression in mice, HEK293 cells transfected with pYL25/pYL30/pYL3 were encapsulated and intraperitoneally injected into OF1 mice (2-3 × 106 cells/mouse). Immediately after implantation, PGN, HK_SA or live S. carnosus was administered intraperitoneally at specific dosages. After 24 hr, mice were sacrificed, blood samples were collected retroorbitally, and SEAP levels were quantified in the serum.
Prevention of foreign body infection
To test the diagnostic and therapeutic efficacy of cell implants against MRSA in mice, C57BL/6 mice were anesthetized followed by the subcutaneous implantation of sterile cylindrical Teflon tissue cages (32 × 10 mm; volume: 1.9 mL) with 130 regularly spaced holes (Angst + Pfister AG, Zurich, Switzerland). Upon complete wound healing, cages were tested for sterility by plating percutaneously aspirated tissue cage fluid from the lumen of the tissue cage on Columbia sheep blood agar plates. Prophylactic cell-based treatment was started at 16 hr prior to infection with an injection of 3-5 × 106 microencapsulated cells contained in 400 μL serum-free DMEM into the lumen of each cage. On the day of infection, 400 μL serum-free DMEM containing 3-5 × 106 microencapsulated cells and 1205 ± 215 CFU of MRSA were injected into each cage. On days 1, 2, 3, 6, 8 post-infection, tissue cage fluid was collected to measure the levels of lysostaphin and to assess planktonic bacterial load by plating on blood agar plates. On day 8, mice were sacrificed, mouse serum was collected via heart puncture, and tissue cage of each mouse was explanted under aseptic conditions.
Curing of foreign body infection
For the acute infection study, mice were infected with 201 ± 58 CFU/cage of MRSA 2 hr before treatment with Infectpro (3 × 107 cells, maintained for 10 hr in medium containing 10 μg/mL PGN), HEKLys (3 × 107 cells), recombinant lysostaphin (5 mg/kg, once intraperitoneal) or vancomycin (200 mg/kg, every 12 hr subcutaneously). A vancomycin treatment schedule of 200 mg/kg every 12 hr was chosen to provide pharmacokinetic-pharmacodynamic parameters exceeding those reached in human treatment schemes: 1 g every 12 hr or a target AUC/MIC value of 400 (Crandon et al., 2010, Rybak et al., 2009). The explanted tissue cages were washed twice with phosphate-buffered saline followed by 30 s vortexing, sonication for 3 min at 130 W and another 30 s vortexing to release adherent bacteria from the biofilm. Quantification of adherent bacteria was performed by plating appropriated dilutions on blood agar plates. The presence of any regrowth of adherent bacteria was examined by further incubation of the cage in tryptic soy broth (TSB) (Becton Dickinson and Company, Allschwil, Switzerland) for 48 hr at 37°C. Any visualization of a positive culture of MRSA was defined as treatment failure. The prevention and cure rate was defined as the percentage of cages without growth in the individual treatment group. Concentrations of lysostaphin in the tissue cage fluid were determined by ELISA as described above.
Quantification and Statistical Analysis
Statistical parameters including the definitions and exact values of n, distributions and deviations, statistical test and statistical significance are reported in the Figures and corresponding Figure Legends. A two-tailed, unpaired Student’s t test and the Mann-Whitney U test were applied to determine the statistical significance of differences among groups using Graphpad Prism. Differences are considered statistically significant at p < 0.05; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; ns, not significant.
Acknowledgments
We thank Clett Erridge (pCD14), Yang Liu (pIRES/hTLR1, pIRES/hTLR6), Trellis Bioscience (TRL1068), and Tobias Strittmatter (pTS1101) for providing plasmids and Leo Scheller, Viktor Haellman, and Ryosuke Kojima for valuable comments on the manuscript. This work was supported by a European Research Council (ERC) advanced grant (ProNet, no. 321381 to M. Fussenegger) and in part by the National Centre of Competence in Research (NCCR) for Molecular Systems Engineering (to M. Fussenegger) and the Swiss National Science Foundation (PZ00P3_142403 to N.K.).
Author Contributions
Y.L., H.Y., and M. Fussenegger designed the project; A.-K.W. and N.K. designed the animal experiments; Y.L., A.-K.W., P.B., M.X., M. Folcher, N.K., and M. Fussenegger analyzed results; Y.L., A.-K.W., G.C.-E.H., and P.B. performed the experimental work; and Y.L., P.B., M.X., A.-K.W., N.K., and M. Fussenegger wrote the manuscript.
Declaration of Interests
The authors declare no competing interests.
Published: June 21, 2018
Footnotes
Supplemental Information includes six figures and one table and can be found with this article online at https://doi.org/10.1016/j.cell.2018.05.039.
A video abstract is available at https://doi.org/10.1016/j.cell.2018.05.039#mmc2.
Contributor Information
Nina Khanna, Email: nina.khanna@usb.ch.
Martin Fussenegger, Email: fussenegger@bsse.ethz.ch.
Supporting Citations
The following references appear in the Supplemental Information: Ausländer et al., 2014, Erridge et al., 2008, Fang et al., 2005, Fang et al., 2007, Ho et al., 2007, Kim et al., 2011, Kowarz et al., 2015, Liu et al., 2012, Loew et al., 2010, Schlabach et al., 2010, Szymczak et al., 2004, Szymczak-Workman et al., 2012, Tannous et al., 2005, Xie et al., 2016, Ye et al., 2017; and Zhang et al. (2005).
Supplemental Information
References
- Akira S., Takeda K. Toll-like receptor signalling. Nat. Rev. Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- Akira S., Uematsu S., Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Ausländer D., Ausländer S., Charpin-El Hamri G., Sedlmayer F., Müller M., Frey O., Hierlemann A., Stelling J., Fussenegger M. A synthetic multifunctional mammalian pH sensor and CO2 transgene-control device. Mol. Cell. 2014;55:397–408. doi: 10.1016/j.molcel.2014.06.007. [DOI] [PubMed] [Google Scholar]
- Barnett B.P., Arepally A., Stuber M., Arifin D.R., Kraitchman D.L., Bulte J.W. Synthesis of magnetic resonance-, X-ray- and ultrasound-visible alginate microcapsules for immunoisolation and noninvasive imaging of cellular therapeutics. Nat. Protoc. 2011;6:1142–1151. doi: 10.1038/nprot.2011.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker S.C., Roach D.R., Chauhan V.S., Shen Y., Foster-Frey J., Powell A.M., Bauchan G., Lease R.A., Mohammadi H., Harty W.J. Triple-acting lytic enzyme treatment of drug-resistant and intracellular Staphylococcus aureus. Sci. Rep. 2016;6:25063. doi: 10.1038/srep25063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjarnsholt T., Alhede M., Alhede M., Eickhardt-Sørensen S.R., Moser C., Kühl M., Jensen P.O., Høiby N. The in vivo biofilm. Trends Microbiol. 2013;21:466–474. doi: 10.1016/j.tim.2013.06.002. [DOI] [PubMed] [Google Scholar]
- Boucher H.W., Corey G.R. Epidemiology of methicillin-resistant Staphylococcus aureus. Clin. Infect. Dis. 2008;46(Suppl 5):S344–S349. doi: 10.1086/533590. [DOI] [PubMed] [Google Scholar]
- Catena R., Santos E., Orive G., Hernández R.M., Pedraz J.L., Calvo A. Improvement of the monitoring and biosafety of encapsulated cells using the SFGNESTGL triple reporter system. J. Control. Release. 2010;146:93–98. doi: 10.1016/j.jconrel.2010.05.018. [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI) 27th ed. Clinical and Laboratory Standards Institute; 2017. Performance Standards for Antimicrobial Susceptibility Testing. [Google Scholar]
- Crandon J.L., Kuti J.L., Nicolau D.P. Comparative efficacies of human simulated exposures of telavancin and vancomycin against methicillin-resistant Staphylococcus aureus with a range of vancomycin MICs in a murine pneumonia model. Antimicrob. Agents Chemother. 2010;54:5115–5119. doi: 10.1128/AAC.00062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czajkowsky D.M., Hu J., Shao Z., Pleass R.J. Fc-fusion proteins: new developments and future perspectives. EMBO Mol. Med. 2012;4:1015–1028. doi: 10.1002/emmm.201201379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaplewski L., Bax R., Clokie M., Dawson M., Fairhead H., Fischetti V.A., Foster S., Gilmore B.F., Hancock R.E.W., Harper D. Alternatives to antibiotics-a pipeline portfolio review. Lancet Infect. Dis. 2016;16:239–251. doi: 10.1016/S1473-3099(15)00466-1. [DOI] [PubMed] [Google Scholar]
- Dajcs J.J., Hume E.B., Moreau J.M., Caballero A.R., Cannon B.M., O’Callaghan R.J. Lysostaphin treatment of methicillin-resistant staphylococcus aureus keratitis in the rabbit(1) Am. J. Ophthalmol. 2000;130:544. doi: 10.1016/s0002-9394(00)00748-0. [DOI] [PubMed] [Google Scholar]
- Dantes R., Mu Y., Belflower R., Aragon D., Dumyati G., Harrison L.H., Lessa F.C., Lynfield R., Nadle J., Petit S., Emerging Infections Program–Active Bacterial Core Surveillance MRSA Surveillance Investigators National burden of invasive methicillin-resistant Staphylococcus aureus infections, United States, 2011. JAMA Intern. Med. 2013;173:1970–1978. doi: 10.1001/jamainternmed.2013.10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darouiche R.O. Treatment of infections associated with surgical implants. N. Engl. J. Med. 2004;350:1422–1429. doi: 10.1056/NEJMra035415. [DOI] [PubMed] [Google Scholar]
- Davies N.W., Smith J.A., Molesworth P.P., Ross J.J. Hydrogen/deuterium exchange on aromatic rings during atmospheric pressure chemical ionization mass spectrometry. Rapid Commun. Mass Spectrom. 2010;24:1105–1110. doi: 10.1002/rcm.4488. [DOI] [PubMed] [Google Scholar]
- Del Pozo J.L., Patel R. Clinical practice. Infection associated with prosthetic joints. N. Engl. J. Med. 2009;361:787–794. doi: 10.1056/NEJMcp0905029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis M.S., Zhang M., Meng Y.G., Kadkhodayan M., Kirchhofer D., Combs D., Damico L.A. Albumin binding as a general strategy for improving the pharmacokinetics of proteins. J. Biol. Chem. 2002;277:35035–35043. doi: 10.1074/jbc.M205854200. [DOI] [PubMed] [Google Scholar]
- Desbois A.P., Sattar A., Graham S., Warn P.A., Coote P.J. MRSA decolonization of cotton rat nares by a combination treatment comprising lysostaphin and the antimicrobial peptide ranalexin. J. Antimicrob. Chemother. 2013;68:2569–2575. doi: 10.1093/jac/dkt243. [DOI] [PubMed] [Google Scholar]
- Di Stasi A., Tey S.K., Dotti G., Fujita Y., Kennedy-Nasser A., Martinez C., Straathof K., Liu E., Durett A.G., Grilley B. Inducible apoptosis as a safety switch for adoptive cell therapy. N. Engl. J. Med. 2011;365:1673–1683. doi: 10.1056/NEJMoa1106152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott R.B., Escobar L., Tan P.L., Muzina M., Zwain S., Buchanan C. Live encapsulated porcine islets from a type 1 diabetic patient 9.5 yr after xenotransplantation. Xenotransplantation. 2007;14:157–161. doi: 10.1111/j.1399-3089.2007.00384.x. [DOI] [PubMed] [Google Scholar]
- Erridge C., Kennedy S., Spickett C.M., Webb D.J. Oxidized phospholipid inhibition of toll-like receptor (TLR) signaling is restricted to TLR2 and TLR4: roles for CD14, LPS-binding protein, and MD2 as targets for specificity of inhibition. J. Biol. Chem. 2008;283:24748–24759. doi: 10.1074/jbc.M800352200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estellés A., Woischnig A.K., Liu K., Stephenson R., Lomongsod E., Nguyen D., Zhang J., Heidecker M., Yang Y., Simon R.J. A high-affinity native human antibody disrupts biofilm from Staphylococcus aureus bacteria and potentiates antibiotic efficacy in a mouse implant infection model. Antimicrob. Agents Chemother. 2016;60:2292–2301. doi: 10.1128/AAC.02588-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J., Qian J.J., Yi S., Harding T.C., Tu G.H., VanRoey M., Jooss K. Stable antibody expression at therapeutic levels using the 2A peptide. Nat. Biotechnol. 2005;23:584–590. doi: 10.1038/nbt1087. [DOI] [PubMed] [Google Scholar]
- Fang J., Yi S., Simmons A., Tu G.H., Nguyen M., Harding T.C., VanRoey M., Jooss K. An antibody delivery system for regulated expression of therapeutic levels of monoclonal antibodies in vivo. Mol. Ther. 2007;15:1153–1159. doi: 10.1038/sj.mt.6300142. [DOI] [PubMed] [Google Scholar]
- Fauci A.S., Marston D. The perpetual challenge of antimicrobial resistance. JAMA. 2014;311:1853–1854. doi: 10.1001/jama.2014.2465. [DOI] [PubMed] [Google Scholar]
- Fauci A.S., Morens D.M. The perpetual challenge of infectious diseases. N. Engl. J. Med. 2012;366:454–461. doi: 10.1056/NEJMra1108296. [DOI] [PubMed] [Google Scholar]
- Finlay B.B., Hancock R.E.W. Can innate immunity be enhanced to treat microbial infections? Nat. Rev. Microbiol. 2004;2:497–504. doi: 10.1038/nrmicro908. [DOI] [PubMed] [Google Scholar]
- Fitzgerald-Hughes D., Devocelle M., Humphreys H. Beyond conventional antibiotics for the future treatment of methicillin-resistant Staphylococcus aureus infections: two novel alternatives. FEMS Immunol. Med. Microbiol. 2012;65:399–412. doi: 10.1111/j.1574-695X.2012.00954.x. [DOI] [PubMed] [Google Scholar]
- Foster T.J., Geoghegan J.A., Ganesh V.K., Höök M. Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat. Rev. Microbiol. 2014;12:49–62. doi: 10.1038/nrmicro3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier B., Philpott D.J. Recognition of Staphylococcus aureus by the innate immune system. Clin. Microbiol. Rev. 2005;18:521–540. doi: 10.1128/CMR.18.3.521-540.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godefroy E., Gallois A., Idoyaga J., Merad M., Tung N., Monu N., Saenger Y., Fu Y., Ravindran R., Pulendran B. Activation of toll-like receptor-2 by endogenous matrix metalloproteinase-2 modulates dendritic-cell-mediated inflammatory responses. Cell Rep. 2014;9:1856–1870. doi: 10.1016/j.celrep.2014.10.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris R.L., Nunnery A.W., Riley H.D., Jr. Effect of lysostaphin on staphylococcal carriage in infants and children. Antimicrob Agents Chemother (Bethesda) 1967;7:110–112. doi: 10.1128/AAC.7.1.110. [DOI] [PubMed] [Google Scholar]
- Hertlein T., Sturm V., Lorenz U., Sumathy K., Jakob P., Ohlsen K. Bioluminescence and 19F magnetic resonance imaging visualize the efficacy of lysostaphin alone and in combination with oxacillin against Staphylococcus aureus in murine thigh and catheter-associated infection models. Antimicrob. Agents Chemother. 2014;58:1630–1638. doi: 10.1128/AAC.01422-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho T.Y., Chen Y.S., Hsiang C.Y. Noninvasive nuclear factor-kappaB bioluminescence imaging for the assessment of host-biomaterial interaction in transgenic mice. Biomaterials. 2007;28:4370–4377. doi: 10.1016/j.biomaterials.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Holmes A.H., Moore L.S.P., Sundsfjord A., Steinbakk M., Regmi S., Karkey A., Guerin P.J., Piddock L.J.V. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet. 2016;387:176–187. doi: 10.1016/S0140-6736(15)00473-0. [DOI] [PubMed] [Google Scholar]
- Jin M.S., Kim S.E., Heo J.Y., Lee M.E., Kim H.M., Paik S.G., Lee H., Lee J.O. Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell. 2007;130:1071–1082. doi: 10.1016/j.cell.2007.09.008. [DOI] [PubMed] [Google Scholar]
- John A.K., Schmaler M., Khanna N., Landmann R. Reversible daptomycin tolerance of adherent staphylococci in an implant infection model. Antimicrob. Agents Chemother. 2011;55:3510–3516. doi: 10.1128/AAC.00172-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J.Y., Nan X., Jin M.S., Youn S.J., Ryu Y.H., Mah S., Han S.H., Lee H., Paik S.G., Lee J.O. Recognition of lipopeptide patterns by Toll-like receptor 2-Toll-like receptor 6 heterodimer. Immunity. 2009;31:873–884. doi: 10.1016/j.immuni.2009.09.018. [DOI] [PubMed] [Google Scholar]
- Kapadia B.H., Berg R.A., Daley J.A., Fritz J., Bhave A., Mont M.A. Periprosthetic joint infection. Lancet. 2016;387:386–394. doi: 10.1016/S0140-6736(14)61798-0. [DOI] [PubMed] [Google Scholar]
- Kerr D.E., Plaut K., Bramley A.J., Williamson C.M., Lax A.J., Moore K., Wells K.D., Wall R.J. Lysostaphin expression in mammary glands confers protection against staphylococcal infection in transgenic mice. Nat. Biotechnol. 2001;19:66–70. doi: 10.1038/83540. [DOI] [PubMed] [Google Scholar]
- Khosravi A., Yáñez A., Price J.G., Chow A., Merad M., Goodridge H.S., Mazmanian S.K. Gut microbiota promote hematopoiesis to control bacterial infection. Cell Host Microbe. 2014;15:374–381. doi: 10.1016/j.chom.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.H., Lee S.R., Li L.H., Park H.J., Park J.H., Lee K.Y., Kim M.K., Shin B.A., Choi S.Y. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS ONE. 2011;6:e18556. doi: 10.1371/journal.pone.0018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiri N., Archer G., Climo M.W. Combinations of lysostaphin with beta-lactams are synergistic against oxacillin-resistant Staphylococcus epidermidis. Antimicrob. Agents Chemother. 2002;46:2017–2020. doi: 10.1128/AAC.46.6.2017-2020.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein E., Smith D.L., Laxminarayan R. Hospitalizations and deaths caused by methicillin-resistant Staphylococcus aureus, United States, 1999-2005. Emerg. Infect. Dis. 2007;13:1840–1846. doi: 10.3201/eid1312.070629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knappskog S., Ravneberg H., Gjerdrum C., Trösse C., Stern B., Pryme I.F. The level of synthesis and secretion of Gaussia princeps luciferase in transfected CHO cells is heavily dependent on the choice of signal peptide. J. Biotechnol. 2007;128:705–715. doi: 10.1016/j.jbiotec.2006.11.026. [DOI] [PubMed] [Google Scholar]
- Kokai-Kun J.F., Walsh S.M., Chanturiya T., Mond J.J. Lysostaphin cream eradicates Staphylococcus aureus nasal colonization in a cotton rat model. Antimicrob. Agents Chemother. 2003;47:1589–1597. doi: 10.1128/AAC.47.5.1589-1597.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokai-Kun J.F., Chanturiya T., Mond J.J. Lysostaphin as a treatment for systemic Staphylococcus aureus infection in a mouse model. J. Antimicrob. Chemother. 2007;60:1051–1059. doi: 10.1093/jac/dkm347. [DOI] [PubMed] [Google Scholar]
- Kokai-Kun J.F., Chanturiya T., Mond J.J. Lysostaphin eradicates established Staphylococcus aureus biofilms in jugular vein catheterized mice. J. Antimicrob. Chemother. 2009;64:94–100. doi: 10.1093/jac/dkp145. [DOI] [PubMed] [Google Scholar]
- Kowarz E., Löscher D., Marschalek R. Optimized Sleeping Beauty transposons rapidly generate stable transgenic cell lines. Biotechnol. J. 2015;10:647–653. doi: 10.1002/biot.201400821. [DOI] [PubMed] [Google Scholar]
- Kusuma C.M., Kokai-Kun J.F. Comparison of four methods for determining lysostaphin susceptibility of various strains of Staphylococcus aureus. Antimicrob. Agents Chemother. 2005;49:3256–3263. doi: 10.1128/AAC.49.8.3256-3263.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Liu Y., Hao W., Wolf L., Kiliaan A.J., Penke B., Rübe C.E., Walter J., Heneka M.T., Hartmann T. TLR2 is a primary receptor for Alzheimer’s amyloid β peptide to trigger neuroinflammatory activation. J. Immunol. 2012;188:1098–1107. doi: 10.4049/jimmunol.1101121. [DOI] [PubMed] [Google Scholar]
- Loew R., Heinz N., Hampf M., Bujard H., Gossen M. Improved Tet-responsive promoters with minimized background expression. BMC Biotechnol. 2010;10:81. doi: 10.1186/1472-6750-10-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston H.D., Dixon D.M., Knisely J.M., Palmore T.N., Fauci A.S. Antimicrobial Resistance. JAMA. 2016;316:1193–1204. doi: 10.1001/jama.2016.11764. [DOI] [PubMed] [Google Scholar]
- McCaig L.F., McDonald L.C., Mandal S., Jernigan D.B. Staphylococcus aureus-associated skin and soft tissue infections in ambulatory care. Emerg. Infect. Dis. 2006;12:1715–1723. doi: 10.3201/eid1211.060190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed N., Wang M.Y., Le Huec J.C., Liljenqvist U., Scully I.L., Baber J., Begier E., Jansen K.U., Gurtman A., Anderson A.S. Vaccine development to prevent Staphylococcus aureus surgical-site infections. Br. J. Surg. 2017;104:e41–e54. doi: 10.1002/bjs.10454. [DOI] [PubMed] [Google Scholar]
- Nilsen N.J., Deininger S., Nonstad U., Skjeldal F., Husebye H., Rodionov D., von Aulock S., Hartung T., Lien E., Bakke O., Espevik T. Cellular trafficking of lipoteichoic acid and Toll-like receptor 2 in relation to signaling: role of CD14 and CD36. J. Leukoc. Biol. 2008;84:280–291. doi: 10.1189/jlb.0907656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowakowska J., Landmann R., Khanna N. Foreign body infection models to study host-pathogen response and antimicrobial tolerance of bacterial biofilm. Antibiotics (Basel) 2014;3:378–397. doi: 10.3390/antibiotics3030378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orive G., Santos E., Poncelet D., Hernández R.M., Pedraz J.L., Wahlberg L.U., De Vos P., Emerich D. Cell encapsulation: technical and clinical advances. Trends Pharmacol. Sci. 2015;36:537–546. doi: 10.1016/j.tips.2015.05.003. [DOI] [PubMed] [Google Scholar]
- Patron R.L., Climo M.W., Goldstein B.P., Archer G.L. Lysostaphin treatment of experimental aortic valve endocarditis caused by a Staphylococcus aureus isolate with reduced susceptibility to vancomycin. Antimicrob. Agents Chemother. 1999;43:1754–1755. doi: 10.1128/aac.43.7.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S.Y., You R.I., Lai M.J., Lin N.T., Chen L.K., Chang K.C. Highly potent antimicrobial modified peptides derived from the Acinetobacter baumannii phage endolysin LysAB2. Sci. Rep. 2017;7:11477. doi: 10.1038/s41598-017-11832-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Placencia F.X., Kong L., Weisman L.E. Treatment of methicillin-resistant Staphylococcus aureus in neonatal mice: lysostaphin versus vancomycin. Pediatr. Res. 2009;65:420–424. doi: 10.1203/PDR.0b013e3181994a53. [DOI] [PubMed] [Google Scholar]
- Proctor R.A. Challenges for a universal Staphylococcus aureus vaccine. Clin. Infect. Dis. 2012;54:1179–1186. doi: 10.1093/cid/cis033. [DOI] [PubMed] [Google Scholar]
- Puhto A.P., Puhto T.M., Niinimäki T.T., Leppilahti J.I., Syrjälä H.P. Two-stage revision for prosthetic joint infection: outcome and role of reimplantation microbiology in 107 cases. J. Arthroplasty. 2014;29:1101–1104. doi: 10.1016/j.arth.2013.12.027. [DOI] [PubMed] [Google Scholar]
- Rybak M.J., Lomaestro B.M., Rotschafer J.C., Moellering R.C., Craig W.A., Billeter M., Dalovisio J.R., Levine D.P. Vancomycin therapeutic guidelines: a summary of consensus recommendations from the infectious diseases Society of America, the American Society of Health-System Pharmacists, and the Society of Infectious Diseases Pharmacists. Clin. Infect. Dis. 2009;49:325–327. doi: 10.1086/600877. [DOI] [PubMed] [Google Scholar]
- Santos E., Larzabal L., Calvo A., Orive G., Pedraz J.L., Hernández R.M. Inactivation of encapsulated cells and their therapeutic effects by means of TGL triple-fusion reporter/biosafety gene. Biomaterials. 2013;34:1442–1451. doi: 10.1016/j.biomaterials.2012.10.076. [DOI] [PubMed] [Google Scholar]
- Schaffner W., Melly M.A., Koenig M.G. Lysostaphin: an enzymatic approach to staphylococcal disease. II. In vivo studies. Yale J. Biol. Med. 1967;39:230–244. [PMC free article] [PubMed] [Google Scholar]
- Schindler C.A., Schuhardt V.T. Lysostaphin: A new bacteriolytic agent for the Staphylococcus. Proc. Natl. Acad. Sci. USA. 1964;51:414–421. doi: 10.1073/pnas.51.3.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlabach M.R., Hu J.K., Li M., Elledge S.J. Synthetic design of strong promoters. Proc. Natl. Acad. Sci. USA. 2010;107:2538–2543. doi: 10.1073/pnas.0914803107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlatter S., Rimann M., Kelm J., Fussenegger M. SAMY, a novel mammalian reporter gene derived from Bacillus stearothermophilus alpha-amylase. Gene. 2002;282:19–31. doi: 10.1016/s0378-1119(01)00824-1. [DOI] [PubMed] [Google Scholar]
- Schleifer K.H., Fischer U. Description of a new species of the genus Staphylococcus: Staphylococcus carnosus. Int. J. Syst. Bacteriol. 1982;32:153–156. [Google Scholar]
- Shah K., McCormack C.E., Bradbury N.A. Do you know the sex of your cells? Am. J. Physiol. Cell Physiol. 2014;306:C3–C18. doi: 10.1152/ajpcell.00281.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen J.L., Rosada C., Serakinci N., Justesen J., Stenderup K., Rattan S.I.S., Jensen T.G., Kassem M. Telomerase expression extends the proliferative life-span and maintains the osteogenic potential of human bone marrow stromal cells. Nat. Biotechnol. 2002;20:592–596. doi: 10.1038/nbt0602-592. [DOI] [PubMed] [Google Scholar]
- Smeds M.R., Duncan A.A., Harlander-Locke M.P., Lawrence P.F., Lyden S., Fatima J., Eskandari M.K., Vascular Low-Frequency Disease Consortium Treatment and outcomes of aortic endograft infection. J. Vasc. Surg. 2016;63:332–340. doi: 10.1016/j.jvs.2015.08.113. [DOI] [PubMed] [Google Scholar]
- Spellberg B., Bartlett J.G., Gilbert D.N. The future of antibiotics and resistance. N. Engl. J. Med. 2013;368:299–302. doi: 10.1056/NEJMp1215093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark F.R., Thornsvard C., Flannery E.P., Artenstein M.S. Systemic lysostaphin in man--apparent antimicrobial activity in a neutropenic patient. N. Engl. J. Med. 1974;291:239–240. doi: 10.1056/NEJM197408012910507. [DOI] [PubMed] [Google Scholar]
- Strauss A., Thumm G., Götz F. Influence of Lif, the lysostaphin immunity factor, on acceptors of surface proteins and cell wall sorting efficiency in Staphylococcus carnosus. J. Bacteriol. 1998;180:4960–4962. doi: 10.1128/jb.180.18.4960-4962.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stryjewski M.E., Corey G.R. Methicillin-resistant Staphylococcus aureus: an evolving pathogen. Clin. Infect. Dis. 2014;58(Suppl 1):S10–S19. doi: 10.1093/cid/cit613. [DOI] [PubMed] [Google Scholar]
- Sutherland I.W. The biofilm matrix--an immobilized but dynamic microbial environment. Trends Microbiol. 2001;9:222–227. doi: 10.1016/s0966-842x(01)02012-1. [DOI] [PubMed] [Google Scholar]
- Szymczak A.L., Workman C.J., Wang Y., Vignali K.M., Dilioglou S., Vanin E.F., Vignali D.A.A. Correction of multi-gene deficiency in vivo using a single ‘self-cleaving’ 2A peptide-based retroviral vector. Nat. Biotechnol. 2004;22:589–594. doi: 10.1038/nbt957. [DOI] [PubMed] [Google Scholar]
- Szymczak-Workman A.L., Vignali K.M., Vignali D.A. Design and construction of 2A peptide-linked multicistronic vectors. Cold Spring Harb. Protoc. 2012;2012:199–204. doi: 10.1101/pdb.ip067876. [DOI] [PubMed] [Google Scholar]
- Tannous B.A., Kim D.E., Fernandez J.L., Weissleder R., Breakefield X.O. Codon-optimized Gaussia luciferase cDNA for mammalian gene expression in culture and in vivo. Mol. Ther. 2005;11:435–443. doi: 10.1016/j.ymthe.2004.10.016. [DOI] [PubMed] [Google Scholar]
- Tong S.Y., Davis J.S., Eichenberger E., Holland T.L., Fowler V.G., Jr. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015;28:603–661. doi: 10.1128/CMR.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trampuz A., Piper K.E., Jacobson M.J., Hanssen A.D., Unni K.K., Osmon D.R., Mandrekar J.N., Cockerill F.R., Steckelberg J.M., Greenleaf J.F., Patel R. Sonication of removed hip and knee prostheses for diagnosis of infection. N. Engl. J. Med. 2007;357:654–663. doi: 10.1056/NEJMoa061588. [DOI] [PubMed] [Google Scholar]
- Vegas A.J., Veiseh O., Doloff J.C., Ma M., Tam H.H., Bratlie K., Li J., Bader A.R., Langan E., Olejnik K. Combinatorial hydrogel library enables identification of materials that mitigate the foreign body response in primates. Nat. Biotechnol. 2016;34:345–352. doi: 10.1038/nbt.3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vegas A.J., Veiseh O., Gürtler M., Millman J.R., Pagliuca F.W., Bader A.R., Doloff J.C., Li J., Chen M., Olejnik K. Long-term glycemic control using polymer-encapsulated human stem cell-derived beta cells in immune-competent mice. Nat. Med. 2016;22:306–311. doi: 10.1038/nm.4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Eiff C., Kokai-Kun J.F., Becker K., Peters G. In vitro activity of recombinant lysostaphin against Staphylococcus aureus isolates from anterior nares and blood. Antimicrob. Agents Chemother. 2003;47:3613–3615. doi: 10.1128/AAC.47.11.3613-3615.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh S., Shah A., Mond J. Improved pharmacokinetics and reduced antibody reactivity of lysostaphin conjugated to polyethylene glycol. Antimicrob. Agents Chemother. 2003;47:554–558. doi: 10.1128/AAC.47.2.554-558.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A., Athan E., Pappas P.A., Fowler V.G., Jr., Olaison L., Paré C., Almirante B., Muñoz P., Rizzi M., Naber C., International Collaboration on Endocarditis-Prospective Cohort Study Investigators Contemporary clinical profile and outcome of prosthetic valve endocarditis. JAMA. 2007;297:1354–1361. doi: 10.1001/jama.297.12.1354. [DOI] [PubMed] [Google Scholar]
- Xie M., Ye H., Wang H., Charpin-El Hamri G., Lormeau C., Saxena P., Stelling J., Fussenegger M. β-cell-mimetic designer cells provide closed-loop glycemic control. Science. 2016;354:1296–1301. doi: 10.1126/science.aaf4006. [DOI] [PubMed] [Google Scholar]
- Ye H., Xie M., Xue S., Charpin-El Hamri G., Yin J., Zulewski H., Fussenegger M. Self-adjusting synthetic gene circuit for correcting insulin resistance. Nat. Biomed. Eng. 2017;1:0005. doi: 10.1038/s41551-016-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Leng Q., Mixson A.J. Alteration in the IL-2 signal peptide affects secretion of proteins in vitro and in vivo. J. Gene Med. 2005;7:354–365. doi: 10.1002/jgm.677. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.