Abstract
Metastasis requires cellular changes related to cell-to-cell and cell-to-matrix adhesion, immune surveillance, activation of growth and survival signalling pathways, and epigenetic modifications. In addition to tumour cells, tumour stroma is also modified in relationship to the primary tumour as well as to distant metastatic sites (forming a metastatic niche). A common denominator of most stromal partners in tumour progression is CD36, a scavenger receptor for fatty acid uptake that modulates cell-to-extracellular matrix attachment, stromal cell fate (for adipocytes, endothelial cells), TGFβ activation, and immune signalling. CD36 has been repeatedly proposed as a prognostic marker in various cancers, mostly of epithelial origin (breast, prostate, ovary, and colon) and also for hepatic carcinoma and gliomas. Data gathered in preclinical models of various cancers have shown that blocking CD36 might prove beneficial in stopping metastasis spread. However, targeting the receptor in clinical trials with thrombospondin mimetic peptides has proven ineffective, and monoclonal antibodies are not yet available for patient use. This review presents data to support CD36 as a potential prognostic biomarker in cancer, its current stage towards achieving bona fide biomarker status, and knowledge gaps that must be filled before further advancement towards clinical practice.
1. Introduction
Metastasis is a rather inefficient process if the number of circulating tumour cells is to be compared with the number of clinically overt metastatic sites [1]. From vascular invasion to secondary site tumour initiation, a cell can go through changes in cell-to-cell and cell-to- matrix adhesion profile, face immune surveillance systems, activate growth and survival signalling pathways, and undergo epigenetic modifications. To be effective, these changes must occur in a time-dependent manner, modifying the cell phenotype for survival in new microenvironments. Under these conditions, how much of the primary tumour is recapitulated in a metastasis? And how can we predict whether or when a primary tumour would seed secondary sites?
Perhaps surprisingly, scrutinizing the less investigated stromal tumour tissue—or the modern and reinvented “stromal metastatic niche”—could provide some answers. The metastatic niche has been defined as “extracellular matrix, nonmalignant cells, and the signalling molecules they produce” [2]. Different in composition and less characterized than its counterpart—the primary tumour niche—the stromal metastatic niche recently underwent a shift in perspective to a “premetastatic niche,” prepared in advance by conditioned infiltrating monocytes. This premetastatic niche, as yet unoccupied by tumour cells, is thought to create a tumour friendly environment to enhance the survival chances of invading cells [3].
A common denominator of most stromal partners playing a role in tumour progression is expression of CD36—a scavenger receptor for fatty acid (FA) uptake that modulates cell-to- extracellular matrix attachment, stromal cell fate (for adipocytes, endothelial cells), TGFβ activation, and immune signalling [4]. Unlike its well-known and better-studied binding partners (thrombospondins (TSPs) 1 and 2) but with controversial involvement in cancer progression, CD36 is increasingly emerging as a prognostic marker associated with the metastatic process. Even more so, its presence seems not to be limited to tumour stroma, as the number of reports describing it on tumour cells is increasing.
Several gaps remain in CD36-related knowledge. Learning from paths travelled in understanding other receptors as cancer-related biomarkers, one can argue that there are still plenty of blank spots on the CD36 cancer-related map. In cancer studies, CD36 is investigated mostly in relationship with TSPs, a family of matrix proteins acting as ligands, and most data are related to TSP interactions and, to a lesser extent, TGFß. Only a handful of papers have been published directly addressing CD36, almost all measuring levels of expression in relationship to tumour growth and metastasis. Furthermore, lessons from myeloproliferative diseases and gliomas suggest that mutated receptors do not require a ligand to be active. In the case of CD36, constitutive activation, regardless of TSP expression within the tumour or tumour niche, could be largely overlooked if the main focus falls on the ligand rather than the receptor.
In addition, a parallel analysis of stromal CD36 versus tumour CD36 is usually missing from the scientific argument in most original articles. Last, but not least, studies in animal models using athymic mice remove a key player from the stroma, the immune cell, yielding an incomplete tumour–stroma interaction panel.
What is the ultimate goal in CD36 research? One size fits all? Probably not. That pattern has not yet been the case for any molecular intervention in diagnostics, prognostics, or therapy. However, the chase for a single but efficient antimetastatic molecular target is justifiable from several perspectives. First and foremost, time is gained for diagnostic and therapeutic intervention. Second, absence of metastases will translate into an absence of cellular polymorphisms derived from environmental change. Thus, molecular therapies addressing the primary tumour could have a greater impact on relapse and survival rates.
How far is CD36 from validated status as a biomarker and what type of biomarker would best fit are the questions the present review will attempt to answer, after analysing the evidence tying CD36 to the metastatic process and the translation into knowledge towards clinical practice.
2. CD36 Distribution and Functions in Normal Tissues
CD36, also known as platelet glycoprotein (GP) 4/, FA translocase (FAT)/, scavenger receptor class B member 3 (SCARB3)/, GP88, GPIIIB, or GPIV [5], is an integral membrane GP encoded by the CD36 gene and belonging to the scavenger receptor family. The intracellular trafficking of the molecule requires different degrees of glycosylation, with the heavy glycosylated protein being exposed on the cell membrane [6, 7]. When exposed, CD36 may associate with other transmembrane proteins, such as integrins (β1, β2, and β5) and tetraspanins (CD9, CD81) [8]. Its intracellular domains associate with members of the src family of tyrosine kinases, such as fyn, lyn, and, yes, a molecular interaction most probably mediated by lipids in the context of lipid rafts [9]. Supramolecular assembly of CD36 into nanoclusters at the plasma membrane, even in the absence of ligands, is important for downstream fyn signalling [10]. The extracellular domain binds to a vast array of ligands, which accounts for the diversity of signal transduction outcomes: (i) adhesive glycoproteins of the TSP family [11]; (ii) collagenic proteins (collagens I and IV) [5]; (iii) lipid ligands (anionic or oxidized phospholypids –PL), native and oxidized lipoproteins [8], FAs); and (iv) peptides such as hexarelin or fibrillar Aβ amyloid species [12].
As a surface protein, CD36 is widely distributed. Found on platelets and monocytes/macrophages, it is involved in cellular activation. It not only mediates the initial binding of platelets on extracellular matrix protein like collagen I [5] and TSP-1 but also triggers signal transduction, inducing an oxidative burst in monocytes [13]. In addition, it is present on erythrocytes, where it mediates adherence of Plasmodium falciparum–infected erythrocytes.
CD36 expression was also detected in differentiated adipocytes [14]. In vitro studies proved that CD36 is located in lipid rafts, along with caveolin-1, mediating FA uptake [15]. Both functional studies with CD36 cross-linking agents and disruption of lipid rafts stop the transport of long-chain FAs [14].
In skeletal muscle, CD36 expression on the cell surface is an important mechanism for FA uptake and short-term regulation through subcellular redistribution [16]. However, CD36 is also found in the mitochondria, where it is responsible for FA oxidation [17]. CD36 expression is regulated by both insulin and contraction, which promotes the translocation of intracellular stored CD36 to the plasma membrane. Increased expression can contribute to lipid accumulation in heart and skeletal muscle [18].
CD36 also has been described on endothelial cells of human dermis, but only in the microvasculature and not in the large vessels [19] and in caveolin-rich membranes isolated from lung endothelium [20]. It is also present on normal mammary epithelial cells [21], which prompted its investigation in breast cancer, as discussed below.
Another type of epithelial cell expressing CD36 is the taste receptor cell within lingual taste buds in tongue of rodents [22], pigs, and humans [23]. CD36 expression is restricted to only the lingual papillae where it has been localized at the apical side of the circumvallate [22] and foliate taste buds [23]. Its expression is lipid-mediated, changing the attraction for fat during a meal [24]. Only lipid discrimination is affected in CD36-null mice [22].
CD36 is expressed in the brush border membrane of duodenal and jejunal enterocytes [25] in both mice and humans [26]. Moreover, early after lipid ingestion, CD36 disappears from the luminal side of intestinal villi [27]. Similar to adipocytes, CD36 is located in lipid rafts, where it colocalizes with caveolin-1. In vitro studies showed that caveolin-1 is required for the transport of CD36 to the apical membrane, thus regulating its surface availability for FA uptake [28]. Moreover, brush border caveolae provide the absorptive surface for dietary FA. Studies performed on caveolin-1 knockout mice proved that FA absorption was compromised and the animals could not gain weight [25].
Also at the intestinal level, CD36 was detected on enteroendocrine cells secreting secretin and cholecystokinin in the mucosa of the duodenum, jejunum, and proximal ileum on both apical membranes and cytosolic granules [29].
In mouse liver, CD36 is expressed on hepatocytes, endothelial cells, and Kupffer cells [30] and the expression is increased by starvation [8] and aging, especially when associated with a high-fat diet. Insulin increases CD36 expression in the liver [31] by activating PPARγ, an upstream regulator of CD36 expression. Enhanced expression and subsequent fat uptake and triglyceride (TG) accumulation may accelerate progression of nonalcoholic fatty liver disease [32], insulin resistance, and type 2 diabetes [33].
In the normal brain, CD36 expression is low, but it is upregulated upon stroke due to monocyte-macrophage infiltration. It appears that CD36 contributes to acute ischemic brain injury during the inflammatory phase and is involved in phagocytosis during the recovery phase [7].
In ovary, CD36 is found on serous ovarian epithelial cells [34], contributing to angiogenesis and folliculogenesis [35].
3. Functions of CD36: Lessons Learned from the CD36 Knockout Mouse Model
CD36 was first identified as platelet GPIV, due to thrombocytes' ability to bind TSP [36]. Later, its overlapping structure with leukocyte differentiation antigen CD36 was demonstrated [37]. In the following years, its role in platelet activation [38] and cell adhesion [39] was investigated. Not long after, its involvement in translocation of long-chain FAs was reported [40].
Febbraio and collaborators have played an important role in understanding CD36 functions by creating mice with a null mutation in the CD36 gene through targeted homologous recombination [41]. The animal model was subsequently used by a large number of researchers, with 528 citations recorded to date (ISI Web of Science Core Collection, searched on March 13, 2018). Some of the significant insights provided by CD36-/- mouse experiments include regulation of CD36 by PPAR-gamma [42] inhibition of angiogenesis in vitro and in vivo; induction of apoptosis by TSP-1 via activation of CD36, p59fyn, caspase- 3–like proteases, and p38 mitogen-activated protein kinases [43]; understanding of atherosclerotic lesion development [44, 45] contribution to uptake and use long-chain FAs [46] diet response [47], serving as an advanced glycation end-products receptor [48] orosensory perception of long-chain FAs [49]. Conversely, mice were also engineered to overexpress CD36 in specific tissues by using the promoter of the muscle creatine kinase gene, resulting in enhanced FA oxidation, reduced plasma TGs and FAs, and increased plasma glucose and insulin [50].
More recent mouse experimental models include double-knockout animals for CD36 and other genes such as leptin [51], tyrosine-protein kinase Mer [52], liver-specific signal transducer and activator of transcription (STAT)5 [53], scavenger receptor-A [54], scavenger receptor class B type I [55], heart-specific lipoprotein lipase [56], apolipoprotein E [57], and liver-type FA-binding protein [58].
The CD36 molecular structure includes two transmembrane domains located near both ends of the molecule, joined by a large extracellular region [59]; the transmembrane domains continue with small intracellular tails that are palmitoylated [60] and are important in localizing CD36 within caveolae and lipid rafts [61]. The N-glycosylated extracellular region has a binding site for TSP-1 (residues 93–120) [62] and one site for competitive binding of FA and oxidized low-density lipoprotein (ox)LDL/oxidized glycerophospholipids (residues: 150–168) [63, 64] that can bind hexarelin, one of several growth hormone–releasing peptides [65], and PfEMP1 proteins of the malaria parasite [66]. Neculai et al. [67] found, through an analogy with the crystal structure of structurally related LIMP-2 that they described, a notable feature of the CD36 extracellular domain: a tunnel, mainly comprising hydrophobic residues, spanning its entire length and apparently able to selectively transfer cholesterol esters from the extracellular environment to the outer leaflet of the cell membrane. Thus, future, more detailed structural studies of CD36 could provide actionable targets for therapies for diseases involving this molecule and its numerous, highly variable binding partners.
Other studies have contributed to identifying and understanding the role of CD36 association with other membrane or intracellular molecules. An interesting example is the discovery of CD36 as a regulator of Toll-like receptors 4 and 6 heterodimer assembly that can subsequently trigger inflammatory signalling in microglia [68]. Such data suggest that CD36 can make a major contribution to sterile inflammation in response to atherogenic lipids and amyloid-beta.
Interesting avenues of research might also be opened by a few recent studies that identified a role for noncoding RNA molecules in CD36 expression regulation with functional consequences. miR-758-5p decreases lipid accumulation in foam cells via regulating CD36-mediated cholesterol uptake [69], long noncoding RNA MALAT1 regulates oxLDL-induced CD36 expression via activating β-catenin [70], and uc.372, an ultraconserved RNA belonging to the class of long noncoding RNAs, regulates expression of genes related to lipid synthesis and uptake, including CD36, via suppression of specific miR molecule maturation [71].
Following data gathered from these models and others not mentioned here, the involvement in lipid metabolism and cell-to-matrix adhesion has been confirmed for various cell types, and other functions that are site-specific have been identified and are presented briefly below.
3.1. Lipid Scavenger Receptor and Subsequent Impact on Lipid Metabolism
CD36 has long been known as a scavenger receptor able to bind oxidized LDL (oxLDL) and HDL (oxHDL) [72, 73] but also native lipoprotein molecules [74]. CD36 is involved in high-affinity FA uptake and processing and eventually lipid accumulation and metabolic dysfunction under excessive supply [8]. First found on monocytes and platelets, CD36 is also responsible for uptake of long-chain FA into muscle and adipose tissues and across the brush border [25]. Ligand binding activates phospholipase C, increases cytosolic Ca concentration, and activates chylomicron production [27]. CD36 also regulates the secretion of hepatic very LDL (VLDL), which may explain the correlation between CD36 protein expression and serum levels of VLDL lipid, particle number, and apolipoprotein B in humans [75]. CD36 deletion decreases VLDL output in vivo by increasing prostaglandins D2, F2, and E2 synthesis in the liver [51].
CD36 is a key factor in acute and adaptive regulation of muscle FA oxidation in response to a chronic metabolic stimulus and for the selection of skeletal muscle fuel under basal conditions, during acute exercise, or after muscle training [76]. In heart muscle, CD36 impacts adaptation of myocardial rhythm to energy deprivation [77]. During fasting, CD36 null mice have abnormal myocardial Ca2+ dynamics, phospholipid composition, and cAMP levels and associated conduction anomalies with a high incidence of sudden death [77]. Moreover, recent data have shown that myocardial CD36-mediated signal transduction activates FA β-oxidation [8].
Pathology Impact. Following the interaction of oxLDL with CD36 on intimal transmigrated macrophages, oxLDLs are internalized. They bind to the nuclear hormone receptor PPARɣ followed by the upregulation of CD36, which amplifies oxLDL uptake and foam cell formation [78]. Moreover, by stimulation of cytokine production, intima is further infiltrated and atherosclerotic lesion is formed [8].
3.2. Cell-to-Matrix Attachment
CD36 can bind to extracellular matrix proteins, such as collagen [5] and thrombospondin 2 in platelets and various cell lines. It also binds to TSP-1, but at concentrations higher than physiological, possibly reached in overdeveloped cancerous stromal tissue. CD47 is required for CD36 activation under TSP-1 ligation [79]. In endothelial cells, TSP binding triggers apoptosis, a mechanism bypassed in cancer, where it favours angiogenesis and tumour growth, as discussed later.
4. CD36 as an Early Biomarker for Metastatic Cancer
The term “biomarker” came into frequent use from the 1970s [80] and is currently defined as a “characteristic that is measured as an indicator of normal biological processes, pathogenic processes, or responses to an exposure or intervention, including therapeutic interventions” [81]. A candidate biomarker should provide measurable features, accuracy in indication for a physiologic or pathogenic mechanism, or pharmacological response to a therapeutic approach. According to the US Food and Drug Administration, an ideal biomarker should be highly sensitive and specific for a certain disease, safe, and easily measured in any biological sample, cost-effective, and able to yield accurate results [82].
Correlated with invasion of tumours and metastasis, CD36 has been repeatedly proposed as a prognostic marker in various types of cancers, mostly of epithelial origin. In the following section, we discuss the results that build up the case for CD36 as an “early prognostic marker in metastasis” and review its progress towards clinical validation.
4.1. Preclinical Studies
To be considered as a potential biomarker of metastatic cancer, CD36 must respect the first rule of biomarkers—to exhibit a constant change in disease versus health, in this case, in metastatic cancer versus normal paired control tissue. Indeed, the expression of CD36 has been demonstrated to be increased in human tumour cell lines, human biopsies from ovarian tumours [83], gastric cancer [84], glioblastoma [85], and oral squamous carcinoma [86]. In contrast, consistent with data from tumour growth mechanism studies, CD36 has been reported to be decreased in endothelial cells, as a bypass program of its antiangiogenic effect [87].
Most preclinical studies address CD36 indirectly, in the context of TSP binding. These studies exploit the antiangiogenic effect of TSP 1 and 2 via CD36 signalling, by using recombinant proteins, or TSR peptides, to compensate for loss of TSP in tumour cells. Controversies arose when migration and invasion of cancer cells seemed to be promoted although the primary tumour responded to treatment. A TSP-1 null/breast cancer mouse model demonstrated reduced pulmonary metastases, although there was no impact on primary tumour growth, indicative of effects on the metastatic [88]. The same group demonstrated that inclusion of the RFK sequence in the TSP recombinant protein impacts positively metastases reduction, but in relation to TGFβ activation [88], and that, to some extent, loss of CD36 binding to TSP is compensated by the RFK sequence, in terms of antitumour effect. Another study on a mouse model of breast cancer, using a TSP-2-derived recombinant protein, reported both inhibition of primary tumour growth and reduction of lymph node and lung metastasis. Although the primary effect was positively correlated with CD36-induced mitochondrial apoptosis in endothelial cells and decreased neoangiogenesis; an antimetastatic effect was correlated with the RFK sequence and TGFβ activation [89].
Results with another mouse model of metastatic breast cancer indicated that although CD36 expression in the whole primary tumour was downregulated, this alteration was related to loss of stromal receptor. This hypothesis was confirmed by normal expression of CD36 on isolated tumour cells [90].
In metastatic prostate cancer, CD36 was activated in tumour cells, which led to increased cell migration and invasion, linked to downstream activation of MAPK [91].
Ovarian tumour cells harvested from ascites of patients also express CD36, which was used by Wang et al. as a target for TSP-1-induced apoptosis and subsequent tumour shrinkage in a mouse xenograft model [92]. A recent study showed upregulation of CD36 in metastatic versus primary human ovarian tumours; moreover, blocking CD36 with monoclonal antibodies resulted in reduced tumour burden in a mouse xenograft model [83]. Furthermore, Russell et al. found that combined therapy with thrombospondin-1 type I repeats (3TSR) and chemotherapy induces regression and improves survival in a mouse model of ovarian cancer[93]. These results suggest that CD36 might offer an interesting therapy target, besides its putative biomarker role.
CD36 has been reported on glioblastoma cells as well, in a specific subset of stem-like cells, with role in stemness preservation and tumour initiation [85].
Recently, a study addressed directly the involvement of CD36 in tumour growth and metastasis, by overexpressing CD36 in oral squamous carcinoma cell lines. Tumour cells were then transplanted into immune-competent mice, showing significantly increased metastatic potential over their wild-type counterparts. Conversely, knocking CD36 down led to zero lymph node invasions. The same antimetastatic effect was obtained with CD36- targeting antibodies. While metastasis was prevented, or if already present, significantly reduced, primary lesions were not affected by the treatment [86]. Unlike previous reports, the work of Pascual et al. highlighted a possible cooperation between adipose tissue and tumour cells via CD36, which favours a predominantly lipidic metabolism. The link between increased lipidic profile and tumour progression was also highlighted in an obese mouse model of breast cancer [87]. The authors reported CD36 expression on some, but not all tumour cells, as well as downregulation of CD36 expression on endothelium of neovessels, presumably due to repressed CD36 gene transcription via PDK1-FOXO1 activation by lysophosphatidic acid.
The model that can be delineated so far is that a high CD36 level correlates with metastatic cancers and thus is poorly prognostic. However, blocking CD36 in a tumour system composed of tumour cells and stromal niche would equally affect both populations. An earlier work of deFillipis et al. [94] proposes that loss of CD36 in the pretumoural breast stroma creates a milieu favourable for tumour initiation or progression. Thus, targeting CD36 to prevent metastasis would have a protumorigenic impact on the surrounding stroma. Add to this the proangiogenic effect on the tumour itself, and the outcome will be, very likely, a thriving primary tumour that is possibly nonmetastatic.
These results highlight the importance of integration of models in a correct spatial context, in which stromal niches and tumour cells interact. From this perspective, which part is more important for metastasis? And, in consequence, which population of cells should be targeted in studies aiming at validation of CD36 as a prognostic biomarker (see Figure 1)? Which population holds greater prognostic value: CD36+ tumour cells, as proposed by Pascual et al., or CD36-depleted stroma, as proposed by deFillipis et al.?
Figure 1.
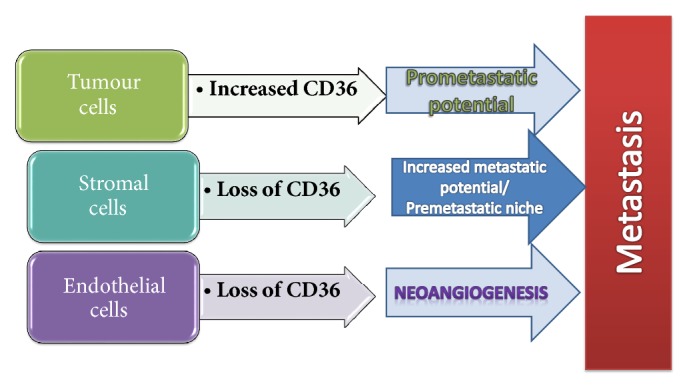
Involvement of CD36 in the metastatic process, related to the main three components of any tumour niche: the tumour cells, stromal cells, and vascularisation. CD36 expression in tumours prone to metastasis is different in each compartment, as demonstrated by animal models and pathology studies in patient samples.
Unfortunately, these aspects have not yet been covered by clinical studies, as discussed in the next section, although CD36 is emerging as a candidate prognostic biomarker in different types of epithelial cancers, alone or in panels with other proteins.
4.2. Human Studies
Data from animal models and in vitro human tumour cell lines point to CD36 as a metastasis-related indicator, prompting investigations on a larger scale in human tumour samples (Table 1). One of the first mentions of CD36 as a possible biomarker for cancer prognostics dates more than 15 years, when it was included in a panel of immunophenotyping for high risk for acute myeloblastic leukaemia [95].
Table 1.
Involvement of CD36 in various types of cancer—data from human sample studies.
| Type of cancer | Location within the tumour | Contribution of CD36 | Refs |
|---|---|---|---|
| Breast cancer | Stroma tissue Decreased endothelial expression Tumour cells |
Angiogenesis | [90] [87] |
| Prostate cancer | Tumour cells | Activation of MAPK signalling, pro-invasion | [91] |
| Ovarian cancer | Tumour cells harvested from patient ascites | Pro-metastatic | [92] |
| Colon cancer | Not specified | Decreased expression in metastatic cancer | [96] |
| Oral squamous cell carcinoma | Tumour cells | Favours lymph node and lung metastasis | [86] |
| Acute myeloid leukaemia | Tumour cells | Part of immunophenotyping panel used for patient stratification | [95] |
| Glioblastoma | Tumour cells | Maintenance of stemness features, tumour initiation | [85] |
| Hepatocellular carcinoma | Not mentioned | Increased CD36 is associated with epithelial-to-mesenchymal transduction | [101] |
In a study of inflammation and cancer, Rachidi et al. used a reductive approach by considering all epithelial cancers as oncoinflammatory events and looking for a common signature. Although CD36 did not meet the criteria for all seven types of cancers studied, a high CD36 gene expression level was proposed as a poor prognostic marker in colon and ovarian cancer when assessed in panel with other proteins [96].
Wang et al. found that 97% of ovarian cancers express CD36, as do 100% of lymph node metastases. Furthermore, the receptor's expression was increased with disease progression [92]. CD36 was also detected in liposarcoma and prostate and breast tumours; in the last type, translocation of the protein from the cytoplasm to cell membrane was related to oestrogen signalling [97].
Firm confirmation of the relationship between CD36 and metastasis came from a study of over 2500 cases of different types of cancers (a “pan-cancer” study) in which genes involved in metabolic rewiring towards aerobic glycolysis and de novo FA synthesis were assessed in metastatic tumours compared to primary tumours. The CD36 gene was frequently amplified in metastatic tumours and survival rates in the high-copy-number group were reduced when compared with low-copy-number patients [98].
5. Clinical Trials
Direct targeting of CD36 in tumour pathology has not yet been addressed in cancer-related clinical trials. Rather, its ability to bind TSP1 and modulate antiangiogenic responses was exploited therapeutically. Several clinical trials tested TSP-1 peptidomimetics specifically binding to CD36, but they were discontinued for lack of response or severe adverse effects [79]. Data gathered in preclinical models of various types of cancers taught that blocking CD36 might prove beneficial in stopping metastases from spreading. Lessons learned from other fields of successful clinical research show that humanized monoclonal antibodies are a valid option (reviewed in [99]). But a recent commentary estimates that development of antibodies against CD36, to be used in clinical trials, would take at least 4 years [100].
Continuing the pipeline of TSP peptidomimetics, in line with the lipid scavenger function of CD36, apolipoprotein A-I–mimetic peptides are being tested in preclinical trials, but mostly for noncancerous pathologies [102–104].
CD36 has been and continues to be investigated as a possible biomarker in metabolic diseases (obesity, insulin resistance, and diabetes type 2), cardiovascular diseases, and autoimmune/inflammatory conditions (Table 2).
Table 2.
Clinical trials investigating CD36 expression.
| No | Conditions | Title | Interventions | Identifier |
|---|---|---|---|---|
| 1 | Healthy Young/Elderly | Effects of Native Whey or Milk Supplementation on Adaptations to 12 Weeks of Strength Training in Young and Elderly | Dietary Supplement: Native whey|Dietary Supplement: Milk|Other: Strength training | NCT03033953 |
| Anabolic Effects of Whey and Casein After Strength Training in Young and Elderly | Strength training|Dietary Supplement: Milk 1%|Dietary Supplement: Whey protein concentrate 80|Dietary Supplement: Native whey | NCT02968888 | ||
|
| ||||
| 2 | Taste Sensitivity|Fatty Acid Type | Fatty Acid Taste Thresholds: Caproic, Lauric, Oleic, Linoleic, Linolenic | NCT01996566 | |
|
| ||||
| 3 | Obesity/Metabolic Syndrome | CD36 and Human Fat Taste Perception | No intervention | NCT02699567 |
| Metabolic and Cardiovascular Impact of CD36 Deficiency in African Americans | NCT02126735 | |||
| Study 8: Fat Perception in Humans (09-0873) | NCT01128400 | |||
| Study 19: Metabolically Normal and Metabolically Abnormal Obesity | Behavioral: overfeeding | NCT01184170 | ||
|
| ||||
| 4 | Insulin Resistance|Endothelial Dysfunction | CD36 in Nutrient Delivery and Its Dysfunction | Sildenafil Citrate (Viagra) | NCT03012386 |
|
| ||||
| 5 | Type 2 Diabetes or Obesity Without Diabetes | Search for Biological Markers of Orosensory Perception of Fatty Acids in Healthy Subjects and Possible Modifications in Patients With Type 2 Diabetes and in Obese Non-diabetic Patients. | Other: Measure the threshold of detection for linoleic acid|Other: Oral stimulation tests|Other: Venous blood samples|Other: Samples for genetic studies (ancillary study) | NCT02028975 |
|
| ||||
| 6 | Moderate Hypertriacylglycerolemic Subjects | Intervention With n3 LC-PUFA-supplemented Yogurt | Dietary Supplement: n3 long chain polyunsaturated fatty acids | NCT01244048 |
|
| ||||
| 7 | Diabetes Mellitus, Type 2|Dyslipidemia Associated With Type II Diabetes Mellitus|Percutaneous Coronary Intervention | Effects of Evolocumab on Platelet Reactivity in Patients With Diabetes Mellitus After Elective Percutaneous Coronary Intervention | Drug: Evolocumab|Drug: Placebo | NCT03258281 |
|
| ||||
| Insulin Resistance|Diabetes|Cancer|Obesity|Inflammation | Study 14: Diet and Metabolic Inflammation | Other: Diet A|Other: Diet B | NCT02539355 | |
|
| ||||
| Renal Failure Chronic Requiring Hemodialysis|Metabolic Syndrome|Diabetes Mellitus Type 2|Hyperlipidemia | Study 16: Salusin-alpha - a New Factor in the Pathogenesis of Lipid Abnormalities in Hemodialysis Patients | Drug: Atorvastatin | NCT01448174 | |
|
| ||||
| Cardiovascular Abnormalities | Study 13: A Study of Immunological Biomarkers as Predictors of Cardiovascular Events | Other: blood tests | NCT02894060 | |
|
| ||||
| Coronary Artery Disease | Polymorphisms in CD36 and STAT3 Genes and Different Dietary Interventions Among Patients With Coronary Artery Disease | Dietary Supplement: Olive oil|Dietary Supplement: Nuts|Dietary Supplement: Control diet | NCT02202265 | |
|
| ||||
| Thrombocytopenia | Study 5: Identification of Donors of CD36-Deficient Platelets Among Japanese Individuals on the NIH Campus | None | NCT00015639 | |
|
| ||||
| Idiopathic Thrombocytopenic Purpura | The Effect of Eltrombopag on the Expression of Platelet Collagen Receptor GPVI in Pediatric ITP. | Drug: Eltrombopag|Drug: conventional | NCT03412188 | |
|
| ||||
| Myelodysplastic Syndromes | Study 22: Azacitidine and Erythropoietin Versus Azacitidine Alone for Patients With Low-Risk Myelodysplastic Syndromes | Azacitidine|Drug: Erythropoietin|Drug: Azacitidine (Monotherapy) | NCT00379912 | |
|
| ||||
| Rheumatoid Arthritis | Study 7: A New Mode of Action of Anti-TNF, Reverse Signaling, in Rheumatoid Arthritis | Diagnostic Test: Blood test | NCT03216928 | |
|
| ||||
| Rheumatoid Arthritis | Atherosclerosis in Rheumatoid Arthritis and Lupus: Restoring Cholesterol Balance | NCT01180361 | ||
|
| ||||
| SLE|Rheumatoid Arthritis|Healthy Subjects | Study 15: Efferocytosis and Genomic Polymorphism in Autoimmune Diseases | NCT00364728 | ||
|
| ||||
| HIV Infections | Study 12: Maraviroc 300 mg Twice Daily in HIV Negative Male Volunteers | Drug: Maraviroc | NCT00771823 | |
The hope is that data gathered from these clinical trials will be highly informative about the pharmacological profile and side-effects of various types of CD-36 related compounds, for further repurposing in cancer therapy.
6. Future Perspectives in CD36-Related Tumour Biology
Plenty of data have been gathered to demonstrate CD36 involvement in metastasis spreading and, yet, novel and exciting avenues are still opening. Along with new reports of CD36 involvement in normal mitochondrial function [105], one could ask how CD36 increase impacts tumour cells energetic metabolism and the effect of CD 36 inhibition on bystander cells.
Further on, based on tumour animal models, the next step would be translation to human pathology. This process has already started, at least at the bioinformatics level, contributing to acknowledgment of CD36 as a possible prognostic biomarker for metastatic cancer, by compiling data from repositories and meta-analyses. So far, high levels of CD36 have been proposed as a poor prognostic marker for colon and ovarian cancer [96] as well as for breast cancer, lung small cell carcinoma, and urinary bladder carcinoma [86].
If the molecule is to be included in further clinical trials, validation in large cohorts remains to be accomplished, along with clarifications in some grey areas, such as site of detection of CD36 (stromal cells versus tumour cells) and proper quantification.
Acknowledgments
This work was partially supported by Core Program, implemented with the support of MRI [Projects nos. 18.21-01.01, 18.21-02.01, and 18.21-01.06].
Contributor Information
Eugen Radu, Email: eugen.radu@umfcd.ro.
Mihail Eugen Hinescu, Email: mhinescu@yahoo.com.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- 1.Massagué J., Obenauf A. C. Metastatic colonization by circulating tumour cells. Nature. 2016;529(7586):298–306. doi: 10.1038/nature17038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sleeman J. P. The metastatic niche and stromal progression. Cancer and Metastasis Reviews. 2012;31(3-4):429–440. doi: 10.1007/s10555-012-9373-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Irmisch A., Huelsken J. Metastasis: new insights into organ-specific extravasation and metastatic niches. Experimental Cell Research. 2013;319(11):1604–1610. doi: 10.1016/j.yexcr.2013.02.012. [DOI] [PubMed] [Google Scholar]
- 4.Silverstein R. L., Febbraio M. CD36, a scavenger receptor involved in immunity, metabolism, angiogenesis, and behavior. Science Signaling. 2009;2(72, article re3):16. doi: 10.1126/scisignal.272re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tandon N. N., Kralisz U., Jamieson G. A. Identification of glycoprotein IV (CD36) as a primary receptor for platelet-collagen adhesion. The Journal of Biological Chemistry. 1989;264(13):7576–7583. [PubMed] [Google Scholar]
- 6.Alessio M., De Monte L., Scirea A., Gruarin P., Tandon N. N., Sitia R. Synthesis, processing, and intracellular transport of CD36 during monocytic differentiation. The Journal of Biological Chemistry. 1996;271(3):1770–1775. doi: 10.1074/jbc.271.3.1770. [DOI] [PubMed] [Google Scholar]
- 7.Woo M. S., Yang J., Beltran C., Cho S. Cell surface CD36 protein in monocyte/macrophage contributes to phagocytosis during the resolution phase of ischemic stroke in mice. The Journal of Biological Chemistry. 2016;291(45):23654–23661. doi: 10.1074/jbc.m116.750018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pepino M. Y., Kuda O., Samovski D., Abumrad N. A. Structure-function of CD36 and importance of fatty acid signal transduction in fat metabolism. Annual Review of Nutrition. 2014;34:281–303. doi: 10.1146/annurev-nutr-071812-161220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thorne R. F., Law E. G., Elith C. A., Ralston K. J., Bates R. C., Burns G. F. The association between CD36 and Lyn protein tyrosine kinase is mediated by lipid. Biochemical and Biophysical Research Communications. 2006;351(1):51–56. doi: 10.1016/j.bbrc.2006.09.156. [DOI] [PubMed] [Google Scholar]
- 10.Githaka J. M., Vega A. R., Baird M. A., Davidson M. W., Jaqaman K., Touret N. Ligand-induced growth and compaction of CD36 nanoclusters enriched in Fyn induces Fyn signaling. Journal of Cell Science. 2016;129(22):4175–4189. doi: 10.1242/jcs.188946. [DOI] [PubMed] [Google Scholar]
- 11.Asch A. S., Silbiger S., Heimer E., Nachman R. L. Thrombospondin sequence motif (CSVTCG) is responsible for CD36 binding. Biochemical and Biophysical Research Communications. 1992;182(3):1208–1217. doi: 10.1016/0006-291X(92)91860-S. [DOI] [PubMed] [Google Scholar]
- 12.Park Y. M. CD36, a scavenger receptor implicated in atherosclerosis. Experimental & Molecular Medicine. 2014;46(6, article e99) doi: 10.1038/emm.2014.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trezzini C., Jungi T. W., Spycher M. O., Maly F. E., Rao P. Human monocytes CD36 and CD16 are signaling molecules. Evidence from studies using antibody-induced chemiluminescence as a tool to probe signal transduction. The Journal of Immunology. 1990;71(1):29–37. [PMC free article] [PubMed] [Google Scholar]
- 14.Pohl J., Ring A., Korkmaz Ü., Ehehalt R., Stremmel W. FAT/CD36-mediated long-chain fatty acid uptake in adipocytes requires plasma membrane rafts. Molecular Biology of the Cell (MBoC) 2005;16(1):24–31. doi: 10.1091/mbc.E04-07-0616. doi: 10.1091/mbc.E04-07-0616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Souto R. P., Vallega G., Wharton J., Vinten J., Tranum-Jensen J., Pilch P. F. Immunopurification and characterization of rat adipocyte caveolae suggest their dissociation from insulin signaling. The Journal of Biological Chemistry. 2003;278(20):18321–18329. doi: 10.1074/jbc.M211541200. [DOI] [PubMed] [Google Scholar]
- 16.Luiken J. J., Dyck D. J., Han X., et al. Insulin induces the translocation of the fatty acid transporter FAT/CD36 to the plasma membrane. American Journal of Physiology-Endocrinology and Metabolism. 2002;282(2):E491–E495. doi: 10.1152/ajpendo.00419.2001. [DOI] [PubMed] [Google Scholar]
- 17.Bezaire V., Bruce C. R., Heigenhauser G. J. F., et al. Identification of fatty acid translocase on human skeletal muscle mitochondrial membranes: Essential role in fatty acid oxidation. American Journal of Physiology-Endocrinology and Metabolism. 2006;290(3):E509–E515. doi: 10.1152/ajpendo.00312.2005. [DOI] [PubMed] [Google Scholar]
- 18.Koonen D. P. Y., Glatz J. F. C., Bonen A., Luiken J. J. F. P. Long-chain fatty acid uptake and FAT/CD36 translocation in heart and skeletal muscle. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 2005;1736(3):163–180. doi: 10.1016/j.bbalip.2005.08.018. [DOI] [PubMed] [Google Scholar]
- 19.Swerlick R. A., Lee K. H., Wick T. M., Lawley T. J. Human dermal microvascular endothelial but not human umbilical vein endothelial cells express CD36 in vivo and in vitro. The Journal of Immunology. 1992;148(1):78–83. [PubMed] [Google Scholar]
- 20.Krieger M. Charting the fate of the “good cholesterol”: identification and characterization of the high-density lipoprotein receptor SR-BI. Annual Review of Biochemistry. 1999;68:523–558. doi: 10.1146/annurev.biochem.68.1.523. [DOI] [PubMed] [Google Scholar]
- 21.Greenwalt D. E., So O. Y., Jiwani N., Watt K. W. K. PAS IV, an Integral Membrane Protein of Mammary Epithelial Cells, Is Related to Platelet and Endothelial Cell CD36 (GP IV) Biochemistry. 1990;29(30):7054–7059. doi: 10.1021/bi00482a015. [DOI] [PubMed] [Google Scholar]
- 22.Laugerette F., Passilly-Degrace P., Patris B., et al. CD36 involvement in orosensory detection of dietary lipids, spontaneous fat preference, and digestive secretions. The Journal of Clinical Investigation. 2005;115(11):3177–3184. doi: 10.1172/JCI25299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simons P. J., Kummer J. A., Luiken J. J. F. P., Boon L. Apical CD36 immunolocalization in human and porcine taste buds from circumvallate and foliate papillae. Acta Histochemica. 2011;113(8):839–843. doi: 10.1016/j.acthis.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 24.Martin C., Passilly-Degrace P., Gaillard D., Merlin J.-F., Chevrot M., Besnard P. The lipid-sensor candidates CD36 and GPR120 are differentially regulated by dietary lipids in mouse taste buds: impact on spontaneous fat preference. PLoS ONE. 2011;6(8) doi: 10.1371/journal.pone.0024014.e24014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siddiqi S., Sheth A., Patel F., Barnes M., Mansbach C. M., II Intestinal caveolin-1 is important for dietary fatty acid absorption. Biochimica et Biophysica Acta. 2013;1831(8):1311–1321. doi: 10.1016/j.bbalip.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lobo M. V. T., Huerta L., Ruiz-Velasco N., et al. Localization of the lipid receptors CD36 and CLA-1/SR-BI in the human gastrointestinal tract: Towards the identification of receptors mediating the intestinal absorption of dietary lipids. Journal of Histochemistry & Cytochemistry. 2001;49(10):1253–1260. doi: 10.1177/002215540104901007. [DOI] [PubMed] [Google Scholar]
- 27.Tran T. T. T., Poirier H., Clément L., et al. Luminal lipid regulates CD36 levels and downstream signaling to stimulate chylomicron synthesis. The Journal of Biological Chemistry. 2011;286(28):25201–25210. doi: 10.1074/jbc.M111.233551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ring A., Le Lay S., Pohl J., Verkade P., Stremmel W. Caveolin-1 is required for fatty acid translocase (FAT/CD36) localization and function at the plasma membrane of mouse embryonic fibroblasts. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 2006;1761(4):416–423. doi: 10.1016/j.bbalip.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 29.Sundaresan S., Shahid R., Riehl T. E., et al. CD36-dependent signaling mediates fatty acid-induced gut release of secretin and cholecystokinin. The FASEB Journal. 2013;27(3):1191–1202. doi: 10.1096/fj.12-217703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malerød L., Juvet L., Gjøen T., Berg T. The expression of scavenger receptor class B, type I (SR-BI) and caveolin-1 in parenchymal and nonparenchymal liver cells. Cell and Tissue Research. 2002;307(2):173–180. doi: 10.1007/s00441-001-0476-9. [DOI] [PubMed] [Google Scholar]
- 31.Steneberg P., Sykaras A. G., Backlund F., Straseviciene J., Söderström I., Edlund H. Hyperinsulinemia enhances hepatic expression of the fatty acid transporter Cd36 and provokes hepatosteatosis and hepatic insulin resistance. The Journal of Biological Chemistry. 2015;290(31):19034–19043. doi: 10.1074/jbc.M115.640292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sheedfar F., Sung M. M. Y., Aparicio-Vergara M., et al. Increased hepatic CD36 expression with age is associated with enhanced susceptibility to nonalcoholic fatty liver disease. AGING. 2014;6(4):281–295. doi: 10.18632/aging.100652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koonen D. P. Y., Jacobs R. L., Febbraio M. Increased hepatic CD36 expression contributes to dyslipidemia associated with diet-induced obesity. Diabetes. 2007;56(12):2863–2871. doi: 10.2337/db07-0907. [DOI] [PubMed] [Google Scholar]
- 34.Petrik J. J., Gentry P. A., Feige J.-J., LaMarre J. Expression and localization of thrombospondin-1 and -2 and their cell-surface receptor, CD36, during rat follicular development and formation of the corpus luteum. Biology of Reproduction. 2002;67(5):1522–1531. doi: 10.1095/biolreprod.102.007153. [DOI] [PubMed] [Google Scholar]
- 35.Osz K., Ross M., Petrik J. The thrombospondin-1 receptor CD36 is an important mediator of ovarian angiogenesis and folliculogenesis. Reproductive Biology and Endocrinology. 2014;12(1, article no. 21) doi: 10.1186/1477-7827-12-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Asch A. S., Barnwell J., Silverstein R. L., Nachman R. L. Isolation of the thrombospondin membrane receptor. The Journal of Clinical Investigation. 1987;79(4):1054–1061. doi: 10.1172/JCI112918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tandon N. N., Lipsky R. H., Burgess W. H., Jamieson G. A. Isolation and characterization of platelet glycoprotein IV (CD36) The Journal of Biological Chemistry. 1989;264(13):7570–7575. [PubMed] [Google Scholar]
- 38.Aiken M. L., Ginsberg M. H., Byers-Ward V., Plow E. F. Effects of OKM5, a monoclonal antibody to glycoprotein IV, on platelet aggregation and thrombospondin surface expression. Blood. 1990;76(12):2501–2509. [PubMed] [Google Scholar]
- 39.Tandon N. N., Ockenhouse C. F., Greco N. J., Jamieson G. A. Adhesive functions of platelets lacking glycoprotein IV (CD36) Blood. 1991;78(11):2809–2813. [PubMed] [Google Scholar]
- 40.Harmon C. M., Abumrad N. A. Binding of sulfosuccinimidyl fatty acids to adipocyte membrane proteins: Isolation and ammo-terminal sequence of an 88-kD protein implicated in transport of long-chain fatty acids. Journal of Membrane Biology. 1993;133(1):43–49. doi: 10.1007/BF00231876. [DOI] [PubMed] [Google Scholar]
- 41.Febbraio M., Abumrad N. A., Hajjar D. P., et al. A null mutation in murine CD36 reveals an important role in fatty acid and lipoprotein metabolism. The Journal of Biological Chemistry. 1999;274(27):19055–19062. doi: 10.1074/jbc.274.27.19055. [DOI] [PubMed] [Google Scholar]
- 42.Chawla A., Barak Y., Nagy L., Liao D., Tontonoz P., Evans R. M. PPAR-γ dependent and independent effects on macrophage-gene expression in lipid metabolism and inflammation. Nature Medicine. 2001;7(1):48–52. doi: 10.1038/83336. [DOI] [PubMed] [Google Scholar]
- 43.Jiménez B., Volpert O. V., Crawford S. E., Febbraio M., Silverstein R. L., Bouck N. Signals leading to apoptosis-dependent inhibition of neovascularization by thrombospondin-1. Nature Medicine. 2000;6(1):41–48. doi: 10.1038/71517. [DOI] [PubMed] [Google Scholar]
- 44.Febbraio M., Podrez E. A., Smith J. D., et al. Targeted disruption of the class B, scavenger receptor CD36 protects against atherosclerotic lesion development in mice. The Journal of Clinical Investigation. 2000;105(8):1049–1056. doi: 10.1172/JCI9259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Young M. P., Febbraio M., Silverstein R. L. CD36 modulates migration of mouse and human macrophages in response to oxidized LDL and may contribute to macrophage trapping in the arterial intima. The Journal of Clinical Investigation. 2009;119(1):136–145. doi: 10.1172/jci35535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Coburn C. T., Knapp F.F. J., Febbraio M., Beets A. L., Silverstein R. L., Abumrad N. A. Defective uptake and utilization of long chain fatty acids in muscle and adipose tissues of CD36 knockout mice. The Journal of Biological Chemistry. 2000;275(42):32523–32529. doi: 10.1074/jbc.M003826200. [DOI] [PubMed] [Google Scholar]
- 47.Hajri T., Han X. X., Bonen A., Abumrad N. A. Defective fatty acid uptake modulates insulin responsiveness and metabolic responses to diet in CD36-null mice. The Journal of Clinical Investigation. 2002;109(10):1381–1389. doi: 10.1172/JCI200214596. doi: 10.1172/JCI200214596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ohgami N., Nagai R., Ikemoto M., et al. CD36, a member of the class B scavenger receptor family, as a receptor for advanced glycation end products. The Journal of Biological Chemistry. 2001;276(5):3195–3202. doi: 10.1074/jbc.M006545200. [DOI] [PubMed] [Google Scholar]
- 49.Gaillard D., Laugerette F., Darcel N., et al. The gustatory pathway is involved in CD36-mediated orosensory perception of long-chain fatty acids in the mouse. The FASEB Journal. 2008;22(5):1458–1468. doi: 10.1096/fj.07-8415com. [DOI] [PubMed] [Google Scholar]
- 50.Ibrahimi A., Bonen A., Blinn W. D., et al. Muscle-specific overexpression of FAT/CD36 enhances fatty acid oxidation by contracting muscle, reduces plasma triglycerides and fatty acids, and increases plasma glucose and insulin. The Journal of Biological Chemistry. 1999;274(38):26761–26766. doi: 10.1074/jbc.274.38.26761. [DOI] [PubMed] [Google Scholar]
- 51.Nassir F., Adewole O. L., Brunt E. M., Abumrad N. A. CD36 deletion reduces VLDL secretion, modulates liver prostaglandins, and exacerbates hepatic steatosis in ob/ob mice. Journal of Lipid Research. 2013;54(11):2988–2997. doi: 10.1194/jlr.M037812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dehn S., Thorp E. B. Myeloid receptor CD36 is required for early phagocytosis of myocardial infarcts and induction of Nr4a1-dependent mechanisms of cardiac repair. The FASEB Journal. 2018;32(1):254–264. doi: 10.1096/fj.201700450R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hosui A., Tatsumi T., Hikita H., et al. Signal transducer and activator of transcription 5 plays a crucial role in hepatic lipid metabolism through regulation of CD36 expression. Hepatology Research. 2016;9:5–15. doi: 10.1111/hepr.12816. [DOI] [PubMed] [Google Scholar]
- 54.Blanchet C., Jouvion G., Fitting C., Cavaillon J.-M., Adib-Conquy M. Protective or deleterious role of scavenger receptors SR-A and CD36 on host resistance to Staphylococcus aureus depends on the site of infection. PLoS ONE. 2014;9(1) doi: 10.1371/journal.pone.0087927.e87927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Truongy T. Q., Aubin D., Falstrault L., Brodeurz M. R., Brissette L. SR-BI, CD36, and caveolin-1 contribute positively to cholesterol efflux in hepatic cells. Cell Biochemistry & Function. 2010;28(6):480–489. doi: 10.1002/cbf.1680. [DOI] [PubMed] [Google Scholar]
- 56.Bharadwaj K. G., Hiyama Y., Hu Y., et al. Chylomicron- and VLDL-derived lipids enter the heart through different pathways: In vivo evidence for receptor- and non-receptor-mediated fatty acid uptake. The Journal of Biological Chemistry. 2010;285(49):37976–37986. doi: 10.1074/jbc.M110.174458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Harb D., Bujold K., Febbraio M., Sirois M. G., Ong H., Marleau S. The role of the scavenger receptor CD36 in regulating mononuclear phagocyte trafficking to atherosclerotic lesions and vascular inflammation. Cardiovascular Research. 2009;83(1):42–51. doi: 10.1093/cvr/cvp081. [DOI] [PubMed] [Google Scholar]
- 58.Xie Y., Cifarelli V., Pietka T., et al. Cd36 knockout mice are protected against lithogenic diet-induced gallstones. Journal of Lipid Research. 2017;58(8):1692–1701. doi: 10.1194/jlr.M077479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Febbraio M., Hajjar D. P., Silverstein R. L. CD36: a class B scavenger receptor involved in angiogenesis, atherosclerosis, inflammation, and lipid metabolism. The Journal of Clinical Investigation. 2001;108(6):785–791. doi: 10.1172/jci200114006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tao N., Wagner S. J., Lublin D. M. CD36 is palmitoylated on both N- and C-terminal cytoplasmic tails. The Journal of Biological Chemistry. 1996;271(37):22315–22320. doi: 10.1074/jbc.271.37.22315. [DOI] [PubMed] [Google Scholar]
- 61.Rios F. J. O., Ferracini M., Pecenin M., et al. Uptake of oxLDL and IL-10 production by macrophages requires PAFR and CD36 recruitment into the same lipid rafts. PLoS ONE. 2013;8(10) doi: 10.1371/journal.pone.0076893.e76893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pearce S. F. A., Wu J., Silverstein R. L. Recombinant GST/CD36 fusion proteins define a thrombospondin binding domain: Evidence for a single calcium-dependent binding site on CD36. The Journal of Biological Chemistry. 1995;270(7):2981–2986. doi: 10.1074/jbc.270.7.2981. [DOI] [PubMed] [Google Scholar]
- 63.Takai M., Kozai Y., Tsuzuki S., et al. Unsaturated long-chain fatty acids inhibit the binding of oxidized low-density lipoproteins to a model CD36. Bioscience, Biotechnology, and Biochemistry. 2014;78(2):238–244. doi: 10.1080/09168451.2014.882750. [DOI] [PubMed] [Google Scholar]
- 64.Tsuzuki S., Yamasaki M., Kozai Y., et al. Assessment of direct interaction between CD36 and an oxidized glycerophospholipid species. The Journal of Biochemistry. 2017;162(3):163–172. doi: 10.1093/jb/mvx019. [DOI] [PubMed] [Google Scholar]
- 65.Demers A., McNicoll N., Febbraio M., et al. Identification of the growth hormone-releasing peptide binding site in CD36: a photoaffinity cross-linking study. Biochemical Journal. 2004;382(2):417–424. doi: 10.1042/bj20040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hsieh F.-L., Turner L., Bolla J. R., Robinson C. V., Lavstsen T., Higgins M. K. The structural basis for CD36 binding by the malaria parasite. Nature Communications. 2016;7 doi: 10.1038/ncomms12837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Neculai D., Schwake M., Ravichandran M., et al. Structure of LIMP-2 provides functional insights with implications for SR-BI and CD36. Nature. 2013;504(7478):172–176. doi: 10.1038/nature12684. [DOI] [PubMed] [Google Scholar]
- 68.Stewart C. R., Stuart L. M., Wilkinson K., et al. CD36 ligands promote sterile inflammation through assembly of a Toll-like receptor 4 and 6 heterodimer. Nature Immunology. 2010;11(2):155–161. doi: 10.1038/ni.1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li B., Xia L., Liu J., et al. miR-758-5p regulates cholesterol uptake via targeting the CD36 3′UTR. Biochemical and Biophysical Research Communications. 2017;494(1-2):384–389. doi: 10.1016/j.bbrc.2017.09.150. [DOI] [PubMed] [Google Scholar]
- 70.Huangfu N., Xu Z., Zheng W., Wang Y., Cheng J., Chen X. LncRNA MALAT1 regulates oxLDL-induced CD36 expression via activating β-catenin. Biochemical and Biophysical Research Communications. 2018;495(3):2111–2117. doi: 10.1016/j.bbrc.2017.12.086. [DOI] [PubMed] [Google Scholar]
- 71.Guo J., Fang W., Sun L., et al. Ultraconserved element uc.372 drives hepatic lipid accumulation by suppressing miR-195/miR4668 maturation. Nature Communications. 2018;9(1) doi: 10.1038/s41467-018-03072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Endemann G., Stanton L. W., Madden K. S., Bryant C. M., White R. T., Protter A. A. CD36 is a receptor for oxidized low density lipoprotein. The Journal of Biological Chemistry. 1993;268(16):11811–11816. [PubMed] [Google Scholar]
- 73.Thorne R. F., Mhaidat N. M., Ralston K. J., Burns G. F. CD36 is a receptor for oxidized high density lipoprotein: Implications for the development of atherosclerosis. FEBS Letters. 2007;581(6):1227–1232. doi: 10.1016/j.febslet.2007.02.043. [DOI] [PubMed] [Google Scholar]
- 74.Calvo D., Gómez-Coronado D., Suárez Y., Lasunción M. A., Vega M. A. Human CD36 is a high affinity receptor for the native lipoproteins HDL, LDL, and VLDL. Journal of Lipid Research. 1998;39(4):777–788. [PubMed] [Google Scholar]
- 75.Love-Gregory L., Sherva R., Schappe T., et al. Common CD36 SNPs reduce protein expression and may contribute to a protective atherogenic profile. Human Molecular Genetics. 2011;20(1):193–201. doi: 10.1093/hmg/ddq449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McFarlan J. T., Yoshida Y., Jain S. S., et al. In vivo, fatty acid translocase (CD36) critically regulates skeletal muscle fuel selection, exercise performance, and training-induced adaptation of fatty acid oxidation. The Journal of Biological Chemistry. 2012;287(28):23502–23516. doi: 10.1074/jbc.M111.315358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pietka T. A., Sulkin M. S., Kuda O., et al. CD36 protein influences myocardial Ca2+ homeostasis and phospholipid metabolism: Conduction anomalies in CD36-deficient mice during fasting. The Journal of Biological Chemistry. 2012;287(46):38901–38912. doi: 10.1074/jbc.M112.413609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rahaman S. O., Lennon D. J., Febbraio M., Podrez E. A., Hazen S. L., Silverstein R. A CD36-dependent signaling cascade is necessary for macrophage foam cell formation. Cell Metabolism. 2006;4(3):211–221. doi: 10.1016/j.cmet.2006.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jeanne A., Schneider C., Martiny L., Dedieu S. Original insights on thrombospondin-1-related antireceptor strategies in cancer. Frontiers in Pharmacology. 2015;6 doi: 10.3389/fphar.2015.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yuhas J. M., Li A. P., Martinez A. O., Ladman A. J. A simplified method for production and growth of multicellular tumor spheroids. Cancer Research. 1977;37(10):3639–3643. [PubMed] [Google Scholar]
- 81.FDA-NIH Biomarker Working Group. BEST (Biomarkers, EndpointS, and other Tools) Resource. Food and Drug Administration (US), Silver Spring (MD); 2016. [PubMed] [Google Scholar]
- 82.Jain K. K. The Handbook of Biomarkers. New York, NY, USA: Springer New York; 2017. Technologies for Discovery of Biomarkers. [DOI] [Google Scholar]
- 83.Ladanyi A., Mukherjee A., Kenny H. A., et al. Adipocyte-induced CD36 expression drives ovarian cancer progression and metastasis. Oncogene. 2018;37(17):2285–2301. doi: 10.1038/s41388-017-0093-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kashihara H., Shimada M., Yoshikawa K., et al. Correlation between thrombospondin-1 expression in non-cancer tissue and gastric carcinogenesis. Anticancer Reseach. 2017;37(7):3547–3552. doi: 10.21873/anticanreS.11724. [DOI] [PubMed] [Google Scholar]
- 85.Hale J. S., Otvos B., Sinyuk M., et al. Cancer stem cell-specific scavenger receptor 36 drives glioblastoma progression. Stem Cells. 2014;32(7):1746–1758. doi: 10.1002/stem.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pascual G., Avgustinova A., Mejetta S., et al. Targeting metastasis-initiating cells through the fatty acid receptor CD36. Nature. 2017;541(7635):41–45. doi: 10.1038/nature20791. [DOI] [PubMed] [Google Scholar]
- 87.Dong L., Yuan Y., Opansky C., et al. Diet-induced obesity links to ER positive breast cancer progression via LPA/PKD-1-CD36 signaling-mediated microvascular remodeling. Oncotarget . 2017;8(14):22550–22562. doi: 10.18632/oncotarget.15123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yee K. O., Connolly C. M., Duquette M., Kazerounian S., Washington R., Lawler J. The effect of thrombospondin-1 on breast cancer metastasis. Breast Cancer Research and Treatment. 2009;114(1):85–96. doi: 10.1007/s10549-008-9992-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Koch M., Hussein F., Woeste A., et al. CD36-mediated activation of endothelial cell apoptosis by an N-terminal recombinant fragment of thrombospondin-2 inhibits breast cancer growth and metastasis in vivo. Breast Cancer Research and Treatment. 2011;128(2):337–346. doi: 10.1007/s10549-010-1085-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Johnstone C. N., Smith Y. E., Cao Y., et al. Functional and molecular characterisation of EO771.LMB tumours, a new C57BL/6-mouse-derived model of spontaneously metastatic mammary cancer. DISEASE MODELS & MECHANISMS. 2015;8(3):237–251. doi: 10.1242/dmm.017830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen P.-C., Tang C.-H., Lin L.-W., et al. Thrombospondin-2 promotes prostate cancer bone metastasis by the up-regulation of matrix metalloproteinase-2 through down-regulating miR-376c expression. Journal of Hematology & Oncology. 2017;10(1, article no. 33) doi: 10.1186/s13045-017-0390-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang S., Blois A., El Rayes T., et al. Development of a prosaposin-derived therapeutic cyclic peptide that targets ovarian cancer via the tumor microenvironment. Science Translational Medicine. 2016;8(329) doi: 10.1126/scitranslmed.aad5653.329ra34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Russell S., Duquette M., Liu J., Drapkin R., Lawler J., Petrik J. Combined therapy with thrombospondin-1 type I repeats (3TSR) and chemotherapy induces regression and significantly improves survival in a preclinical model of advanced stage epithelial ovarian cancer. The FASEB Journal. 2015;29(2):576–588. doi: 10.1096/fj.14-261636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.DeFilippis R. A., Chang H., Dumont N., et al. CD36 repression activates a multicellular stromal program shared by high mammographic density and tumor tissues. Cancer Discovery. 2012;2(9):826–839. doi: 10.1158/2159-8290.CD-12-0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Casasnovas R. O., Slimane F. K., Garand R., et al. Immunological classification of acute myeloblastic leukemias: relevance to patient outcome. Leukemia. 2003;17(3):515–527. doi: 10.1038/sj.leu.2402821. [DOI] [PubMed] [Google Scholar]
- 96.Rachidi S. M., Qin T., Sun S., Zheng W. J., Li Z. Molecular profiling of multiple human cancers defines an inflammatory cancer-associated molecular pattern and uncovers KPNA2 as a uniform poor prognostic cancer marker. PLoS ONE. 2013;8(3) doi: 10.1371/journal.pone.0057911.e57911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kuemmerle N. B., Rysman E., Lombardo P. S., et al. Lipoprotein lipase links dietary fat to solid tumor cell proliferation. Molecular Cancer Therapeutics. 2011;10(3):427–436. doi: 10.1158/1535-7163.MCT-10-0802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nath A., Chan C. Genetic alterations in fatty acid transport and metabolism genes are associated with metastatic progression and poor prognosis of human cancers. Scientific Reports. 2016;6 doi: 10.1038/srep18669.18669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pento J. T. Monoclonal antibodies for the treatment of cancer. Anticancer Reseach. 2017;37(11):5935–5939. doi: 10.21873/anticanres.12040. [DOI] [PubMed] [Google Scholar]
- 100.Ledford H. Fat fuels cancer’s spread in mice. Nature. 2016 doi: 10.1038/nature.2016.21092. [DOI] [Google Scholar]
- 101.Nath A., Li I., Roberts L. R., Chan C. Elevated free fatty acid uptake via CD36 promotes epithelial-mesenchymal transition in hepatocellular carcinoma. Scientific Reports. 2015;5 doi: 10.1038/srep14752.14752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Souza A. C. P., Bocharov A. V., Baranova I. N., et al. Antagonism of scavenger receptor CD36 by 5A peptide prevents chronic kidney disease progression in mice independent of blood pressure regulation. Kidney International. 2016;89(4):809–822. doi: 10.1016/j.kint.2015.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yokoi H., Yanagita M. Targeting the fatty acid transport protein CD36, a class B scavenger receptor, in the treatment of renal disease. Kidney International. 2016;89(4):740–742. doi: 10.1016/j.kint.2016.01.009. [DOI] [PubMed] [Google Scholar]
- 104.Bocharov A. V., Wu T., Baranova I. N., et al. Synthetic amphipathic helical peptides targeting CD36 attenuate lipopolysaccharide-induced inflammation and acute lung injury. The Journal of Immunology. 2016;197(2):611–619. doi: 10.4049/jimmunol.1401028. [DOI] [PubMed] [Google Scholar]
- 105.Anderson C. M., Kazantzis M., Wang J., et al. Dependence of brown adipose tissue function on CD36-mediated coenzyme Q uptake. Cell Reports. 2015;10(4):505–515. doi: 10.1016/j.celrep.2014.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]


