Abstract
The correlation between miR-200 family overexpression and cancer prognosis remains controversial. Therefore, we conducted a systematic review and meta-analysis by searching PubMed, Embase, Cochrane Library, China Biology Medicine disc (CBM), and China National Knowledge Infrastructure (CNKI) to identify eligible studies. Hazard ratios (HRs) with 95% confidence intervals (CIs) were calculated to evaluate the strength of the correlations. Additionally, different subgroup analyses and publication bias test were performed. Eventually, we analyzed 23 articles that included five tumor types and 3038 patients. Consequently, high expression of miR-200 family in various tumors was associated with unfavorable overall survival (OS) in both univariate (HR = 1.32, 95% CI: 1.14–1.54, P < 0.001) and multivariate (HR = 1.32, 95% CI: 1.16–1.49, P < 0.001) analyses. Likewise, a similar result was found in different subgroups of the patient source, cancer type, test method, sample source, miR-200 component, and sample size. However, no association of miR-200 family was detected with recurrence- or relapse-free survival (RFS) (univariate: HR = 1.02, 95% CI: 0.96–1.09, P = 0.47; multivariate: HR = 1.07, 95% CI: 1.00–1.14, P = 0.07), progression-free survival (PFS) (univariate: HR = 0.96, 95% CI: 0.54–1.70, P = 0.88; multivariate: HR = 1.17, 95% CI: 0.86–1.61, P = 0.32), and disease-free survival (DFS) (univariate: HR = 0.90, 95% CI: 0.74–1.09, P = 0.29; multivariate: HR = 0.98, 95% CI: 0.68–1.41, P = 0.90). Our findings have provided convincing evidence that miR-200 family overexpression suggested poor prognosis of various cancer types, which efforts may raise the potential use of miR-200 family for cancer prognosis in clinical practice.
1. Introduction
MicroRNAs (miRNAs) are evolutionarily conserved, endogenous small noncoding, and single-stranded RNAs of 18–22 nucleotides in length. They often negatively regulate gene targets by translational inhibition or mRNA degradation [1, 2]. It has been revealed that the posttranscriptional regulation could influence various biological processes including apoptosis, differentiation, proliferation, stress response, and metabolism [3, 4]. miRNAs could also be able to predict cancer prognosis due to their crucial roles in cancer progression and metastasis. Previous studies have explored that deregulated miRNAs with aberrant expression levels were closely correlated with cancer prognosis and even could be a novel kind of biomarkers for various cancer types [5, 6].
Interestingly, the miR-200 family is a typical and most extensively studied example in functional miRNAs. The miR-200 family, composed of five miRNA sequences (miR-141, miR-200a, miR-200b, miR-200c, and miR-429) and located in two clusters in the genome, is involved in the epithelial to mesenchymal transition (EMT) through regulation of E-cadherin expression via suppression of ZEB1 and ZEB2 [7, 8]. Recent studies have reported that miR-200 cluster is overexpressed in different tumors and played a critical role in mRNA degradation or inhibition through targeted binding to the relevant 3′-untranslated region (UTR) [9]. miR-200 family has been shown to offer a great potential in both cancer diagnosis and prognosis. Despite the potential roles of miR-200 family high expression in prognosis for cancer patients that have been attempted, no definite conclusions have been drawn so far. Meta-analysis can explore the authentic and comprehensive results through incorporating all available evidences to get a relatively precise and accurate estimation by using statistical analyses [10]. Thus, we have performed the current meta-analysis to explore the potential associations between miR-200 family and cancer prognosis, which efforts should hold great promise in verifying the potential of miRNAs as biomarkers for evaluating therapeutic efficacy and prognosis of various cancers.
2. Methods
2.1. Ethics Statement
The PRISMA statement was used to conduct the current meta-analysis [11]. No patient's privacy or clinical samples were involved in this study; hence, the ethical approval was not required.
2.2. Search Strategy
Literature resources including PubMed, Cochrane Library, Embase, CBM, and CNK were introduced to search eligible studies, by using the terms “microRNA OR miRNA OR miR-200 OR miR-141 OR miR-429 OR miR-200 family OR miR-200 cluster,” “survival OR prognosis OR prognostic,” and “cancer OR tumor OR tumour OR neoplasm OR neoplasma OR neoplasia OR carcinoma OR cancers OR tumors OR tumours OR neoplasms OR neoplasmas OR neoplasias OR carcinomas.” Last search of current investigation was updated on November 25th, 2017. Additionally, the publication language was only limited to English and Chinese. In case of omission, we identified the reference lists of the relevant articles and reviewed articles to seek for the potentially relevant studies. Conventionally, we have not contacted the corresponding authors even if the relevant data were unavailable.
2.3. Inclusion and Exclusion Criteria
Studies complied with the following criteria could be identified: (1) clinical study about the association of miR-200 family with cancer prognosis and (2) relevant data of the hazard ratios (HRs) and their corresponding 95% confidence intervals (CIs) to evaluate its associations were available. Studies which met the following four criteria were excluded: (1) the available data regarding associations was absent; (2) similar or duplicate study (when the same or similar cohort was applied, after careful examination, the most complete information was included); (3) other types of articles such as reviews or abstracts; and (4) studies involved with cell lines or animal models.
2.4. Data Extraction
In the light of inclusion and exclusion criteria, we extracted the relevant data from each eligible study. If disagreements were noticed, we are clearly open to discussion by each other (Wen Liu and Kaiping Zhang) or reviewed by a third author (Pengfei Wei). The data on first author, publication year, study country, age, cancer type, miRNA category, sample source, sample size, follow-up time, test method, survival outcome, analysis method, HR (95% CI), and the cut-off value were extracted. We have not contacted any author of the original researches even if the essential information could not be available. Besides, patient sources came from Asia, Europe, and North America. Sample sources were stratified into tissue, blood, formalin-fixed and paraffin-embedded (FFPE), and tissue microarray (TMA). Test methods included TaqMan, in situ hybridization (ISH), and reverse transcription polymerase chain reaction (RT-PCR). Sample sizes were separated into ≥100 and <100. Cancer types included epithelial ovarian cancer (EOC), breast cancer (BC), nonsmall cell lung cancer (NSCLC), gastric cancer (GC), and colorectal cancer (CRC). Analyses methods were divided into univariate analysis and multivariate analysis. Patients' prognostic outcomes included overall survival (OS), relapse-free survival (RFS), progression-free survival (PFS), and disease-free survival (DFS).
3. Statistical Analysis
We have explored the association of miR-200 family with cancer prognosis by applying Review Manager software (RevMan 5, The Cochrane Collaboration, Oxford, UK) and Stata software (Version 12.0, Stata Corporation, College Station, TX). HR and 95% CI were collected for assessing the prognostic value of high expression of miR-200 family in various cancers. Meanwhile, the heterogeneity has been assessed via chi-square-based Q and I2 test across studies (no heterogeneity I2 < 25%, moderate heterogeneity I2 = 25%–50%, extreme heterogeneity I2 > 50%) [12]. In case of extreme heterogeneity (I2 > 50% or P < 0.01 for Q test), we used random-effects (DerSimonian and Laird method) model [13]. Otherwise, fixed-effects (Mantel-Haenszel method) model was introduced [14]. One-way sensitivity analyses which individually removed publications in meta-analysis were conducted to assess results' stability. It mainly explores the impact of specific study upon mixed HR. In Begg's funnel plots, logHR was plotted against SE. P value less than 0.05 indicated that there was a bias of the study [15]. Additionally, different subgroups consisted of patient source, cancer type, test method, sample source, sample size, and miR-200 component were conducted.
4. Results
4.1. Characteristics of the Studies
Consequently, 23 studies consisted of 3038 samples satisfied the eligible criteria [16–38] (Figure 1).
Figure 1.
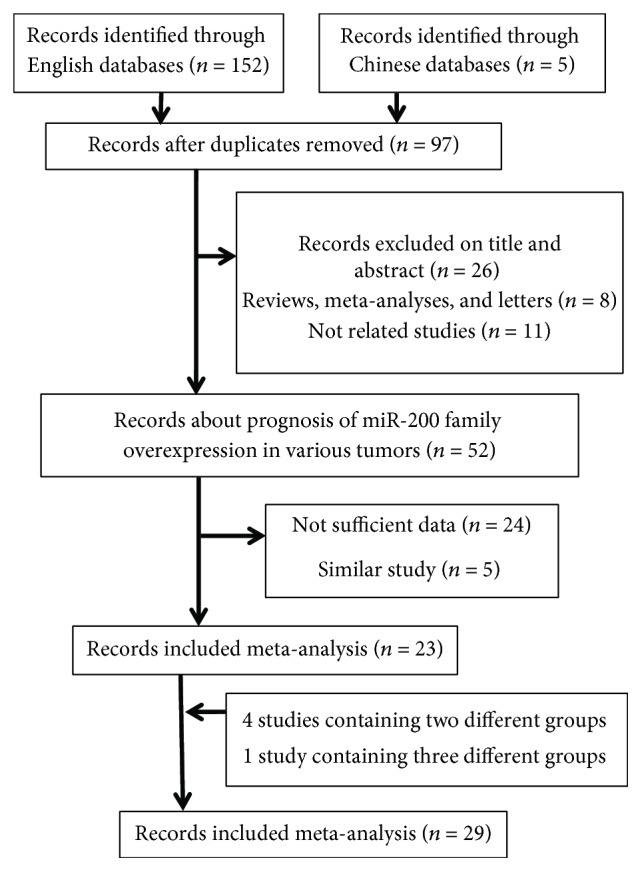
Flow diagram of the study selection process in the meta-analysis.
The principal characteristics of the eligible studies were summarized in Table 1.
Table 1.
Main characteristics of the eligible studies.
| First author | Year | Country | Age | Cancer type | MicroRNA | Sample size | Follow-up, median (range) | Outcome |
|---|---|---|---|---|---|---|---|---|
| Zou J. [16] | 2017 | China | NA | EOC | miR-429 | 72 | NA | OS/PFS |
| Han Y. [17] | 2017 | China | NA | CRC | miR-429 | 71 | 34.2 | OS |
| Maierthaler M. [18] | 2017 | Germany | 70 (33–92) 68.0 (36–92) |
CRC | miR-200a, miR-200b, miR-200c, miR-141, miR-429 | 527 | NA | OS/RFS |
| Si L. [19] | 2017 | China | 60.5 (41–78) | NSCLC | miR-200c | 110 | NA | OS/DFS |
| Meng X. [20] | 2016 | Germany | 60 (23–91) | EOC | miR-200a, miR-200b, miR-200c | 163 | 20 (1–136) | OS/RFS |
| Dong S. J. [21] | 2016 | China | 56 (31–79) | CRC | miR-429 | 116 | NA | OS |
| Antolín S. [22] | 2015 | Spain | 54.8 (29–73) | BC | miR-200c, miR-141 | 57 | 74.6 (74.2–77.7) | OS/PFS |
| Gao Y. C. [23] | 2015 | China | NA | EOC | miR-200c, miR-141 | 93 | NA | OS |
| Lu Y. B. [24] | 2015 | China | NA | GC | miR-141 | 95 | NA | OS |
| Liu J. Y. [25] | 2015 | China | 57.48 | EOC | miR-200a | 44 | 26 (5–49) | OS/PFS |
| Cao Q. [26] | 2014 | China | 58 (26–88) | EOC | miR-200a, miR-200b, miR-200c | 100 | 36.8 (6–56) | OS |
| Kim M. K. [27] | 2014 | Korea | 64 (26–77) | NSCLC | miR-200c | 72 | 31 (1–135) | OS |
| Zhu W. [28] | 2014 | China | 59 | NSCLC | miR-429 | 70 | NA | OS |
| Song F. [29] | 2014 | China | 60.5 | GC | miR-200a, miR-200b, miR-200c | 385 | 35 (1–112) | OS/PFS |
| Tejero R. [30] | 2014 | Spain | 65 (35–85) | NSCLC | miR-200c/141 | 155 | 43 (2–160) | OS |
| Toiyama Y. [31] | 2014 | Japan | 67 | CRC | miR-200c | 182 | NA | OS |
| Sun Q. [32] | 2014 | China | NA | EOC | miR-200a | 53 | 56.79 (11–98) | OS |
| Liu X. G. [33] | 2012 | China | NA | NSCLC | miR-200c, miR-141 | 70 | 24 | OS |
| Chao A. [34] | 2012 | China | NA | EOC | miR-200a | 176 | 40 (3–109) | OS/RFS |
| Marchini S. [35] | 2011 | Italy | 52 (21–82) | EOC | miR-200b, miR-200c | 144 | 110.4 (82.8–139.2) | OS/PFS |
| Cheng H. [36] | 2011 | USA | NA | CRC | miR-141 | 156 | NA | OS |
| Leskelä S. [37] | 2010 | Spain | 57 (35–85) | EOC | miR-200a, miR-200b, miR-200c, miR-141, miR-429 | 72 | NA | OS/PFS/RFS |
| Hu X. [38] | 2009 | USA | 58.3 | EOC | miR-200a | 55 | NA | OS/PFS |
NA: not available; EOC: epithelial ovarian cancer; BC: breast cancer; NSCLC: nonsmall cell lung cancer; GC: gastric cancer; CRC: colorectal cancer; OS: overall survival; DFS: disease-free survival; PFS: progression-free survival; RFS: recurrence- or relapse-free survival; HR: hazard ratio; CI: confidence interval.
Among these studies, Cheng's study was involved with three different cohorts of Tianjin cohort, TexGen cohort, and all cohort [36]. Zhu et al. designed a study to detect tissue and serum miRNA expression [28]. Tejero et al. analyzed the role of members of the miR-200 family from NSCLC patients after surgery both in the entire cohort and adenocarcinoma cohort [30]. Maierthaler et al. explore miRNA expression in two different cohorts of nonmetastatic and metastatic CRC [18]. Toiyama et al. conducted a study to detect the prognostic value of the miR-200 family in CRC from blood and FFPE samples. As mentioned above, we treated them independently into meta-analysis [31]. Eventually, this meta-analysis was established based on 29 studies (Table 2). Among these 29 studies, 28 were written in English while one was published in Chinese. The sample sizes ranged from 44 to 527. The cancer types contained ten EOC, one BC, seven NSCLC, two GC, and nine CRC. Meanwhile, one ISH, 24 RT-PCR, and four TaqMan in test methods were applied. According to the sample sources, there were seven FFPE, ten tissue, ten blood, and two TMA. For the survival outcomes, 29 eligible studies were divided into 42 datasets: 29 for OS, six for PFS, five for RFS, and two for DFS. However, the cut-off value for the miR-200 family was inconsistent among these included studies (Table 2).
Table 2.
MicroRNA evaluation and survival data of the selected studies.
| First author | Year | Country | Test method | Cancer type | MicroRNA | Sample source | Outcome | HR (95% CI) | Cut-off value |
|---|---|---|---|---|---|---|---|---|---|
| Zou J. | 2017 | China | RT-PCR | EOC | miR-429 | Tissue | OS | (U) 0.641 (0.412–0.996)/(M) 0.763 (0.458–1.270) | >0.532 |
| Zou J. | 2017 | China | RT-PCR | EOC | miR-429 | Tissue | PFS | (U) 0.661 (0.478–0.915)/(M) 0.710 (0.504–1.001) | |
|
| |||||||||
| Han Y. | 2017 | China | RT-PCR | CRC | miR-429 | Tissue | OS | (M) 1.852 (1.019–3.326) | Median |
|
| |||||||||
| Maierthaler M.-1 | 2017 | Germany | TaqMan | CRC | miR-200a | Blood | OS | (U) 0.929 (0.707–1.211)/(M) 1.053 (0.791–1.401) | Median |
| Maierthaler M.-1 | 2017 | Germany | TaqMan | CRC | miR-200b | Blood | OS | (U) 0.704 (0.524–0.945)/(M) 0.772 (0.570–1.045) | |
| Maierthaler M.-1 | 2017 | Germany | TaqMan | CRC | miR-200c | Blood | OS | (U) 0.808 (0.646–1.010)/(M) 0.840 (0.659–1.070) | |
| Maierthaler M.-1 | 2017 | Germany | TaqMan | CRC | miR-141 | Blood | OS | (U) 0.925 (0.713–1.200)/(M) 1.038 (0.785–1.374) | |
| Maierthaler M.-1 | 2017 | Germany | TaqMan | CRC | miR-429 | Blood | OS | (U) 0.951 (0.734–1.235)/(M) 0.968 (0.721–1.300) | |
| Maierthaler M.-2 | 2017 | Germany | TaqMan | CRC | miR-200a | Blood | OS | (U) 1.198 (0.986–1.456)/(M) 1.227 (1.008–1.495) | |
| Maierthaler M.-2 | 2017 | Germany | TaqMan | CRC | miR-200b | Blood | OS | (U) 1.172 (0.946–1.453)/(M) 1.208 (0.975–1.497) | |
| Maierthaler M.-2 | 2017 | Germany | TaqMan | CRC | miR-200c | Blood | OS | (U) 1.117 (0.947–1.318)/(M) 1.152 (0.975–1.362) | |
| Maierthaler M.-2 | 2017 | Germany | TaqMan | CRC | miR-141 | Blood | OS | (U) 1.071 (0.877–1.305)/(M) 1.105 (0.904–1.350) | |
| Maierthaler M.-2 | 2017 | Germany | TaqMan | CRC | miR-429 | Blood | OS | (U) 1.010 (0.853–1.196)/(M) 1.006 (0.845–1.198) | |
| Maierthaler M.-1 | 2017 | Germany | TaqMan | CRC | miR-200a | Blood | RFS | (U) 0.929 (0.718–1.203)/(M) 1.031 (0.786–1.353) | |
| Maierthaler M.-1 | 2017 | Germany | TaqMan | CRC | miR-200b | Blood | RFS | (U) 0.714 (0.539–0.947)/(M) 0.750 (0.561–1.005) | |
| Maierthaler M.-1 | 2017 | Germany | TaqMan | CRC | miR-200c | Blood | RFS | (U) 0.819 (0.657–1.019)/(M) 0.835 (0.658–1.060) | |
| Maierthaler M.-1 | 2017 | Germany | TaqMan | CRC | miR-141 | Blood | RFS | (U) 0.910 (0.705–1.175)/(M) 0.999 (0.760–1.312) | |
| Maierthaler M.-1 | 2017 | Germany | TaqMan | CRC | miR-429 | Blood | RFS | (U) 0.954 (0.743–1.227)/(M) 1.076 (0.716–1.618) | |
| Maierthaler M.-2 | 2017 | Germany | TaqMan | CRC | miR-200a | Blood | RFS | (U) 1.175 (0.973–1.420)/(M) 1.200 (0.989–1.456) | |
| Maierthaler M.-2 | 2017 | Germany | TaqMan | CRC | miR-200b | Blood | RFS | (U) 1.109 (0.893–1.377)/(M) 1.143 (0.919–1.422) | |
| Maierthaler M.-2 | 2017 | Germany | TaqMan | CRC | miR-200c | Blood | RFS | (U) 1.076 (0.911–1.272)/(M) 1.100 (0.930–1.302) | |
| Maierthaler M.-2 | 2017 | Germany | TaqMan | CRC | miR-141 | Blood | RFS | (U) 1.057 (0.871–1.284)/(M) 1.085 (0.890–1.321) | |
| Maierthaler M.-2 | 2017 | Germany | TaqMan | CRC | miR-429 | Blood | RFS | (U) 1.080 (0.916–1.272)/(M) 1.078 (0.910–1.277) | |
|
| |||||||||
| Si L. | 2017 | China | RT-PCR | NSCLC | miR-200c | Tissue | OS | (M) 2.095 (1.241–3.536) | The 2−ΔΔCq |
| Si L. | 2017 | China | RT-PCR | NSCLC | miR-200c | Tissue | DFS | (M) 1.647 (1.049–2.585) | |
|
| |||||||||
| Meng X. | 2016 | Germany | RT-PCR | EOC | miR-200a | Blood | OS | (U) 1.7 (0.8–3.5) | Median |
| Meng X. | 2016 | Germany | RT-PCR | EOC | miR-200b | Blood | OS | (U) 2.7 (1.3–5.7)/(M) 2.8 (1.1–6.8) | |
| Meng X. | 2016 | Germany | RT-PCR | EOC | miR-200c | Blood | OS | (U) 2.4 (1.2–4.9)/(M) 2.5 (1.1–6.1) | |
| Meng X. | 2016 | Germany | RT-PCR | EOC | miR-200a | Blood | RFS | (U) 1.1 (0.6–1.9) | |
| Meng X. | 2016 | Germany | RT-PCR | EOC | miR-200b | Blood | RFS | (U) 1.6 (0.9–2.8) | |
| Meng X | 2016 | Germany | RT-PCR | EOC | miR-200c | Blood | RFS | (U) 2.0 (1.1–3.6)/(M) 1.7 (0.8–3.6) | |
|
| |||||||||
| Dong S. J. | 2016 | China | RT-PCR | CRC | miR-429 | Tissue | OS | (M) 2.296 (1.105–4.528) | Median |
|
| |||||||||
| Antolín S. | 2015 | Spain | RT-PCR | BC | miR-200c | Blood | OS | (U) 1.38 (1.11–1.71)/(M) 2.79 (1.01–7.7) | >1.29 relative expression value |
| Antolín S. | 2015 | Spain | RT-PCR | BC | miR-200c | Blood | PFS | (U) 1.37 (1.09–1.71)/(M) 3.33 (1.22–9.07) | |
| Antolín S. | 2015 | Spain | RT-PCR | BC | miR-141 | Blood | OS | (M) 0.986 (0.942–1.032) | |
| Antolín S. | 2015 | Spain | RT-PCR | BC | miR-141 | Blood | PFS | (M) 0.987 (0.95–1.025) | |
|
| |||||||||
| Gao Y. C. | 2015 | China | RT-PCR | EOC | miR-200c | Blood | OS | (U) 3.14 (1.67–5.93) | −ΔCt method with 95% CI |
| Gao Y. C. | 2015 | China | RT-PCR | EOC | miR-141 | Blood | OS | (U) 1.83 (1.00–3.33) | |
|
| |||||||||
| Lu Y. B. | 2015 | China | RT-PCR | GC | miR-141 | Tissue | OS | (M) 2.972 (1.297–10.001) | Median |
|
| |||||||||
| Liu J. Y. | 2015 | China | RT-PCR | EOC | miR-200a | Tissue | OS | (M) 0.354 (0.149–0.840) | Log 2−ΔΔCt |
| Liu J. Y. | 2015 | China | RT-PCR | EOC | miR-200a | Tissue | PFS | (M) 0.395 (0.210–0.742) | |
|
| |||||||||
| Cao Q | 2014 | China | ISH | EOC | miR-200a | Tissue | OS | (U) 22.69 (1.32–50.53)/(M) 17.26 (1.36–36.98) | Median |
| Cao Q. | 2014 | China | ISH | EOC | miR-200b | Tissue | OS | (U) 20.28 (1.20–42.28)/(M)15.41 (1.13–31.36) | |
| Cao Q. | 2014 | China | ISH | EOC | miR-200c | Tissue | OS | (U) 21.42 (1.26–48.33)/(M) 16.22 (1.27–33.81) | |
|
| |||||||||
| Kim M. K. | 2014 | Korea | RT-PCR | NSCLC | miR-200c | FFPE | OS | (M) 3.67 (1.17–11.45) | Median |
|
| |||||||||
| Zhu W.-1 | 2014 | China | RT-PCR | NSCLC | miR-429 | Tissue | OS | (U) 1.686 (0.570–4.984)/(M) 2.749 (0.706–10.707) | Mean |
| Zhu W.-2 | 2014 | China | RT-PCR | NSCLC | miR-429 | Blood | OS | (U) 6.458 (1.409–29.593)/(M) 12.875 (2.295–72.23) | |
|
| |||||||||
| Song F. | 2014 | China | RT-PCR | GC | miR-200a | TMA | OS | (U) 0.82 (0.57–1.20)/(M) 0.72 (0.47–1.13) | Median |
| Song F. | 2014 | China | RT-PCR | GC | miR-200b | TMA | OS | (U) 0.87 (0.60–1.26)/(M)0.93 (0.63–1.41) | |
| Song F. | 2014 | China | RT-PCR | GC | miR-200c | TMA | OS | (U) 1.19 (0.80–1.77)/(M) 1.32 (0.82–2.12) | |
| Song F. | 2014 | China | RT-PCR | GC | miR-200a | TMA | DFS | (U) 0.81 (0.58–1.14)/(M) 0.67 (0.45–0.99) | |
| Song F. | 2014 | China | RT-PCR | GC | miR-200b | TMA | DFS | (U) 0.84 (0.60–1.18)/(M) 0.82 (0.56–1.19) | |
| Song F. | 2014 | China | RT-PCR | GC | miR-200c | TMA | DFS | (U) 1.08 (0.76–1.54)/(M) 1.06 (0.70–1.60) | |
|
| |||||||||
| Tejero R.-1 | 2014 | Spain | TaqMan | NSCLC | miR-200c/141 | FFPE | OS | (M) 2.787 (1.087–7.148) | Mean |
| Tejero R.-2 | 2014 | Spain | TaqMan | NSCLC | miR-200c/141 | FFPE | OS | (M) 10.649 (2.433–46.608) | |
|
| |||||||||
| Toiyama Y.-1 | 2014 | Japan | RT-PCR | CRC | miR-200c | Blood | OS | (U) 2.43 (1.26–4.68)/(M)2.67 (1.28–5.67) | Median |
| Toiyama Y.-2 | 2014 | Japan | RT-PCR | CRC | miR-200c | FFPE | OS | (U) 0.56 (0.28–1.10) | |
|
| |||||||||
| Sun Q. | 2014 | China | RT-PCR | EOC | miR-200a | TMA | OS | (U) 0.58 (0.08–4.05) | Median (≥12.623) |
|
| |||||||||
| Liu X. G. | 2012 | China | RT-PCR | NSCLC | miR-200c | Tissue | OS | (U) 6.020 (1.344–26.971) | 2−ΔΔCt > 2.0 |
| Liu X. G. | 2012 | China | RT-PCR | NSCLC | miR-141 | Tissue | OS | (U) 4.135 (0.467–36.597) | |
| Chao A. | 2012 | China | RT-PCR | EOC | miR-200a | FFPE | OS | (M) 1.466 (0.786–2.734) | Log ratio > 1.3 |
| Chao A. | 2012 | China | RT-PCR | EOC | miR-200a | FFPE | RFS | (M) 1.213 (0.70–2.101) | |
|
| |||||||||
| Marchini S. | 2011 | Italy | RT-PCR | EOC | miR-200b | Tissue | OS | (U) 2.137 (0.801–5.701)/(M) 2.051 (0.640–6.570) | >25% |
| Marchini S. | 2011 | Italy | RT-PCR | EOC | miR-200b | Tissue | PFS | (U) 3.197 (1.417–7.213)/(M) 2.335 (0.857–6.363) | |
| Marchini S. | 2011 | Italy | RT-PCR | EOC | miR-200c | Tissue | OS | (U) 0.309 (0.112–0.850)/(M) 0.244 (0.076–0.785) | |
| Marchini S. | 2011 | Italy | RT-PCR | EOC | miR-200c | Tissue | PFS | (U) 0.392 (0.174–0.885)/(M) 0.419 (0.146–1.204) | |
|
| |||||||||
| Cheng H.-1 | 2011 | USA | RT-PCR | CRC | miR-141 | Blood | OS | (U) 3.80 (1.46–9.91)/(M) 1.36 (0.45–4.14) | 2−ΔΔCt |
| Cheng H.-2 | 2011 | USA | RT-PCR | CRC | miR-141 | Blood | OS | (U) 4.83 (2.06–11.35)/(M) 3.41 (1.36–8.56) | |
| Cheng H.-3 | 2011 | USA | RT-PCR | CRC | miR-141 | Blood | OS | (U) 3.61 (1.96–6.65)/(M) 2.40 (1.18–4.86) | |
|
| |||||||||
| Leskelä S. | 2010 | Spain | RT-PCR | EOC | miR-200a | FFPE | PFS | (M) 1.22 (0.57–2.58) | 75% of positive cells |
| Leskelä S. | 2010 | Spain | RT-PCR | EOC | miR-200b | FFPE | PFS | (M) 1.35 (0.62–2.93) | |
| Leskelä S. | 2010 | Spain | RT-PCR | EOC | miR-200c | FFPE | PFS | (M) 2.24 (1.00–5.03) | |
| Leskelä S. | 2010 | Spain | RT-PCR | EOC | miR-141 | FFPE | PFS | (M) 2.35 (0.98–5.59) | |
| Leskelä S. | 2010 | Spain | RT-PCR | EOC | miR-429 | FFPE | PFS | (M) 2.10 (0.92–4.79) | |
| Leskelä S. | 2010 | Spain | RT-PCR | EOC | miR-429 | FFPE | RFS | (M) 2.01 (1.11–3.66) | |
| Leskelä S. | 2010 | Spain | RT-PCR | EOC | miR-429 | FFPE | OS | (M) 2.08 (1.03–4.20) | |
|
| |||||||||
| Hu X. | 2009 | USA | RT-PCR | EOC | miR-200a | FFPE | OS | (U) 0.70 (0.03–14.29) | >11 |
| Hu X. | 2009 | USA | RT-PCR | EOC | miR-200a | FFPE | PFS | (U) 0.64 (0.22–1.81) | |
EOC: epithelial ovarian cancer; BC: breast cancer; NSCLC: nonsmall cell lung cancer; NMIBC: nonmuscle-invasive bladder cancer; GC: gastric cancer; CRC: colorectal cancer; OS: overall survival; DFS: disease-free survival; PFS: progression-free survival; RFS: recurrence- or relapse-free survival; HR: hazard ratio; CI: confidence interval; U: univariate analysis; M: multivariate analysis; ISH: in situ hybridization; RT-PCR: reverse transcription-polymerase chain reaction; FFPE: formalin-fixed and paraffin-embedded; TMA: tissue microarray; OS: overall survival; DFS: disease-free survival; PFS, progression-free survival; RFS: recurrence- or relapse-free survival.
4.2. Meta-Analysis of OS
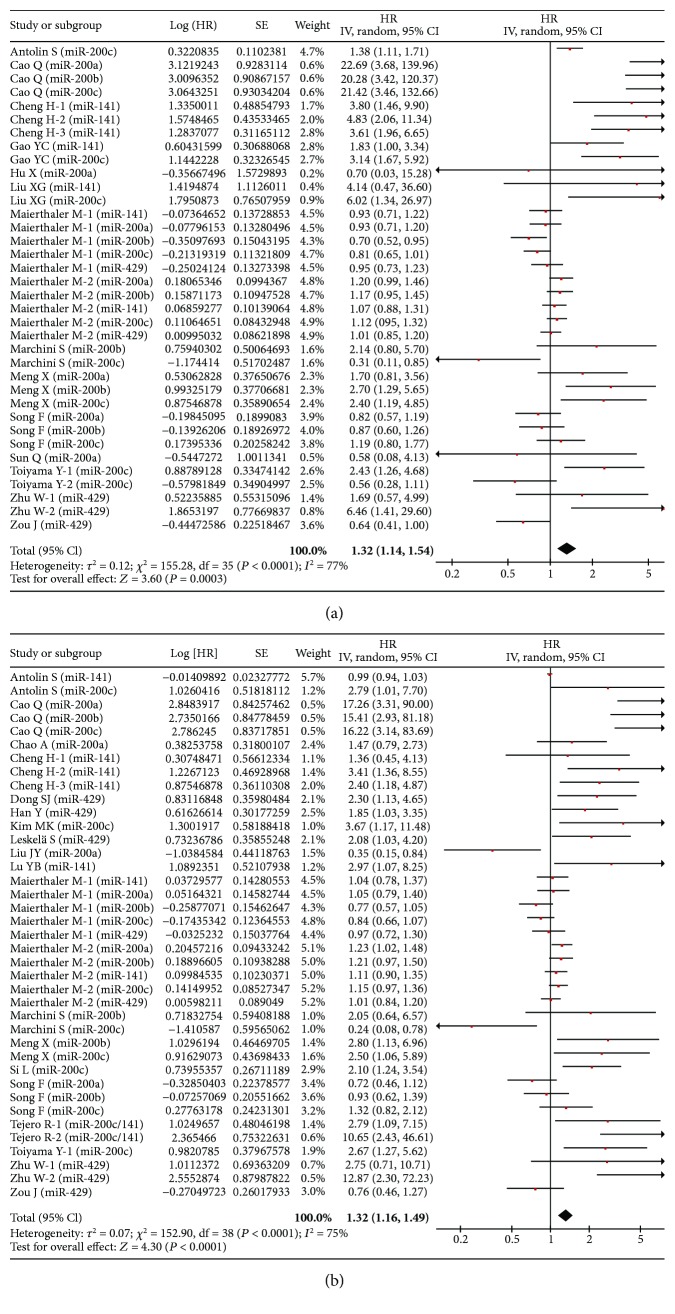
In univariate analysis, 19 studies were involved in current meta-analysis to assess the prognosis of miR-200 family overexpression in various cancers. High expression of miR-200 family was found to be associated with unfavorable OS (HR = 1.32, 95% CI: 1.14–1.54, P < 0.001) (Figure 2(a)). Besides, it indicated that there were certain associations via subanalyses regarding patient source, cancer type, test method, sample source, sample size, and miR-200 component (Table 3).
Figure 2.
Forest plot of the association between high expression of the miR-200 family in various tumors and OS under different types of analysis. (a) Univariate analysis; (b) multivariate analysis. The squares and horizontal lines correspond to the study-specific HR and 95% CI. The area of the squares reflects the weight. The diamond represents the summary HR and 95% CI. CI = confidence interval, HR = hazard ratio.
Table 3.
Stratified analysis of the high expression of the miR-200 family and overall survival.
| Categories | Subgroups | Univariate analyses | Multivariate analyses | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Number of datasets | HR (95% CI) | P value | I 2 | Ph | Number of datasets | HR (95% CI) | P value | I 2 | Ph | ||
| All | 19 | 1.32 (1.14–1.54) | <0.001 | 77.50% | <0.001 | 24 | 1.32 (1.16–1.49) | <0.001 | 75.10% | <0.001 | |
|
| |||||||||||
| Patient source | Asia | 10 | 1.91 (1.26–2.92) | 0.003 | 80.10% | <0.001 | 13 | 1.98 (1.34–2.90) | 0.001 | 78.20% | <0.001 |
| Europe | 5 | 1.07 (0.95–1.21) | 0.286 | 66.80% | <0.001 | 8 | 1.11 (0.99–1.24) | 0.071 | 67.10% | <0.001 | |
| North America | 4 | 3.81 (2.46–5.90) | <0.001 | 0.00% | 0.685 | 3 | 2.37 (1.44–3.91) | 0.001 | 0.00% | 0.457 | |
|
| |||||||||||
| Cancer type | EOC | 7 | 2.18 (1.23–3.86) | 0.008 | 79.90% | <0.001 | 7 | 1.98 (1.03–3.80) | 0.039 | 81.80% | <0.001 |
| CRC | 7 | 1.12 (0.96–1.31) | 0.140 | 77.70% | <0.001 | 8 | 1.15 (1.02–1.30) | 0.026 | 60.10% | 0.001 | |
| NSCLC | 3 | 3.36 (1.64–6.89) | 0.001 | 0.00% | 0.411 | 6 | 2.91 (1.99–4.26) | <0.001 | 33.40% | 0.185 | |
| GC | 1 | 0.94 (0.75–1.17) | 0.565 | 1.90% | 0.361 | 2 | 1.10 (0.72–1.68) | 0.669 | 62.30% | 0.047 | |
| BC | 1 | 1.38 (1.11–1.71) | 0.003 | / | / | 1 | 1.46 (0.54–3.91) | 0.454 | 75.10% | 0.045 | |
|
| |||||||||||
| Test method | RT-PCR | 16 | 1.64 (1.24–2.16) | 0.001 | 75.30% | <0.001 | 19 | 1.57 (1.23–1.99) | <0.001 | 75.10% | <0.001 |
| ISH | 1 | 21.42 (7.54–60.83) | <0.001 | 0.00% | 0.996 | 1 | 16.28 (6.28–42.24) | <0.001 | 0.00% | 0.995 | |
| TaqMan | 2 | 1.01 (0.95–1.08) | 0.686 | 47.70% | 0.046 | 4 | 1.07 (0.95–1.20) | 0.249 | 58.80% | 0.005 | |
|
| |||||||||||
| Sample source | FFPE | 2 | 0.57 (0.29–1.10) | 0.095 | 0.00% | 0.890 | 5 | 2.27 (1.56–3.32) | <0.001 | 43.10% | 0.135 |
| Tissue | 5 | 3.19 (1.19–8.52) | 0.021 | 84.40% | <0.001 | 9 | 2.04 (1.13–3.68) | 0.017 | 80.70% | <0.001 | |
| Blood | 10 | 1.34 (1.15–1.57) | <0.001 | 79.00% | <0.001 | 9 | 1.14 (1.02–1.28) | 0.019 | 68.30% | <0.001 | |
| TMA | 2 | 0.93 (0.75–1.16) | 0.527 | 0.00% | 0.519 | 1 | 0.94 (0.73–1.21) | 0.649 | 40.90% | 0.184 | |
|
| |||||||||||
| Sample size | ≧100 | 11 | 1.25 (1.06–1.47) | 0.007 | 78.90% | <0.001 | 14 | 1.29 (1.11–1.49) | 0.001 | 71.60% | <0.001 |
| <100 | 8 | 1.74 (1.10–2.75) | 0.018 | 68.50% | 0.001 | 10 | 1.84 (1.17–2.90) | 0.008 | 79.60% | <0.001 | |
|
| |||||||||||
| miR-200 component | miR-200a | 7 | 1.14 (0.81–1.61) | 0.438 | 64.80% | 0.009 | 6 | 1.07 (0.72–1.59) | 0.723 | 78.30% | <0.001 |
| miR-200b | 6 | 1.38 (0.88–2.16) | 0.166 | 82.10% | <0.001 | 6 | 1.36 (0.89–2.08) | 0.158 | 76.70% | 0.001 | |
| miR-200c | 11 | 1.38 (1.01–1.89) | 0.040 | 82.40% | <0.001 | 10 | 1.62 (1.12–2.33) | 0.010 | 79.30% | <0.001 | |
| miR-141 | 7 | 2.01 (1.26–3.21) | 0.003 | 83.50% | <0.001 | 7 | 1.24 (0.99–1.56) | 0.060 | 68.00% | 0.005 | |
| miR-429 | 5 | 0.99 (0.73–1.34) | 0.953 | 62.20% | 0.032 | 8 | 1.41 (1.01–1.98) | 0.043 | 70.30% | 0.001 | |
EOC: epithelial ovarian cancer; BC: breast cancer; NSCLC: nonsmall cell lung cancer; GC: gastric cancer; CRC: colorectal cancer; RT-PCR: reverse transcription-polymerase chain reaction; ISH: in situ hybridization; FFPE: formalin-fixed and paraffin-embedded; TMA: tissue microarray; HR: hazard ratio; CI: confidence interval; Ph: P value of the heterogeneity test.
In multivariate analysis, 24 studies were included in meta-analysis to explore the prognostic value of the miR-200 family. As a result, high expression of the miR-200 family in various cancers was associated with unfavorable overall survival (HR = 1.32, 95% CI: 1.16–1.49, P < 0.001) (Figure 2(b)). Likewise, a similar result was found in different subgroups (Table 3).
4.3. Meta-Analysis of RFS/PFS/DFS
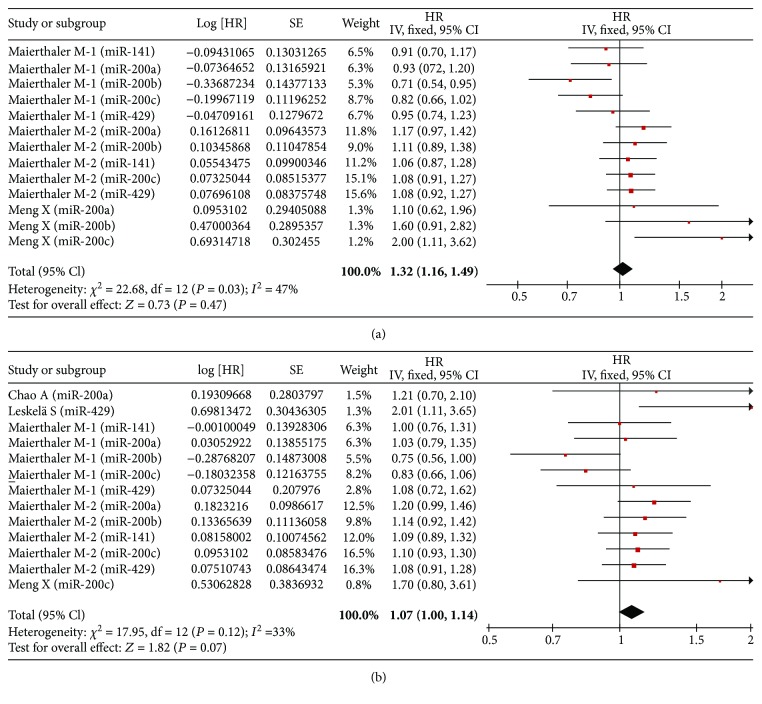
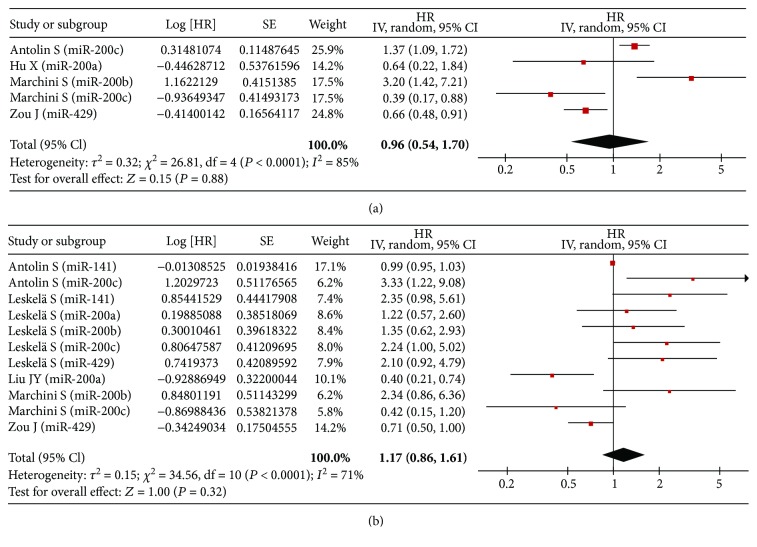
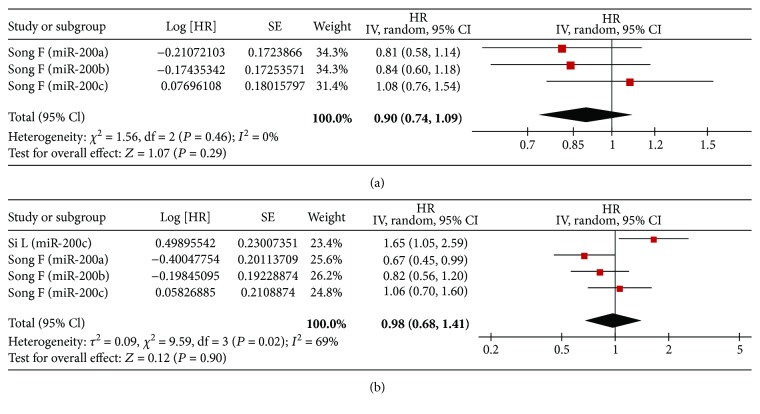
In univariate analysis, there were three studies, four studies, and one study involved with RFS, PFS, and DFS, respectively. Correspondingly, five studies, five studies, and two studies were collected in multivariate analysis, respectively. Ultimately, we found that no association of high expression of the miR-200 family was detected with RFS (univariate: HR = 1.02, 95% CI: 0.96–1.09, P = 0.47; multivariate: HR = 1.07, 95% CI: 1.00–1.14, P = 0.07) (Figure 3), PFS (univariate: HR = 0.96, 95% CI: 0.54–1.70, P = 0.88; multivariate: HR = 1.17, 95% CI: 0.86–1.61, P = 0.32) (Figure 4), and DFS (univariate: HR = 0.90, 95% CI: 0.74–1.09, P = 0.29; multivariate: HR = 0.98, 95% CI: 0.68–1.41, P = 0.90) (Figure 5).
Figure 3.
Forest plot of the association between high expression of the miR-200 family in various tumors and RFS under different types of analysis. (a) Univariate analysis; (b) multivariate analysis. The squares and horizontal lines correspond to the study-specific HR and 95% CI. The area of the squares reflects the weight. The diamond represents the summary HR and 95% CI. CI = confidence interval, HR = hazard ratio.
Figure 4.
Forest plot of the association between high expression of the miR-200 family in various tumors and PFS under different types of analysis. (a) Univariate analysis; (b) multivariate analysis. The squares and horizontal lines correspond to the study-specific HR and 95% CI. The area of the squares reflects the weight. The diamond represents the summary HR and 95% CI. CI = confidence interval, HR = hazard ratio.
Figure 5.
Forest plot of the association between high expression of the miR-200 family in various tumors and DFS under different types of analysis. (a) Univariate analysis; (b) multivariate analysis. The squares and horizontal lines correspond to the study-specific HR and 95% CI. The area of the squares reflects the weight. The diamond represents the summary HR and 95% CI. CI = confidence interval, HR = hazard ratio.
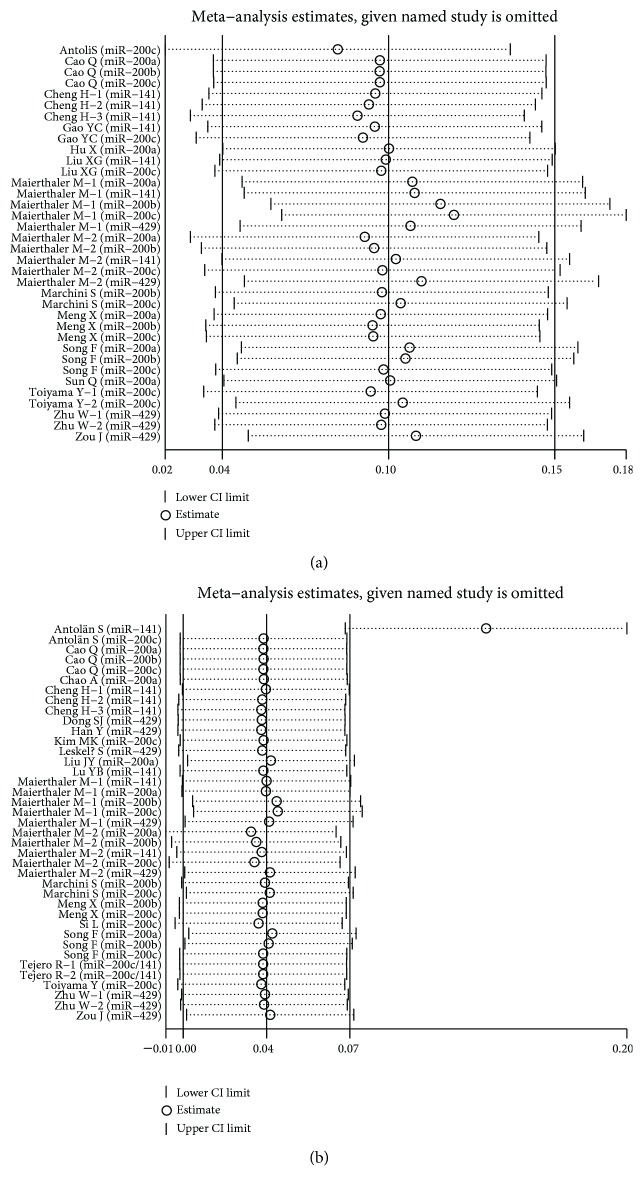
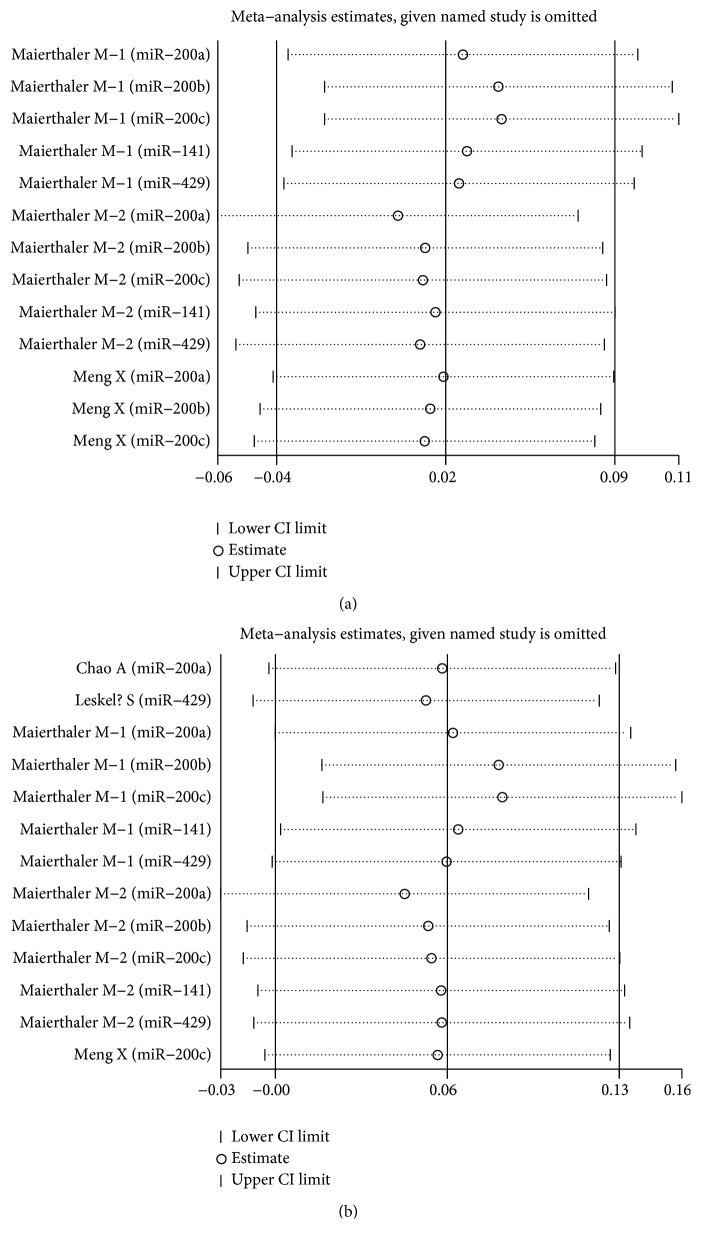
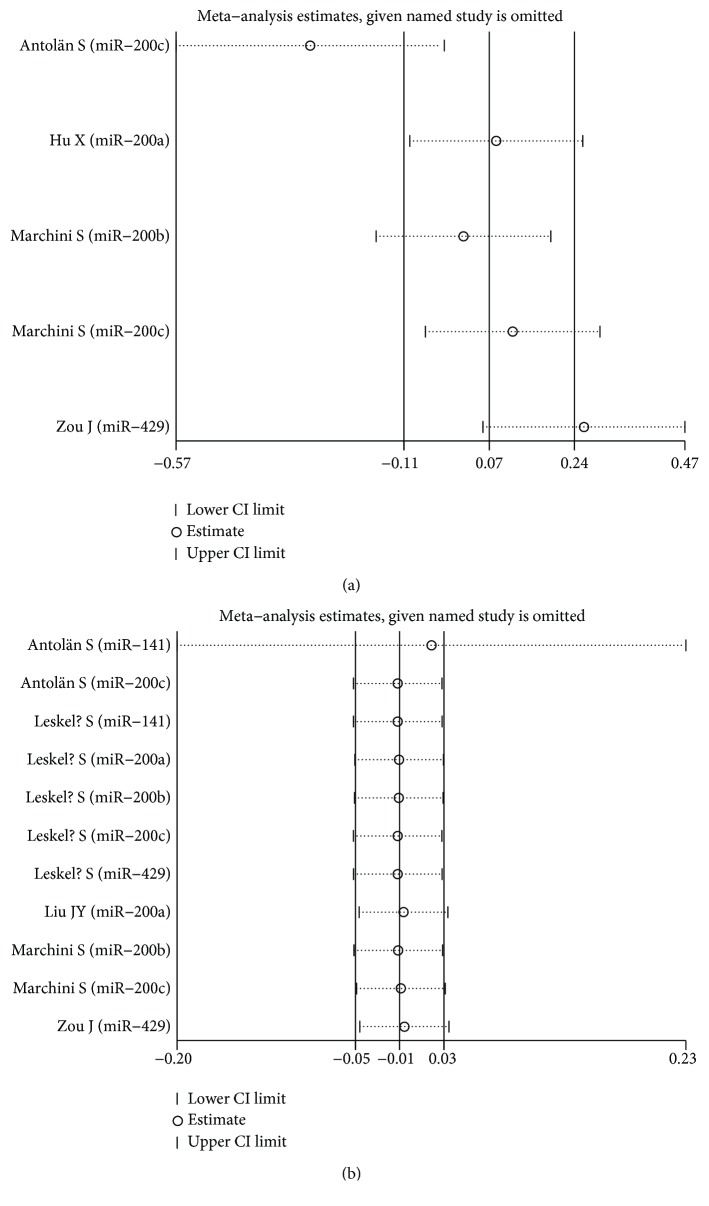
4.4. Sensitivity Analysis
Each single included study was deleted at a time to assess the specific effect of the individual data on the pooled HRs, and one-way sensitivity analysis suggested that most pooled results were relatively stable. Among them, the pooled results of OS, RFS, and PFS in both univariate analysis and multivariate analysis were shown in Figures 6(a), 6(b), Figures 7(a), 7(b), and Figures 8(a), 8(b), respectively. As shown in Figure 6(b), after excluding the study conducted by Antolín et al. [22], heterogeneity was slightly reduced between miR-200 family overexpression and OS under multivariate analysis (I2 from 75.1% to 73.3%), while the pooled results remained unchanged (multivariate: HR = 1.40, 95% CI: 1.21–1.63, P < 0.001). Likewise, as shown in Figure 8(a), the similar result was found between miR-200 family overexpression and PFS under univariate analysis (I2 from 85.1% to 80.4%), and the pooled results remained unchanged (univariate: HR = 0.85, 95% CI: 0.38–1.88, P = 0.684) after excluding the aforementioned study [22].
Figure 6.
One-way sensitivity analysis of high expression of the miR-200 family in various tumors with OS under different types of analysis. (a) Univariate analysis; (b) multivariate analysis. Individually removed the studies and suggested that the results of this meta-analysis were relatively stable.
Figure 7.
One-way sensitivity analysis of high expression of the miR-200 family in various tumors with RFS under different types of analysis. (a) Univariate analysis; (b) multivariate analysis. Individually removed the studies and suggested that the results of this meta-analysis were stable.
Figure 8.
One-way sensitivity analysis of high expression of the miR-200 family in various tumors with PFS under different types of analysis. (a) Univariate analysis; (b) multivariate analysis. Individually removed the studies and suggested that the results of this meta-analysis were relatively stable.
4.5. Publication Bias Evaluation
Begg's funnel plot indicated that there was a significant publication bias in meta-analysis of OS under both univariate analysis (P = 0.028) and multivariate analysis (P < 0.001). However, no publication bias was found in meta-analysis of RFS (univariate: P = 0.760; multivariate: P = 0.855), PFS (univariate: P = 1.000; multivariate: P = 0.087), and DFS (univariate: P = 0.296; multivariate: P = 0.308).
5. Discussion
Generally, cancer progression and blood-borne metastasis are the primary factors contributed to the great majority of cancer deaths. The specific biomarkers of metastatic phenotype hold great promise in individualized therapy and improved prognosis prediction in several neoplastic diseases [39]. In recent decades, to explore the clinically useful cancer signatures remains to be research hotpot due to the complexity of cancer. Gene expression signatures of carcinomas have led to new classifications of cancer subgroups and also carried prognostic and predictive information [40]. miRNAs are small noncoding RNAs that regulate human protein-coding gene expression of specific mRNAs by either translational repression or degradation. miRNA expression signatures have distinct functions in controlling the cell cycle, proliferation, invasion, and metastasis [41], which could thus be developed into a potential prognostic signature [42]. The latest miRBase release contains 24,521 miRNA loci from 206 species, further processed to produce 30,424 mature miRNA products [43]. To date, significant miRNA expression changes have been observed in multiple cancers analyzed by profiling and next generation sequencing technologies [44].
The miR-200 family of miRNAs consists of five members grouped into two independent transcriptional clusters: miR-200a, miR-200b, and miR-429 on chromosome 1 (1p36.33), and miR-141 and miR-200c on chromosome 12 (12p13.31). Deregulation of the miR-200 family of microRNAs has been involved in cell plasticity, apoptosis, molecular subtype, oestrogen regulation, control of the growth and function of stem cells, and regulation of the downstream transcriptional program that mediate distant metastasis [45]. Cancer progression is associated with a dynamic process of epithelial-to-mesenchymal transition (EMT), during which epithelial cells lose their cell polarity and cell-cell adhesion and gain migratory as well as invasive properties by downregulating E-cadherin and upregulating vimentin expression [46, 47]. The miR-200 family members may play a major role in the suppression of EMT and metastasis [48]. Deregulation of miR-200 in cancer cell lines caused upregulation of E-cadherin and reduced motility of cancer cells. Conversely, inhibition of miR-200 reduced E-cadherin expression, increased expression of vimentin, and induced EMT [49]. In addition, the miR-200 family is known as a key transcriptional regulator of EMT and the maintenance of a less invasive and aggressive epithelial phenotype by targeting ZEB1 and ZEB2, two important transcriptional repressors of the E-cadherin gene [48]. ZEB was inhibited by miR-200 members at the posttranscriptional level by binding to highly conserved target sites in their 3′-UTR; the functional link of ZEB factors with the miR-200 family in a double negative feedback loop is known as the ZEB/miR-200 feedback loop [50]. It also has been reported that several tumor suppressor genes, including BRD7, BAP1, GATA, CLOCK, and PTPN12, might be potential targets of the miR-200 family [51, 52].
To date, studies focused on the association of high expression of the miR-200 family with cancer prognosis have yielded conflicting results. Notably, small sample-sized studies lacking statistical power often have resulted in apparently contradicting conclusions. Meta-analysis is a useful tool for providing convincing evidence as it could present inconsistent results from different studies to get a relatively precise result. As far as we know, the current meta-analysis is the first try to comprehensively assess the correlation of miR-200 cluster high expression with cancer prognosis. We have explored the potential associations in overall population and the corresponding subgroups. Consequently, of particular interest is the finding of significant correlation between high expression of miR-200 cluster and poor OS by two different statistical methods. Likewise, a similar result was found in different subgroups. However, no association of miR-200 family was detected with RFS/PFS/DFS.
In the current meta-analysis, significant heterogeneity was found, which required careful interpretation and searched for influencing factors by further subgroup analyses. Consequently, impact of ethnicity, detection methods, cancer types, sample size, and sample source on prognosis in patients was considerable, which should be taken into consideration when evaluating the prognosis of cancer for patients. Some potential or undiscovered factors including adjustment for surgery, radiation, chemotherapy, socioeconomic status, and tumor characteristics should not be ignored. Moreover, there was a significant publication bias in meta-analysis of OS under both univariate analysis and multivariate analysis, suggesting that only published studies in English and Chinese might not provide so sufficient evidences. As for RFS/PFS/DFS, we did not perform subgroup analyses due to relatively fewer eligible studies. Although the studies regarding various tumors without a consistent cut-off value may influence the ultimate results and the heterogeneity suggested that potential or undiscovered factors might be ignored, a certain relationship of high expression of the miR-200 family in cancer prognosis was found in the current study.
6. Conclusion
In summary, the current study is the first original meta-analysis to address the correlation between miR-200 family expression and prognosis for cancer patients. A significant correlation was explored in overall population as well as the corresponding subgroups. Concretely, it presented that miR-200 family overexpression might be associated with poor OS to some extent, while no association was detected between high miR-200 family expression and RFS/PFS/DFS. In the future, detailed investigations comprising large cohort size from multicenter are required to confirm our conclusions.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (81601600) and the China Postdoctoral Science Foundation (2016M590576, 2017T100455).
Contributor Information
Min Chao, Email: cm0654@sina.com.
Jing Wang, Email: ahwangjing1968@126.com.
Data Availability
All data have been shared in the figures and tables.
Conflicts of Interest
The authors have no conflict of interests to declare.
Authors' Contributions
Wen Liu, Kaiping Zhang, Min Chao, and Jing Wang conceived and designed the study. Wen Liu, Kaiping Zhang, and Yue Hu conducted the eligible study collection, quality assessment, and data extraction. Pengfei Wei and Yaqin Peng analyzed the data. Wen Liu, Xiang Fang, and Guoping He interpreted the results. Limin Wu and Min Chao prepared the tables and figures. Wen Liu and Kaiping Zhang wrote the manuscript; Pengfei Wei, Min Chao, and Jing Wang revised it. Wen Liu, Kaiping Zhang, and Pengfei Wei contributed equally to this work. All authors read and approved the final manuscript.
References
- 1.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 2.Calin G. A., Sevignani C., Dumitru C. D., et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(9):2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Niu J., Sun Y., Guo Q., Niu D., Liu B. miR-1 inhibits cell growth, migration, and invasion by targeting VEGFA in osteosarcoma cells. Disease Markers. 2016;2016:8. doi: 10.1155/2016/7068986.7068986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grosshans H., Filipowicz W. Molecular biology: the expanding world of small RNAs. Nature. 2008;451(7177):414–416. doi: 10.1038/451414a. [DOI] [PubMed] [Google Scholar]
- 5.Wu Y., Jia Z., Cao D., et al. Predictive value of miR-219-1, miR-938, miR-34b/c, and miR-218 polymorphisms for gastric cancer susceptibility and prognosis. Disease Markers. 2017;2017:9. doi: 10.1155/2017/4731891.4731891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Halvorsen A. R., Kristensen G., Embleton A., et al. Evaluation of prognostic and predictive significance of circulating microRNAs in ovarian cancer patients. Disease Markers. 2017;2017:9. doi: 10.1155/2017/3098542.3098542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi P.-W., Ng S.-W. The functions of microRNA-200 family in ovarian cancer: beyond epithelial-mesenchymal transition. International Journal of Molecular Sciences. 2017;18(6):p. 1207. doi: 10.3390/ijms18061207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koutsaki M., Libra M., Spandidos D. A., Zaravinos A. The miR-200 family in ovarian cancer. Oncotarget. 2017;8(39):66629–66640. doi: 10.18632/oncotarget.18343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zidar N., Boštjančič E., Jerala M., et al. Down-regulation of microRNAs of the miR-200 family and up-regulation of snail and slug in inflammatory bowel diseases-hallmark of epithelial-mesenchymal transition. Journal of Cellular and Molecular Medicine. 2016;20(10):1813–1820. doi: 10.1111/jcmm.12869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Munafo M. R., Flint J. Meta-analysis of genetic association studies. Trends in Genetics. 2004;20(9):439–444. doi: 10.1016/j.tig.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 11.Liberati A., Altman D. G., Tetzlaff J., et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. PLoS Medicine. 2009;6(7, article e1000100) doi: 10.1371/journal.pmed.1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Higgins J. P. T., Thompson S. G. Quantifying heterogeneity in a meta-analysis. Statistics in Medicine. 2002;21(11):1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 13.DerSimonian R., Laird N. Meta-analysis in clinical trials. Controlled Clinical Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 14.Mantel N., Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. Journal of the National Cancer Institute. 1959;22(4):719–748. [PubMed] [Google Scholar]
- 15.Begg C. B., Berlin J. A. Publication bias and dissemination of clinical research. Journal of the National Cancer Institute. 1989;81(2):107–115. doi: 10.1093/jnci/81.2.107. [DOI] [PubMed] [Google Scholar]
- 16.Zou J., Liu L., Wang Q., et al. Downregulation of miR-429 contributes to the development of drug resistance in epithelial ovarian cancer by targeting ZEB1. American Journal of Translational Research. 2017;9(3):1357–1368. [PMC free article] [PubMed] [Google Scholar]
- 17.Han Y., Zhao Q., Zhou J., Shi R. miR-429 mediates tumor growth and metastasis in colorectal cancer. American Journal of Cancer Research. 2017;7(2):218–233. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Maierthaler M., Benner A., Hoffmeister M., et al. Plasma miR-122 and miR-200 family are prognostic markers in colorectal cancer. International Journal of Cancer. 2017;140(1):176–187. doi: 10.1002/ijc.30433. [DOI] [PubMed] [Google Scholar]
- 19.Si L., Tian H., Yue W., et al. Potential use of microRNA-200c as a prognostic marker in non-small cell lung cancer. Oncology Letters. 2017;14(4):4325–4330. doi: 10.3892/ol.2017.6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meng X., Müller V., Milde-Langosch K., Trillsch F., Pantel K., Schwarzenbach H. Diagnostic and prognostic relevance of circulating exosomal miR-373, miR-200a, miR-200b and miR-200c in patients with epithelial ovarian cancer. Oncotarget. 2016;7(13):16923–16935. doi: 10.18632/oncotarget.7850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong S. J., Cai X. J., Li S. J. The clinical significance of miR-429 as a predictive biomarker in colorectal cancer patients receiving 5-fluorouracil treatment. Medical Science Monitor. 2016;22:3352–3361. doi: 10.12659/MSM.900674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Antolín S., Calvo L., Blanco-Calvo M., et al. Circulating miR-200c and miR-141 and outcomes in patients with breast cancer. BMC Cancer. 2015;15(1):p. 297. doi: 10.1186/s12885-015-1238-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao Y. C., Wu J. MicroRNA-200c and microRNA-141 as potential diagnostic and prognostic biomarkers for ovarian cancer. Tumour Biology. 2015;36(6):4843–4850. doi: 10.1007/s13277-015-3138-3. [DOI] [PubMed] [Google Scholar]
- 24.Lu Y. B., Hu J. J., Sun W. J., Duan X. H., Chen X. Prognostic value of miR-141 down-regulation in gastric cancer. Genetics and Molecular Research. 2015;14(4, article 17305):17311. doi: 10.4238/2015.December.16.31. [DOI] [PubMed] [Google Scholar]
- 25.Liu J. Y., Zhao Y. R., Zhang L. N., Yan Y., Zheng H. Expression and clinical significance of four miRNAs in epithelial ovarian cancer. Tianjin Medical Journal. 2015;43(9):996–999. [Google Scholar]
- 26.Cao Q., Lu K., Dai S., Hu Y., Fan W. Clinicopathological and prognostic implications of the miR-200 family in patients with epithelial ovarian cancer. International Journal of Clinical and Experimental Pathology. 2014;7(5):2392–2401. [PMC free article] [PubMed] [Google Scholar]
- 27.Kim M. K., Jung S. B., Kim J.-S., et al. Expression of microRNA miR-126 and miR-200c is associated with prognosis in patients with non-small cell lung cancer. Virchows Archiv. 2014;465(4):463–471. doi: 10.1007/s00428-014-1640-4. [DOI] [PubMed] [Google Scholar]
- 28.Zhu W., He J., Chen D., et al. Expression of miR-29c, miR-93, and miR-429 as potential biomarkers for detection of early stage non-small lung cancer. PLoS One. 2014;9(2, article e87780) doi: 10.1371/journal.pone.0087780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song F., Yang D., Liu B., et al. Integrated microRNA network analyses identify a poor-prognosis subtype of gastric cancer characterized by the miR-200 family. Clinical Cancer Research. 2014;20(4):878–889. doi: 10.1158/1078-0432.CCR-13-1844. [DOI] [PubMed] [Google Scholar]
- 30.Tejero R., Navarro A., Campayo M., et al. miR-141 and miR-200c as markers of overall survival in early stage non-small cell lung cancer adenocarcinoma. PLoS One. 2014;9(7, article e101899) doi: 10.1371/journal.pone.0101899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toiyama Y., Hur K., Tanaka K., et al. Serum miR-200c is a novel prognostic and metastasis-predictive biomarker in patients with colorectal cancer. Annals of Surgery. 2014;259(4):735–743. doi: 10.1097/SLA.0b013e3182a6909d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun Q., Zou X., Zhang T., Shen J., Yin Y., Xiang J. The role of miR-200a in vasculogenic mimicry and its clinical significance in ovarian cancer. Gynecologic Oncology. 2014;132(3):730–738. doi: 10.1016/j.ygyno.2014.01.047. [DOI] [PubMed] [Google Scholar]
- 33.Liu X. G., Zhu W. Y., Huang Y. Y., et al. High expression of serum miR-21 and tumor miR-200c associated with poor prognosis in patients with lung cancer. Medical Oncology. 2012;29(2):618–626. doi: 10.1007/s12032-011-9923-y. [DOI] [PubMed] [Google Scholar]
- 34.Chao A., Lin C. Y., Lee Y. S., et al. Regulation of ovarian cancer progression by microRNA-187 through targeting disabled homolog-2. Oncogene. 2012;31(6):764–775. doi: 10.1038/onc.2011.269. [DOI] [PubMed] [Google Scholar]
- 35.Marchini S., Cavalieri D., Fruscio R., et al. Association between miR-200c and the survival of patients with stage I epithelial ovarian cancer: a retrospective study of two independent tumour tissue collections. Lancet Oncology. 2011;12(3):273–285. doi: 10.1016/S1470-2045(11)70012-2. [DOI] [PubMed] [Google Scholar]
- 36.Cheng H., Zhang L., Cogdell D. E., et al. Circulating plasma miR-141 is a novel biomarker for metastatic colon cancer and predicts poor prognosis. PLoS One. 2011;6(3, article e17745) doi: 10.1371/journal.pone.0017745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leskelä S., Leandro-García L. J., Mendiola M., et al. The miR-200 family controls β-tubulin III expression and is associated with paclitaxel-based treatment response and progression-free survival in ovarian cancer patients. Endocrine Related Cancer. 2010;18(1):85–95. doi: 10.1677/ERC-10-0148. [DOI] [PubMed] [Google Scholar]
- 38.Hu X., Macdonald D. M., Huettner P. C., et al. A miR-200 microRNA cluster as prognostic marker in advanced ovarian cancer. Gynecologic Oncology. 2009;114(3):457–464. doi: 10.1016/j.ygyno.2009.05.022. [DOI] [PubMed] [Google Scholar]
- 39.Montani F., Bianchi F. Circulating cancer biomarkers: the macro-revolution of the micro-RNA. eBioMedicine. 2016;5:4–6. doi: 10.1016/j.ebiom.2016.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prat A., Ellis M. J., Perou C. M. Practical implications of gene-expression-based assays for breast oncologists. Nature Reviews Clinical Oncology. 2011;9(1):48–57. doi: 10.1038/nrclinonc.2011.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Z., Guo Y., Pu X., Li M. Dissecting the regulation rules of cancer-related miRNAs based on network analysis. Scientific Reports. 2016;6(1):p. 34172. doi: 10.1038/srep34172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jay C., Nemunaitis J., Chen P., Fulgham P., Tong A. W. miRNA profiling for diagnosis and prognosis of human cancer. DNA and Cell Biology. 2007;26(5):293–300. doi: 10.1089/dna.2006.0554. [DOI] [PubMed] [Google Scholar]
- 43.Kozomara A., Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Research. 2014;42(D1):D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang L., Wei P., Shen X., et al. MicroRNA expression profile in penile cancer revealed by next-generation small RNA sequencing. PLoS One. 2015;10(7, article e0131336) doi: 10.1371/journal.pone.0131336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uhlmann S., Zhang J. D., Schwäger A., et al. miR-200bc/429 cluster targets PLCγ1 and differentially regulates proliferation and EGF-driven invasion than miR-200a/141 in breast cancer. Oncogene. 2010;29(30):4297–4306. doi: 10.1038/onc.2010.201. [DOI] [PubMed] [Google Scholar]
- 46.Yu M., Bardia A., Wittner B. S., et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339(6119):580–584. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gregory P. A., Bert A. G., Paterson E. L., et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nature Cell Biology. 2008;10(5):593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 48.Davalos V., Moutinho C., Villanueva A., et al. Dynamic epigenetic regulation of the microRNA-200 family mediates epithelial and mesenchymal transitions in human tumorigenesis. Oncogene. 2012;31(16):2062–2074. doi: 10.1038/onc.2011.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen D., Dang B. L., Huang J. Z., et al. MiR-373 drives the epithelial-to-mesenchymal transition and metastasis via the miR-373-TXNIP-HIF1α-TWIST signaling axis in breast cancer. Oncotarget. 2015;6(32):32701–32712. doi: 10.18632/oncotarget.4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wellner U., Schubert J., Burk U. C., et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nature Cell Biology. 2009;11(12):1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 51.Park Y. A., Lee J. W., Choi J. J., et al. The interactions between microRNA-200c and BRD7 in endometrial carcinoma. Gynecologic Oncology. 2012;124(1):125–133. doi: 10.1016/j.ygyno.2011.09.026. [DOI] [PubMed] [Google Scholar]
- 52.Iorio M. V., Visone R., di Leva G., et al. MicroRNA signatures in human ovarian cancer. Cancer Research. 2007;67(18):8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data have been shared in the figures and tables.