Abstract
The function of the poly(A) signal in spleen necrosis virus (SNV) is dependent upon the distance between the cap site and the poly(A) site, while the function of the SV40 late poly(A) signal is independent of the distance. Deletions in the SNV poly(A) sequence do not alter the distance-dependent function. SNV/SV40 chimeric poly(A) signals show intermediate behavior between the SNV and SV40 poly(A) signals. These results indicate that multiple sequence elements are involved in the functions of either the SNV or SV40 poly(A) signals. This intermediate behavior is also observed with poly(A) signals from the mouse α-globin and herpes simplex virus thymidine kinase genes.
Retroviruses are RNA viruses that replicate through a DNA intermediate, the provirus. The proviral DNA is synthesized by the virus-encoded reverse transcriptase. During reverse transcription, long terminal repeat (LTR) sequences are created at both ends of the proviral DNA. The LTR sequences contain cis-regulatory elements necessary for replication of the retro-virus: sequences for integration, transcription initiation, transcription enhancement, and RNA 3′ end formation (for review, see Weiss et al., 1985; Coffin 1990). Because of the duplication of the LTR sequences, signals for transcription initiation and RNA 3′ end formation are located at both the 5′ and 3′ ends of the provirus.
For proper RNA 3′ end formation, retro-viruses, like cellular genes, require two sequence elements: one is the AAUAAA sequence; and the other is the GU- or U-rich sequence, the so-called downstream element, which is located downstream from the poly(A) site (for review, see Birnstiel et al., 1985; Humphrey and Proud-foot, 1988; Manley, 1988; Wickens, 1990; Proudfoot, 1991). In addition to the requirement for these elements, retroviruses use at least three different ways to inactivate the 5′ poly(A) signal or to activate the 3′ poly(A) signal for RNA 3′ end formation (for review, see Proudfoot, 1991).
In some retroviruses, such as Rous sarcoma virus and human T cell leukemia virus-1 (Ahmed et al., 1991), the AAUAAA sequence is located upstream of the cap site and is not transcribed into RNA at the 5′ end (for review, see Proud-foot, 1991).
In another retrovirus, human immunodeficiency virus-1 (Brown et al., 1991; DeZazzo et al., 1991; Valsamakis et al., 1991), and a pararetro-virus, hepatitis B virus (Russnak and Ganem, 1990), an upstream element(s) is required for RNA 3′ end formation in addition to the AAUAAA and downstream elements. Since this upstream element is not transcribed into RNA at the 5′ end but is transcribed near the 3′ end, the second poly(A) site is processed, while the first poly(A) site remains unprocessed.
In spleen necrosis virus (Iwasaki and Temin, 1990) and cauliflower mosaic virus (Sanfacon and Hohn, 1990), the poly(A) signals require a distance from the cap site to be active. The poly(A) signal in human immunodeficiency virus-1 also requires a distance from the cap site for RNA 3′ end formation (Weichs an der Glon et al., 1991), in addition to the upstream elements.
Previously, we reported that in SNV the distance between the cap site and the poly(A) site is critical: if it is shorter than 500 bases, the poly(A) signal is inactive; if it is longer than 1000 bases, the poly(A) site is used efficiently. In contrast, the poly(A) signal for the SV40 late genes functions independent of its distance from the cap site (Iwasaki and Temin, 1990).
In this report we asked what features of the SNV poly(A) signal sequences are responsible for distance-dependent RNA 3′ end formation by making (1) deletions in the SNV sequence and (2) SNV/SV40 chimeras. Our experiments indicated that multiple sequence elements are involved in the functions of either the SNV or SV40 poly(A) signal.
Materials and methods
Nomenclature
Plasmids have the letter “p” before their names [e.g., pR/U5], whereas transcripts made from these plasmids do not [e.g., R/U5].
Cells
D17 cells (a dog osteosarcoma cell line) were grown as previously described (Watanabe and Temin, 1983). Transfection of D17 cells was performed as described by Kawai and Nishizawa (1984).
Plasmids
Recombinant DNA techniques were as described by Sambrook et al. (1989). The L construct containing the R/U5 region of SNV was identical to pR/U5(1423) in our previous report (Iwasaki and Temin, 1990). The 5′S and 3′S constructs containing the R/U5 region of SNV were identical to pR/U5(497) and pR/U5(47l), respectively, in our previous report (Iwasaki and Temin, 1990). We created a Bgl II site at the 3′ end of the U5 region in these vectors by adding two nucleotides, GA, using polymerase chain reaction (PCR) mutagenesis. Addition of these two nucleotides did not change expression of RNA compared with the original sequences (data not shown).
This Bgl II site and an Xba I site (see Fig. 1A) were used to replace the R/U5 region with all the deletions, replacement, chimeric, and other poly(A) signal sequences.
Figure 1.
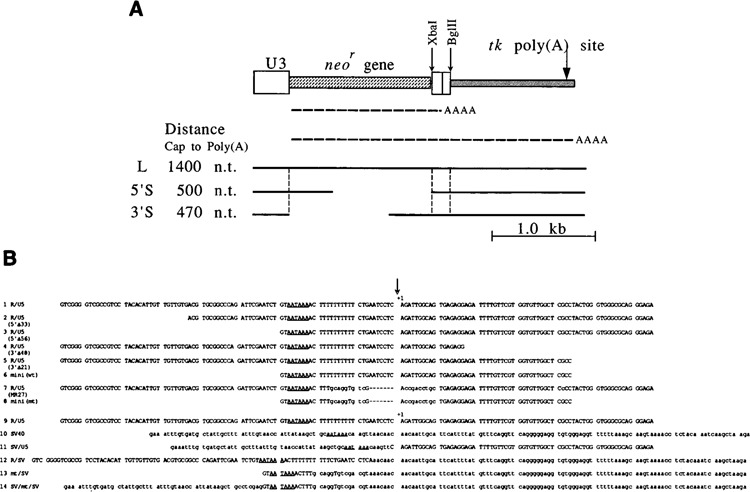
Structures of vectors and sequences used as the poly(A) signals. A. Vectors used. These vectors contained the U3 region of SNV for a promoter ( ), the neomycin resistance gene [neo
r] (
), the neomycin resistance gene [neo
r] ( ) to extend the distance from the cap site to the first poly(A) site, the first poly(A) signal to be tested (
) to extend the distance from the cap site to the first poly(A) site, the first poly(A) signal to be tested ( ), and the herpes simplex virus thymidine kinase [tk] gene to provide the second poly(A) site (
), and the herpes simplex virus thymidine kinase [tk] gene to provide the second poly(A) site ( ). The RNAs expressed from these vectors are shown by (----AA); one RNA species is polyadenylated at the first poly(A) site, and the other RNA species is polyadenylated at the tk poly(A) site. Tests for RNA 3′ end formation were performed using three types of vectors: L, 5′S, and 3′S. Gaps in the 5′S and 3′S constructs indicate deleted regions in these vectors. The distances from the cap site to the first poly(A) site in the L, 5′S, and 3′S constructs were approximately 1400, 500, and 470 nucleotides, respectively. By measuring levels of the two RNA species, the efficiency of RNA 3′ end formation was evaluated for the first poly(A) signals. A probe to detect RNA species transcribed from L and 5′S was the 5′ neor gene fragment. A probe to detect RNA species transcribed from L and 3′S was the 3′ neor gene fragment. (See Materials and Methods.) Other symbols: Xba I and Bgl II, sites of restriction enzymes used; vertical broken lines, boundaries of regions; thick lines, structures of vectors. B. Mutations of the SNV R/U5 region used to identify additional sequences necessary for RNA 3′ end formation. The R region is upstream from the poly(A) site, and the U5 region is downstream from the poly(A) site. The AATAAA poly (A) signal sequences are underlined. The poly(A) sites are marked by vertical arrows and +1. All the sequences were aligned at the poly(A) sites. The lowercase letters in the sequences indicate the nucleotides different from the parental sequence (R/U5). The broken lines in the sequences indicate the deleted nucleotides compared to the parental sequence. This R/U5 sequence contained two extra nucleotides at the 3′ end, GA, to create a Bgl II site, compared with the original R/U5 sequence. The addition of these nucleotides did not influence RNA 3′ end formation at the SNV poly(A) site (data not shown). The remainder of the vector sequences was identical for all constructs, except for pR/U5(3′Δ48). pR/U5(3′Δ48) contained 6 extra bases, CAGGTA BspM I site, at the junction between the U5 and tk sequences. These extra nucleotides did not influence RNA 3′ end formation (data not shown).
). The RNAs expressed from these vectors are shown by (----AA); one RNA species is polyadenylated at the first poly(A) site, and the other RNA species is polyadenylated at the tk poly(A) site. Tests for RNA 3′ end formation were performed using three types of vectors: L, 5′S, and 3′S. Gaps in the 5′S and 3′S constructs indicate deleted regions in these vectors. The distances from the cap site to the first poly(A) site in the L, 5′S, and 3′S constructs were approximately 1400, 500, and 470 nucleotides, respectively. By measuring levels of the two RNA species, the efficiency of RNA 3′ end formation was evaluated for the first poly(A) signals. A probe to detect RNA species transcribed from L and 5′S was the 5′ neor gene fragment. A probe to detect RNA species transcribed from L and 3′S was the 3′ neor gene fragment. (See Materials and Methods.) Other symbols: Xba I and Bgl II, sites of restriction enzymes used; vertical broken lines, boundaries of regions; thick lines, structures of vectors. B. Mutations of the SNV R/U5 region used to identify additional sequences necessary for RNA 3′ end formation. The R region is upstream from the poly(A) site, and the U5 region is downstream from the poly(A) site. The AATAAA poly (A) signal sequences are underlined. The poly(A) sites are marked by vertical arrows and +1. All the sequences were aligned at the poly(A) sites. The lowercase letters in the sequences indicate the nucleotides different from the parental sequence (R/U5). The broken lines in the sequences indicate the deleted nucleotides compared to the parental sequence. This R/U5 sequence contained two extra nucleotides at the 3′ end, GA, to create a Bgl II site, compared with the original R/U5 sequence. The addition of these nucleotides did not influence RNA 3′ end formation at the SNV poly(A) site (data not shown). The remainder of the vector sequences was identical for all constructs, except for pR/U5(3′Δ48). pR/U5(3′Δ48) contained 6 extra bases, CAGGTA BspM I site, at the junction between the U5 and tk sequences. These extra nucleotides did not influence RNA 3′ end formation (data not shown).
All the deletion and replacement sequences in the R/U5 region were constructed by PCR mutagenesis. These sequences were confirmed by dideoxy sequencing. Primers used in PCR and details of the methods will be request.
The chimeric sequences, R/SV and SV/U5, were constructed as follows: The upstream and downstream sequences of SNV were separated at the Dra I site that is located at its poly(A) site and made blunt-ended by Klenow treatment. The SV40 DNA fragment has been previously described (Iwasaki and Temin, 1990) and is shown in Figure 1B. The upstream and downstream sequences of SV40 were separated at the Hpa I site that is located 6 bases upstream from its poly(A) site. These half sequences were ligated to make the chimeric signals and were cloned into L, 5′S, and 3′S constructs.
The fragment used in mt/SV was made by elongation by Klenow enzyme of two partially complementary oligonucleotides. The oligonucleotides used were:
5′-CTCTAGACTCGAGTAATAAAACTTTGCAGGTG-3′,
and
5′GGTACGTCGACACCTGCAAAGT-3′.
The first oligonucleotide was identical to a primer used to construct mini(mt). After elongation by the Klenow enzyme, the fragment was self-ligated, digested with Xba I and Rsa I, and purified by gel electrophoresis in 3% low point melting agarose. The purified fragment was li-gated with the Hpa I-Bgl II fragment of the SV40 sequence and cloned into L, 5′S, and 3′S constructs. The sequence of mt/SV was confirmed by dideoxy-sequencing. The construction of the SV/mt/SV sequence was essentially similar to the construction of the mt/SV sequence. Details for this construction will be supplied upon request.
The herpes simplex virus thymidine kinase (tk) poly(A) signal that we used was previously described (Iwasaki and Temin, 1990).
The 140 bases of the mouse a-globin poly(A) signal were cloned from pBR322/genomic α-globin (Nishioka and Leder, 1979), using the Bal I site located 74 bases upstream and the Nco I site located 63 bases downstream from the poly(A) site. The Bal I site was converted to an Xba I site by ligation with an Xba I linker. The Nco I site was converted to a Bgl II site by using Klenow enzyme and ligating with a Bgl II linker. This Xba I-Bgl II fragment was used to make the α-globin constructs.
The 150 bases of the rat P450 gene poly(A) signal were generated by PCR. For this PCR λor15 (Porter et al., 1990) was used as template DNA and primers were as follows:
5′-CCTCGAGCTGGGGTGCAGCCCCCA-3′,
and
5′-CAGATCTTGTCTAATTGAAGC-3′.
The PCR-generated DNA fragment was cloned as described as above and was confirmed by dideoxy sequencing.
Plasmids for probes used in RNase protection assays were constructed by cloning the Ava II-Bgl II fragments (approximately 400 bases of the 3′ neo r fragment and the poly(A) sequence) of the L constructs having the R/U5, R/U5(MR27), R/SV, SV/U5 sequences into a plasmid modified from pBluescript+, using the Xho I and Bgl II sites. This Ava II site was converted to an Xho I site by treatment with Klenow enzyme and ligation with an Xho I linker. This Bgl II site was created at the original Cla I site of pBluescript+ by insertion of a Bgl II linker.
RNA analyses
Cellular RNA was isolated as described elsewhere (Berger and Birkenmeier, 1979; Iwasaki and Temin, 1990). Isolated RNA was treated with RQ1 DNase (Promega) in the presence of RNasin (Promega) as described by the supplier.
Northern blot analysis was performed with DNase-treated RNA (20 μg) as described by Sambrook et al. (1989). Labeling of the DNA fragments to make radioactive probes was performed with a random priming procedure (Fein-berg and Vogelstein, 1984). The 5′ and 3′ neo r probes were fragments from the 5′ end to the Eag I site, and from the Nco I site to the 3′ end of the neo r gene fragment, respectively. These neo r fragments were used for the 5′S and 3′S constructs.
RNase protection assays were performed as described by Farnham et al. (1985). RNA probes were synthesized in vitro using phage T7 RNA polymerase in the presence of 32P(α) UTP.
Results
Experimental design
To identify sequences – other than the AAU-AAA and GU-rich sequences – that are required for RNA 3′ end formation in the R/U5 region of spleen necrosis virus (SNV), the L vector was used (Fig. 1A). The L vector contained a transcriptional promoter derived from SNV (the U3 region in the LTR), the 1.3 kilobase (kb) fragment of the neomycin phosphotransferase gene (neo r) to extend the distance from the cap site to the poly(A) site, the R/U5 region of SNV for the first poly(A) signal (the R region is upstream of the poly(A) site and the U5 region is downstream of the poly(A) site), and the 2.2 kb fragment of the herpes simplex virus thymidine kinase gene fragment (tk) for the second poly(A) signal. The R/U5 region in this vector was mutated to identify additional sequences for RNA 3′ end formation in SNV (Fig. 1B). The efficiency of RNA 3′ end formation was evaluated by measuring the levels of RNAs processed at the first and second poly(A) sites. Since the tk poly(A) signal was used as the second poly(A) signal in all constructs, efficiency of RNA 3′ end formation means the relative efficiency for the first poly(A) signal under competition with the second tk poly(A) signal. Previously, we showed that addition of the tk fragment did not change the stability of steady state RNA (Iwasaki and Temin, 1990). Therefore, measuring the levels of two RNA species transcribed from a vector should accurately reflect the relative efficiency of RNA 3′ end formation in the context of the vectors.
To identify sequences responsible for the distance-dependent function, two additional vectors were used, 5′S and 3′S (Fig. 1A). The distances from the cap site to the first poly(A) site in the 5′S and 3′S vectors were approximately 500 and 470 bases, respectively. At these distances, the SNV poly(A) signal is inactive for RNA 3′ end formation (Iwasaki and Temin, 1990). 5′S contained the 5′ one-third fragment of the neomycin phosphotransferase gene, whereas 3′S contained the 3′ one-third fragment of the neomycin phosphotransferase gene. The reason we used these two different vectors was to test for effects of any specific sequences on distance-dependent RNA 3′ end formation.
D17 cells were transfected (Kawai and Nishizawa, 1984) with DNAs of the L, 5′S, or 3′S vectors, and pCMV-CAT (Stinski and Roehr, 1985). Forty-eight hours after transfection, RNA was isolated (Berger and Birkenmeier, 1979; Iwasaki and Temin, 1990) and was subjected to Northern blot hybridization (Sambrook et al., 1989). At the same time a fraction of cells was subjected to CAT assay to normalize the efficiency of transfection (Gorman et al., 1982). To confirm the reproducibility of data, each construct was tested two or more times.
Additive effects on RNA 3′ end formation by the upstream and downstream sequences in the R/U5 region of spleen necrosis virus
Deletions of 33 bases and 56 bases from the 5′ end of the R region in the L construct decreased the amount of RNA processed at these SNV poly(A) sites [Fig. 2A, constructs 2 and 3; Fig. 2B, lane 3 (1400 n.t. RNA), and Northern data not shown for R/U5(5′Δ33)]. Likewise, the deletion of 21 bases from the 3′ end of the U5 region in the L construct decreased processing [Fig. 2A, construct 5; Fig. 2B, lane 7 (1400 n.t. RNA)]. Therefore, deletions upstream and downstream of the poly(A) site slightly decreased usage of that site from 70% to 40–51%.
Figure 2.
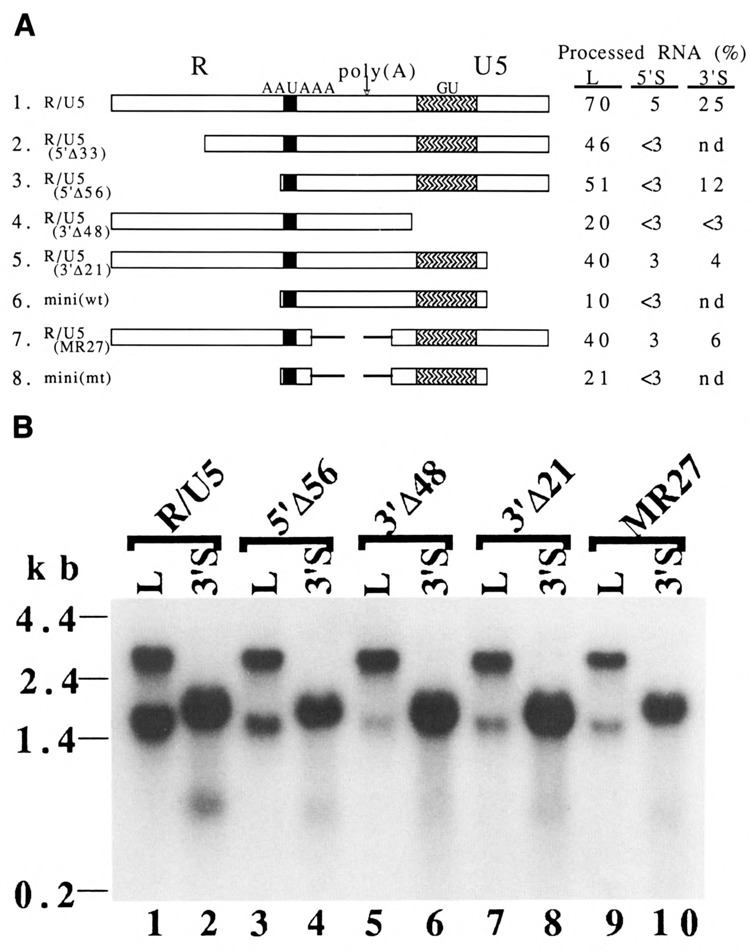
Deletion mutants to identify the upstream and downstream elements in the R/U5 region for RNA 3′ end formation. A. Structures of the deletion mutants and the efficiency of RNA 3′ end formation at the SNV poly(A) site. The sequence of the R/U5 region is shown by  . The AAUAAA and an 18-base stretch of GU-rich sequences (bases +20 to +38 in Fig. 1B) are indicated by
. The AAUAAA and an 18-base stretch of GU-rich sequences (bases +20 to +38 in Fig. 1B) are indicated by  and
and  . The poly(A) site is indicated by a vertical arrow. The replaced sequences at the middle of the R/U5 region are shown by thin lines in constructs 7 and 8. Names of the poly(A) sequences are shown at the left side. L, 5′S, and 3′S indicate which constructs were used (see Fig. 1A). The percentages of RNA processed at the SNV poly(A) sites are shown at the right side of each construct, nd = not determined. The percentages of processed RNA were obtained by microdensi-tometry scanning of Northern bands. Variation was the largest, 40% ± 11 (mean ± s.d.), for R/U5(3′Δ21); and the smallest, 20% ± 6, for R/U5(3′Δ48). Some data were mean values of two experiments. The probe to detect RNA species transcribed from L and 5′S was the 5′ neo
r gene fragment. The probe to detect RNA species transcribed from L and 3′S was the 3′ neo
r gene fragment. (See Materials and Methods.) B. Northern blot analysis. The sizes of marker RNAs are shown at the left. RNAs processed at the first poly(A) site were approximately 1.4 kb for L constructs and 0.5 kb for the 3′S constructs. RNAs processed at the tk poly(A) site were approximately 2.7 kb for L constructs and 1.8 kb for the 3′S constructs. D17 cells were transfected (Kawai and Nishizuwa, 1984) with the indicated constructs, and RNA was isolated (Berger and Birkenmeier, 1979) at 48 hours after transfection and was subjected to Northern blot analysis (Iwasaki and Temin, 1990). The DNA fragment used as the probe was the 3′ neo
r fragment (see Materials and Methods). Transfection efficiency was normalized to levels of chloramphenicol acetyltransferase activity (Gorman et al., 1982) resulting from cotransfection with pCMV-CAT (Stinski and Roehr, 1985) (data not shown).
. The poly(A) site is indicated by a vertical arrow. The replaced sequences at the middle of the R/U5 region are shown by thin lines in constructs 7 and 8. Names of the poly(A) sequences are shown at the left side. L, 5′S, and 3′S indicate which constructs were used (see Fig. 1A). The percentages of RNA processed at the SNV poly(A) sites are shown at the right side of each construct, nd = not determined. The percentages of processed RNA were obtained by microdensi-tometry scanning of Northern bands. Variation was the largest, 40% ± 11 (mean ± s.d.), for R/U5(3′Δ21); and the smallest, 20% ± 6, for R/U5(3′Δ48). Some data were mean values of two experiments. The probe to detect RNA species transcribed from L and 5′S was the 5′ neo
r gene fragment. The probe to detect RNA species transcribed from L and 3′S was the 3′ neo
r gene fragment. (See Materials and Methods.) B. Northern blot analysis. The sizes of marker RNAs are shown at the left. RNAs processed at the first poly(A) site were approximately 1.4 kb for L constructs and 0.5 kb for the 3′S constructs. RNAs processed at the tk poly(A) site were approximately 2.7 kb for L constructs and 1.8 kb for the 3′S constructs. D17 cells were transfected (Kawai and Nishizuwa, 1984) with the indicated constructs, and RNA was isolated (Berger and Birkenmeier, 1979) at 48 hours after transfection and was subjected to Northern blot analysis (Iwasaki and Temin, 1990). The DNA fragment used as the probe was the 3′ neo
r fragment (see Materials and Methods). Transfection efficiency was normalized to levels of chloramphenicol acetyltransferase activity (Gorman et al., 1982) resulting from cotransfection with pCMV-CAT (Stinski and Roehr, 1985) (data not shown).
A deletion of 48 bases from the 3′ end of the U5 region, which included the GU-rich sequence (Fig. 1B), decreased processing more dramatically, to 20% [Fig. 2A, construct 4; Fig. 2B, lane 5, (1400 n.t. RNA)]. Therefore, this GU-rich sequence is part of the downstream element necessary for efficient RNA 3′ end formation in SNV.
To test further the effects of the upstream 56 base and downstream 21 base sequences on RNA 3′ end formation, we constructed a minimal poly(A) signal (Figs. 1B and 2A, construct 6). Deletion of both upstream and downstream sequences in the R/U5 region decreased the processed RNA to 10% (Northern data not shown), indicating that the modest requirement for sequences upstream and downstream are additive, as exhibited by constructs R/U5(5′Δ56) and R/U5(3′Δ21) (Fig. 2A, constructs 3 and 5).
A 27-base region located between the AAU-AAA and GU-rich sequences was replaced with a sequence not related to SNV (Fig. 1B, 7). This replacement slightly decreased processing at the poly(A) site [Fig. 2A, construct 7; Fig. 2B, lane 9 (1400 n.t. RNA)].
To test further the effects on RNA 3′ end formation of the middle region in addition to the upstream and downstream sequences, another minimal poly(A) signal was constructed (Figs. 1B and 2A, 8). The mini(mt) sequence gave 21 % of processed RNA (Nothern data not shown); the positive effect of the middle region on RNA 3′ end formation was observed only in the presence of the upstream and downstream sequences.
We observed 1.2–2.2 times reduction of total RNA levels in the L constructs having deletion mutants compared with the wild-type R/U5 sequence. We do not know if this reduction of RNA levels results from differences in the RNA stability among the deletion sequences. However, since we always estimated the efficiency of RNA 3′ end formation by comparing the levels of two RNA species, RNA processed at the first poly(A) site and RNA processed at the second poly(A) site, the slight variation of total RNA, levels is unlikely to influence our results greatly.
No specific sequences are responsible for distance-dependent RNA 3′ end formation
To determine sequences involved in distance-dependent RNA 3′ end formation, the mutant SNV poly(A) signals in the 5′S and 3′S constructs were examined by Northern analysis. We hypothesized that, if specific sequences in the R/U5 region are responsible for distance-dependent RNA 3′ end formation, these deletions would activate RNA 3′ end formation near the cap site.
First, we describe results obtained with the 3′S constructs. The parental R/U5 region in the 3′S construct gave reduced processing at this poly(A) site [Fig. 2A, construct 1; Fig. 2B, lane 2 (500 n.t. RNA)]. Thus, this R/U5 poly(A) signal showed distance-dependent RNA 3′ end formation. The mutant poly(A) signals of R/U5(5′Δ56), R/U5(3′Δ48), R/U5(3′Δ21), and R/ U5(MR27) reduced processing a little more than the parental signal (Fig. 2A, constructs 3–6). Therefore, all the mutant poly(A) signals maintained the distance-dependent phenomenon. Furthermore, the reduction observed among mutant sequences in the 3′S constructs roughly correlated with the reduction observed in the L constructs (Fig. 2A and 2B). Since none of these mutant poly(A) signals activated RNA 3′ end formation near the cap site, it appeared that no specific sequences within the R/U5 region of SNV were responsible for the distance-dependent function.
In the 5′S construct, these poly(A) signals gave <3–5% of the RNA processed at their poly(A) sites (Fig. 2A; Northern data not shown). These results indicated again that no specific sequences in the R/U5 region of SNV were responsible for the distance-dependent function.
In the context of the 5′S and 3′S constructs, we observed almost no difference in total RNA levels among all the constructs including the wild-type R/U5 constructs.
Upstream and downstream sequences in the SV40 poly(A) signal separately increase the efficiency of RNA 3′ end formation
We previously reported two differences between the SNV and SV40 poly(A) signals (Iwasaki and Temin, 1990): (1) the SV40 poly(A) signal was more efficient than the SNV poly(A) signal (the R/U5 region) when both were located 1400 bases or more from the cap site; and (2) the SV40 poly(A) signal was active independent of the distance from the cap site, whereas the efficiency of the SNV poly(A) signal correlated with the distance from the cap site.
To determine what sequences cause these differences between the SNV and SV40 poly(A) signals, we constructed SNV/SV40 chimeric poly(A) signals (Fig. 1B). One chimeric signal, SV/U5, contained SV40 sequence upstream from the poly(A) site and the SV40 sequence downstream from the poly(A) site. These chimeric poly(A) signals were analyzed in the L, 5′S, and 3′S constructs (Fig. 1A).
We first asked what sequences are responsible for the high efficiency of the SV40 signals at 1400 bases or more from the cap site. In the L constructs the SV40 poly(A) signal consistently gave higher processing than SNV [Fig. 3A, constructs 1 and 2; Fig. 3B, lanes 1, 3, 5, and 7 (1400 n.t. RNA)]. The SV/U5 and R/SV chimeric signals increased processing at these poly(A) sites, compared to the R/U5 signal [Fig. 3A, constructs 3 and 4; Fig. 3B, lanes 9, 11, 13, and 15 (1400 n.t. RNA)]. This result suggested that the sequences upstream and downstream from the poly(A) site in SV40 independently enhance RNA 3′ end formation compared with the R/U5 region of SNV.
Figure 3.
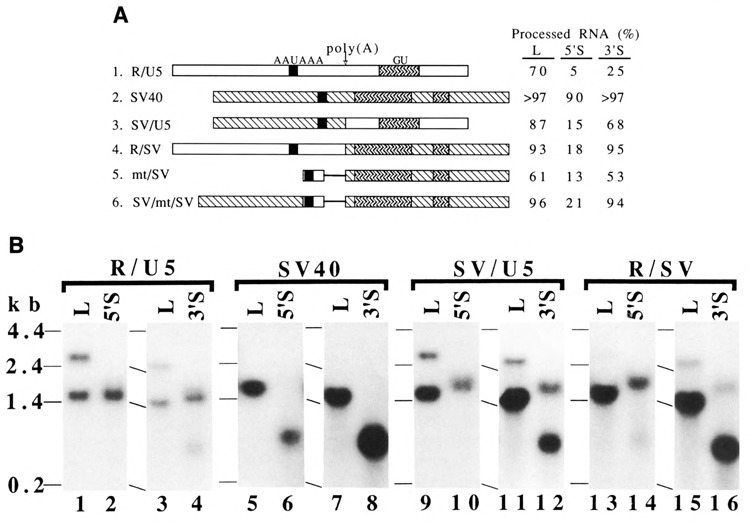
Chimeric poly(A) signal sequences to test distance-dependent RNA 3′ end formation. A. Structures of chimeric poly(A) signals and the efficiency of RNA 3′ end formation at the poly(A) sites. The R/U5 region and the SV40 late poly(A) signal sequences are shown by  and
and  , respectively. The downstream elements of SV40 shown in this figure were defined by others (Conway and Wickens, 1985; Sadofsky et al., 1985). Variation was the largest, 61% ± 13 (mean ± s.d.), for R/SV; and the smallest, 93% ± 2, for R/SV. When the level of total RNA from the L construct was taken as 1.0 in each set, the relative levels of total RNA for other constructs were as follows: R/U5 (L: 5′S: 3′S = 1: 0.9 :1.2), SV40 (L: 5′S; 3′S = 1: 0.3 :1.4), SV/U5 (L: 5′S: 3′S = 1: 0.6 :0.4), R/SV (L: 5′S: 3′S = 1: 0.4 :1.1), mt/SV (L: 5′S: 3′S = 1: 0.8 :0.8), and SV/mt/SV (L: 5′S: 3′S = 1: 1: 0.6). See the legend of Figure 2A for other symbols. B. Northern blot analyses. D17 cells were transfected with the indicated constructs. See the legend of Figure 2B for for other descriptions.
, respectively. The downstream elements of SV40 shown in this figure were defined by others (Conway and Wickens, 1985; Sadofsky et al., 1985). Variation was the largest, 61% ± 13 (mean ± s.d.), for R/SV; and the smallest, 93% ± 2, for R/SV. When the level of total RNA from the L construct was taken as 1.0 in each set, the relative levels of total RNA for other constructs were as follows: R/U5 (L: 5′S: 3′S = 1: 0.9 :1.2), SV40 (L: 5′S; 3′S = 1: 0.3 :1.4), SV/U5 (L: 5′S: 3′S = 1: 0.6 :0.4), R/SV (L: 5′S: 3′S = 1: 0.4 :1.1), mt/SV (L: 5′S: 3′S = 1: 0.8 :0.8), and SV/mt/SV (L: 5′S: 3′S = 1: 1: 0.6). See the legend of Figure 2A for other symbols. B. Northern blot analyses. D17 cells were transfected with the indicated constructs. See the legend of Figure 2B for for other descriptions.
The effect of the SV40 upstream sequence was also seen by removing it. The construct mt/SV contained no SV40 upstream sequence, but it contained the AAUAAA sequence, a sequence unrelated to either SNV or SV40, and the downstream sequence of SV40 (Fig. 1B, 13). The construct mt/SV reduced processing at this poly(A) site, compared with SV40 itself (Fig. 3A, construct 5).
Returning part of the SV40 upstream sequence restored high processing. SV/mt/SV contained the SV40 upstream sequence from the AAUAAA sequence and a sequence identical to the mt/SV construct downstream from the AAUAAA sequence (Fig. 1B, 14). SV/mt/SV gave processing almost as high as SV40 (Fig. 3A, construct 6; Northern data not shown). This result again suggested that the upstream sequence of SV40 enhanced RNA 3′ end formation. Furthermore, comparison between the R/SV and mt/SV poly(A) signals (Fig. 3A, constructs 4 and 5) indicated that the upstream sequence in the R region of SNV also contained elements that enhanced RNA 3′ end formation. This result was consistent with the results of the experiment in Figure 2A.
No single sequence element is fully responsible for distance-dependent and distance-independent RNA 3′ end formation
We then asked what sequences cause the difference between the SNV and SV40 poly(A) signals with respect to distance-dependence. RNA 3′ end formation at 500 bases from the cap site was tested in the 5′S and 3′S constructs. We assumed that: (1) if any sequences derived from SNV in these chimeric poly(A) signals are responsible for the distance-dependent function, either (or both, if the SNV sequence has redundant signals for the distance-dependent function) of the chimeric signals should behave similarly to the SNV poly(A) signal; or (2) if sequences derived from the SV40 poly(A) signal are responsible for distance-independent RNA 3′ end formation, either or both of the chimeric signals should behave similarly to the SV40 poly(A) signal.
As expected, the R/U5 region of SNV gave low processing in the 5′S construct (500 n.t. RNA) and in the 3′S construct (470 n.t. RNA) (Fig. 3A, construct 1; Fig. 3B, lanes 2 and 4). These levels of processed RNA were significantly lower than that of the processed RNA in the L construct (Fig. 3A, construct 1). The SV40 poly(A) signal gave very high processing in the L, 5′S, and 3′S constructs (Fig. 3A, construct 2; Fig. 3B, lanes 5–8).
The chimeric SNV/SV40 poly(A) signals behaved differently in the two different constructs. In the 5′S constructs, these chimeric signals, SV/U5 and R/SV, gave low processing at these sites (500 n.t. RNA) (Fig. 3A, constructs 3 and 4; Fig. 3B, lanes 10 and 14). However, in the 3′S constructs, these chimeric signals resulted in high processing (470 n.t. RNA) (Fig. 3A, constructs 3 and 4; Fig. 3B, lanes 12 and 16). Therefore, one part of either the SNV or SV40 poly (A) signal is not sufficient to restore completely the distance-dependent or distance-independent function in these different contexts.
The mt/SV and SV/mt/SV synthetic signals also showed context-dependent behavior similar to the chimeric poly(A) signals (Fig. 3A, constructs 5 and 6; Northern data not shown). The result with SV/mt/SV suggested that the SV40 sequence downstream from the poly(A) site and the SV40 sequence upstream from the AAUAAA sequence were still not sufficient for full activity of RNA 3′ end formation in the 5′S construct. This observation indicated that strength of a poly(A) signal is not the determinant of the distance-independent function. The strength of processing at a poly(A) signal in the L construct correlated with that in the short constructs (5′S and 3′S), but it did not correlate with the distance-independent function.
One might speculate that the low percentages of processed RNA at these chimeric poly(A) sites in the 5′S constructs were the result of an inhibitory effect of the 5′ neo r gene fragment on RNA 3′ end formation, not just the result of distance-dependent RNA 3′ end formation. To test this hypothesis, we inserted the 5′ two-thirds of the neo r gene fragment just upstream of the 5′ neo r gene fragment in the 5′S constructs; therefore, these new vectors contained a duplication of the 5′ one-third of the neo r gene fragment and none of the 3′ one-third of the neo r gene fragment. The SV/U5 and R/SV poly(A) signals gave 76% and 80% of processed RNA at the poly(A) sites in these constructs (data not shown). This result suggested that the 5′ one-third of the neor gene fragment did not have an inhibitory effect on RNA 3′ end formation.
To confirm the results of the Northern analyses, we performed RNA protection assays with some of the same RNA preparations used in the Northern analyses (R/U5 in the L and 3′S constructs, SV/U5 in the L and 3′S constructs, and R/SV in the L and 3′S constructs). The RNA protection assays produced results essentially consistent with the Northern analyses (Fig. 4). Two species of protected fragments appeared in each lane (Fig. 4). Smaller fragments (approximately 420 bases, depending upon RNA samples) corresponded with the sizes of RNA processed at the first poly(A) sites and larger fragments (approximately 490 bases, depending upon RNA samples) corresponded with the sizes of RNA processed at the second poly(A) site. Relative intensities of these bands in each lane were consistent with the results from the Northern analyses (see Fig. 3).
Figure 4.
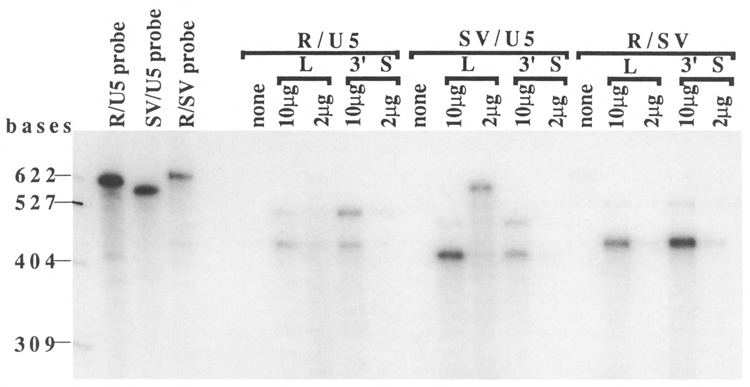
RNA protection assay. The sizes of marker RNAs are shown at the left. The input probes are shown in the left three lanes in this gel: R/U5, SV/U5, and R/SV probes. The names of probes and the names and amounts of the RNA samples are indicated at the top. The names of probes used were the same as the RNA samples (e.g., the R/U5 probe was used for RNA samples with R/U5 in the L and 3′S constructs). The sizes of the input RNA probes were 588 bases for the R/U5 probe, 560 bases for the SV/U5 probe, and 608 bases for the R/SV probe. Protected fragments were 433 bases (polyadenylated RNA) and 497 bases (pass-through RNA) at the R/U5 poly(A) site, 405 bases (polyadenylated RNA) and 469 bases (pass-through RNA) at the SV/U5 poly(A) site, and 440 bases (polyadenylated RNA) and 517 bases (pass-through RNA) at the R/SV poly(A) site. The RNA protection assay was performed as described by Farnham et al. (1985). Total RNA in reactions was adjusted to 10 μg with tRNA. “None” indicates reactions with only tRNA. RNA probes were synthesized in vitro using T7 RNA polymerase.
In conclusion, since none of the chimeric or synthetic poly(A) signals fully restored distance-dependent or distance-independent RNA 3′ end formation, it is likely that no single sequence element is solely responsible for the distance-dependent or distance-independent function.
The efficiencies of RNA 3′ end formation in other poly(A) signals are reduced more in the 5′S constructs than in the 3′S constructs
We then asked whether other poly(A) signals show distance-dependent or distance-independent RNA 3′ end formation. We again constructed three kinds of vectors, L, 5′S, and 3′S, with other poly(A) signals. The poly(A) signals used in this experiment were derived from the mouse α-globin (Nishioka and Leder, 1979), the herpes simplex virus thymidine kinase (tk) (Wagner et al., 1981), and the rat NADPH-cytochrome P450 oxidoreductase genes (Porter et al., 1990) (Fig. 5A).
Figure 5.
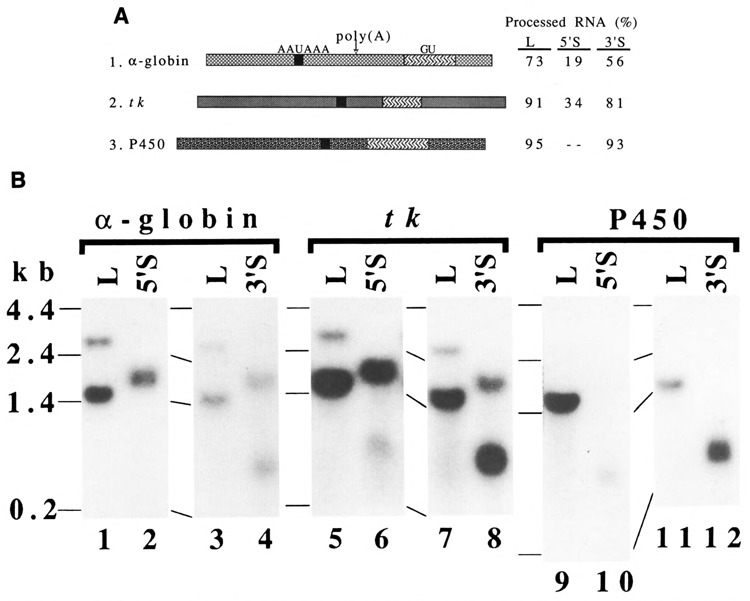
Other poly(A) signals for distance-dependent RNA 3′ end formation. A. Structures of poly(A) signals and the efficiency of RNA 3′ end formation at the poly(A) sites. The poly(A) signals for the mouse α-globin gene (Nishioka and Leder, 1979), the herpes simplex virus thymidine kinase (tk) gene (Wagner et al., 1981), and the rat NADPH-cytochrome P450 oxidoreductase gene (Porter et al., 1990) are shown by  ,
,  , and
, and  , respectively. The downstream elements of the tk poly(A) signal were defined by others (Zhang et al., 1986). The GU-rich sequences downstream from the poly(A) sites in the α-globin and P450 oxidoreductase genes were determined by sequence analysis. Variation was the largest, 34% ± 13 (mean ± s.d.), for tk; and the smallest, 91% ± 5, for tk. Some data were mean values of two experiments. The percentage of processed RNA at the P450 poly(A) signal in the 5′S construct was not determined, because of low expression of RNA. When the level of total RNA from the L construct was taken as 1.0 in each set, relative levels of total RNA for other constructs were as follows: α-globin (L: 5′S: 3′S = 1: 1.2 :1.3), tk (L: 5′S: 3′S = 1: 0.6 :1.3), and P450 (L: 3′S = 1: 0.7). See the legend of Figure 2A for other symbols B. Northern blot analyses. D17 cells were transfected with the indicated constructs. See the legend of Figure 2B for other descriptions.
, respectively. The downstream elements of the tk poly(A) signal were defined by others (Zhang et al., 1986). The GU-rich sequences downstream from the poly(A) sites in the α-globin and P450 oxidoreductase genes were determined by sequence analysis. Variation was the largest, 34% ± 13 (mean ± s.d.), for tk; and the smallest, 91% ± 5, for tk. Some data were mean values of two experiments. The percentage of processed RNA at the P450 poly(A) signal in the 5′S construct was not determined, because of low expression of RNA. When the level of total RNA from the L construct was taken as 1.0 in each set, relative levels of total RNA for other constructs were as follows: α-globin (L: 5′S: 3′S = 1: 1.2 :1.3), tk (L: 5′S: 3′S = 1: 0.6 :1.3), and P450 (L: 3′S = 1: 0.7). See the legend of Figure 2A for other symbols B. Northern blot analyses. D17 cells were transfected with the indicated constructs. See the legend of Figure 2B for other descriptions.
The poly(A) signals for the α-globin, tk, and P450 oxidoreductase genes gave processing as high as 73–95% in the L constructs (Fig. 5A; Fig. 5B, L lanes). In the 5′S construct, the a-globin and tk poly(A) signals resulted in low processing at their poly(A) sites (Fig. 5A, constructs 1 and 2; Fig. 5B, lanes 2 and 6). However, in the 3′S construct, the α-globin and tk poly(A) signals resulted in high processing at the sites (Fig. 5A, constructs 1 and 2; Fig. 5B, lanes 4 and 8). Thus, the α-globin and tk poly(A) signals behaved differently in different contexts, just like the SNV/SV40 chimeric poly(A) signals. The P450 poly(A) signal in the 3′S construct gave processing as high as that in the L construct (Fig. 5A, construct 3; Fig. 5B, lane 12). In the 5′S construct, the level of steady state RNA was extremely low (Fig. 5B, lane 10), so it was difficult to draw any firm conclusions.
In conclusion, the poly(A) signals for the α-globin and tk genes did not consistently show either distance-dependent or distance-independent RNA 3′ end formation in the different contexts. Furthermore, these observations again support the hypothesis that the strength of a poly(A) signal is not the major determinant for the distance-independent function. For example, the α-globin poly(A) signal had a similar strength of RNA 3′ end formation to the strength of the R/U5 poly(A) signal in the L construct. However, this signal showed low processing in the 5′S construct and high processing in the 3′S construct, while the R/U5 poly(A) signals showed consistently low processing in the 5′S and 3′S constructs.
Discussion
We analyzed the R/U5 region in spleen necrosis virus (SNV) for RNA 3′ end formation and found multiple sequence elements that increase processing at the SNV poly(A) site. These elements are located in (1) the first 33 bases in the R region, (2) the 27 bases in the middle of the U5 region, and (3) the last 21 bases in the U5 region. The second element contained an 18-base stretch of a GU-rich sequence, and the third element also contained a small stretch of a GU-rich sequence. Therefore, we speculate that the downstream element for RNA 3′ end formation in SNV consists of the second and third elements. The primary sequences in these two elements were different from each other, but the effects of these elements on RNA 3′ end formation seemed to be additive.
The additive effects of the downstream elements in the SNV poly(A) signal are different from the downstream elements in some other poly(A) signals, especially the SV40 late poly(A) signal. In the SV40 poly(A) signal the downstream element is functionally redundant: small deletions and base substitutions in the SV40 downstream element did not reduce processing efficiency (Zarkower and Wickens, 1988). This difference between the SNV and SV40 elements might be a part of these functional differences.
The necessity of the upstream sequences for efficient RNA 3′ end formation has been reported in other viruses: the poly(A) signals for the SV40 late (Carswell and Alwine, 1989), the adenovirus late (DeZazzo and Imperiale, 1989), hepatitis B virus (Russnak and Ganem, 1990), cauliflower mosaic virus (Sanfacon et al., 1991), and human immunodeficiency virus–1 (Brown et al., 1991; DeZazzo et al., 1991; Valsamakis et al., 1991). The distances of these upstream elements from the poly(A) site or the AAUAAA sequence varies among these poly(A) signals. In some cases, the upstream elements have an important role in activating only the second poly(A) site in an RNA that contains two copies of the poly(A) signals (for review, see Coffin and Moore, 1990; Proudfoot, 1991).
In the case of SNV, the U3 region seems to contain another upstream element for RNA 3′ end formation, but the distance from the cap site to the poly(A) site is more important than the U3 upstream element to determine the efficiency of RNA 3′ end formation at the SNV poly(A) site (Iwasaki and Temin, 1990). It has also been reported that the distance from the cap site to the poly(A) site is important for RNA 3′ end formation in some other viruses (Sanfacon and Hohn, 1990; Weichs an der Glon et al., 1991).
Therefore, activation of RNA 3′ end formation at the 3′ poly(A) signal appears to be different in different virus species. We interpret these observations as follows: The AAUAAA, upstream (in the R region), and downstream (in the U5 region) elements are necessary for efficient RNA 3′ end formation. The short distance from the cap site to the poly(A) site in the 5′ LTR inactivates RNA 3′ end formation at the 5′ poly(A) site, even in the presence of all these elements. Increasing the distance from the cap site to the poly(A) site activates a poly(A) signal which contains a complete set of these elements. In the case of SNV, an upstream element in the U3 region further enhances RNA 3′ end formation at the 3′ poly(A) site in addition to the effects of the other elements in the R/U5 region and the distance. It is possible that some other retro-viruses contain upstream elements only in the U3 region. These could be equivalent to the upstream elements in the R region and the U3 region in SNV. In such cases, the deletion of the U3 region itself may have profound effects on RNA 3′ end formation.
Deletions in the SNV poly(A) signal sequences indicated that no specific sequence elements were responsible for distance-dependent RNA 3′ end formation in SNV. We took another approach by making SNV/SV40 chimeras to identify what sequences cause the difference between the SNV and SV40 poly(A) signals. The SNV/ SV40 chimeric poly(A) signals showed different behavior in different vectors. None of the chimeric or synthetic poly(A) signals had full distance-dependent or distance-independent RNA 3′ end formation. Therefore, it is likely that no single sequence element is solely responsible for the distance-dependent or distance-independent functions.
The entire sequence of the SV40 poly(A) signal appears to be required for distance-independent RNA 3′ end formation, and no specific SNV sequences are involved in distance-dependent RNA 3′ end formation. The following two observations support these hypotheses: (1) Deletion analysis of the R/U5 region in SNV did not reveal any sequence elements solely responsible for distance-dependent RNA 3′ end formation. If such elements existed in the R/U5 region of SNV, at least some of the deletion mutants should have behaved similarly to the chimeric signals. (2) The mt/SV and SV/mt/SV synthetic poly(A) signals had almost no sequences related to SNV, but they also showed behavior similar to the chimeric signals.
We used the term “distance-independent” RNA 3′ end formation based on our observations. However, it is unlikely that the SV40 poly(A) signal has evolved to gain a distance-independent function, because the distance from the cap site to the poly(A) signal in the SV40 genome is fixed. We speculate that the phenomena of distance-dependent and distance-independent RNA 3′ end formation reflect an unknown aspect of poly(A) signal functions, and that retroviruses take advantage of this function.
Acknowledgments
We thank P. Bell for a gift of λorl5 DNA; T. Jacoby, B. Pietz, and J. Schoening for technical assistance; M. Sheets and M. Wickens for constructive discussion during this work and for comments on the manuscript; and P. Bernstein, J. Dahlberg, E. Freed, P. Lambert,J. Jones, O. Niwa, R. Kollmar, J. Ross, and K. Takimoto for helpful comments on the manuscript. This work was supported by grants CA-07175 and CA-22443 from the Public Health Service. H.M.T is an American Cancer Society Research Professor.
The costs of publishing this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC Section 1734 solely to indicate this fact.
Kouichi Iwasaki is currently at the Department of Genetics, Washington University School of Medicine, 4566 Scott Avenue, St. Louis, MO 63110.
References
- Ahmed Y. F., Gilmartin G. M., Hanly S. M., Nevins J. R., and Greene W. C. (1991), Cell 64, 727–737. [DOI] [PubMed] [Google Scholar]
- Berger S. L. and Birkenmeier C. S. (1979), Biochemistry 18, 5143–5149. [DOI] [PubMed] [Google Scholar]
- Benz E. W. Jr., Wydro R. M., Nadal-Ginard B., and Dina D. (1980), Nature 288, 665–669. [DOI] [PubMed] [Google Scholar]
- Birnstiel M. L., Busslinger M., and Strub K. (1985), Cell 41, 349–359. [DOI] [PubMed] [Google Scholar]
- Brown P. H., Tiley L. S., and Cullen B. R. (1991), J Virol 65, 3340–3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carswell S. and Alwine J. C. (1989), Mol. Cell. Biol. 9, 4248–4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffin J. M. (1990), in Fields Virology, 2nd ed., Raven Press Ltd., New York. [Google Scholar]
- Coffin J. M. and Moore C. (1990), Trends Genet 6, 276–277. [DOI] [PubMed] [Google Scholar]
- Colbere-Garapin F., Chousterman S., Horodniceanu F., Kourilsky P., and Garapin A.-C. (1979), Proc Natl Acad Sci USA 76, 3755–3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway L. and Wickens M. (1985), Proc Natl Acad Sci USA 82, 3949–3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeZazzo J. D., Kilpatrick J. E., and Imperiale M. J. (1991), Mol. Cell. Biol. 11, 1624–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeZazzo J. D. and Imperiale M. J. (1989), Mol. Cell. Biol. 9, 4951–4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty J. P. and Temin H. M. (1986), Mol Cell Biol 6, 4387–4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnham P. J., Abrams J. M., and Schimke R. T. (1985), Proc Natl Acad Sci USA 82, 3978–3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P. and Vogelstein B. (1984), Anal Biochem 137, 266–267. [DOI] [PubMed] [Google Scholar]
- Gorman C. M., Moofat L. F., and Howard B. H. (1982), Mol Cell Biol 2, 1044–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey T. and Proudfoot N. J. (1988), Trends Genet 4, 243–245. [DOI] [PubMed] [Google Scholar]
- Iwasaki K. and Temin H. M. (1990), Genes Dev 4, 2299–2307. [DOI] [PubMed] [Google Scholar]
- Kawai S. and Nishizawa M. (1984), Mol Cell Biol 4, 1172–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley J. L. (1988), Biochem Biophys Acta 950, 1–12. [DOI] [PubMed] [Google Scholar]
- Nishioka Y. and Leder P. (1979), Cell 18, 875–882. [DOI] [PubMed] [Google Scholar]
- Porter T. D., Beck T. W., and Kasper C. B. (1990), Biochemistry 29, 9814–9818. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. (1991), Cell 64, 671–674. [DOI] [PubMed] [Google Scholar]
- Russnak R. and Ganem D. (1990) Genes Dev 4, 764–776. [DOI] [PubMed] [Google Scholar]
- Sadofsky M., Connelly S., Manley J. L., and Alwine J. C. (1985), Mol Cell Biol 5, 2713–2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F., and Maniatis T. (1989), in Molecular Cloning: A Laboratory Manual, 2nd ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Sanfacon H. and Hohn T. (1990), Nature 346, 81–84. [DOI] [PubMed] [Google Scholar]
- Sanfacon H., Brodmann P., and Hohn T. (1991), Genes Dev 5, 141–149. [DOI] [PubMed] [Google Scholar]
- Stinski M. F. and Roehr T. J. (1985) J Virol 55, 431–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valsamakis A., Zeichner S., Carswell S. and Alwine J. C. (1991), Proc Natl Acad Sci USA 88, 2108–2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M. J., Sharp J. A., and Summers W. C. (1981), Proc Natl Acad Sci USA 78, 1441–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S. and Temin H. M. (1983), Mol Cell Biol 3, 2241–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichsan der Glon C., Monks J., and Proudfoot N. J. (1991), Genes Dev 5, 244–253. [DOI] [PubMed] [Google Scholar]
- Weiss R., Teich N., Varmus H., and Coffin J. (1985), in RNA Tumor Viruses, 2nd ed., supplements and appendices, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Wickens M. (1990), Trends Biochem Sci 15, 277–281. [DOI] [PubMed] [Google Scholar]
- Zarkower D. and Wickens M. (1988), J Biol Chem 263, 5780–5788. [PubMed] [Google Scholar]
- Zhang F, Denome R. M., and Cole C. N. (1986), Mol Cell Biol 6, 4611–4623. [DOI] [PMC free article] [PubMed] [Google Scholar]


