Abstract
A rapid method for defining novel steroid-responsive elements has been developed. Large libraries of degenerate oligonucleotides were analyzed using a yeast-based screen to identify estrogen-responsive DNA sequences. From a library of 40,000 recombinants, seven estrogen-responsive clones were identified. When sequenced, these elements showed remarkable diversity and were different from the consensus vitellogenin A2 ERE. One surprising result was the presence of the two half sites as direct repeats in some of the clones. This implies that in vivo estrogen receptor can bind and transactivate yeast genes through response elements in which the two half sites align as direct repeats. This protocol requires no purified protein and specifically selects for functional response elements. It has a wide application in the study of any transcription factor/DNA interaction.
Steroid hormones are potent modulators of the transcriptional events that regulate homeostosis and development. These hormones mediate their effects by binding to specific intracellular receptor proteins. Such binding leads to a conversion of the protein from an inactive to a transcriptionally active form. The activated receptor is then capable of regulating transcription rates of responsive genes by interacting with specific regulatory sequences near target genes (Ringold, 1985; Yamamoto, 1985; Strahle et al., 1987; Evans, 1989; Beato, 1989; O’Malley, 1990).
A number of DNA recognition sequences (steroid response elements, SREs) for the steroid hormone receptor super-family have been defined. A comparison of these sequences has indicated that they are similar, though distinct (Beato, 1989; O’Malley, 1990). Classically, SREs have been defined by deletion analysis of steroid-responsive promoters, and by a determination of the minimal sequences required to transfer steroid responsiveness upon a heterologous promoter (Druege et al., 1986). Such approaches have identified SREs for the estrogen (Klock et al., 1987; Glass et al., 1987; Kumar and Chambon, 1988), progesterone/glucocorticoid (Tsai et al., 1988; Pfahl, 1982; Giesse et al., 1982), thyroid hormone (Evans, 1989; Glass et al., 1987), retinoic acid, and vitamin D receptors (Walker et al., 1984; Kerner et al., 1989). Sequence comparisons reveal that these SREs are remarkably similar, and with the exception of SREs for the vitamin D receptor and retinoic acid receptor, they generally represent short palindromic sequences that have a dyad axis of symmetry (Kerner et al., 1989). The contribution of each of these bases in the sequences has been analyzed extensively; this allows definition of critical contact points of the receptor and DNA (Strahle et al., 1987; Kumar and Chambon, 1988; Umesono and Evans, 1989). Such studies rely on the identification of a few distinct SREs, with additional elements defined by mutational modifications of these original sequences (Strahle et al., 1987).
We wished to develop a random and rapid approach to identify novel steroid response elements using synthetic response elements derived from random-sequence oligonucleotides. Previously, random-sequence oligonucleotides have been used to define consensus sequences of E. coli promoter elements (Oliphant and Struhl, 1988), yeast GCN4 protein-binding sites (Oliphant et al., 1989), different classes of yeast TATA elements (Singer et al., 1990), and the binding sites of MyoD and E2A proteins (Blackwell and Weintraub, 1990). These methods require transcription factors free of other DNA-binding proteins. For this study we chose to develop a screen that would allow the identification of DNA sequences from libraries of degenerate oligonucleotides which would allow a specific transcriptional response to receptor. During the course of our studies, a similar approach was reported to define an NGFI-B binding site (Wilson et al., 1991). This approach utilized the DNA-binding domain of NGFI-B expressed as part of aLexA-NGFI-B-Gal4 chimeric activator protein.
We and others have previously reported on the successful reconstitution of an estrogen-responsive transcription unit in yeast (Metzger et al., 1988; Fuqua et al., 1991; Pham et al., 1991). This yeast-based technology was used to develop a screen that would identify novel steroid response elements for intact receptors among large libraries of degenerate oligonucleotides. Our goal was to enhance the understanding of estrogen action at its cognate DNA response elements and would provide a simple model that could be used to identify response elements for authentic members of the steroid hormone receptor superfamily.
Materials and methods
Materials
DNA-manipulating enzymes were obtained from Promega, Boehringer Mannheim, or New England Biolabs. [125I] protein A was obtained from ICN. The immobilon-P (PVDF) transfer membranes (IPVM 30uRo) were purchased from Millipore, and rabbit anti-rat antibody (IgG) was purchased from Zymed. Estradiol-l7β, adenine sulfate, o-nitrophenyl β-d-galactopyranoside, and uracil were from Sigma. Casamino acids, yeast nitrogen base without amino acids, and dextrose were obtained from Difco.
Buffers
Homogenization buffer for analysis of receptor contained 10mM Tris-HCl, 2mM EDTA, 5mM dithiothreitol, and 10% glycerol (TEDG) and 300mM KC1 (TEDG with salt). Transcriptional buffer for β-galactosidase assays contained 0.12M Na2HPO4, 0.04M NaH2PO4, 10mM KC1, 1mM MgSO4, and 0.27% 2-mercaptoethanol, pH 7.0.
Yeast strains
The S. cerevisiae strain BJ3505 (mat a, pep4::His3, Prb61-Δ 1.6R, His3, Lys 2-208, Trpl-Δ 101, Ura 3-52, Gal2 (CUP 1)) was used throughout this study (Moerle et al., 1986). All yeast transformations were carried out using the lithium acetate transformation protocol (Ito et al., 1983).
DNA constructions
The construction of YEPE10, a high copy number yeast expression vector, has been described previously (McDonnell et al., 1991). The expression vector YEPE10 was transformed into BJ3505, and transformants were isolated by tryptophan auxotrophy. The construction of the estrogen-responsive reporter YRPE2 has been described previously (Fuqua et al., 1991; Pham et al., 1991). To construct the reporter plasmid YRPD2, in which the two receptor binding half sites are aligned as direct repeats, two copies of an oligonucleotide were inserted into the unique Bgl II site of plasmid PC3 (Pham et al., 1991). The oligonucleotide sequence is as follows: GATCCTAGAGGTCACAGGGTCATACGA. The copy number and orientation of the inserted sequences were determined by sequencing.
Preparation of yeast extracts
S. cerevisiae cells containing YEPE10 were grown overnight in minimal media containing 2% glucose and essential amino acids at 30°C. The yeast were then subcultured in fresh medium and allowed to grow until early mid-log phase (OD600 = 1.0). Induction of receptor was initiated by the addition of 100μM copper sulfate to the culture, which was then allowed to grow for another 4–6 hours. Cells were harvested by centrifugation and washed twice with TEDG. This and all subsequent steps were done at 4°C. The pellet was resuspended in TEDG buffer containing 300mM KC1. The cell suspension was mixed with an equal volume of glass beads (0.5 mm; Braun Instruments) and disrupted by vortexing in a microfuge tube. The homogenate was centrifuged at 12,000g for 10 minutes. The supernatant was collected, and protein concentration was estimated by the method of Bradford (1976) using bovine serum albumin as a standard.
Western immunoblotting
Proteins from yeast were resolved on a 10% SDS-polyacrylamide gel and transferred to immobilon membranes as described previously (Carson et al., 1987). Solid phase radioimmuno-assay was performed as described (Carson et al., 1987) using monoclonal antibodies (D-75) directed against human ER (kindly provided by Dr. Geoffrey Greene, University of Chicago).
Transcriptional assays
Yeast strains containing the receptor expression plasmid and reporter plasmid were grown in minimal medium minus uracil and tryptophan in the presence and absence of hormone (17 β-estradiol) and copper sulfate overnight at 30 °C. Yeast extracts were prepared from these cells and assayed for β-galactosidase activity as described previously (Miller, 1972).
Construction and propagation of (SRE)n library
Single-stranded oligonucleotides corresponding to the sequences below were synthesized. Degeneracy was achieved by using a mix of all four bases at each random position (N). The primary oligonucleotide sequence was:
5′ GCATGCATAGATCTNNNGNNCNNNNNGNNCNNNGGATCCAAGCTTCG 3′
The degenerate oligonucleotides were converted to double-stranded DNA (Oliphant et al., 1986; Oliphant and Struhl, 1987) using a 14-mer primer complementary to the 3′ end of the degenerate oligonucleotides. A library of 40,000 clones was prepared in E. coli by cloning the double-stranded oligonucleotides into the unique Bgl II site of yeast reporter plasmid YRPC3 (Pham et al., 1991), yielding YRP(SRE)n. DNA was prepared from the E. coli library and then transformed into yeast strain BJ3505 carrying the receptor expression plasmid. Transformants were identified by tryptophan and uracil auxotrophy on plates containing X-gal and 17β-estradiol.
Transformants developing blue color in the presence of estrogen were picked, purified, and analyzed for hormone-dependent transcriptional activation, as described above. Reporter plasmids were rescued from transformants exhibiting hormone-dependent transcriptional activity (Hoffman and Winston, 1987), and the DNA inserts were sequenced by the dideoxy chain termination method (Sanger et al., 1977).
Results
Development of the screen
The yeast expression plasmid carrying the human estrogen receptor cDNA(YEPE10) was transformed into a protease-deficient strain of Saccharomyces cerevisiae (BJ3505) by the standard techniques (Ito et al., 1983). The yeast cells carrying YEPE10 were grown overnight in minimal media at 30°C. The yeast cells were then subcultured in fresh medium and allowed to grow until early mid-log phase (OD600 = 1). Induction of receptor was then initiated by the addition of 100μM copper sulfate to culture, which was then allowed to grow for another 4–6 hours. Following induction, cells were harvested and lysed, and the resultant extracts were analyzed by immunoblot using D-75 ER antibody. As shown in Figure 1, the human estrogen receptor expressed in yeast is intact and migrates as a 65 kDa protein. A subsequent analysis of this material showed that the hormone-binding properties were identical to wild-type receptor (McDonnell et al., 1991). Furthermore, the transcriptional activity of hER expressed in yeast was similar to that observed in transfected mammalian cells (Fuqua et al., 1991; Pham et al., 1991; McDonnell et al., 1991).
Figure 1.

Western immunoblot analysis of hER expressed in yeast. Yeast extract (50 μg of protein) from YEPE10 was analyzed by immunoblot as described in Materials and Methods, using D-75 ER antibody.
Development of a system for identification of SREs
Much of the existing methodology (Strahle et al., 1987; Beato, 1989; Druege et al., 1986; Kumar and Chambon, 1988; Walker et al., 1984; Kerner et al., 1989) for identification of steroid response elements is laborious and limited to comparative analysis of sequences from a few steroid-responsive genes. To overcome these problems, we utilized the method shown in Figure 2. A degenerate oligonucleotide was designed following a comparison of the known steroid response elements (Table 1). Nucleotides common to all response elements are boxed and were left constant in our otherwise random oligonucleotide (Strahle et al., 1986; Klock et al., 1987). Where choices were involved, we left all nucleotides constant if they were previously reported to be receptor contact points. A pattern evident from this comparison of different response elements was the variable nature of the spacer region between the two receptor binding half sites. For the estrogen receptor we chose a spacer of 3 oligonucleotides, all of which were randomized. The oligonucleotides were converted to double-stranded DNA by using a specific primer complementary to the 3′ end of the degenerate oligonucleotide (Oliphant et al., 1986; Oliphant and Struhl, 1987). The double-stranded oligonucleotides were cloned in the Bgl II site of the YRPC3 vector (Pham et al., 1991) and propagated in E. coli, generating a library of approximately 40,000 clones.
Figure 2.

Identification of estrogen response elements using a yeast-based screen. A library of degenerate oligonucleotides YRP(SRE)n was created by cloning the double-stranded oligonucleotides into the unique Bgl II site of yeast reporter plasmid YRPC3. CYC1 is the yeast iso-1-cytochrome C promoter fused to the structural gene of E. coli Lac Z. URA3 is the selective marker for uracil. 2 Micron is the replicating DNA of yeast. The library was introduced into yeast cells carrying the estrogen receptor expression plasmid (YEPE10), and the cells were selected for tryptophan and uracil auxotrophy on plates containing X-gal and 17β-estradiol. Transformants developing blue color were picked, and functional estrogen response elements were selected.
Table 1.
Sequence of degenerate oligonucleotide and comparison with other SREs.
| The sequence of the degenerate oligonucleotide is indicated and aligned to the response elements for estrogen glucocorticoid/progesterone and thyroxine. The sequences found in all response elements are boxed and were left constant in our oligonucleotide. |

|
To assay for functional steroid response elements, the library was introduced into yeast cells carrying an estrogen receptor expression plasmid, and transformants were selected for tryptophan and uracil auxotrophy on selective plates containing estradiol and X-gal. The plates were incubated at 30°C for several days. A total of 157 blue colonies were picked, purified, and reassayed in the presence and absence of estradiol. Ninety-six percent of blue colonies exhibited hormone-independent transcriptional activity. We attributed these to the generation of cryptic transcription factor binding sites in our library. However, 4% of the blue colonies exhibited hormone-dependent transcriptional activity. Reporter plasmids were rescued from hormone-dependent transformants, and the DNA inserts were sequenced. The sequence and transcriptional activities of hormone-dependent transformants are shown in Table 2.
Table 2.
Sequence and transcriptional activities of hormone-dependent response elements.
| Reporter plasmids were rescued from hormone-dependent transformants, and the DNA inserts were sequenced. The sequence and transcriptional activities of hormone-dependent transformants were aligned to the reference reporter (YRPE2), carrying the vitellogenin A2 ERE and YRPC3. |

|
The transcriptional activity of YRPE2, the reference reporter carrying the vitellogenin A2 ERE, and transformant #12 were absolutely hormone-dependent. Transformants #1, #4, #9, #13, #39, and #45 exhibited a low basal activity which was inducible by hormone. With one exception, the basal activity was <10% of the inducible activity. Induction varied 7- to 23-fold from the basal level. To demonstrate that the transcriptional activity exhibited by these transformants was receptor-dependent, the reporter plasmids containing synthetic EREs were transformed into a yeast strain that did not contain estrogen receptor, and transformants were selected by uracil auxotrophy. These transformants were grown in the presence or absence of estradiol and copper sulfate. The cytosols were prepared and analyzed for β-galactosidase activity. As expected, neither the reference reporter, YRPE2, nor the reporter plasmids containing synthetic EREs stimulated induction of β-galactosidase activity under any conditions (data not shown).
Sequence analysis of hormone-dependent transformants suggests that the sequences of all the transformants were different from the consensus sequence of the estrogen response element. In the latter response element, the two half sites are organized as inverted repeats. Surprisingly, the two half sites of clones #9 and #45 consisted of direct repeats. In the clone #45, direct repeats are misaligned by one base. These results might suggest that the estrogen receptor can function in yeast from a response element in which the two half sites are present as either direct or indirect repeats. In the clone #39, the conserved C in the first half is changed to A, while in the second half both conserved G and C are changed to C and T respectively. These changes in the conserved nucleotides might have originated during oligonucleotide synthesis. This suggests that these bases are not critically important for functional interaction of estrogen receptor with its response elements.
To confirm that the estrogen receptor can bind and transactivate transcription from a response element in which the two receptor binding half sites are aligned as direct repeats, an oligonucleotide was constructed containing the receptor binding sites as perfect direct repeats. This oligonucleotide was cloned into the Bgl II site of the plasmid PC3 (Pham et al., 1991), (YRPD2). This plasmid was transformed into the yeast cells containing the receptor expression plasmid. The transcriptional activity of estrogen receptor through this response element was compared to those of a perfect palindrome under varying copper concentrations in the presence and absence of hormone. Cytosols were prepared, and induction of lacZ activity was measured (Fig. 3). These data suggest that in the absence of hormone, the receptor cannot stimulate induction of transcription from either response elements. Transcriptional activity was observed only in the presence of hormone. Both reporters demonstrated a similar transcriptional activity. These results confirmed that the estrogen receptor can transactivate transcription from yeast response elements in which receptor binding half sites are aligned in direct or indirect repeats. These results are in contrast with other published results (Naar et al., 1991).
Figure 3.
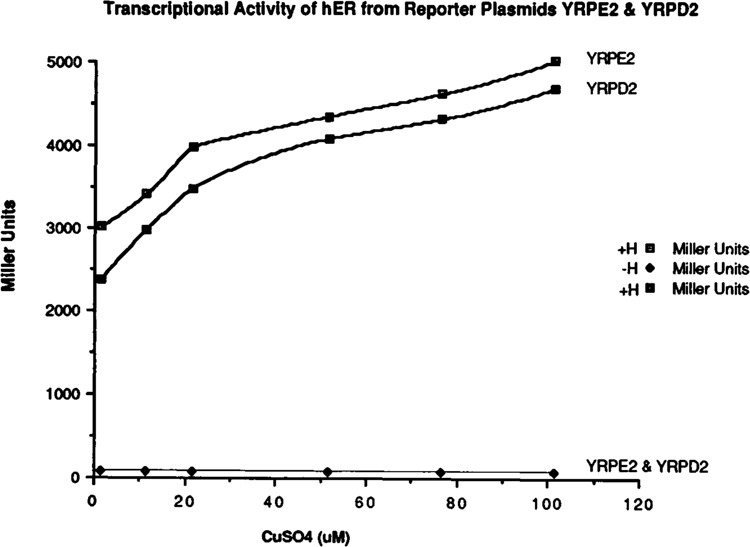
Transcriptional activity of hER from reporter plasmids containing receptor binding sites as direct or indirect repeats. The yeast cells containing YEPE10 and YRPE2 plasmids and the cells containing YEPE10 and YRPD2 plasmids were grown in minimal media in the absence and presence of estradiol with increasing concentrations of copper sulfate (CuS04). Cytosols were prepared, and induction of lacZ was measured. The β-galactosidase activity measured in yeast extracts is expressed as Miller units per mg protein.
The EREs identified in this study were compared to the mammalian gene bank by computer scanning analysis (Table 3) to see if novel estrogen responsive genes could be identified. This analysis revealed that homologous sequences were present in promoters of several mammalian genes. A review of the literature indicated that most of these genes had been reported to be under estrogen regulation, but these reports did not identify the sequences responsible for this effect. For example, the ERE in transformant #4 is homologous to the sequences found in the promoter region of human phosphoglycerate kinase gene. The ERE sequences of transformant #12 are homologous to the sequences present in the promoter of human serum retinol binding protein. The ERE of transformant #13 is similar to sequences found in the promoter regions of human elastin, muscle creatin kinase, complement C3, and HMG CoA reductase gene, as well as those found in the promoter regions of mouse muscle creatin kinase gene and chicken apo VLDL II genes.
Table 3.
Comparison of synthetic ERE sequences with the mammalian gene bank.
| The estrogen response elements identified from large libraries of degenerate oligonucleotides were compared to the mammalian gene bank. The genes containing homologous sequences and regulated by estrogen were identified. | |
| Sequences of estrogen response elements identified from degenerate oligonudeotide libraries | Genes containing homologous sequences in the promoter regions and regulated by estrogen |
|---|---|
| # 4 GGTTCGNNNCGTGC | Human phosphoglycerate kinase gene |
| #12 GCACANNNGGACC | Human serum retinol binding protein |
| #13 GGACANNNGGCCC | Human elastin gene |
| Human muscle creatin kinase gene | |
| Human complement C3 gene | |
| Human HMG CoA reductase gene | |
| Mouse muscle creatin kinase gene | |
| Chicken apo VLDL II gene | |
Using this identical strategy, we have also identified several progesterone-responsive elements. Some of the progesterone-responsive elements are also present in direct repeats (data not shown).
Discussion
Most current methods for the identification of the steroid response elements rely on a deletion analysis of steroid-responsive promoters to define the minimal amount of sequence required to transfer hormonal regulation from one gene to another (Druege et al., 1986). This analysis is often followed by mutagenesis of the core element. We describe here an alternative method that utilizes a reconstituted estrogen response system in yeast to screen large libraries of possible steroid response elements. Using this approach, we have identified and sequenced seven estrogen response elements. A comparison of these sequences suggests that most are similar but nonidentical to the consensus vitellogenin ERE (Klein-Hitpass et al., 1988). One surprising result was the presence of the two half sites arranged as direct repeats in some of the clones. This suggests that the estrogen receptor can transactivate genes in yeast via response elements in which the two half sites align as direct repeats. To confirm this hypothesis, a synthetic response element containing two receptor binding sites as direct repeats was constructed, and induction of gene transcription from this response element was examined. This study confirmed that estrogen receptor can activate gene transcription through response elements in which receptor recognition sites are present as direct or indirect repeats.
It has been suggested that thyroid hormone receptor, retinoic acid receptor, and vitamin D3 receptor can selectively bind and transactivate genes from direct repeat response elements (Umesono et al., 1991). Notably, our results contradict those of Naar et al. (1991), who suggest that estrogen receptor cannot induce gene transcription from response elements in which receptor binding sites are aligned as direct repeats. We are in general agreement with this observation from our previous studies in the same animal cells (data not shown). Nevertheless, it is quite possible that these differences may be due to cell-specific factors which modulate gene induction via protein-protein interaction and should be tested in a wide variety of eucaryotic cells.
Comparison of the estrogen response elements identified in this study with the mammalian gene bank revealed that homologous sequences were present in the promoters of various mammalian genes. Most of these genes were reported to be regulated by estrogen, but their response elements have not yet been identified.
The system described here is not limited to a single receptor, and it can be used to identify the response element for any DNA-binding protein. To confirm this we have also used this system to identify progesterone receptor response elements (PREs). This system could also be useful for identifying the response elements for the “orphan” members of the steroid receptor superfamily. Identification of such sequences is required for eventual functional analysis of these receptors. In view of the expanding number of sequences in the gene bank, it is possible that this approach may be useful in identifying new steroid-responsive genes.
The system developed in this study has major advantages over in vitro methods (Oliphant et al., 1989; Singer et al., 1990; Blackwell and Weintraub, 1990). This system does not require purified protein, high affinity antibodies, or a heterologous expression system. Our system also has limited advantages over the previously developed in vivo system which was employed to identify the NGFI-B DNA-binding sites (Wilson et al., 1991). The system utilized the DNA-binding domain of NGFI-B expressed as part of a LexA-NGFI-B-Gal4 chimeric protein. The LexA DNA-binding domain may interfere with the selection of target sequences, whereas our method does not require fused protein. Also, our system requires one-step selection, not two-step.
This system also has several other advantages. Using a random oligo approach, all possible response elements can be identified, not just those based upon a known sequence. The screen can be designed to rank elements relative to activity. The system is rapid and requires only the expression of the receptor in yeast. No purified receptor is necessary for this assay. The same oligo library can be used for most steroid superfamily members. Finally, by comparing the sequences of the identified elements, a more informative consensus binding sequence can be derived.
Acknowledgments
We thank Geoffrey Greene for the ER cDNA and D75 antibodies. We also thank Paula Howard for technical assistance and Lisa Gamble for typing the manuscript. This research was supported by ACS funding.
The costs of publishing this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC Section 1734 solely to indicate this fact.
References
- Beato M. (1989), Cell 56, 335–344. [DOI] [PubMed] [Google Scholar]
- Blackwell T. K. and Weintraub H. (1990), Science 250, 1104–1110. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. (1976), Anal Biochem 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Carson M. A., Tsai M-J., Conneely O. M., Maxwell B. L., Clarck J. M., Dobson A. D. W., Elbrecht A., Toft D. O., Schrader W T., and O’Malley B. W. (1987), Mol Endocrinol 1, 791–801. [DOI] [PubMed] [Google Scholar]
- Druege P. M., Klein-Hitpass L., Green S., Stack G., Chambon P., and Ryffel G. U. (1986), Nucl Acids Res 23, 9329–9337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans R. A. (1989), Science 240, 889–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuqua S. A. W., Fitzgerald S. D., Tandon A. K., McDonnell D. P., Nawaz Z., O’Malley B. W., and McGuire W. L. (1991), Cancer Res 51, 105–109. [PubMed] [Google Scholar]
- Giesse S., Scheidereit C., Westphal H. M., Hynes N. E., Gronerand B., and Beato M. (1987), EMBO J 1, 1613–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass C. K., Franco R., Weinberger C., Albert V. R., Evans R. M., and Rossnfeld M. G. (1987), Nature 329, 734–736. [DOI] [PubMed] [Google Scholar]
- Hoffman C. S. and Winston F. (1987), Gene 57, 267–272. [DOI] [PubMed] [Google Scholar]
- Ito M., Fukuda Y., Murata K., and Kimura A. (1983), J Bacteriol 153, 163–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerner S. A., Scott R. A., and Pike J. W. (1989) Proc Natl Acad Sci USA 86, 4455–4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein-Hitpass L., Ryffel G. V., Heithinger E., and Cato A. C. B. (1988), Nucl Acids Res 16, 647–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klock G., Strahle U., and Schutz G. (1987), Nature 329, 734–736. [DOI] [PubMed] [Google Scholar]
- Kumar V. and Chambon P. (1988), Cell 55, 361–369. [DOI] [PubMed] [Google Scholar]
- McDonnell D. P., Nawaz Z., Densmore C., Weigel N. L., Pham T. A., Clarck J. H., and O’Malley B. W. (1991), J Steroid Biochem Mol Biol 39, 291–297. [DOI] [PubMed] [Google Scholar]
- Metzger D., White J. H., and Chambon P. (1988), Nature 334, 31–36. [DOI] [PubMed] [Google Scholar]
- Miller J. M. (1972), in Experiments in Molecular Genetics (Miller J. M., ed.), Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 352–355. [Google Scholar]
- Moerle C. M., Ayanardi M. W., Kolodny M. R., Park F. J., and Jones E. W. (1986), Genetics 115, 255–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naar A. M., Boutin J-M., Lipkin S. M., Yu V. C., Holloway J. M., Glass C. K., and Rosenfeld M. G. (1991), Cell 65, 1267–1279. [DOI] [PubMed] [Google Scholar]
- Oliphant A. R., Hill D. E., Nussbaum A. L., and Struhl K. (1986), Gene 44, 177. [DOI] [PubMed] [Google Scholar]
- Oliphant A. R., Brandl C. J., and Struhl K. (1989), Mol Cell Biol 9, 2944–2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliphant A. R. and Struhl K. (1987), Methods Enzymol 155, 568–582. [DOI] [PubMed] [Google Scholar]
- Oliphant A. R. and Struhl K. (1988), Nucl Acids Res 16, 7673–7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley B. W. (1990), Mol Endocrinol 4, 3263–3269. [Google Scholar]
- Pfahl M. (1982), Cell 31, 475–482. [DOI] [PubMed] [Google Scholar]
- Pham T. A., Elliston J. F., Nawaz Z., McDonnell D. P., Tsai M-J., and O’Malley B. W. (1991), Proc Natl Acad Sci USA 88, 3125–3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringold G. A. (1985), Annu Rev Pharmacol Toxicol 25, 529–566. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., and Coulson A. R. (1977), Proc Natl Acad Sci USA 74, 5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer V. L., Wobbe R., and Struhl K. (1990), Genes Dev 4, 636–645. [DOI] [PubMed] [Google Scholar]
- Strahle U., Kloch G., and Schutz G. (1987), Proc Natl Acad Sci USA 84, 7871–7875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai S. Y, Carlstedt-Duke J., Weigel N. L., Dahlman K., Gustafsson J. A., Tsai M-J., and O’Malley B. W. (1988), Cell 55, 361–369. [DOI] [PubMed] [Google Scholar]
- Umesono K., Murakami K. K., Thompson C. C., and Evans R. A. (1991), Cell 65, 1255–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umesono K. and Evans R. A. (1989), Cell 57, 1139–1146. [DOI] [PubMed] [Google Scholar]
- Walker P., Germond J. E., Brown-Luedi M., Givel F., and Wahli W. (1984), Nucl Acids Res 12, 8611–8626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson T. E., Fahrner T. J., Johnston M., and Milbrandt J. (1991), Science 252, 1296–1300. [DOI] [PubMed] [Google Scholar]
- Yamamoto K. (1985), Annu Rev Genet 19, 209–252. [DOI] [PubMed] [Google Scholar]


