Abstract
This study included 168 and 85 mother–infant dyads from Asian and United States of America cohorts to examine whether a genomic profile risk score for major depressive disorder (GPRSMDD) moderates the association between antenatal maternal depressive symptoms (or socio-economic status, SES) and fetal neurodevelopment, and to identify candidate biological processes underlying such association. Both cohorts showed a significant interaction between antenatal maternal depressive symptoms and infant GPRSMDD on the right amygdala volume. The Asian cohort also showed such interaction on the right hippocampal volume and shape, thickness of the orbitofrontal and ventromedial prefrontal cortex. Likewise, a significant interaction between SES and infant GPRSMDD was on the right amygdala and hippocampal volumes and shapes. After controlling for each other, the interaction effect of antenatal maternal depressive symptoms and GPRSMDD was mainly shown on the right amygdala, while the interaction effect of SES and GPRSMDD was mainly shown on the right hippocampus. Bioinformatic analyses suggested neurotransmitter/neurotrophic signaling, SNAp REceptor complex, and glutamate receptor activity as common biological processes underlying the influence of antenatal maternal depressive symptoms on fetal cortico-limbic development. These findings suggest gene–environment interdependence in the fetal development of brain regions implicated in cognitive–emotional function. Candidate biological mechanisms involve a range of brain region-specific signaling pathways that converge on common processes of synaptic development.
Keywords: amygdala, cortical thickness, depression, morphological shape, polygenic risk score
Introduction
Antenatal maternal depression, independent of postnatal maternal mood, influences fetal neurodevelopment, in particular the amygdala, where alterations associate with affective disorders (Rifkin-Graboi et al. 2013; Qiu et al. 2015a; Wen et al. 2017). Since depression is a familial disorder with a significant estimate of heritability (Sullivan et al. 2000), biological pathways linking maternal mood during pregnancy to offspring neurodevelopment are likely to vary as a function of genetic vulnerability for depression. Studies of candidate genes or of family history suggest the influence of “depressogenic” environmental conditions, such as childhood maltreatment and socio-economic status (SES), is moderated by genotype (Moffitt and Caspi 2006), reflecting gene–environment interdependence. However, candidate gene approaches bear weaknesses (Duncan and Keller 2011) and gene products operate in networks, such that alterations in neural systems that enhance vulnerability for psychopathology derive from genomic variants at multiple sites and may converge to influence common biological systems. Indeed, the genetic contribution to susceptibility for depression appears to reflect the cumulative influence of multiple genetic variants (Lubke et al. 2012). This idea led to the use of methods of genomic risk profiling to examine the influence of genetic burden as reflected by a set of “risk” alleles for specific psychiatric disorders (Ripke et al. 2013). The risk alleles and effect sizes of single-nucleotide polymorphisms (SNPs) are established from existing genome-wide association studies (GWAS) for depression using relevant “discovery” samples based on their P-values below a defined threshold. A genomic profile risk score (GPRS) is calculated for each individual in the target sample as the sum of the count of risk alleles weighted by the effect size in the discovery sample. The GPRS for major depressive disorder (GPRSMDD) predicts the risk for depression (P < 10−6) (Ripke et al. 2013) and moderates the influence of childhood maltreatment on depressive symptoms (Peyrot et al. 2014). The target phenotype can also differ from that examined in the discovery sample allowing cross-phenotype analyses and studies of individual differences in endophenotypes. For example, a higher GPRSMDD associates with cortical thinning in the medial prefrontal cortex (Holmes et al. 2012), a region strongly implicated in the pathophysiology of depression (Kupfer et al. 2012).
Given the evidence on the relationship between antenatal maternal depression and the amygdala development during the fetal period (Rifkin-Graboi et al. 2013; Qiu et al. 2015a), we examined whether such relationship was moderated by infant GPRSMDD, controlling for maternal GPRSMDD using a longitudinal Asian birth cohort sample. We then identified candidate genetic pathways and biological processes that might mediate the influence of environment conditions associated with the genetic risk for MDD on the amygdala. We repeated the analysis on the amygdala using a US cohort of mother–infant dyads with measures of antenatal maternal depressive symptoms, neonatal neuroimaging data, and measures of GPRSMDD (see the description of the US cohort in the Supplementary Material). In this study, we computed GPRSMDD based on the SNPs selected from a Caucasian GWAS (Ripke et al. 2013). Given the fact on population differences in genotype frequency and risk genetic variants for MDD (Woo et al. 2004; Serretti et al. 2007; Porcelli et al. 2012; Sheikh et al. 2013; Ming et al. 2013, 2015), we noted that there may be a potential limitation of using a Caucasian sample as a discovery sample for studying the GPRSMDD in the Asian sample. Nevertheless, to the best of our knowledge, the discovery sample used in this study is from one of the most robust GWAS for MDD with the largest sample size (Ripke et al. 2013) at the current stage. Despite its limitation, it is informative in the sense that individual SNPs have opposing effects between Asian and Caucasian populations but they are still risk SNPs for MDD. For instance, Caucasians with S allele of the serotonin transporter polymorphism (5-HTTPLR) show higher amygdala activation in response to emotional stimuli (e.g., Hariri et al. 2002, 2005; Murphy et al. 2013), whereas in Asians, L allele is associated with amygdala hyperactivation (Li et al. 2012). Hence, we expected that if Asian neonates with high GPRSMDD show the positive association between antenatal maternal depressive symptoms and the amygdala volume, whereas Caucasian neonates with high GPRSMDD show the opposite association or vice versa.
We further performed an exploratory analysis on whole brain morphology, including cortical thickness and hippocampal morphology, and examined whether GPRSMDD also modulates the association of antenatal maternal depression and other brain regions, particularly in the cortico-limbic system. Due to the compatibility in the magnetic resonance imaging (MRI) acquisition and the anatomical definitions of the brain structures, this study only performed the exploratory analysis using the Asian cohort. In particular, we employed large deformation diffeomorphic metric mapping (LDDMM) (Du et al. 2011) to perform detailed analysis of the morphological shape of cortical and subcortical brain regions. This approach allows for precise anatomical examination of these structures, well beyond that examined in brain volumetric studies, and reveals specific-regional shape changes associated with disease status (Qiu et al. 2009, 2010). Based on genetic contribution to MDD (Lubke et al. 2012) and the evidence that antenatal maternal depression influences fetal cortico-limbic development (Qiu et al. 2015a), we hypothesized that neonates with high GPRSMDD may show the negative relationship of antenatal maternal depression with fetal development of the cortico-limbic structures, including the hippocampus and prefrontal cortex. To further explore the moderating influence of the GPRSMDD, we examined the effects of SES, which influences the risk for a broad range of mental disorders, including MDD (e.g., Gilman et al. 2003), and cortico-limbic development (Farah et al. 2010; Noble et al. 2015).
This study, to the best of our knowledge, is the first kind to provide a strategy for the analysis of gene–environment interdependence and identify candidate biological processes that might mediate the influence of antenatal maternal depressive symptoms on fetal brain development. Because of challenges in imaging neonates shortly after birth, the sample size of this study was modest for the study of gene–environment interaction and hence our findings are preliminary but may suggest biologically plausible candidate processes for transgenerational transmission of depression from mother to child.
Materials and Methods
Subjects
One-hundred and eighty-nine infants of mothers who participated in the prospective Growing Up in Singapore Towards healthy Outcomes (GUSTO) birth cohort study (Soh et al. 2012) were recruited for neuroimaging (Asian cohort). The Asian cohort recruited pregnant women attending an antenatal ultrasound scan clinic who were Singapore citizens or Permanent Residents of Chinese, Malay, or Indian ethnic background, and had no history of psychotropic medication.
We included the 168 neonates with genetic and neuroimaging data, gestational age at birth ≥35 weeks, birth weight >2500 g, and a 5-min Appearance, Pulse, Grimace, Activity, and Respiration score ≥9 and whose mothers had genetic data and returned the questionnaires stated below. The Asian study was approved by the National Healthcare Group Domain Specific Review Board (NHG DSRB) and the Sing Health Centralized Institutional Review Board (CIRB). Written consent was obtained from mothers.
Scales of Maternal Depressive Symptoms and SES
In GUSTO, the Edinburgh Postnatal Depression Scale (EPDS) questionnaire was administered to mothers at 26 weeks of pregnancy to quantify depressive symptomatology. The EPDS (Cox et al. 1987) is a widely used 10-item self-report scale designed as a screening instrument for postnatal depression and valid for use in gestation. Each item of the EPDS is scored on a 4-point scale (0–3), and items 3 and 5–10 are reverse scored. The reliability of the EPDS was 0.82 assessed using Cronbach's analysis for our cohort. SES was measured using monthly household income (Table 1) and was collected at 26 weeks gestation via questionnaire.
Table 1.
Demographics of the Asian cohort
| Mean | SD | |
|---|---|---|
| Gestational age (week) | 38.69 | 1.13 |
| Birth weight (gram) | 3117.85 | 391.76 |
| Birth weight adjusted by gestational age (gram) | 3118.14 | 379.01 |
| Postconceptual age at the MRI visit (week) | 40.08 | 1.21 |
| Antenatal maternal depression | 8.59 | 4.46 |
| N | % | |
| Sex, % | ||
| Male | 168 | 53.6 |
| Ethnicity, % | ||
| Chinese | 168 | 44.0 |
| Malay | 39.3 | |
| Indian | 16.7 | |
| Monthly household income (US$), % | ||
| ≤768 | 168 | 3.0 |
| 769–1538 | 13.7 | |
| 1538–3076 | 39.3 | |
| 3077–4615 | 23.2 | |
| ≥4616 | 13.7 | |
| Upreported | 7.1 | |
Note: SD, standard deviation.
MRI Acquisition and Analysis
In the Asian cohort, axial fast spin-echo T2-weighted MRI (repetition time = 3500 ms; echo time = 110 ms; field of view = 256 mm × 256 mm; matrix size = 256 × 256; 50 axial slices with 2.0 mm thickness) were acquired in infants 5–14 days of age using a 1.5-Tesla general electric scanner at the Department of Diagnostic and Interventional Imaging of the KK Women's and Children's Hospital. The detailed acquisition and image quality check procedure were previously reported (Qiu et al. 2013, 2015b). No sedation was used and precautions taken to reduce exposure to the MRI scanner noise. A neonatologist was present during scanning. A pulse oximeter was used to monitor heart rate and oxygen saturation throughout the entire scans. All brain scans were reviewed by a neuroradiologist (M.V.F). To determine image motion, all axial slices of the T2-weighted MRI data were visually inspected to ensure no cross-slice motion and checkerboard patterns.
Within individual subjects, when possible, 2 T2-weighted MRI acquisitions were first rigidly aligned and averaged to increase signal-to-noise ratio. In cases where only one scan was acquired, data from one scan were used in lieu of the average axial image. The skull of the averaged axial image was removed using Brain Extraction Tool (Smith 2002). A Markov random field model was used to automatically delineate amygdala, hippocampus, cortical gray matter, white matter, and cerebrospinal fluid (CSF) from the neonatal T2-weighted MRI data. The validation of this method in our sample was reported elsewhere (Qiu et al. 2013, 2015b; Rifkin-Graboi et al. 2013). Against the manual segmentation of the 20 brain images randomly selected in our data set, the segmentation accuracy rates of the amygdala, hippocampus, cortical gray matter, and white matter were respectively 0.75, 0.71, 0.79, and 0.86.
Based on the above tissue segmentation, a cortical surface was constructed at the boundary between gray and white matter using a graph-based topology correction algorithm. Cortical thickness was measured as the distance between cortical surfaces at the boundary between gray matter and CSF and at the boundary of gray matter and white matter. We employed a LDDMM algorithm to align individual cortical surfaces to the Asian atlas that was generated based on the cortical anatomy of the 20 subjects of this study and transferred the thickness of each individual subject to the atlas (Bai et al. 2012). The cortical thickness was smoothed using the Laplace–Beltrami basis functions on the cortical surface (Qiu et al. 2006).
Amygdala and hippocampal morphological shapes were represented using surfaces contoured at the structure boundary. The LDDMM-surface mapping (Qiu and Miller 2008; Zhong and Qiu 2010; Tan and Qiu 2016) was then used to take surfaces as objects and provided transformation that deformed the Asian atlas to be similar to subjects (Bai et al. 2012). Shape variations of individual subjects relative to the Asian atlas were characterized by the Jacobian determinant of the transformation in the logarithmic scale, which represents the ratio of structural volume to the atlas volume. Positive values correspond to expansion, while negative values correspond to compression of the structure relative to the atlas at each anatomical location.
SNP Genotyping and GPRS Computation for MDD
In the Asian cohort, genomic DNA was extracted from frozen umbilical cord specimens for infants and from blood for mothers according to standard procedures. The samples were genotyped on Illumina Omni express arrays, shown to perform well and with better coverage than alternatives in Asian populations (Jiang et al. 2013), and on Illumina Exome arrays, following the manufacturer's instructions (Expression Analysis Inc). Further quality control on the genotyping calls were previously described (Qiu et al. 2015b). SNPs were verified for a genotyping rate ≥95% and no deviation from Hardy–Weinberg equilibrium (P < 0.001), and minor allele frequency ≥0.05, in PLINK (Purcell et al. 2007).
The GPRSMDD of infants and mothers was created using PLINK based on their SNPs and meta-analysis results from the Psychiatric Genomics Consortium (PGC; European ancestry, 9240 MDD cases, and 9519 controls) (Ripke et al. 2013). The GPRSMDD was a cumulative summary score computed as the sum across SNPs of the number of risk alleles (0, 1, and 2) weighted by odd ratio. Odd ratios were obtained from the meta-analysis on MDD from the Psychiatric Genomics Consortium website (http://www.med.unc.edu/pgc) (Ripke et al. 2013) and represented the allele-specific risk for MDD. We selected the SNPs with low linkage disequilibrium to each other (r2 < 0.25 within 500 kb window, filtering for significance; PLINK-command -clump-p1 1 -clump-p2 1 -clump-r2 0.25 -clump-kb 500) (Peyrot et al. 2014) and that survived at P-value thresholds (0.05, 0.1, and 0.2) to incorporate the proportion of variation in disease risk explained through their additive effects (Purcell et al. 2009; Holmes et al. 2012; Peyrot et al. 2014). The resulting numbers of selected SNPs for infant GPRSMDD were 2766, 5081, and 8935. The numbers of SNPs for maternal GPRSMDD were 2769, 5078, and 8902. This GPRSMDD was then standardized to a mean of zero and standard deviation of one to aid interpretation of results.
Assume that genetic risks for rheumatoid arthritis (RA) should not contribute to the risk for MDD and hence should not modulate the relationship between maternal depressive symptoms and neonatal morphology. In this study, we computed GPRS scores for RA (GPRSRA) as a negative control. We anticipated that GPRSRA would not modulate the relationship between GPRSMDD and neonatal brain morphology.
Statistical Analysis
Linear regressions were used to examine interaction effects of infant GPRSMDD with antenatal maternal depressive symptoms (or SES) on amygdala and hippocampus volumes and shapes, as well as cortical thickness of the whole brain in the Asian sample. The EPDS scores (or SES) and the GPRSMDD of the infant were included as main factors, which were entered into the first block of linear regressions along with covariates of gender, maternal ethnicity, birth weight adjusted by gestational age, postconceptual age on the MRI day. The interaction of the 2 main factors (EPDS score/SES and GPRSMDD) was then entered into the second block of the regression. For statistical analysis on shapes and thickness, the data were smoothed on the atlas surfaces using a heat kernel with 30-mm full-width-at-half-maximum (Chung et al. 2005). Statistical results were illustrated with P-value < 0.001 at the vertex level and with the cluster level of significance (P < 0.05) after correction for multiple comparisons based on random field theory (Chung et al. 2005, 2010). The correction of multiple comparisons was applied to one interaction model at a time, but not across genomic polygenic risk scores (GPRSMDD and GPRSRA) and different brain measures (volume, shape, and thickness).
If the interaction of the EPDS score (or SES) and GPRSMDD was significant, we performed post hoc analyses where the sample was then divided into 2 groups, one with a GPRSMDD greater than its mean value, 0 (high GPRSMDD group) and the other ≥0 (low GPRSMDD group). We examined associations of antenatal maternal depressive symptoms (or SES) and brain measures in each GPRSMDD group using regression with EPDS (or SES) as a main factor and covariates of gender, maternal ethnicity, adjusted birth weight, and postconceptual age on the MRI day. The brain measures used in this post hoc analysis were computed as average values of thickness, or shape deformation of the brain regions (colored regions shown in Fig. 2A and Fig. 3A) with the significant interaction of GPRSMDD with antenatal maternal depressive symptoms (or SES). The same analysis was repeated while additionally controlling for maternal GPRSMDD. For the interaction of GPRSMDD and antenatal maternal depressive symptoms, the same analysis was also repeated while additionally controlling for SES and vice versa.
Figure 2.
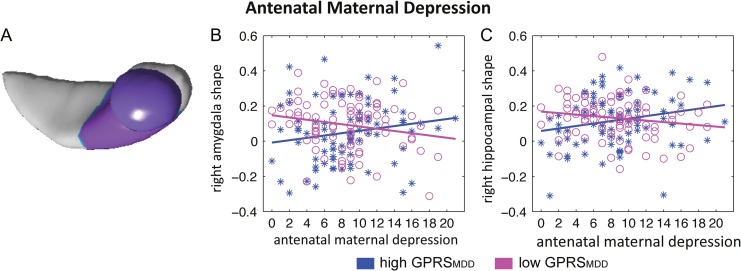
Statistical maps of the interaction effects of the infant GPRSMDD with antenatal maternal depressive symptoms (A) on the amygdala and hippocampal shapes. Panels (B, C) illustrate the relationship of antenatal maternal depressive symptoms with right amygdala shape (left panel) and right hippocampal shape (right panel) in neonates with high GPRS for major depressive disorder (GPRSMDD, blue astericks) and those with low GPRSMDD (magenta circles). Antenatal maternal depressive symptoms are represented by the score of EPDS, with a higher score indicating more severe of depressive symptoms.
Figure 3.
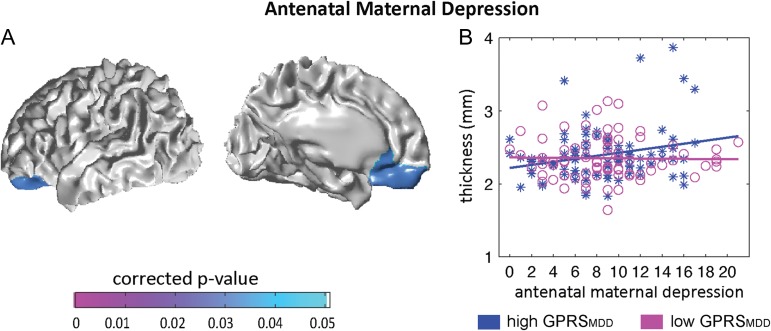
Statistical maps for interaction of the infant GPRSMDD with antenatal maternal depressive symptoms (A) on cortical thickness. Panel (B) illustrates the relationship of antenatal maternal depressive symptoms with thickness of left OFC and vmPFC in neonates with high GPRSMDD, (blue asterisks) and those with low GPRSMDD (magenta circles). Antenatal maternal depressive symptoms are represented by the score of EPDS, with a higher score indicating more severe of depressive symptoms.
Genetic Pathway Annotation Analysis
Genetic pathway enrichment was used to explore the underlying biological processes associated with genes involved in the GPRSMDD computation. We first selected the top SNPs that moderated the relationship of antenatal maternal depressive symptoms with individual brain regions. These SNPs were mapped to genes using the batch query function in UCSC Genome Bioinformatics (http://genome.ucsc.edu). PANTHER (http://pantherdb.org) was then used to identify pathways that were statistically over-represented against the reference genes involved in the same genetic pathways based on fold enrichment analysis.
We further explored biological functions of each genetic pathway using GeneMania (Mostafavi et al. 2008: http://Genemania.org). GeneMania was developed as a heuristic algorithm, derived from ridge regression, to integrate multiple functional association networks and predict biological functions from a single process-specific pathway. GeneMania was used to assess candidate biological functions associated with the genes in the pathways identified by PANTHER using multiple functional association data sets, including co-expression analysis, protein–protein interactions, co-localization, gene–gene interactions and protein domain similarity.
Results
Demographics
Table 1 and Supplementary Table S1 list the demographic information of the Asian and US cohorts. In the Asian cohort, antenatal maternal depressive symptoms did not vary as a function of infant gender (t166 = 0.11, P = 0.92) and ethnicity (F2165 = 2.26, P = 0.11; Table 1). Antenatal maternal depressive symptoms did not vary as a function of infant or maternal GPRSMDD based on the 3 thresholds (all P > 0.2). But, SES was significantly correlated with antenatal maternal depressive symptoms (r = −0.273; P = 0.001) and significantly differed among the 3 maternal ethnicity (F2155 = 4.726, P = 0.01).
Among 168 neonates, 85 had 1 good T2-weighted MRI scan and 83 had 2 T2-weighted MRI scans. Between these 2 groups, we did not observe differences in birth weight (t166 = −0.833, P = 0.406), postconceptual age on the MRI day (t166 = 0.180, P = 0.857), gender P = 0.216), maternal ethnicity P = 0.662), and antenatal maternal depressive symptoms (t166 = 1.140, P = 0.256).
Interaction of GPRSMDD with Antenatal Maternal Depressive Symptoms on Neonatal Brain Morphology in the Asian cohort
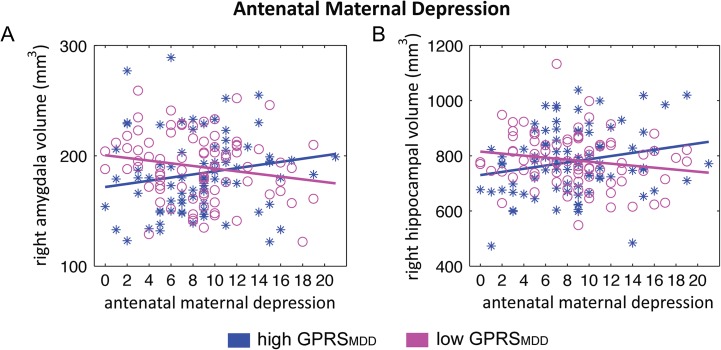
Regression analysis revealed a significant interaction effect between antenatal maternal depressive symptoms and infant GPRSMDD on the right, but not left amygdala and hippocampal volumes (Table 2) after adjusting for gender, maternal ethnicity, adjusted birth weight, and postconceptual age on the MRI day. The interaction effect was apparent at all 3 threshold levels of the GPRSMDD, with the largest interaction effect found using the infant GPRSMDD at P = 0.2. These findings remained significant when additionally controlling for the mother GPRSMDD (the Supplementary Material; Table 2) and also the finding for the right amygdala remained significant when additionally controlling for SES (the Supplementary Material). Subsequent post hoc analysis revealed a significant positive association between antenatal maternal depressive symptoms and right hippocampal volume (t78 = 2.04, P = 0.045; Fig. 1B) and the marginal, positive association between antenatal maternal depressive symptoms and right amygdala volume (t78 = 1.80, P = 0.076; Fig. 1A) in neonates with high GPRSMDD (>0). No associations were found between antenatal maternal depressive symptoms and right amygdala volume (t80 = −1.62, P = 0.109) and between antenatal maternal depressive symptoms and right hippocampal volume (t80 = −1.62, P = 0.109) in neonates with low GPRSMDD (<0).
Table 2.
Statistical results for amygdala and hippocampal volumes in the Asian sample
| Structure | Mean (SD) (mm3) | Interaction effects |
|---|---|---|
| Left amygdala | 211.41 (34.48) | F1161 = 2.96, P = 0.087 |
| Right amygdala | 187.09 (33.77) | F1161 = 6.88*,a, P = 0.009 |
| Left hippocampus | 779.40 (108.18) | F1161 = 0.002, P = 0.964 |
| Right hippocampus | 781.76 (109.11) | F1161 = 4.15*,a, P = 0.043 |
Note: F-values indicate the interaction effects of infant GPRSMDD with antenatal maternal depressive symptoms on the structural volumes. * P-value < 0.05.
aThe result remains significant when maternal GPRSMDD was entered in regression analysis as an additional covariate.
Figure 1.
Interaction of the infant's GPRSMDD with antenatal maternal depressive symptoms on the amygdala and hippocampal volumes. Scatter plots in panels (A) and (B) illustrate the relationship of antenatal maternal depressive symptoms with right amygdala volume and right hippocampal volume in neonates with high GPRS for major depressive disorder (GPRSMDD, blue asterisks) and those with low GPRSMDD (magenta circles).
Similarly, we found significant interaction effects between infant GPRSMDD and antenatal maternal depressive symptoms on the right amygdala shape (corrected cluster P = 7.86e-5; Fig. 2A) and anterior right hippocampal shape (corrected cluster P = 0.016; Fig. 2A), with the largest interaction effect at GPRSMDD of P = 0.2 among the 3 thresholds (0.05, 0.1, and 0.2). This interaction effect was apparent for GPRSMDD of all 3 threshold levels. These findings remained significant when controlling for maternal GPRSMDD and the same finding on the right amygdala shape remained significant when controlling for SES (the Supplementary Material). The post hoc analysis revealed a significant positive association between antenatal maternal depressive symptoms and right anterior hippocampal shape (t78 = 2.03, P = 0.046; Fig. 2C) and a marginal, positive association between antenatal maternal depressive symptoms and right amygdala shape (t78 = 1.84, P = 0.070; Fig. 2B) in neonates with high infant GPRSMDD (>0). No associations were found between antenatal maternal depressive symptoms and right amygdala shape (t80 = −1.77, P = 0.08) and between antenatal maternal depressive symptoms and right anterior hippocampal shape (t80 = −1.63, P = 0.108) in neonates with low infant GPRSMDD (<0).
We found a significant interaction effect between antenatal maternal depressive symptoms and infant GPRSMDD on thickness of left orbitofrontal cortex (OFC) and ventromedial prefrontal cortex (vmPFC) (corrected cluster P = 0.031; Fig. 3A), with the largest interaction effect using the GPRSMDD at P = 0.1 among the 3 thresholds (0.05, 0.1, and 0.2); the interaction was also apparent using the GPRSMDD at P = 0.2. These findings remained significant when additionally controlling for the maternal GPRSMDD (the Supplementary Material). These findings became a trend of significance after additionally controlling for SES (the Supplementary Material). The post hoc analysis further revealed the significant positive association between antenatal maternal depressive symptoms and cortical thickness of left OFC and vmPFC (t85 = 2.63, P = 0.010) in neonates with high infant GPRSMDD (>0), suggesting that a greater maternal depressive score was associated with thicker cortex in left OFC and vmPFC (see Fig. 3B). No association was found between antenatal maternal depressive symptoms and cortical thickness of left OFC and vmPFC (t80 = −0.054, P = 0.957) in neonates with low infant GPRSMDD (<0).
In contrast to the GPRSMDD, GPRSRA did not moderate the association of antenatal maternal depressive symptoms with the right amygdala and hippocampus at all the 3 thresholds (0.05, 0.1, and 0.2) of the GPRSRA computation (Table 3). Moreover, the GPRSRA did not affect the association between cortical thickness in the vmPFC and OFC and antenatal maternal depressive symptoms (cluster P-value > 0.05).
Table 3.
Statistical results for amygdala and hippocampal volumes in the Asian sample.
| Threshold for the GPRSRA computation | Right amygdala | Right hippocampus |
|---|---|---|
| Antenatal maternal depression | ||
| P < 0.05 | F1159 = 0.867, P = 0.353 | F1159 = 1.457, P = 0.229 |
| P < 0.1 | F1159 = 1.662, P = 0.199 | F1159 = 1.267, P = 0.262 |
| P < 0.2 | F1159 = 1.662, P = 0.199 | F1159 = 1.267, P = 0.262 |
Note: F-values indicate the interaction effects of the infant's GPRSRA with antenatal maternal depression on the structural volumes.
Interaction of GPRSMDD with SES on Neonatal Brain Morphology in the Asian Cohort
Likewise, a significant interaction effect between SES and infant GPRSMDD on the right, but not left amygdala and hippocampal volumes and shapes (Supplementary Table S2) after adjusting for gender, maternal ethnicity, adjusted birth weight, and postconceptual age on the MRI day. Such interaction effects remained significant for the right hippocampus but not the right amygdala when additionally controlling for antenatal maternal depressive symptoms. No interaction effect between SES and infant GPRSMDD was observed on cortical thickness.
Brain Region-Specific Gene Networks and Biological Processes
Among the SNPs involved in the GPRSMDD, the top 1551 showed the most significant interaction with antenatal maternal depressive symptoms (P < 0.2) on the right amygdala, 1170 SNPs on the right hippocampus, and 1236 SNPs on the left OFC and vmPFC thickness, which were mapped to 629, 508, and 503 genes, respectively. PANTHER analysis revealed biological pathways for the 3 structures. These pathways were statistically over-representative in relation to those in the PANTHER pathway database (Table 4). For the right amygdala, 78 genes could not be found in the PANTHER database. The remaining 551 genes formed one statistically over-represented pathway (P = 0.003) labeled by PANTHER as “metabotropic glutamate receptor group III”. For the left OFC and vmPFC, 61 genes could not be found in the PANTHER database. The remaining 442 genes formed 5 statistically over-represented pathways, including “heterotrimeric G-protein signaling pathway-Gq alpha and Go alpha” (P = 0.004), “endothelin signaling” (P = 0.007), “histamine H1 receptor” (P = 0.005), 5-HT2 type receptor (P = 0.013), and “oxytocin receptor” (P = 0.044). For the right hippocampus, 69 genes could not be found in the PANTHER database. The remaining 439 genes formed 2 statistically over-represented genetic pathways, including “axon guidance mediated by Slit/Robo” (P = 0.027) and “Alzheimer's disease-amyloid secretase” (P = 0.047). Supplementary Table S3 lists the genes involved in each pathway.
Table 4.
Genetic pathways associated with interaction of GPRSMDD and antenatal maternal depressive symptoms in the Asian sample
| Structure | Genetic pathways | Fold enrichment | P-valuea |
|---|---|---|---|
| Interaction of GPRSMDD and antenatal maternal depression | |||
| Right amygdala | Metabotropic glutamate receptor group III | >5 | 0.003 |
| Left OFC and vmPFC | Heterotrimeric G-protein signaling pathway-Gq alpha and Go alpha mediated pathway |
>5 | 0.004 |
| Histamine H1 receptor-mediated signaling | >5 | 0.005 | |
| Endothelin signaling | >5 | 0.007 | |
| 5-HT2 type receptor-mediated signaling | >5 | 0.013 | |
| Oxytocin receptor-mediated signaling | >5 | 0.044 | |
| Right hippocampus | Axon guidance mediated by Slit/Robo | >5 | 0.027 |
| Alzheimer's disease-amyloid secretase | >5 | 0.047 | |
| Interaction of GPRSMDD and SES | |||
| Right hippocampus | CCKR signaling map | 4.36 | 0.009 |
| 5-HT2 type receptor medicated signaling pathway | >5 | 0.021 | |
| Beta1 adrenergic receptor signaling pathway | >5 | 0.021 | |
| Beta2 adrenergic receptor signaling pathway | >5 | 0.021 | |
Notes: The Fold Enrichment is defined as the ratio of the proportion of our input genes involved in a particular genetic pathway to the proportion of the reference genes involved in the same genetic pathway.
aBonferroni corrected P-value.
The same analysis for SNPs in infant GPRSMDD that nominally moderate the association between SES and fetal neurodevelopment did not reveal any significantly enriched pathways for the right amygdala. For the right hippocampus, 40 genes could not be found in the PANTHER database and the remaining 311 genes formed 4 statistically over-represented pathways (Table 4), including “CCK receptor signaling” (P = 0.009), “Beta1” (P = 0.021) and “Beta2 (P = 0.021) adrenergic receptor signaling”, and “5-HT2 type receptor” mediated signaling pathways (P = 0.021). Supplementary Table S4 lists the genes involved in each pathway.
We then entered the genes from the individual pathways identified by PANTHER into GeneMania and identified candidate biological processes (Table 5). GeneMania identified “Glutamate receptor activity” as the primary function of the metabotropic glutamate receptor group III pathway, which included genes that encode for subunits of the N-methyl-D-aspartate (GRIN1, GRIN2A, GRIN2B, GRIN2C, and GRIN2D), Kainate (GRK1-5), and α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (GRIA1-4) receptors, as well as for the metabotropic receptors (GRM1-8). This same pathway was also significantly associated with the SNAp REceptor (SNARE) complex function. Likewise, the SNARE complex emerged as the primary biological function associated with both the 5-HT2-type receptor- and the oxytocin receptor-mediated signaling pathway identified as candidate moderators of the effect of antenatal maternal depressive symptoms on cortical thickness (Table 5).
Table 5.
Biological functions of genetic pathway in the Asian sample
| Genetic pathway | Biological function | FDR corrected P-value | Coverage |
|---|---|---|---|
| Metabotropic glutamate receptor group III | Glutamate receptor activity | 7.3e–46 | 20/20 |
| SNARE complex | 3.2e–18 | 12/32 | |
| G-protein signaling | G-protein coupled receptor signaling | 1.2e–32 | 29/125 |
| Histamine H1 receptor-mediated signaling | Inositol phosphate metabolic process | 6.0e–19 | 13/55 |
| Endothelin signaling | Fibroblast growth factor signaling | 1.5e–43 | 35/170 |
| 5-HT2 type receptor-mediated signaling | SNARE complex | 3.1e–25 | 15/32 |
| Oxytocin receptor-mediated signaling | SNARE complex | 5.0e–26 | 15/32 |
| Axon guidance mediated by Slit/Robo | Small GTPase mediated signal transduction | 3.9e–8 | 12/291 |
| Amyloid secretase | Neurotrophic signaling | 2.4e–20 | 23/277 |
| Beta1 adrenergic receptor signaling | SNARE complex | 1.6e–27 | 15/32 |
| Beta2 adrenergic receptor signaling | SNARE complex | 1.6e–27 | 15/32 |
| CCKR signaling | Intracellular ligand-gated calcium channel activity | 3.1e–14 | 7/10 |
The Findings from the US Cohort
We also examined the neonatal amygdala and hippocampus structures using the US sample of 85 dyads with available neonatal brain imaging data of the amygdala and hippocampal volumes, SNP data genotyped using the Illumina Omni express arrays, and measures of maternal depressive symptoms assessed using the Center for Epidemiologic Studies Depression scale during the third trimester (the Supplementary Material). We used the same statistical model as that employed with the Asian cohort data and found a statistically significant interaction effect between infant GPRSMDD and antenatal maternal depressive symptoms on the right (F1,77 = 4.09, P = 0.047), but not left amygdala volume (F1,77 = 1.21, P = 0.275), with the largest interaction effect at GPRSMDD of P = 0.1 among the 3 thresholds (0.05, 0.1, and 0.2). Interestingly, the US cohort showed the significant positive association between antenatal maternal depressive symptoms and right amygdala volume (t36 = 2.21, P = 0.034) in neonates with low infant GPRSMDD (<0) and the marginal negative association between antenatal maternal depressive symptoms and right amygdala volume (t37 = −1.66, P = 0.105) in neonates with high infant GPRSMDD (>0), which is opposite to that seen in the Asian sample. This could be due to differences in populations, which has been reported in other studies on Asian and Caucasian populations based on individual SNPs (Sheikh et al. 2013) (Woo et al. 2004; Serretti et al. 2007; Porcelli et al. 2012; Ming et al. 2013, 2015). Nevertheless, interaction effects between infant GPRSMDD and antenatal maternal depressive symptoms were not observed on bilateral hippocampal volumes (left: F1,77 = 0.064, P = 0.801; right: F1,77 = 0.903, P = 0.345).
Among the SNPs involved in the GPRSMDD for the US sample, the top 747 SNPs with the most significant interaction with antenatal maternal depressive symptoms (P < 0.1) on the right amygdala volume were mapped to 594 genes. For the right amygdala, 65 genes could not be found in the PANTHER database. The remaining 529 genes formed 2 statistically over-represented pathway labeled by PANTHER as “metabotropic glutamate receptor group III” (P = 0.002) and “metabotropic glutamate receptor group I” (P = 0.003). Thus, despite the differences in sample size and ethnicity, these biological pathways were strikingly similar to those identified in the Asian cohort for the interaction between antenatal maternal depressive symptoms and GPRSMDD on the right amygdala volume.
Discussion
This study provides evidence for the role of infant genotype as a moderator of the association of antenatal maternal mental health with fetal development of cortico-limbic structures, including structures such as the amygdala, hippocampus, OFC, and vmPFC that are implicated in affective disorders. These interaction effects were apparent using measures of depressive symptoms across a community sample and remained significant even after controlling for maternal GPRSMDD thus revealing selective moderation by infant genotype. This study further revealed over-representative genetic pathways and biological processes formed by the genes included in the GPRSMDD computation for which variations best contributed to modulating the relationship of antenatal maternal depressive symptoms and the morphology of the aforementioned brain structures.
The amygdala, hippocampus, OFC, and vmPFC are cortico-limbic structures implicated in mood disorders and central to emotion and social regulation. Abnormal morphology and activity in these regions are reported in children (Thomas et al. 2001; Pagliaccio et al. 2014) and adults with MDD (Johnstone et al. 2007). Smaller hippocampal volume and greater activity in response to sad faces predict increased depressive symptoms in children (Suzuki et al. 2013). In depressed children severity correlates with activity of the amygdala–hippocampal complex and medial PFC during sad mood elaboration (Pagliaccio et al. 2012) and amygdala reactivity during emotional face viewing (Gaffrey et al. 2011). Likewise, children with early-life stress show smaller OFC (Hanson et al. 2010). Children of parents with MDD also show altered amygdala activity in the emotional facial expression (Monk et al. 2008). Increased amygdala activation in response to novelty or threatening faces in children is associated with measures of negative emotionality (Perez-Edgar et al. 2007), which in turn predicts a greater risk for mood disorders (Bruder-Costello et al. 2007). Moreover, altered functional connectivity between the amygdala and medial PFC, including OFC and vmPFC, is observed in 6-month-old infants born to mothers with increased antenatal maternal depressive symptoms (Qiu et al. 2015a). These findings suggest that the cortico-limbic systems may be particularly vulnerable to maternal influences during fetal development and reflect, in part, the neural basis for the familial transmission of phenotypes associated with depression.
Our study revealed an interaction effect between maternal depressive symptoms and infant GPRSMDD that was apparent in the right amygdala and hippocampus. Individual differences in stress reactivity associate with morphology and function of the prefrontal cortex, the hippocampus and the amygdala, as well as the connections between these regions (Pruessner et al. 2008; van Marle et al. 2009) and there is compelling evidence implicating the right hemisphere, notably the right amygdala and right hippocampus in the regulation of stress responses (Abercrombie et al. 1998; Carrion et al. 2007; Hamilton and Gotlib 2008). The risk for depression is linked to heightened stress reactivity, which associates with a familial risk for depression: individuals with increased genetic vulnerability for depression show greater negative affect in response to daily life stress, which then predicts depression (Wichers et al. 2009). Our findings suggest the familial transmission of individual differences in neural systems that underlie stress reactivity may occur during fetal development.
We explored gene networks and their associated biological processes as candidate mediators of the influence of clinically relevant environmental conditions on fetal neurodevelopment. The gene pathways (metabotropic glutamate receptor group pathways) derived from SNPs that best mediated the interaction of antenatal maternal depressive symptoms and infant GPRSMDD on the right amygdala in both the Asian and US cohorts were strongly associated with glutamate receptor activity. Multiple approaches, including genome-wide analyses, implicate glutamatergic synaptic transmission in the etiology of depression (Skolnick et al. 2009; Lee et al. 2012) and patients with MDD exhibit abnormal glutamate concentrations in the amygdala (Michael et al. 2003). Postmortem studies with MDD suggest altered glutamatergic synaptic signaling (Hashimoto et al. 2007) and ketamine, a glutamatergic intervention, is a treatment for MDD (Caddy et al. 2014). The antidepressant, tianeptine, blocks stress-induced increases in glutamate release in the amygdala (Piroli et al. 2013) and moderates behavioral responses to stress. This glutamate receptor activity included the GRM1 and GRM8 genes, which are nominally associated with MDD (Terracciano et al. 2010). Knockout mouse models of genes coding for metabotropic glutamate receptor proteins, which modulate postsynaptic glutamate signaling, associate with increased anxiety (Bures et al. 2005). The “G-protein signaling” network also identified as a candidate mediator for the effects of maternal depression on cortical structure (Supplementary Table S3) includes the GRM1, GRM4, GRM 7, and GRM8 genes.
The genetic pathway analysis in the right amygdala identified the “SNARE complex” biological process, which is a fundamental component of neural connectivity and neurotransmitter release comprised of syntaxin, SNAP-25, and VAMP. Indeed, perhaps the most compelling finding from our analysis was the broad implication of the SNARE complex as a biological process mediating the impact of antenatal maternal depressive symptoms on multiple brain structures (Table 5). Postmortem studies revealed abnormal expression of SNARE proteins in the hippocampus and frontal cortex of subjects with MDD (Scarr et al. 2006). Likewise, proteomic analyses implicate synaptic dysfunction in MDD and identify altered expression of SNARE complex proteins (Martins-de-Souza et al. 2012). Administration of ketamine as well as a broad range of antidepressant medications alters the accumulation of SNARE complexes in synaptic membranes (Bonanno et al. 2005), which then associates with altered capacity for stimulated glutamate release (Bonanno et al. 2005). Two SNP's (rs3787283 and rs3746544) in the SNAP-25 gene have been associated with MDD in a Han Chinese sample (Wang et al. 2015), an ethnic group that accounted for the majority of the Asian subjects in our study.
Our findings suggest that the influence of antenatal maternal depressive symptoms on the cortico-limbic structure is mediated by brain-specific signaling pathways (oxytocin, 5-HT2, and glutamate receptor pathways). These pathways are linked to processes underlying synaptic development. In addition to association with the glutamate receptor activity, the SNARE complex was the top biological process for the oxytocin and 5-HT2 gene networks (Table 5). There is strong evidence for the importance of serotonin, including 5-HT2 receptor activation, in perinatal brain structure (see Lesch and Waider 2012 for a review). While oxytocin receptor activation is not extensively described in terms of brain development, there is a period of transient neocortical expression of oxytocin receptor binding in rodent brain that suggests a developmental influence (Hammock and Levitt 2013). Finally, our analyses identified neurotrophic signaling systems as candidate mediators of the effects of antenatal maternal depression on hippocampus (Table 4). The Fibroblast growth factor signaling system is particularly interesting to consider in light of the findings with rodent models that early postnatal administration of FGF2 decreases fear behavior in animals genetic predisposed to increased expression of anxiety-like behaviors (Eren-Kocak et al. 2011). Sustained FGF2 effects on genes expression included targets involved in cell survival, an effect that is consistent with the role for neurotrophic signaling pathways as a mediator of the effects of antenatal maternal depression. Neurotrophic signaling pathway was the top biological process emerging from the amyloid secretase network and includes genes coding for proteins that mediate the effects of brain-derived neurotrophic factor signaling, implicated in the pathophysiology of MDD and associated with the activation of inositol phosphate metabolism (Duman and Voleti 2012), which was also a top biological process identified in our analysis. Taken together, our findings are consistent with the hypothesis that environmental conditions that increase the risk for MDD associate with alterations in mechanisms controlling synaptic plasticity in cortico-limbic brain regions (Duman and Aghajanian 2012).
Interestingly, the direction of the interaction between maternal depressive symptoms and infant genotype on right amygdala volume in the US sample is in the opposite direction to that observed in the Asian sample. This is not surprising as there is now considerable evidence for opposing effects of individual SNPs between Asian and Caucasian populations. For instance, Sheikh et al. 2013 found that during childhood Caucasian children homozygous for the Catechol-O-methyltransferase (COMT) val allele show higher levels of internalizing symptoms compared to children with at least one copy of the COMT met allele. In contrast, Baumann et al. 2013 found that in Asians homozyges of the COMT met allele with adverse childhood experiences show higher anxiety sensitivity. Similar findings are apparent in studies of the serotonin transporter polomorphism (5-HTTPLR). Ming et al. 2015 report a replication of the well-established interaction between the 5-HTTPLR and stressful life events on emotional health in Han Chinese, the principal ethnic group in our Asian sample. However, in this study it was the L and not the S allele that conferred greater sensitivity to stress; studies with Caucasian samples commonly report that the S allele confers sensitivity to adversity. This study was also a replication of a previous, similar finding with the 5-HTTPLR variant (Ming et al. 2013). Meta-analyses likewise reveal differential effects of 5-HTTLPR on selective serotonin reuptake inhibitors efficacy between Caucasians and Asians (Serretti et al. 2007; Porcelli et al. 2012). Importantly, functional MRI studies show a link between S allele and higher amygdala activation in response to emotional stimuli in Caucasians (e.g., Hariri et al. 2002, 2005; Murphy et al. 2013), whereas in Asians L allele is associated with amygdala hyperactivation (Li et al. 2012). These findings imply that the same risk allele may operate in the opposite fashion as a function of ethnic background. Allele frequency may be a factor. The frequency of the L allele is much lower than that of S allele in Chinese population and Japan populations, but higher in Caucasians or African–Americans (Lesch et al. 1996; He et al. 2010). Importantly, despite the directional difference, the pathway analyses revealed the same underlying biological processes for the amygdala effect in both samples. Indeed, PANTHER identified the same primary pathway for both samples using independent analyses. These findings are consistent with the studies noted above, that reveal effects of polymorphisms in the same genetic regions, consistent with common underlying biological processes, but with polymorphisms operating in an ethnicity-specific manner.
The relationship between antenatal maternal depression and children's outcomes is often observed as a function of gender. Recent evidence suggests that girls born to depressed mothers may be more likely to exhibit increased cortisol reactivity in childhood and subsequently altered amygdala–prefrontal cortex activity in adolescence (Burghy et al. 2012). Nevertheless, our study did not observe the gender effect, which may partly because of the age of children in this study. It remains possible that gender differences could be observable at later stages in development.
This study is best considered as exploratory and intended to provide a strategy for the analysis of gene–environment interdependence and identify candidate biological processes that might mediate the influence of environment conditions associated with the risk for MDD on fetal brain development. While preliminary, our findings do suggest biologically plausible candidate processes in two independent cohorts. While the neonatal neuroimaging dataset in this study is highly unique in its timing of acquisition and number of subjects, it is nevertheless a modest sample size for genomic analyses. Due to the time limit for scanning of neonates and brain immaturity, the neonatal brain MRI, in general, has relatively low contrast between gray and white matter, which could be challenging for imaging and the brain segmentation. The contrast between gray matter and white matter is relatively clear on T2-weighted MRI at the neonatal stage but is diminished around 4–9 months of age. We reported the brain segmentation accuracy in the Materials and Methods section to indicate our constraint with the neonatal brain images. An additional limitation is the use of GWAS datasets derived from Caucasians for the definition of a GPRS to study within an Asian sample. While this is an important caveat, we note that the Wang et al. (2015) study identified multiple SNPs in the SNAP-25 gene, which is a member of the SNARE complex, in the pathophysiology of MDD in Asian population. Moreover, our assessment of maternal depressive symptoms was based on a common screening tool intended to elicit a subjective report of emotional well-being, but does not constitute clinical assessment. The brain variations in the offspring are thus best considered as being associated with self-reported depressive symptoms and not to clinical depression, per se. Furthermore, other factors, such as marital discord, paternal mental illness, medication during pregnancy, maternal substance use, or breastfeeding, could also influence the fetal brain development over and beyond antenatal maternal depression since the brain develops rapidly in early life (Uematsu et al. 2012). In our study, SES was highly correlated with antenatal maternal depressive symptoms and both showed its interaction with GPRSMDD and influence the right amygdala and hippocampus (Supplementary Table S2 and Figures S1 and S2). After controlling for each other, the interaction effect of antenatal maternal depression and GPRSMDD was mainly shown on the right amygdala, while the interaction effect of SES and GPRSMDD was mainly shown on the right hippocampus (the Supplementary Material). These findings suggested that environment conditions associated with the risk for MDD interplay with genetic risks for MDD on fetal brain development. Finally, comparing brain anatomical measures across different cohorts, particularly, on neonates, is challenging. This is partly because of different strength of the MRI scanners and imaging acquisition protocols. More importantly, the challenge also lies in the definition of the anatomy of individual brain structures. In this study, we obtained the comparable volume measure for the amygdala from the two cohorts. Hence, this study incorporated the amygdala findings from both cohorts where we could be certain of comparable anatomical localization. Furthermore, the US cohort was lack of maternal GPRSMDD information and hence our reported results for the US cohort could not count for the contribution of maternal GPRSMDD.
Conclusion
We present a hypothesis-generating approach to the study of gene × environment interaction that uses GPRS together with bioinformatic approaches to identify candidate biological processes mediating the influence of environmental conditions on brain development and function. This approach is considered as hypothesis-generating. For example, in larger samples, with sufficient statistical power, we predict that a multi-gene locus score, reflecting polymorphisms in genes involved in the biological processes of glutamate receptor activity, SNARE complex, 5-HT2 receptor signaling, should moderate brain region-specific effects of “depressogenic” environmental conditions during fetal development.
Supplementary Material
Notes
We thank the GUSTO study group and all clinical and home visit staff involved. The voluntary participation of all participants is greatly appreciated. The GUSTO study group includes Pratibha Agarwal, Arijit Biswas, Choon Looi Bong, Shirong Cai, Jerry Kok Yen Chan, Yiong Huak Chan, Cornelia Yin Ing Chee, Yin Bun Cheung, Audrey Chia, Amutha Chinnadurai, Chai Kiat Chng, Mary Foong-Fong Chong, Shang Chee Chong, Mei Chien Chua, Chun Ming Ding, Eric Andrew Finkelstein, Doris Fok, Keith M. Godfrey, Anne Eng Neo Goh, Yam Thiam Daniel Goh, Joshua J. Gooley, Wee Meng Han, Mark Hanson, Christiani Jeyakumar Henry, Joanna D. Holbrook, Chin-Ying Hsu, Hazel Inskip, Jeevesh Kapur, Ivy Yee-Man Lau, Bee Wah Lee, Yung Seng Lee, Ngee Lek, Sok Bee Lim, Yen-Ling Low, Iliana Magiati, Lourdes Mary Daniel, Cheryl Ngo, Krishnamoorthy Naiduvaje, Wei Wei Pang, Boon Long Quah, Victor Samuel Rajadurai, Mary Rauff, Salome A. Rebello, Jenny L. Richmond, Lynette Pei-Chi Shek, Allan Sheppard, Borys Shuter, Leher Singh, Shu-E Soh, Walter Stunkel, Lin Lin Su, Kok Hian Tan, Oon Hoe Teoh, Mya Thway Tint, Hugo P S van Bever, Rob M. van Dam, Inez Bik Yun Wong, P. C. Wong, Fabian Yap, George Seow Heong Yeo. Conflict of Interest: None declared.
Supplementary Material
Funding
The National Medical Research Council (NMRC; NMRC/TCR/004-NUS/2008 and NMRC/CBRG/0039/2013), the Singapore Ministry of Education Academic Research Fund Tier 2 (MOE2012-T2-2-130) and funding from the Hope for Depression Research Foundation, the Sackler Foundation, the National Institute of Health (R01 MH091351, R01 HD060628), and ERA-NET NEURON 01EW1407A and the Canadian Institutes for Advanced Research.
References
- Abercrombie HC, Schaefer SM, Larson CL, Oakes TR, Lindgren KA, Holden JE, Perlman SB, Turski PA, Krahn DD, Benca RM, et al. 1998. Metabolic rate in the right amygdala predicts negative affect in depressed patients. Neuroreport. 9:3301–3307. [DOI] [PubMed] [Google Scholar]
- Bai J, Abdul-Rahman MF, Rifkin-Graboi A, Chong YS, Kwek K, Saw SM, Godfrey KM, Gluckman PD, Fortier MV, Meaney MJ, et al. 2012. Population differences in brain morphology and microstructure among Chinese, Malay, and Indian Neonates. PLoS One. 7:e47816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann C, Klauke B, Weber H, Domschke K, Zwanzger P, Pauli P, Deckert J, Reif A. 2013. The interaction of early life experiences with COMT val158met affects anxiety sensitivity. Genes Brain Behav. 12:821–829. [DOI] [PubMed] [Google Scholar]
- Bonanno G, Giambelli R, Raiteri L, Tiraboschi E, Zappettini S, Musazzi L, Raiteri M, Racagni G, Popoli M. 2005. Chronic antidepressants reduce depolarization-evoked glutamate release and protein interactions favoring formation of SNARE complex in hippocampus. J Neurosci. 25:3270–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruder-Costello B, Warner V, Talati A, Nomura Y, Bruder G, Weissman M. 2007. Temperament among offspring at high and low risk for depression. Psychiatry Res. 153:145–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bures M, Swanson CJ, Linden A-M, Schoepp DD, Johnson MP, Monn JA. 2005. Metabotropic glutamate receptors as novel targets for anxiety and stress disorders. Nat Rev Drug Discov. 4:131–144. [DOI] [PubMed] [Google Scholar]
- Burghy CA, Stodola DE, Ruttle PL, Molloy EK, Armstrong JM, Oler JA, Fox ME, Hayes AS, Kalin NH, Essex MJ, et al. 2012. Developmental pathways to amygdala-prefrontal function and internalizing symptoms in adolescence. Nat Neurosci. 15:1736–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caddy C, Giaroli G, White TP, Shergill SS, Tracy DK. 2014. Ketamine as the prototype glutamatergic antidepressant: pharmacodynamic actions, and a systematic review and meta-analysis of efficacy. Ther Adv Psychopharmacol. 4:75–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrion VG, Weems CF, Reiss AL. 2007. Stress predicts brain changes in children: a pilot longitudinal study on youth stress, posttraumatic stress disorder, and the hippocampus. Pediatrics. 119:509–516. [DOI] [PubMed] [Google Scholar]
- Chung MK, Robbins SM, Dalton KM, Davidson RJ, Alexander AL, Evans AC. 2005. Cortical thickness analysis in autism with heat kernel smoothing. Neuroimage. 25:1256–1265. [DOI] [PubMed] [Google Scholar]
- Chung MK, Worsley KJ, Nacewicz BM, Dalton KM, Davidson RJ. 2010. General multivariate linear modeling of surface shapes using SurfStat. Neuroimage. 53:491–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox JL, Holden JM, Sagovsky R. 1987. Detection of postnatal depression. Development of the 10-item Edinburgh postnatal depression scale. Br J Psychiatry. 150:782–786. [DOI] [PubMed] [Google Scholar]
- Du J, Younes L, Qiu A. 2011. Whole brain diffeomorphic metric mapping via integration of sulcal and gyral curves, cortical surfaces, and images. Neuroimage. 56:162–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duman RS, Aghajanian GK. 2012. Synaptic dysfunction in depression: potential therapeutic targets. Science. 338:68–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duman RS, Voleti B. 2012. Signaling pathways underlying the pathophysiology and treatment of depression: novel mechanisms for rapid-acting agents. Trends Neurosci. 35:47–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan LE, Keller MC. 2011. A critical review of the first 10 years of candidate gene-by-environment interaction research in psychiatry. Am J Psychiatry. 168:1041–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren-Kocak E, Turner CA, Watson SJ, Akil H. 2011. Short-hairpin RNA silencing of endogenous fibroblast growth factor 2 in rat hippocampus increases anxiety behavior. Biol Psychiatry. 69:534–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farah MJ, Hackman DA, Meaney MJ. 2010. Socioeconomic status and the brain: mechanistic insights from human and animal research. Nat Rev Neurosci. 11:651–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffrey MS, Luby JL, Belden AC, Hirshberg JS, Volsch J, Barch DM. 2011. Association between depression severity and amygdala reactivity during sad face viewing in depressed preschoolers: an fMRI study. J Affect Disord. 129:364–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilman SE, Kawachi I, Fitzmaurice GM, Buka SL. 2003. Family disruption in childhood and risk of adult depression. Am J Psychiatry. 160:939–946. [DOI] [PubMed] [Google Scholar]
- Hamilton JP, Gotlib IH. 2008. Neural substrates of increased memory sensitivity for negative stimuli in major depression. Biol Psychiatry. 63:1155–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammock EA, Levitt P. 2013. Oxytocin receptor ligand binding in embryonic tissue and postnatal brain development of the C57BL/6 J mouse. Front Behav Neurosci. 7:195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson JL, Chung MK, Avants BB, Shirtcliff EA, Gee JC, Davidson RJ, Pollak SD. 2010. Early stress is associated with alterations in the orbitofrontal cortex: a tensor-based morphometry investigation of brain structure and behavioral risk. J Neurosci. 30:7466–7472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariri AR, Drabant EM, Munoz KE, Kolachana BS, Mattay VS, Egan MF, Weinberger DR. 2005. A susceptibility gene for affective disorders and the response of the human amygdala. Arch Gen Psychiatry. 62:146–152. [DOI] [PubMed] [Google Scholar]
- Hariri AR, Mattay VS, Tessitore A, Kolachana B, Fera F, Goldman D, Egan MF, Weinberger DR. 2002. Serotonin transporter genetic variation and the response of the human amygdala. Science. 297:400–403. [DOI] [PubMed] [Google Scholar]
- Hashimoto K, Sawa A, Iyo M. 2007. Increased levels of glutamate in brains from patients with mood disorders. Biol Psychiatry. 62:1310–1316. [DOI] [PubMed] [Google Scholar]
- He Q, Xue G, Chen C, Lu Z, Dong Q, Lei X, Ding N, Li J, Li H, Chen C, et al. 2010. Serotonin transporter gene-linked polymorphic region (5-HTTLPR) influences decision making under ambiguity and risk in a large Chinese sample. Neuropharmacology. 59:518–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes AJ, Lee PH, Hollinshead MO, Bakst L, Roffman JL, Smoller JW, Buckner RL. 2012. Individual differences in amygdala-medial prefrontal anatomy link negative affect, impaired social functioning, and polygenic depression risk. J Neurosci. 32:18087–18100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Willner D, Danoy P, Xu H, Brown MA. 2013. Comparison of the performance of two commercial genome-wide association study genotyping platforms in Han Chinese samples. G3 (Bethesda). 3:23–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone T, van Reekum CM, Urry HL, Kalin NH, Davidson RJ. 2007. Failure to regulate: counterproductive recruitment of top-down prefrontal-subcortical circuitry in major depression. J Neurosci. 27:8877–8884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupfer DJ, Frank E, Phillips ML. 2012. Major depressive disorder: new clinical, neurobiological, and treatment perspectives. Lancet. 379:1045–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PH, Perlis RH, Jung JY, Byrne EM, Rueckert E, Siburian R, Haddad S, Mayerfeld CE, Heath AC, Pergadia ML, et al. 2012. Multi-locus genome-wide association analysis supports the role of glutamatergic synaptic transmission in the etiology of major depressive disorder. Transl Psychiatry. 2:e184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesch K-P, Waider J. 2012. Serotonin in the modulation of neural plasticity and networks: implications for neurodevelopmental disorders. Neuron. 76:175–191. [DOI] [PubMed] [Google Scholar]
- Lesch KP, Bengel D, Heils A, Sabol SZ, Greenberg BD, Petri S, Benjamin J, Muller CR, Hamer DH, Murphy DL. 1996. Association of anxiety-related traits with a polymorphism in the serotonin transporter gene regulatory region. Science. 274:1527–1531. [DOI] [PubMed] [Google Scholar]
- Li S, Zou Q, Li J, Li J, Wang D, Yan C, Dong Q, Zang YF. 2012. 5-HTTLPR polymorphism impacts task-evoked and resting-state activities of the amygdala in Han Chinese. PLoS One. 7:e36513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubke GH, Hottenga JJ, Walters R, Laurin C, de Geus EJ, Willemsen G, Smit JH, Middeldorp CM, Penninx BW, Vink JM, et al. 2012. Estimating the genetic variance of major depressive disorder due to all single nucleotide polymorphisms. Biol Psychiatry. 72:707–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins-de-Souza D, Guest PC, Harris LW, Vanattou-Saifoudine N, Webster MJ, Rahmoune H, Bahn S. 2012. Identification of proteomic signatures associated with depression and psychotic depression in post-mortem brains from major depression patients. Transl Psychiatry. 2:e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael N, Erfurth A, Ohrmann P, Arolt V, Heindel W, Pfleiderer B. 2003. Neurotrophic effects of electroconvulsive therapy: a proton magnetic resonance study of the left amygdalar region in patients with treatment-resistant depression. Neuropsychopharmacology. 28:720–725. [DOI] [PubMed] [Google Scholar]
- Ming Q, Zhang Y, Yi J, Wang X, Zhu X, Yao S. 2015. Serotonin transporter gene polymorphism (5-HTTLPR) L allele interacts with stress to increase anxiety symptoms in Chinese adolescents: a multiwave longitudinal study. BMC Psychiatry. 15:248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming QS, Zhang Y, Chai QL, Chen HY, Hou CJ, Wang MC, Wang YP, Cai L, Zhu XZ, Yi JY, et al. 2013. Interaction between a serotonin transporter gene promoter region polymorphism and stress predicts depressive symptoms in Chinese adolescents: a multi-wave longitudinal study. BMC Psychiatry. 13:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt TE, Caspi A. 2006. Gene-environment interactions in psychiatry: joining forces with neuroscience. Nat Rev Neurosci. 7:583–590. [DOI] [PubMed] [Google Scholar]
- Monk CS, Klein RG, Telzer EH, Schroth EA, Mannuzza S, Moulton JL 3rd, Guardino M, Masten CL, McClure-Tone EB, Fromm S, et al. 2008. Amygdala and nucleus accumbens activation to emotional facial expressions in children and adolescents at risk for major depression. Am J Psychiatry. 165:90–98. [DOI] [PubMed] [Google Scholar]
- Mostafavi S, Ray D, Warde-Farley D, Grouios C, Morris Q. 2008. GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 9(Suppl 1):S4 10.1186/gb-2008-9-s1-s4 Epub 2008 Jun 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy SE, Norbury R, Godlewska BR, Cowen PJ, Mannie ZM, Harmer CJ, Munafo MR. 2013. The effect of the serotonin transporter polymorphism (5-HTTLPR) on amygdala function: a meta-analysis. Mol Psychiatry. 18:512–520. [DOI] [PubMed] [Google Scholar]
- Noble KG, Houston SM, Brito NH, Bartsch H, Kan E, Kuperman JM, Akshoomoff N, Amaral DG, Bloss CS, Libiger O, et al. 2015. Family income, parental education and brain structure in children and adolescents. Nat Neurosci. 18:773–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliaccio D, Luby J, Gaffrey M, Belden A, Botteron K, Gotlib IH, Barch DM. 2012. Anomalous functional brain activation following negative mood induction in children with pre-school onset major depression. Dev Cogn Neurosci. 2:256–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliaccio D, Luby JL, Luking KR, Belden AC, Barch DM. 2014. Brain-behavior relationships in the experience and regulation of negative emotion in healthy children: implications for risk for childhood depression. Dev Psychopathol. 26:1289–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Edgar K, Roberson-Nay R, Hardin MG, Poeth K, Guyer AE, Nelson EE, McClure EB, Henderson HA, Fox NA, Pine DS, et al. 2007. Attention alters neural responses to evocative faces in behaviorally inhibited adolescents. Neuroimage. 35:1538–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyrot WJ, Milaneschi Y, Abdellaoui A, Sullivan PF, Hottenga JJ, Boomsma DI, Penninx BW. 2014. Effect of polygenic risk scores on depression in childhood trauma. Br J Psychiatry. 205:113–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piroli GG, Reznikov LR, Grillo CA, Hagar JM, Fadel JR, Reagan LP. 2013. Tianeptine modulates amygdalar glutamate neurochemistry and synaptic proteins in rats subjected to repeated stress. Exp Neurol. 241:184–193. [DOI] [PubMed] [Google Scholar]
- Porcelli S, Fabbri C, Serretti A. 2012. Meta-analysis of serotonin transporter gene promoter polymorphism (5-HTTLPR) association with antidepressant efficacy. Eur Neuropsychopharmacol. 22:239–258. [DOI] [PubMed] [Google Scholar]
- Pruessner JC, Dedovic K, Khalili-Mahani N, Engert V, Pruessner M, Buss C, Renwick R, Dagher A, Meaney MJ, Lupien S. 2008. Deactivation of the limbic system during acute psychosocial stress: evidence from positron emission tomography and functional magnetic resonance imaging studies. Biol Psychiatry. 63:234–240. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, Sullivan PF, Sklar P. 2009. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 460:748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu A, Bitouk D, Miller MI. 2006. Smooth Functional and Structural Maps on the Neocortex via Orthonormal Bases of the Laplace-Beltrami Operator. IEEE Trans Med Imaging. 25(10):1296–1306. [DOI] [PubMed] [Google Scholar]
- Qiu A, Miller MI. 2008. Multi-Structure Network Shape Analysis via Normal Surface Momentum Maps. NeuroImage. 42(4):1430–1438. [DOI] [PubMed] [Google Scholar]
- Qiu A, Adler M, Crocetti D, Miller MI, Mostofsky SH. 2010. Basal ganglia shapes predict social, communication, and motor dysfunctions in boys with autism spectrum disorder. J Am Acad Child Adolesc Psychiatry. 49:539–551 e531-534. [DOI] [PubMed] [Google Scholar]
- Qiu A, Anh TT, Li Y, Chen H, Rifkin-Graboi A, Broekman BF, Kwek K, Saw SM, Chong YS, Gluckman PD, et al. 2015. a. Prenatal maternal depression alters amygdala functional connectivity in 6-month-old infants. Transl Psychiatry. 5:e508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu A, Crocetti D, Adler M, Mahone EM, Denckla MB, Miller MI, Mostofsky SH. 2009. Basal ganglia volume and shape in children with attention deficit hyperactivity disorder. Am J Psychiatry. 166:74–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu A, Rifkin-Graboi A, Chen H, Chong YS, Kwek K, Gluckman PD, Fortier MV, Meaney MJ. 2013. Maternal anxiety and infants’ hippocampal development: timing matters. Transl Psychiatry. 3:e306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu A, Tuan TA, Ong ML, Li Y, Chen H, Rifkin-Graboi A, Broekman BF, Kwek K, Saw SM, Chong YS, et al. 2015. b. COMT haplotypes modulate associations of antenatal maternal anxiety and neonatal cortical morphology. Am J Psychiatry. 172:163–172. [DOI] [PubMed] [Google Scholar]
- Rifkin-Graboi A, Bai J, Chen H, Hameed WB, Sim LW, Tint MT, Leutscher-Broekman B, Chong YS, Gluckman PD, Fortier MV, et al. 2013. Prenatal maternal depression associates with microstructure of right amygdala in neonates at birth. Biol Psychiatry. 74:837–844. [DOI] [PubMed] [Google Scholar]
- Ripke S, Wray NR, Lewis CM, Hamilton SP, Weissman MM, Breen G, Byrne EM, Blackwood DH, Boomsma DI, Cichon S, et al. 2013. A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry. 18:497–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarr E, Gray L, Keriakous D, Robinson PJ, Dean B. 2006. Increased levels of SNAP-25 and synaptophysin in the dorsolateral prefrontal cortex in bipolar I disorder. Bipolar Disord. 8:133–143. [DOI] [PubMed] [Google Scholar]
- Serretti A, Kato M, De Ronchi D, Kinoshita T. 2007. Meta-analysis of serotonin transporter gene promoter polymorphism (5-HTTLPR) association with selective serotonin reuptake inhibitor efficacy in depressed patients. Mol Psychiatry. 12:247–257. [DOI] [PubMed] [Google Scholar]
- Sheikh HI, Kryski KR, Smith HJ, Dougherty LR, Klein DN, Bufferd SJ, Singh SM, Hayden EP. 2013. Catechol-O-methyltransferase gene val158met polymorphism and depressive symptoms during early childhood. Am J Med Genet B Neuropsychiatr Genet. 162B:245–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skolnick P, Popik P, Trullas R. 2009. Glutamate-based antidepressants: 20 years on. Trends Pharmacol Sci. 30:563–569. [DOI] [PubMed] [Google Scholar]
- Smith SM. 2002. Fast robust automated brain extraction. Hum Brain Mapp. 17:143–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soh SE, Lee SS, Hoon SW, Tan MY, Goh A, Lee BW, Shek LP, Teoh OH, Kwek K, Saw SM, et al. 2012. The methodology of the GUSTO cohort study: a novel approach in studying pediatric allergy. Asia Pac Allergy. 2:144–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, Neale MC, Kendler KS. 2000. Genetic epidemiology of major depression: review and meta-analysis. Am J Psychiatry. 157:1552–1562. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Botteron KN, Luby JL, Belden AC, Gaffrey MS, Babb CM, Nishino T, Miller MI, Ratnanather JT, Barch DM. 2013. Structural-functional correlations between hippocampal volume and cortico-limbic emotional responses in depressed children. Cogn Affect Behav Neurosci. 13:135–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Qiu A. 2016. Large Deformation Multiresolution Diffeomorphic Metric Mapping for Multiresolution Cortical Surfaces: A Coarse-to-Fine Approach. IEEE Trans Image Process. 25(9):4061–4074. [DOI] [PubMed] [Google Scholar]
- Terracciano A, Tanaka T, Sutin AR, Sanna S, Deiana B, Lai S, Uda M, Schlessinger D, Abecasis GR, Ferrucci L, et al. 2010. Genome-wide association scan of trait depression. Biol Psychiatry. 68:811–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas KM, Drevets WC, Dahl RE, Ryan ND, Birmaher B, Eccard CH, Axelson D, Whalen PJ, Casey BJ. 2001. Amygdala response to fearful faces in anxious and depressed children. Arch Gen Psychiatry. 58:1057–1063. [DOI] [PubMed] [Google Scholar]
- Uematsu A, Matsui M, Tanaka C, Takahashi T, Noguchi K, Suzuki M, Nishijo H. 2012. Developmental trajectories of amygdala and hippocampus from infancy to early adulthood in healthy individuals. PLoS One. 7:e46970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Marle HJ, Hermans EJ, Qin S, Fernandez G. 2009. From specificity to sensitivity: how acute stress affects amygdala processing of biologically salient stimuli. Biol Psychiatry. 66:649–655. [DOI] [PubMed] [Google Scholar]
- Wang Q, Wang Y, Ji W, Zhou G, He K, Li Z, Chen J, Li W, Wen Z, Shen J, et al. 2015. SNAP25 is associated with schizophrenia and major depressive disorder in the Han Chinese population. J Clin Psychiatry. 76:e76–e82. [DOI] [PubMed] [Google Scholar]
- Wen D, Poh J, Soe N, Chong Y, Chen H, Kwek K, Shek L, Gluckman P, Fortier M, Meaney M, et al. 2017. Influences of prenatal and postnatal maternal depression on amygdala volume and microstructure in young children. Transl Psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichers M, Geschwind N, Jacobs N, Kenis G, Peeters F, Derom C, Thiery E, Delespaul P, van Os J. 2009. Transition from stress sensitivity to a depressive state: longitudinal twin study. Br J Psychiatry. 195:498–503. [DOI] [PubMed] [Google Scholar]
- Woo JM, Yoon KS, Choi YH, Oh KS, Lee YS, Yu BH. 2004. The association between panic disorder and the L/L genotype of catechol-O-methyltransferase. J Psychiatr Res. 38:365–370. [DOI] [PubMed] [Google Scholar]
- Zhong J, Qiu A. 2010. Multi-Manifold Diffeomorphic Metric Mapping for Aligning Cortical Hemispheric Surfaces. NeuroImage. 49(1):355–365. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.