Abstract
In appropriate mammalian cells, interferon regulatory factor-1 (IRF-1) can activate the virus-responsive element of the IFN-β promoter (VREβ″) or the synthetic oligonucleotide (GAAAGT)4. The latter contains two copies of the functional equivalent of PRDI, one of the regulatory domains of VREP′. We prepared yeast strains containing an IRF-1 expression plasmid under the control of the galactose-inducible Gal1 promoter and a reporter plasmid with either (GAAAGT)4, VREβ″, or other test sequences placed upstream of a minimal promoter linked to the β-galactosidase coding sequence. Upon induction of IRF-1 expression, the (GAAAGT)4-containing promoter was activated, but VREβ″ and all other sequences tested were inactive. Our results showed that IRF-1 belongs to a class of higher eukaryotic transcription factors that can interact with the yeast transcriptional machinery. Our findings also raised the question why the duplicate PRDI-like sequences in (GAAAGT)4 can be activated by IRF-1 synthesized in yeast, but not VREβ″, which also contains at least two PRDI-like sequences.
Regulation of the IFN-β promoter is mediated by at least four sequence elements (Goodbourn et al., 1985; Fujita et al., 1987; Dinter and Hauser, 1987; Goodbourn and Maniatis, 1988; Keller and Maniatis, 1988; Fan and Maniatis, 1989; Leblanc et al., 1990) contained within a 125-nucleotide segment preceding the cap site (Fujita et al., 1985; Harada et al., 1990), designated in this paper as VREβ″ (Fig. 1E). PRDII is a binding site for the activating factor NF-κB (Clark and Hay, 1989; Fujita et al., 1989; Hiscott et al., 1989; Lenardo et al., 1989; Visvanathan and Goodbourn, 1989), and PRDI and PRDIII bind the activating factor IRF-1 (Fujita et al., 1988; Miyamoto et al., 1988; Harada et al., 1989; Xanthoudakis et al., 1989; Reis et al., 1992). PRDIV is a binding site for a protein of the cAMP response element binding protein (ATF/CREB) family of transcription factors (Du and Maniatis, 1992). PRDI and PRDII, and two additional domains, NRDI and NRDII, are believed to bind negative regulatory factors as well (Goodbourn and Maniatis, 1988; Keller and Maniatis, 1988; Whittemore and Maniatis, 1990). In particular, IRF-2, which binds to PRDI (Harada et al., 1990), and the zinc finger protein PRDI-BF1 (Keller and Maniatis, 1991; Keller and Maniatis, 1992) have been shown to act as transcriptional repressors. Additional factors binding to PRDI have been described (Whiteside et al., 1992). Virus induction is thought to come about on the one hand by relief of repression and on the other by activation of pre-existing NK-κB and de novo synthesis as well as activity enhancement of IRF-1, which then act cooperatively on the IFN-β promoter (Fujita et al., 1989, Lenardo et al., 1989; Visvanathan and Goodbourn, 1989; Leblanc et al., 1990; Watanabe et al., 1991). Reporter constructs containing PRDI supported by an SV40 enhancer, or (GAAAGT)4, which contains the functional equivalent of dimeric PRDI (Naf et al., 1991), are activated not only by viruses but also by over-expression of IRF-1 (MacDonald et al., 1990; Näf et al., 1991). However, the role of IRF-1 in virus-induced activation of the IFN-β promoter has been questioned (Pine et al., 1990; Whiteside et al., 1992).
Figure 1.

Yeast expression plasmids. A. Map of the IRF-1 expression plasmid YEpGIR. B. Map of the generic reporter plasmid YEpΔUAS. C. Map of the generic reporter plasmid pUMS-UAS-GH. Restriction sites in square brackets were destroyed during construction. The UAS insert is flanked by Cla I and Hind III sites in all cases except that of (GAAAGT)4, where a fortuitous point mutation obliterated the Hind III site. “RβG” is a rabbit β-globin promoter segment extending from −56 to −10 (Dierks et al., 1983). “pBR” is a pBR322 segment extending from 4172 to 28 (Watson, 1988). D. Nucleotide sequences of the regions (a) upstream and (b) downstream of the UAS cloning site in YEpΔUAS and pUMS-UAS-GH. E. Nucleotide sequence of VREβ” and (GAAAGT)4 including the linker sequences (lower case letters). The binding sites are as defined by Whiteside et al. (1992), Näf et al. (1991), Leblanc et al. (1990), Visvanathan and Goodbourn (1989), and Du and Maniatis (1992).
The IFN-αl promoter is induced differently: it lacks an NF-κB site and binds IRF-1 at least an order of magnitude less tightly than does the IFN-(3 promoter (Miyamoto et al., 1988; MacDonald et al., 1990). It contains a domain designated as TG sequence, which, when supported by an SV40 enhancer, mediates virus inducibility but shows little response to IRF-1 (Näf et al., 1991). Multimers of GAAATG, a sequence in the TG element, mediate virus inducibility when supported by an enhancer, but do not respond to IRF-1 (MacDonald et al., 1990; Näf et al., 1991).
Several mammalian transcription factors were shown to be active in yeast (Metzger et al., 1988; Hanes and Brent, 1989; Lambert et al., 1989; Samson et al., 1989; Schena et al., 1989; Struhl et al., 1989; Ding et al., 1991; Scharer and Iggo, 1992), and vice versa (Kakidani and Ptashne, 1988; Webster et al., 1988; Carey et al., 1990; Lin et al., 1990), or to cooperate in vitro with mammalian factors (Buratowski et al., 1988; Cavallini et al., 1988; Lue et al., 1989). Some mammalian factors, however, seem not to function in yeast (W. Schaffner and G. Braus, personal communication).
We now report that IRF-1 is active in yeast. An inducible IRF-1 expression plasmid and a reporter plasmid containing a potential activating sequence (UAS) upstream of a minimal promoter were introduced into S. cerevisiae, and expression of the reporter sequence was determined. Whereas (GAAAGT)4 mediated activation by IRF-1, other sequences of the type (GAAANN)4, as well as VREβ″ and VREαl (the virus-responsive element of the IFN-αl promoter), did not give a detectable response. This shows that IRF-1, as synthesized in yeast, is sufficient to activate (GAAAGT)4-mediated transcription in conjunction with the yeast transcriptional machinery, but that activation of VREβ″ requires either modification of IRF-1 and/or the cooperation of other factors present in mammalian cells but not in yeast.
Materials and methods
Strains
The strains SS328 (MATα ade2-101 his3Δ200 ura3-52 lys2-801) and SS330 (MATa ade2-101 his3Δ200 ura3-52 tyrl) (Vijayraghavan et al., 1989) were used as hosts for the Gal1/IRF-1 expression plasmid and the lacZ-reporter plasmids, respectively
Plasmids
Plasmids were constructed by standard procedures (Maniatis et al., 1982). To construct the yeast IRF expression plasmid YRpGIR, plasmid IRF1-L (Miyamoto et al., 1988) was cleaved at the Bal I site (position +86, 120 nucleotides upstream of the translation initiation site), and Sal I linkers were added. The linear plasmid DNA was cleaved with Xba I downstream of the IRF-1 coding region within the vector CDM8 (Aruffo and Seed, 1987), and the 5′ overhang was blunted with Klenow DNA polymerase. The resulting IRF-1-containing fragment was cleaved with Sal I and ligated into Sal I- and Sma I-cleaved plasmid YGRA2, resulting in plasmid YRpGIR. Plasmid YGRA2 (Omari, 1990; obtained from S. Omari and F. Thoma) contains the His3 gene, an ARS sequence, and the Gal1/10 promoter. For the construction of YEpGIR (Fig. 1A), the 2.2 kb EcoR I fragment of pLG669-Z containing the 2 μm plasmid origin (Guarente and Ptashne, 1981) was inserted into the Gal1-proximal EcoR I site of YRpGIR. For the construction of reporter plasmids, the starting plasmid was pLGABS (obtained from K. Harshman and C. Parker), a derivative of pLG669-Z (Guarente and Ptashne, 1981) in which the 0.85 kb Sal I-Xho I fragment of the CYC1 promoter was replaced by a Bgl II linker. Different UAS elements were inserted into the resulting Bgl II site. Insertion of the Ssp I-Pvu II fragment of plasmid 42P-(GAAAGT)4 gave plasmid YEpΔGT; analogous Ssp I-Pvu II fragments, containing (GAAAGC)4, (GAAATG)4, and VREαl, yielded YEpΔGC, YEpΔTG, and YEpΔVREαl, respectively. For the construction of YEpΔVREβ″ and YEpΔAA, the EcoR I-Pvu II fragments of plasmid 42P-VREβ″ and 42P-(GAAAAA)4 were ligated to a Bgl 11-Nhe I fragment of pLGΔFBS and a Nhe I-EcoR I fragment of YEpΔVREαl. Plasmids of the 42P series were constructed by introducing the desired test sequence, carrying a Cla I-compatible overhang (5′-CGAT) on the 5′ end of the top strand and a Hind III-compatible overhang (TTCGA-5′) on the 3′ end of the bottom strand, into the large Hind Ill-Cla I fragment of plasmid 42P (Kuhl et al., 1987). The orientation of the UAS sequences relative to the CYC1 transcription initiation site is identical to the orientation used in the mammalian test system (Kuhl et al., 1987; MacDonald et al., 1990). All UAS inserts were confirmed by sequencing. Partial digestion of YEpAGT with EcoR I and religation yielded YIpAGT. To construct YCpAGT, YCp50 (Rose et al., 1987) was cleaved with Nde I and, after blunting the ends, cleaved with Apa I; the resulting CEN/ARS-containing fragment was ligated to a Sma I/Apa I-cleaved YIpΔGT vector.
To construct the human growth hormone reporter plasmids, the upstream mouse sequence (UMS), which contains transcriptional stop site(s), was excised from plasmid pUMS-SB1 (Heard et al., 1987; a gift from M. Yaniv) as a 1025 bp Sal I fragment and inserted into the Sal I site of p0GH (Selden et al., 1986) to give pUMS0GH. The short EcoR I/Pvu II fragment of plasmid 12P (Kuhl et al., 1987) comprising VREαl and the rabbit β-globin minimal promoter was blunted with Klenow DNA polymerase and ligated into the filled-in BamH I site of pUMS0GH to create pUMSVREαlGH, the Hind III site within the pUC polylinker (upstream of UMS) was made resistant, and the test between the Cla I and Hind III sites flanking VREαl.
Yeast genetic techniques
Yeast media were as described by Sherman et al. (1983). Yeast was transformed according to Ito et al. (1983). To obtain different combinations of reporter and IRF-1 expression plasmids, strain SS330, harboring a reporter plasmid, and strain SS328, containing an expression plasmid, were mated, and diploid cells containing both the expression and the reporter plasmids were selected on minimal medium lacking histidine, uracil, tyrosine, and lysine. To carry out mass mating, about 30–50 colonies each of a haploid reporter and the IRF-1 expression strain were mixed on YPD plates and grown overnight. Diploid cells were selected in glucose-containing liquid minimal medium for at least 14 hours.
Yeast cell growth, induction, and β-galactosidase assay
For induction by galactose, cells were grown under selective conditions in glucose-containing medium, washed once in water, and used to inoculate minimal medium containing either glucose or galactose as sole carbon source. The inoculum was about 10–15 times heavier for the galactose-containing than for the glucose-containing culture. β-Galactosidase activity was measured (Guarente, 1983) after 16–24 hours in the case of the glucose-grown samples and after 36–48 hours for the galactose-grown samples, at a cell density of about 107/ml. The assay values were normalized to the OD600 of the culture at the time of assay.
Transient transfection of P19 cells and hGH assay
Undifferentiated P19 cells (obtained from Dr. T Taniguchi) were transfected essentially as described by Harada et al. (1990). The cells were seeded into 6-well plates (Falcon, No. 3046) and transfected with 2.5 μg reporter DNA and either 2.5 μg IRF-1 expression plasmid pAct-1 (Harada et al., 1990) for IRF-induced, or 2.5 μg pBlue-script for mock-induced samples. Samples for hGH assay were harvested 48 hours after transfection and assayed for human growth hormone (hGH) with the Allégro radioimmune assay kit as described by the manufacturer (Nichols Institute Diagnostics, San Juan Capistrano, CA).
Results
The transactivation system
In a preliminary experiment IRF-1 was expressed in S. cerevisiae from IRF-1 cDNA (Miyamoto et al., 1988) under the control of the inducible PH05 promoter (Miyanohara et al., 1983). Trans-activation was monitored using a reporter plasmid with a truncated CYC1 promoter directing expression of the lacZ gene (Guarente and Ptashne, 1981). Insertion of (GAAAGT)4 but not of (GAAATG)4 upstream of the truncated promoter gave 6- to 30-fold more β-galactosidase activity in phosphate-free than in high phosphate medium, showing that IRF-1 could specifically transactivate a promoter containing an IRF-1-responsive sequence (data not shown).
Because it is technically simpler to induce by galactose addition than by phosphate deprivation, we switched to the Gal1 promoter (Johnston and Davis, 1984) to express IRF-1 in yeast cells. We placed different replicons into the IRF-1 expression and reporter plasmids and tested the effect on transactivation.
The haploid yeast strain SS328 was transfected with an IRF-1 expression plasmid (Fig. 1A) containing the autonomous replication sequence ARS (YRpGIR) either alone or in conjunction with the 2 μm plasmid origin (YEpGIR). An ARS-containing vector devoid of the IRF-1 coding sequence (YGRA2) served as a negative control. A (GAAAGT)4 reporter plasmid (Fig. 1B) containing either the CEN/ARS sequence (YCPΔGT), the 2 μm plasmid replicon (YEpΔGT), or no replicon (YIpAGT) was introduced into the haploid yeast strain SS330. Diploid strains containing different combinations of expression and reporter plasmids were obtained by mating the appropriate parents and selecting diploid progeny on a suitable medium. Yeast strains were grown in galactose- and glucose-containing media to induce or repress formation of IRF-1, respectively. Figure 2 shows that the presence or absence of a replicon on the reporter plasmid had no significant effect on the inducibility of β-galactosidase activity by IRF-1. However, the nature of the replicon in the IRF-1 expression plasmid had a dramatic influence on β-galactosidase expression; the presence of the 2 μm plasmid origin resulted in a strong activation of the reporter, while there was little activity when an ARS sequence was used as replication origin.
Figure 2.

Effect of different origins of replication on the activation of a (GAAAGT)4-containing reporter by an IRF-1 expression vector. The reporter plasmid contained (GAAAGT)4 as UAS element and either the CEN/ARS sequences (YCpΔGT), the 2 μm plasmid origin (YEpAGT), or no replicating sequences (YIpΔGT). The IRF-1 expression plasmid contained the ARS sequences either alone (YRpGIR) or in combination with the 2 μm plasmid replicon (YEpGIR). Cells were grown for 24 hours in glucose medium (GLU) or 48 hours in galactose medium (GAL). (β-Galactosidase (β-GAL) activity was measured in duplicate for three individual transformants and averaged.
It is believed that in the presence of the 2 nm plasmid origin, a stable high plasmid copy number is maintained, while in its absence plasmid maintenance is unstable, resulting in a variable copy number (Rose and Broach, 1991). Quite likely the expression of IRF-1 is the limiting factor for the induction of the reporter plasmid. In all further experiments both the IRF-1 expression plasmid and the reporter plasmid were equipped with the 2 μm plasmid origin.
The specificity of IRF-1-driven expression in yeast
It was shown earlier that multimerization of certain hexamers of the type GAAANN gives rise to three types of elements that are virus-inducible in mammalian cells (MacDonald et al., 1990). Type I oligonucleotides, such as (GAAAGT)4 or (GAAAGC)4, respond to IRF-1. The type II oligonucleotide (GAAATG)4 requires an enhancer for virus inducibility and does not respond to IRF-1. The type III oligonucleotide (GAAACG)4 does not respond to IRF-1 and in contrast to the other two types shows considerable constitutive activity, which was ascribed by MacDonald et al. (1990) to its response to the ubiquitous GA factor characterized by LaMarco and McKnight (1989). (GAAAAA)4 was inert.
Using the assay system described above, we tested the response to IRF-1 expression of different promoter elements. As shown in Figure 3, transactivation by IRF-1 was observed solely with (GAAAGT)4, while (GAAACG)4, (GAAATG)4, (GAAAAA)4, and the virus-responsive elements of the IFN-αl and IFN-β promoters (VREα and VREβ″, respectively) were inactive. Figure 4 shows that in the presence of galactose, but not of glucose, IRF-1 mRNA is expressed to an equal extent both in yeast containing the (GAAAGT)4 and the VREP″ reporter constructs.
Figure 3.
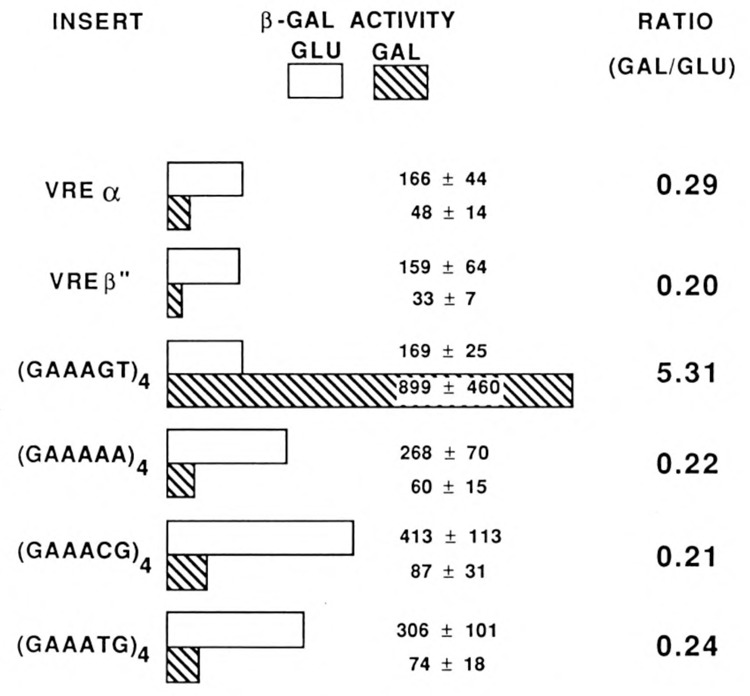
Response of different promoter elements to IRF-1 expression. Diploid cells contained the IRF-1 expression plasmid YEpGIR and the YEpUAS reporter (Fig. 1B) with the UAS indicated. Cells were grown for 24 hours in glucose medium (GLU) or for 40 hours in galactose medium (GAL). The values for β-galactosidase (β-GAL) activity are the averages of seven to nine independent experiments, resulting from mass mating of the appropriate strains.
Figure 4.

IRF-1 transcript level in uninduced and galactose-induced diploid yeast cells containing an IRF-1 expression plasmid. A–C: Diploid cells contained the IRF-1 expression plasmid YEpGIR and the YEpUAS reporter (Fig. 1B) with (A) VREαl, (B) VREβ”, (C) (GAAAGT)4. In the control sample (D), the diploid cells contained Z669 and YGRA2. Cells were grown for 24–38 hours in galactose (GAL) or glucose (GLU) medium, and the RNA extracted according to Vijayraghavan et al. (1989). Samples (5 μg) were run through a 1 % agarose gel. Endogenous ribosomal RNA was used as molecular weight marker. The samples were transferred to Gene Screen nylon membranes (DuPont) and hybridized as described (Büeler et al., 1992). The probe was the 1141 bp Nco I fragment of IRF-1 cDNA (Miyamoto et al., 1988) labeled by random priming (Feinberg and Vogelstein, 1983) and used at 3.5 × 105 cpm/ml. Autoradiography was for 1 hour.
The relative efficiencies of (GAAAGT)4 and VREβ″ as promoter elements in P19 cells
In L cells both (GAAAGT)4 and VREβ″ mediate activation by virus, but only (GAAAGT)4 is significantly activated by overexpression of IRF-1 (MacDonald et al., 1990). Harada et al. (1990) showed that in P19 cells VREβ″ is activated by overexpression of IRF-1; however, a comparison with (GAAAGT)4 was not reported. In view of the transactivation experiments in yeast, we determined the relative responses of (GAAAGT)4 and VREβ″ to IRF-1 overexpression. We prepared a reporter plasmid in which the promoter elements used in the yeast expression plasmids were linked to the human growth hormone coding sequence. P19 cells were cotransfected with these reporter plasmids and an IRF-1 expression plasmid. Table 1 shows that the inducibility of the VREβ″ promoter by IRF-1 was 11 times higher than that of the (GAAAGT)4 element, while the absolute induced expression level with (GAAAGT)4 was 3.7 times higher than with VREβ″. In yeast, inducibility of the VREβ″ promoter by IRF-1 was at least 26 times lower than that of the (GAAAGT)4 element, while the expression level with (GAAAGT)4 after induction was 15 times higher than with VREβ″. Thus, in yeast the response of VREP″ to IRF-1 relative to that of (GAAAGT)4, if any, is far lower than in P19 cells.
Table 1.
Response of different promoter elements to IRF-1 in P19 cells.
| P19 cells were co-transfected with the growth hormone reporter plasmid pUMS-UAS-GH containing the UAS sequence indicated and either the constitutive IRF-1 expression plasmid pAct-1 (Harada et al., 1990) (“IRF-1”) or pBluescript (“MOCK”). After 48 hours growth hormone was determined in the supernatant. Values were determined in duplicate from 3 independent experiments except for the values marked with *, which were from a single experiment. | |||
| Promoter insert (UAS) | hGH conc. (ng/ml) | Inducibility | |
|---|---|---|---|
| MOCK | IRF-1 | ||
| (GAAAGT)4 | 36 ± 4 | 2500 ± 500 | 70 |
| VREβ″ | 0.9 ± 0.4 | 683 ± 210 | 760 |
| (GAAATG)4 | 0.6* | 0.8* | – |
Discussion
(GAAAGT)4 – but none of the other oligonucleotides tested—mediated the response of a reporter plasmid to the mammalian transcription factor IRF-1 in S. cerevisiae. (GAAAGT)4 contains at least two copies of GAAAGTGAAAGT, which functionally mimics the IFN-β promoter element PRDI (AGAAGTGAAAGT; Näf et al., 1991). IRF-1 binds in vitro to PRDI and to the related sequence PRDIII (Miyamoto et al., 1988) as well as to (GAAAGT)4 (MacDonald et al., 1990).
We expected that VREβ″ would also mediate activation by IRF-1, because it contains two IRF-1 binding sites and because it efficiently mediates expression in response to IRF-1 in P19 cells, albeit not in L cells (Harada et al., 1990). Harada et al. (1990) explain this difference by their finding that L cells – but not P19 cells – contain substantial levels of IRF-2, which binds to PRDI and prevents activation by IRF-1. It is still not clear, however, why (GAAAGT)4 responds to IRF-1 in L cells (MacDonald et al., 1990) despite its binding capacity for IRF-2, which abounds in these cells.
Although yeast most likely contains no IRF-2, VREβ″ was not activated by IRF-1 in these cells. This may simply reflect the fact that IRF-1 has a higher binding affinity for (GAAAGT)4 than for VRE β″, and that the level of IRF-1 in yeast is too low to mediate activation by VREβ″. However, other explanations can be considered. (1) IRF-1 by itself may not be able to activate VREβ″ but may need to cooperate with an additional DNA-binding factor, such as NF-κB (Leblanc et al., 1990). P19 cells, in which both VREβ″ (Harada et al., 1990) and (GAAAGT)4 are active (Table 1), contain constitutive NF-κB-like activity (Harada et al., 1989), but yeast likely does not. (2) To function with VREβ″ –but not with (GAAAGT)4-IRF-1 must be modified either covalently or by association with other component(s) present in mammalian cells (Whiteside et al., 1992) but not in yeast. One report suggests that a posttranslational modification, likely phosphorylation of IRF-1, may play an important role in viral induction of the IFN-β gene (Watanabe et al., 1991). (3) Yeast contains a protein which fortuitously binds to the IFN-β promoter–but not to (GAAAGT)4 – and prevents activation by IRF-1.
Several exogenous transcription factors, such as mammalian nuclear receptors (Metzger et al., 1988; Schena et al., 1989), the mammalian p53 protein (Schärer and Iggo, 1992), the transcription factor Pit-1 (Ding et al., 1991), the bicoid (Hanes and Brent, 1989; Struhl et al., 1989), ubx, and abd (Samson et al., 1989) proteins from Drosophila, and the papilloma virus E2 trans-activator (Lambert et al., 1989) have been shown to function in yeast, presumably by cooperating with an endogenous target protein, perhaps yeast RNA polymerase II. However, it would seem that not all mammalian transcription factors are active in yeast (W. Schaffner and G. Braus, personal communication). It is interesting that IRF-1, which is involved in activation of genes pertaining to the lymphokine system, can also interact with the yeast transcriptional machinery. This most likely means that IRF-1 has an activating domain functionally similar to a domain represented on a yeast transcription factor(s) whose DNA binding domain does not recognize (GAAAGT)4.
Finally, we would mention that the yeast trans-activation system we have described could be used for expression cloning of mammalian transactivating factors. In a reconstruction experiment, a library of L929 cDNA under the control of the Gal1 promoter in a plasmid homologous to YEpGIR was spiked with YEpGIR and introduced into S. cerevisiae containing the (GAAAGT)4 reporter plasmid YEpΔGT. After replica plating on X-Gal/glucose and X-Gal/galactose plates, colonies exhibiting strong blue color on the galactose but not the glucose plates were found to contain YEpGIR (K. Nagata, unpublished results).
Acknowledgments
We thank Dr. T. Taniguchi for unpublished information, Dr. T. Nomura for plasmid pUMSVREβ″GH, Drs. S. Omari and F. Thoma for plasmid YGRA2, Mr. J. Ecsoedi for synthetic oligonucleotides, and Mr. R. Beerli and Mr. K. Lüthi for technical assistance.
This work was supported by the Schweizerische Nationalfonds and the Kanton of Zürich.
The costs of publishing this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC Section 1734 solely to indicate this fact.
Kiyoshi Nagata is currently at Shionogi & Co., Ltd., Research Laboratories, 12-4, Sagisu 5-chome, Fukushima-ku, Osaka 553, Japan.
References
- Aruffo A. and Seed B. (1987), Proc Natl Acad Sci USA 84, 8573–8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S., Hahn S., Sharp P. A., and Guarente L. (1988), Nature 334, 37–42. [DOI] [PubMed] [Google Scholar]
- Büeler H., Fischer M., Lang Y., Bluethmann H., Lipp H.-P., DeArmond J., Prusiner S. B., Aguet M., and Weissmann C. (1992), Nature 356, 577–582. [DOI] [PubMed] [Google Scholar]
- Carey M., Lin Y. S., Green M. R., and Ptashne M. (1990), Nature 345, 361–364. [DOI] [PubMed] [Google Scholar]
- Cavallini B., Huet J., Plassat J. L., Sentenac A., Egly J. M., and Chambon P. (1988), Nature 334, 77–80. [DOI] [PubMed] [Google Scholar]
- Clark L. and Hay R. T. (1989), Nucleic Acids Res 17, 499–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierks P., van Ooyen A., Cochran M. D., Dobkin C., Reiser J., and Weissmann C. (1983), Cell 32, 695–706. [DOI] [PubMed] [Google Scholar]
- Ding Y., Roberson M. S., Moye-Rowley W. S., and Maurer R. A. (1991), Mol Endocrinol 5, 1239–1245. [DOI] [PubMed] [Google Scholar]
- Dinter H. and Hauser H. (1987), Eur J Biochem 166, 103–109. [DOI] [PubMed] [Google Scholar]
- Du W. and Maniatis T. (1992), Proc Natl Acad Sci USA 89, 2150–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C. M. and Maniatis T. (1989), EMBO J 8, 101–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P. and Vogelstein B. (1983), Anal Biochem 132, 6–13. [DOI] [PubMed] [Google Scholar]
- Fujita T., Ohno S., Yasumitsu H., and Taniguchi T. (1985), Cell 41, 489–496. [DOI] [PubMed] [Google Scholar]
- Fujita T., Shibuya H., Hotta H., Yamanishi K., and Taniguchi T. (1987), Cell 49, 357–367. [DOI] [PubMed] [Google Scholar]
- Fujita T., Sakakibara J., Sudo Y., Miyamoto M., Kimura Y., and Taniguchi T. (1988), EMBO J 7, 3397–3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita T., Miyamoto M., Kimura Y., Hammer J., and Taniguchi T. (1989), Nucleic Acids Res 17, 3335–3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodbourn S., Zinn K., and Maniatis T. (1985), Cell 41, 509–520. [DOI] [PubMed] [Google Scholar]
- Goodbourn S. and Maniatis T. (1988), Proc Natl Acad Sci USA 85, 1447–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. and Ptashne M. (1981), Proc Natl Acad Sci USA 78, 2199–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. (1983), Methods Enzymol 101, 181–191. [DOI] [PubMed] [Google Scholar]
- Hanes S. D. and Brent R. (1989), Cell 57, 1275–1283. [DOI] [PubMed] [Google Scholar]
- Harada H., Fujita T., Miyamoto M., Kimura Y., Maruyama M., Furia A., Miyata T., and Taniguchi T. (1989), Cell 58, 729–739. [DOI] [PubMed] [Google Scholar]
- Harada H., Willison K., Sakakibara J., Miyamoto M., Fujita T., and Taniguchi T. (1990), Cell 63, 303–312. [DOI] [PubMed] [Google Scholar]
- Heard J.-M., Herbomel P., Ott M.-O., Mottura-Rollier A., Weiss M., and Yaniv M. (1987), Mol Cell Biol 7, 2425–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscott J., Alper D., Cohen L., Leblanc J. F., Sportza L., Wong A., and Xanthoudakis S. (1989), J Virol 63, 2557–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., and Kimura A. (1983), J Bacteriol 153, 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M. and Davis R. W. (1984), Mol Cell Biol 4, 1440–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakidani H. and Ptashne M. (1988), Cell 52,161–167. [DOI] [PubMed] [Google Scholar]
- Keller A. D. and Maniatis T. (1988), Proc Natl Acad Sci USA 85, 3309–3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A. D. and Maniatis T. (1991), Genes Dev 5, 868–879. [DOI] [PubMed] [Google Scholar]
- Keller A. D. and Maniatis T. (1992), Mol Cell Biol 12, 1940–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhl D., de la Fuente J., Chaturvedi M., Parimoo S., Ryals J., Meyer F., and Weissmann C. (1987), Cell 50, 1057–1069. [DOI] [PubMed] [Google Scholar]
- LaMarco K. L. and McKnight S. L. (1989), Genes Dev 3, 1372–1383. [DOI] [PubMed] [Google Scholar]
- Lambert P. F., Dostatni N., McBride A. A., Yaniv M., Howley P. M., and Arcangioli B. (1989), Genes Dev 3, 38–48. [DOI] [PubMed] [Google Scholar]
- Leblanc J. F., Cohen L., Rodrigues M., and Hiscott J. (1990), Mol Cell Biol 10, 3987–3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenardo M. J., Fan C. M., Maniatis T., and Baltimore D. (1989), Cell 57, 287–294. [DOI] [PubMed] [Google Scholar]
- Lin Y S., Carey M., Ptashne M., and Green M. R. (1990), Nature 345, 359–361. [DOI] [PubMed] [Google Scholar]
- Lue N. F., Flanagan P. M., Sugimoto K., and Kornberg R. D. (1989), Science 246, 661–664. [DOI] [PubMed] [Google Scholar]
- MacDonald N. J., Kuhl D., Maguire D., Näf D., Gallant P., Goswamy A., Hug H., Büeler H., Chaturvedi M., de la Fuente J., Ruffner H., Meyer F., and Weissmann C. (1990), Cell 60, 767–779. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Fritsch E. F., and Sambrook J. (1982), Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Metzger D., White J. H., and Chambon P. (1988), Nature 334, 31–36. [DOI] [PubMed] [Google Scholar]
- Miyamoto M., Fujita T., Kimura Y, Maruyama M., Harada H., Sudo Y, Miyata T, and Taniguchi T. (1988), Cell 54, 903–913. [DOI] [PubMed] [Google Scholar]
- Miyanohara A., Toh A., Nozaki C., Hamada F., Ohtomo N., and Matsubara K. (1983), Proc Natl Acad Sci USA 80, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Näf D., Hardin S. E., and Weissmann C. (1991), Proc Natl Acad Sci USA 88, 1369–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omari S. (1990), Dissertation No. 9131: Chromatin Structure during Transcription in the Yeast S. cerevisiae, Federal Institute of Technology, Zürich, Switzerland. [Google Scholar]
- Pine R., Decker T., Kessler D. S., Levy D E., and Darnell J. E. (1990), Mol Cell Biol 10, 2448–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis L. F. L., Harada H., Wolchok J. D., Taniguchi T., and Vilcek J. (1992), EMBO J 11, 185–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M. D. and Broach J. R. (1991), Methods Enzymol 194, 195–229. [DOI] [PubMed] [Google Scholar]
- Rose M. D., Novick P., Thomas J. H., Botstein D., and Fink G. R. (1987), Gene 60, 237–243. [DOI] [PubMed] [Google Scholar]
- Samson M. L., Jackson-Grusby L., and Brent R. (1989), Cell 57, 1045–1052. [DOI] [PubMed] [Google Scholar]
- Schena M., Freedman L. P., and Yamamoto K. R. (1989), Genes Dev 3, 1590–1601. [DOI] [PubMed] [Google Scholar]
- Schärer E. and Iggo R. (1992), Nucleic Acids Res 20, 1539–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selden R. F., Burke-Howie K., Rowe M. E., Goodman H. M., and Moore D. D. (1986), Mol Cell Biol 6, 3173–3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F., Fink G. R., and Hicks J. B. (1986), Laboratory Course Manual for Methods in Yeast Genetics, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Struhl G., Struhl K., and Macdonald P. M. (1989), Cell 57, 1259–1273. [DOI] [PubMed] [Google Scholar]
- Vijayraghavan U., Company M., and Abelson J. (1989), Genes Dev 3, 1206–1216. [DOI] [PubMed] [Google Scholar]
- Visvanathan K. V. and Goodbourn S. (1989), EMBO J 8, 1129–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe N., Sakakibara J., Hovanessian A. G., Taniguchi T., and Fujita T. (1991), Nucleic Acids Res 19, 4421–4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson N. (1988), Gene 70, 399–403. [DOI] [PubMed] [Google Scholar]
- Webster N., Jin J. R., Green S., Hollis M., and Chambon P. (1988), Cell 52, 169–178. [DOI] [PubMed] [Google Scholar]
- Whittemore L. A. and Maniatis T. (1990), Proc Natl Acad Sci USA 87, 7799–7803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteside S. T., Visvanathan K. V., and Goodbourn S. (1992), Nucleic Acids Res 20, 1531–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood T. G., McGeady M. L., Baroudy B. M., Blair D. G., and Van de Woude G. F. (1984), Proc Natl Acad Sci USA 81, 7817–7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xanthoudakis S., Cohen L., and Hiscott J. (1989), J Biol Chem 264, 1139–1145. [PubMed] [Google Scholar]


