Abstract
The effects of human alternative splicing factor, ASF, on in vitro splicing of adenovirus E1A pre-mRNA were examined. E1A pre-mRNA is a complex substrate, and splicing in HeLa cell nuclear extracts produces six different RNAs using three alternative 5′ splice sites and two 3′ splice sites. Addition of excess ASF to splicing reactions produced a simplified splicing pattern, in which only one spliced product, 13S RNA, was detected. Inhibition of 12S and 9S splicing, which use 5′ splice sites upstream of the 13S 5′ splice site, extends previous observations that when multiple 5′ splice sites compete for the same 3′ splice site, ASF causes preferential selection of the proximal 5′ splice site. However, inhibition of the other splices, which use a different upstream 3′ splice site, represents a novel activity of ASF, as competition between 5′ splice sites is not involved. The effect of ASF on 12S splicing was found to depend on its position relative to competing 5′ splice sites, indicating that the ability of ASF to activate proximal 5′ splice sites is position- but not sequence-dependent. Finally, addition of small amounts of ASF to ASF-lacking S100 extract was able to activate distal as well as proximal 5′ splice sites in two of three pre-mRNAs tested, indicating that in these cases changes in the concentration of ASF alone can be sufficient to modulate alternative 5′ splice site selection.
Alternative splicing of pre-mRNAs is an important step in regulating the expression of many cellular and viral genes, allowing a single primary transcript to produce different protein products in different cell types, or at different developmental stages (see Smith et al., 1989, for recent review). In Drosophila, the expression of several genes, including those for sex determination, are regulated by alternative splicing, and trans-acting factors that control this process have recently been identified (reviewed by Baker, 1989). Genetic analysis of the genes controlling sex determination has identified three genes encoding proteins that directly or indirectly modulate splice site selection. These are transformer (Boggs et al., 1987), Sex-lethal (Bell et al., 1988), and transformer-2 (Amrein et al., 1988; Goralski et al., 1989). The Sex-lethal gene product appears to regulate the splicing of its own pre-mRNA and that of the transformer gene by suppressing a male-specific default 3′ splice site in female flies (Sosnowski et al., 1989; Inoue et al., 1990). The transformer and transformer-2 products then activate a female-specific 3′ splice site in double-sex pre-mRNA (Hedley and Maniatis, 1991; Hoshijima et al., 1991; Ryner and Baker, 1991). The product of another Drosophila gene, suppressor-of-white-apricot, can inhibit splicing of the first intron of its own pre-mRNA (reviewed in Bingham et al., 1988).
Alternative splicing factor (ASF), a human protein that regulates alternative splicing in vitro, has recently been purified (Ge and Manley, 1990). ASF was initially characterized based on its ability to increase the ratio of small t to large T mRNAs produced when an SV40 pre-mRNA was spliced in vitro, which represents an increase in selection of the 5′ splice site proximal to the common 3′ splice site. The activity was shown to reside in a protein of ∼32kD (Ge and Manley, 1990). At the same time, it was reported that the splicing factor SF2, apparently essential for all splicing in vitro, could affect 5′ splice site selection by enhancing the use of the proximal 5′ splice in β-globin pre-mRNAs containing multiple cryptic 5′ splice sites or duplicated 5′ splice sites (Krainer et al., 1990a;1990b). Based on similarities in their physical properties and activities, and most recently, the sequence of cDNA clones, it is now clear that ASF and SF2 are identical (Ge et al., 1991; Krainer et al., 1991). The primary structure of the ASF/SF2 protein reveals significant similarities to the Drosophila splicing regulators transformer and transformer-2, as well as to the U1 snRNP 70K protein (Ge et al., 1991; Krainer et al., 1991). In addition, the discovery by Ge et al. (1991) that multiple forms of ASF cDNAs are generated by alternative 3′ splice site selection, raises the intriguing possibility that a family of splicing regulators with different activities might be produced by the ASF gene. However, little is known about how ASF works, and additional insights could come from examination of its effects on additional pre-mRNAs.
In this paper, adenovirus E1A RNA splicing has been used to examine the effects of ASF on in vitro splicing of a very complex splicing substrate. The E1A gene produces at least six different mRNAs by alternative splicing using three 5′ splice sites and two 3′ splice sites (see Fig. 1A). With the exception of one species (12.5S RNA), which has only been detected in vitro, all of these RNAs have been identified both in vivo (Berk and Sharp, 1978; Chow et al., 1979; Stephens and Harlow, 1987; Ulfendahl et al., 1987) and in vitro (Schmitt et al., 1988; Gattoni et al., 1988). In adenovirus infected cells, E1A mRNA splicing is regulated so that at early times after infection, 13S RNA is the major species, with five-fold lower levels of 12S RNA and very low levels of 9S, 10S, or 11S RNAs detected, while at late times, all other RNAs increase relative to 13S RNA (Berk and Sharp, 1978; Spector et al., 1978; Chow et al., 1979; Svensson et al., 1983; Stephens and Harlow, 1987; Ulfendahl et al., 1987). This in vivo regulation of E1A splicing suggested that E1A pre-mRNA splicing was a good candidate for examining regulation of pre-mRNA splicing by ASF in vitro. In this report, we show that in addition to modulating selection of competing 5′ splice sites, ASF can affect splicing of an intron that does not involve competing 5′ splice sites, possibly by influencing 3′ splice site selection.
Figure 1.
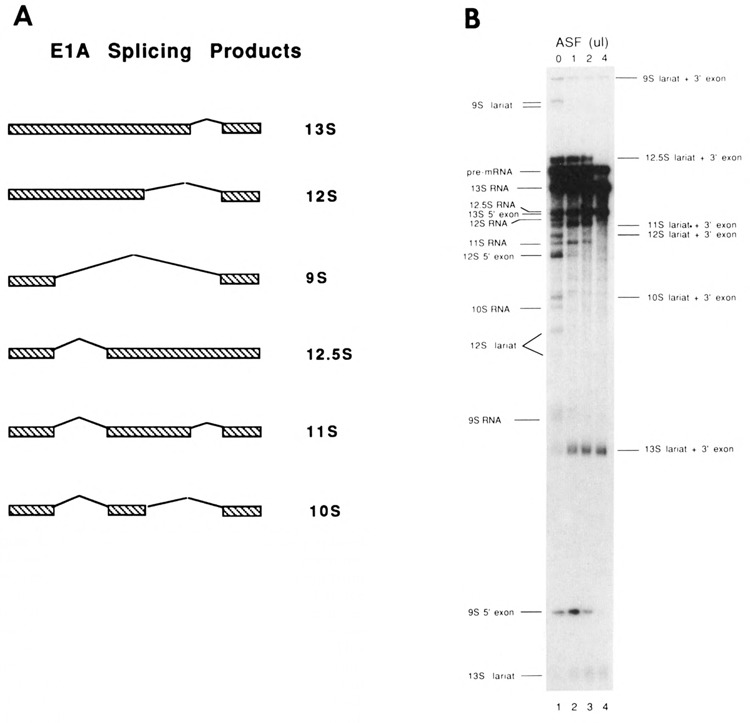
A. Structure of the six alternatively spliced E1A mRNAs produced during in vitro splicing reactions. 13S, 12S, and 9S RNAs are produced by splicing from one of 3 alternative 5′ splice sites to a common 3′ splice site. Additional RNAs are produced by splicing an intron between the 9S 5′ splice site and a 3′ splice site 216 nucleotides downstream of the 9S 5′ splice site. This produces 12.5S RNA in the absence of additional splicing, US RNA when the 13S intron is also removed, and 1 0S RNA when the 12S intron is also removed. B. Products of splicing E1A pre mRNA in the absence of additional ASF (lane 1) and in the presence of 1 μl (lane 2), 2μl (lane 3), or 4μl (lane 4) of purified ASF (see Materials and Methods for purification details).
Materials and methods
Materials
Restriction enzymes, SP6 RNA polymerase, DNA ligase, and m7GpppG cap structure analogue are from New England Biolabs. DEAE cellulose was from Whatman. DNaseI and RNAsin were from Promega Biotech. Centricon 10 centrifugal concentration units were from Amicon.
Plasmid constructions
The construction of pSP64-E1A has been described previously (Harper and Manley, 1991). Briefly, plasmids contain the adenovirus 2 E1A gene from nt 498–1572. DNA fragments carrying mutated 5′ splice sites were substituted for wild-type fragments. The E1A mutations dl1500 and pm975 were obtained from the plasmids pEK dl1500 and pEK pm975, respectively, which were provided by A. Berk. These mutations were cloned into pSP64 by replacing the fragment of the E1A gene from the Cla I site (nt.917) to the Xba I site (nt.1339) in pSP64-E1A with the corresponding fragment from each of the mutants. The SV40 plasmid, derived from the plasmid pSTER-i66 (12), was provided by H. Ge.
Preparation of splicing extracts, pre-mRNA synthesis, and in vitro splicing
Nuclear extracts and cytoplasmic S100 were prepared from suspension HeLa cells by the method of Dignam et al. (1983), and dialyzed for 8–12 hours against a modified Dignam buffer D, containing 20 mM HEPES pH 7.9, 0.1M KCl, 0.2 mM EDTA, 5% glycerol and 1 mM DTT. Insoluble material was removed by centrifugation.
E1A substrate RNAs were made by SP6 RNA polymerase transcription of pSP64-E1A, or appropriate mutant plasmid DNAs, linearized at the Xba I site (Ad2 nt. #1339). SV40 substrate RNA was transcribed from a plasmid containing the SV40 early region carrying a 66 base-pair insertion in the small t intron, derived from the plasmid pSTER-i66 (Ge and Manley, 1990). In vitro transcription was carried out such that the final concentration of GTP was 0.1mM; alpha 32P-GTP was added to give a specific activity of 6 Ci/mmole; and 2mM m7GpppG was included to cap transcripts. RNAs were purified on denaturing polyacrylamide gels.
In vitro splicing reactions were carried out essentially as previously described (Harper and Manley, 1991). Standard splicing reactions were performed in 25 μl containing 10 μl of extract or S100, and 5 μl of extract dialysis buffer containing various concentrations of ASF. Final concentrations in the reactions were 2.25mM MgCl2, 500μM ATP, 20mM creatine phosphate, 3% polyvinyl alcohol, 10ng substrate RNA, 12mM HEPES (pH 7.9), 4% glycerol, 0.12mM EDTA, 1 Unit/μl RNAsin, 5mM DTT, and 60mM KCl. For reactions in which KCl concentration was varied, varying concentrations of KCl were included in the 5 μl fo extract dialysis buffer added, to give the correct KCl concentration in the splicing reaction.
The alternative splicing factor ASF/SF2 used in these experiments was purified from Hela cell nuclear extracts by the method of Ge and Manley (1990) through the Mono Q step, or to approximately 50% homogeneity, except in the experiment shown in Figure 3, in which the ASF was purified only through the Superose 6 step of the procedure.
Figure 3.
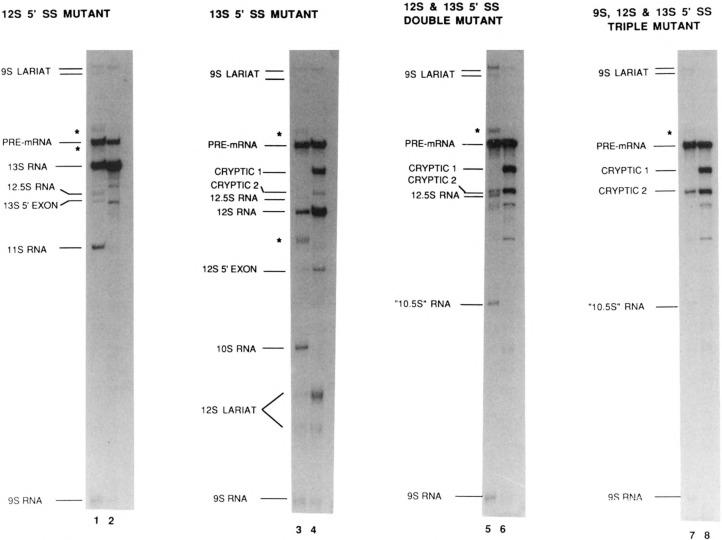
Influence of ASF on the splicing of mutant E1A pre-mRNAs. Products of splicing the 12S 5′ splice site mutant (lanes 1 and 2), 13S 5′ splice site mutant (lanes 3 and 4), 12S, 13S double 5′ splice site mutant (lanes 5 and 6), and 13S, 12S, 9S triple 5′ splice site mutant (lanes 7 and 8) pre-mRNAs; in the absence (lanes 1, 3, 5, and 7) or presence (lanes 2, 4, 6, and 8) of excess ASF. In addition to the authentic E1A splicing products, the positions of two cryptic splicing products are indicated. These are the only products of splicing the triple mutant RNA in the presence of excess ASF (lane 8); two other bands seen in this lane are the 5′ exons for cryptic 1 and cryptic 2 splicing. In this experiment 4 μl of partially purified ASF (see Materials and Methods for details of ASF purification) were added to the reactions shown in even-numbered lanes. Asterisks indicate the positions of the 12.5S, 11S, and 10S lariat + 3′ exon intermediates, which are in different positions relative to the linear products in the gel shown here, compared to that shown in Figure 1B, because the two gels were not run under identical conditions.
Analysis of splicing products
Products of splicing 32P-labeled RNAs were separated by electrophoresis on 4% polyacrylamide gels containing 7M urea, 0.1M Tris-Borate (pH 8.3), 10mM EDTA, and visualized by auto-radiography on Kodak X-Omat AR5 film using an intensifying screen. Initial characterization of the E1A splicing products has been described (Harper and Manley, 1991). Novel splicing products were identified based on their linear size compared to RNA size markers, and the appropriate effects of various mutations in the splicing substrates.
Results
ASF simplifies the alternative splicing pattern of E1A pre-mRNA in vitro
E1A pre-mRNA can be alternatively spliced in vitro using three 5′ splice sites and two 3′ splice sites, to produce the six RNAs shown in Figure 1A (Schmitt et al., 1987; Gattoni et al., 1988; Harper and Manley, 1991). To determine the effects of elevated concentrations of ASF on splicing of the E1A pre-mRNA, increasing amounts of ASF were added to E1A splicing reactions carried out in HeLa cell nuclear extracts (Fig. 1B). In the absence of added ASF, all of the possible splicing products were detected (lane 1). Addition of increasing amounts of ASF (lanes 2–4) caused a gradual simplification of the splicing pattern. At relatively low ASF concentrations, 9S and 12S (and therefore also 10S) splicing was inhibited (lane 2), while 11S and 12.5S splicing was also inhibited, but only at higher levels of ASF (lanes 3–4). 13S splicing was progressively enhanced, until at the highest concentration of ASF tested, only 13S products could be detected (lane 4).
The above results indicate that ASF can affect E1A splice site selection in two distinct ways. First, ASF causes a shift in 5′ splice site selection from use of all three 5′ splice sites to preferential selection of the 13S 5′ splice site. This finding is consistent with previous reports showing that when multiple 5′ splice sites are competing for a common 3′ splice site, addition of ASF favors selection of the proximal 5′ splice site (Ge and Manley, 1990; Krainer et al., 1990a), and inhibition of 12S, 10S, and 9S was likely due to this activity of ASF. The second effect of ASF on E1A splice site selection was to inhibit splicing of 11S and 12.5S RNAs. 12S and 13S 5′ splice sites do not compete with the 9S 5′ splice site for the upstream 3′ splice site used in these splices. Thus the inhibition of 12.5S and 11S RNA was unexpected, as these splices do not involve 5′ splice site competition. It is noteworthy that inhibition of 12.5S and 11S splicing occurred only at higher concentrations of ASF than were required for inhibition of 12S, 10S, and 9S splicing (Fig. 2, lanes 2–4). Indeed, the amount of several detectable 12.5S and 11S products and intermediates appeared to increase at the lowest concentration of ASF added (lane 2) before decreasing at higher concentrations. When a pre-mRNA that contained only this first intron (truncated upstream of the 12S 5′ splice site) was spliced in vitro, ASF also inhibited its splicing (data not shown), indicating that this effect was not due to the presence of downstream competing splice sites.
Figure 2.
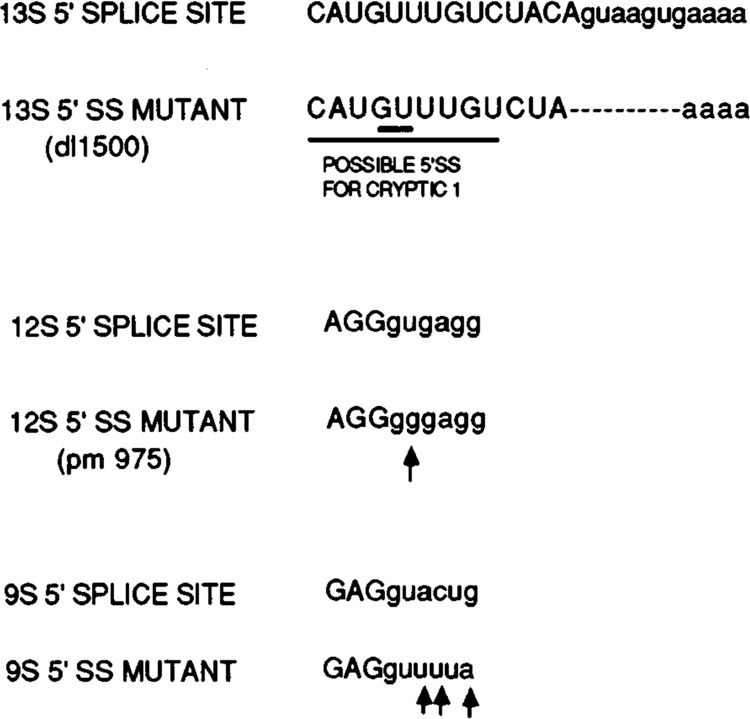
Sequences of E1A 5′ splice sites. The sequences of the 13S, 12S, and 9S 5′ splice sites are shown, with the sequences of mutants in the splice site shown below the wild-type sequence. Uppercase letters indicate exon sequences, and lowercase letters indicate intron sequences. For the 13S 5′ splice site mutant, dl1500 (Montell et al., 1984), the dashed line indicates the position of a 9 nt. deletion. The 12S 5′ splice site mutation is point mutant pm975 (Montell et al., 1982). Also the probable sequence of the cryptic-1 5′ splice site is indicated by underlining in the dl1500 sequence.
The effect of ASF on 5′ splice site selection is position-dependent and sequence-independent
The inhibition by ASF of splicing the E1A first intron raised the possibility that the effect of ASF on a particular 5′ splice site might depend on its sequence or surrounding environment, rather than on its proximity to the common 3′ splice site. This possibility could not be ruled out by the small number of pre-mRNAs shown previously to be affected by ASF (Ge and Manley, 1990; Krainer et al., 1990a). To address this issue, we examined the effect of ASF on in vitro splicing of a series of E1A pre-mRNAs carrying 5′ splice site mutations. The sequences of the 5′ splice mutations are shown in Figure 2, and the results are shown in Figure 3. Splicing of a pre-mRNA carrying a point mutation in the 12S 5′ splice site was similar to that of the wild-type substrate in the absence (lane 1) as well as the presence of added ASF (lane 2), except for the expected lack of 12S and 10S RNAs. Splicing of a pre-mRNA containing a small deletion that removes the 13S 5′ splice site resulted in the production of 12S RNA as the predominant product in the absence of added ASF (lane 3), and addition of ASF increased 12S splicing while eliminating 10S splicing and slightly decreasing 9S splicing (lane 4). A novel cryptic 5′ splice site located near the position of the 13S 5′ splice site (denoted cryptic 1; see Fig. 2) was also activated strongly by addition of ASF. These results show that the effect of increased ASF concentration on 12S 5′ splice site selection depends on its position relative to competing 5′ splice sites, rather than its sequence or its immediate environment. That is, when the proximal 13S 5′ splice site was present, ASF inhibited 12S splicing (Fig. 1B), while in the absence of a functional 13S 5′ splice site, ASF enhanced 12S splicing (Fig. 3, lanes 3 and 4). It is noteworthy that use of the cryptic-1 5′ splice site was absolutely dependent on elevated ASF concentration.
When a pre-mRNA carrying both the 12S and 13S 5′ splice mutations was used, the level of 9S and 12.5S splicing increased, and two additional novel products were also detected (Fig. 3, lane 5). The new products were the RNA-labeled cryptic 2, which results from activation of a cryptic 5′ splice site between the 12S and 13S 5′ splice sites (approximately 50 nucleotides 3′ to the 12S site), and the product labeled 10.5S RNA, which results from cryptic 2 splicing combined with removal of the first intron. Splicing of cryptic 2 RNA has been observed previously in vivo in HeLa cells transfected with a 12S/13S double mutant E1A gene (Zhuang and Weiner, 1986). Addition of ASF inhibited 9S, 12.5S, and 10.5S splicing, and activated splicing of cryptic 1 as well as cryptic 2 (lane 6), consistent with a position-dependent effect of ASF. When the 12S and 13S 5′ splice site mutations were combined with a triple point mutation at the 9S 5′ splice site (that reduces but does not eliminate 9S splicing), the amounts of 9S, 12.5S, and 10.5S RNAs were all reduced (lane 7). Addition of ASF eliminated the small amount of 9S, 12.5S, and 10.5S splicing, and again increased splicing of cryptic 2 RNA and activated cryptic 1 RNA splicing (lane 8). The most striking result of the experiments with this series of mutant pre-mRNAs was that cryptic 1 splicing was never observed in the absence of excess ASF, even when all three natural 5′ splice sites were mutated as in the reaction shown in lane 7. This suggests that the 5′ splice site used to produce cryptic 1 RNA (see Figure 2) is very weak, even in the absence of competing 5′ splice sites, but that it can be strongly activated by increased ASF concentration, apparently due to its proximity to the 3′ splice site.
The effects of ASF are distinct from those of increasing ionic strength in the splicing reaction
Variations in the ionic conditions of in vitro splicing reactions have been shown to affect the ratios of alternative splicing products of E1A pre-mRNA (Schmitt et al., 1987) and other complex substrates (Helfman et al., 1988; Mayeda and Ohshima, 1988). We have previously shown that these effects on E1A splicing reflect changes in cis-competition between 5′ splice sites rather than the properties of a particular splice site per se (Harper and Manley, 1991). In general, with pre-mRNAs that contain competing 5′ splice sites, increasing the ionic strength in splicing reactions increases selection of the proximal 5′ splice site, which is similar to the effect of adding ASF. In order to examine whether or not the influence of ASF on splice site selection was identical to that caused by increasing salt concentrations, we compared the effects of ASF addition to those of increasing ionic strength in splicing of E1A wild-type (Fig. 4, lanes 1–10) and the 13S 5′ splice site mutant (lanes 11–20) pre-mRNAs. Splicing of the wild-type El A substrate in the presence of increasing amounts of ASF (lanes 1–5) again resulted in the gradual simplification of the splicing pattern, until at the highest concentration of ASF, 13S RNA was the only major splicing product detected (lane 5). Increasing the KCl concentration in the splicing reaction also enhanced 13S splicing (although not as efficiently as did ASF) and reduced 12S splicing, but in addition increased 9S splicing until a concentration of 110mM, above which all splicing was somewhat inhibited. These results show that the effect of ASF on 9S splicing are distinct from those of increasing ionic strength, and further show that elevated salt concentrations do not always lead to preferential use of proximal 5′ splice sites.
Figure 4.
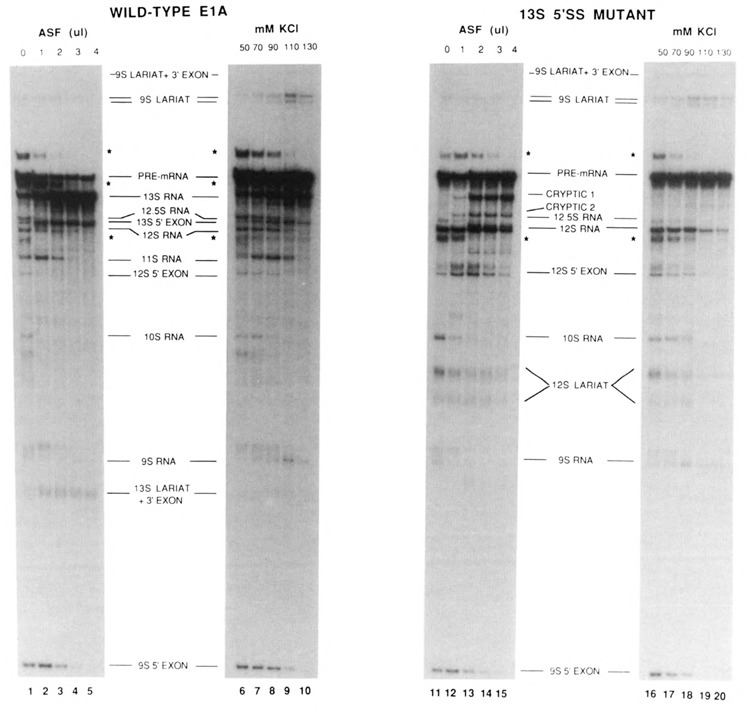
Comparison of the effects of ASF and increasing ionic strength in splicing reactions. Wild-type E1A (lanes 1–10) or the 13S 5′ splice site mutant (lanes 11–20) pre-mRNAs were spliced in under standard splicing condition as described in Materials and Methods (lanes 1 and 11); in reactions to which 1 μl (lanes 2 and 12), 2 μl (lanes 3 and 13), 3 μl (lanes 4 and 14), or 4 μl (lanes 5 and 15) of ASF were added; or in reactions containing increasing concentrations of KCl as indicated (lanes 6–10 and 16–20). Asterisks indicate the positions of the 12.5S, US, and 10S lariat + 3′ exon intermediates (see legend to Figure 3).
When the 13S 5′ splice site mutant pre-mRNA was used (lanes 11–20), more dramatic differences were observed. Increasing amounts of ASF in the splicing reaction resulted in strong inhibition of 9S and 10S splicing, and a gradual shift from 12S splicing to splicing of cryptic RNAs 1 and 2 (lanes 11–15). Increasing the ionic strength of the splicing reaction resulted in a slight activation of 9S and cryptic 2 splicing, but no detectable activation of cryptic 1 splicing, even at the highest KCl concentration (130mM) tested (lanes 16–20). Together, these results demonstrate that the effects of ASF on E1A splicing are distinct from those of increasing ionic strength.
Very low levels of ASF can activate distal as well as proximal 5′ splice sites
The finding that increasing the concentrations of ASF in nuclear extract preferentially activated proximal 5′ splice sites raised the question of what effect reduction of ASF levels might have on alternative splice site selection. To examine this, we made use of the fact that ASF/SF2 is the only essential splicing factor missing from cytoplasmic S100 extracts, and its addition is sufficient to activate splicing (Krainer et al., 1990a,b; Ge and Manley, 1991). The concentration of ASF in splicing reactions containing S100 rather than nuclear extract can thus be easily varied, from very small amounts barely adequate to activate splicing up to the higher concentrations required to switch splicing to the proximal 5′ splice site.
We have used this system to examine the effects of low concentrations of ASF on splicing of SV40 early pre-mRNA, in addition to the wild-type and 13S 5′ splice site mutant E1A pre-mRNAs. The SV40 early region produces two mRNAs, coding for the large T and small t tumor antigens, by utilization of alternative 5′ splice sites and a common 3′ splice site. In vitro splicing of the SV40 early pre-mRNA in HeLa cell nuclear extracts produces predominantly large T RNA, but small t splicing can be activated by addition of excess ASF (Ge and Manley, 1990 and references therein). Because the level of small t splicing is very low in the absence of excess ASF, we have used an SV40 pre-mRNA containing a 66 nucleotide insertion in the small t intron, called SVi66, which gives increased small t splicing both in vivo (Fu and Manley, 1987) and in vitro (Ge and Manley, 1990).
The results of titrating ASF into S100 extracts containing the E1A pre-mRNAs are shown in Figure 5, and the SV40 pre-mRNA in Figure 6. The effect of limiting ASF concentrations was similar for the E1A 13S 5′ splice site mutant (Fig. 5, lanes 6–10) and SV40 pre-mRNAs (Fig. 6). In both cases, low levels of ASF activated distal as well as proximal 5′ splice sites. Splicing of 12S RNA as well as cryptic RNAs 1 and 2 were observed when the 13S 5′ splice site mutant pre-mRNA was used (Fig. 5, lanes 7 and 8). A switch to selective use of the proximal 5′ splice site occurred at high ASF concentrations, resulting in production of high levels of cryptic 1 RNA (and large amounts of cryptic 1 5′ exon, which migrates slightly more slowly that 12S RNA), and the elimination of 12S and cryptic 2 RNA splicing (Fig. 5, lanes 9 and 10). In reactions containing SV40 pre-mRNA, both small t and large T RNA splicing was activated at low ASF concentrations (Fig. 6, lanes 2–4) and splicing switched to exclusively small t splicing when high concentrations of ASF were used (lane 6). However, a significantly different pattern of activation was observed with the wild-type E1A pre-mRNA (Fig. 5, lanes 1–5). At all concentrations tested, only the proximal, 13S 5′ splice site was utilized efficiently. Splicing from the distal 12S and 9S 5′ splice sites was barely detectable. Thus, with this pre-mRNA even low concentrations of ASF led to preferential activation of the proximal 5′ splice site. Together, these results indicate that variations in the concentration of ASF alone are able to modulate selection of 5′ splice sites in some pre-mRNAs effectively, but not in others.
Figure 5.
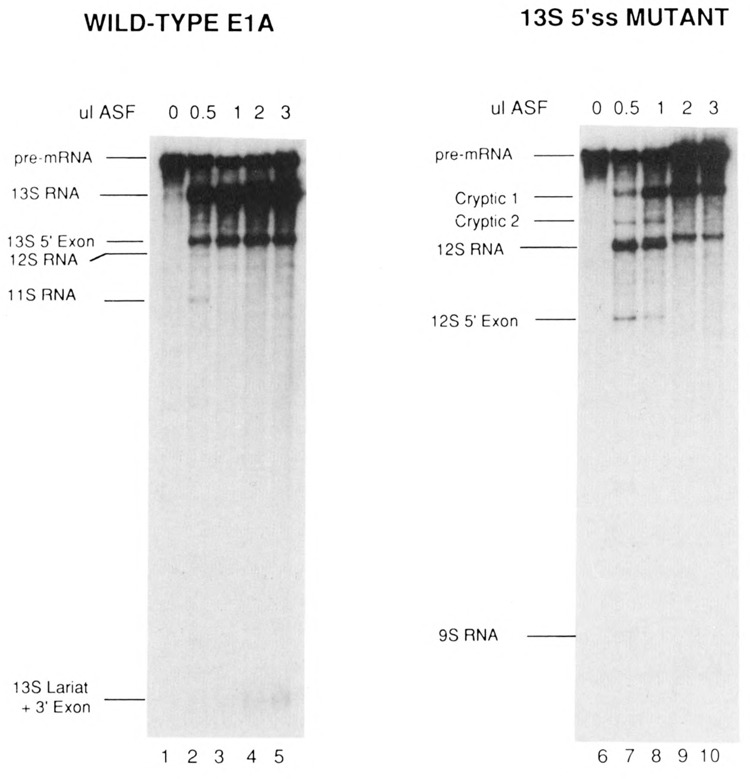
Effect of low ASF concentrations on alternative splicing of E1A pre-mRNAs. Wild-type E1A (lanes 1–5) and the 13S 5′ splice site mutant (lanes 6–10) were processed in S100 extract in the absence of ASF (lanes 1 and 6), or in the presence of 0.5 μl (lanes 2 and 7), 1 μl (lanes 3 and 8), 2 μl (lanes 4 and 9), or 4 μl (lanes 5 and 10) of ASF.
Figure 6.
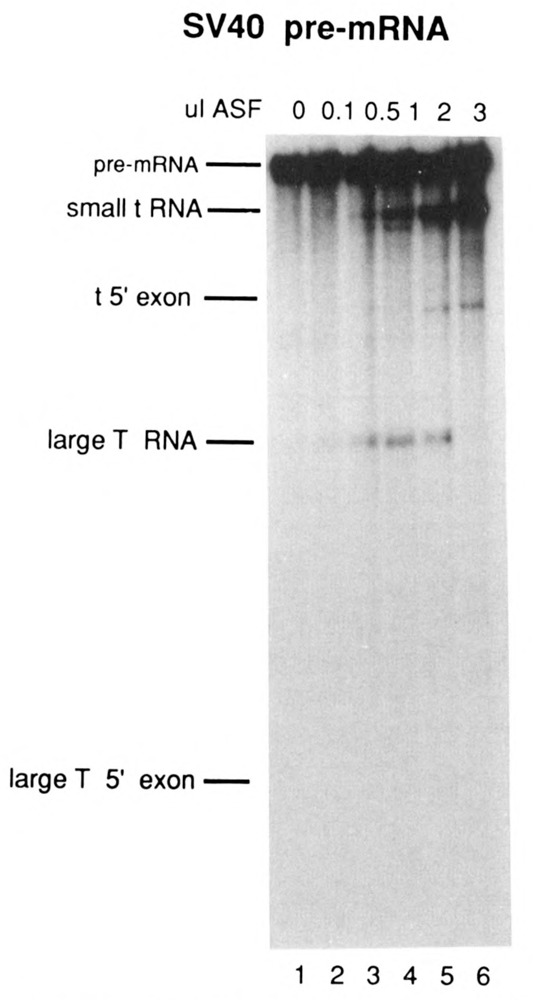
Effect of low ASF concentrations on alternative splicing of SV40 early region pre-mRNA. SV40-i66 pre-mRNA was processed in S100 extract in the absence of ASF (lane 1), or in the presence of 0.1 μl (lane 2), 0.5 μl (lane 3), 1 μl (lane 4), 2 μl (lane 5), or 3 μl (lane 6) of ASF.
Discussion
The experiments presented here have demonstrated that the alternative splicing factor ASF has two apparently distinct activities that result in dramatic shifts in the adenovirus E1A in vitro splicing pattern, reducing the number of major spliced products from six to just one. The first activity, which switches selection of alternative 5′ splice sites to favor the use of the site most proximal to a common 3′ splice site, has been observed with SV40 early pre-mRNA (Ge and Manley, 1990) and cryptic or duplicated 5′ splice sites in β-globin pre-mRNA (Krainer et al., 1990a). The possibility that the effect of ASF on 5′ splice site switching was dependent on the particular sequence context could not be excluded on the basis of results from these two pre-mRNA splicing substrates alone. In this paper, we have shown that 12S splicing is inhibited when ASF is added to reactions containing the wild-type E1A pre-mRNA, but it is enhanced when ASF is added to reactions containing the 13S 5′ splice site mutant pre-mRNA. The opposite effects of ASF on 12S splicing, in the context of wild-type and mutant pre-mRNA s, provides compelling evidence that this property indeed reflects activation of the proximal 5′ splice site and repression of distal sites in a position-dependent manner, and that the sequence and/or context of the splice site is not a critical determinant.
The second activity of ASF inhibits splicing of the upstream intron required to produce the 12.5S/11S/10S E1A RNAs, even though this splicing reaction does not involve competing 5′ splice sites. This finding indicates that in some cases ASF can be involved in modulating 3′ splice site selection. Regardless of whether the inhibition of E1A first intron splicing is due to inhibition of the 9S 5′ splice site or direct inhibition of the first intron 3′ splice, this activity represents a novel function for ASF in modulating selection of alternative splice sites, as it occurs in the absence of 5′ splice site competition. Although the mechanism(s) by which ASF functions to modulate splice site selection is unknown, it is Intriguing that higher concentrations of ASF were required to block upstream splicing than to enhance 13S 5′ splice site selection. Perhaps inhibition of upstream splicing requires more molecules of ASF, or reflects a lower affinity interaction, than does activation of the downstream 5′ splice site. ASF was originally observed to inhibit all splicing at very high concentrations (Ge and Manley, 1990; Krainer et al., 1990a,b). However, the concentration of ASF required to inhibit E1A first intron splicing is well below the amount needed to detect general inhibition of splicing; in fact, it is below the level necessary to obtain maximal 13S RNA splicing.
The E1A first intron has some unusual features, and it may be that these play a role in its response to ASF. It contains a long distance between the 3′ splice site and the branch point adenosines, located 51, 55, and 59 nucleotides upstream of the 3′ splice site (Gattoni et al., 1988), as well as the potential for significant secondary structure between the branch point and the 3′ splice site, which appears to be important for efficient splicing of the intron (Chebli et al., 1989). Perhaps these features are in some way important for ASF function. Although these properties of the E1A first intron are not commonly found, similar features have been detected in alternatively spliced introns of the chicken β-tropomyosin (D’Orval et al., 1991) and the rat tropomyosin 1 (Helfman and Ricci, 1989; Helfman et al., 1990). It is possible that ASF in some way recognizes these unusual structural features, and participates in regulating splicing of these and other similar introns.
The position-dependent, sequence-independent nature of proximal 5′ splice site activation by ASF is similar to that of a newly recognized splicing factor, called DSF for distal splicing factor, which activates distal 5′ splice sites when multiple 5′ splice sites are competing for a common 3′ splice site (Harper and Manley, 1991). DSF was found to be required to activate 12S and 9S splicing from the wild-type E1A pre-mRNA, and large T splicing from an SV40 pre-mRNA, in a fractionated nuclear extract. In addition, DSF was required to activate 12S and 9S splicing in S100 extract complemented with ASF. The opposite activities of the two alternative splicing factors on selection of competing 5′ splice sites is particularly interesting in light of the similar (position-dependent) manner in which they appear to function. Indeed it is possible that they function together to modulate alternative 5′ splice site selection in different tissues or developmental stages.
We have shown that ASF can activate distal 5′ splice sites when very low levels of the factor are added to splicing reactions carried out in ASF-lacking cytoplasmic S100 extracts. This activation of distal 5′ splice sites by low levels of ASF was observed with SV40 early or the E1A 13S 5′ splice site mutant pre-mRNAs, but not with the wild-type E1A pre-mRNA. This may reflect differences in the relative strength of the competing 5′ splice sites. As shown in Figure 2, the 13S 5′ splice site is an excellent match to the consensus 5′ splice site sequence (7/9 overall, and 6/6 within the intron), while the 12S 5′ splice site is somewhat poorer (7/9 overall, but 5/6 within the intron), and the probable cryptic-1 5′ splice site is poorer still (6/9 overall and 4/6 within the intron). Our results are consistent with the notion that when the proximal 5′ splice site is very strong, reduced concentrations of ASF are not sufficient to activate competing distal 5′ splice sites, but when the proximal 5′ splice site is relatively weak, low levels of ASF can activate both distal and proximal 5′ splice sites. A possible explanation for this could be that at low ASF concentrations, productive U1 snRNP binding to each 5′ splice site is determined principally by relative splice site “strength” (e.g., complimentarity to the 5′ end of U1 snRNA), and this alone determines the relative utilization of competing 5′ splice sites. At higher, perhaps saturating, ASF concentrations, interactions between U1 snRNP and different 5′ splice sites becomes equivalent, and proximity to the 3′ splice site is now the predominant determinant. The results obtained with SV40 pre-mRNA at low ASF concentrations are not entirely consistent with this idea, since the proximal small 15′ splice site is an excellent match to the consensus (8/9 overall and 5/6 within the intron). However, this site is nonetheless very weak in in vitro splicing reactions, indicating that some other feature, such as short intron length, reduces its efficiency and thus its ability to compete with large T splicing.
Whatever the mechanism, our results show that when a distal 5′ splice site is competing with a relatively weak proximal site, as is the case in the SV40 early pre-mRNA, and when the E1A 12S 5′ splice site competes with cryptic splice sites 1 and 2, increasing concentrations of ASF can sequentially activate and then inhibit utilization of the distal 5′ splice site(s), indicating that in some instances changes in the concentration or activity of ASF alone can modulate 5′ splice site selection. However, when the proximal 5′ splice is very strong, as is the case with the E1A 13S splice site, distal 5′ splice sites cannot be activated by ASF alone. This suggests that in such instances an additional factor is required for use of distal sites. Indeed, we have recently shown that DSF is required for activation of 12S splicing from a wild-type E1A pre-mRNA (Harper and Manley, 1991), and it thus may be that the activities of ASF and DSF are together necessary for alternative 5′ splice selection in this type of pre-mRNA.
Acknowledgments
We thank H. Ge for useful discussions and for providing a sample of purified ASF for use in preliminary experiments; and M. Wang for technical assistance. This work was supported by NIH grants CA46121 and CA33620.
The costs of publishing this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC Section 1734 solely to indicate this fact.
References
- Amrein H., Gorman M., and Nothiger R. (1988), Cell 55, 1025–1035. [DOI] [PubMed] [Google Scholar]
- Baker B. S. (1989), Nature 340, 521–524. [DOI] [PubMed] [Google Scholar]
- Berk A. J. and Sharp P. A. (1978), Cell 14, 695–711. [DOI] [PubMed] [Google Scholar]
- Bell L. R., Maine E. M., Schedl P., and Cline T. W. (1988), Cell 55, 1037–1046. [DOI] [PubMed] [Google Scholar]
- Bingham P. M., Chou T.-B., Mims I., and Zachar Z. (1988), Trends Genet 4, 134–138. [DOI] [PubMed] [Google Scholar]
- Boggs R. T., Gregor P., Idriss S., Belote J. M., and McKeown M. (1987), Cell 50, 739–747. [DOI] [PubMed] [Google Scholar]
- Chebli K., Gattoni R., Schmitt P., Hildwein G., and Stevenin J. (1989), Mol Cell Biol 9, 4852–4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow L. T., Broker T. R., and Lewis J. B. (1979), J Mol Biol 134, 265–303. [DOI] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., and Roeder R. G. (1983), Nucl Acids Res 11, 1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Orval C., d’Aubenton C., Sirand-Pugnet P., Gallego M., Brody E., and Marie J. (1991), Science 252, 1823–1828. [DOI] [PubMed] [Google Scholar]
- Fu X.-Y. and Manley J. L. (1987), Mol Cell Biol 7, 738–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattoni R., Schmitt P., and Stevenin J. (1988), Nucl Acids Res 16, 2389–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge H. and Manley J. L. (1990), Cell 62, 25–34. [DOI] [PubMed] [Google Scholar]
- Ge H., Zuo P., and Manley J. L. (1991), Cell 66, 373–382. [DOI] [PubMed] [Google Scholar]
- Goralski T. J., Edström J.-E., and Baker B. S. (1989), Cell 56, 1011–1018. [DOI] [PubMed] [Google Scholar]
- Harper J. E. and Manley J. L. (1991), Mol Cell Biol, 11, 5945–5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedley M. L. and Maniatis T. (1991), Cell 65, 579–586. [DOI] [PubMed] [Google Scholar]
- Helfman D. M., Ricci W. M., and Finn L. A. (1988), Genes Dev 2, 1627–1638. [DOI] [PubMed] [Google Scholar]
- Helfman D. M. and Ricci W. M. (1989), Nucl Acids Res 17, 5633–5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfman D. M., Roscigno R. F., Mulligan G. J., Finn L. A., and Weber K. S. (1990), Genes Dev 4, 98–110. [DOI] [PubMed] [Google Scholar]
- Hoshijima K., Inoue K., Higuchi I., Sakamoto H., and Shimura Y. (1991), Science 252, 833–836. [DOI] [PubMed] [Google Scholar]
- Inoue K., Hishijima K., Sakamoto H., and Shimura Y. (1990), Nature 344, 461–463. [DOI] [PubMed] [Google Scholar]
- Krainer A. R., Conway G. C., and Kozak D. (1990a), Cell 62, 35–42. [DOI] [PubMed] [Google Scholar]
- Krainer A. R., Conway G. C., and Kozak D. (1990b), Genes Dev 4, 1158–1171. [DOI] [PubMed] [Google Scholar]
- Krainer A. R., Mayeda A., Kozak D., and Binns G. (1991), Cell 66, 383–394. [DOI] [PubMed] [Google Scholar]
- Mayeda A. and Ohshima Y. (1988), Mol Cell Biol 8, 4484–4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montell C., Fisher E. F., Caruthers M. H., and Beck A. J. (1982), Nature 295, 380–384. [DOI] [PubMed] [Google Scholar]
- Montell C., Courtois G., Eng C., and Berk A. (1984), Cell 36, 951–961. [DOI] [PubMed] [Google Scholar]
- Ryner L. C. and Baker B. S. (1991), Genes Dev 5, 2071–2085. [DOI] [PubMed] [Google Scholar]
- Schmitt P., Gattoni R., Keoharong P., and Stevinin J. (1987), Cell 50, 31–39. [DOI] [PubMed] [Google Scholar]
- Smith C. W.J., Patton J. G., and Nadal-Ginard B. (1989), Annu Rev Genet 23, 527–577. [DOI] [PubMed] [Google Scholar]
- Sosnowski B. A., Belote J. M., and McKeown M. (1989), Cell 58, 449–459. [DOI] [PubMed] [Google Scholar]
- Spector D. J., McGrogan M., and Raskas H. J. (1978), J Mol Biol 126, 395–414. [DOI] [PubMed] [Google Scholar]
- Stephens C. and Harlow E. (1987), EMBO J 6, 2027–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson C., Pettersson U., and Akusjarvi G. (1983), J Mol Biol 165, 475–499. [DOI] [PubMed] [Google Scholar]
- Ulfendahl P. J., Linder S., Kreivi J. P., Nordqvist K., Svensson C., Hultberg H., and Akusjarvi G. (1987), EMBO J 6, 2037–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y. and Weiner A. M. (1986), Cell 46, 827–835. [DOI] [PubMed] [Google Scholar]