Abstract
All genes encoding proteins in eukaryotes are transcribed by RNA polymerase II. The first step in analyzing transcriptional regulation requires understanding the general mechanisms of RNA polymerase II-specific gene transcription. The basal promoter, a template containing a TATA box devoid of upstream regulatory sequences, has been used to identify and characterize the factors which, together with RNA polymerase II, govern transcription in mammalian systems: TFIIA, TFIIB, TFIID, TFIIE, TFIIF, TFIIG, TFIIH, and TFIIJ. Interactions between regulatory transcription factors and basal elements of the transcriptional machinery affect the transcriptional rate in a positive or negative fashion. As these multiple proteins are purified, and their coding sequences are isolated, we come closer to reproducing these processes in vitro with pure components, and thus to elucidating the complex interactions among them.
Present understanding of eukaryotic transcriptional mechanisms derives in great part from mammalian systems. In documenting current knowledge in the field, this review will emphasize mammalian transcription factors. Yeast and Drosophila will be mentioned briefly if complementary information is available, and other organisms will be excluded entirely. Likewise, several controversial issues are beyond the scope of this review. (For recent comprehensive reviews, see Saltzman and Weinmann, 1989; Mermelstein et al., 1989; Sawadogo and Sentenac, 1990; and Stone et al., 1991.)
Figure 1 summarizes our current understanding of transcription in mammalian cell systems. It represents schematically the basic reactions occurring during transcription initiation by RNA polymerase II, together with the function of each transcription factor. While detailed knowledge of some of these processes has lagged because of difficulties in obtaining purified factors, considerable progress has been made recently through the cloning of specific factors and an abundance of cloned gene products in bacteria. The availability of these proteins, free of other contaminant polypeptides, allows us to assign specific functions unequivocally. At the same time, novel requirements are revealed each time new, recombinant, or purified transcription factors—the latter free of other mammalian proteins—are used in reconstituted in vitro transcription systems (see for example Dynlacht et al., 1991; Meisterernst et al., 1991). Some of the genes encoding transcription factors described below are now cloned, and active, bacterially expressed versions of their proteins can be easily obtained in the lab in large amounts, while others must still be purified from cell extracts.
Figure 1.
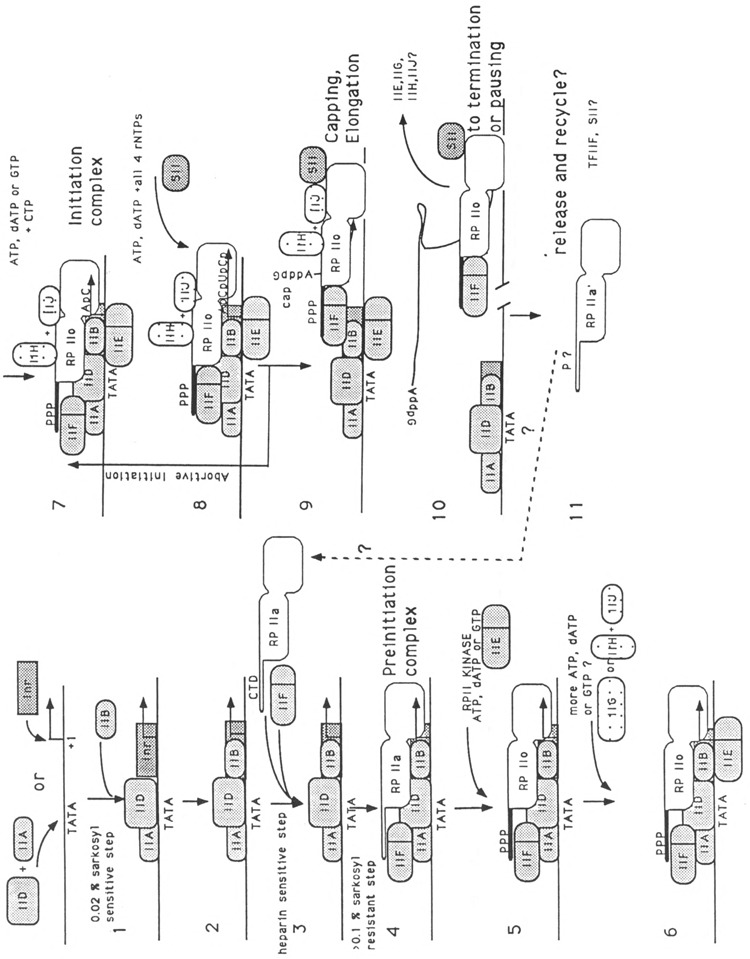
Schematic representation of transcription initiation on the adenovirus 2 major late promoter (Ad2 MLP). The diagram summarizes data available as of October 10, 1991, and contains many modifications which are not fully described or attributed in the text, since not all of the data have been published. The location and role of some required factors, such as BTF-3 (Zheng et al., 1990), TFIIG (Sumimoto et al., 1990), or TFIIH (Flores et al., 1991), are less well characterized. The inverted G in step 9 indicates the 5′-5′ CAP dinucleotide formation, catalyzed by a guanosyltransferase that may also be associated with the transcription complex (Reinberg et al., 1987). Interactions with elongation factor SII or TFIIS (Rappaport et al., 1987, 1988) and the abortive initiation process (Luse and Jacob, 1987; Rougvie and Lis, 1988) or pausing (Resnekov and Aloni, 1989) are not discussed in this review. They do, however, point to areas of further study, and to the fate of transcription factors and RNA polymerase in recycling.
The earlier fractionation of transcriptionally competent HeLa cell extracts after phosphocellulose chromatography (Matsui et al., 1980) resulted in fractions eluted at 0.1 M KC1, 0.3 M KC1,0.5 M KCl, and 1.0 M KC1. In transcription reconstitution experiments, only fractions eluting at 0.1, 0.5, and 1 M KC1 were required (Matsui et al., 1980). The fraction eluting at 1 M KCl contained two activities, a poly-ADP-ribopolymerase binding at DNA nicks (Slattery et al., 1983) and TFIID, while the fraction eluting at 0.5 M KC1 contained TFIIB and TFIIE (Reinberg and Roeder, 1987).
The process of transcription initiation begins by the binding of protein TFIID to the template at the TATA box (step 1). The TATA box is an important promoter sequence element present in a large number of mammalian genes; it is located approximately 30 nucleotides upstream of the initiation start site. Binding of the IID protein is the earliest event in the process of transcription initiation and determines template commitment (Davison et al., 1983; Fire et al., 1984; Reinberg et al., 1987). When binding to the TATA box by a partially purified TFIID fraction precedes the assembly of chromatin on those same DNA fragments, the corresponding promoter is active in an in vitro transcription system (Workman and Roeder, 1987); in contrast, when chromatin assembly precedes TFIID binding, the promoter is inactive. Thus, the presence of the TATA box binding protein (TBP) is an essential determinant for fulfilling the potential for gene modulation by other transcription factors. Housekeeping genes and some viral genes (adenovirus E2e and IVa2) have AT-rich sequences which are distinct from typical TATA boxes. However, the protein fraction containing factor TFIID is still required for transcription of the adenovirus IVa2 gene and artificial SP1 TATA-less constructs in vitro (Carcamo et al., 1989; Pugh and Tjian, 1991). In the case of IVa2, the functional TATA box appears to be located downstream of the transcription start site, and the direction of transcription may be determined by the initiator (Smale and Baltimore, 1989; Carcamo et al., 1990, 1991). The initiator element, whose presence was suggested by the effect of point mutants around the start site (Lee et al., 1988), was demonstrated by the experiments of Smale and Baltimore (1989) on the terminal deoxynucleotidyltransferase promoter, and confirmed by the experiments of Carcamo et al. (1989, 1990, 1991). The initiator element will be discussed more fully below.
The TATA box binding protein (TBP)
TFIID, purified from the fraction eluting with 1 M salt from phosphocellulose, participates in the initial steps of template commitment. This protein’s instability has been a major roadblock for further purification of other factors. The differential thermal sensitivity of TFIID (Nakajima et al., 1988) and the fact that a yeast protein fraction can complement the human TFIID-depleted extracts (Buratowski et al., 1988) have allowed several groups to purify the yeast TFIID polypeptide and clone the yeast gene encoding for it. The cloned gene predicts a protein of 27 kDa for yeast (Cavallini et al., 1989; Hahn et al., 1989; Horikoshi et al., 1989; Schmidt et al., 1989; Eisenmann et al., 1989; Fikes et al., 1990). Using the sequence of the yeast TFIID gene, the human TBP gene was cloned (Kao et al., 1990; Peterson et al., 1990; Hoffman et al., 1990), and the open reading frame predicts a protein of 37 kDa. TBP contain a very highly conserved carboxy-terminal 160-amino acid domain with tandem repeats and homology to bacterial o factor (Horikoshi et al., 1989; Hoffman et al., 1990). Limited homology to bacterial o has also been found in RAP 30 (the small subunit of TFIIF), the fourth subunit of yeast RNA polymerase II, and mitochondrial RNA polymerase (reviewed by Jaehning, 1991). A single gene appears to encode for the human, Drosophila, and yeast TBP, but two TBP genes are present in Arabidopsis (Gash et al., 1990; Hoey et al., 1990; Hoffman et al., 1990). The human TBP contains an amino-terminal domain with unique features, such as a run of 38 glutamines in a row. Although the yeast and human proteins appear to function interchangeably in a mammalian in vitro transcription system, the human TFIID is unable to substitute for the yeast TFIID in vivo. A region in the carboxy-terminal domain (CTD) of the yeast protein seems to be essential for this species-specific restriction (Gill and Tjian, 1991; McCormack et al., 1991). Binding specificity to modified TATA boxes (TGTAAA) correlates to amino acid changes in the second tandem repeat of the conserved domain (Strubin and Struhl, 1992). The purified yeast TBP induces DNA bending when bound to DNA (Horikoshi et al., 1992). Activation by upstream regulatory factors may require additional proteins (Peterson et al., 1990; Pugh and Tjian, 1990; Kelleher et al., 1990; Flanagan et al., 1991; Lewin, 1990). Most importantly, the yeast and mammalian TBP overproduced in bacteria are active in the in vitro transcriptional systems, insuring a steady source of this important reagent.
TBP-associated factors
Although in in vitro transcription experiments recombinant TBP is able to support transcription from basal promoter in a highly purified system, no response of upstream regulatory factors, such as SP1, could be detected, unless the less pure mammalian cell TFIID fraction was used (Peterson et al., 1990; Pugh and Tjian, 1990). The nomenclature we follow, in accord with these authors, is to call the multicomponent complex purified from cells TFIID, and to call the recombinant 37-kDa protein TBP, for TATA box binding protein. Purification or immunoprecipitation of TBP from cell extracts has revealed the presence of tightly associated proteins important for transactivation. They form what has been called the TFIID fraction or complex (Inostroza et al., 1991; Dynlacht et al., 1991). Some or all of the six cellular proteins coprecipitating with TBP-specific antibodies are required for activation by upstream factors (Dynlacht et al., 1991). Other proteins, like TFIID repressor (Dr), co-purifying with the TBP, might be negative regulators, since they displace the TBP-IIA DNA complex to form TBP-Dr complexes with distinct gel mobilities (J. Inostroza, F, Mermelstein, I. Ha, W. S. Lane, and D Reinberg, unpublished data). A fraction containing upstream stimulatory activity (USA) responsible for stimulation by upstream SP1 and USF contains both positive and negative regulatory elements (Meisterernst et al., 1991) and may contain some of the same proteins found in the TFIID complex.
Transactivation
The exact mechanism by which TFIID-mediated transactivation occurs remains unclear. Promoter activity may be stimulated by direct interaction of acidic transactivator proteins with TFIID. The viral transactivator VP16 is able to interact with yeast TFIID (Stringer et al., 1990). Point mutations in VP16 that affect its transactivation potential also affect its interaction with TFIID (Ingles et al., 1991). Others have suggested that VP16 interaction with TFIID is weak, while interaction with TFIIB is strong (Lin and Green, 1991). Experiments in a yeast-reconstituted transcription system suggest that “mediator” proteins are required to detect gal4-VP16 effects (Kelleher et al., 1990). We and others have recently shown that human and yeast TFIIDs are able to interact directly with adenovirus E1 A, a potent pleiotropic transactivator (Horikoshi et al., 1991; Lee et al., 1991). These interactions occur between the first repeat and the basic domain of TBP and the Zn++ finger of E1A (Lee et al., 1991; N. Horikoshi, U. Graeven, and R. Weinmann, unpublished data). In contrast to VP16 (Lin and Green, 1991), the interactions between E1A and TBP are strong, while those between E1A and TFIIB are weak (I. Ha, N. Horikoshi, D. Reinberg, and R. Weinmann, unpublished data). Recent studies have revealed negative regulators involved in basal and activated transcription. For example, in the case of the retinoblastoma protein whose association with transcription factor E2F represses its DNA-binding activity, dissociation by ElA of E2F/Rb complexes results in DNA binding and possibly transcriptional activation (Bagchi et al, 1991; Hiebert et al., 1992). A 19-kDa repressor protein (Dr) bound to TBP has recently been purified and its encoding sequences isolated (Inostroza and Reinberg, 1991). Properties of the negative component of USA (NCI; Meisterernst et al., 1991) appear similar to the negative regulator Dr. By analogy, dissociation of this Dr transcriptional inhibitor from TFIID by E1 A may correlate with E1A-mediated activation via TATA boxes (see discussion in Horikoshi et al., 1991).
Transcription initiation factor (Inr or TFII I)
The TATA motif is not the only promoter element required for the assembly of transcription competent complexes. Natural promoters without apparent TATA motifs or mutations which alter the sequence reduce but do not completely obliterate transcription. The sequences around the start site and their importance as promoter elements were first pointed out for the terminal deoxynucleotidyl-transferase initiator (Inr; Smale and Baltimore, 1989). Similar motifs with a common  sequence (where the A is the transcriptional start) have been described for the adenovirus major late and IVa2 promoters (Carcamo et al., 1991).
sequence (where the A is the transcriptional start) have been described for the adenovirus major late and IVa2 promoters (Carcamo et al., 1991).
The initiator protein bound to DNA can substitute for the TATA box as nucleation center for the transcription initiation complex (Carcamo et al., 1991). The DNA-bound initiator element is directly and weakly recognized by RNA polymerase II. This process is greatly enhanced by transcription factors IID, IIB, and IIF, which selectively stimulate correct start site utilization (Carcamo et al., 1991). It appears that strong initiators are required with weak TATA boxes, while strong TATA boxes tolerate weak initiators. Thus, cooperative interactions between the two elements appear important (Lee et al., 1988; Carcamo et al., 1991). Moreover, the initiator motif alone is able to nucleate all the other transcription factors around itself in a complex containing IID, IIB, RNA polymerase II, and IIF in the absence of a TATA box (Carcamo et el., 1991). This explains the TBP requirement of TATA-less promoters, since TBP can be recruited to the promoter by the other proteins of the initiation complex (Pugh and Tjian, 1991). The gene encoding the sequence for the transcription factor YY-1, a Krüppel-like 55-kDa polypeptide with four Zn++ fingers, was recently cloned by several laboratories (Shi et al., 1991; Hariharan et al., 1991; Park and Atchison, 1991; Flanagan et al., 1991). This protein is able to function in an in vitro transcription system as an initiator, either alone or in the presence of transcription factor SP1 (Seto et al., 1991). A novel 120-kDa helix-loop-helix initiator protein, which binds at the initiation site of the adenovirus major late promoter and is able to interact functionally with the upstream factor USF, has been described by Roy et al. (1991) as TFII I.
Transcription factor IIA (TFIIA)
The visualization of the sequential steps in transcription complex assembly was greatly aided by the development of a gel mobility shift assay in which sequential binding of the transcription factors could be readily followed (Buratowski et al., 1989). The presence of TFIIA is required for formation of gel-shifted complexes between TFIID and promoter DNA, but this requirement can be overcome in the presence of excess Mg++ (Buratowski et al., 1989; Horikoshi et al., 1989). Some report transcription factor IIA as a 43-kDa polypeptide (Egly et al., 1984), or as a 13- and 17-kDa protein complex (Samuels and Sharp, 1986); others have purified a TFIIA 35-kDa active protein from wheat (Burke et al., 1990). The protein purified from calf thymus has full activity in the in vitro system (Samuels and Sharp, 1986). Using eight chromatographic steps to purify this TFIIA factor, it appears that it contains two activities (TFIIA and TFIIJ; Cortes et al., 1992). The TFIIA activity was further purified by bacterially produced TDB-affinity chromatography and contains polypeptides of 35, 19, and 14 kDa, corresponding to the previously described TFIIA activity (Samuels and Sharp, 1986; Cortes et al., 1992). It stimulates transcription but is not required when TBP from recombinant sources is used in reconstitution experiments. In contrast, TFIIJ is required and apparently acts at a later stage in the initiation process (step 6; Cortes et al., 1992). A more complete understanding of the role of the different forms of TFIIA and TFIIJ will have to await characterization and cloning of the encoding genes, and further study of the proteins. The requirement of TFIIA for the formation of the TBP promoter complex appears controversial, and depends on the source of TBP and binding conditions.
Transcription factor IIB (TFIIB)
After the TFIID-TFIIA complex is formed on promoter DNA, the factor TFIIB enters into the transcription complex (step 3). TFIIA stimulates the formation of the DAB complex significantly if D and B alone are provided but does not affect later stages (Maldonado et al., 1990). The TFIIB 33-kDa protein appears distinct from the recently reported BTF-3 (Zheng et al., 1990). The gene encoding TFIIB has been cloned by Ha et al. (1991) and Malik et al. (1991). It contains two 76 amino acid repeats and exhibits some homology with bacterial σ factors. Evidence for interaction of VP16-derived acidic activators with TFIIB has been recently presented (Lin and Green, 1991; Lin et al., 1991). This interaction appears to be stronger than the VP16-TBP interaction, in contrast to what occurs with ElA (see above).
Transcription factor IIF (TFIIF)
The use of more purified factors in transcription reconstitution has permitted the identification of other activities in the phosphocellulose fraction eluting at 0.5 M KCl: TFIIF and TFIIE (Reinberg and Roeder, 1987). The two-subunit transcription factor TFIIF is probably the same as RAP 74-30 kDa, a bimolecular complex binding to RNA polymerase II (Sopta et al., 1985; Flores et al., 1988; Burton et al., 1988), and also possibly similar to βγ (Conaway et al., 1989) and FC (Kitajima et al., 1990). The native protein sediments as a dimer of 220 kDa, containing two subunits of 30 kDa and two subunits of 78 kDa (Flores et al., 1990). The latter is phosphorylated in vitro by casein kinase II (O. Flores, R. Weinmann, and D. Reinberg, unpublished data). The role of TFIIF is to mediate the entry of RNA polymerase IIa into the complex of TFIID, TFIIB, and promoter DNA (Flores et al., 1991; step 3). Moreover, it appears that the small subunit of TFIIF is sufficient to insure entry of RNA polymerase II into the transcriptional complex (Flores et al., 1991). It is interesting to note that the sequence homology found between the 30-kDa subunit of TFIIF and the Escherichia coli σ 70 protein (Sopta et al., 1989) may thus extend to their functional role in transcription (Flores et al., 1991). Although some reports suggested that RAP 74 may be a helicase, no ATP-dependent helicase activity or ATPase has been detected in extensively purified human TFIIF (Flores et al., 1990). The final word will come from recombinant proteins. The small subunit of this two-subunit protein has been cloned by Sopta et al. (1989). The large subunit has recently been cloned by Aso et al. (1992), and by Finkelstein et al. (1992). The recombinant large and small TFIIF subunits associate and stimulate transcriptional activity of depleted extract, but lack ATPase or helicase activities (Aso et al., 1992; Finkelstein et al., 1992).
Transcription factor IIE (TFIIE)
TFIIE, also purified from phosphocellulose fraction eluting at 0.5 M KC1, can be separated from TFIIF on DEAE-5PW columns (Flores et al., 1990). TFIIE is also composed of two subunits, one 56 kDa and one 34 kDa, and behaves in sizing columns as a 200-kDa tetramer (Inostroza et al., 1991). The similar molecular weight of the polypeptides of the transcription factors TFIIE and TFIIF has generated a great deal of confusion. A factor purified from rat liver with activities equivalent to TFIIE is ε (Conaway et al., 1991b). The two subunits of the TFIIE protein have been recently cloned (M. G. Peterson et al., 1991; Ohkuma et al., 1991; Sumimoto et al., 1991). The 34-kDa subunit contains a consensus nucleotide binding site, and the 56-kDa contains a pocket with a sequence similar to all kinases, in addition to a Zn finger domain possibly involved in DNA binding (Peterson et al., 1991). Although an ATPase activity was found associated with TFIIE, it is apparently absent in TFIIE preparations purified to homogeneity (Inostroza et al., 1991). The purification of this factor was greatly facilitated by the finding that it leaks to the cytoplasmic fraction (S100) during extract preparation.
Other transcription factors
The recently described TFIIG protein fraction (Sumimoto et al., 1990) is separated from the TFIID fraction on further purification. Because different laboratories use complementary fractions with varying degrees of purification, the exact nature of this factor will require purification to homogeneity to ascertain whether it is indeed a new factor or part of others previously described.
Recent purification results of Flores and Reinberg (1991) and Cortes et al. (1992) suggest that the fraction described as TFIIG may be composed of other proteins. After chromatography on SP-Sepharose, S-Sepharose, and phenyl-Superose, two activities, called TFIIJ and TFIIH, were separated (Flores and Reinberg, 1991). TFIIH could also be purified from the TFIIF fraction, while TFIIJ also co-purified through seven steps with TFIIA (Flores and Reinberg, 1991). TFIIH activity co-purified with polypeptides of 33, 40, 42, 60, and 90 kDa, while TFIIJ has not yet been purified to homogeneity (Flores and Reinberg, 1991). Both factors enter the transcription complex after DBPolFE are assembled on the promoter (steps 6 and 7 in the diagram). TFIIJ was required only when bacterially produced TBP was used, but was dispensable with TFIID purified from cells (Cortes et al., 1991). The small subunit of TFIIF or TFIIH does not cross-react with anti-BTF3 antibodies (Flores and Reinberg, 1990). BTF3 is a 30-kDa polypeptide which binds to RNA polymerase II and is required in the transcription system from which it was purified and cloned (Zheng et al., 1990). Transcription factor BTF2 is also composed of 5 subunits of 90, 60, 43, 41, and 35 kDa and appears homologous to TFIIH and the rat liver δ (Gerard et al., 1991). The kinase involved in conversion of RNA polymerase IIa to IIo in the transcription complex appears to be associated with TFIIH (D. Reinberg, personal communication). A δ factor isolated from rat liver consists of at least 6 poly-peptides and has a native molecular mass of 390 kDa (Conaway et al., 1989, 1991b). It is interesting to note that rat liver δ also contains polyeptides of 95, 85, 68, 46, 43, 38, and 35 kDa (Conaway et al., 1991b), which may be related to TFIIH. The native molecular weight of this δ complex and its polypeptide composition also appear to be related to the TFIID complex found in Drosophila (Dynlacht et al., 1991). This liver transcription factor has an ATPase activity associated with TATA box DNA binding (Conaway et al., 1989). The exact relationship between BTF2, the δ multisubunit complex, TFIIG, TFIIH, and TFIIJ remains to be determined.
RNA polymerase II
This enzyme, well characterized and purified by different laboratories, is at the moment the reagent which even within the next few years will continue to be purified from cells or tissues. For recent reviews see Sawadogo and Sentenac (1990), Corden (1990), Woychik and Young (1990), and Young (1991). This is a large enzyme with 10 sub-units, the largest of which is 215 kDa in size. Most subunits of the yeast RNA polymerase II enzyme have been cloned by the laboratories of Sentenac and Young (see Sawadogo and Sentenac, 1990; Kolodziej et al., 1990; Young, 1991). Some subunits of the human enzyme have also been cloned recently (Saltzman and Weinmann, 1989; Pati and Weissman, 1989, 1990). Although most of the subunits may be cloned in the next few years, the reassembly of active enzyme from these components seems to present a difficult problem. Two-dimensional crystals of yeast RNA polymerase II are powerful analytical tools that have been used to obtain structural information at a resolution of 16Å (Edwards et al., 1990; Darst et al., 1991). Structural analysis has revealed a 25Å-diameter channel through which the DNA may be threaded. This major 25Å-wide groove then bifurcates into a narrow groove, where single-stranded nucleic acids may be interacting (nascent RNA; Darst et al., 1991). A finger of protein projecting outward from the structure may represent the CTD (Darst et al., 1991).
Phosphorylation of the CTD of RNA polymerase II
Three forms of RNA polymerase II are normally purified from tissues: IIo, IIa, and IIb. These differ in the apparent size of the largest subunit, which migrates in gels as 240 kDa (IIo), 215 kDa (Ha), and 180 kDa (IIb). From the results of Corden et al. (1985), it is clear that IIb is a proteolyzed form of Ha, lacking the CTD multiheptapeptide repeated sequence of the large subunit A unique feature of the 215 kDa polypeptide is the presence at the CTD of a heptapeptide which is repeated 52 times in the mouse enzyme (Ahearn et al., 1987) and 27 times in yeast, with variable numbers in other organisms (reviewed in Corden, 1990). The IIo form of RNA polymerase differs from IIa in the phosphorylation of this multiheptapeptide repeated sequence and is the only form found to crosslink with RNA in transcription complexes (Bartholomew et al., 1986). Although the IIb proteolyzed form has been discounted as a contributor to transcription in vivo, the transcriptional activity of IIb in an in vitro system (Zehring et al., 1988; Kim and Dahmus, 1989) suggests that this problem has to be reexamined. Pure RNA polymerase can be efficiently obtained by using nuclear pellets that are byproducts of the preparation of nuclear extracts (Lu et al., 1991). In the in vitro transcription system, reconstituted with highly purified factors devoid of endogenous phosphatase or kinase activities, the IIa form is 8–10 times more active in specific initiation than the IIo form, and forms stable complexes with promoter DNA, TFIID, TFIIB, and TFIIF (Lu et al., 1991).
The interconversion of the two forms apparently occurs in the transition from initiation to elongation (Laybourn and Dahmus, 1989, 1990; Payne et al., 1989; Lu et al., 1991; step 5 in the diagram) and is accompanied by phosphorylation of the CTD. The kinase responsible for phosphorylation remains to be identified.
We have established an ATP-hydrolysis requirement (Bunick et al., 1982) and a 5,6-diCl-1-β-D ribofuranosyl-benzimidazole (DRB)-sensitive process (Zandomeni et al., 1983) before formation of the first dinucleotide bond (step 6). The ATP hydrolysis suggested involvement of a kinase, a DNA-dependent gyrase, or an ATPase (Bunick et al., 1982), as did the identification of DRB sensitivity at an early transcriptional step (Zandomeni et al., 1983). We have identified casein kinase II as a DRB-sensitive kinase (Zandomeni and Weinmann, 1984; Zandomeni et al., 1986), although only a single phosphorylation site for this enzyme is present in the unique region of the CTD (Corden, 1990). It is important to establish which kinase(s) is responsible for the phosphorylation of the large subunit associated with the enzyme in the transcription complexes. One of the questions that remains unanswered is the nature of the ATP-hydrolysis requirement at transcription initiation (Bunick et al., 1982).
The steps in the formation of the initiation complexes have been defined by their sensitivities to sarkosyl, heparin, and salt, and the challenge by second templates (see Saltzman and Weinmann, 1989, for references). Clarification of the order of this process was recently provided by the analysis of the assembly intermediates of these complexes in a native gel system (Buratowski et al., 1989). The phosphorylation step required to pass from the preinitiation to the initiation complex results in the conversion of the RpIIa to RpIIo. The CTD domain of RpII contains 52 heptamer repeats, and it has been reported that from 15 to 200 phosphates are added to the CTD of RPIIa (Zhang and Corden, 1991; Arias et al., 1991). This amount of incorporated phosphate may suffice to produce the mobility shift to IIo and may explain, at least in part, the ATP requirement at initiation (steps 4–5). Since RNA polymerase Ha, Ho, and IIb have similar ATP hydrolysis requirements for transcription initiation, in addition to the phosphorylation of the CTD, other ATP-requiring steps may be involved (Laybourn and Dahmus, 1990). Several kinases have been implicated in this step of the initiation process, and it is not yet clear which one is involved. We have provided evidence for casein kinase II or a DRB-sensitive kinase (Zandomeni et al., 1986), while others suggest that a cell-cycle related kinase is involved in this process (Cisek and Corden, 1989). Interestingly, these latter kinases are also sensitive to DRB (Cisek and Corden, 1991). Kinases purified from Aspergillus (Stone and Reinberg, 1992) and yeast (Lee and Greenleaf, 1989) appear as single-subunit of 57 kDa or three-subunit enzymes of 58 kDa, 38 kDa, and 32 kDa. The large subunit of the yeast CTD kinase (CTK1) has been cloned and is required for normal growth of yeast (Lee and Greenleaf, 1991). One of the mammalian kinases recently purified contains polypeptides of 58 and 34 kDa. Its small subunit is related to the CDC2-CDC28 cell cycle genes of yeast, and the activity of the enzyme is sensitive to DRB (Cisek and Corden, 1989, 1991). A variant enzyme contains subunits of 62 kDa (equivalent to cyclin B) and 34 kDa (cdc2), implying that it is a major cell-cycle specific kinase (Cisek and Corden, 1991). The enzyme purified from yeast appears to be quite distinct in substrate specificity, amino acid sequence, and chromatographic properties (Lee and Greenleaf, 1989). An enzyme which is enhanced under heat-shock conditions and able to phosphorylate a CTD peptide appears to be more similar to the yeast-Aspergillus enzyme (Legagneux et al., 1990). A template-associated kinase has been implicated in CTD phosphorylation, but the enzyme has not been purified yet (Arias et al., 1991).
The substrate for the RpII-kinase able to efficiently convert the RNA polymerase IIa to IIo in the transcription complex can be either ATP, dATP, or GTP (Lu et al., 1991). DRB acts as a mixed-type inhibitor of casein kinase II (which can also use ATP, GTP, or dATP as substrates) and of in vivo and in vitro transcription at similar DRB concentrations (Zandomeni et al., 1986; Zandomeni, 1989). Thus, the kinase involved in phosphorylation of RpII would also be sensitive to DRB and, if different from CKII, sequence-related in the active site. Finally, phosphorylation by one kinase may be required to induce changes that allow phosphorylation by a second kinase. The difficulty here is that these enzymes act catalytically rather than stoichiometrically, like the transcription factors described above. This problem will be resolved when more purified components or recombinant proteins are used in transcription reconstitution. Phosphorylation of the CTD and utilization of up to 200 molecules of ATP per initiation event do not exclude the possibility that other ATP-dependent processes, such as unwinding, or other ATPase and kinase activities, are associated with the transcription initiation process.
Future directions
The availability of an increasing number of mammalian transcription factors from recombinant sources will greatly facilitate the purification, characterization, and cloning of the remaining transcription factors. Although studies have focused on the adenovirus major late promoter as a template and the HeLa cell as a source of proteins, application to other promoters and systems will permit fuller understanding of the principles of promoter selection, the mechanics of transcription initiation, and the effects of regulators on this process.
Other steps in the transcriptional process, such as elongation (which can be regulated by multiple factors, including TFIIS or SII and TFIIF), may play an important role in the control of gene expression. Moreover, it seems that some elements of the transcriptional machinery may be reutilized directly, because they remain bound to the promoter after initiation of RNA synthesis, like TFIID or TFIIA. Others, like the RpIIo, have to be dephosphorylated so that the RpIIa can re-enter the transcription process, while the fate of such factors as IIE, IIG, IIH, and IIJ is unknown. Little is known about how this recycling may occur and which enzymes are involved. The tools provided by studying the basal transcriptional machinery will also help to clarify the multiple ways in which promoter activity is regulated, as well as the multiple sites of regulation used by different effectors. Finally, we have addressed only those proteins directly interacting with naked DNA. In higher eukaryotes, cellular DNA is organized into chromatin structures, and these are also important determinants of gene expression (for a recent review, see Felsenfeld, 1992).
References
- Ahearn J. M., Bartolomei M. S., West M. L., Cisek L. J., and Corden J. L. (1987), J Biol Chem 265,425–437. [PubMed] [Google Scholar]
- Arias J. A., Peterson S. R., and Dynan W. S. (1991), J Biol Chem 266, 8055–8061. [PubMed] [Google Scholar]
- Aso T., Vasavada H. A., Kawaguchi T., Germino F. J., Ganguly S., Kitajima S., Weissman S., and Yasukochi Y. (1992), Nature 355, 461–464. [DOI] [PubMed] [Google Scholar]
- Bagchi S., Weinmann R., and Raychaudhuri P. (1991), Cell 65, 1063–1072. [DOI] [PubMed] [Google Scholar]
- Bartholomew B., Dahmus M. E., and Meares C. F. (1986), J Biol Chem 261, 14226–14231. [PubMed] [Google Scholar]
- Bunick D., Zandomeni R., Ackerman S., and Weinmann R. (1982), Cell 29, 877–886. [DOI] [PubMed] [Google Scholar]
- Buratowski S., Hahn S., Sharp P. A., and Guarente L., (1988), Nature 334, 37–42. [DOI] [PubMed] [Google Scholar]
- Buratowski S., Hahn S., Guarente L., and Sharp P. A. (1989), Cell 56, 549–561. [DOI] [PubMed] [Google Scholar]
- Burke C., Xu-Bu Y., Marchitell L., Davis E. A., and Ackerman S. (1990), Nucl Acids Res 18, 3611–3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton Z. F., Killeen M., Sopta M., Ortolan L. G., and Greenblatt J. (1988), Mol Cell Biol 8, 1602–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carcamo J., Lobos S., Merino A., Buckbinder L., Weinmann R., Natarajan V., and Reinberg D. (1989), J Biol Chem 264, 7704–7714. [PubMed] [Google Scholar]
- Carcamo J., Maldonado E., Cortes P., Ahn M. H., Ha I., Kasai Y., Flint J., and Reinberg D. (1990), Genes Dev 4, 1611–1622. [DOI] [PubMed] [Google Scholar]
- Carcamo J., Buckbinder L., and Reinberg D. (1991), Proc Natl Acad Sci USA 88, 8052–8056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallini B., Fans I., Mathes H., Chipoulet J. M., Egly J. M., and Chambon P. (1989), Proc Natl Acad Sci USA 86, 9803–9807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisek L. J. and Corden J. L. (1989), Nature 339, 679–684. [DOI] [PubMed] [Google Scholar]
- Cisek L. J. and Corden J. L. (1991), Methods Enzymol 200, 301–324. [DOI] [PubMed] [Google Scholar]
- Conaway J. W. and Conaway R. C. (1989), J Biol Chem 264, 2357–2362. [PubMed] [Google Scholar]
- Conaway J. W., Hanley J. P., Garrett K. P., and Conaway R. C. (1991a), J Biol Chem 266, 7804–7811. [PubMed] [Google Scholar]
- Conaway R. C., Garrett K. P., Hanley J. P., and Conaway J. W. (1991b), Proc Natl Acad Sci USA 88, 6205–6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corden J., Cadena D. L., Ahearn J. M., and Dahmus M. E. (1985), Proc Natl Acad Sci USA 82, 7934–7938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corden J. L. (1990), Trends Biochem Sci 15, 387–389. [DOI] [PubMed] [Google Scholar]
- Cortes P., Flores O., and Reinberg D. (1992), Mol Cell Biol 12, 413–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darst S. A., Edwards A. M., Kubalek E. W, and Kornberg R. D. (1991), Cell 66, 121–128. [DOI] [PubMed] [Google Scholar]
- Davison B. L., Egly J. M., Mulvihill E. R., and Chambon P. (1983), Nature 301, 680–686. [DOI] [PubMed] [Google Scholar]
- Dynlacht B. D., Hoey T., and Tjian R. (1991), Cell 66, 563–576. [DOI] [PubMed] [Google Scholar]
- Edwards A. M., Darst S. A., Feaver W. J., Thompson N. E., Burgess R. R., and Kornberg R. D. (1990), Proc Natl Acad Sci USA 87, 2122–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egly J. M., Miyamoto N. G., Moncollin V., and Chambon P. (1984), EMBO J 3, 2363–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenmann D. M., Dollard C., and Winston R. (1989), Cell 58, 1183–1191. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G. (1992), Nature 355, 219–224. [DOI] [PubMed] [Google Scholar]
- Fikes J. D., Becker D. M., Winston F., and Guarente L. (1990), Nature 346, 291–294. [DOI] [PubMed] [Google Scholar]
- Finkelstein A., Kostrub C. F., Li J., Chavez D. P., Wang B. Q., Fang S. M., Greenblatt J., and Burton Z. F. (1992), Nature 355, 464–467. [DOI] [PubMed] [Google Scholar]
- Fire A., Samuels S., and Sharp P. A. (1984), J Biol Chem 259, 2509–2516. [PubMed] [Google Scholar]
- Flanagan P. M., Kelleher R. J. III, Sayre M. H., Tschochner H., and Kornberg R. D. (1991), Nature 350, 436–438. [DOI] [PubMed] [Google Scholar]
- Flores O., Ha I., and Reinberg D. (1990), J Biol Chem 265, 5629–5634. [PubMed] [Google Scholar]
- Flores O., Lu H., Killen M., Greenblatt J., Burton Z. F., and Reinberg D. (1991), Proc Natl Acad Sci USA 88, 9999–10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores O., Maldonado E., Burton Z., Greenblatt J., and Reinberg D. (1988), J Biol Chem 263,10812–10816. [PubMed] [Google Scholar]
- Flores O., Maldonado E., and Reinberg D. (1989), J Biol Chem 264, 8913–8921. [PubMed] [Google Scholar]
- Gash A., Hoffman A., Horikoshi M., Roeder R. G., and Chua N.-H. (1990), Nature 346, 390–394. [DOI] [PubMed] [Google Scholar]
- Gerard M., Fischer L., Moncollin V., Chipoulet J.-M., Chambon P., and Egly J.-M. (1991), J Biol Chem 226, 20940–20945. [PubMed] [Google Scholar]
- Gill G. and Tjian R. (1991), Cell 65, 333–340. [DOI] [PubMed] [Google Scholar]
- Gregor P. D., Sawadogo M., and Roeder R. G. (1990), Genes Dev 4, 1730–1740. [DOI] [PubMed] [Google Scholar]
- Ha I., Lane W. J., and Reinberg D. (1991), Nature 352, 689–695. [DOI] [PubMed] [Google Scholar]
- Hahn S., Buratowski S., Sharp P. A., and Guarente L. (1989), Cell 58, 1173–1181. [DOI] [PubMed] [Google Scholar]
- Hariharan N., Kelley D. E., and Perry R. P. (1991), Proc Natl Acad Sci USA 88, 9799–9803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiebert S. W., Chellappan S. P., Horowitz J. M., and Nevins J. R. (1992), Genes Dev 6, 177–185. [DOI] [PubMed] [Google Scholar]
- Hoey T., Dynlacht B. D., Peterson M. G., Pugh B. F., and Tjian R. (1990), Cell 61, 1179–1186. [DOI] [PubMed] [Google Scholar]
- Hoffman A., Sinn E., Yamamoto T., Wang J., Roy A., Horikoshi M., and Roeder R. G. (1990), Nature 346, 387–390. [DOI] [PubMed] [Google Scholar]
- Horikoshi M., Hai T., Lin Y.-S., Green M. R., and Roeder R. G. (1988), Cell 54, 1033–1042. [DOI] [PubMed] [Google Scholar]
- Horikoshi M., Wang C. K., Fuji H., Cromlish J. A., Weil P. A., and Roeder R. G. (1989), Nature 341, 299–303. [DOI] [PubMed] [Google Scholar]
- Horikoshi N., Maguire K., Kralli A., Maldonado E., Reinberg D., and Weinmann R. (1991), Proc Natl Acad Sci USA 88, 5124–5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikoshi M., Beruccioli C., Takeda R., Wang J., Yamamoto T., and Roeder R. G. (1992), Proc Natl Acad Sci USA 89, 1060–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingles C. J., Shales M., Cress W. D., Triezenberg S. J., and Greenblatt J. (1991), Nature 351, 588–590. [DOI] [PubMed] [Google Scholar]
- Inostroza J., Flores O., and Reinberg D. (1991), J Biol Chem 266, 9304–9308. [PubMed] [Google Scholar]
- Jaehning J. A. (1991), Science 253, 859. [DOI] [PubMed] [Google Scholar]
- Kao C. C., Lieberman P. M., Schmidt M. C., Zhou Q., Pei R., and Berk A. J. (1990), Science 248, 1646–1650. [DOI] [PubMed] [Google Scholar]
- Kelleher R. J. III, Flanagan P. M., and Kornberg R. D. (1990), Cell 61, 1209–1215. [DOI] [PubMed] [Google Scholar]
- Kim W. Y. and Dahmus M. E. (1989), J Biol Chem 264, 3169–3176. [PubMed] [Google Scholar]
- Kitajima S., Tanaka Y, Kawaguchi T., Nagaska T., Weissman S. M., and Yasukochi Y. (1990), Nucl Acids Res 18, 4843–4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodziej P. A., Woychik N., Liao S.-M., and Young R. A. (1990), Mol Cell Biol 10, 1915–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laybourn P. J. and Dahmus M. E. (1989), J Biol Chem 264, 6693–6698. [PubMed] [Google Scholar]
- Laybourn P. J. and Dahmus M. E. (1990), J Biol Chem 265, 13165–13173. [PubMed] [Google Scholar]
- Lee J. M. and Greenleaf A. L. (1989), Proc Natl Acad Sci USA 86, 3624–3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. M. and Greenleaf A. L. (1991), Gene Expr 1, 149–167. [PMC free article] [PubMed] [Google Scholar]
- Lee R. F., Concino M. F., and Weinmann R. (1988), Virology 165, 51–56. [DOI] [PubMed] [Google Scholar]
- Lee W S., Kao C. C., Bryant G. O., Liu X., and Berk A. J. (1991), Cell 67, 365–376. [DOI] [PubMed] [Google Scholar]
- Legagneux V., Morange M., and Besaude O. (1990), Eur J Biochem 193, 121–126. [DOI] [PubMed] [Google Scholar]
- Lewin B. (1990), Cell 61, 1161–1164. [DOI] [PubMed] [Google Scholar]
- Lin Y.-S. and Green M. R. (1991), Cell 64, 971–981. [DOI] [PubMed] [Google Scholar]
- Lin Y. S., Ha I., Maldonado E., Reinberg D., and Green M. R. (1991), Nature 353, 569–571. [DOI] [PubMed] [Google Scholar]
- Lu H., Flores Q, Weinmann R., and Reinberg D (1991), Proc Natl Acad Sci USA 88, 10004–10008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luse D. S. and Jacob G. A. (1987), J Biol Chem 262, 14990–14997. [PubMed] [Google Scholar]
- Maldonado E., Ha I., Cortes P., Weis L., and Reinberg D. (1990), Mol Cell Biol 10, 6335–6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S., Hisatake K., Sumimoto H., Horikoshi M., and Roeder R. G. (1991), Proc Nad Acad Sci USA 88, 9553–9557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T., Segall J., Weil A. P., and Roeder R. G. (1980), J Biol Chem 255, 1192–1196. [PubMed] [Google Scholar]
- McCormack B. P., Strubin M., Ponticelli A. S., and Struhl S. K. (1991), Cell 65, 341–348. [DOI] [PubMed] [Google Scholar]
- Meisterernst M., Roy A. L., Liev H. M., and Roeder R. G. (1991), Cell 66, 981–993. [DOI] [PubMed] [Google Scholar]
- Mermelstein F. H., Flores O., and Reinberg D. (1989), Biochim Biophys Acta 1009, 1–10. [DOI] [PubMed] [Google Scholar]
- Nakajima N., Horikoshi M., and Roeder R. (1988), Mol Cell Biol 8, 4028–4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkuma Y., Sumimoto H., Hoffman A., Shimasaki S., Horikoshi M., and Roeder R. G. (1991), Nature 354, 401–404. [DOI] [PubMed] [Google Scholar]
- Park K. and Atchison M. (1991), Proc Natl Acad Sci USA 88 9804–9808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pati U. K. and Weissman S. M. (1989), J Biol Chem 264, 13114–13121. [PubMed] [Google Scholar]
- Pati U. K. and Weissman S. M. (1990), J Biol Chem 265, 8400–8403. [PubMed] [Google Scholar]
- Payne J. M., Laybourn P. J., and Dahmus M. E. (1989), J Biol Chem 264, 19621–19629. [PubMed] [Google Scholar]
- Peterson M. G., Inostroza J., Maxon M. E., Flores O., Admon A., Reinberg D, and Tjian R. (1991), Nature 354, 369–373. [DOI] [PubMed] [Google Scholar]
- Peterson M. G., Tanese N., Pugh B. F., and Tjian R. (1990), Science 248, 1625–1630. [DOI] [PubMed] [Google Scholar]
- Pugh B. F. and Tjian R. (1990), Cell 61, 1187–1197. [DOI] [PubMed] [Google Scholar]
- Pugh B. F. and Tjian R. (1991), Genes Dev 5, 1935–1945. [DOI] [PubMed] [Google Scholar]
- Rappaport J., Cho K., Saltzman A., Prenger J., Golomb M., and Weinmann R. (1988), Mol Cell Biol 8, 3136–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappaport J., Zandomeni R., Reinberg D., and Weinmann R. (1987), J Biol Chem 262, 5227–5232. [PubMed] [Google Scholar]
- Reinberg D., Horikoshi M., and Roeder R. G. (1987), J Biol Chem 262, 3322–3330. [PubMed] [Google Scholar]
- Reinberg D. and Roeder R. G. (1987), J Biol Chem 262, 3331–3337. [PubMed] [Google Scholar]
- Resnekov O. and Aloni Y. (1989), Proc Natl Acad Sci USA 86, 12–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougvie A. E. and Lis J. T. (1988), Cell 54, 795–804. [DOI] [PubMed] [Google Scholar]
- Roy A. L., Meisterernst M., Pognonec P., and Roeder R. G. (1991), Nature 354, 245–248. [DOI] [PubMed] [Google Scholar]
- Saltzman A. and Weinmann R. (1989), FASEB J 3, 1723–1728. [DOI] [PubMed] [Google Scholar]
- Samuels M. and Sharp P. A. (1986), J Biol Chem 261, 2003–2013. [PubMed] [Google Scholar]
- Sawadogo M. and Roeder R. G. (1985), Cell 43,165–175. [DOI] [PubMed] [Google Scholar]
- Sawadogo M. and Sentenac A. (1990), Annu Rev Biochem 59, 711–754. [DOI] [PubMed] [Google Scholar]
- Schmidt M. C., Kao C. C., Pei R., and Berk A. J. (1989), Proc Natl Acad Sci USA 86, 7785–7789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto E., Shi Y., and Shenk T. (1991), Nature 354, 241–245. [DOI] [PubMed] [Google Scholar]
- Shi Y., Seto E., Chang L., and Shenk T. (1991), Cell 67, 377–388. [DOI] [PubMed] [Google Scholar]
- Slattery E., Dignam J. D., Matsui T., and Roeder R. G. (1983), J Biol Chem 258, 5955–5959. [PubMed] [Google Scholar]
- Smale S. T. and Baltimore D. (1989), Cell 57,103–113. [DOI] [PubMed] [Google Scholar]
- Sopta M., Burton Z. F., and Greenblatt J. (1989), Nature 341, 410–414. [DOI] [PubMed] [Google Scholar]
- Sopta M., Carthew R. W., and Greenblatt J. (1985), J Biol Chem 260, 10353–10360. [PubMed] [Google Scholar]
- Stevens A. and Maupin M. K. (1989), Biochem Biophys Res Commun 159, 508–515. [DOI] [PubMed] [Google Scholar]
- Stone N., Flores O., and Reinberg D. (1991), Pharm Technol 15, 36–46 and 128. [Google Scholar]
- Stone N. and Reinberg D. (1992), J Biol Chem 267, 6353–6360. [PubMed] [Google Scholar]
- Stringer F. F., Ingles C. J., and Greenblatt J. (1990), Nature 345, 783–785. [DOI] [PubMed] [Google Scholar]
- Strubin M. and Struhl K. (1992), Cell 68, 721–730. [DOI] [PubMed] [Google Scholar]
- Sumimoto H., Ohkuma Y., Yamamoto T., Horikoshi M., and Roeder R. G. (1990), Proc Natl Acad Sci USA 87, 9158–9162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumimoto H., Ohkuma Y, Sinn E., Kato H., Shimasaki S., Horikoshi M., and Roeder R. G. (1991), Nature 354, 398–401. [DOI] [PubMed] [Google Scholar]
- Van Dyke M. W., Roeder R. G., and Sawadogo M. (1988), Science 241, 1335–1338. [DOI] [PubMed] [Google Scholar]
- Workman J. L. and Roeder R. G. (1987), Cell 51, 613–630. [DOI] [PubMed] [Google Scholar]
- Woychik N. A. and Young R. A. (1990), Trends Biochem Sci 15, 347–351. [DOI] [PubMed] [Google Scholar]
- Young R. A. (1991), Annu Rev Biochem 60, 689–715. [DOI] [PubMed] [Google Scholar]
- Zandomeni R., Bunick D., Ackerman S., Mittleman B. S., and Weinmann R. (1983), J Mol Biol 167, 561–574. [DOI] [PubMed] [Google Scholar]
- Zandomeni R. and Weinmann R. (1984), J Biol Chem 259, 14804–14811. [PubMed] [Google Scholar]
- Zandomeni R., Zandomeni M. C., Shugar D., and Weinmann R. (1986), J Biol Chem 261, 3414–3419. [PubMed] [Google Scholar]
- Zhang J. and Corden J. L. (1991), J Biol Chem 266, 2290–2296. [PubMed] [Google Scholar]
- Zheng X. M., Black D., Chambon P., and Egly J. M. (1990), Nature 344, 556–559. [DOI] [PubMed] [Google Scholar]
- Zehring W. A., Lee J. M., Weeks J. R., Jokerst R. S., and Greenleaf A. L. (1988), Proc Natl Acad Sci USA 85, 3698–3702. [DOI] [PMC free article] [PubMed] [Google Scholar]


