Mammalian gene expression can be regulated by cell type, developmental stage, and numerous extereal stimulti Regulation is brought about by a complex interaction of cis-acting DNA sequences and trans-acting factors. Considerable effort has been expended to identify and characterize regulatory DNA sequences and to understand the basic mechanisms by which they exert their actions. Traditionally, the transcriptional activities of putative promoters and regulatory sequences have been measured by transfections into cultured cells of various DNA constructs consisting of these sequences fused to a reporter gene and subsequent quantitation of reporter gene expression. This approach has provided a great deal of information concerning the sequences mediating the expression of genes in various cell types in response to physiologic perturbations such as hormones—and, to a lesser extent, during specific developmental stages, such as the fusion of myoblasts to form myotubes.
Given the complexity of biological processes in vivo, however, a question arises whether regulatory elements identified in cultured cells accurately explain the regulation of gene expression in vivo. This becomes particularly important in considering the use of immortalized cell lines or primary cultures, whose patterns of gene expression often differ significantly from those of the intact tissue.
One way to evaluate the relevance of in vitro transfection studies to gene regulation in vivo is to create transgenic animals using constructs that have also been tested in cultured cells. This approach requires the generation of several independent mouse lines for each construct under study, an expensive and time-consuming undertaking compared with transfections into cultured cells. One alternative to transgenic mice that may approximate in vivo gene regulation more closely than cell culture transfections is the injection of DNA sequences linked to reporters into cardiac and skeletal muscle tissue in vivo (Kitsis et al., 1991). However, this gene transfer technique appears to be limited to striated muscle cells (Wolff et al., 1991).
Regardless of whether gene regulation is studied in vitro or in vivo, the state of the foreign DNA may influence the regulation of its expression. With transient transfections into cultured cells or injections of DNA into muscle in vivo (Wolff, et al., 1990), the construct being tested exists in an episomal state. The geometric configuration of the regulatory sequences in the construct may differ markedly from its natural configuration in the endogenous locus. This, in turn, may influence regulation of the expression of the construct. In addition, an episome will not be subject to regulatory information that may reside in the chromatin configuration. On the other hand, with stably transfected cells and transgenic animals, the construct undergoes integration into random chromosomal sites whose chromatin configuration may differ from that of the endogenous locus. In fact, the importance of long-range chromosome effects is supported by the existence of locus control regions (LCR), such as those seen in the β globin gene complex (see Grosveld, et al., 1987; Forrester et al., 1987), which act at a distance to confer copy number-dependent, position-independent expression onto a gene. Although it would be optimal to study the regulation of a construct at its endogenous locus, or at least within its LCR, this is usually not possible. Therefore, studies should be performed on multiple independent cell or transgenic lines to separate effects which result from the cis-acting elements under study from those which are due to the position of integration.
In addition to transcriptional regulation, posttranscriptional control plays an important role in the control of mammalian gene expression. Expression of a test construct consisting of promoter and regulatory elements linked to a reporter gene will not reflect posttranscriptional processes that may be relevant to the regulation of the gene under study, since sequences specifying the endogenous pattern of RNA splicing and mRNA stability have been replaced with heterologous sequences. Some investigators have used constructs containing a minigene, rather than a reporter gene, to address some of these issues (for example, see Dente et al., 1988). Though obviously important, the regulatory role played by posttranscriptional events is beyond the scope of the present review.
Although in vivo experiments have often yielded results identical to those obtained in cultured cells (for example, see Leask et al., 1990), there are multiple examples of discordance between gene regulation in vitro and in vivo. The purpose of this review is to discuss some of the examples in which transcriptional regulation conferred by a given cis-acting DNA sequence element differs significantly between cultured cells and in vivo. In addition, possible mechanisms for the discordance will be discussed.
Gene regulation in vitro and in vivo
In a relatively small number of cases, some of the same DNA constructs tested in transfections have also been evaluated in transgenic mice (Table 1). This section will compare results that have been obtained using both approaches. Discordance between gene regulation in vitro and in vivo can take the form of “promiscuous” expression of the DNA construct in transfections into different cell types with appropriate expression in transgenics. In the case of the liver-specific α1-acid glycoprotein gene (AGP-A), ∼1.2 kb of upstream sequence and ∼2 kb of 3′-flanking sequence were able to direct transcription with similar efficiencies, both in HeLa cells, where the gene “should not” be expressed, and in Hep3B hepatoma cells, which express the endogenous AGP-A gene. In contrast, in transgenic mice, the same sequences resulted in appropriate liver-restricted expression (Dente et al., 1988). This type of discordance is also exhibited by a human N-myc minigene, which was expressed promiscuously following transient and stable transfections into mouse 3T3 cells, where the endogenous gene is not expressed, but was expressed concordantly with the endogenous gene in transgenic animals (Zimmerman et al., 1990). The endogenous rat α-cardiac myosin heavy chain gene is expressed in cardiac but not skeletal muscle. Despite this, ∼0.6 kb of 5′-flanking sequence was sufficient to direct transcription in both myoblasts and myotubes of the L6E9 rat skeletal muscle cell line (Tsika et al., 1990). This sequence was not transcriptionally active in HeLa cells or 3T3 fibroblasts, demonstrating limited—but not entirely specific—expression of this construct in vitro. In contrast, the same upstream sequence directed cardiac-specific expression in transgenic mice (Katz et al., 1992) and following injection into the hearts of adult rats in vivo (Kitsis et al., 1991).
Table 1.
Discordance between gene regulation in vitro and in vivo
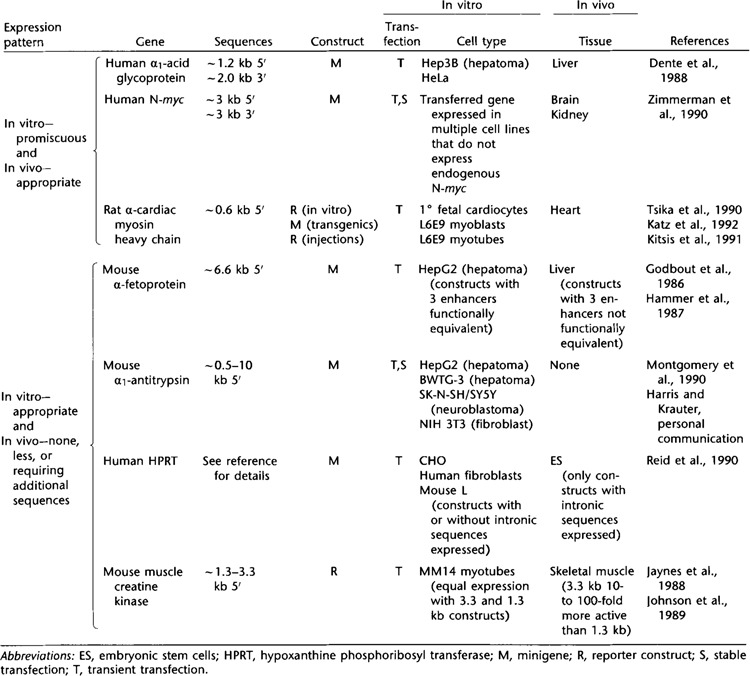
Another form of discordance is observed when cis-acting elements are sufficient to direct expression in transfected cells but are inadequate to drive transgene expression in the analogous cell type in vivo or in embryonic stem cells. For example, transfection of an α-feto-protein minigene into hepatoma cells demonstrated the functional equivalence of three enhancers located in the 5′-flanking region (Godbout et al., 1986). In contrast, constructs containing various combinations of these elements differed in their capacities to direct transcription in the livers of transgenic animals (Hammer et al., 1987). Another example is the mouse α1-antitrypsin gene. Approximately 0.5–10 kb of 5′-flanking sequence results in expression of a human growth-releasing factor minigene in transiently and stably transfected hepatoma cells and primary hepatocyte cultures, but not in 3T3 or neuroblastoma cells (Montgomery et al., 1990). In contrast, identical constructs lacked sufficient information to be expressed in multiple lines of transgenic animals (T. Harris and K. Krauter, personal communication). Discordance between gene expression in somatic and embryonic stem (ES) cells has also been observed with a hypoxanthine phosphoribosyl transferase (HPRT) minigene, which was expressed following transfection into multiple somatic cell types but not into ES cells, which express the endogenous HPRT gene (Reid et al., 1990). Addition of HPRT intronic sequences to the minigene, however, resulted in its expression in ES cells. Gene expression in cultured cells can be quantitatively discordant from that in transgenic animals, even if expression is limited to the appropriate tissues in both systems. For example, ∼1.3 kb of mouse muscle creatine kinase (MCK) upstream sequence was as efficient as a ∼3.3 kb construct in driving reporter gene expression in differentiated MM14 cells, a mouse myoblast line (Jaynes et al., 1988). In several lines of transgenic animals, however, the longer construct was 10- to 100-fold more transcriptionally active than the shorter one (Johnson et al., 1989).
Cell culture models for studying gene regulation
Given the discordance between gene regulation in vitro and in vivo, it is worthwhile to examine mechanisms that might explain the phenomenon. One place to begin is to compare quantitative and qualitative patterns of endogenous gene expression in cultured cells and intact tissues. For example, many hepatoma cell lines have been shown to express liver-specific genes at very low levels or not at all (Clayton et al., 1985). The H4AZC2 rat hepatoma cell line, for instance, expresses (among others) one liver-specific gene, glutathione S-transferase, but only at 5% of the level seen in the intact liver. Although culture conditions can be manipulated to induce expression of this gene several fold, it remains substantially lower than that observed in vivo (Gatmaitan et al., 1983). In addition, culture conditions designed to promote liver-specific gene expression have failed to activate the liver-specific α1-antitrypsin gene, which is silent in H4AZC2 cells. Muscle cell lines also exhibit differences in gene expression relative to normal muscle tissue. This is exemplified by the L6J1 myoblast line, which—although capable of fusing into myotubes and initiating a program of muscle-specific gene expression—does not express the entire program of myogenic regulators, since it does not express MyoD (Hinterberger, et al., 1991). Additionally, the normal temporal and quantitative program of myosin heavy chain gene expression does not occur in muscle cell lines (see Weydert et al., 1987). In summary, the phenotype of many cell lines diverges significantly from that of cells in the intact organ.
There are several potential mechanisms for altered gene expression in cell lines. First, the genotype of immortalized cells may be altered, which may affect the expression of specific genes. Second, cells in culture lack many of the cues that trigger physiological and developmental processes. For example, paracrine and mechanical signals from other cell types with which they are in contact in a tissue undoubtedly influence gene expression. Other stimuli that may be different (or lacking) in cell culture relate to the physical substrate on which cells are grown and the media in which they are bathed. Of note, using a chemical and/or a second cell type to model the extracellular matrix and hormonally defined media to replicate the extracellular milieu has been shown to modulate gene expression in cultured cells to mimic more closely that in vivo (see Jefferson et al., 1984; Enat et al., 1984). In addition, cultured cells do not experience the myriad of developmental stimuli to which cells that are formed and grow in an embryo are subjected. It is possible that the absence of this developmental history results in alterations in the subsequent program of gene expression.
Primary cell cultures, which may more accurately reflect gene expression in vivo, are an alternative to immortalized cell lines for gene regulation studies. In some cases, such as cardiac myocytes, primary cultures must be used, since there are as yet no permanent cell lines. Unfortunately, gene expression in primary cultures can also be discordant with that in the same cell type in the environment of the intact organ. This can be the result of the cell dissociation procedure, cell culture conditions (see Jefferson et al., 1984), or the absence of the appropriate developmental stimuli, as discussed above. When hepatocytes are disaggregated from the liver and cultured on a plastic substrate in serum-containing media, the cells assume a flat morphology, and liver-specific gene expression decreases dramatically, while the expression of housekeeping genes such as tubulin remains constant (see Clayton and Darnell, 1983; Clayton et al., 1985). Albumin transcription decreases to 50% of baseline after 4 hours and to 10% after 24 hours in culture (Xanthopoulas et al., 1989). Similarly, levels of skeletal α-actin mRNA specifically decline in cultured neonatal cardiac myocytes compared with the intact neonatal heart (Bishopric et al., 1987).
In other cases, the discordance between gene expression in vitro and in vivo may reflect the phenotype of the cells at the time they are placed in culture. For example, it has been possible to use only fetal and neonatal cardiocytes in transfection studies. The phenotype of these cells differs significantly from that of adult cardiocytes, thereby limiting the applicability of the results of gene regulation experiments in these primary cultures. In support of this idea, it has recently been shown that DNA constructs that are inactive in cultured fetal cardiocytes are active when injected into the adult heart in vivo (P. M. Buttrick et al., unpublished data). Despite the limitations of extrapolating results obtained with myocytes from an earlier developmental stage to adults, neonatal cardiac myocyte cultures have been useful for analyzing some complex physiological processes, such as cellular hypertrophy, in which patterns of gene expression in cultured neonatal cells mimic those in vivo (Waspe et al., 1990).
Bases of discordance
Clues to the molecular basis for discordance between gene expression in cultured cells and the same cell type in the intact tissue have been provided by analyses of trans-acting factors. Steady-state levels of mRNAs encoding several transcription factors have been observed to decrease in cultured liver cells. For example, the sequence-specific DNA binding protein C/EBP is expressed at levels that are at least 10-fold lower in cultured hepatoma cells compared with the intact liver (Friedman et al., 1989). Further, it has been demonstrated that transcription of this gene decreases almost 10-fold within 24 hours of placing hepatocytes into culture (Xanthopoulos et al., 1989). In fact, recent studies suggest that decreases in C/EBP occur in response to activation of cell proliferation, thereby suggesting that expression of high levels of C/EBP might be incompatible with most rapidly growing cell lines (Mischoulon et al., 1992). Another example is provided by repression of the expression of HNF-4, a member of the steroid hormone receptor superfamily, in dedifferentiated hepatomas and in hepatocyte–somatic cell hybrids in which the liver phenotype was extinguished (Kuo et al., 1992). This alteration, in turn, resulted in absence of HNF-1α, a homeoprotein that requires HNF-4 to transactivate its expression. In these systems, expression of other transcription factors, such as C/EBP, was unchanged, demonstrating that the observed changes in gene expression were not the result of general decreases in the abundance of all transcription factors. The implication of such findings for the study of gene regulation is significant. If certain trans-acting factors are required to achieve appropriate tissue- or developmental stage-specific or stimulus-responsive expression, then gene regulation in cultured cells, which express different levels of these factors compared with the intact tissue, may be discordant with that in vivo. It seems likely that some of the cases of discordance discussed in this review can be attributed to altered expression of trans-acting factors in cultured cells.
In summary, the regulation of many mammalian genes is different in cultured cells than in intact tissues. Future studies directed at defining the differences in cellular milieu in these two states will undoubtedly extend our understanding of this discordance and of the underlying basis for gene regulation in vivo.
Acknowledgments
We thank Drs. Kenneth Krauter and Peter Buttrick for helpful discussions.
This work was supported by National Institutes of Health grants HL 02699 (R. N. Kitsis), GM 29090, and HL 57412 (L. A. Leinwand); American Heart Association, New York City Affiliate Grants-in-Aid (R. N. Kitsis and L. A. Leinwand); and the Irma T. Hirschl Scholar Award (L. A. Leinwand).
The costs of publishing this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC Section 1734 solely to indicate this fact.
References
- Acsadi G., Jiao S., Jani A., Duke D., Williams P., Chong W., and Wolff J. A. (1991), New Biol 3, 71–81. [PubMed] [Google Scholar]
- Bishopric N. H., Simpson P. C., and Ordahl C. P. (1987), J Clin Invest 80, 1194–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton D. F., Weiss M., and Darnell J. E. Jr. (1985) Mol Cell Biol 5, 2633–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton D. F. and Darnell J. E. (1983) Mol Cell Biol 3, 1552–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dente L., Ruther U., Tripodi M., Wagner E., and Cortese R. (1988), Genes Dev 2, 259–266. [DOI] [PubMed] [Google Scholar]
- Enat R., Jefferson D. M., Ruiz-Opazo N., Gatmaitan Z., Leinwand L. A., and Reid L. M. (1984), Proc Natl Acad Sci USA 81, 1411–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester W. C., Takegawa S., Papayannopoulou T., Stamatoyannopoulos G., and Groudine M. (1987), Nucleic Acids Res 15, 10159–10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman A. D., Landschultz W. H., and McKnight S. L. (1989), Genes Dev 3, 1314–1322. [DOI] [PubMed] [Google Scholar]
- Gatmaitan Z., Jefferson D., Ruiz-Opazo N., Biempica L., Arias I., Dudas G., Leinwand L., and Reid L. M. (1983), J Cell Biol 97, 1179–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godbout R., Ingram R., and Tilghman S. M. (1986) Mol Cell Biol 6, 477–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosveld F., Blom van Alssndelft G., Greavess G. and Kollias G. (1987), Cell 51, 975–985. [DOI] [PubMed] [Google Scholar]
- Hammer R. H., Krumlauf R., Cramper S. A., Brinster R. L., and Tilghman S. M. (1987), Science 235, 53–58. [DOI] [PubMed] [Google Scholar]
- Hinterberger T. J., Sassoon D. A., Rhodes S. J., and Konieczny S. F. (1991), Dev Biol 147, 144–156. [DOI] [PubMed] [Google Scholar]
- Jaynes J. B., Johnson J. E., Bushin J. N., Gartside C. L., and Hauschka S. D. (1988), Mol Cell Biol 8, 62–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson D. M., Clayton D. F., Darnell J. E. Jr., and Reid L. M. (1984), Mol Cell Biol 4, 1929–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. E., Wold B. J., and Hauschka S. D. (1989), Mol Cell Biol 9, 3393–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz E. B., Steinhelper M. E., Delcarpio J. B., Daud A. J., Claycomb W. C., and Field L. J. (1992), Am J Physiol 31, H1867–H1876. [DOI] [PubMed] [Google Scholar]
- Kitsis R. N., Buttrick P. M., McNally E. M., Kaplan M. L., and Leinwand L. A. (1991), Proc Natl Acad Sci USA 88, 4138–4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo C. J., Conley P. B., Chen L., Sladek E M., Darnell J. E. Jr., and Crabtree G. R. (1992), Nature 355, 457–461. [DOI] [PubMed] [Google Scholar]
- Leask A., Rosenberg M., Vassar R., and Fuchs E. (1990), Genes Dev 4, 1985–1998. [DOI] [PubMed] [Google Scholar]
- Mischoulon D., Rana B., Bucher N. L. R., and Farmer S. R. (1992), Mol Cell Biol 12, 2553–2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery K. T., Tardiff J., Reid L. M., and Krauter K. S. (1990), Mol Cell Biol 10, 2625–2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid L. H., Gregg R. G., Smithies O., and Koller B. H. (1990), Proc Natl Acad Sci USA 87, 4299–4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsika R. W., Bahl J. J., Leinwand L. A., and Morkin E. (1990), Proc Natl Acad Sci USA 87, 379–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waspe L. E., Ordhal C. P., and Simpson P. C. (1990), J Clin Invest 85, 1206–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weydert A., Barton P., Harris A. J., Pinset C., and Buchingham M. (1987), Cell 49, 121–129. [DOI] [PubMed] [Google Scholar]
- Wolff J. A., Malone R. W., Williams P., Chong W., Acsadi G., Jani A., and Feigner P. L. (1990), Science 247, 1465–1468. [DOI] [PubMed] [Google Scholar]
- Xanthopoulos K. G., Mirkovitch J., Decker T., Kuo C. F., and Darnell J. E. Jr. (1989), Proc Natl Acad Sci USA 86, 4117–4121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman K., Legouy E., Stewart V., DePinho R., and Alt F. W. (1990), Mol Cell Biol 10, 2096–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]


