Abstract
Volatile molecules in exhaled breath represent potential biomarkers in the setting of infectious diseases, particularly those affecting the respiratory tract. In particular, Pseudomonas aeruginosa is a critically-important respiratory pathogen in specific subsets of the population, such as those with cystic fibrosis. Infections caused by P. aeruginosa can be particularly problematic when co-infection with respiratory syncytial virus (RSV) occurs, as this is correlated with the establishment of chronic P. aeruginosa infection. In the present study, we evaluate the volatile metabolites produced by P. Aeruginosa (PAO1)-infected, RSV-infected, co-infected, or uninfected cystic fibrosis bronchial epithelial (CFBE) cells, in vitro. We identified a volatile metabolic signature that could discriminate between P. aeruginosa-infected and non-P. aeruginosa-infected CFBE with an area under the receiver operating characteristic curve (AUROC) of 0.850, using the machine learning algorithm Random Forest (RF). Although we could not discriminate between RSV-infected and non-RSV-infected CFBE (AUROC = 0.431), we note that sample classification probabilities for RSV-infected cell, generated using RF, were between those of uninfected CFBE and P. aeruginosa-infected CFBE, suggesting that RSV infection may result in a volatile metabolic profile that shares attributes with both of these groups. To more precisely elucidate the biological origins of the volatile metabolites that were discriminatory between P. aeruginosa-infected and non-P. aeruginosa-infected CFBE, we measured the volatile metabolites produced by P. aeruginosa grown in the absence of CFBE. Our findings suggest that the discriminatory metabolites produced likely result from the interaction of P. aeruginosa with the CFBE cells, rather than the metabolism of media components by the bacterium. Taken together, our findings support the notion that P. aeruginosa interacting with CFBE yields a particular volatile metabolic signature. Such a signature may have clinical utility in the monitoring of individuals with cystic fibrosis.
Keywords: Cystic fibrosis, respiratory syncytial virus (RSV), Pseudomonas aeruginosa, VOCs, metabolomics, comprehensive two-dimensional gas chromatography (GC×GC), mass spectrometry
1. Introduction
Respiratory tract infections are among the leading causes of death worldwide, resulting in 3.2 million deaths in 2015 alone [1], and pneumonia remains one of the world’s leading causes of death for children under the age of five [2]. According to the Etiology of Pneumonia in the Community (EPIC) study conducted by Centers for Disease Control and Prevention (CDC), human rhinovirus (9%), influenza virus (6%), and Streptococcus pneumoniae (5%) are the most prevalent pathogenic agents in the adult population, while respiratory syncytial virus (28%) and human rhinovirus (27%) are the predominant pathogens in children [3,4]. In this study, polymicrobial infections (i.e., infections caused by more than one pathogen) were identified in 36% of adults and 81% of children, with the occurrence of polymicrobial infections resulting in substantially worse morbidity and mortality [5–8]. Of particular interest are viral-bacterial polymicrobial infections, as viral infections have been shown to promote bacterial colonization of the airway up to several weeks following initial infection [9–13].
Polymicrobial infections involving influenza virus and S. pneumoniae are commonly encountered in the human population [10]. However, other viral-bacterial interactions may occur in specific subsets of the population, such as in the setting of cystic fibrosis (CF). In CF, a complex and dynamic respiratory microbiota interacts and evolves in a way that is not fully characterized to-date [14]. In this population, co-infection of respiratory viruses and Pseudomonas aeruginosa is correlated with the establishment and exacerbation of chronic P. aeruginosa lung infection [15][16–20], with respiratory syncytial virus (RSV) as the etiological agent in 9–58% of viral infections in these patients [21]. Chronic P. aeruginosa infection has dire health consequences in these individuals, including decreased quality of life and reduced life expectancy [22]. It has been demonstrated that the antiviral interferon host response to RSV, along with the release of transferrin-bound iron that occurs during viral infection, promotes P. aeruginosa biofilm formation, and this, in turn, can be correlated with disease exacerbation [23].
Volatile molecules have been investigated as potential biomarkers for the diagnosis of both acute and chronic respiratory infections [24]. Numerous studies have focused on the volatile metabolic fingerprints generated by bacterial infections in vivo [25–31], but none have evaluated the ability of this approach to discriminate between virally- and bacterially-derived volatile compounds, nor deconvolute bacterial- and viral-associated signatures in the setting of a polymicrobial environment. Nevertheless, several transcriptomics studies have demonstrated that viral and bacterial infections induce distinct responses by host cells [32–42]. Such findings provide the rationale behind our hypothesis that volatile metabolic signatures can be used to differentiate between virally-infected, bacterially-infected, and co-infected cells. The aim of this study is thus to investigate the ability of volatile metabolites to discriminate between respiratory epithelium infected, in vitro, with P. aeruginosa and RSV singularly, and to evaluate the polymicrobial fingerprint generated by the infection of P. aeruginosa and RSV simultaneously. For this, we employ solid-phase microextraction (SPME) followed by comprehensive two-dimensional gas chromatography (GC×GC) with a time-of-flight mass spectrometer (ToF MS), a powerful technique for the analysis of complex mixtures.
2. Materials and Methods
2.1. Viral and bacterial infection sample preparation
Cell culture: Immortalized homozygous CFTR ΔF508 CFBE41o- human bronchial epithelial cells (CFBE) were maintained in a humidified incubator at 37°C 5% CO2 in Minimum Essential Medium (MEM) containing phenol red (Gibco, Waltham, MA, United States) supplemented with 10% fetal bovine serum (FBS, Gemini Bio-Products, West Sacramento, CA, United States), 0.5 μg/mL Plasmocin prophylactic (InvivoGen, San Diego, CA, United States), 2 mM L-glutamine, 5 U/mL penicillin, and 5 μg/mL streptomycin (Sigma-Aldrich, St. Louis, MO, United States) (Hendricks PNAS 2016, 113:1642–1647). Immortalized STATE−/− NY3.2 mouse fibroblasts were maintained in MEM containing phenol red supplemented with 5% FBS, 5 U/mL penicillin, and 5 μg/mL streptomycin. Both cell types are not on the commonly misidentified list, and cells were tested quarterly for mycoplasma using a Southern Biotech mycoplasma detection kit. CFBE41o- cells were seeded at near confluency on 12 mm diameter, 0.4 μm pore size Transwell permeable polycarbonate membrane supports (Costar, St. Louis, MO, United States). After attachment and confluency, epithelial cells were differentiated at air-liquid interface (ALI) for one week. Respiratory syncytial virus (RSV) line A2 was propagated in NY3.2 cells. RSV was extracted and stored in high salt media (HSM) at −80°C. Plaque-forming units were determined by immunoreactive staining assay in NY3.2 cells. P. aeruginosa strain PAOl was grown in Lysogeny Broth (LB) at 37°C on a roller.
P. aeruginosa-only samples: Bacteria were prepared by washing an overnight culture in MEM supplemented with 2 mM L-glutamine and culturing in MEM supplemented with 2 mM L-glutamine in a 25 cm2 cell culture flask (Coming, Coming, NY, United States) at 37°C 5% CO2 for 24 h. Media was then transferred to a tube and centrifuged to remove bacterial aggregates. Clarified media was filtered through a 0.22 μm filter (Merck Millipore, Burlington, MA, United States) and stored at −80°C. The same set up was used without P. aeruginosa inoculation for media-only controls.
Bacterial-epithelial co-cultures: ALI-differentiated CFBE41o- cells were washed with MEM lacking phenol red (Gibco) supplemented with 2 mM L-glutamine, and basolateral media was replaced with MEM containing phenol red supplemented with 10% FBS and 2 mM L-glutamine. Cells were inoculated with RSV at a multiplicity of infection (MOI) of 1 in MEM supplemented with 2 mM L-glutamine in the apical compartment. Control cells were inoculated with MEM containing an equal volume of vehicle (HSM). Apical media was removed after infection and cells were returned to ALI and maintained in antibiotic-free basolateral media. RSV-infected cells were then inoculated with P. aeruginosa that was prewashed in MEM lacking phenol red supplemented with 2 mM L-glutamine at an MOI of approximately 25. After 1 h attachment, non-attached bacteria were removed and apical media was adjusted to 0.4% L-arginine. After an additional 5 h, apical media was transferred to a tube and epithelium-associated biofilms were disrupted in 0.1% Triton X-100 (Bio-Rad, Hercules, CA, United States) and analyzed by serial dilution and plating on LB agar for colony-forming units. Apical media was centrifuged to remove bacterial aggregates, passed through a 0.22 μm filter, and stored at −80°C. RNA was collected from a separate set of Transwells by RNeasy with QIAshredder columns (Qiagen, Hilden, Germany) according to manufacturer recommendations. Transcript levels for GAPDH, the RSV N gene, and IFNL1 were assessed by converting RNA into cDNA with iScript cDNA Synthesis Kit and performing RT-qPCR using iQ SYBR Green Supermix (Bio-Rad) on a StepOne Real-Time PCR System (ThermoFisher Scientific, Waltham, MA, United States). Primers for GAPDH (5’- CGACCACTTTGTCAAGCTCA-3’, 5’-AGGGGAGATTCAGTGTGGTG-3’), for RSV N gene (5’-CGCCTTGGAAGAGTCACTCA-3′, 5’-GAAGCCTCAGGTCCCAATTC-3’), and for IFNL1 (5‘-GCTCTTAGCAAAGTCAAGTTGAATGA-3‘, 5‘-TGCTCCGTTGGATGGTGTATT-3‘). Conditions analyzed were epithelial cells alone, RSV-infected epithelial cells, P. aeruginosa-infected epithelial cells, and co-infected epithelial cells.
2.2. Sample preparation for volatile metabolites analysis
Five-hundred μL of filtered apical media was collected into a 10 mL air-tight glass vial sealed with a PTFE/silicone cap (Sigma-Aldrich) frozen at −20°C until analysis (within one month of collection). Samples were incubated for 15 min (the equilibration phase) at 37°C before fiber exposure for 45 min at the same temperature (the extraction phase). Samples were agitated at 250 rpm during both the equilibration and extraction phases. A divinylbenzene/carboxen/polydimethylsiloxane (DVB/CAR/PDMS) df 50/30 μm, 2 cm length fiber (Supelco, Bellefonte, PA, USA) was used to pre-concentrate the volatile molecules. The fiber was then introduced into the GC injector for thermal desorption for 1 min at 250°C in splitless mode. The fiber was conditioned before use and baked-out after each run for 7 minutes at 250°C. Absence of carryover was verified by desorbing the fiber into the GC injector after the baking time.
2.3. Analytical instrumentation
The analyses of volatile molecules were carried out using a Pegasus 4D (LECO Corp., St. Joseph, MI, United States) GC×GC time-of-flight (TOF) MS instrument with an Agilent 6890 GC, and equipped with an MPS autosampler (Gerstel, Linthicum Heights, MD, United States). The first dimension column was an Rxi-624Sil (60 m × 250 μm × 1.4 μm (length × internal diameter × film thickness)) connected in series with a Stabilwax secondary column (1 m × 250 μm × 1.4 μm), both from Restek (Bellefonte, PA, USA). The carrier gas was helium, at a flow rate of 2 mL/min. The primary oven temperature program was 35°C (hold 1 min) ramped to 230°C at a rate of 3.5°C/min. The secondary oven and the thermal modulator were offset from the primary oven by +5°C and +25°C, respectively. A modulation period of 2.0 s (alternating 0.5 s hot and 0.5 s cold pulses) was used. The transfer line temperature was set at 250°C. A mass range of m/z 30 to 500 was collected at a rate of 200 spectra/s following a 2.5 min acquisition delay. The ion source was maintained at 200°C. Data acquisition and analysis were performed using ChromaTOF software, version 4.50 (LECO Corp).
2.4. Processing and analysis of chromatographic data
Chromatographic data were processed and aligned using ChromaTOF. A signal-to-noise (S/N) cutoff was set at 50:1 in at least one chromatogram and a minimum of 20:1 S/N ratio in all others. For the alignment of peaks across chromatograms, maximum first and second-dimension retention time deviations were set at 6 s and 0.2 s, respectively, and the inter-chromatogram spectral match threshold was set at 600 (out of 1000). Compounds eluting prior to 4 min and artifacts were removed prior to statistical analysis. Artifacts were identified by running a series of instrument blanks (i.e., runs without SPME fiber injections) and fiber blanks (i.e., runs with SPME fiber injections but without fiber headspace exposure) to identify the compounds deriving from column and fiber bleeding (e.g., siloxane, silanol, etc.). Furthermore, the most common and ubiquitous plasticizers (i.e. phthalates) were removed as well.
A mixture of normal alkanes (C6-C20) was analyzed every 20 runs to calculate the linear retention index [43] and evaluate the instrument performance. The SPME and GC methods are as described above, except for the SPME exposition time, which was shorter (5 min) to avoid excessive overload of the fiber.
In agreement with the minimum reported standard guidelines suggested by the Metabolomics Standard Initiative, the most discriminatory features were putative annotated (Level 2) or putatively assigned to a chemical class (Level 3) [44] based on mass spectral similarities to the NIST 2011 mass spectral library, with a match score ≥ 800 (out of 1000) required for putative identifications. In addition to the MS similarity, the putative identification was also supported by the linear retention index (LRI) in agreement (i.e., in the ±15 range), with data reported using the same stationary phase [45] (if available). Unless confirmed by the LRI value, hydrocarbons were generally assigned as “alkylated hydrocarbons”, as only the chemical class of these compounds can be assigned considering their mass spectral fragmentation pattern and their location in the two-dimensional chromatogram.
2.5. Statistical analysis
All statistical analyses were performed using R v3.3.2 (R Foundation for Statistical Computing, Vienna, Austria). Prior to statistical analyses, the chromatographic area of compounds across chromatograms was normalized using Probabilistic Quotient Normalization [46], Variables were log-transformed, mean-centered, and unit-scaled prior to all statistical analyses. Random forest (RF) [47] was used to identify the most highly discriminatory volatile molecules and predict the class to which a leave-one-out cross validation (LOOCV) sample belonged. LOOCV was repeated 31 times, leaving out each sample (e.g., 8 PA+/RSV+, 8 PA−/RSV+, 8 PA+/RSV−, and 7 PA−/RSV−) once. The class probabilities were used to generate receiver operating characteristic (ROC) curves, and from these ROC curves, sensitivities, specificities, and area under the ROC curve (AUROC) were calculated. The optimal thresholds for class probabilities were calculated using Youden’s J statistic [48], rather than the 0.5 cutoff that is traditionally applied to two-class classification problems. Mean decrease in accuracy (MDA) was used as the measure of variable importance. The Mann-Whitney U test [49] was used to compare the normalized area of volatile molecules between experimental groups (Table 1), while a Student’s t-test was used to compare class probabilities between groups.
Table 1.
List of volatile metabolites significantly different between experimental groups, along with their putative identification. The direction of the arrow indicate the group in which the compound was significantly more abundant, “↑” indicates that the compound was significantly more abundant in the first group listed in the column heading, and “↓” indicates that the compound was significantly more abundant in the second group.
| # | Compound | Class | Formula | CAS | Similarity | LRIEXP | LRILit | 1tR (min:s) | 2tR (S) | Reference | PA−/RSV−vs. PA+/RSV− |
PA−/RSV− vs. PA−/RSV+ |
PA−/RSV− vs. PA+/RSV+ |
PA+/RSV− vs. PA−/RSV+ |
PA+/RSV− vs. PA+/RSV+ |
PA−/RSV+ vs. PA+/RSV+ |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2-Propenal | Ald | C3H4O | 107–02-8 | 855 | 538 | 523 | 6:00 | 1.0 | ↓ | ↓ | |||||
| 2* | Acetone | Ket | C3H6O | 67–64-1 | 978 | 545 | 530 | 6:15 | 0.8 | 51,60 | ↓ | ↓ | ↓ | ↑ | ↓ | |
| 3 | 2-methyl-pentane | Hyd | C6H14 | 107–83-5 | 941 | 570 | 564 | 7:12 | 0.6 | 58 | ↑ | ↑ | ||||
| 4 | Unknown | Aid | 595 | 8:08 | 0.7 | ↓ | ||||||||||
| 5 | Alkylated hydrocarbons |
Hyd | 627 | 9:27 | 0.7 | ↑ | ||||||||||
| 6 | Butanal | Aid | C4H8O | 123–72-8 | 863 | 627 | 629 | 9:28 | 0.8 | 51,57,60 | ↓ | |||||
| 7* | 2-Butanone | Ket | C4H8O | 78–93-3 | 965 | 629 | 637 | 9:32 | 1.00 | 51–57,59,60 | ↓ | ↓ | ↓ | |||
| 8 | Unknown | S-Com | 650 | 10:28 | 1.5 | ↑ | ||||||||||
| 9 | Tetrahydrofuran | Het-Cyc | C4H8O | 109–99-9 | 651 | 655 | 10:31 | 0.7 | 58 | ↓ | ↓ | |||||
| 10* | 3-methylbutanal | Aid | C5H10O | 590–86-3 | 924 | 693 | 694 | 12:19 | 0.8 | 54–56 | ↓ | ↑ | ↓ | |||
| 11* | 2-methylbutanal | Aid | C5H10O | 96–17-3 | 850 | 701 | - | 12:41 | 0.7 | 54,55,57 | ↓ | ↑ | ||||
| 12 | Alkylated hydrocarbons | Hyd | 753 | 15:24 | 0.6 | ↑ | ||||||||||
| 13 | Alkylated hydrocarbons | Hyd | 760 | 15:46 | 0.6 | ↑ | ||||||||||
| 14 | Alkylated hydrocarbons | Hyd | 767 | 16:06 | 0.6 | ↑ | ↑ | ↑ | ||||||||
| 15 | 4-methyl-2-pentanone | Ket | C6H12O | 108–10-1 | 850 | 780 | 783 | 16:46 | 1.00 | |||||||
| 16 | Alkylated hydrocarbons | Hyd | 791 | 17:21 | 0.8 | ↓ | ||||||||||
| 17 | Alkylated hydrocarbons | Hyd | 822 | 19:02 | 0.6 | ↑ | ↑ | ↑ | ||||||||
| 18 | Alkylated hydrocarbons | Hyd | 846 | 20:17 | 0.7 | ↑ | ↑ | ↑ | ||||||||
| 19 | Alkylated hydrocarbons | Hyd | 858 | 20:56 | 0.6 | ↑ | ↑ | ↑ | ||||||||
| 20 | 2-methyl-octane | Hyd | C9H20 | 3221–61-2 | 862 | 864 | 873 | 21:14 | 0.6 | 58 | ↑ | ↑ | ↑ | |||
| 21 | Alkylated hydrocarbons | Hyd | 874 | 21:48 | 0.6 | ↑ | ↑ | ↑ | ||||||||
| 22 | Cyclohexanone | Ket | C6H10O | 108–94-1 | 910 | 955 | 957 | 25:58 | 0.9 | ↓ | ↑ | |||||
| 23 | Alkylated hydrocarbons | Hyd | 977 | 27:05 | 0.6 | ↑ | ↑ | |||||||||
| 24* | 2-ethylhexanal | Ald | C8H16 | 123–05-7 | 918 | 993 | - | 27:54 | 0.8 | 29 | ↓ | |||||
| 25 | Alkylated hydrocarbons | Hyd | 1088 | 32:35 | 0.6 | ↓ | ||||||||||
| 26 | Alkylated hydrocarbons | Hyd | 1091 | 32:44 | 0.6 | ↓ | ||||||||||
| 27 | Alkylated hydrocarbons | Hyd | 1097 | 33:02 | 0.6 | ↑ | ↑ | ↑ | ||||||||
| 28* | Undecane | Hyd | C11H24 | 1120–21-4 | 872 | 1102 | 1100 | 33:14 | 0.7 | 56 | ↑ | |||||
| 29* | Alkylated hydrocarbons | Hyd | 1115 | 33:50 | 0.7 | ↑ | ||||||||||
| 30 | Alkylated hydrocarbons | Hyd | 1121 | 34:08 | 0.9 | ↓ | ↓ | |||||||||
| 31 | Unknown | Ket | 1137 | 34:50 | 1.3 | ↓ | ↓ | ↓ | ||||||||
| 32 | Alkylated hydrocarbons | Hyd | 1159 | 35:51 | 0.7 | ↓ | ||||||||||
| 33* | Alkylated hydrocarbons | Hyd | 1175 | 36:33 | 0.6 | ↓ | ↑ | |||||||||
| 34 | Alkylated hydrocarbons | Hyd | 1214 | 38:17 | 0.6 | ↑ | ↓ | |||||||||
| 35 | Alkylated hydrocarbons | Hyd | 1294 | 41:42 | 0.6 | ↑ | ↑ | |||||||||
| 36 | Alkylated hydrocarbons | Hyd | 1301 | 42:02 | 0.6 | ↑ | ↑ | |||||||||
| 37 | Alkylated hydrocarbons | Hyd | 1319 | 42:44 | 0.6 | ↓ | ||||||||||
| 38 | Alkylated hydrocarbons | Hyd | 1325 | 42:58 | 0.6 | ↑ | ↑ | |||||||||
| 39 | Alkylated hydrocarbons | Hyd | 1335 | 43:21 | 0.7 | ↑ | ||||||||||
| 40* | Alkylated hydrocarbons | Hyd | 1340 | 43:34 | 0.6 | ↑ | ||||||||||
| 41 | Alkylated hydrocarbons | Hyd | 1496 | 49:32 | 0.6 | ↑ | ||||||||||
| 42 | Alkylated hydrocarbons | Hyd | 1541 | 51:08 | 0.6 | |||||||||||
| 43 | Alkylated hydrocarbons | Hyd | 1567 | 52:04 | 0.7 | ↑ |
MDA>0.01 for the comparison of P. aeruginosa-infected versus non-P. aeruginosa-infected cells
3. Results and Discussion
3.1. Respiratory syncytial virus and P. aeruginosa infections
Infections with RSV and/or P. aeruginosa were confirmed by RT-qPCR and dilution plating, respectively (Figure 1). RSV transcripts were readily detected in both RSV-infected and co-infected respiratory epithelium (Figure 1A), indicating that a productive RSV infection was achieved. In addition to RSV transcript detection, we also observed a strong antiviral response by the RSV-infected and co-infected respiratory epithelium, evidenced by upregulation of the gene encoding IFN-λΙ (IFNL1, Figure 1B). Similar to previous studies [23,50], RSV co-infection of the respiratory epithelium enhanced P. aeruginosa biofilm biogenesis, as compared to P. aeruginosa alone (Figure 1C). Together, these results indicate that the samples processed for volatile fingerprinting were suitable to serve as representative uninfected (PA−/RSV−), P. aeruginosa- infected (PA+/RSV−), RSV-infected (PA−/RSV+), and co-infected (PA+/RSV+) respiratory epithelium.
Figure 1.
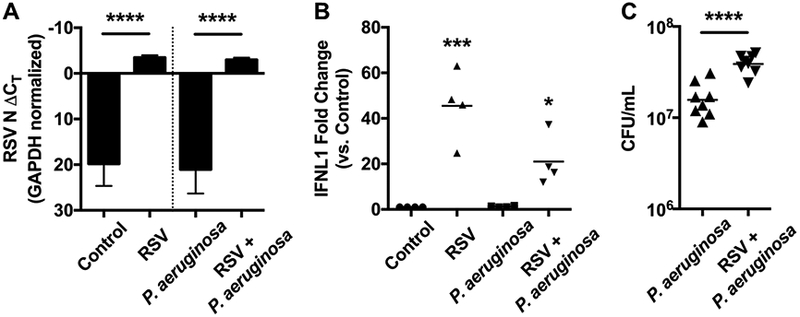
RSV and P. aeruginosa infection of CF respiratory epithelial cells. A) ΔCT values for RT-qPCR of infected CFBE cells with RSV N gene primers, normalized to GAPDH. Y axis inverted to represent that lower numbers (including more negative) indicate greater abundance of RNA. Data represent the mean and standard deviation, statistical comparison by paired ANOVA with Sidak correction for multiple comparisons. B) Fold change in IFNL1 expression for infected CFBE cells as determined by RT-qPCR, normalized to GAPDH and compared to uninfected control cells. Data represent the mean, statistical comparison to uninfected control by paired ANOVA with Dunnett correction for multiple comparisons. C) CFU of P. aeruginosa growing in mucosal biofilms associated with infected CFBE cells. Data represent the mean, statistical comparison by paired t-test. *: p < 0.05, ***: p < 0.001, ****: p < 0.0001.
3.2. Volatile fingerprints of P. aeruginosa and respiratory syncytial virus infections
Raw chromatographic data consisting of 762 features across the 31 samples of interest (8 PA+/RSV+, 8 PA−/RSV+, 8 PA+/RSV−, and 7 PA−/RSV−) were pre-processed to remove artifacts, resulting in a final data matrix consisting of 230 compounds plausibly derived from the biological specimens themselves.
We first sought to determine whether there was a subset of volatile metabolites that were significantly different in abundance between experimental groups. Forty-three features were identified as significantly different in at least one comparison and are reported in Table 1, along with their putative identification (if possible). The abundance of each feature in the different pairwise comparisons is reported in Table 1. Arrow directions indicate the group in which the compound was more abundant, with “↑” indicating that the compound was significantly more abundant in the first group listed in the column heading, and “↓,” indicating that the compound was significantly more abundant in the second group. Thirty-four of these 43 features were significantly different between PA+/RSV− and PA−/RSV−, 19 between PA−/RSV+ and PA−/RSV−, and 16 between PA+/RSV+ and PA−/RSV−, of which only three (all alkylated hydrocarbons) were not overlapped with the features reported from the comparison between PA+/RSV− and PA−/RSV−. Only six features were significantly different between PA+/RSV− and PA−/RSV+, all more expressed in the former group, except one alkylated hydrocarbon. Notably, only one feature was significantly more expressed in PA+/RSV− versus PA+/RSV+ (cyclohexanone), whereas five were significantly different in the comparison of PA−/RSV+ versus PA+/RSV+, all more expressed in the latter group Seven of the putatively identified metabolites have been previously identified in the headspace of P. aeruginosa grown under in vitro conditions [51–57], three metabolites were previously identified in the headspace of cell line infected with human respiratory viruses [58], and one, 2-ethylhexanal, has been detected in the headspace of several cancer cell cultures [28].
3.3. Discriminatory ability of the volatile metabolite signature
We next sought to determine whether volatile metabolic fingerprints could discriminate between infected and uninfected cystic fibrosis bronchial epithelial cells (CFBE). Random forest (RF) was used to assess the capability of volatile fingerprints to discriminate between: I) P. aeruginosa-infected (including both PA+/RSV− and PA+/RSV+) versus non-P. aeruginosa-infected (PA−/RSV− and PA−/RSV+) cells, and II) RSV-infected (including both PA−/RSV+ and PA+/RSV+) versus non-RSV-infected (PA+/RSV− and PA−/RSV−) cells. Receiver operating characteristic (ROC) curves were generated using class probabilities for leave-one-out cross-validation (LOOCV) samples (Figure 2).
Figure 2.
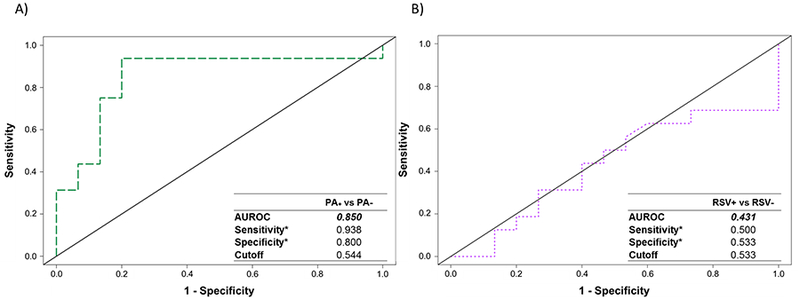
Receiver operating characteristic (ROC) curves for the discrimination between: A) P. aeruginosa-infected (PA+/RSV− and PA+/RSV+) versus non-P. aeruginosa-infected (PA−/RSV− and PA−/RSV+) cells, and B) RSV-infected (PA−/RSV+ and PA+/RSV+) versus non-RSV-infected (PA−/RSV− and PA+/RSV−), generated using Random Forest. *Optimal sensitivity and specificity of each statistical model are calculated at the given class prediction cutoff (ranging from 0 to 1).
The results obtained from the first model (P. aeruginosa versus non-P. aeruginosa) yielded an AUROC of 0.850, with a sensitivity of 0.938 and specificity of 0.800 at an optimal class probability cutoff of 0.544 (2a, left). In contrast, the ability to discriminate between RSV-infected and non-RSV-infected cells was approximately random, with an AUROC of 0.431, and optimal sensitivity and specificity of 0.500 and 0.533, respectively (2b, right). Taken together, these findings suggest that under the experimental conditions employed in the present study, infection of CFBE with P. aeruginosa produces a detectable volatile molecular fingerprint, while infection of CFBE with RSV does not.
The class probabilities for each sample in the comparison of P. aeruginosa versus non-P. aeruginosa infected CFBE were plotted, with 0 indicating non-P. aeruginosa-infected, and 1 indicating P. aeruginosa-infected (Figure 3). Within the P. aeruginosa-infected group, there was no significant difference in class probabilities between PA+/RSV− and PA+/RSV+ (p = 0.71). It is interesting, however, that the class probabilities for PA−/RSV+ were significantly different from both PA−/RSV− (p = 0.03) as well as PA+/RSV− (p = 0.02). This finding suggests that RSV singular infection results in a volatile molecular fingerprint that shares attributes with both PA−/RSV− as well as PA+/RSV−, possibly resultant from a general host response to infection. In contrast to the comparison of P. aeruginosa-infected versus non-P. aeruginosa-infected cells, there were no significant differences in class probabilities between groups for the comparison of RSV-infected versus non-RSV-infected cells (Supplementary Figure S1).
Figure 3.
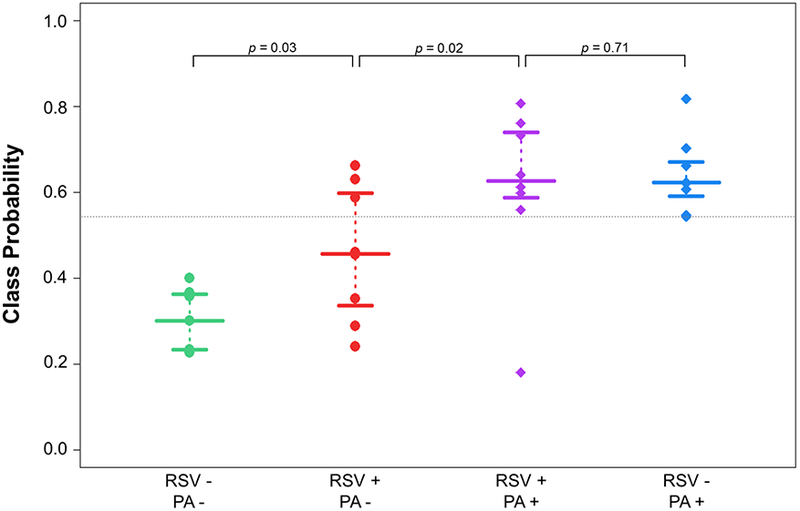
Class probabilities for LOOCV samples (n = 31) for the comparison of P. aeruginosa-infected versus non-P. aeruginosa-infected cells. Points towards the top of the plot indicate a higher probability of classifying as P. aeruginosa-infected, while points towards the bottom of the plot indicate a higher probability of classifying as non-P. aeruginosa-infected. The horizontal dotted line corresponds to optimal sensitivity/specificity cutoff (0.544). Wide bars represent median probability for each class, while narrow bars represent 25th and 75th percentiles.
3.4. Most discriminatory metabolites and their possible origin
For the comparison of P. aeruginosa-infected versus non-P. aeruginosa-infected cells, volatile molecules were ranked according to their discriminatory ability, as defined by their mean decrease in accuracy (MDA, a measure of variable importance). The intensity profile of the most discriminatory molecules (MDA > 0.01) are visualized in Figure 4. Six of these nine were putatively identified [two ketones (acetone and 2-butanone), three aldehydes (3-methylbutanal, 2-methylbutanal, and 2-ethylhexanal), one hydrocarbon (undecane)] and three compounds were generally assigned as alkylated hydrocarbons (Table 1).
Figure 4.
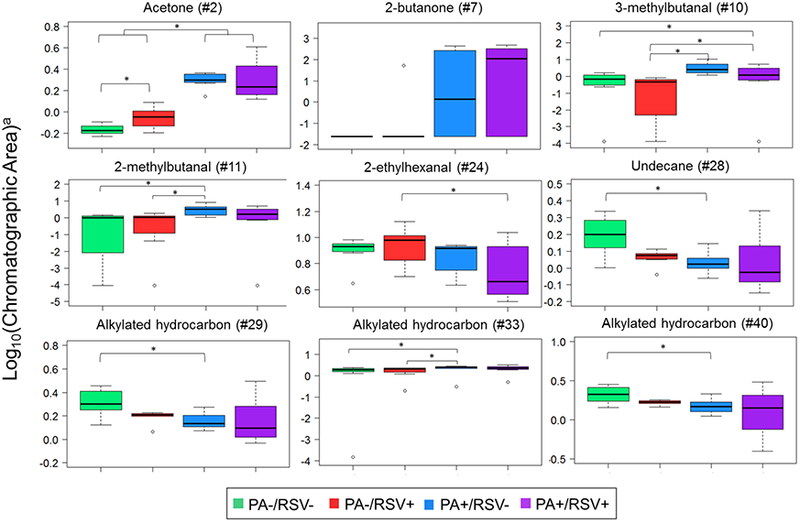
Boxplot of the most discriminatory features (MDA > 0.01) for discrimination between P. aeruginosa-infected versus non-P. aeruginosa-infected cells. *: p < 0.05. a: Data were mean centered and scaled before plotting. Identification as per Table 1.
The chemical classes (ketones, aldehydes, hydrocarbons) reported herein in culture headspace can be plausibly attributed to lipid oxidation pathways. Acetone and 2-butanone derive from the decarboxylation of ß-keto acids, and have been reported as important markers of P. aeruginosa elsewhere [51–57,59,60]. Aldehydes have been related to lipid peroxidation during inflammation [61], and 3-methylbutanal and 2-methylbutanal have been also reported as key intermediate in the catabolism of amino acids, such as leucine [62,63]. Aliphatic hydrocarbons (e.g. undecane) may be derived from the metabolism of fatty acids, which are converted to aldehydes due to the activity of acyl-CoA reductase, and enzymatically decarboxylated to hydrocarbons [64]. A preponderance of hydrocarbons were also highlighted in the headspace of human and murine epithelial cells infected with RSV and Influenza A, respectively [58]. Indeed, it has been shown that viral infection increases the production of reactive oxygen species, in part through a NAD(P)H oxidase-dependent mechanism [65], which is associated with the formation of hydrocarbons.
3.4. Analysis of the headspace of P. aeruginosa during growth in MEM
Finally, we hypothesized that a subset of the nine metabolites identified as highly discriminatory between P. aeruginosa-infected versus non-P. aeruginosa-infected CFBE resulted from either the consumption or production of metabolites by the bacterium itself. To test this, P. aeruginosa was grown as a biofilm in MEM in the absence of respiratory epithelium. Of the nine compounds selected, six were found in the headspace of P. aeruginosa grown alone, namely acetone, 2-butanone, 2-methylbutanal, undecane, and two hydrocarbons (#29 and #40). However, none of these differed significantly in abundance between P. aeruginosa in MEM and sterile media (MEM alone), suggesting that the selected metabolites likely result from the interaction of P. aeruginosa with the CFBE cells, or are produced in higher quantity during the interaction between P. aeruginosa and CFBE cells. Three compounds, 3-methylbutanal, 2-ethylhexanal and a hydrocarbon (#33), were not found in the headspace of P. aeruginosa grown only in media, suggesting that they originate from the interaction of P. aeruginosa with CFBE cells, or from the response of CFBE cells to the presence of P. aeruginosa.
3.4. Study strengths and limitations
The present study represents a first attempt at evaluating the volatile metabolic signatures of both singular P. aeruginosa and RSV infection, as well as P. aeruginosa-RSV co-infection, in the setting of in vitro CFBE cell co-culture. The use of CFBE cell culture represents a more clinically-significant growth environment relative to growth in nutrient-rich in vitro environments. This, in turn, can provide insight for future in vivo experimental models and inform translational studies in patient populations. Furthermore, our evaluation of P. aeruginosa grown in MEM allows us to hypothesize about the origins of these metabolites, although further experiments are needed to determine whether these discriminatory metabolites arise from metabolism of alternate substrates by P. aeruginosa in the presence of CFBE, and/or the response of CFBE to the presence of P. aeruginosa.
With respect to study limitations, we acknowledge that we consider only a single, reference strain (PAO1), which may not be metabolically representative of all possible P. aeruginosa strains. Furthermore, we were able to analyze only a small volume (500 μL) per replicate, and that this may have contributed to a relatively weak signal intensity, possibly below the detection limit for a subset of analytes. This, in turn, may have contributed to our inability to identify a robust volatile metabolic signature associated with RSV infection. Moreover, the volatile profiles obtained do not necessarily mirror the real profile present in the headspace of the sample, since it is mediated by the specific selectivity of the SPME fiber (PDMS/Car/DVB) used. The identification of the discriminatory compounds was putative and the confirmation of their identity with analytical standards should be confirmed in future studies. Finally, it is important to consider that the CFBE cell system models the initial interaction (6 h) of P. aeruginosa with the respiratory epithelium. It is therefore possible that a longer interaction time might yield to a more robust signal and altered metabolite detection as the infection persists.
Conclusions
In the present study, we demonstrate that volatile metabolites can be used to discriminate between P. aeruginosa (PAO1) infected and non-P. aeruginosa infected respiratory epithelium. Although weak and non-discriminatory, a signature related to the RSV singular infection can be observed (see Figure 3), while the volatile signature of co-infected samples (PA+/RSV+) is dominated by the signature produced by P. aeruginosa. The contribution of the host response to the abundance of some of these discriminatory metabolites can be hypothesized, since their abundances were significantly different from the control when RSV singular infection was present (acetone, undecane, and a hydrocarbon (#29)). Furthermore, the formation of 2-butanone, 3-methylbutanal, and 2-ethylhexanal can be associated with the specific interaction between P. aeruginosa and the CFBE cells, as these metabolites were not detected in the headspace of P. aeruginosa grown as a biofilm in the absence of CFBE.
Supplementary Material
Acknowledgements
Financial support for this work was provided by Hitchcock Foundation and the National Institute of Health (NIH, Project # 1R21AI12107601). CAR was supported by the Burroughs Wellcome Fund Institutional Program Unifying Population and Laboratory Based Sciences, awarded to Dartmouth College (Grant#1014106), and a T32 training grant (T32LM012204, PI: Christopher I Amos). JAM was supported by the Cystic Fibrosis Foundation (MELVIN15F0) and a T32 training grant (T32AI049820, PI: Neal A. DeLuca). JMB was supported by the Cystic Fibrosis Foundation (BOMBER14G0) and National Institutes of Health (R01HL123771).
References
- [1].WHO 2014. Media centre The top 10 causes of death Fact sheet N°310 (Updated May 2014) 2012–4
- [2].UNICEF 2015. Levels and trends in child mortality New York: UNICEF; 1–30 [Google Scholar]
- [3].Jain S, Williams DJ, Arnold SR, Ampofo K, Bramley AM, Reed C, Stockmann C, Anderson EJ, Grijalva CG, Self WH, Zhu Y, Patel A, Hymas W, Chappell JD, Kaufman RA, Kan JH, Dansie D, Lenny N, Hillyard DR, Haynes LM, Levine M, Lindstrom S, Winchell JM, Katz JM, Erdman D, Schneider E, Hicks LA, Wunderink RG, Edwards KM, Pavia AT, McCullers JA and Finelli L 2015. Community-Acquired Pneumonia Requiring Hospitalization among U.S. Children N. Engl. J. Med 372 835–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Jain S, Self WH, Wunderink RG, Fakhran S, Balk R, Bramley AM, Reed C, Grijalva CG, Anderson EJ, Courtney DM, Chappell JD, Qi C, Hart EM, Carroll F, Trabue C, Donnelly ΗK, Williams DJ, Zhu Y, Arnold SR, Ampofo K, Waterer GW, Levine M, Lindstrom S, Winchell JM, Katz JM, Erdman D, Schneider E, Hicks LA, McCullers JA, Pavia AT, Edwards KM and Finelli L 2015. Community-Acquired Pneumonia Requiring Hospitalization among U.S. Adults N. Engl. J. Med 373 415–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Peters BM, Jabra-Rizk MA, O’May GA, William Costerton J and Shirtliff ΜE 2012. Polymicrobial interactions: Impact on pathogenesis and human disease Clin. Microbiol. Rev 25 193–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Bosch AATM, Biesbroek G, Trzcinski K, Sanders EAM and Bogaert D 2013. Viral and Bacterial Interactions in the Upper Respiratory Tract PLoS Pathog 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Melvin JA and Bomberger JM 2016. Compromised Defenses: Exploitation of Epithelial Responses During Viral-Bacterial Co-Infection of the Respiratory Tract PLoS Pathog 12 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Caliendo AM, Gilbert DN, Ginocchio CC, Hanson KE, May L, Quinn TC, Tenover FC, Alland D, Blaschke AJ, Bonomo RA, Carroll KC, Ferraro MJ, Hirschhom LR, Joseph WP, Karchmer T, MacIntyre AT, Reller LB and Jackson AF 2013. Better Tests, Better Care: Improved Diagnostics for Infectious Diseases Clin. Infect. Dis 57 S139–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hament JM, Aerts PC, Fleer A, Van Dijk H, Harmsen T, Kimpen JLL and Wolfs TFW 2005. Direct binding of respiratory syncytial virus to pneumococci: A phenomenon that enhances both pneumococcal adherence to human epithelial cells and pneumococcal invasiveness in a murine model Pediatr. Res 58 1198–203 [DOI] [PubMed] [Google Scholar]
- [10].McCullers JA 2006. Insights into the interaction between influenza virus and pneumococcus Clin. Microbiol. Rev 19 571–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Avadhanula V, Rodriguez CA, DeVincenzo JP, Wang Y, Webby RJ, Ulett GC and Adderson EE 2006. Respiratory Viruses Augment the Adhesion of Bacterial Pathogens to Respiratory Epithelium in a Viral Species- and Cell Type-Dependent Manner J. Virol 80 1629–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Stark JM, Stark MA, Colasurdo GN and LeVine AM 2006. Decreased Bacterial Clearance From the Lungs of Mice Following Primary Respiratory Syncytial Virus Infection James J. Med. Virol 78 829–38 [DOI] [PubMed] [Google Scholar]
- [13].van der Sluijs KF, van Elden LJR, Nijhuis M, Schuurman R, a JM, Florquin S, Goldman M, Jansen ΗM, Lutter R and van der Poll T 2004. IL-10 Is an Important Mediator of the Enhanced Susceptibility to Pneumococcal Pneumonia after Influenza Infection J. Immunol 172 7603–9 [DOI] [PubMed] [Google Scholar]
- [14].Frayman KB, Armstrong DS, Grimwood K and Ranganathan SC 2017. The airway microbiota in early cystic fibrosis lung disease Pediatr. Pulmonol 1–21 [DOI] [PubMed] [Google Scholar]
- [15].Wat D, Gelder C, Hibbitts S, Cafferty F, Bowler I, Pierrepoint M, Evans R and Doull I 2008. The role of respiratory viruses in cystic fibrosis. J. Cyst. Fibros 7 320–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Smyth AR, Smyth RL, Tong CYW, Hart CA and Heaf DP 1995. Effect of respiratory virus infections including rhinovirus on clinical status in cystic fibrosis Arch. Dis. Child 73 117–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Collinson J, Nicholson KG, Cancio E, Ashman J, Ireland DC, Hammersley V, Kent J and Callaghan CO 1996. Effects of upper respiratory tract infections in patients with cystic fibrosis Thorax 51 1115–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hiatt PW, Grace SC, Kozinetz CA, Raboudi SH, Treece DG, Taber LH, Piedra PA and Objective A 2018. Effects of Viral Lower Respiratory Tract Infection on Lung Function in Infants With Cystic Fibrosis Pediatrics 103 619–26 [DOI] [PubMed] [Google Scholar]
- [19].Brownlee JW and Turner RB 2008. New developments in the epidemiology and clinical spectrum of rhinovirus infections Curr. Opin. Pediatr 20 67–71 [DOI] [PubMed] [Google Scholar]
- [20].Petersen NT, Hoiby N, Mordhorst CH, Lind K, Flensborg EW and Bruun B 1981. Respiratory infections in cystic fibrosis patients caused by virus, chlamydia and mycoplasma-possible synergism with Pseudomonas aeruginosa Acta Paediatr. Scand 70 626–8 [DOI] [PubMed] [Google Scholar]
- [21].Van Ewijk BE, Van Der Zalm ΜM, Wolfs TFW and Van Der Ent CK 2005. Viral respiratory infections in cystic fibrosis J. Cyst. Fibros 4 31–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Courtney JM, Bradley J, Mccaughan J, Ο TM, Shortt C, Bredin CP, Bradbury I and Elborn JS 2007. Predictors of Mortality in Adults With Cystic Fibrosis Pediatr. Pulmonol 42 525–32 [DOI] [PubMed] [Google Scholar]
- [23].Hendricks MR, Lashua LP, Fischer DK, Flitter BA, Eichinger KM, Durbin JE, Sarkar SN, Coyne CB, Empey KM and Bomberger JM 2016. Respiratory syncytial virus infection enhances Pseudomonas aeruginosa biofilm growth through dysregulation of nutritional immunity Proc. Natl. Acad. Sci 113 1642–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Sethi S, Nanda R and Chakraborty T 2013. Clinical application of volatile organic compound analysis for detecting infectious diseases Clin. Microbiol. Rev 26 462–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zhu J, Bean HD, Jimenez-Diaz J and Hill JE 2013. Secondary electrospray ionization-mass spectrometry (SESI-MS) breathprinting of multiple bacterial lung pathogens, a mouse model study J. Appl. Physiol 114 1544–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Zhu J, Bean HD, Wargo MJ, Leclair LW and Hill JE 2014. Detecting Bacterial Lung Infections: in vivo Evaluation of in vitro Volatile Fingerprints J Breath Res 7 16003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zhu J, Jiménez-Díaz J, Bean HD, Daphtary NA, Aliyeva ΜI, Lundblad LKA and Hill JE 2013. Robust detection of P. aeruginosa and S. aureus acute lung infections by secondary electrospray ionization-mass spectrometry (SESI-MS) breathprinting: from initial infection to clearance J. Breath Res 7 37106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Filipiak W, Mochalski P, Filipiak A, Ager C, Cumeras R, Davis CE, Agapiou A, Unterkofler K and Troppmair J 2016. A Compendium of Volatile Organic Compounds (VOCs) Released By Human Cell Lines. Curr. Med. Chem 23 2112–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Filipiak W, Beer R, Sponring A, Filipiak A, Ager C, Schiefecker A, Lanthaler S, Helbok R, Nagl M, Troppmair J and Amann A 2015. Breath analysis for in vivo detection of pathogens related to ventilator-associated pneumonia in intensive care patients: a prospective pilot study Breath analysis for in vivo detection of pathogens related to ventilator-associated pneumonia in intensive J. Breath Res 9 16004. [DOI] [PubMed] [Google Scholar]
- [30].Fowler SJ, Basanta-sanchez M, Xu Y, Goodacre R and Dark PM 2015. Surveillance for lower airway pathogens in mechanically ventilated patients by metabolomic analysis of exhaled breath: a case-control study Thorax 70 320–5 [DOI] [PubMed] [Google Scholar]
- [31].Schnabel R, Fijten R, Smolinska A, Dallinga J and Van Schooten FJ 2015. Analysis of volatile organic compounds in exhaled breath to diagnose ventilator-associated pneumonia Sci. Rep 17179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Sweeney TE, Shidham A, Wong HR, Khatri P, Alto P and Alto P 2016. A comprehensive time-course–based multicohort analysis of sepsis and sterile inflammation reveals a robust diagnostic gene set. Sci. Transl. Med 7 1–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Sweeney T and Purvesh K 2015. Comprehensive validation of the FAIM3:PLAC8 ratio in time matched public gene expression data Am. J. Respir. Crit. Care Med 192 1260–1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].McHugh L, Seldon TA, Brandon RA, Kirk JT, Rapisarda A, Sutherland AJ, Presneill JJ, Venter DJ, Lipman J, Thomas MR, Klein Klouwenberg PMC, van Vught L, Scicluna B, Bonten M, Cremer OL, Schultz MJ, van der Poll T, Yager TD and Brandon RB 2015. A Molecular Host Response Assay to Discriminate Between Sepsis and Infection-Negative Systemic Inflammation in Critically Ill Patients: Discovery and Validation in Independent Cohorts PLoS Med 12 1–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Scicluna BP, Klein Klouwenberg PMC, Van Vught LA, Wiewel MA, Ong DSY, Zwinderman AH, Franitza M, Toliat MR, Nürnberg P, Hoogendijk AJ, Horn J, Cremer OL, Schultz MJ, Bonten MJ and Van Der Poll T 2015. A molecular biomarker to diagnose community-acquired pneumonia on intensive care unit admission Am. J. Respir. Crit. Care Med 192 826–35 [DOI] [PubMed] [Google Scholar]
- [36].Hu X, Yu J, Crosby SD and Storch GA 2013. Gene expression profiles in febrile children with defined viral and bacterial infection Proc. Natl. Acad. Sci 110 12792–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Zaas AK, Burke T, Chen M, Mcclain M, Nicholson B, Veldman T, Tsalik EL, Fowler V, Rivers EP, Kingsmore SF, Voora D, Lucas J, Hero AO, Carin L, Woods CW and Ginsburg GS 2013. A Host-Based RT-PCR Gene Expression Signature to Identify Acute Respiratory Viral Infection Sci Transl Med 5203ra126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Suarez NM, Bunsow E, Falsey AR, Walsh EE, Mejias A and Ramilo O 2015. Superiority of transcriptional profiling over procalcitonin for distinguishing bacterial from viral lower respiratory tract infections in hospitalized adults. J. Infect. Dis 212 213–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Delneste Y, Beauvillain C and Jeannin P 2007. Innate immunity: structure and function of TLRs Med Sci 23 67–73 [DOI] [PubMed] [Google Scholar]
- [40].Medzhitov R and Janeway CAJ 1997. Innate immunity: Minireview the virtues of a nonclonal system of recognition Cell 91 295–8 [DOI] [PubMed] [Google Scholar]
- [41].Medzhitov R and Janeway CJ 2000. Innate immune recognition: mechanisms and pathways Immunol. Rev 173 89–97 [DOI] [PubMed] [Google Scholar]
- [42].Sung RY, Hui SH, Wong CK, Lam CW and Yin J 2001. A comparison of cytokine responses in respiratory syncytial virus and influenza A infections in infants. Eur. J. Pediatr 160 117–22 [DOI] [PubMed] [Google Scholar]
- [43].d’Acampora Zellner B, Bicchi C, Dugo P, Rubiolo P, Dugo G and Mondello L 2008. Linear retention indices in gas chromatographic analysis: a review Flavour Fragr. J 23 297–314 [Google Scholar]
- [44].Sumner LW, Samuel T, Noble R, Gmbh SD, Barrett D, Beale ΜH and Hardy N 2007. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI) Metabolomics 3 211–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Schallschmidt K, Becker R, Jung C, Bremser W, Walles T, Neudecker J, Leschber G, Frese S and Nehls I 2016. Comparison of volatile organic compounds from lung cancer patients and healthy controls-challenges and limitations of an observational study. J. Breath Res 10 46007. [DOI] [PubMed] [Google Scholar]
- [46].Dieterle F, Ross A, Schlotterbeck G and Senn H 2006. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics Anal. Chem 78 4281–90 [DOI] [PubMed] [Google Scholar]
- [47].Breiman L 2001. Random forests Mach. Learn 45 5–32 [Google Scholar]
- [48].Ruopp MD, Perkins NJ, Whitcomb BW and Schisterman enrique F 2008. Youden Index and Optimal Cut-Point Estimated from Observations Affected by a Lower Limit of Detection Biometrical J 50 419–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Mann ΗB and Whitney DR 1947. On a Test of Whether one of Two Random Variables is Stochastically Larger than the Other Author (s): Mann ΗB and Whitney DR Source: The Annals of Mathematical Statistics, Vol. 18, No. 1 (Mar., 1947), pp. 50–60 Published by: Institute Ann. Math. Stat. 18 50–60 [Google Scholar]
- [50].Melvin JA, Lashua LP, Kiedrowski MR, Yang G, Deslouches B, Montelaro RC and Bomberger JM 2016. Simultaneous Antibiofilm and Antiviral Activities of an Engineered Antimicrobial mSphere 1 e00083–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Shestivska V, Spanel P, Dryahina K, Sovova K, Smith D, Musilek M and Nemec A 2012. Variability in the concentrations of volatile metabolites emitted by genotypically different strains of Pseudomonas aeruginosa J. Appl. Microbiol 113 701–13 [DOI] [PubMed] [Google Scholar]
- [52].Bean HD, Rees CA and Hill JE 2016. Comparative analysis of the volatile metabolomes of Pseudomonas aeruginosa clinical isolates J. Breath Res 10 47102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Labows JN, Mcginley KJ, Webster GUYF Leyden JJ 1980. Headspace Analysis of Volatile Metabolites of Pseudomonas aeruginosa and Related Species by Gas Chromatography-Mass Spectrometry J. Clin. Microbiol 12 521–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Filipiak W, Sponring A, Baur ΜM, Filipiak A, Ager C, Wiesenhofer H, Nagl M, Troppmair J and Amann A 2012. Molecular analysis of volatile metabolites released specifically by staphylococcus aureus and pseudomonas aeruginosa BMC Microbiol 113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Boots AW. Identification of microorganisms based on headspace analysis of volatile organic compounds by gas chromatography – mass spectrometry. J. Breath Res. 2014;8:27106. doi: 10.1088/1752-7155/8/2/027106. [DOI] [PubMed] [Google Scholar]
- [56].Zechman M, Aldinger S and Labows J 1986. Characterization of pathogenic bacteria by automated headspace concentration-gas chromatography J. Chromatogr 377 49–57 [DOI] [PubMed] [Google Scholar]
- [57].Neerincx AH, Geurts BP, Habets MFJ, Booij JA, Van Loon J, Jansen JJ, Buydens LMC, Van Ingen J, Mouton JW, Harren FJM, Wevers RA, Merkus PJFM, Cristescu SM and Kluijtmans LAJ 2016. Identification of Pseudomonas aeruginosa and Aspergillus fumigatus mono- and co-cultures based on volatile biomarker combinations Identification of Pseudomonas aeruginosa and Aspergillus fumigatus mono- and co-cultures based on volatile biomarker combinat J. Breath Res 10 16002. [DOI] [PubMed] [Google Scholar]
- [58].Purcaro G, Rees CA, Wieland-alter WF, Schneider MJ, Wang X, Stefanuto P, Wright PF, Enelow RI and Hill JE 2018. Volatile fingerprinting of human respiratory viruses from cell culture J. Breath Res 12 26015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Rudzinski CM, Herzig-marx R, Lin J, Szpiro A, Johnson B and Street W 2004. Pathogen detection using headspace analysis Scientific Conference on Chemical and Biological Defense Research [Google Scholar]
- [60].Dryahina K, Sovová K, Nemec A and Španěl P 2016. Differentiation of pulmonary bacterial pathogens in cystic fibrosis by volatile metabolites emitted by their in vitro cultures: Pseudomonas aeruginosa, Staphylococcus aureus, Stenotrophomonas maltophilia and the Burkholderia cepacia complex Differentia J. Breath Res 10 37102. [DOI] [PubMed] [Google Scholar]
- [61].Forman HJ 2010. Reactive oxygen species and alpha, beta-unsaturated aldehydes as second messengers in signal transduction. Ann. N. Y. Acad. Sci 1203 35–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Schulz S and Dickschat JS 2007. Bacterial volatiles: the smell of small organisms. Nat. Prod. Rep 24 814–42 [DOI] [PubMed] [Google Scholar]
- [63].Marilley L and Casey MG 2004. Flavours of cheese products: metabolic pathways, analytical tools and identification of producing strains Int. J. Food Microbiol 90 139–59 [DOI] [PubMed] [Google Scholar]
- [64].Ladygina N, Dedyukhina EG and Vainshtein ΜB 2006. A review on microbial synthesis of hydrocarbons Process Biochem 41 1001–14 [Google Scholar]
- [65].Hosakote YM, Liu T, Castro SM, Garofalo RP and Casola A 2009. Respiratory syncytial virus induces oxidative stress by modulating antioxidant enzymes Am. J. Respir. Cell Mol. Biol 41 348–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


