Abstract
ADP-ribosylation is a post-translational modification of proteins found in organisms from all kingdoms of life which regulates many important biological functions including DNA repair, chromatin structure, unfolded protein response, and apoptosis. Several cellular enzymes, such as macrodomain containing proteins PARG and TARG1, reverse protein ADP-ribosylation. Here we show that human Nudix-Type Motif 16 (hNUDT16) represents a new enzyme class that can process protein ADP-ribosylation in vitro, converting it into ribose-5’-phosphate tags covalently attached to the modified proteins. Furthermore, our data show that hNUDT16 enzymatic activity can be used to trim ADP-ribosylation on proteins in order to facilitate analysis of ADP-ribosylation sites in proteins by mass spectrometry.
Keywords: Post-translational modification (PTM), ADP-ribosylation, PARP, Nudix hydrolases, NUDT16
Introduction
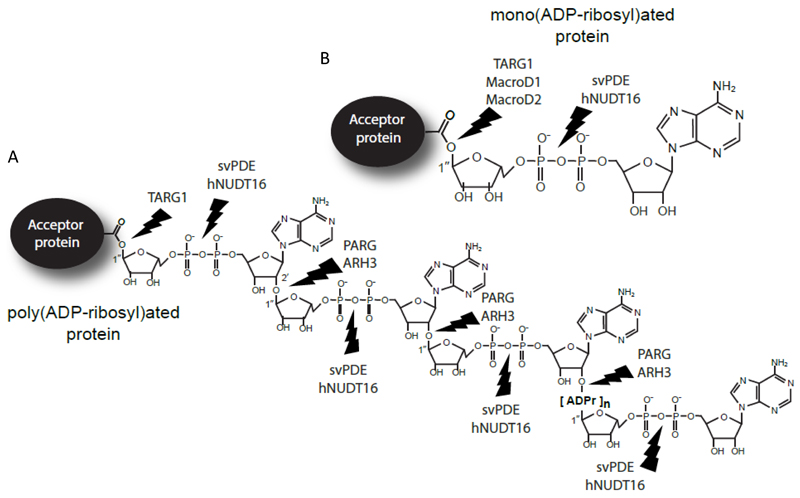
ADP-ribosylation is a post-translational modification (PTM) of proteins found in organisms from all kingdoms of life that regulates many important cellular processes [1,2]. The poly(ADP-ribose) polymerases (PARPs) are the major family of intracellular enzymes which transfer ADP-ribose (ADPr) from donor NAD+ onto target proteins, primarily the glutamate and aspartate residues [2–8]. While some PARPs transfer only a single ADPr unit (mono-ADPr; MAR), other PARPs are able to attach repeating units of ADPr via a unique 2’,1”-O-glycosydic ribose-ribose bond to generate long chains of poly-ADPr (PAR), [5–7]. Poly(ADP-ribosyl)ation (PARylation) is a highly dynamic PTM with an estimated half-life of only 1-6 min [9]. Poly(ADP-ribose) glycohydrolase (PARG) is the main enzyme in humans for PAR hydrolysis, which specifically cleaves the ribose-ribose bonds between the ADPr subunits of the PAR chains using its catalytic macrodomain [10,11,12,13] (Fig. 1A). Another enzyme that can act on these bonds is ADP-ribosylhydrolase 3 (ARH3) [14]. However, these two enzymes are unable to cleave MAR attached to a protein [11], which is in turn the substrate for three other macrodomain proteins: terminal ADP-Ribose protein glycohydrolase (TARG1), MacroD1 and MacroD2 [15–17] (Fig. 1B). Together, this makes ADP-ribosylation a dynamic, reversible PTM that has been linked to many important cellular processes such as the DNA damage response, Wnt signalling and NF-kB signaling [4,18]. Intriguingly, other enzymes may have the potential to process ADP-ribose chains in yet another way. For example, accumulation of glutamyl ribose-5’-phosphate (glutamyl-R5P) has been detected in the tissue extracts from a patient that died from neurological deterioration and renal failure [19], indicating the existence of an unknown cellular phosphodiesterase that can hydrolyse the pyrophosphate bond within an ADPr molecule attached to a glutamate group in proteins (thereby releasing adenine monophosphate (AMP) and peptidyl-R5P, which is subsequently degraded by cellular proteases). More recently, R5P sites have been also identified on arginine residues of several cellular proteins [20]. Nucleoside diphosphate linked moiety X (Nudix) hydrolases class of enzymes could be responsible for such reaction products [21,22]. Nudix hydrolases are found in Bacteria, Viruses, Eukaryotes, and Archaea and some of their family members have been shown to act on free ADPr [21,22]. Nudix proteins are characterized by a highly conserved 23-amino-acid Nudix motif, GX5EX7REUXEEXGU, where U is an aliphatic or hydrophobic residue [21,22]. Nudix enzymes have protective, regulatory, and signalling functions in metabolism through their ability to remove a wide range of organic pyrophosphates from the cellular environment [22]. Although prokaryotic Nudix enzymes, like E. coli Orf209 and Thermus thermofilus HB8 (TtADPRase), and human NUDT5 and NUDT9 are active as ADPr pyrophosphatase on free ADPr [21–23], no Nudix proteins were tested for their capacity to hydrolyse ADP-ribosylation directly linked to proteins. In the present study, we investigated 11 human Nudix enzymes and tested them for the ability to cleave protein PARylation and MARylation. Among the Nudix members tested in our study, we found human Nudix-Type Motif 16 (hNUDT16) as the only one able to remove both PAR- and MARylation (Fig. 1A and B). Our study represents the first evidence that intracellular phosphodiesterases can process protein ADP-ribosylation and convert it into another PTM, namely phosphoribosylation (the functionality of the R5P protein tags in vivo is currently unknown). We also show that hNUDT16 can be used as a tool to trim the complex protein ADP-ribosylation into a simple mark easily detectable by mass spectrometry and may represent a better alternative to the commonly used snake venom Phosphodiesterase I (svPDE) [24–26].
Figure 1. Schematic representation of the enzymes that can hydrolyse the protein ADP-ribosylation.
A and B Schematic representation showing PARylated (A) and MARylated (B) proteins and the chemical bonds cleaved by hNUDT16, svPDE, PARG, ARH3, TARG1, MacroD1 and MacroD2.
Experimental Procedures
Plasmids and Proteins
PARP1 wild type protein was from Trevigen (high specific activity). PARP1-E988Q was expressed from pET28a(+) and purified as previously described [15]. pET28a His6-Herpetosiphon aurantiacus PARP was purified as previously described [11]. hNUDT1 was expressed from the pET28a(+) vector in E.coli BL21 DE3, hNUDT4 and hNUDT9 were amplified from cDNA prepared from HL60 cells using PCR and subcloned into the pET28a(+) vector. Bacterial expression constructs of hNUDT3, hNUDT5, hNUDT10-12 and hNUDT14-16 in pNIC28-Bsa4 were kind gifts from the Protein Science Facility (PSF) at Karolinska Institutet. hNUDT5 was expressed in-house as earlier described [27]. hNUDT3-4, hNUDT9-12 and hNUDT14-16 were initially expressed and purified by PSF at Karolinska Institutet using HisTrap HP (GE Healthcare) followed by gel filtration on HiLoad 16/60 Superdex 75 (GE Healthcare). The purity of all protein preparations were verified by SDS-PAGE followed by Coomassie staining. For expression and purification of hNUDT16, 1.5 L of transformed Rosetta2 (DE) cells were grown in TB medium in presence of 100 μg/mL Kanamycin and 34 μg/mL Chloramphenicol at 37°C. When the optical density (OD) reached 2.0, the temperature was decreased to 18°C. When the OD reached 3.0, the expression of recombinant protein was induced with isopropyl-thiogalactopyranoside (IPTG) 0.2 mM and continued over the night. Bacteria were then lysed fusing BugBuster protein extraction reagent (Novagen) and Benzonase (Sigma) in a buffer composed by 20 mM Tris-HCl pH 8.0, 500 mM NaCl, 10 mM Imidazole, 5 μM β-mercaptoethanol, 10% glycerol and supplemented with Complete Protease Inhibitor (Roche). After 1 hour incubation, the lysate was clarified by centrifugation and supernatant applied on 5 mL HisTrap HP (GE Healthcare) affinity chromatography, the His tagged protein was eluted in 500 mM Imidazole and further purified by size-exclusion chromatography on a Superdex 200 column (GE Healthcare) using ÄKTA pure (GE Healthcare) dialysing the protein in a buffer composed by 25 mM Tris pH 8.0, 300 mM NaCl, 10% Glycerol and 1mM DTT. Protein were then concentrated up to 9 mg/mL using 10 kDa Amicon-Ultra centrifugal filter (Millipore).
Purification of snake venom Phosphodiesterase
Phosphodiesterase I (svPDE) from Crotalus adamanteus venom was purified as previously described [24–26], with slightly modifications. Briefly, approximately 2,52 mg dried weight of partially purified svPDE (Whorthigton) were dissolved in 1 mL of loading buffer (10 mM Tris-HCl pH 7.5, 50 mM NaCl, 10% Glycerol) and loaded onto a pre-equilibrated 1 mL HiTrap blue HP (GE Healthcare). The column was washed with 5 column volumes (CV) of loading buffer followed by an increasing gradient of KPO4 pH 8.0 up to 150 mM. The svPDE protein was eluted using 1M KPO4 buffer. Desired fractions were pulled and loaded onto analytical size-exclusion chromatography Superdex 200 (GE Healthcare) using ÄKTA pure (GE Healthcare) in a buffer composed of 10 mM Tris pH 8.0, 50 mM NaCl, 15 mM MgCl2 and 1% Glycerol. The fractions were assayed separately for PAR-hydrolytic and proteolytic activity. Concentration of desired fractions (~97 kDa molecular weight) was measured using Nanodrop (Thermo) and stored at -80 °C.
Assay for hydrolysis of free ADP-ribose
Hydrolysis of the free ADP-ribose was assayed for 20 minutes at 22 °C in assay buffer (100 mM TrisAcetate pH 8.0, 40 mM NaCl, 10 mM MgAc, 1 mM DTT and 0,005% Tween 20) in quadruplicate. Briefly, 50 μM ADP-ribose was incubated together with Nudix enzyme in buffer containing an excess of the coupled enzyme Calf Intestinal Phosphatase (Sigma) (5U/ml), converting formed R5P to inorganic-phosphate (Pi) and ribose. Released Pi was detected using a colorimetric method using a malachite green reagent [28] and measurement of the absorbance at 630 nm. The concentrations of Nudix enzymes in the reaction were: hNUDT1, 250 nM; hNUDT5, 6 nM; hNUDT9, 25 nM; hNUDT12, 250 nM; hNUDT14, 4 nM; hNUDT15, 75 nM; hNUDT16 250 nM. The signal was converted to formed Pi per μM Nudix enzyme per minute using a standard curve (A630=0.01743*[Pi] (μM)).
Nudix activity assays on ADP-ribosylated proteins
Poly(ADP-ribosyl)ated and mono(ADP-ribosylated) PARP1 proteins were prepared as described [15,29] in a reaction buffer containing 50 mM Tris-HCl (pH 8.0), 4 mM MgCl2, 50 mM NaCl, 0.2 mM DTT, 200 mM NAD+ (Trevigen) and 130 ng activated DNA (BPS Bioscience). For the PAR hydrolysis activity assays 0.5 μM PARylated PARP1 substrate was used in 10 μL reaction. We estimate that the average number of the ADP-ribose units in chains linked to a PARP1 protein prepared this way is 40 units per protein, giving an approximate concentration of 20 µM monomeric units in the reaction. For the MARylated PARP1, 2 μM of PARP1-E988Q was used as a substrate. Reactions were stopped by the addition of PARP1 inhibitor Olaparib (1 μM). The MgCl2 (Sigma) concentration was adjusted to 15 mM to allow full Nudix hydrolase activity. Automodified PARP1 was then incubated for 3 hours at 30°C with hydrolytic enzymes in 10 μL reaction. Concentrations of hydrolytic enzymes used are as indicated in figures. Reactions were stopped by addition of Laemmli loading buffer, samples boiled at 90°C for 1.5 minutes and fractionated by NuPAGE Novex Bis-Tris 4-12% Gel using MOPS buffer (Invitrogen). PARP1-E988Q experiments were visualized by autoradiography, experiments using wild-type PARP1 were visualized by Western blot using a specific anti-PAR antibody.
Thin layer Chromatography (TLC)
The TLC was performed as previously described [11]. PARylated PARP1 was automodified in presence of [32P]-labelled NAD as described above. The product of reaction was then cleaned by G25 desalting columns, MgCl2 was added to get 15 mM and 10 μL reaction samples were processed by 18 μM of hNUDT16 or 0.5 μM of svPDE as described above. 1 μL of reaction was spotted onto polyethyleneimine (PEI)-cellulose plates (Macherey-Nagel, Polygram CEL 300 PEI/UV254) and developed in 0.15 M LiCl and 0.15 M formic acid. Dried plates were exposed on X-ray film or visualized by UV254 shadowing.
Immunoblotting
Fractionated proteins on gradient gels were transferred onto nitrocellulose membranes using Trans-Blot Turbo Transfer System (Biorad) at 1.3A/25V for 20 minutes. Membranes were blocked in 5% Non-Fat Dry Milk (NFDM; Biorad) diluted in 0.1% Tween 20-PBS and subsequently incubated with the following primary antibodies: rabbit polyclonal anti-PAR (1:2000; Trevigen), mouse monoclonal anti-6xHis (1:5000; Clontech), rabbit polyclonal anti-PARP1 (1:1000, Abcam). Primary antibody incubations were then followed by incubation with secondary antibodies as indicated and developed with ECL Western blotting detection reagent (GE Healthcare).
Preparation samples for Mass Spectrometry analysis
Reactions were performed as above described with slightly modifications. 0.5 μM PARP1 was automodified and incubated with or without 18 μM of recombinant hNUDT16 or 1 μM of human PARG for 3 hours at 30°C in presence of 15 mM MgCl2. Samples were processed either by in-gel or in-solution tryptic digestion. For samples processed by in-gel digestion, reactions were stopped by addition of Laemmli loading buffer, samples were not boiled. 3 μL aliquots of reaction were loaded on SDS-PAGE and probed using anti-PAR antibody. The remaining samples were resolved on SDS-PAGE and stained using SYPRO-Ruby (Invitrogen) following manufacturer’s instructions. Gels were visualized and scanned by Pharos FX plus (Biorad). Proteins were visualized by UV trans-illumination and excised from the gel for the LC/MS analysis. Protein samples processed by in-solution tryptic digestion were processed in an alternative way. Briefly, after enzymatic reactions proteins were precipitated by cold acetone to get the protein mix rid of glycerol. Protein pellets were then processed for in-solution tryptic digestion.
In-gel Trypsin Digestion
Samples were de-stained in 50 mM ammonium bicarbonate (Sigma) and 50% acetonitrile (HPLC grade, Fisher), reduced with TCEP (Pierce) and alkylated with chloroacetamide (Sigma). Proteins were digested using 200 ng trypsin (Promega) for 16 hours at 37°C. Samples were desalted prior to mass spectrometry using an in-house manufactured C18 reverse-phase tip.
In-solution Trypsin Digestion
After enzymatic reactions, proteins were precipitated by cold acetone. Pellets were dissolved in 6M Guanidium-chloride (Sigma), reduced with TCEP (Pierce) and alkylated with chloroacetamide (Sigma). After a 10-fold dilution with 50 mM ammonium bicarbonate (Fluka), proteins were digested using 200 ng trypsin Gold (Promega) for 16 hours at 37°C. Samples were desalted prior to mass spectrometry using an in-house manufactured C18 reverse-phase tip.
Mass Spectrometry
Samples from in-gel tryptic digestion were analyzed by LC-MS/MS using a Q Exactive mass spectrometer (Thermo, Hemel Hempstead) coupled to a Dionex Ultimate 3000 RSLCnano system. Samples were resolved on a 90 min gradient on a home-packed 25 cm long, by 75 µm inner-diameter column, at a flow-rate of 300 nL min-1 run in direct injection. The Q Exactive was operated in a “Top20” Data Dependent Acquisition mode using HCD fragmentation. Samples from in solution tryptic digestion were analyzed by LC-MS/MS using a Q Exactive Plus mass spectrometer (Thermo), essentially as described in [6], with a following modifications: samples were resolved during a 90 min gradient on a Picofrit C18 column from New Objective (75 μm inner diameter x 25 cm, 1.9 μm particle size, 120 A pore size) and peptides were injected into the mass spectrometer through a Thermo Scientific Nanospray Flex ion source. In addition to the data dependent acquisition described in [6], a mass inclusion list was used to improve the quality of the MS/MS spectra.
Data Analysis
.raw files were converted to .mgf using msconvert V2.1 [30]. .mgf files were searched using Mascot V2.5 (MatrixScience, London) against human UniProt database. Trypsin enzyme specificity and up to 2 missed cleavages were allowed. The precursor mass tolerance was 20 ppm and fragment ion tolerance 0.02 Da. The fixed modification was defined as carbamidomethyl (C) and the variable modifications as oxidation (M), and phosphoribosylation (+ 212.01 Da) (DE). Neutral losses of H3PO4 (phosphoric acid, -97.98 Da) and C5H9PO7 (phosphoribosylation, -212.01 Da) were defined in Mascot for the phosphoribosylation. Spectra were manually inspected and annotated using Mascot annotation backbone. Raw files were also analysed with Andromeda-based MaxQuant (version 1.5.0.30). MaxQuant was set up essentially as in [20]. The reported spectrum was manually validated using stringent criteria.
Results
Screening of human Nudix enzymes for PAR hydrolysing activity
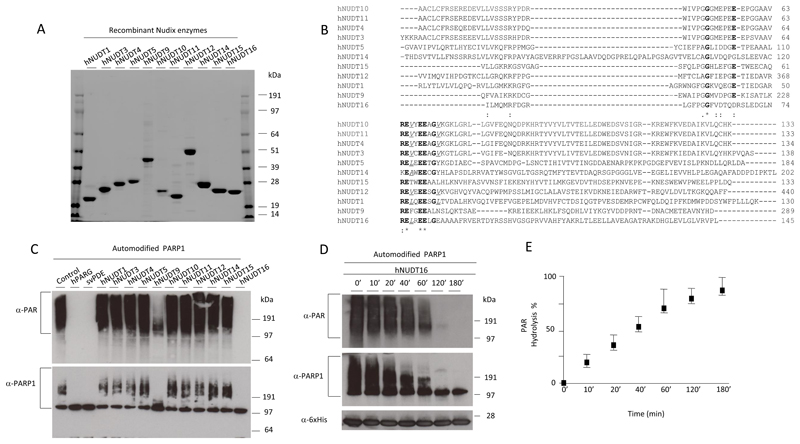
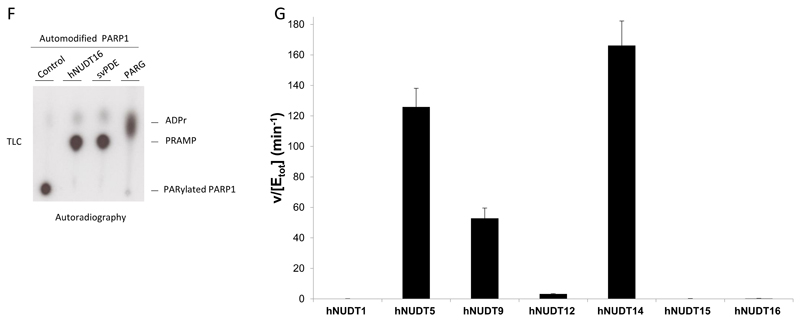
The cDNAs encoding the following human members of Nudix proteins: hNUDT1, hNUDT3-5, hNUDT9-12 and hNUDT14-16 were cloned into bacterial expression vectors and proteins subsequently purified (Fig. 2A) [27]. Sequence alignment of the Nudix domains from proteins taken under consideration in the present study showed high divergence, but conservation of their main catalytic residues known to be important for phosphodiesterase activity is still retained (Fig. 2B). To test whether any of human Nudix enzymes exhibit phosphodiesterase activity against PARylated proteins, we incubated 20 μM of purified Nudix proteins with PAR covalently linked to PARP1 protein as a substrate for 3 hours. The concentration of PAR used in this assay (defined in monomeric ADP-ribose units that are potential substrates for Nudix enzymes) was equimolar relative to the Nudix hydrolase proteins tested. PAR was visualized by Western blot using an antibody specifically recognizing poly and oligo chains of ADPr but not MAR. Strikingly, the recombinant hNUDT9 and especially hNUDT16 showed significant ability to remove the PAR signal (Fig. 2C, upper panel). hNUDT16 was capable of a quantitative reaction even at shorter time points and collapsed the PARP1 signal into a single band visualised by the SDS-PAGE (Fig. 2C-E). Furthermore, the analysis of the hNUDT16 reaction products using radiolabelled PAR as a substrate showed that the main released product was phosphoribosyl-AMP (PRAMP), as observed for svPDE previously [25] (Fig 2F). All the other Nudix enzymes tested were incapable of cleaving PAR (Fig 2C), but the activity assays against the free ADP-ribose revealed that some of them (e.g. hNUDT5, hNUDT12 and hNUDT14) can hydrolyse this substrate (Fig. 2G).
Figure 2. Screening of human Nudix enzymes for PAR-hydrolysing activity.
A 2μg of human recombinant Nudix (hNUDT) enzymes were loaded on SDS-PAGE and gel stained with Coomassie-blue. B ClustualW2 alignment of the Nudix domain belonging to hNUDT enzymes taken under consideration in the present study. The highly conserved residues of characteristic Nudix motif GX5EX7REUXEEXGU are in bold in the figure, while the variable aliphatic/hydrophobic residues (U) are highlighted. C Hydrolysis of the protein PARylation. 0.5 μM of human recombinant PARP1 was automodified to produce approximately 20 μM PAR substrate (defined in monomeric ADP-ribose units) and incubated with buffer only (control), 1 μM human recombinant PARG, 0.5 μM of purified svPDE or 20 μM of recombinant hNUDT proteins in 10 μL of reaction for 3 hours at 30°C. Samples were fractionated on SDS-PAGE and probed using anti-PAR (upper panel), and anti-PARP1 antibodies (lower panel). D Automodified PARP1 was incubated with 18 μM of recombinant hNUDT16 for the indicated times and analysed by anti-PAR, anti-PARP1 and anti-6xHis antibodies. E Percentage of PAR hydrolysis calculated quantifying anti-PAR signal of three separate experiments such as that showed in Fig. 2D. Quantification was made using ImageJ. F 0.5 μM of wild type human recombinant PARP1 was automodified in presence of [32P]-NAD+ and then incubated with buffer (Control), 18 μM of recombinant hNUDT16, 0.45 μM of purified svPDE and 1 μM of PARG for 3 hours at 30°C. The products of enzymatic reaction were then assayed by the thin layer chromatography (TLC). G Hydrolysis of the free ADP-ribose by Nudix enzymes measured by the release of inorganic phosphate (Pi) upon coupling the reaction with Calf Intestinal Phoshatase. Pi was detected using a colorimetric assay using malachite green reagent. Data is presented as mean of duplicates of hydrolysed ADP-ribose (in µM) per µM enzyme per minute.
hNUDT16 is able to efficiently hydrolase PARylated and oligo(ADP-ribosyl)ated proteins
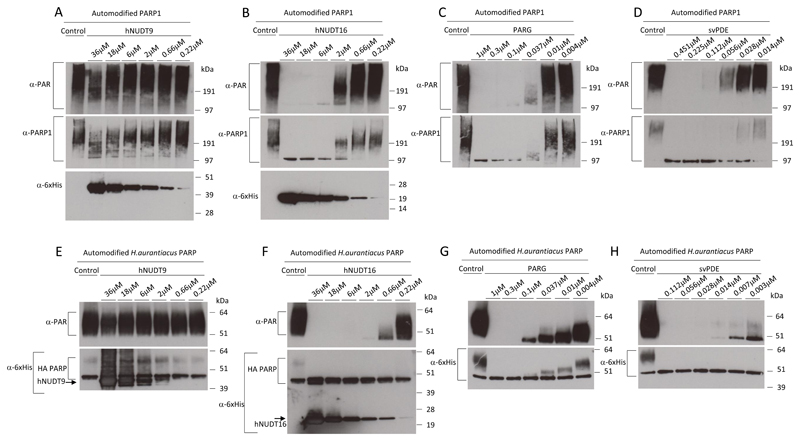
We further tested the range of hNUDT9 and hNUDT16 protein concentrations by PAR hydrolysis assay and the PAR-removing activity of the Nudix enzymes was compared to PARG and snake venom phosphodiesterase (svPDE) that served as positive controls (Fig. 3). As showed in Fig. 3A, hNUDT9 has a very low activity towards PAR. On the other hand, hNUDT16 hydrolysed PAR more efficiently (Fig. 3B), but this activity was nevertheless still considerably lower than observed for PARG (Fig. 3C) and svPDE (Fig. 3D). To test whether hNUDT16 exhibits a phosphodiesterase activity against oligoADP(ribosyl)ated proteins, His6-tagged Herpetosiphon aurantiacus PARP was automodified and used as substrate for hNUDT16 [11]. This bacterial PARP was shown previously to produce only short ADPr chains, up to ten repeats. We observed similar activities of Nudix enzymes against oligo(ADPribosyl)ation, as seen with the PARylated PARP1 substrate (Fig. 3 E-H). hNUDT9 did not show significant activity against the oligo(ADP-ribose) (Fig. 3E), while hNUDT16 was active in a similar range of protein concentrations that were necessary to remove PAR from PARP1 (Fig. 2F). Again, PARG (Fig. 3G) and svPDE (Fig. 3H) were notably faster than NUDT16 in hydrolysing short ADP-ribose chains. Altogether, these data show the ability of hNUDT16 to trim ADPr chains of various lengths covalently linked to a modified protein.
Figure 3. hNUDT16 is able to hydrolase PAR- and oligo(ADP-ribosyl)ated proteins.
A-D Automodified PARP1 (prepared as in Fig 2) was incubated with buffer only (control) or decreasing concentrations of recombinant hNUDT9 (A), hNUDT16 (B), PARG (C) or svPDE (D) and analysed with an anti-PAR, anti-PARP1 and anti-6xHis antibodies. Note, the His6-tag in PARG recombinant protein was previously cleaved and, therefore, not visualizable by anti-6xHis antibody. E-F 1 µM of Herpetosiphon aurantiacus PARP (HA PARP) was automodified and incubated with buffer only (control) or decreasing concentrations of the recombinant hNUDT9 (E), NUDT16 (F), PARG (G) or svPDE (H), and analysed with anti-PAR and anti-6xHis antibodies. Due to the proximity of His-tagged HA PARP and His-tagged hNUDT9, hNUDT9 was indicated by an arrow in anti-6xHis western blot (lower panel).
hNUDT16 processes MAR from modified proteins
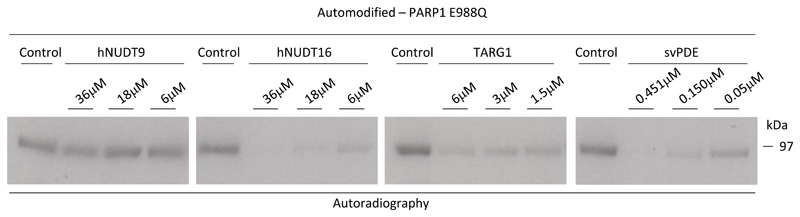
In order to investigate whether hNUDT16 is able to remove MAR from proteins, the recombinant PARP1-E988Q, a PARP1 mutant catalysing MARylation instead of PARylation, was incubated with decreasing concentrations of hNUDT9 and hNUDT16, as well as with TARG1 and svPDE that served as positive controls (Fig. 4A). hNUDT16, was able to remove PARP1-linked MAR, whereas hNUDT9 was not active under the same conditions. These data suggest that hNUDT16 is able to hydrolyse protein MARylation, in addition to oligo(ADP-ribosyl)ation and PARylation.
Figure 4. hNUDT16 is able to efficiently hydrolase MARylated proteins.
A 2 μM of recombinant PARP1-E988Q mutant was automodified using 32P-labeled NAD+ and then incubated with buffer only (control), and decreasing concentration of recombinant hNUDT9, hNUDT16, TARG1 and decreasing concentrations of purified svPDE. Samples were loaded on SDS-PAGE and [32P]-NAD+ incorporation was detected by autoradiography.
hNUDT16 leaves ribose-5’-phosphate (5RP) tags upon acting on PARylated proteins
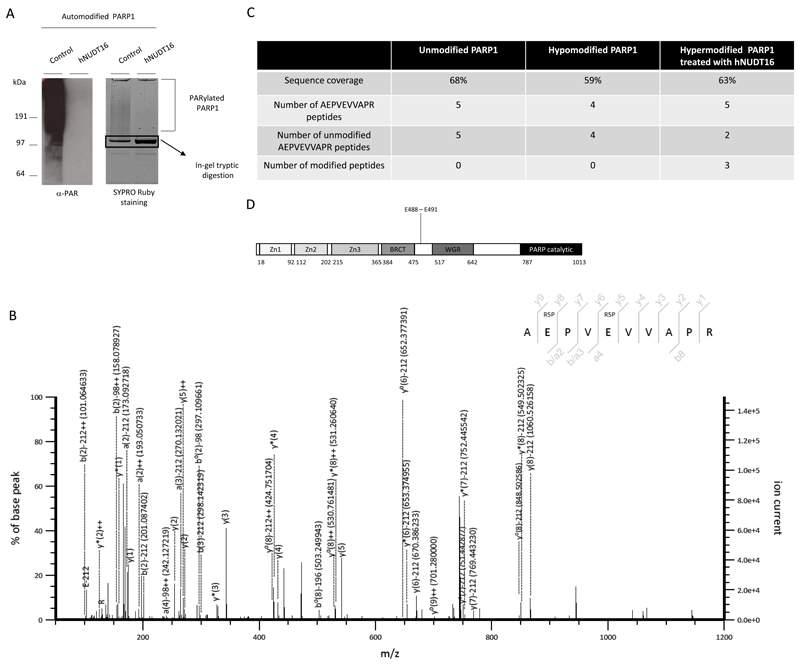
As Nudix proteins readily hydrolyse nucleotides at the pyrophosphate bonds [21–23], the action of hNUDT16 on PARylated proteins should produce a protein covalently linked to a R5P moiety (leading to a mass shift of 212.01 Da), while releasing the PRAMP as a by-product (Fig. 1A-B and 2F) [25–26]. To confirm the conversion of PAR to R5P, the peptide reaction products were analysed by high performance liquid chromatography (HPLC) coupled to electrospray ionization tandem mass spectrometry (ESI-LC/MS) [31]. PARP1 was first automodified and next incubated with hNUDT16 under the conditions described above. The protein was then resolved on SDS-PAGE. The efficiency of hNUDT16 reaction was analyzed using anti-PAR immunoblot (Fig. 4A, left panel). As a result of hNUDT16-hydrolazing activity, a compacted PARP1 band could be visualised using SYPRO-Ruby staining and the corresponding gel band excised for the analysis (Fig. 5A, right panel). The protein was then digested using trypsin and analyzed by LC/MS. Although MS-induced fragmentation produced a large number of the neutral losses of the full modification (212.01 Da loss) or just the phosphate group (97.98 Da loss) [26], we could also observe several peptides where the two glutamate residues were covalently linked to the R5P, providing a direct evidence that a canonical Nudix reaction mechanism took place (Fig. 5B and C). Importantly, the R5P-modified peptides could be observed only in the hNUDT16-treated sample and not in the untreated samples (Fig. 5C). By contrast, no ADPr sites were detected, confirming the efficiency of hNUDT16-dependent PAR hydrolysis. On the other hand, the ADP-ribosylated site on the same peptide was detected in the control PARylated sample treated with the PARG enzyme that is expected to leave the mono(ADP-ribose) mark (Supplementary Fig. 1). The two R5P sites detected on automodified PARP1 following hNUDT16 or PARG treatment are in the automodification domain and are known to be major ADP-ribosylation sites of PARP1 described in all the studies identifying ADP-ribosylation sites on PARP1 published so far (Fig 5D) [15,25,26,32,33].
Figure 5. hNUDT16 leaves the ribose-5’-phosphate tags on PARylated proteins.
A Recombinant wild-type PARP1 was auto-modified and then incubated with or without (control) human hNUDT16. Reactions were stopped by addition of Laemmli loading buffer. A small aliquot of the reaction was loaded on SDS-PAGE and probed using anti-PAR antibody (left panel). The remaining sample was resolved on the SDS-PAGE and stained using SYPRO-Ruby. The proteins were visualized by UV trans-illumination and the compacted band corresponding to PARP1 was excised from the gel for the LC/MS analysis (panel on the right). B Representative MS/MS spectrum of HCD-induced fragmentation of the main modified PARP1 peptide (AEPVEVVAPR) carrying the double R5P modification on the two adjacent glutamate residues (labelled as R5P). The spectrum was annotated using an in house annotation system. The observed m/z ratio was indicated in brackets only for the fragmented ions including the Glutamate residues. Legend of ion types: * ion minus NH3, ° ion minus H2O, + peptide charge. C Incidence of modified AEPVEVVAPR PARP1 peptide in unreacted PARP1, hypo modified PARP1 and hyper modified PARP1 treated with hNUDT16. D Schematic representation of the PARP1 domain including the position of the modification sites.
Discussion
A neurodegenerative disorder characterized by an uncontrolled activity of an unknown enzyme able to cleave ADPr on proteins to AMP and peptidyl-R5P and leading to a lysosomal accumulation of proteins carrying glutamic covalently blocked by R5P, has been described previously [19]. This disorder was thought to be due to deficiency of the main protein deADP-ribosylation enzymes and the subsequent action of an endogenous phosphodiesterase type of protein on the persisting non-recycled protein ADP-ribosylation. Another study identified the R5P sites on arginine residues of several cellular vertebrate proteins [20]. Therefore we sought to discover the cellular phoshodiesterases that could act on protein ADP-ribosylation. In particular, we focused on enzymes belonging to the Nudix hydrolase family and identified hNUDT16 as the enzyme able to in vitro cleave the PAR and MAR linked to modified proteins, leaving the R5P mark (Fig. 5). hNUDT16 is a nuclear and nucleoplasmic protein firstly described as m(7)G decapping enzyme highly conserved during evolution [34]. Later on, it was shown that hNUDT16 has a relatively relaxed specificity and has hydrolytic activity on several purine triphosphates/diphosphates nucleosides, converting them in corresponding nucleoside monophosphates [35]. The nuclear localization of hNUDT16 means that in addition to free nucleotides, it also encounters high levels of protein ADP-ribosylation, especially after cellular stress. Thus, it is possible that ADP-ribosylation cleaving ability of hNUDT16 observed in vitro may be significant under certain conditions and it may represent an alternative way of removing ADP-ribosylation signals in cells. Alternatively, the processing of protein ADP-ribosylation into bound ribose-phosphate may represent an unfavourable, toxic event that may become pronounced in pathological conditions (such as already mentioned lysosome storage disease) when the activity of protein ADP-ribosyl macrodomain proteins is compromised or when hNUDT16 is overexpressed [15,19]. The progress in the ADP-ribosylation field has been held back significantly by the lack of reliable mass spectrometry methods to study modification sites [31]. One of the main issues has been the lack of general biochemical tools to reduce the size and heterogeneity of the protein PARylation. One recent study employed hydroxamic acid to reduce sample complexity [32], however, this method can only capture acidic residues and would not identify e.g. the modified lysines, cysteines or arginines identified in other studies [6,8,25,26]. Other studies employed the svPDE, a protein purified from venom of the rattle-snake Crotalus adamanteus to reduce PARylation into small and readily MS-detectable R5P sites [24,25,26,31]. However, the commercially available svPDE is costly. More importantly, it contains variable amounts of contaminants (including 5’nucleotidase, phosphatase and protease), so commercial svPDE preparations readily require additional purification steps before they can be used [24,25,26 and this study]. Finally, svPDE cannot be expressed in bacteria and predictably requires glycosylation for its activity [36]. In this work, we have established the first recombinant source for an enzyme that can be used as an alternative to svPDE for in vitro conversion of PARylated and MARylated proteins into proteins containing the R5P mark. Indeed, hNUDT16 enzyme can be produced recombinantly in large amounts from E. coli with minimal costs, so our data also provide an important technical development for the mass spectrometry analyses of the protein ADP-ribosylation.
Supplementary Material
HCD MS/MS spectrum of the ADP ribose modified peptide on E491 (m/z 598.260408; (3+); mass error 0.65 ppm).
Summary statement.
NUDT16 cleaves protein ADP-ribosylation in vitro, converting it into ribose-5’-phosphate; This novel enzymatic activity suggests an alternative pathway for protein ADP-ribosylation removal. NUDT16 activity can also be used as an in vitro tool to improve mass spectrometry-based identification of protein ADP-ribosylation sites.
Acknowledgements
The authors would thank all members of IA laboratory and Oreste Acuto (Sir William Dunn School of Pathology, University of Oxford) for their helpful suggestions. We thank PSF for protein purification and Central Proteomic Facility at Dunn School of Pathology (University of Oxford).
Funding
The work in IA laboratory is supported by the Wellcome Trust and the European Research Council. LP is supported by an Italian Cancer Research Foundation (FIRC) fellowship for abroad. K.F. is supported by an EMBO fellowship. T.H. laboratory is mainly supported by the Torsten and Ragnar Söderberg foundation. IM acknowledges funding from the DFG (CECAD).
Abbreviations footnote
- PTM
post-translational modification
- ADPr
ADP-ribose
- MAR
Mono-ADP-ribose
- PAR
Poly(ADP-ribose)
- MARylation
Mono(ADP-ribosyl)ation
- PARylation
Poly(ADP-ribosyl)ation
- R5P
Ribose-5’-phosphate
- PRAMP
Phosphoribosyl-AMP
- svPDE
snake venom Phosphodiesterase
- HCD
Higher-energy Collision Dissociation
- MS
Mass Spectrometry
Footnotes
Author contribution
LP performed biochemical experiments. LP, BT, OLe, TM and IM performed mass-spectrometry experiments and data analysis. LP, A-SJ, OLo, JCP and TH purified recombinant proteins. RZ and KF performed supporting studies. LP and IA conceived the study and wrote the manuscript.
The authors declare no competing financial interests.
References
- 1.Hassa PO, Haenni SS, Elser M, Hottiger MO. Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going? Microbiol Mol Biol Rev. 2006;70:789–829. doi: 10.1128/MMBR.00040-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perina D, Mikoč A, Ahel J, Cetković H, Zaja R, Ahel I. Distribution of protein poly(ADP-ribosyl)ation systems across all domains of life. DNA Repair (Amst) 2014;23:4–16. doi: 10.1016/j.dnarep.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ame JC, Spenlehauer C, de Murcia G. The PARP superfamily. Bioessays. 2004;26:882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- 4.Gibson BA, Kraus WL. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol. 2012;13:411–24. doi: 10.1038/nrm3376. [DOI] [PubMed] [Google Scholar]
- 5.Barkauskaite E, Jankevicius G, Ladurner AG, Ahel I, Timinszky G. The recognition and removal of cellular poly(ADP-ribose) signals. FEBS J. 2013;280:3491–3507. doi: 10.1111/febs.12358. [DOI] [PubMed] [Google Scholar]
- 6.Vyas S, Matic I, Uchima L, Rood J, Zaja R, Hay RT, Ahel I, Chang P. Family-wide analysis of poly(ADP-ribose) polymerase activity. Nat Commun. 2014;5:4426. doi: 10.1038/ncomms5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D'Amours D, Desnoyers S, D'Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J. 1999;342:249–268. [PMC free article] [PubMed] [Google Scholar]
- 8.Altmeyer M, Messner S, Hassa PO, Fey M, Hottiger MO. Molecular mechanism of poly(ADP-ribosyl)ation by PARP1 and identification of lysine residues as ADP-ribose acceptor sites. Nucleic Acids Res. 2009;37:3723–3738. doi: 10.1093/nar/gkp229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alvarez-Gonzales R, Althaus FR. Poly(ADP-ribose) catabolism in mammalian cells exposed to DNA-damaging agents. Mutat Res. 1989;218:47–74. doi: 10.1016/0921-8777(89)90012-8. [DOI] [PubMed] [Google Scholar]
- 10.Lin W, Amé JC, Aboul-Ela N, Jacobson EL, Jacobson MK. Isolation and characterization of the cDNA encoding bovine poly(ADP-ribose) glycohydrolase. J Biol Chem. 1997;272:11895–11901. doi: 10.1074/jbc.272.18.11895. [DOI] [PubMed] [Google Scholar]
- 11.Slade D, Dunstan M, Barkauskaite E, Weston R, Lafite P, Dixon N, Ahel M, Leys D, Ahel I. The structure and catalytic mechanism of a poly(ADP-ribose) glycohydrolase. Nature. 2011;477:616–620. doi: 10.1038/nature10404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tucker JA, Bennett N, Brassington C, Durant ST, Hassall G, Holdgate G, McAlister M, Nissink JW, Truman C, Watson M. Structures of the human poly (ADP-ribose) glycohydrolase catalytic domain confirm catalytic mechanism andexplain inhibition by ADP-HPD derivatives. PLoS One. 2012;7:e50889. doi: 10.1371/journal.pone.0050889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lambrecht MJ, Brichacek M, Barkauskaite E, Ariza A, Ahel I, Hergenrother PJ. Synthesis of Dimeric ADP-Ribose and Its Structure with Human Poly(ADP-ribose) Glycohydrolase. J Am Chem Soc. 2015 doi: 10.1021/ja512528p. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oka S, Kato J, Moss J. Identification and characterization of a mammalian 39-kDa poly(ADP-ribose) glycohydrolase. J Biol Chem. 2006;281:705–713. doi: 10.1074/jbc.M510290200. [DOI] [PubMed] [Google Scholar]
- 15.Sharifi R, Morra R, Appel CD, Tallis M, Chioza B, Jankevicius G, Simpson MA, Matic I, Ozkan E, Golia B, Schellenberg MJ, et al. Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease. EMBO J. 2013;32:1225–1237. doi: 10.1038/emboj.2013.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosenthal F, Feijs KL, Frugier E, Bonalli M, Forst AH, Imhof R, Winkler HC, Fischer D, Caflisch A, Hassa PO, Lüscher B, et al. Macrodomain-containing proteins are new mono-ADP-ribosylhydrolases. Nat Struct Mol Biol. 2013;20:502–507. doi: 10.1038/nsmb.2521. [DOI] [PubMed] [Google Scholar]
- 17.Jankevicius G, Hassler M, Golia B, Rybin V, Zacharias M, Timinszky G, Ladurner AG. A family of macrodomain proteins reverses cellular mono-ADP-ribosylation. Nat Struct Mol Biol. 2013;20:508–14. doi: 10.1038/nsmb.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feijs KL, Verheugd P, Lüscher B. Expanding functions of intracellular resident mono-ADP-ribosylation in cell physiology. FEBS J. 2013;280:3519–3529. doi: 10.1111/febs.12315. [DOI] [PubMed] [Google Scholar]
- 19.Williams JC, Chambers JP, Liehr JG. Glutamyl ribose 5-phosphate storage disease. A hereditary defect in the degradation of poly(ADP-ribosylated) proteins. J Biol Chem. 1984;259:1037–1042. [PubMed] [Google Scholar]
- 20.Matic I, Ahel I, Hay RT. Reanalysis of phosphoproteomics data uncovers ADP-ribosylation sites. Nat Methods. 2012;9:771–772. doi: 10.1038/nmeth.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mildvan AS, Xia Z, Azurmendi HF, Saraswat V, Legler PM, Massiah MA, Gabelli SB, Bianchet MA, Kang LW, Amzel LM. Structures and mechanisms of Nudix hydrolases. Arch Biochem Biophys. 2005;433:129–143. doi: 10.1016/j.abb.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 22.McLennan AG. The Nudix hydrolase superfamily. Cell Mol Life Sci. 2006;63:123–143. doi: 10.1007/s00018-005-5386-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zha M, Guo O, Zhang Y, Yu B, Ou Y, Zhong C, Ding J. Molecular mechanisms of ADP-ribose hydrolysis by human NUDT5 from structural and kinetic studies. J Mol Biol. 2008;379:568–578. doi: 10.1016/j.jmb.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 24.Oka J, Ueda K, Hayaishi O. Snake venom phosphodiesterase: simple purification with Blue Sepharose and its application to poly(ADP-ribose) study. Biophys Res Commun. 1978;80:841–848. doi: 10.1016/0006-291x(78)91321-9. [DOI] [PubMed] [Google Scholar]
- 25.Chapman JD, Gagné JP, Poirier GG, Goodlett DR. Mapping PARP-1 auto-ADP-ribosylation sites by liquid chromatography-tandem mass spectrometry. J Proteome Res. 2013;12:1868–1880. doi: 10.1021/pr301219h. [DOI] [PubMed] [Google Scholar]
- 26.Daniels CM, Ong SE, Leung AK. Phosphoproteomic approach to characterize protein mono- and poly(ADP-ribosyl)ation sites from cells. J Proteome Res. 2014;13:3510–3522. doi: 10.1021/pr401032q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gad H, Koolmeister T, Jemth AS, Eshtad S, Jacques SA, Ström CE, Svensson LM, Schultz N, Lundbäck T, Einarsdottir BO, Saleh A, et al. MTH1 inhibition eradicates cancer by preventing sanitation of the dNTP pool. Nature. 2014;508:215–221. doi: 10.1038/nature13181. [DOI] [PubMed] [Google Scholar]
- 28.Baykov AA, Evtushenko OA, Avaeva SM. A malachite green procedure for orthophosphate determination and its use in alkaline phosphatase-based enzyme immunoassay. Anal Biochem. 1988;171:266–270. doi: 10.1016/0003-2697(88)90484-8. [DOI] [PubMed] [Google Scholar]
- 29.Barkauskaite E, Brassington A, Tan ES, Warwicker J, Dunstan MS, Banos B, Lafite P, Ahel M, Mitchison TJ, Ahel I, Leys D. Visualization of poly(ADP-ribose) bound to PARG reveals inherent balance between exo- and endo-glycohydrolase activities. Nat Commun. 2013;4:2164. doi: 10.1038/ncomms3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chambers MC, MacLean B, Burke R, Amode D, Ruderman DL, Neumann S, Gatto L, Fischer B, Pratt B, Egertson J, Hoff K, et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat Biotechnol. 2012;30:918–920. doi: 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vivelo CA, Leung AK. Proteomics approaches to identify mono(ADP-ribosyl)ated and poly(ADP-ribosyl)ated proteins. Proteomics. 2014 doi: 10.1002/pmic.201400217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, Wang J, Ding M, Yu Y. Site-specific characterization of the Asp- and Glu-ADP-ribosylated proteome. Nat Methods. 2013;10:981–984. doi: 10.1038/nmeth.2603. [DOI] [PubMed] [Google Scholar]
- 33.Tao Z, Gao P, Liu HW. Identification of the ADP-ribosylation sites in the PARP-1 automodification domain: analysis and implications. J Am Chem Soc. 2009;131:14258–14260. doi: 10.1021/ja906135d. [DOI] [PubMed] [Google Scholar]
- 34.Peculis BA, Reynolds K, Cleland M. Metal determines efficiency and substrate specificity of the nuclear NUDIX decapping proteins X29 and H29K (Nudt16) J Biol Chem. 2007;282:24792–24805. doi: 10.1074/jbc.M704179200. [DOI] [PubMed] [Google Scholar]
- 35.Iyama T, Abolhassani N, Tsuchimoto D, Nonaka M, Nakabeppu Y. NUDT16 is a (deoxy)inosine diphosphate, and its deficiency induces accumulation of single-strand breaks in nuclear DNA and growth arrest. Nucleic Acids Res. 2010;38:4834–4843. doi: 10.1093/nar/gkq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bollen M, Gijsbers R, Ceulemans H, Stalmans W, Stefan C. Nucleotide pyrophosphatases/phosphodiesterases on the move. Crit Rev Biochem Mol Biol. 2000;35:393–432. doi: 10.1080/10409230091169249. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
HCD MS/MS spectrum of the ADP ribose modified peptide on E491 (m/z 598.260408; (3+); mass error 0.65 ppm).