Abstract
Cardiovascular disease (CVD) is the leading cause of morbidity and mortality in the United States. Human epidemiological studies provide challenges for understanding mechanisms that regulate initiation and progression of CVD due to variation in lifestyle, diet, and other environmental factors. Studies describing metabolic and physiologic aspects of CVD, and those investigating genetic and epigenetic mechanisms influencing CVD initiation and progression, have been conducted in multiple Old World nonhuman primate (NHP) species. Major advantages of NHPs as models for understanding CVD are their genetic, metabolic, and physiologic similarities with humans, and the ability to control diet, environment, and breeding. These NHP species are also genetically and phenotypically heterogeneous, providing opportunities to study gene by environment interactions that are not feasible in inbred animal models. Each Old World NHP species included in this review brings unique strengths as models to better understand human CVD. All develop CVD without genetic manipulation providing multiple models to discover genetic variants that influence CVD risk. In addition, as each of these NHP species age, their age-related comorbidities such as dyslipidemia and diabetes are accelerated proportionally 3 to 4 times faster than in humans.
In this review, we discuss current CVD-related research in NHPs focusing on selected aspects of CVD for which nonprimate model organism studies have left gaps in our understanding of human disease. We include studies on current knowledge of genetics, epigenetics, calorie restriction, maternal calorie restriction and offspring health, maternal obesity and offspring health, nonalcoholic steatohepatitis and steatosis, Chagas disease, microbiome, stem cells, and prevention of CVD.
Keywords: caloric restriction, Chagas disease, epigenetics, genetics, maternal nutrition, non-alcoholic fatty liver disease, stem cells
Introduction
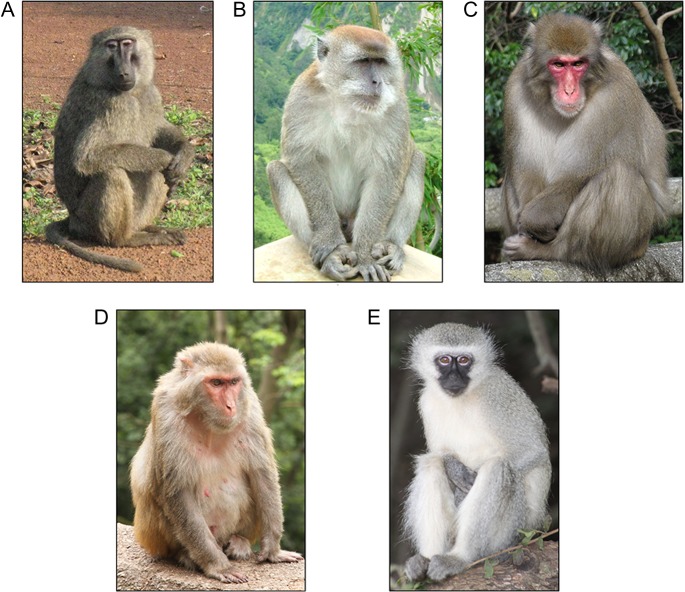
Cardiovascular disease (CVD) is the leading cause of morbidity and mortality in the United States (Mozaffarian et al. 2016). Human epidemiological studies provide significant challenges for understanding mechanisms underlying initiation and progression of CVD due to extensive variation in lifestyle and other environmental factors. Nonhuman primates (NHPs) have been studied for decades as a model to understand the pathogenesis and progression of CVD. Studies describing metabolic and physiologic aspects of CVD, and studies investigating genetic and epigenetic mechanisms influencing CVD initiation and progression, have been conducted in baboons (Papio hamadryas), rhesus macaques (Macaca mulatta), cynomologous macaques (Macaca fascicularis), snow monkeys (Macaca fuscata), and vervet monkeys (Chlorocebus aethiops sabaeus) (Figure 1). Major advantages of studying CVD in NHPs are their genetic, metabolic, and physiologic similarities with humans, and equally important is the ability to control diet, environment, and breeding. In addition, these NHP species are genetically and phenotypically heterogeneous, providing opportunities to study gene by environment interactions that cannot be feasibly studied in inbred animal models.
Figure 1.
Old World monkeys (Cercopithecidae) included in studies to understand mechanisms and pathology of cardiovascular disease: (A) Olive baboon, (B) Cynomolgus macaque, (C) Japanese macaque, (D) Rhesus macaque, and (E) Vervet monkey. References for figures: A. Olive baboon: Photograph by Charles J Sharp. https://en.wikipedia.org/wiki/Baboon#/media/File:Baboons_on_rock.jpg B. Cynomolgus macaque: Photograph by Sakurai Midori. https://en.wikipedia.org/wiki/Crab-eating_macaque#/media/File:Ngarai_Sianok_sumatran_monkey.jpg C. Japanese macaque: Photograph by Kenpei. https://en.wikipedia.org/wiki/Japanese_macaque#/media/File:Macaca_fuscata_fuscata1.jpg D. Rhesus macaque: Photograph by Einar Fredriksen. https://en.wikipedia.org/wiki/Rhesus_macaque#/media/File:Macaca_mulatta_in_Guiyang.jpg E. Vervet monkey: Photograph by Derek Keats. https://en.wikipedia.org/wiki/Vervet_monkey#/media/File:Vervet_monkey_Krugersdorp_game_reserve_(5657678441).jpg
Each of the Old World NHP species listed above brings unique strengths as models of human CVD. All develop CVD without genetic manipulation, providing multiple models to discover genetic variants that influence CVD risk. As these species age, their age-related comorbidities such as dyslipidemia and diabetes are accelerated proportionally 3 to 4 times faster than in humans (Cox et al. 2013). In addition, due to their large body size it is feasible to conduct longitudinal studies, collecting biopsy tissue samples in healthy and diseased animals at multiple time points. While it is feasible to collect various biopsy samples in an NHP model, advanced imaging technologies are increasingly being applied. Among the advantages of the NHP model already noted, the overall anatomical similarity and general larger body size allow for the direct application of imaging techniques and equipment already being used with humans. As an example, molecular resonance imaging (MRI) has been utilized in several studies focused on cardiometabolic function and health across a range of NHP species (Kochunov et al. 2017; Kuo et al. 2017).
Among NHP species, lipid metabolism in baboons more strongly correlates with humans than does lipid metabolism of rhesus macaques, cynomolgus macaques, or vervet monkeys (Eggen 1974). Initial observations of lesions in baboons were first published by McGill et al. in 1960 (McGill et al. 1960) and subsequent studies have characterized atherosclerosis in baboons (McGill et al. 1981). Baboon plasma cholesterol and triglyceride (TG) concentrations respond to dietary cholesterol, fat, and carbohydrates similar to those of humans (Higgins et al. 2010; Kushwaha and McGill 1998; Kushwaha et al. 1994). Chronic feeding of a high-fat diet leads to fatty streaks and atherosclerotic plaque (McGill et al. 1981). In addition, a diet high in simple carbohydrates and fat causes baboons to develop increased body fat and TG concentrations, altered adipokine concentrations, and altered glucose metabolism, consistent with observations in humans (Higgins et al. 2010). Even on a normal chow diet, some animals develop insulin resistance, dyslipidemia, atherosclerosis, and CVD, recapitulating metabolic complications seen in humans (Higgins et al. 2014; Kamath et al. 2011; Kulkarni et al. 2014).
Rhesus macaques, cynomologous macaques, vervet monkeys, and snow monkeys also develop atherosclerosis when fed a high-fat diet long term (Eggen 1974; Rudel et al. 1990), and their smaller size compared with baboons makes housing and husbandry more amenable for some research facilities.
NHPs also bring challenges to studies of CVD. Compared with mice and rats, NHPs have longer lifespans and longer health spans; and due to their larger body sizes, they are more expensive to maintain, and require greater amounts of experimental reagents for testing of therapies and drugs. In addition, NHP genomes are less well annotated than mouse and rat genomes, which poses challenges for identifying and validating genetic variants that influence CVD initiation and progression. Some of these challenges can be overcome by using comparative genomic analysis and annotation tools, such as those available through the UCSC Genome Browser (Rosenbloom et al. 2015), and will be overcome in the near future as annotation of NHP genomes improves.
In this article, we review current CVD-related research in NHPs focusing on selected aspects of CVD for which nonprimate model organism studies have left gaps in our understanding of human disease. This review includes genetics, epigenetics, calorie restriction, maternal calorie restriction and offspring health, maternal obesity and offspring health, nonalcoholic steatohepatitis (NASH) and steatosis, Chagas disease, microbiome, stem cells, and prevention. This review does not include some other clinically important topics to which CVD research with NHPs has made vital contributions, including vascular aging, hypertension, diastolic function, social status, and stress, among others.
Genetics
Influence of Genetic Variation on CVD Risk
Studies to identify genetic loci and genetic variation that are correlated with CVD risk have been conducted with the pedigreed baboon colony at the Southwest National Primate Research Center (SNPRC). Initial studies identified correlations of variation in apolipoprotein(a) isoform frequencies with lipoprotein(a) serum concentrations (Williams-Blangero and Rainwater 1991); and serum concentrations of lipoprotein(a) correlate with risk of CVD (Kronenberg et al. 1996). The SNPRC baboon colony has also been used to map genetic loci related to hypertension (Kammerer et al. 2003), loci related to low density lipoprotein (LDL) size phenotypes (Rainwater et al. 2003), and loci regulating plasma levels of gamma glutamyl transferase and albumin—quantitative traits that correlate with CVD (Bose et al. 2009). Baboons from the SNPRC colony were also used to identify functional genetic variants that regulate high density lipoprotein cholesterol (HDL-C) plasma concentrations (Cox et al. 2007) and to identify novel candidate genes that regulate LDL-C plasma concentrations (Karere et al. 2013).
Studies at the Oregon National Primate Research Center have shown that plasma HDL-C concentrations are also heritable in rhesus macaques (Vinson et al. 2013), providing another NHP model for research on the genetics of this CVD-related quantitative trait. Addition of whole genome sequence data for the SNPRC pedigreed baboon colony, as well as a pedigreed snow monkey colony (Oregon National Primate Research Center) and pedigreed rhesus macaque colonies at the other NPRCs, will accelerate the identification of functional genetic variants that influence CVD initiation and progression.
Gene by Diet Interactions and CVD Risk
The expression of an organism's genes occurs within an environmental context in which almost all gene expression is affected by gene by environment interactions. Perhaps the most profound and pervasive form of gene by environment interaction is that of gene by diet. As food becomes available on a continuous basis, the nutrient composition of an organism's diet can have a chronic and significant impact on various physiological and metabolic processes, all of which are mediated to varying degrees by the genetic makeup of the organism. While the nutritional environment represents a potent factor impacting gene action, the clear detection of such effects is difficult in free-living populations of humans due to the pronounced diversity in individual diets. However, NHP models such as the baboon, with their strong similarity in dietary adaption with humans (both being highly omnivorous) as well as the large extent of genetic conservation and the ability to control diet and breeding, presents a unique opportunity to investigate gene by diet interactions under tightly controlled experimental conditions. Investigators at the SNPRC have extensive experience utilizing a colony of pedigreed baboons to understand the interaction of saturated fat and cholesterol with genes involved in lipid metabolism and the corresponding impact on risk for CVD. Since the early 1980s, more than 3000 pedigreed baboons have been challenged with a high-cholesterol, high-fat (HCHF) diet (containing 41% of the energy as fat by the addition of lard) and 6.37 mg cholesterol/g (Wang et al. 2004) for varying periods of time to assess the impact of this diet exposure on a wide range of phenotypes associated with lipid metabolism and cardiovascular health. The results documented many significant gene by diet interactions, including diet-specific effects of two major loci on apo-A1 levels in response to a high-fat diet (Blangero et al. 1990), and a pattern of shared genetic effects (i.e., pleiotropy) among three subfractions of HDL (HDL1-C, HDL2-C, and HDL3-C) varied depending on dietary exposure (Mahaney et al. 1999). The colony has also been used to assess the interactions of vitamin E supplementation on risk factors for CVD (PMID: 17823422) and the impact of a chronic (2-year) HCHF diet on blood lipids and lipoproteins, lipoprotein-related enzymes, biomarkers of inflammation and oxidative stress, blood pressure, arterial compliance (stiffness), and extent of atherosclerotic lesions (Mahaney et al. 2017). The results of that and many previous studies clearly established that experimental dietary manipulations of NHPs have biological impacts that closely resemble those of humans whose diets have high levels of cholesterol and fat and that there is considerable individual variation in response to these dietary components.
MicroRNA by Diet Interactions
MicroRNAs (miRNAs) are endogenous, small non-protein coding RNAs (approximately 22 nts) that posttranscriptionally regulate gene expression by degradation of mRNAs or translational silencing (Ambros 2004). MiRNAs exist in virtually all organisms and are highly conserved evolutionarily (Ambros 2001), suggesting an essential role in biological processes. Expression of these small RNAs is highly tissue and cell specific. Circulating extracellular miRNAs are present in body fluids, including urine, saliva, breast milk, and plasma, where they are protected against ribonucleases by forming complexes with proteins, lipids, exosomes, and/or microvesicles. Thus, miRNAs are useful biomarkers for some diseases (Hunter et al. 2008; Valadi et al. 2007). MiRNAs also play roles in cell-cell communication, where they are loaded in carriers such as HDL-C particles and extracellular vesicles and transported to and taken up by distant tissues to regulate gene expression (Nolte-‘t Hoen et al. 2015; Vickers et al. 2011). MiRNAs have been implicated in a plethora of biological pathways, including cholesterol metabolism, and contribute to the progression of a number of diseases (Rayner et al. 2011; Sun et al. 2014). Because many genes can be regulated by a single miRNA and different miRNAs can target one gene, miRNAs fine-tune gene expression and coordinate genetic networks underlying common complex human diseases, such as CVD.
De novo cholesterol synthesis is tightly regulated to maintain homeostasis, and diet-derived LDL-C that perturbs cholesterol homeostasis is a major risk factor for CVD. Investigation into the role of miRNAs on LDL-C variation in half-sib baboons discordant for serum LDL-C concentrations (n = 6, low LDL-C; n = 6, high LDL-C), challenged with HCHF diet for 7 weeks, revealed 226 differentially expressed miRNAs (66 upregulated and 160 downregulated) (Karere et al. 2012). Overlaying these miRNAs onto genetic networks to identify molecular mechanisms that may regulate variation in LDL-C revealed seven candidate miRNAs (Karere et al. 2013). These findings demonstrated that liver miRNAs are responsive to diet, that miRNA response to a HCHF diet challenge differs among baboons with different LDL-C serum concentrations, and that miRNAs may regulate LDL-C variation.
Serum HDL-C concentrations are inversely associated with CVD, and HDL-C particles are essential for sequestering cholesterol from organs and macrophages. MiR-33 and miR-27b have been implicated in cholesterol synthesis and fatty acid oxidation (Rayner et al. 2011; Vickers et al. 2013). Ouimet et al. (Ouimet et al. 2016) identified oxysterol-binding protein-like 6 (OSBPL6) as a target of miR-33 and miR-27b. OSBPL6 is localized on a region of chromosome 2 linked to premature CVD (Nsengimana et al. 2007) and linked to variation in HDL-C plasma concentrations in women (North et al. 2003). In addition, OSBPL6 is an LXR-responsive gene that is upregulated in response to feeding vervet monkeys a high-fat, high-simple carbohydrate diet. In mice, knockdown of OSBPL6 was associated with reduced cholesterol esterification, and overexpression was associated with increased cholesterol trafficking and efflux in macrophages and hepatocytes. A study of 200 healthy individuals showed a positive correlation between OSBPL6 hepatic expression and plasma HDL-C concentrations. These findings suggest that miR-33 and miR-27b regulate variation in HDL-C metabolism (Ouimet et al. 2016).
Calorie Restriction and CVD
The majority of studies investigating the impact of caloric restriction (CR) on CVD risk has been performed using mice and rats, and more recently naked mole rats. Numerous studies have shown that CR has beneficial effects on health and longevity including reduction of CVD risk factors in these short-lived species (reviewed in Stenvinkel et al. 2016). In NHPs, only three studies have prospectively investigated the effects of prolonged CR on metabolism, physiology, and CVD incidence: studies of rhesus macaques at the Wisconsin National Primate Research Center (WNPRC) (Colman et al. 2009, 2014) and the National Institute of Aging (NIA) at the National Institutes of Health (Lane et al. 1992, 1999; Mattison et al. 2012; Verdery et al. 1997), and a study of cynomolgus macaques at the Wake Forest School of Medicine (Cefalu et al. 1999).
In the rhesus macaque study at the WNPRC, young adult male rhesus macaques of Indian origin were fed a low-fat, low-cholesterol diet ad libitum from weaning until the study began. During the first 6 weeks of the study, animals were housed singly with food consumption monitored. During the subsequent 3 months, the amount of food for CR animals was reduced by 10% per month compared with ad libitum consumption. For the NIA study, male rhesus macaques of Indian and Chinese origin, in three age groups (6–12 months, 3–5 years, and 18–25 years), were included. Six years after the start of the study, female rhesus macaques in three age groups (1–3 years, 6–14 years, and 16–21 years) were added to the study (Colman et al. 2009, 2014). As with the WNPRC study, after an initial assessment of ad libitum food consumption amounts, amounts of food available for the CR animals were reduced by 10% per month compared with ad libitum consumption (Lane et al. 1992, 1999; Mattison et al. 2012; Verdery et al. 1997).
In both of these studies, monkeys showed a decrease in body weight and body fat, especially abdominal visceral fat. CR correlated with improved insulin sensitivity and glucose tolerance, and increased insulin-stimulated glucose uptake when measured using the clamp technique compared with age-matched ad libitum-fed controls. In the NIA study, CR monkeys did not have a beneficial effect on extent of atherosclerotic lesions (Mattison et al. 2012). In the WNPRC study, CR monkeys showed improvement of plasma triglyceride levels, lipoprotein levels, blood pressure, and extent of atherosclerotic lesions compared with controls (Colman et al. 2014).
In the Wake Forest School of Medicine study, young adult male cynomolgus macaques were initially fed a moderately atherogenic diet. Then over a 3-month period, 30% CR was gradually implemented. After 4 years, the CR cynomolgus macaques showed a decrease in body fat and an increase in insulin sensitivity compared with controls. Similar to the NIA CR rhesus macaque study, the CR cynomolgus macaques did not show improved lipid profiles or extent of atherosclerotic lesions compared with controls (Cefalu et al. 1999).
In these three experimental paradigms using Indian-origin rhesus, Chinese-origin rhesus, and cynomolgus macaques, CR consistently positively impacted insulin sensitivity. Although diabetes and CVD are tightly linked, CR did not consistently show a similar positive impact on lipoprotein and triglyceride profiles or extent of atherosclerotic lesions (reviewed in (Kemnitz 2011)). It is possible that genetic variation plays a role in these differences, supporting the need for further studies to understand genetic mechanisms central to cardiovascular health.
Maternal Nutrition and Offspring CVD
Maternal CR During Pregnancy and Offspring CVD Risk
It is well established that suboptimal maternal nutrition during pregnancy alters fetal development (Langley-Evans 2015), with greater risk of adult-onset diseases including CVD (Barker et al. 1989; Hanson and Gluckman 2015). Human epidemiological and experimental animal studies have shown that programming affects the risk of developing diseases during adulthood and correlates with maternal diet and availability of nutrients during development (McMullen and Mostyn 2009; Ozanne et al. 2004). Effects of the in utero environment on health span are termed “developmental programming.” Maternal under-nutrition during pregnancy, which has been studied more extensively than other programming challenges, typically results in fetal intrauterine growth restriction (IUGR). IUGR is an important obstetric complication that affects 4% to 8% of babies in developed countries and 6% to 30% in developing countries, resulting in long-term health complications in the offspring during adulthood.
One of the first human epidemiological studies to address programming by IUGR was the Nurses’ Health Study at Harvard Medical School. This study of more than 120 000 women showed that low birth weight was associated with increased incidence of CVD (Rich-Edwards et al. 1997). These findings raised questions about the influence of programming on the genome, influencing gene by environment interactions in IUGR offspring; for example, does programming modify the epigenome and offspring resiliency (Rich-Edwards et al. 1997; Sun et al. 2013; Tarrade et al. 2015)?
A baboon model of IUGR has been developed to study tissue, cellular, and molecular changes in the developing fetus (Cox et al. 2013; Li et al. 2009) and the impact on offspring health (Choi et al. 2011). Fetal studies showed that maternal CR (MCR, 30% reduction of controls fed ad libitum) influences the fetal kidney transcriptome and renal tubule structure (Cox et al. 2006), with mTOR signaling central to this response (Nijland et al. 2007). In addition, MCR impacts the fetal liver transcriptome and energy storage (Li et al. 2009), and fetal liver changes correlate with epigenetic changes in the energy management enzyme PEPCK1 (Nijland et al. 2010). Similar findings have not been reported in nonprimate models of IUGR, suggesting fundamental molecular and cellular differences between primate and nonprimate response to a suboptimal in utero environment. Furthermore, IUGR juvenile offspring in this model show signs of hypertension, insulin resistance, and metabolic syndrome (Choi et al. 2011).
In addition to IUGR being associated with increased incidence of dyslipidemia and hypertension, cardiac dysfunction has been detected in IUGR fetuses and neonates (Fouzas et al. 2014). However, the mechanisms by which IUGR programs fetal heart and postnatal cardiac function are poorly understood. Some measures of cardiac function are inversely correlated with birth weight (Jones et al. 2008; Ward et al. 2004), and human IUGR offspring as adults have higher systolic blood pressure and smaller aortic dimension, suggesting impairment of future left ventricular performance (Bjarnegård et al. 2013). A better understanding of the underlying pathogenesis will allow development of imaging biomarkers for diagnosis and offer more timely treatment options.
A recent study by Kuo et al. (2016) investigated the effects of IUGR on young adult baboons (age 5.7 ± 1.3 years). They used cardiac magnetic resonance imaging, an established noninvasive method to quantify cardiac changes indicative of subclinical heart disease in humans and found systolic dysfunction, diastolic derangement, and morphological remodeling of the left ventricle in these young adult baboons. Results from this study showed that the effects of cardiac function on IUGR persist postnatally and that baboons exhibit cardiac abnormalities similar to humans. The investigators found differences in cardiac dysfunction by comparison with nonprimate models, indicating either variation in IUGR protocols or differences between primate and nonprimate cardiac physiology (Kuo et al. 2016). Use of this imaging modality where animals can be monitored longitudinally may provide foundation data for discovery of biomarkers early in the disease process. In addition, because this modality is established for human cardiac functional assessment, findings in baboons can be directly translated to humans.
Maternal Obesity During Pregnancy and Offspring CVD Risk
The incidence of obesity and overweight has reached epidemic proportions in the developed world, with approximately 64% of women of childbearing age in the United States being overweight (BMI ≥ 25 kg/m2 and <30 kg/m2) or obese (BMI ≥ 30 kg/m2) (Wilson and Messaoudi 2015). It is well established that an obesogenic nutritional environment and a sedentary lifestyle contribute to the risk of developing obesity. A growing body of evidence links early-life nutritional adversity to the development of long-term metabolic disorders (Li et al. 2011). Therefore, early-life exposure of offspring to environmental stimuli, including altered nutrition during critical periods of development, can program alterations in organogenesis, tissue development, and metabolism, predisposing offspring to obesity, metabolic disease, and CVD in later life (Dong et al. 2013; Reynolds et al. 2013; Segovia et al. 2014). Supporting the programming hypothesis is the increasing prevalence of maternal obesity and excess maternal weight gain associated with increased risk of obesity in offspring (Nathanielsz et al. 2007) and metabolic-related diseases such as diabetes and CVD (Alfaradhi and Ozanne 2011).
Animal studies suggest that inappropriate energy metabolism during pregnancy has an adverse effect on fetal development and is an important factor in metabolic programming (Rees et al. 2008). Maternal obesity-induced developmental programming has been validated in mice, rats, sheep, and NHPs (Maloyan et al. 2013; McCurdy et al. 2009). McCurdy et al. (McCurdy et al. 2009) showed lipid accumulation in the livers of Japanese macaque fetuses that were approaching birth. In addition, a baboon model of maternal obesity revealed fetal hepatic lipid accumulation (Puppula et al. unpublished data) as well as dysregulation of fetal cardiac miRNA expression and early signs of fibrosis in the fetal heart (Maloyan et al. 2013). Because the liver is a central metabolic regulator, the observed accumulation of lipid in NHP fetal livers may be an upstream event that influences CVD and metabolic disease risk in offspring of obese pregnancies. Also of interest is the observed disruption of the methionine cycle in obese pregnant baboons, suggesting an epigenetic mechanism by which obesity during pregnancy may impact fetal development (Nathanielsz et al. 2015).
NASH and Steotosis
Nonalcoholic fatty liver disease (NAFLD), the most common cause of chronic liver disease in the United States, progresses to NASH, which can progress to end-stage liver disease. NAFLD describes a spectrum of liver pathologies ranging from simple steatosis (>5% TG accumulation in hepatocytes) to NASH, including lobular and portal vein inflammation, hepatocellular ballooning, and fibrosis. NAFLD has been shown to confer increased CVD-related deaths due to the high prevalence of dyslipidemia in these individuals (Cohen et al. 2011). In addition, resolution of NASH is associated with improved lipoprotein profiles and triglycerides, suggesting that NASH directly impacts CVD progression (Corey et al. 2015). Over 50% of obese adults in the United States develop NAFLD. Of these, about one-half will progress to NASH, and 20% to 30% will develop additional liver health complications such as cirrhosis or hepatocellular carcinomas within 10 years. With the increasing prevalence of obesity and diabetes, there has been a steep rise in the incidence of nonalcoholic NAFLD in the developed world (Cohen et al. 2011).
The primary challenge in developing a suitable animal model for NAFLD and NASH has been the need for animals to develop all characteristic hepatic abnormalities together with the metabolic systemic complications such as dyslipidemia, insulin resistance, obesity, and characteristic cytokine profiles. In rodent models, it has been challenging to recreate all NAFLD-related health complications effectively and reproducibly. Genetic models such as the Zucker rat or the Ob/Ob mouse develop obesity, dyslipidemia, and insulin resistance (Phillips et al. 1996; Tilg and Diehl 2000) but fail to develop the entire complement of liver pathologies, even on a challenge diet. Challenging these mice with lipopolysaccharides accelerates the development of steatosis and hepatic injury. Similarly, rodents fed a methionine- and choline-deficient diet develop steatosis and some aspects of the hepatic inflammation seen in human NAFLD, but fail to develop insulin resistance and obesity (Dela Peña et al. 2005). Rodent studies also have suggested that fructose and high-carbohydrate diets potently promote the development of NAFLD. However, rodents have higher lipogenesis rates compared to humans, and the primary sites of lipogenesis, the inflammatory responses, and toll-like receptor expression patterns, all hallmarks of human NAFLD, are different. Also, despite the effects of challenge diets, rodents do not readily develop fibrosis (Bergen and Mersmann 2005; Ketloy et al. 2008; Sanches et al. 2015; Seok et al. 2013).
Other mammalian species have been proposed as NAFLD models, such as Ossabaw (Bell et al. 2010) or Lee-Sung (Li et al. 2016) mini-pigs and laboratory opossums (Chan et al. 2012), but to date, they have not been widely used for NAFLD-related studies, and no long-term data are available to validate the entire complement of characteristic molecular and histological abnormalities seen in human patients.
Recent studies suggest that NHPs may develop NAFLD-like abnormalities. A small percentage of captive marmosets, a small New World monkey species, developed NAFLD-like pathologies, potentially triggered by exposure to excess dietary iron. The animals developed hepatomegaly, hepatic inflammation, and obesity, but no overt signs of liver fibrosis or insulin resistance (Kramer et al. 2015). In contrast, recent long-term studies suggest that vervet monkeys developed liver fibrosis, in addition to other hallmarks of NAFLD, obesity, and insulin resistance, after seven years on a high-fructose diet (Cydylo et al. 2017). These findings suggest that NHP species develop NAFLD-like symptoms similar to humans. A NHP model would have benefits over current rodent models due to extensive human-NHP genetic, physiologic, and nutritional behavior similarities.
Baboons also develop significant liver steatosis, initial signs of hepatic inflammation, and insulin resistance on a HCHF diet. A HCHF diet also causes baboons to develop increased body fat and TG concentrations, altered adipokine concentrations, and altered glucose metabolism, consistent with observations in humans (Higgins et al. 2010). Steatosis can be observed within a few weeks, much more rapidly than reported in other NHP species. The rate of steatosis is variable among animals, suggesting that genetic factors also influence the development of steatosis, similar to humans. Therefore, it is likely that baboons develop all characteristic features of NAFLD and NASH, including the hepatic pathologies and the systemic complications when exposed to a HCHF challenge diet for extended periods. Due to baboon body size (which facilitates repeated collection of biopsy samples without harm to the animal), its similarity to humans genetically and physiologically, and its susceptibility to diet-induced metabolic abnormalities, the baboon may be an ideal animal model for research on NAFLD and NASH.
Chagas Disease
Overview of Chagas Disease
Chagas disease is caused by a protozoan parasite, Trypanosoma cruzi, which is typically transmitted by blood-sucking triatomine bugs. In general (although perhaps with rare exceptions), once a person is infected, he or she is always infected; the immune system is quite effective in eliminating parasites from the blood and maintaining blood parasitemia at low levels after the acute phase, but it cannot eliminate parasites from the heart or other tissues. After many years or several decades, approximately 30% of infected people develop cardiac pathologies. Many infected people die of sudden cardiac arrest or complications of congestive heart failure. There is no vaccine or prophylactic drug for Chagas disease, and therapeutic drugs have been tested in clinical trials for half a century without leading to validation of sufficient efficacy and safety to be widely marketed for use in chronically infected adults.
Chagas Disease in NHPs
Like many other wild mammals, monkeys serve as a reservoir for T. cruzi. Although T. cruzi in nature is confined to the Americas, ranging from central and the southern half of the United States to the deep south of South America, Old World monkeys also can be experimentally infected with the parasite (see Seah et al. 1974), and they exhibit disease progression and outcomes similar to those of humans (see, for example, Carvalho et al. 2003; Sathler-Avelar et al. 2016; Vitelli-Avelar et al. 2017; Zabalgoitia et al. 2003a, 2003b, 2004). Old World monkeys, which are phylogenetically and physiologically more similar to humans than are New World monkeys, are the species of choice for translational research on Chagas disease. The species that are most frequently used are baboons, rhesus macaques, and cynomolgus macaques. These and other NHP species are prone to natural infection with T. cruzi when they are maintained in outdoor or indoor-outdoor housing in areas where T. cruzi infection is endemic in wild mammal reservoirs.
Fundamental Questions that can be Addressed with Old World Monkeys in Translational Research on Chagas Disease
There are at least four fundamental questions pertaining to prevention and treatment of Chagas disease that can be best answered by conducting research with Old World monkeys, in concert with research in mice and human subjects.
Which candidate drugs, which dosages and durations of treatment, and which combinations of those drugs are most efficacious as therapies for Chagas disease?
Which candidate vaccines, and which dosages and vaccination regimens, are most efficacious for prophylaxis against Chagas disease?
Which candidate vaccines, which dosages and vaccination regimens, and in which combinations with candidate drugs are most efficacious for treating patients?
Does reduction in blood parasitemia and/or cardiac parasite load in response to therapy lead to a reduction in the rate of disease progression; and does elimination of all parasites form the body in response to therapy lead to prevention of disease progression or reduction in the rate of disease progression?
Each of these questions involves many variables that may affect the answers; therefore, it is appropriate that the many candidate drugs and vaccines undergo extensive testing in mouse models to identify those that exhibit the greatest potential. However, clinical therapeutic drug trials have been conducted with great optimism after stunningly successful results from experiments with mice, and none has produced in humans the favorable results that were anticipated (see for example Pecoul et al. 2016). Apparently, the combined metabolic and immunologic systems of mice do not sufficiently resemble those of humans for the results from mice to be predictive of results for humans.
Posaconazole is an example of a candidate drug that resulted in sterile cure of mice infected with T. cruzi (reviewed by Urbina 2009) and failed to eradicate parasites from infected humans (Molina et al. 2014). The same drug, administered to 12 chronically infected baboons in a dosing regimen that yielded similar blood levels of the drug over the same duration of time as is achieved in humans, failed to completely clear the parasites from any of the animals (J. L. VandeBerg, unpublished data). The experiment was overpowered; if only three or four of the baboons had been used in the experiment, the same conclusion would have been reached—not to proceed with a clinical trial. This result indicates that NHP drug (or vaccine) trials can be conducted economically by using a small number of animals per group.
Impediments to Therapeutic Clinical Trials for Chagas Disease
The results of clinical trials on Chagas disease are particularly difficult to interpret, because it is not possible to determine with certainty if the parasites have been eliminated from heart tissues. Even many untreated people (or monkeys) who are known to be infected with T. cruzi do not have detectable levels of parasites in the blood in spite of repeated serial PCR assays in which the limit of detection is less than one parasite per milliliter. Therefore, the inability to detect parasites in blood after serial sampling and PCR posttreatment does not necessarily reflect the absence of parasites in tissues. Only in animal models can the absence of parasites in tissues be determined; first, by immunosuppression, which often leads to increased blood parasitemia, and then by direct examination of tissues by PCR after euthanasia. This strategy has been used extensively with mice (for example, see Khare et al. 2015), and it also has been used in an active study with NHPs (J. L. VandeBerg, R. L. Tarleton, and I. Ribeiro, unpublished data).
Moreover, the course of the disease in humans is gradual over many years, so it is not possible to ascertain in a follow-up of only a few years if candidate therapies lead to a reduction in disease progression or not. However, Old World monkeys can provide reliable information in regard to disease progression in an experiment of only a few years of duration, for two reasons. First, while the average duration from the time of infection to the development of clinically detectable symptoms might be many years, the accumulation of lymphocytes in the heart and consequent inflammation (myocarditis) occur continuously as the immune system responds to the presence of parasites. Therefore, in the NHP model, euthanasia and histological examination of the heart can determine if disease is progressing, long before clinical abnormalities could be detected. Second, just as Old World monkeys and their immune systems age about three times faster than humans (i.e., three monkey years is approximately equivalent to one human year), so also Chagas disease progresses about three times faster in Old World monkeys than in humans (as do many other chronic diseases). Therefore, while it might take six to nine years to detect significant clinical differences in posttreatment clinical outcomes of candidate therapies between groups of humans, the same magnitude of group differences would probably be observed in two to three years in Old World monkeys. These factors, along with the ability to determine at necropsy if each posttreatment monkey is or is not still infected with T. cruzi by PCR assays of tissues, will enable the fourth question posed above to be answered rigorously in a practical time frame.
Microbiome—Diet Responsiveness
The microbiota is a complex community of microorganisms composed of bacteria, archaea, anaerobic fungi, protozoa, and viruses that influences host health and disease. Recent studies have shown that dysbiosis (or microbial imbalance) is associated with a wide range of diseases, including CVD (Aron-Wisnewsky and Clément 2016; Griffin et al. 2015; Haghikia and Landmesser 2015; Mell et al. 2015; Meyer and Bennett 2016; Miller 2013; Tang and Hazen 2014; Tuohy et al. 2014). Previous studies examining dietary patterns suggest a strong association between gut dysbiosis and Western diet, which tends to be high in fat and animal protein (e.g., red meat), high in sugar, and low in plant-based fiber (Amato et al. 2015; Hold 2014; Manzel et al. 2014; Miele et al. 2015). Therefore, it is not surprising that even though alteration of the microbiota has been linked to a number of genetic and environmental factors (e.g., antibiotic use, stress, infection, geography, and race), most research to date has focused on the association of diet and gut dysbiosis (Aron-Wisnewsky and Clément 2016; Bhatnagar 2015; Clayton et al. 2016; Del Chierico et al. 2014; do Rosario et al. 2016; Ferguson et al. 2016; Meyer and Bennett 2016; Miller 2013; Sonnenburg and Bäckhed 2016; Vos 2014).
The gastrointestinal (GI) tract is inhabited by trillions of commensal bacteria that directly impact host nutrition, as they are essential mediators of metabolism and obesity in mammals (Ma et al. 2014; Pacheco and Sperandio 2015). These bacteria encode enzymatic pathways that enable metabolism and synthesis of fatty acids and vitamins (Janiak 2016; Ma et al. 2014). For example, they allow the host to extract calories from otherwise indigestible complex carbohydrates and plant polysaccharides via enzymes that are not encoded within the host genome (Janiak 2016; Ma et al. 2014; Miele et al. 2015; Pacheco and Sperandio 2015), so alteration of the type or amount of microbes in the gut influences metabolic processes and host nutrition (Amato 2016; Flint et al. 2007; Turnbaugh et al. 2006). Metagenomic studies have shown that the species composition of the gut microbiota is very dynamic and that the majority of changes are a result of the selective pressure that diet exerts on the microbial community (Amato 2016; David et al. 2014; Turnbaugh et al. 2006). For example, most microbes inhabiting the gut belong to the Bacteroidetes (Gram-negative) and Firmicutes (Gram-positive) phyla (Pacheco and Sperandio 2015). However, obese individuals consuming a Western diet tend to have a decreased ratio of Bacteroidetes to Firmicutes. When switched to a lean diet, obese individuals lose weight and regain Bacteroidetes (Ma et al. 2014).
Most animal studies of the gut microbiota have been done in a rodent model. However, NHPs are known to be important model systems for understanding human health, so there is growing interest in the NHP gut microbiota. In agreement with previous findings in humans, recent studies have shown that diet has a profound impact on the microbial composition of the NHP gut microbiota. In a recent paper, Mareike Janiak (Janiak 2016) discusses the relationship between diet and gut adaption that allows primates to maximize the energy obtained from food and allows them to exploit food sources that were previously difficult to digest. For example, metagenomic analysis of black howler monkeys (Alouatta pigra) revealed how diet influences gut microbiota composition to enable more efficient extraction of energy and nutrients during periods of low fruit intake by producing more short chain fatty acids (Amato et al. 2015). Similarly, Sun et al. (Sun et al. 2016) identified variation between winter and spring gut microbiota in free-ranging Tibetan macaques (Macaca thibetana) that allowed proper nutrition in diverse climates. A few studies have used infant rhesus macaques to compare the gut microbiota following formula-feeding and breast-feeding (Ardeshir et al. 2014; Narayan et al. 2015; O'sullivan et al. 2013). Findings from these studies indicate that metabolic and gut microbiome development is different in formula-fed infants from breast-fed infants and that the choice of infant feeding may have future health consequences, such as an increased susceptibility to CVD. Previous studies have shown that maternal obesity contributes to an increased risk of lifelong morbidity and mortality of the offspring (Boney et al. 2005; Chu et al. 2016; Dong et al. 2013; Drake and Reynolds 2010; Harris et al. 2016; Ramsay et al. 2002; Reynolds et al. 2013), but the molecular mechanisms underlying these risks are still unclear. Using a Japanese macaque model of maternal obesity, Ma et al. (Ma et al. 2014) demonstrated that a high-fat, caloric-dense maternal diet structures the offspring's gut microbiota and that the resultant dysbiosis is only partially corrected by a low-fat control diet after weaning.
Other studies involving captive or wild NHPs (Amaral et al. 2017; Bo et al. 2010; Degnan et al. 2012; Frey et al. 2006; Kisidayová et al. 2009; Ley et al. 2008; McCord et al. 2014; Moeller and Ochman 2013; Ochman et al. 2010; Szekely et al. 2010; Uenishi et al. 2007; Yildirim et al. 2010) indicate that the NHP microbiota is species-specific and altered by environmental factors (e.g., diet, geography, and social factors). They also reveal that the abundance of Firmicutes and Bacteroidetes in the NHP microbiota is similar to that of healthy humans (Bo et al. 2010; Frey et al. 2006; Kisidayová et al. 2009; Szekely et al. 2010; Uenishi et al. 2007; Yildirim et al. 2010). Furthermore, studies show that chimpanzees (Pan troglodytes), our closest living relative, have similar gut microbiota (Degnan et al. 2012; Ellis et al. 2013; Moeller et al. 2012; Ochman et al. 2010). Despite differences in host diet, findings from these studies indicate that the gut microbial communities within humans and chimpanzees overlap at broad taxonomic levels and assort into similar enterotypes based on the relative abundances of bacterial genera.
Although these findings make a compelling case for using NHPs to study how diet affects the gut microbiota of humans and influences CVD, a recent study by Amato et al. (Amato et al. 2015) suggests that such research should be done with caution. In this study, the authors compared the gut microbiota of humans and vervet monkeys after consuming both a Western diet (high in animal fat and protein and low in fiber) and a non-Western diet (low in animal fat and protein and high in fiber). Their findings indicate that host-gut microbe interactions differ in humans and vervets. For example, they found that humans had an increased relative abundance of Firmicutes and a reduced relative abundance of Prevotella on a Western diet, while vervets showed the opposite pattern. Furthermore, they identified an increased relative abundance of genes associated with carbohydrate metabolism in the microbiome of humans consuming a Western diet but not in vervets. In addition to the differences identified in the gut microbiota, they also found that the physiological responses to the Western diet differed between humans and vervets. Humans gained weight on the Western diet, while most of the vervets did not. Therefore, the authors state that “NHP models of host-gut microbe relationships may be less ideal than assumed for addressing questions regarding human diet and physiology in the context of the gut microbiota” and speculate that the human gut microbiota may provide increased susceptibility to obesity and metabolic disorders, particularly when hosts are consuming a high-protein, high-fat diet. Even though this study suggests that the vervet gut microbiota may not provide an ideal model for understanding the effect of the human gut microbiota on host metabolism and nutrition in the context of a Western diet, the authors state that it may help identify novel therapeutics to improve human resistance to obesity via the gut microbiota if the vervet gut microbiota truly possesses properties that make it resistant to obesity when subjected to a Western diet (Amato et al. 2015). Furthermore, it should be noted that this is only one study focusing on one NHP species. Given the similarities between human and chimpanzee gut microbiota and similarities between human and baboon omnivorous diets, other NHP species may be appropriate models for studying the effects of diet on the human gut microbiota and how these host-microbe interactions influence CVD risk.
Stem Cells—Therapies and Interventions
Stem cell therapies are being developed to treat CVD by two fundamental mechanisms: the stimulation of angiogenesis to develop improved vascularization of damaged heart or vascular tissue, and the stimulation of myogenesis to repair damaged cardiac muscle. Mouse models have pioneered preclinical research, but the immunological and physiological differences between mice and humans limit the capacity to translate many results obtained from mice directly to human subjects. Moreover, it is impossible to scale many of the results obtained from mice to human applications. For example, some approximate measures are as follows (mouse/human): heart weight, 0.165 vs. 250 g (1500-fold difference); coronary artery diameter, 0.16 vs. 4.5 mm (28-fold difference) (Oberhoffer et al. 1989; Thüroff et al. 1984; Wiedemann 1962). For these and perhaps other reasons, efficacious results obtained in research with mice have not translated well to clinical trials, which have had disappointing outcomes. Consequently, NHPs are becoming more widely used for late stage preclinical stem cell research aimed at developing cardiovascular therapies and interventions. The most widely used NHPs for this purpose are rhesus and cynomolgus macaques and baboons, which, as Old World monkeys, are phylogenetically and physiologically closer to humans than New World monkeys and are larger than New World monkeys used in laboratory research. Male baboons can weigh as much as 30 to 40 kg, so this species among all NHPs used in research is the most appropriate for investigations where scaling to human size is an important factor. Pluripotent embryonic stem cell (ESC) lines, as well as induced pluripotent stem cell (iPSC) lines, have been developed from all three of these species, and these species are all used in research aimed at developing stem cell therapies and interventions. Old World NHPs also are invaluable models for research on multipotent adult stem cells, which exist throughout the body after development.
Potential Therapy Involving Bone Marrow Stem Cells (BMSCs)
BMSCs are continuously released from bone marrow and serve to repair damage to the vascular endothelium, and they are released in large quantities in response to acute, severe damage such as occurs during myocardial infarction. They also are released in large numbers in response to some cytokines, particularly granulocyte colony-stimulating factor (G-CSF), which has been extensively used clinically to harvest BMSCs for autologous transplantation to treat patients immediately after myocardial infarction. However, treatment of patients with BMSCs released in response to G-CSF has had inconsistent outcomes and modest success, at best (Fisher et al. 2015; Gyöngyösi et al. 2016). Shi et al. (Shi et al. 2004) hypothesized that BMSCs mobilized by G-CSF may not have the same therapeutic capacity as those mobilized naturally in response to acute, severe vascular damage. They used a baboon model to compare the characteristics of BMSCs isolated from the circulation of five baboons treated with G-CSF to those of BMSCs isolated from the circulation of five baboons from which a section of femoral artery had been removed. They observed that the BMSC populations released in response to the two experimental interventions had different characteristics and that the cells released in response to femoral artery ligation have higher capacity for vascular differentiation, suggesting that alternative strategies for inducing the release of BMSCs for clinical purposes might be more therapeutically effective than administration of G-CSF. This result illustrates the value of NHP models for developing applications of adult stem cell therapies for cardiovascular therapies.
Potential Therapy for Repair of Vascular Endothelium
Damage to the arterial endothelium is a critical early event in atherogenesis and peripheral vascular disease, and deterioration of the vascular endothelium occurs naturally throughout life. It follows, then, that stem cell therapies might be used in treating disorders in which the endothelium has been damaged or is deteriorating (see Chong et al. 2016). Toward that end, Shi et al. (Shi et al. 2012, 2013) induced baboon ESCs to differentiate into CD34+, CD31−, and CD146− endothelial progenitor cells (multi-potential hematopoietic stem cells), labelled them with a fluorescent tag, and inoculated them into an arterial segment that had been denuded of endothelial cells in an ex vivo system. By 14 days postinoculation, the cells had attached and integrated into the denuded surface and had undergone maturation events that led to the expression of CD31 and CD146 (antigens expressed on mature endothelial cells). This experiment provided proof-of-concept and paves the way for in vivo experiments that will be conducted with baboons in which the arterial endothelium has been damaged by a balloon catheter. Although the inoculated stem cells will be broadly distributed throughout the vasculature (unlike in the ex vivo model, which involved only a segment of an artery), it is expected that the stem cells will home to and preferentially attach to the injured site, as occurs when BMSCs are released into the circulation after vascular injury.
Construction of Bioengineered Arterial Segments
A much more ambitious potential use of pluripotent stem cells is the construction of bioengineered arterial segments that could be used to replace diseased sections of arteries. The concept is to develop separate lineages of endothelial progenitor cells and smooth muscle progenitor cells, which can be used to seed the inside and the outside, respectively, of a segment of tubular scaffold placed in a bioreactor in which natural conditions on the inside and the outside of an artery wall are simulated. The scaffold can be constructed of a biodegradable material, with a lumen and wall thickness of the artery intended to be replaced. As endothelial cells differentiate on the interior surface and smooth muscle cells differentiate on the exterior surface, the scaffold degrades at the rate for which it was engineered, leaving a bioengineered arterial segment that could be surgically implanted in a patient (or the scaffold could be bioengineered to degrade after implantation and in vivo remodeling of the arterial segment). Progress toward this goal has been made by Q. Shi and J. L. VandeBerg (unpublished data) using baboon ESCs, although the segments produced in initial in vitro experiments were not sufficiently robust for transitioning to in vivo experiments.
Regeneration of Heart Tissue After Myocardial Infarction
Another potential use of stem cell therapy is the regeneration of heart tissue after myocardial infarction to prevent or to treat heart failure as a consequence of the death of myocardial cells. Toward that goal, Shiba et al. (Shiba et al. 2016) induced cynomolgus macaque iPSCs with a transfected fluorescent marker to develop into cardiomyocytes, and they injected the cells into the hearts of four other monkeys after myocardial infarction had been experimentally induced. The monkeys were immunosuppressed with a clinically relevant regimen to protect against immunological rejection. Twelve weeks after transplantation, when the monkeys were killed at the experimental endpoint, the fluorescent cells had contributed to partial remuscularization of the scar area, and the regenerated tissue was well vascularized. Moreover, contractile function of the hearts was improved at the endpoint and was already improved by 4 weeks after transplantation, by comparison with control animals. Although transient ventricular tachycardia was significantly increased in the treated animals, the results of this experiment clearly establish the potential for the therapeutic use of pluripotent stem cells for treating myocardial infarction patients.
IPSCs Versus ESCs for Cardiovascular Therapies
On the surface, it might seem that iPSCs would be superior to ESCs for developing therapies for CVD, because autologous cells are likely to be less prone to histocompatibility problems (even though there may be some histocompatibility differences between iPSC-derived cells and natural host cells). However, histocompatibility problems can be overcome relatively easily, as they are for every type of organ transplant. The fundamental problems in using iPSCs rather than ESCs are the financial and temporal costs. Developing an iPSC line from every patient would be financially costly given that each line must be characterized in detail for many properties, including chromosomal aberrations and oncogenic potential. Even more important is the temporal cost when the intent is to treat an acute condition such as myocardial infarction. However, ESCs from well-characterized lines that have been rigorously established to be safe could be used to develop standardized off-the-shelf reagents (e.g., cardiomyocytes, endothelial progenitor cells, and arterial segments of various lengths and diameters) that could be administered immediately to patients who experience an acute cardiovascular problem. While histo-incompatibility could be judiciously managed by standard immunosuppression regimens alone, the problems of histo-incompatiblity could be further diminished by creating the reagents from a panel of ESCs derived from different embryos selected to have a broad representation of the more common major histocompatibility complex haplotypes. Of course, such a panel of well-characterized and safe pluripotent stem cells representing many different histocompatibility complex haplotypes could be created from iPSCs, but the concept of treating each patient with autologous iPSC-derived cells appears not to be a practical goal. Therefore, it is suggested that researchers focus on ESCs in NHP research aimed at developing CVD therapies.
Detection and Prevention of CVD
Despite recent scientific advancements, treatment options, including statins, angiotensin-converting enzyme inhibitors, beta blockers, and other drugs, the prevalence of CVD continues to increase, underscoring the need for new therapeutic strategies (van Rooij and Olson 2007). In this section we discuss scientific advancements in discovery of therapies for preventing CVD and the fundamental role of NHP models.
HMG-CoA Reductase Inhibitor (Statins)
Statins are synthetic molecules that bind and decrease the activity of HMG-CoA reductase, a rate-limiting enzyme in cholesterol biosynthesis pathway. Statins are the most widely used therapy for lowering plasma LDL-C (Ramkumar et al. 2016). There is a strong body of evidence that indicates statin therapy has significant mortality and morbidity benefits for both primary and secondary prevention from CVD. Moreover, studies have demonstrated that statins are potent in regressing atherosclerosis and improving endothelium function by decreasing LDL oxidation, reducing smooth muscle cells migration and proliferation, activation of monocytes to macrophages, inhibition of cytokine production, and reduction in adhesion of monocytes to arterial walls (Cerda et al. 2015; Koh 2000; Treasure et al. 1995). Despite these intriguing findings, the direct mechanistic assessment of statin potency beyond lipid lowering presents considerable obstacles, because it is not feasible to obtain target tissues from healthy humans. Previous studies confirmed that statins improve endothelial function in cynomolgus macaques similarly to humans (Sukhova et al. 2002; Williams et al. 1998). One difference between human and cynomolgus macaques was the effect of statins on smooth muscle cell content in plaques; smooth muscle cells are a source of extracellular matrices and cytokine production. The study in cynomolgus macaques revealed increased smooth muscle cell content in plaques, while human studies suggested that statin therapy decreased proliferation of smooth muscle cell in plaques (Corpataux et al. 2005; Sukhova et al. 2002). This apparent difference between humans and cynomolgus macaques merits further investigation.
Despite the positive effect of statins on cardiovascular end-points, there are concerns regarding the adverse effects of statin therapy, including myalgia, rhabdomyolysis, liver toxicity, and diabetes (Rha et al. 2016; Russo et al. 2014; Yokote et al. 2011). NHP models of atherosclerosis provide an opportunity to comprehensively investigate the side effects of statins and the underlying mechanisms.
PCSK9 Inhibitor (Evolocumab)
The proprotein convertase subtilisin/kexin type 9 (PCSK9) inhibitor Evolocumab is a recently discovered monoclonal antibody therapy for lowering plasma LDL-C. Interest in PCSK9 emerged after it was found that individuals with a nucleotide variant in PCSK9 had low plasma LDL-C (Horton et al. 2007). PCSK9 asserts its function by binding to and degrading the LDL receptor (LDLR), impacting the LDLR recycling process necessary for cholesterol absorption in hepatocytes. Evolocumab binds to the PCSK9 domain, preventing LDLR-PCSK9 interaction and subsequent degradation of LDLR. Evolocumab reduces LDL-C by nearly 50% and >60% when combined with statins. This therapy costs approximately $1400 per patient per year, so it is recommended only for individuals at high risk who cannot tolerant statins or statins are not effective in lowering LDL-C.
The potency of Evolocumab has been evaluated in NHPs, it was shown to lower LDL-C in healthy and in diet-induced hypercholesterolemic cynomolgus macaques (Liang et al. 2012). In addition, Evolocumab therapy significantly lowered LDL-C in monkeys treated with statins. Currently there is inadequate information about PCSK9 therapy, including the long-term CVD effects, the age range for treatment, and interaction with other therapies. NHP models provide an opportunity to evaluate the age range of treatment, drug interaction, toxicity, and effects on atherosclerosis. Studies with NHPs may be helpful in supporting the new paradigm of time-limited but very intensive LDL-C lowering by PCSK9 inhibitor. Studies evaluating plaque morphology after the discontinuation of aggressive lipid-lowering therapy while maintained on background statin therapy will be an important area for future investigation.
Other lipid-lowering therapies that have not been evaluated in NHPs include apoB and microsomal triglyceride transfer protein and NPC1L1 inhibitor (Ezetimibe). These therapies are designed to target genes involved in lipid metabolic pathways.
The Potential of Genome Editing to Validate and Understand Mechanisms of Functional Genetic Variants
Development of programmable nuclease-based genome-editing technologies, among which CRISPR (clustered regularly interspaced short palindromic repeats)/Cas9 is currently the most prevalent, enables targeted and efficient modification of almost any genome. This approach provides the means to genetically manipulate cells and whole organisms to test the function of genetic variants. Recent studies by Guo and Li (Guo and Li 2015) demonstrate feasibility of gene knockout by CRISPR/Cas9 in rhesus macaques, indicating that this system can be used to test candidate functional variants in NHPs, overcoming one of the major limitations of working with NHP models by comparison with mice. However, the quality of NHP genome annotations currently limits the ability to design guide RNAs for genetic manipulation. With improved quality of NHP genome annotation, NHP genomic manipulation studies will likely progress rapidly, providing the means to assess mechanisms by which functional genetic variants impact cell and organism phenotype (Luo et al. 2016).
Conclusions
This is an exciting time in biomedical research with availability of technologies to quantify molecular changes at the cellular level, bioinformatics tools to provide biological frameworks to understand the impact of molecular activities, the ability to genetically manipulate cells to test functional variants and functional networks, and the capacity to generate complex tissues and organs from pluripotent stem cells. In addition, progress in the field of organ/tissue growth in the laboratory provides the means to move from in vitro cell culture experiments to more complex systems where cell-cell interactions and communication can be studied. NHP research completes the overarching research paradigm by allowing investigation of these complex systems in a whole animal that closely resembles humans in metabolism, physiology, genetics, and CVD risk factors. The NHP whole-animal model provides the opportunity to develop a better understanding of biological mechanisms involved in CVD by unraveling the complexities of system-wide organization and communication.
References
- Alfaradhi M, Ozanne S. 2011. Frontiers: Developmental programming in response to maternal overnutrition. Front Genet 2:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral WZ, Lubach GR, Proctor A, Lyte M, Phillips GJ, Coe CL. 2017. Social influences on prevotella and the gut microbiome of young monkeys. Psychosom Med. doi:10.1097/PSY.0000000000000454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato KR. 2016. Incorporating the gut microbiota into models of human and non-human primate ecology and evolution. Am J Phys Anthropol 159:S196–S215. [DOI] [PubMed] [Google Scholar]
- Amato KR, Leigh SR, Kent A, Mackie RI, Yeoman CJ, Stumpf RM, Wilson BA, Nelson KE, White BA, Garber PA. 2015. The gut microbiota appears to compensate for seasonal diet variation in the wild black howler monkey (Alouatta pigra). Microb Ecol 69:434–443. [DOI] [PubMed] [Google Scholar]
- Amato KR, Yeoman CJ, Cerda G, Schmitt CA, Cramer JD, Miller ME, Gomez A, Turner TR, Wilson BA, Stumpf RM, Nelson KE, White BA, Knight R, Leigh SR. 2015. Variable responses of human and non-human primate gut microbiomes to a western diet. Microbiome 3:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V. 2001. MicroRNAs: Tiny regulators with great potential. Cell 107:823–826. [DOI] [PubMed] [Google Scholar]
- Ambros V. 2004. The functions of animal micrornas. Nature 431:350–355. [DOI] [PubMed] [Google Scholar]
- Ardeshir A, Narayan NR, Méndez-Lagares G, Lu D, Rauch M, Huang Y, Van Rompay KK, Lynch SV, Hartigan-O'Connor DJ. 2014. Breast-fed and bottle-fed infant rhesus macaques develop distinct gut microbiotas and immune systems. Sci Transl Med 6:252ra120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aron-Wisnewsky J, Clément K. 2016. The gut microbiome, diet, and links to cardiometabolic and chronic disorders. Nat Rev Nephrol 12:169–181. [DOI] [PubMed] [Google Scholar]
- Barker DJ, Winter PD, Osmond C, Margetts B, Simmonds SJ. 1989. Weight in infancy and death from ischaemic heart disease. Lancet 2:577–580. [DOI] [PubMed] [Google Scholar]
- Bell LN, Lee L, Saxena R, Bemis KG, Wang M, Theodorakis JL, Vuppalanchi R, Alloosh M, Sturek M, Chalasani N. 2010. Serum proteomic analysis of diet-induced steatohepatitis and metabolic syndrome in the ossabaw miniature swine. Am J Physiol Gastrointest Liver Physiol 298:G746–G754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergen WG, Mersmann HJ. 2005. Comparative aspects of lipid metabolism: Impact on contemporary research and use of animal models. J Nutr 135:2499–2502. [DOI] [PubMed] [Google Scholar]
- Bhatnagar D. 2015. Gut flora, diet and intestinal metabolism on cardiovascular risk. Curr Opin Lipidol 26:148–149. [DOI] [PubMed] [Google Scholar]
- Bjarnegård N, Morsing E, Cinthio M, Länne T, Brodszki J. 2013. Cardiovascular function in adulthood following intrauterine growth restriction with abnormal fetal blood flow. Ultrasound Obstet Gynecol 41:177–184. [DOI] [PubMed] [Google Scholar]
- Blangero J, MacCluer JW, Kammerer CM, Mott GE, Dyer TD, McGill HC. 1990. Genetic analysis of apolipoprotein A-I in two dietary environments. Am J Hum Genet 47:414–428. [PMC free article] [PubMed] [Google Scholar]
- Bo X, Zun-xi H, Xiao-yan W, Run-chi G, Xiang-hua T, Yue-lin M, Yun-Juan Y, Hui S, Li-da Z. 2010. Phylogenetic analysis of the fecal flora of the wild pygmy loris. Am J Primatol 72:699–706. [DOI] [PubMed] [Google Scholar]
- Boney CM, Verma A, Tucker R, Vohr BR. 2005. Metabolic syndrome in childhood: Association with birth weight, maternal obesity, and gestational diabetes mellitus. Pediatrics 115:e290–e296. [DOI] [PubMed] [Google Scholar]
- Bose T, Voruganti VS, Tejero ME, Proffitt JM, Cox LA, VandeBerg JL, Mahaney MC, Rogers J, Freeland-Graves JH, Cole SA, Comuzzie AG. 2009. Quantitative loci regulating plasma levels of gamma glutamyl transferase and albumin and their genetic correlations with cardiovascular risk factors. Exp Biol Med (Maywood) 234:vi, 1519–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho CM, Andrade MC, Xavier SS, Mangia RH, Britto CC, Jansen AM, Fernandes O, Lannes-Vieira J, Bonecini-Almeida MG. 2003. Chronic chagas’ disease in rhesus monkeys (Macaca mulatta): Evaluation of parasitemia, serology, electrocardiography, echocardiography, and radiology. Am J Trop Med Hyg 68:683–691. [PubMed] [Google Scholar]
- Cefalu WT, Wagner JD, Bell-Farrow AD, Edwards IJ, Terry JG, Weindruch R, Kemnitz JW. 1999. Influence of caloric restriction on the development of atherosclerosis in nonhuman primates: Progress to date. Toxicol Sci 52:49–55. [DOI] [PubMed] [Google Scholar]
- Cerda A, Rodrigues AC, Alves C, Genvigir FD, Fajardo CM, Dorea EL, Gusukuma MC, Pinto GA, Hirata MH, Hirata RD. 2015. Modulation of adhesion molecules by cholesterol-lowering therapy in mononuclear cells from hypercholesterolemic patients. Cardiovasc Ther 33:168–176. [DOI] [PubMed] [Google Scholar]
- Chan J, Sharkey FE, Kushwaha RS, VandeBerg JF, VandeBerg JL. 2012. Steatohepatitis in laboratory opossums exhibiting a high lipemic response to dietary cholesterol and fat. Am J Physiol Gastrointest Liver Physiol 303:G12–G19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Li C, McDonald TJ, Comuzzie A, Mattern V, Nathanielsz PW. 2011. Emergence of insulin resistance in juvenile baboon offspring of mothers exposed to moderate maternal nutrient reduction. Am J Physiol Regul Integr Comp Physiol 301:R757–R762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong MS, Ng WK, Chan JK. 2016. Concise review: Endothelial progenitor cells in regenerative medicine: Applications and challenges. Stem Cells Transl Med 5:530–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu DM, Antony KM, Ma J, Prince AL, Showalter L, Moller M, Aagaard KM. 2016. The early infant gut microbiome varies in association with a maternal high-fat diet. Genome Med 8:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton JB, Vangay P, Huang H, Ward T, Hillmann BM, Al-Ghalith GA, Travis DA, Long HT, Tuan BV, Minh VV, Cabana F, Nadler T, Toddes B, Murphy T, Glander KE, Johnson TJ, Knights D. 2016. Captivity humanizes the primate microbiome. Proc Natl Acad Sci USA 113:10376–10381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JC, Horton JD, Hobbs HH. 2011. Human fatty liver disease: Old questions and new insights. Science 332:1519–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman RJ, Anderson RM, Johnson SC, Kastman EK, Kosmatka KJ, Beasley TM, Allison DB, Cruzen C, Simmons HA, Kemnitz JW, Weindruch R. 2009. Caloric restriction delays disease onset and mortality in rhesus monkeys. Science 325:201–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman RJ, Beasley TM, Kemnitz JW, Johnson SC, Weindruch R, Anderson RM. 2014. Caloric restriction reduces age-related and all-cause mortality in rhesus monkeys. Nat Commun 5:3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corey KE, Vuppalanchi R, Wilson LA, Cummings OW, Chalasani N NASH Consortium . 2015. NASH resolution is associated with improvements in HDL and triglyceride levels but not improvement in LDL or non-HDL-C levels. Aliment Pharmacol Ther 41:301–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpataux JM, Naik J, Porter KE, London NJ. 2005. The effect of six different statins on the proliferation, migration, and invasion of human smooth muscle cells. J Surg Res 129:52–56. [DOI] [PubMed] [Google Scholar]
- Cox LA, Birnbaum S, Mahaney MC, Rainwater DL, Williams JT, VandeBerg JL. 2007. Identification of promoter variants in baboon endothelial lipase that regulate high-density lipoprotein cholesterol levels. Circulation 116:1185–1195. [DOI] [PubMed] [Google Scholar]
- Cox LA, Comuzzie AG, Havill LM, Karere GM, Spradling KD, Mahaney MC, Nathanielsz PW, Nicolella DP, Shade RE, Voruganti S, VandeBerg JL. 2013. Baboons as a model to study genetics and epigenetics of human disease. ILAR J 54:106–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox LA, Nijland MJ, Gilbert JS, Schlabritz-Loutsevitch NE, Hubbard GB, McDonald TJ, Shade RE, Nathanielsz PW. 2006. Effect of 30% maternal nutrient restriction from 0.16 to 0.5 gestation on fetal baboon kidney gene expression. J Physiol 572:67–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cydylo MA, Davis AT, Kavanagh K. 2017. Fatty liver promotes fibrosis in monkeys consuming high fructose. Obesity (Silver Spring) 25:290–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. 2014. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505:559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan PH, Pusey AE, Lonsdorf EV, Goodall J, Wroblewski EE, Wilson ML, Rudicell RS, Hahn BH, Ochman H. 2012. Factors associated with the diversification of the gut microbial communities within chimpanzees from Gombe National Park. Proc Natl Acad Sci USA 109:13034–13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dela Peña A, Leclercq I, Field J, George J, Jones B, Farrell G. 2005. NF-kappaB activation, rather than TNF, mediates hepatic inflammation in a murine dietary model of steatohepatitis. Gastroenterology 129:1663–1674. [DOI] [PubMed] [Google Scholar]
- Del Chierico F, Vernocchi P, Dallapiccola B, Putignani L. 2014. Mediterranean diet and health: Food effects on gut microbiota and disease control. Int J Mol Sci 15:11678–11699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- do Rosario VA, Fernandes R, Trindade EB. 2016. Vegetarian diets and gut microbiota: Important shifts in markers of metabolism and cardiovascular disease. Nutr Rev 74:444–454. [DOI] [PubMed] [Google Scholar]
- Dong M, Zheng Q, Ford SP, Nathanielsz PW, Ren J. 2013. Maternal obesity, lipotoxicity and cardiovascular diseases in offspring. J Mol Cell Cardiol 55:111–116. [DOI] [PubMed] [Google Scholar]
- Drake AJ, Reynolds RM. 2010. Impact of maternal obesity on offspring obesity and cardiometabolic disease risk. Reproduction 140:387–398. [DOI] [PubMed] [Google Scholar]
- Eggen DA. 1974. Cholesterol metabolism in rhesus monkey, squirrel monkey, and baboon. J Lipid Res 15:139–145. [PubMed] [Google Scholar]
- Ellis RJ, Bruce KD, Jenkins C, Stothard JR, Ajarova L, Mugisha L, Viney ME. 2013. Comparison of the distal gut microbiota from people and animals in Africa. PLoS One 8:e54783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson JF, Allayee H, Gerszten RE, Ideraabdullah F, Kris-Etherton PM, Ordovás JM, Rimm EB, Wang TJ, Bennett BJ and American Heart Association Council on Functional Genomics and Translational Biology, Council on Epidemiology and Prevention, and Stroke Council . 2016. Nutrigenomics, the microbiome, and gene-environment interactions: New directions in cardiovascular disease research, prevention, and treatment: A scientific statement from the American Heart Association. Circ Cardiovasc Genet 9:291–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher SA, Doree C, Mathur A, Martin-Rendon E. 2015. Meta-analysis of cell therapy trials for patients with heart failure. Circ Res 116:1361–1377. [DOI] [PubMed] [Google Scholar]
- Flint HJ, Duncan SH, Scott KP, Louis P. 2007. Interactions and competition within the microbial community of the human colon: Links between diet and health. Environ Microbiol 9:1101–1111. [DOI] [PubMed] [Google Scholar]
- Fouzas S, Karatza AA, Davlouros PA, Chrysis D, Alexopoulos D, Mantagos S, Dimitriou G. 2014. Neonatal cardiac dysfunction in intrauterine growth restriction. Pediatr Res 75:651–657. [DOI] [PubMed] [Google Scholar]
- Frey JC, Rothman JM, Pell AN, Nizeyi JB, Cranfield MR, Angert ER. 2006. Fecal bacterial diversity in a wild gorilla. Appl Environ Microbiol 72:3788–3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin JL, Wang X, Stanley E. 2015. Does our gut microbiome predict cardiovascular risk? A review of the evidence from metabolomics. Circ Cardiovasc Genet 8:187–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Li XJ. 2015. Targeted genome editing in primate embryos. Cell Res 25:767–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyöngyösi M, Wojakowski W, Navarese EP, Moye LÀ and ACCRUE Investigators . 2016. Meta-Analyses of human cell-based cardiac regeneration therapies: Controversies in meta-analyses results on cardiac cell-based regenerative studies. Circ Res 118:1254–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghikia A, Landmesser U. 2015. Emerging role of the gut microbiome for cardiovascular disease. Eur Heart J 36:3130–3132. [PubMed] [Google Scholar]
- Hanson MA, Gluckman PD. 2015. Developmental origins of health and disease—global public health implications. Best Pract Res Clin Obstet Gynaecol 29:24–31. [DOI] [PubMed] [Google Scholar]
- Harris RA, Alcott CE, Sullivan EL, Takahashi D, McCurdy CE, Comstock S, Baquero K, Blundell P, Frias AE, Kahr M, Suter M, Wesolowski S, Friedman JE, Grove KL, Aagaard KM. 2016. Genomic variants associated with resistance to high fat diet induced obesity in a primate model. Sci Rep 6:36123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins PB, Bastarrachea RA, Lopez-Alvarenga JC, Garcia-Forey M, Proffitt JM, Voruganti VS, Tejero ME, Mattern V, Haack K, Shade RE, Cole SA, Comuzzie AG. 2010. Eight week exposure to a high sugar high fat diet results in adiposity gain and alterations in metabolic biomarkers in baboons (Papio hamadryas sp.). Cardiovasc Diabetol 9:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins PB, Rodriguez PJ, Voruganti VS, Mattern V, Bastarrachea RA, Rice K, Raabe T, Comuzzie AG. 2014. Body composition and cardiometabolic disease risk factors in captive baboons (Papio hamadryas sp.): Sexual dimorphism. Am J Phys Anthropol 153:9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hold GL. 2014. Western lifestyle: A ‘master’ manipulator of the intestinal microbiota? Gut 63:5–6. [DOI] [PubMed] [Google Scholar]
- Horton JD, Cohen JC, Hobbs HH. 2007. Molecular biology of PCSK9: Its role in LDL metabolism. Trends Biochem Sci 32:71–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter MP, Ismail N, Zhang X, Aguda BD, Lee EJ, Yu L, Xiao T, Schafer J, Lee ML, Schmittgen TD, Nana-Sinkam SP, Jarjoura D, Marsh CB. 2008. Detection of microrna expression in human peripheral blood microvesicles. PLoS One 3:e3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janiak MC. 2016. Digestive enzymes of human and nonhuman primates. Evol Anthropol 25:253–266. [DOI] [PubMed] [Google Scholar]
- Jones A, Beda A, Osmond C, Godfrey KM, Simpson DM, Phillips DI. 2008. Sex-specific programming of cardiovascular physiology in children. Eur Heart J 29:2164–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath S, Chavez AO, Gastaldelli A, Casiraghi F, Halff GA, Abrahamian GA, Davalli AM, Bastarrachea RA, Comuzzie AG, Guardado-Mendoza R, Jimenez-Ceja LM, Mattern V, Paez AM, Ricotti A, Tejero ME, Higgins PB, Rodriguez-Sanchez IP, Tripathy D, DeFronzo RA, Dick EJ Jr, Cline GW, Folli F. 2011. Coordinated defects in hepatic long chain fatty acid metabolism and triglyceride accumulation contribute to insulin resistance in non-human primates. PLoS One 6:e27617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammerer CM, Rainwater DL, Schneider JL, Cox LA, Mahaney MC, Rogers J, VandeBerg JF. 2003. Two loci affect angiotensin i-converting enzyme activity in baboons. Hypertension 41:854–859. [DOI] [PubMed] [Google Scholar]
- Karere GM, Glenn JP, Birnbaum S, Rainwater DL, Mahaney MC, VanderBerg JL, Cox LA. 2013. Identification of candidate genes encoding an LDL-C QTL in baboons. J Lipid Res 54:1776–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karere GM, Glenn JP, VandeBerg JL, Cox LA. 2012. Differential microrna response to a high-cholesterol, high-fat diet in livers of low and high LDL-C baboons. BMC Genomics 13:320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemnitz JW. 2011. Calorie restriction and aging in nonhuman primates. ILAR J 52:66–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketloy C, Engering A, Srichairatanakul U, Limsalakpetch A, Yongvanitchit K, Pichyangkul S, Ruxrungtham K. 2008. Expression and function of toll-like receptors on dendritic cells and other antigen presenting cells from non-human primates. Vet Immunol Immunopathol 125:18–30. [DOI] [PubMed] [Google Scholar]
- Khare S, Liu X, Stinson M, Rivera I, Groessl T, Tuntland T, Yeh V, Wen B, Molteni V, Glynne R, Supek F. 2015. Antitrypanosomal treatment with benznidazole is superior to posaconazole regimens in mouse models of Chagas disease. Antimicrob Agents Chemother 59:6385–6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisidayová S, Váradyová Z, Pristas P, Piknová M, Nigutová K, Petrzelková KJ, Profousová I, Schovancová K, Kamler J, Modrý D. 2009. Effects of high- and low-fiber diets on fecal fermentation and fecal microbial populations of captive chimpanzees. Am J Primatol 71:548–557. [DOI] [PubMed] [Google Scholar]
- Kochunov P, Wey HY, Fox PT, Lancaster JL, Davis MD, Wang DJ, Lin AL, Bastarrachea RA, Andrade MC, Mattern V, Frost P, Higgins PB, Comuzzie AG, Voruganti VS. 2017. Changes in cerebral blood flow during an alteration in glycemic state in a large non-human primate (Papio hamadryas sp.). Front Neurosci 11:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh KK. 2000. Effects of statins on vascular wall: Vasomotor function, inflammation, and plaque stability. Cardiovasc Res 47:648–657. [DOI] [PubMed] [Google Scholar]
- Kramer JA, Grindley J, Crowell AM, Makaron L, Kohli R, Kirby M, Mansfield KG, Wachtman LM. 2015. The common marmoset as a model for the study of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Vet Pathol 52:404–413. [DOI] [PubMed] [Google Scholar]
- Kronenberg F, Steinmetz A, Kostner GM, Dieplinger H. 1996. Lipoprotein(a) in health and disease. Crit Rev Clin Lab Sci 33:495–543. [DOI] [PubMed] [Google Scholar]
- Kulkarni H, Meikle PJ, Mamtani M, Weir JM, Almeida M, Diego V, Peralta JM, Barlow CK, Bellis C, Dyer TD, Almasy L, Mahaney MC, Comuzzie AG, Göring HH, Curran JE, Blangero J. 2014. Plasma lipidome is independently associated with variability in metabolic syndrome in mexican american families. J Lipid Res 55:939–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo AH, Li C, Li J, Huber HF, Nathanielsz PW, Clarke GD. 2016. Cardiac remodelling in a baboon model of intrauterine growth restriction mimics accelerated ageing. J Physiol 595:1093–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo AH, Li C, Li J, Huber HF, Nathanielsz PW, Clarke GD. 2017. Maternal nutrient restriction during pregnancy and lactation leads to impaired right ventricular function in young adult baboons. J Physiol 595:4245–4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushwaha RS, McGill HC. 1998. Diet, plasma lipoproteins and experimental atherosclerosis in baboons (papio sp.). Hum Reprod Update 4:420–429. [DOI] [PubMed] [Google Scholar]
- Kushwaha RS, Reardon CA, Getz GS, Lewis DS, Rice KS, Carey KD, McGill HC. 1994. Metabolic mechanisms for responses to dietary cholesterol and fat in high and low LDL responding baboons (papio sp.). J Lipid Res 35:633–643. [PubMed] [Google Scholar]
- Lane MA, Ingram DK, Cutler RG, Knapka JJ, Barnard DE, Roth GS. 1992. Dietary restriction in nonhuman primates: Progress report on the NIA study. Ann NY Acad Sci 673:36–45. [DOI] [PubMed] [Google Scholar]
- Lane MA, Ingram DK, Roth GS. 1999. Calorie restriction in nonhuman primates: Effects on diabetes and cardiovascular disease risk. Toxicol Sci 52:41–48. [DOI] [PubMed] [Google Scholar]
- Langley-Evans SC. 2015. Nutrition in early life and the programming of adult disease: A review. J Hum Nutr Diet 28Suppl 1:1–14. [DOI] [PubMed] [Google Scholar]
- Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, Gordon JI. 2008. Evolution of mammals and their gut microbes. Science 320:1647–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Schlabritz-Loutsevitch NE, Hubbard GB, Han V, Nygard K, Cox LA, McDonald TJ, Nathanielsz PW. 2009. Effects of maternal global nutrient restriction on fetal baboon hepatic insulin-like growth factor system genes and gene products. Endocrinology 150:4634–4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Sloboda DM, Vickers MH. 2011. Maternal obesity and developmental programming of metabolic disorders in offspring: Evidence from animal models. Exp Diabetes Res 2011:592408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SJ, Ding ST, Mersmann HJ, Chu CH, Hsu CD, Chen CY. 2016. A nutritional nonalcoholic steatohepatitis minipig model. J Nutr Biochem 28:51–60. [DOI] [PubMed] [Google Scholar]
- Liang H, Chaparro-Riggers J, Strop P, Geng T, Sutton JE, Tsai D, Bai L, Abdiche Y, Dilley J, Yu J, Wu S, Chin SM, Lee NA, Rossi A, Lin JC, Rajpal A, Pons J, Shelton DL. 2012. Proprotein convertase substilisin/kexin type 9 antagonism reduces low-density lipoprotein cholesterol in statin-treated hypercholesterolemic nonhuman primates. J Pharmacol Exp Ther 340:228–236. [DOI] [PubMed] [Google Scholar]
- Luo X, Li M, Su B. 2016. Application of the genome editing tool CRISPR/cas9 in non-human primates. Dongwuxue Yanjiu 37:214–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Prince AL, Bader D, Hu M, Ganu R, Baquero K, Blundell P, Alan Harris R, Frias AE, Grove KL, Aagaard KM. 2014. High-fat maternal diet during pregnancy persistently alters the offspring microbiome in a primate model. Nat Commun 5:3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahaney MC, Blangero J, Rainwater DL, Mott GE, Comuzzie AG, MacCluer JW, VandeBerg JL. 1999. Pleiotropy and genotype by diet interaction in a baboon model for atherosclerosis: A multivariate quantitative genetic analysis of HDL subfractions in two dietary environments. Arterioscler Thromb Vasc Biol 19:1134–1141. [DOI] [PubMed] [Google Scholar]
- Mahaney MC, Karere GM, Rainwater DL, Voruganti VS, Dick EJ Jr, Owston MA, Rice KS, Cox LA, Comuzzie AG, VandeBerg JL. 2017. Diet-induced early-stage atherosclerosis in baboons: Lipoproteins, atherogenesis, and arterial compliance. J Med Primatol. doi:10.1111/jmp.12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloyan A, Muralimanoharan S, Huffman S, Cox LA, Nathanielsz PW, Myatt L, Nijland MJ. 2013. Identification and comparative analyses of myocardial mirnas involved in the fetal response to maternal obesity. Physiol Genomics 45:889–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzel A, Muller DN, Hafler DA, Erdman SE, Linker RA, Kleinewietfeld M. 2014. Role of “Western diet” in inflammatory autoimmune diseases. Curr Allergy Asthma Rep 14:404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattison JA, Roth GS, Beasley TM, Tilmont EM, Handy AM, Herbert RL, Longo DL, Allison DB, Young JE, Bryant M, Barnard D, Ward WF, Qi W, Ingram DK, de Cabo R. 2012. Impact of caloric restriction on health and survival in rhesus monkeys from the NIA study. Nature 489:318–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCord AI, Chapman CA, Weny G, Tumukunde A, Hyeroba D, Klotz K, Koblings AS, Mbora DN, Cregger M, White BA, Leigh SR, Goldberg TL. 2014. Fecal microbiomes of non-human primates in western Uganda reveal species-specific communities largely resistant to habitat perturbation. Am J Primatol 76:347–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCurdy CE, Bishop JM, Williams SM, Grayson BE, Smith MS, Friedman JE, Grove KL. 2009. Maternal high-fat diet triggers lipotoxicity in the fetal livers of nonhuman primates. J Clin Invest 119:323–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill HC, McMahan CA, Kruski AW, Kelley JL, Mott GE. 1981. Responses of serum lipoproteins to dietary cholesterol and type of fat in the baboon. Arteriosclerosis 1:337–344. [DOI] [PubMed] [Google Scholar]
- McGill HCJ, McMahan CA, Kruski AW, Mott GE. 1981. Relationship of lipoprotein cholesterol concentrations to experimental atherosclerosis in baboons. Arteriosclerosis 1:3–12. [DOI] [PubMed] [Google Scholar]
- McGill HC, Strong JP, Holman RL, Werthessen NT. 1960. Arterial lesions in the kenya baboon. Circ Res 8:670–679. [Google Scholar]
- McMullen S, Mostyn A. 2009. Animal models for the study of the developmental origins of health and disease. Proc Nutr Soc 68:306–320. [DOI] [PubMed] [Google Scholar]
- Mell B, Jala VR, Mathew AV, Byun J, Waghulde H, Zhang Y, Haribabu B, Vijay-Kumar M, Pennathur S, Joe B. 2015. Evidence for a link between gut microbiota and hypertension in the Dahl rat. Physiol Genomics 47:187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KA, Bennett BJ. 2016. Diet and gut microbial function in metabolic and cardiovascular disease risk. Curr Diab Rep 16:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miele L, Giorgio V, Alberelli MA, De Candia E, Gasbarrini A, Grieco A. 2015. Impact of gut microbiota on obesity, diabetes, and cardiovascular disease risk. Curr Cardiol Rep 17:120. [DOI] [PubMed] [Google Scholar]
- Miller MJ. 2013. Risk factors for cardiovascular disease: A cautionary tale of diet-microbiome interactions. J Am Coll Nutr 32:75–78. [DOI] [PubMed] [Google Scholar]
- Moeller AH, Degnan PH, Pusey AE, Wilson ML, Hahn BH, Ochman H. 2012. Chimpanzees and humans harbour compositionally similar gut enterotypes. Nat Commun 3:1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller AH, Ochman H. 2013. Factors that drive variation among gut microbial communities. Gut Microbes 4:403–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina I, Gómez i Prat J, Salvador F, Treviño B, Sulleiro E, Serre N, Pou D, Roure S, Cabezos J, Valerio L, Blanco-Grau A, Sánchez-Montalvá A, Vidal X, Pahissa A. 2014. Randomized trial of posaconazole and benznidazole for chronic chagas’ disease. N Engl J Med 370:1899–1908. [DOI] [PubMed] [Google Scholar]
- Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, Das SR, de Ferranti S, Després J-P, Fullerton HJ. 2016. Executive summary: Heart disease and stroke statistics-2016 update: A report from the american heart association. Circulation 133:447. [DOI] [PubMed] [Google Scholar]
- Narayan NR, Méndez-Lagares G, Ardeshir A, Lu D, Van Rompay KK, Hartigan-O'Connor DJ. 2015. Persistent effects of early infant diet and associated microbiota on the juvenile immune system. Gut Microbes 6:284–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathanielsz PW, Poston L, Taylor PD. 2007. In utero exposure to maternal obesity and diabetes: Animal models that identify and characterize implications for future health. Clin Perinatol 34:515–526, v. [DOI] [PubMed] [Google Scholar]
- Nathanielsz PW, Yan J, Green R, Nijland M, Miller JW, Wu G, McDonald TJ, Caudill MA. 2015. Maternal obesity disrupts the methionine cycle in baboon pregnancy. Physiol Rep 3:e12564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijland MJ, Mitsuya K, Li C, Ford S, McDonald TJ, Nathanielsz PW, Cox LA. 2010. Epigenetic modification of fetal baboon hepatic phosphoenolpyruvate carboxykinase following exposure to moderately reduced nutrient availability. J Physiol 588:1349–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijland MJ, Schlabritz-Loutsevitch NE, Hubbard GB, Nathanielsz PW, Cox LA. 2007. Non-human primate fetal kidney transcriptome analysis indicates mammalian target of rapamycin (mtor) is a central nutrient-responsive pathway. J Physiol 579:643–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte-‘t Hoen EN, Van Rooij E, Bushell M, Zhang CY, Dashwood RH, James WP, Harris C, Baltimore D. 2015. The role of microrna in nutritional control. J Intern Med 278:99–109. [DOI] [PubMed] [Google Scholar]
- North KE, Martin LJ, Dyer T, Comuzzie AG, Williams JT and Framingham Heart Study . 2003. HDL cholesterol in females in the framingham heart study is linked to a region of chromosome 2q. BMC Genet 4Suppl 1:S98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nsengimana J, Samani NJ, Hall AS, Balmforth AJ, Mangino M, Yuldasheva N, Maqbool A, Braund P, Burton P, Bishop DT, Ball SG, Barrett JH and British Heart Foundation Family Heart Study Research Group . 2007. Enhanced linkage of a locus on chromosome 2 to premature coronary artery disease in the absence of hypercholesterolemia. Eur J Hum Genet 15:313–319. [DOI] [PubMed] [Google Scholar]
- Oberhoffer R, Lang D, Feilen K. 1989. The diameter of coronary arteries in infants and children without heart disease. Eur J Pediatr 148:389–392. [DOI] [PubMed] [Google Scholar]
- Ochman H, Worobey M, Kuo CH, Ndjango JB, Peeters M, Hahn BH, Hugenholtz P. 2010. Evolutionary relationships of wild hominids recapitulated by gut microbial communities. PLoS Biol 8:e1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Sullivan A, He X, McNiven EM, Haggarty NW, Lönnerdal B, Slupsky CM. 2013. Early diet impacts infant rhesus gut microbiome, immunity, and metabolism. J Proteome Res 12:2833–2845. [DOI] [PubMed] [Google Scholar]
- Ouimet M, Hennessy EJ, van Solingen C, Koelwyn GJ, Hussein MA, Ramkhelawon B, Rayner KJ, Temel RE, Perisic L, Hedin U, Maegdefessel L, Garabedian MJ, Holdt LM, Teupser D, Moore KJ. 2016. MiRNA targeting of oxysterol-binding protein-like 6 regulates cholesterol trafficking and efflux. Arterioscler Thromb Vasc Biol 36:942–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozanne SE, Fernandez-Twinn D, Hales CN. 2004. Fetal growth and adult diseases. Semin Perinatol 28:81–87. [DOI] [PubMed] [Google Scholar]
- Pacheco AR, Sperandio V. 2015. Enteric pathogens exploit the microbiota-generated nutritional environment of the gut. Microbiol Spectr 3. doi:10.1128/microbiolspec.MBP-0001-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecoul B, Batista C, Stobbaerts E, Ribeiro I, Vilasanjuan R, Gascon J, Pinazo MJ, Moriana S, Gold S, Pereiro A, Navarro M, Torrico F, Bottazzi ME, Hotez PJ. 2016. The BENEFIT trial: Where do we go from here? PLoS Negl Trop Dis 10:e0004343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips MS, Liu Q, Hammond HA, Dugan V, Hey PJ, Caskey CJ, Hess JF. 1996. Leptin receptor missense mutation in the fatty zucker rat. Nat Genet 13:18–19. [DOI] [PubMed] [Google Scholar]
- Rainwater DL, Kammerer CM, Mahaney MC, Rogers J, Cox LA, Schneider JL, VandeBerg JL. 2003. Localization of genes that control LDL size fractions in baboons. Atherosclerosis 168:15–22. [DOI] [PubMed] [Google Scholar]
- Ramkumar S, Raghunath A, Raghunath S. 2016. Statin therapy: Review of safety and potential side effects. Acta Cardiol Sin 32:631–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay JE, Ferrell WR, Crawford L, Wallace AM, Greer IA, Sattar N. 2002. Maternal obesity is associated with dysregulation of metabolic, vascular, and inflammatory pathways. J Clin Endocrinol Metab 87:4231–4237. [DOI] [PubMed] [Google Scholar]
- Rayner KJ, Esau CC, Hussain FN, McDaniel AL, Marshall SM, van Gils JM, Ray TD, Sheedy FJ, Goedeke L, Liu X, Khatsenko OG, Kaimal V, Lees CJ, Fernandez-Hernando C, Fisher EA, Temel RE, Moore KJ. 2011. Inhibition of mir-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature 478:404–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner KJ, Sheedy FJ, Esau CC, Hussain FN, Temel RE, Parathath S, van Gils JM, Rayner AJ, Chang AN, Suarez Y, Fernandez-Hernando C, Fisher EA, Moore KJ. 2011. Antagonism of mir-33 in mice promotes reverse cholesterol transport and regression of atherosclerosis. J Clin Invest 121:2921–2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees WD, McNeil CJ, Maloney CA. 2008. The roles of ppars in the fetal origins of metabolic health and disease. PPAR Res 2008:459030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds RM, Allan KM, Raja EA, Bhattacharya S, McNeill G, Hannaford PC, Sarwar N, Lee AJ, Bhattacharya S, Norman JE. 2013. Maternal obesity during pregnancy and premature mortality from cardiovascular event in adult offspring: Follow-up of 1 323 275 person years. BMJ 347:f4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rha SW, Choi BG, Seo HS, Park SH, Park JY, Chen KY, Park Y, Choi SY, Shim MS, Kim JB, Park T, Park J, Lee JJ, Park EJ, Park SH, Choi JY, Lee S, Na JO, Choi CU, Lim HE, Kim JW, Kim EJ, Park CG, Oh DJ. 2016. Impact of statin use on development of new-onset diabetes mellitus in asian population. Am J Cardiol 117:382–387. [DOI] [PubMed] [Google Scholar]
- Rich-Edwards JW, Stampfer MJ, Manson JE, Rosner B, Hankinson SE, Colditz GA, Willett WC, Hennekens CH. 1997. Birth weight and risk of cardiovascular disease in a cohort of women followed up since 1976. BMJ 315:396–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbloom KR, Armstrong J, Barber GP, Casper J, Clawson H, Diekhans M, Dreszer TR, Fujita PA, Guruvadoo L, Haeussler M, Harte RA, Heitner S, Hickey G, Hinrichs AS, Hubley R, Karolchik D, Learned K, Lee BT, Li CH, Miga KH, Nguyen N, Paten B, Raney BJ, Smit AF, Speir ML, Zweig AS, Haussler D, Kuhn RM, Kent WJ. 2015. The UCSC genome browser database: 2015 update. Nucleic Acids Res 43:D670–D681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudel LL, Haines JL, Sawyer JK. 1990. Effects on plasma lipoproteins of monounsaturated, saturated, and polyunsaturated fatty acids in the diet of african green monkeys. J Lipid Res 31:1873–1882. [PubMed] [Google Scholar]
- Russo MW, Hoofnagle JH, Gu J, Fontana RJ, Barnhart H, Kleiner DE, Chalasani N, Bonkovsky HL. 2014. Spectrum of statin hepatotoxicity: Experience of the drug-induced liver injury network. Hepatology 60:679–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanches SC, Ramalho LN, Augusto MJ, da Silva DM, Ramalho FS. 2015. Nonalcoholic steatohepatitis: A search for factual animal models. Biomed Res Int 2015:574832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathler-Avelar R, Vitelli-Avelar DM, Mattoso-Barbosa AM, Perdigão-de-Oliveira M, Costa RP, Elói-Santos SM, Gomes Mde S, Amaral LR, Teixeira-Carvalho A, Martins-Filho OA, Dick EJ, Hubbard GB, VandeBerg JF, VandeBerg JL. 2016. Phenotypic features of circulating leukocytes from non-human primates naturally infected with Trypanosoma cruzi resemble the major immunological findings observed in human Chagas disease. PLoS Negl Trop Dis 10:e0004302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seah SK, Marsden PD, Voller A, Pettitt LE. 1974. Experimental Trypanosoma cruzi infection in rhesus monkeys—the acute phase. Trans R Soc Trop Med Hyg 68:63–69. [DOI] [PubMed] [Google Scholar]
- Segovia SA, Vickers MH, Gray C, Reynolds CM. 2014. Maternal obesity, inflammation, and developmental programming. Biomed Res Int 2014:418975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu W, Richards DR, McDonald-Smith GP, Gao H, Hennessy L, Finnerty CC, López CM, Honari S, Moore EE, Minei JP, Cuschieri J, Bankey PE, Johnson JL, Sperry J, Nathens AB, Billiar TR, West MA, Jeschke MG, Klein MB, Gamelli RL, Gibran NS, Brownstein BH, Miller-Graziano C, Calvano SE, Mason PH, Cobb JP, Rahme LG, Lowry SF, Maier RV, Moldawer LL, Herndon DN, Davis RW, Xiao W, Tompkins RG Inflammation and Host Response to Injury, Large Scale Collaborative Research Program . 2013. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci USA 110:3507–3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Q, Hodara V, Simerly C, Schattan G, VandeBerg JL. 2012. Ex vivo reconstitution of arterial endothelium by embryonic-stem-cell-derived endothelial progenitor cells in baboons. Stem Cells Dev 22:631–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Q, Schatten G, Hodara V, Simerly C, VandeBerg JL. 2013. Endothelial reconstitution by CD34+ progenitors derived from baboon embryonic stem cells. J Cell Mol Med 17:242–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Q, Wang J, Wang XL, VandeBerg JL. 2004. Comparative analysis of vascular endothelial cell activation by tnf-alpha and LPS in humans and baboons. Cell Biochem Biophys 40:289–303. [DOI] [PubMed] [Google Scholar]
- Shiba Y, Gomibuchi T, Seto T, Wada Y, Ichimura H, Tanaka Y, Ogasawara T, Okada K, Shiba N, Sakamoto K, Ido D, Shiina T, Ohkura M, Nakai J, Uno N, Kazuki Y, Oshimura M, Minami I, Ikeda U. 2016. Allogeneic transplantation of ips cell-derived cardiomyocytes regenerates primate hearts. Nature 538:388–391. [DOI] [PubMed] [Google Scholar]
- Sonnenburg JL, Bäckhed F. 2016. Diet-microbiota interactions as moderators of human metabolism. Nature 535:56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenvinkel P, Kooman JP, Shiels PG. 2016. Nutrients and ageing: What can we learn about ageing interactions from animal biology? Curr Opin Clin Nutr Metab Care 19:19–25. [DOI] [PubMed] [Google Scholar]
- Sukhova GK, Williams JK, Libby P. 2002. Statins reduce inflammation in atheroma of nonhuman primates independent of effects on serum cholesterol. Arterioscler Thromb Vasc Biol 22:1452–1458. [DOI] [PubMed] [Google Scholar]
- Sun B, Wang X, Bernstein S, Huffman MA, Xia DP, Gu Z, Chen R, Sheeran LK, Wagner RS, Li J. 2016. Marked variation between winter and spring gut microbiota in free-ranging tibetan macaques (macaca thibetana). Sci Rep 6:26035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Burgner DP, Ponsonby AL, Saffery R, Huang RC, Vuillermin PJ, Cheung M, Craig JM. 2013. Effects of early-life environment and epigenetics on cardiovascular disease risk in children: Highlighting the role of twin studies. Pediatr Res 73:523–530. [DOI] [PubMed] [Google Scholar]
- Sun X, Luo S, He Y, Shao Y, Liu C, Chen Q, Cui S, Liu H. 2014. Screening of the mirnas related to breast cancer and identification of its target genes. Eur J Gynaecol Oncol 35:696–700. [PubMed] [Google Scholar]
- Szekely BA, Singh J, Marsh TL, Hagedorn C, Werre SR, Kaur T. 2010. Fecal bacterial diversity of human-habituated wild chimpanzees (pan troglodytes schweinfurthii) at mahale mountains national park, western tanzania. Am J Primatol 72:566–574. [DOI] [PubMed] [Google Scholar]
- Tang WH, Hazen SL. 2014. The contributory role of gut microbiota in cardiovascular disease. J Clin Invest 124:4204–4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarrade A, Panchenko P, Junien C, Gabory A. 2015. Placental contribution to nutritional programming of health and diseases: Epigenetics and sexual dimorphism. J Exp Biol 218:50–58. [DOI] [PubMed] [Google Scholar]
- Thüroff JW, Hort W, Lichti H. 1984. Diameter of coronary arteries in 36 species of mammalian from mouse to giraffe. Basic Res Cardiol 79:199–206. [DOI] [PubMed] [Google Scholar]
- Tilg H, Diehl AM. 2000. Cytokines in alcoholic and nonalcoholic steatohepatitis. N Engl J Med 343:1467–1476. [DOI] [PubMed] [Google Scholar]
- Treasure CB, Klein JL, Weintraub WS, Talley JD, Stillabower ME, Kosinski AS, Zhang J, Boccuzzi SJ, Cedarholm JC, Alexander RW. 1995. Beneficial effects of cholesterol-lowering therapy on the coronary endothelium in patients with coronary artery disease. N Engl J Med 332:481–487. [DOI] [PubMed] [Google Scholar]
- Tuohy KM, Fava F, Viola R. 2014. ‘The way to a man's heart is through his gut microbiota’—dietary pro- and prebiotics for the management of cardiovascular risk. Proc Nutr Soc 73:172–185. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. 2006. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444:1027–1031. [DOI] [PubMed] [Google Scholar]
- Uenishi G, Fujita S, Ohashi G, Kato A, Yamauchi S, Matsuzawa T, Ushida K. 2007. Molecular analyses of the intestinal microbiota of chimpanzees in the wild and in captivity. Am J Primatol 69:367–376. [DOI] [PubMed] [Google Scholar]
- Urbina JA. 2009. Ergosterol biosynthesis and drug development for Chagas disease. Mem Inst Oswaldo Cruz 104Suppl 1:311–318. [DOI] [PubMed] [Google Scholar]
- Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. 2007. Exosome-mediated transfer of mrnas and micrornas is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9:654–659. [DOI] [PubMed] [Google Scholar]
- van Rooij E, Olson EN. 2007. MicroRNAs put their signatures on the heart. Physiol Genomics 31:365–366. [DOI] [PubMed] [Google Scholar]
- Verdery RB, Ingram DK, Roth GS, Lane MA. 1997. Caloric restriction increases HDL2 levels in rhesus monkeys (Macaca mulatta). Am J Physiol 273:E714–E719. [DOI] [PubMed] [Google Scholar]
- Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. 2011. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol 13:423–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers KC, Shoucri BM, Levin MG, Wu H, Pearson DS, Osei-Hwedieh D, Collins FS, Remaley AT, Sethupathy P. 2013. MicroRNA-27b is a regulatory hub in lipid metabolism and is altered in dyslipidemia. Hepatology 57:533–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinson A, Mitchell AD, Toffey D, Silver J, Raboin MJ. 2013. Sex-specific heritability of spontaneous lipid levels in an extended pedigree of Indian-origin rhesus macaques (Macaca mulatta). PLoS One 8:e72241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitelli-Avelar DM, Sathler-Avelar R, Mattoso-Barbosa AM, Gouin N, Perdigão-de-Oliveira M, Valério-Dos-Reis L, Costa RP, Elói-Santos SM, Gomes MS, Amaral LR, Teixeira-Carvalho A, Martins-Filho OA, Dick EJ, Hubbard GB, VandeBerg JF, VandeBerg JL. 2017. Cynomolgus macaques naturally infected with Trypanosoma cruzi-i exhibit an overall mixed pro-inflammatory/modulated cytokine signature characteristic of human Chagas disease. PLoS Negl Trop Dis 11:e0005233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos MB. 2014. Nutrition, nonalcoholic fatty liver disease and the microbiome: Recent progress in the field. Curr Opin Lipidol 25:61–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XL, Rainwater DL, Mahaney MC, Stocker R. 2004. Cosupplementation with vitamin E and coenzyme Q10 reduces circulating markers of inflammation in baboons. Am J Clin Nutr 80:649–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward AM, Moore VM, Steptoe A, Cockington RA, Robinson JS, Phillips DI. 2004. Size at birth and cardiovascular responses to psychological stressors: Evidence for prenatal programming in women. J Hypertens 22:2295–2301. [DOI] [PubMed] [Google Scholar]
- Wiedemann MP. 1962. Lengths and diameters of peripheral arterial vessels in the living animal. Circ Res 10:686–690. [DOI] [PubMed] [Google Scholar]
- Williams JK, Sukhova GK, Herrington DM, Libby P. 1998. Pravastatin has cholesterol-lowering independent effects on the artery wall of atherosclerotic monkeys. J Am Coll Cardiol 31:684–691. [DOI] [PubMed] [Google Scholar]
- Williams-Blangero S, Rainwater DL. 1991. Variation in lp(a) levels and apo(a) isoform frequencies in five baboon subspecies. Hum Biol 63:65–76. [PubMed] [Google Scholar]
- Wilson RM, Messaoudi I. 2015. The impact of maternal obesity during pregnancy on offspring immunity. Mol Cell Endocrinol 418Pt 2:134–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim S, Yeoman CJ, Sipos M, Torralba M, Wilson BA, Goldberg TL, Stumpf RM, Leigh SR, White BA, Nelson KE. 2010. Characterization of the fecal microbiome from non-human wild primates reveals species specific microbial communities. PLoS One 5:e13963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokote K, Shimano H, Urashima M, Teramoto T. 2011. Efficacy and safety of pitavastatin in japanese patients with hypercholesterolemia: LIVES study and subanalysis. Expert Rev Cardiovasc Ther 9:555–562. [DOI] [PubMed] [Google Scholar]
- Zabalgoitia M, Ventura J, Anderson L, Carey KD, Williams JT, VandeBerg JL. 2003. a. Morphologic and functional characterization of chagasic heart disease in non-human primates. Am J Trop Med Hyg 68:248–252. [PubMed] [Google Scholar]
- Zabalgoitia M, Ventura J, Anderson L, Williams JT, Carey KD, VandeBerg JL. 2003. b. Electrocardiographic findings in naturally acquired chagasic heart disease in nonhuman primates. J Electrocardiol 36:155–160. [DOI] [PubMed] [Google Scholar]
- Zabalgoitia M, Ventura J, Lozano JL, Anderson L, Carey KD, Hubbard GB, Williams JT, VandeBerg JL. 2004. Myocardial contrast echocardiography in assessing microcirculation in baboons with chagas disease. Microcirculation 11:271–278. [DOI] [PubMed] [Google Scholar]