Abstract
Muscle strength is a functional measure of quality of life in humans. Declines in muscle strength are manifested in diseases as well as during inactivity, aging, and space travel. With conserved muscle biology, the simple genetic model C. elegans is a high throughput platform in which to identify molecular mechanisms causing muscle strength loss and to develop interventions based on diet, exercise, and drugs. In the clinic, standardized strength measures are essential to quantitate changes in patients; however, analogous standards have not been recapitulated in the C. elegans model since force generation fluctuates based on animal behavior and locomotion. Here, we report a microfluidics-based system for strength measurement that we call ‘NemaFlex’, based on pillar deflection as the nematode crawls through a forest of pillars. We have optimized the micropillar forest design and identified robust measurement conditions that yield a measure of strength that is independent of behavior and gait. Validation studies using a muscle contracting agent and mutants confirm that NemaFlex can reliably score muscular strength in C. elegans. Additionally, we report a scaling factor to account for animal size that is consistent with a biomechanics model and enables comparative strength studies of mutants. Taken together, our findings anchor NemaFlex for applications in genetic and drug screens, for defining molecular and cellular circuits of neuromuscular function, and for dissection of degenerative processes in disuse, aging, and disease.
Graphical abstract
I. Introduction
Skeletal muscle provides the contractile system critical for animal locomotion across phyla. The conserved biology of muscle includes sarcomere composition and organization, the calcium-initiated contractile mechanism, and the ATP-driven translocation of actin and myosin for force generation1, 2. A powerful indicator of muscle health is strength —a measure of maximum force that can be voluntarily exerted for a given task. Diagnosis of muscle degenerative disorders such as muscular dystrophy relies on strength and physical performance tests in the clinic3, 4. Similar diagnostic evaluation is crucial for, sarcopenia and dynapenia, the age-related progressive loss of muscle mass and muscle strength, respectively5, 6. Additionally, muscle atrophy and strength declines are a major impediment for manned deep space exploration7.
Given conservation of basic muscle biology, simple genetic models such as Drosophila melanogaster8 and the 959-celled Caenorhabditis elegans9, 10 constitute attractive platforms with which to decipher the molecular mechanisms of age-10,11, disease-12, or microgravity-13 associated muscle decline. For example, studies in C. elegans have led to important new insights into the assembly, maintenance, and regulation of striated muscle10, 14–16, with the loss-of-function mutants in sarcomeric proteins showing reduced capacity to swim or crawl17–19. These model organisms also offer a powerful means with which to investigate how diet, exercise protocols, and drug interventions may combat muscle dysfunction. Still, the notion of muscle strength and its measurement in these simple models has remained elusive.
A technical challenge in reporting muscle strength in invertebrate models is that its definition crucially depends on system geometry and measurement protocol. As a result, even though forces on the order of microNewtons have been reported to be exerted by C. elegans20–26; the approaches used yield different values of forces due to worm locomotion, body posture, and behavior. Without a measurement protocol that is reproducible and system geometry that is validated, fluctuating forces cannot be used to extract a meaningful metric for strength. Additionally, the influence of animal size on force production has not been established, even though it is well recognized that animals from the same culture as well as mutants can have different body sizes. Thus, force measures that reveal muscular strength independent of animal behavior and approaches to compensate forces for variations in animal size, have not been established in the C. elegans model.
In humans, precisely defined strength metrics such as maximum voluntary force (MVF) and one repetition maximum (1RM) are absolutely essential to quantitate and compare subjects, which constitutes the clinical basis of muscle physiology investigations27 (Fig. 1a). Here, MVF is defined as the peak force recorded for a particular set of muscle (leg extensor, elbow extensor etc.) during one trial28, 29, and 1RM is defined as the maximum weight that a person can lift for one repetition30. It is imperative to recapitulate analogous metrics and standardize them in the C. elegans model for the field to move forward with genetic studies on problems ranging from Duchenne muscular dystrophy to muscle wasting due to aging to metabolic disorders.
Figure 1. Muscle strength measurement in humans and the NemaFlex system for force measurement in C. elegans.
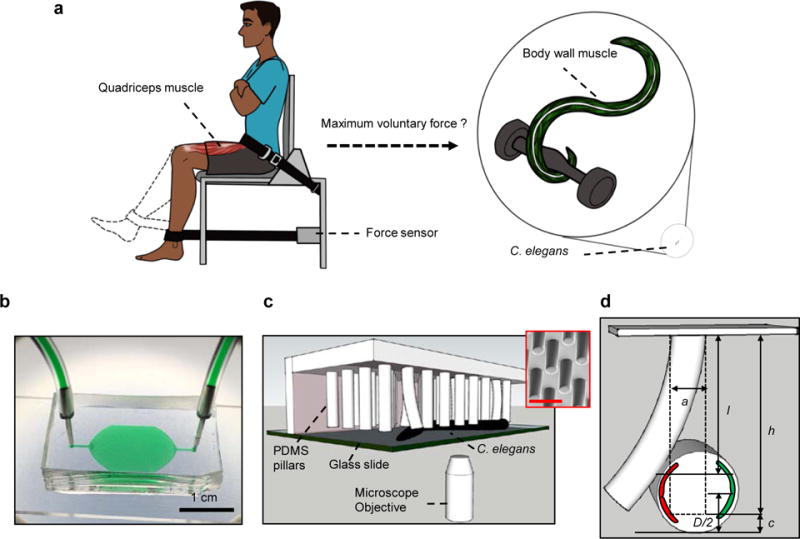
(a) Human muscle function is measured in terms of maximum voluntary force (MVF) during a standardized knee extensor test, where the peak force exerted by the quadriceps muscle is recorded using a force sensor. An equivalent measure of maximum exertable force in C. elegans is currently unavailable. (b) Image of the NemaFlex device filled with green food dye for highlighting the arena and the ports. Scale bar, 1 cm. (c) Schematic showing the C. elegans strength measurement apparatus including the chamber for housing worms, deformable pillar arrays, microscope objective for visualizing pillar deflection, and crawling nematode. Scanning electron microscope (SEM) picture of the pillars (inset). Scale bar, 100 μm. (d) Schematic showing interaction with a pillar by the worm body (exaggerated view). The pillar is deflected due to the action of the body wall muscles (shown in red and green). Parameters defined in Eqn. (1) are shown with actual values being a = 38.3 ± 0.4, h = 71.8 ± 2.9, c = 27.6 ± 2.9 and D = 50 -70 μm (for WT, age = 60 – 84 hrs).
Here we define a microfluidics-based system for strength measurement in nematodes by pillar deflection that we call “NemaFlex” (Fig. 1b-d). We study the effects of micropillar forest design and animal behavior to identify robust measurement conditions that define maximum exertable force (MEF) as a measure that is reproducible and insensitive to animal locomotion and behavior. Our force-velocity analysis in animals experiencing different confinements and validation studies using a muscle contracting agent mutants indeed show that MEF is analogous to MVF measured in humans. We also report a scaling factor to correct MEF for animal size, thus establishing NemaFlex as a quantitative and reproducible system for measurement of muscle strength among individual worms, mutants, and/or prospectively in an individual worm with age or experimental intervention. Our findings establish the means to exploit drug, genetic, and experiential outcomes that may define molecular and cellular circuits of muscle use and maintenance, which in turn might inspire therapies for extended muscle healthspan.
II. Experimental
Strains and worm preparation
C. elegans were maintained on E. coli OP50 bacterial lawns using standard protocol31. Wild-type (WT) animals were Bristol isolate (N2). Other strains used in this study were unc-52(e669), unc-112(r367ts)V, unc-17(e245), and lon-2(e678)X. Age synchronized, well-fed young adult animals were used for all force measurement experiments unless otherwise indicated. Age synchronized populations were obtained using a standard protocol. Approximately 20 gravid adult animals were transferred to a NGM plate (previously seeded with bacteria E. coli OP50) by hand picking and allowed to lay eggs on the bacteria lawn for 3 hours. After 3 hours, gravid adults were removed from the plate. Hatched worms were cultured at 20°C. The age of animals whose strength was evaluated is mentioned in either the main text or figure captions and is given as the time from hatching.
Microfluidic device fabrication
The micropillar-based NemaFlex force measurement device, shown in Fig. 1b, was fabricated using soft lithography32. Two master molds were used in this study. The first is a composite arena design that contains three concentric circular regions (A1, A2, and A3) with varying pillar diameters and spacing. This mold was used to optimize arena geometry for evaluation of maximum exertable forces produced by a crawling worm. The measured dimensions of the pillars in the A1, A2, and A3 regions are provided in Table 1. The second mold referred to as “NemaFlex” in Table 1 is an arena containing pillar matrix with geometrical dimensions very similar to region A3. We used this mold in all the post-optimization studies.
Table 1.
Geometrical details of the composite arena and measured gait and velocity in different regions of the arena. Here D = 65.1 ± 3.2 μm. Age of the worm is 84 hours after hatching. The height of the pillars in the composite arena is 83.6 ± 1.8 μm.
| Composite Arena | Pillar diameter, a (μm) | Confinement, D/s | Crawling amplitude, A (μm) | Crawling wavelength, λ (μm) | Speed, v (μm/s) |
|---|---|---|---|---|---|
| A1 | 63.5 ± 1.0 | 0.67 ± 0.03 | 88.1 ± 4.6 | 579.4 ± 25.6 | 264.1 ± 147.1 |
| A2 | 53.8 ± 1.2 | 0.85 ± 0.04 | 92.6 ± 3.5 | 526.0± 19.4 | 177.4 ± 66.0 |
| A3 | 44.1 ± 0.9 | 1.16 ± 0.06 | 85.0 ± 3.5 | 508.2 ± 32.6 | 127.7 ± 45.6 |
All the master molds used in this study were fabricated using the following procedure. A two-layer mold was fabricated in SU-8 2050 negative photoresist (Microchem) on a 3" silicon wafer as substrate (University Wafer)22. First, a 25-μm-tall, oval-shaped flat layer was fabricated, which forms the boundary of the arena. On top of this layer, a second layer of 75-μm height was fabricated with cylindrical holes that form the micropillars. This two-layer approach provides a total chamber depth of approximately 100 μm and creates dangling (deformable) pillars of height 75 μm (Fig. 1c,d).
Polydimethylsiloxane (PDMS) devices were casted (Sylgard 184 Part A (base) and Part B (curing agent) 10:1 by weight; Dow Corning) over the SU-8 mold by curing for ~2 hours at 75°C. The PDMS replica was then treated in an air-plasma cleaner (Harrick Plasma, Ithaca, NY) for 1 minute and bonded to a 1×3" glass slide. Bonding was done ensuring the pillars did not collapse or deform. Inlet and outlet holes were cored with a 1mm hole puncher (Accuderm) before bonding.
Device preparation and worm loading into the device
Bonded and cleaned microfluidic pillar arenas were prepared by surface treating with 5 wt% Pluronic F127 solution. Pluronic solution was loaded into the device through one of the inlets and incubated for 30 minutes, followed by washing with M9 buffer. The pluronic treatment minimizes protein/bacterial fouling and also helps to reduce bubble formation during worm loading33.
Immediately before each strength experiment, plates were flooded with 3-4 mL of M9 solution (approximately 100 animals/mL). For individual worm loading, the worm solution from the M9-flooded culture plate was diluted to 10X (approximately 10 animals/mL). Single animals were loaded into the device by using a syringe connected to tubing. Individual loading can also be done by hand picking a single worm. Experiments involving the composite arena, levamisole drug treatment, and body size effects were carried out with individuals. Muscle strength of mutants was evaluated at a population level, in which case the identity of the worm is not preserved during imaging. For population studies, approximately 25-30 animals were gently injected into the NemaFlex using a 1 mL syringe. While retaining animals between pillar obstacles, residual bacteria were removed from the device by flow of M9 buffer. In both the individual and population experiments, animals were allowed to habituate in the arena for approximately 5 minutes before imaging in the food-free environment.
Image acquisition
The imaging was conducted on two sets of microscopes and camera combinations. Most of the initial trials and strength measurement of mutants were performed on an inverted microscope (IX71, Olympus, Center Valley, PA) with image acquisition from a CCD camera (ImageEM, Hamamatsu, Japan). The magnification used was 6.4X with an image resolution of 2.52 μm/pixel. Imaging for experiments dealing with arena design optimization, forced contraction assay with levamisole, and influence of body size on strength were carried out using another microscope (IX70, Olympus, Center Valley, PA) instrumented with a CCD camera (Retiga R6, QImaging, Canada). The magnification used was 6X with an image resolution of 0.7 μm/pixel. Images were acquired at 5 fps for a duration of 30-120 s of one single worm with a field of view of 1880×1540 μm2. All imaging was conducted at a temperature of 20 ± 1°C.
Image processing for quantitation of pillar displacements
Movies were processed offline using custom routines written in MATLAB (Mathworks, R2014b). Worm diameters were measured manually using ImageJ 1.48v. Below, we outline the key steps in the procedure for quantitating pillar displacement from the original images of a crawling worm in the pillar arena (Fig. 2a). Pillar tracking algorithm and image processing steps are described in detail in Supplementary Note 1.
Figure 2. Illustration of the steps in image processing to quantify pillar displacements.
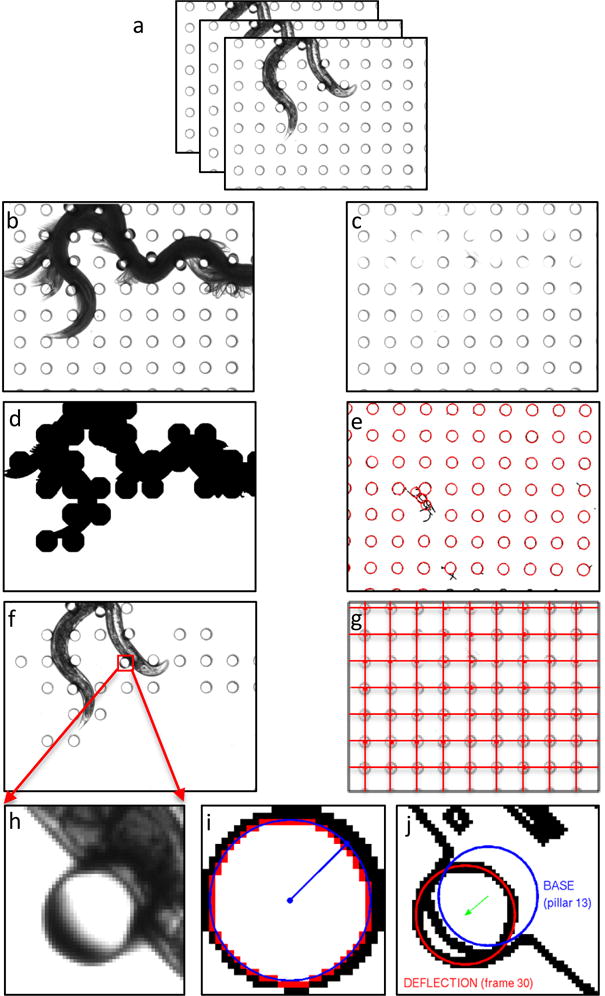
(a) Original images of a crawling worm. (b) Foreground image and (c) background image from a stack of images in the preprocessing step. (d) Mask generation from the foreground. (e) Identification of all pillars by applying circular Hough transform. (f) Identification of the candidate pillar for tracking using the mask. (g) Grid verification and validation of pillar location. (h) A candidate pillar selected from a frame (see red arrows) for illustration of deflection measurement. (i) Determination of pillar base location and radius when the worm is not touching the pillar. (j) Measurement of pillar displacement.
A median filter is applied to each image (Fig. 2a) to eliminate the outlier pixels. The maximum pixel value (i.e. brightest) at each pixel location across all frames provides the background image. Likewise, the minimum pixel value (i.e. darkest) at each pixel location yields the foreground image. As shown in Fig. 2b, these operations make the worm’s entire trajectory visible in the foreground image and absent from the background image. The background contains mostly pillars (see Fig. 2c) and extraneous objects not part of the worm’s trajectory.
A “mask” is created from the foreground, which contains the worm trajectory with contiguous pillars only (Fig. 2b). The mask (Fig. 2d) is then applied to the background image to identify the candidate pillar (Fig. 2f) and candidate frames for tracking. In parallel, the background image (Fig. 2c) is used to identify pillars and their base locations (Fig. 2e) by using circular Hough-transform (CHT). A grid connecting the pillar locations (Fig. 2g) is used to eliminate spurious pillars due to dirt, air bubble etc. Verified candidate pillars are then subject to refined CHT, each one at a time (Fig. 2h-j), and displacements of the pillars are calculated from their base locations. Supplementary Movie 1 shows the output of our pillar tracking algorithm.
Experimental error in the measurement of forces
The error in measurement of forces results from uncertainties associated with quantifying the pillar deflection. Uncertainty in determining pillar deflection can result from several sources including improper fitting of the boundaries of the pillar (e.g. due to day-to-day variations in light intensity), minor heterogeneity in the size distribution of pillars during microfabrication, and mechanical drift in the microscope stage. To account for these different sources of error cumulatively, we tracked a single undeflected pillar from twenty different movies (of duration ≈ 30s) acquired during the study and binned the putative forces to produce a probability distribution (Fig. 3). This maximal force distribution has a mean at 0.9 μN, a standard deviation of 0.8 μN, and the force corresponding to 95% cumulative probability, f95 value, of 2.2 μN (see inset of Fig. 3). Given that the force values due to the stationary pillar can reach as high as 2.2 μN, we use ± 2.5 μN as a conservative estimate of the error in our force measurement. This error is less than 10% of the population-averaged strength of 60-hour-old wild-type animals, as shown in the inset of Fig. 3. There is also zero overlap between the two force distributions, allowing clear distinction between true forces and errors.
Figure 3. Estimation of error in force measurement.
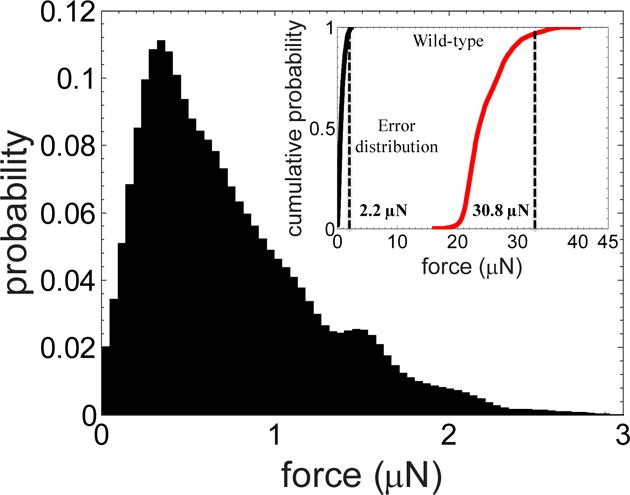
False positive force detections in the system were determined by tracking a single undeflected pillar from twenty movies of wild-type worms (age = 60 hrs). The error distribution has a mean at 0.9 μN, a standard deviation of 0.8 μN, and an f95 value of 2.2 μN. Inset compares error distribution relative to the strength of the population. The f95 value of the error distribution is less than 10% of the f95 value for the worm population. There is also zero overlap between the two force distributions, allowing clear distinction between true forces and errors.
Additional bias in strength measurement could be potentially introduced when the worm body diameter is larger than the pillar spacing, as in the region A3 in the composite arena where the worm confinement D/s = 1.16 (Table 1). Here D is the worm body diameter at mid-length and s is the edge-to-edge pillar spacing. In this case, a default force will be registered even when the worms are not active. These default forces are smaller than the maximal forces exerted by the worm. Since our strength calculation analysis only selects the maximal forces, the measured value is not affected by these default weak forces induced by higher confinement of the worm body in the pillar arena.
To estimate the magnitude of the default force, we choose hypothetical worm body diameters that produce confinement above 1 and calculate the corresponding passive deflections (Δ) and forces (F). As shown in Supplementary Table 1, for all the cases considered, Δ < 5 μm and F < 12 μN. These values are smaller than the maximal deflections and forces experimentally observed for a wild-type animal of similar body size that is actively pushing the pillars (see for example, Supplementary Movie 2 which shows the large pillar deflections for a worm with confinement level of 1.07). Moreover, we observe that when the worm pushes the pillars on one side of the body, sufficient gap is created and therefore pillars on the other side of the body do not experience any push. Thus small forces due to passive deflections occur rarely in an actively crawling worm.
Animal velocity and gait analysis
To calculate the crawling speed of C. elegans, the displacement of the midpoint of the body skeleton was tracked in time. For force-velocity correlation, instantaneous velocity from two consecutive frames was calculated, and the correlation was plotted using the maximal force in the later frame. For characterizing the mean crawling speed, the entire episode was used, and displacements of the midpoint were measured in time. Gait analysis included measuring the wavelength and amplitude of the undulatory forward motion by determining the distance between two peaks in the body wave and the peak height, respectively. Data from ten animals was measured and averaged.
Muscle contraction (levamisole) assay
20-30 animals were loaded into the micropillar chambers housing individual worms. Since the loading process can take 15-20 min, bacteria suspension of 100 mg E. coli OP50/mL of S-complete was added to each chamber after worm loading so that the individuals did not suffer from food deprivation. Imaging of each animal before exposure to levamisole was carried out after removing food with S-complete and allowing the animal to habituate for 5 minutes in the food-free environment. The muscle contraction experiment was carried out with 1 mM levamisole solution in S-complete. The levamisole solution was added to the individual chambers using a 1 mL syringe after the first round of imaging. Muscle contraction induced by levamisole (worm body length contraction) was captured 30 seconds after addition of levamisole solution and continued for 3 minutes or until the worm was paralyzed. ImageJ 1.48v was used to measure the length of the worm before and after the levamisole exposure.
Force characterization during long-time variation in behavior
Individual wild-type young adult worms were loaded into the pillar chambers. The first set of images was captured 2 minutes after loading, which is denoted as t = 0 hr. Food (100 mg E. coli OP50/mL of S-complete) was added at the 1-hour mark to avoid food limitation. The food was then removed at the 2-hour time point, and then 2 minutes after food removal the worm was imaged to obtain strength data at t = 2 hr and 2.5 hr time points.
Data analysis
All the data analysis was conducted in MATLAB (Mathworks, R2014b). Cumulative probability distributions were generated using Kernel Density Estimate (KDE). The bin size is estimated using the built-in asymptotic approximation. The force value at a cumulative probability of 95% (f95) is taken as the strength of the animal. A two-sample t-test was used to determine significance of data from muscle contraction assays. The Wilcoxon rank-sum test (WRST) was used to compare genetic mutants.
III. Results and Discussion
Basic principle of NemaFlex and force analysis
At the core of the NemaFlex technology is a liquid-filled microfluidic chamber containing elastic micropillars dangling from the chamber roof that can be deformed by a nematode push (Fig. 1c, d). As C. elegans threads through the pillars, the individual pillar deflections Δ are quantified using a microscope-camera system and image analysis (see Supplementary Movie 2). The force F causing the observed pillar deflection is calculated using the Timoshenko theory for an elastic rod22, 34,
| (1) |
In Eqn. (1), a, h, and I are the diameter, height, and moment of inertia of the cylindrical pillar respectively. E and γ are the elastic modulus and Poisson’s ratio of the polydimethylsiloxane (PDMS) pillar respectively. The point of contact of the worm with the pillar edge is denoted by l = h – (D-c)/2, which is calculated knowing the worm diameter D at mid-length and the clearance c between the pillar edge and the bottom surface of the chamber. The values used for the different parameters in Eqn. (1) as well as its validity for force measurement are discussed in Supplementary Note 2.
In general, we find that the magnitude of the pillar forces depends on the contact point along the nematode body, body configuration, and behavioral characteristics of C. elegans. Given this probabilistic nature of pillar forces exerted by the worm, we sought to establish a robust and reproducible procedure for quantifying animal strength. Fig. 4a-d shows this procedure in which we typically record a 30 second behavioral episode per animal. In every image of the acquired video, we identify the pillar with the maximal deflection (labeled red in Fig. 4a among all the candidate pillars), allowing us to extract an instantaneous maximal force value from each frame (Fig. 4b). We bin all the instantaneous maximal force values and generate a cumulative force distribution curve that defines a probability for exerting a maximal force lower than or equal to a given value (Fig. 4c). From this distribution, we define the maximal force corresponding to 95% cumulative probability, f95, as the maximum exertable force (MEF) of C. elegans (Fig. 4d). Our error in force detection, measured from pillars that are not in animal contact, is markedly lower than the maximal forces exerted by the nematode (Fig. 4b), with a cumulative measurement error of 2.5 μN in the f95 value (c.f. Fig. 3).
Figure 4. Data analysis workflow for NemaFlex strength measurement.
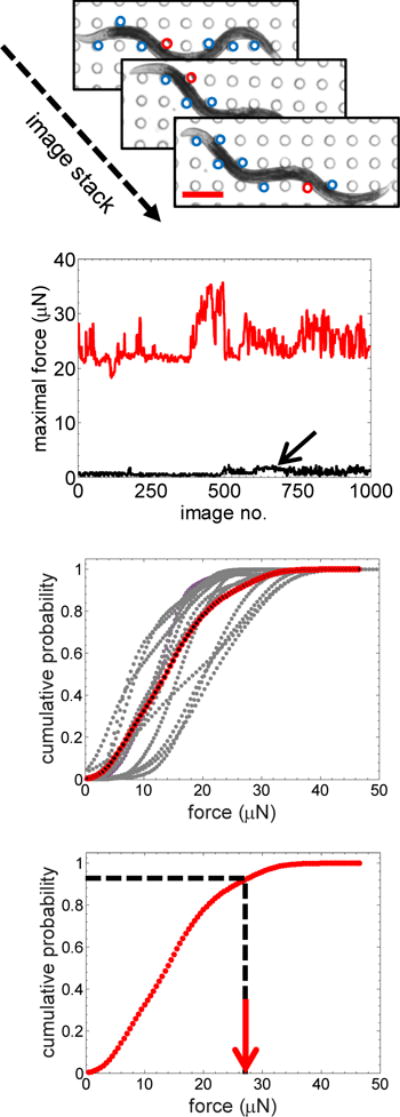
(a) Stack of images showing the worm interacting with different pillars during a locomotory episode. The deflected pillars are circled in blue, and the red circle denotes the pillar that experiences the maximum force. Scale bar, 200 μm. (b) Variation of maximal force over time due to a worm interacting with pillars (in red). The black curve shows force variation from the pillars that are not in contact with the worm – giving an estimate of error in our force measurement. (c) Cumulative probability distribution curves of maximal forces for different worms (age = 60 hrs) showing the variability between individuals (n = 14). The red trace represents the cumulative force distribution curve for the population. (d) From the cumulative force distribution curve, the maximum exertable force, f95, is defined as the maximal force at 95% cumulative probability.
Our approach allows defining the MEF at the level of an individual or a population, depending on whether the data constitute maximal forces from a single worm or a collection of worms. The cumulative force distribution curve for individuals typically consists of 200 data points, and for a population at least 2000. Fig. 4c shows the cumulative force distribution curve for wild-type (WT, age = 60 hrs) individuals. We find variability in the f95 values between individuals suggesting that some animals are naturally stronger than others, even though their body diameters are very similar (5% variation of mean body diameter).
Resistance to locomotion in the pillar arena determines maximal forces
The purpose of our study is to establish a reliable measure for C. elegans strength that is equivalent to maximum voluntary force in humans. To achieve this, we sought to identify micropillar forest designs that create high mechanical resistance to locomotion. Analogous to human strength evaluation where strong forces are registered when resistance to motion is high (c.f. Fig. 1a), we expect that animals in tight pillar arenas experience strong resistance to locomotion and produce large forces. Below we show results supporting that animals produce strong forces in highly resistive arenas and discuss the existence of different regimes of resistance to locomotion based on the size of nematode relative to gap between pillars.
Highly resistive pillar environment is characterized by low velocity and strong forces
To identify an arena geometry that induces high resistance to locomotion and makes C. elegans produce maximal forces frequently we fabricated a composite arena (Fig. 5a) that contained three regions (A1, A2, and A3) with distinct pillar diameters (in the range of a ≈ 40 – 60 μm) and gap between pillars (s ≈ 100 – 60 μm) (Supplementary Note 3). Regions A1, A2 and A3 produced confinement levels of D/s ≈ 0.67, 0.85, and 1.16 for wild type adults respectively (Fig. 5b). The composite arena is unique as it allowed us to investigate the dependence of force-velocity correlation and gait parameters on animal confinement.
Figure 5. Resistance to locomotion determines maximal forces.
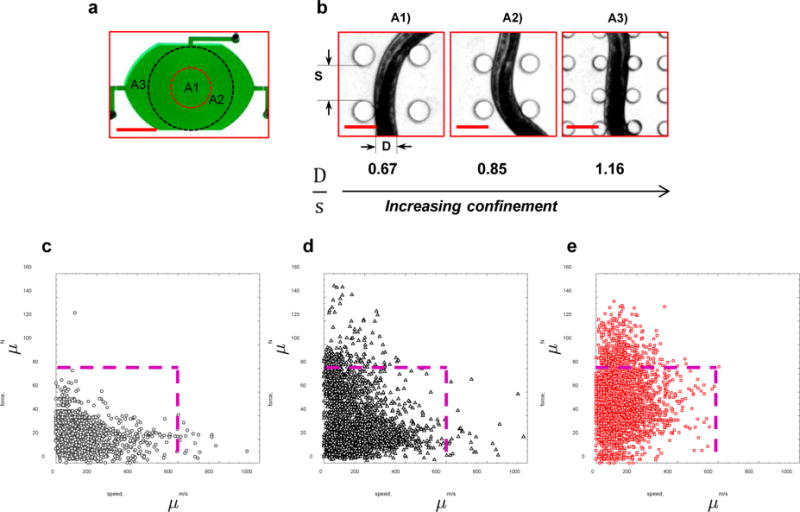
(a) A composite micropillar arena containing sections A1, A2, and A3 with different pillar spacings to investigate the influence of animal confinement on force generation. Scale bar, 5 mm. (b) The same nematode shown in the three different arenas. The level of confinement (D/s) increases as the worm crawls from arena A1 to A3. Scale bar, 100 μm. The force-velocity data for animals in (c) A1, (d) A2, and (e) A3 arenas. The lines indicate force-velocity cut-offs of 80 μN and 600 μm/s. The data correspond to 17 WT individuals of age 84 hours.
Pooling data from individuals (WT, age = 84 hrs), in Fig. 5c-e we show the maximal pillar force (from each image) versus the instantaneous animal velocity in the three arenas. We observe that in the moderately confined A1 arena, animals move at velocities as large as 600 μm/s and rarely exert forces more than 80 μN (Fig. 5c). In contrast, animals in the strongly confining A3 arena move at much slower velocities and often exert forces > 80 μN (Fig. 5e). Such large forces occur less frequently in arena A2 (Fig. 5d). Scoring the forward crawling locomotion, we find that between A1 and A3 arenas, the body wavelength reduces by 12% and amplitude varies by 3%, suggesting that in the moderate-strong confinement regime the force-velocity correlation is much more strongly affected than the forward crawling gait.
Frequent exertion of maximal forces in A3 arena can be understood from the high mechanical resistance to locomotion provided by this tight pillar arena where the nematode muscles push against the pillar, but its body cannot adjust its position in response to reaction forces due to the body being constrained simultaneously by multiple pillars. Evidence for the strong mechanical resistance is exemplified by the high frequency of turns and reversals in the highly resistive A3 and A2 arenas compared to A1 (Fig. 6).
Figure 6. Behavioral phenotyping of C. elegans in pillar arenas with different confinements.
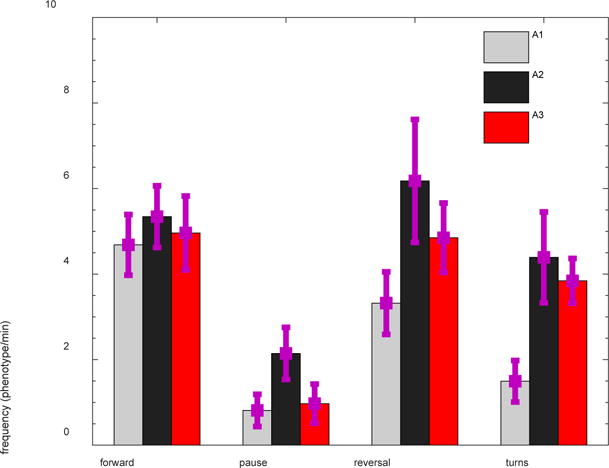
The frequency of reversals and turns is higher under tighter confinement due to strong mechanical resistance of the pillar environment. The data correspond to 17 WT individuals of age 84 hours.
Identification of different regimes of resistance to locomotion
Our experimental results thus far and previous work in pillar environments22–24, 35–38 (see Supplementary Table 2 for description of pillar geometries used in prior works), paint the following general picture; depending on the body size and pillar forest geometry, C. elegans locomotion between microfluidic pillars can be classified as non-resistive, moderately resistive or highly resistive. The existence of such regimes has been previously unrecognized.
In the non-resistive regime, the submerged nematode is typically in contact with one or two pillars (Fig. 7a). The forces would be low here because the nematode translates and rotates in response to a body contact with a pillar, exhibiting swimming-like behavior. An example of this regime was shown by Majmudar et al., where for D/s = 0.17, nematode swim trajectories were observed to be diverted by mechanical interactions with the pillars38.
Figure 7. Schematic illustrating the effect of animal confinement on mechanical resistance and force generation in pillar environments.
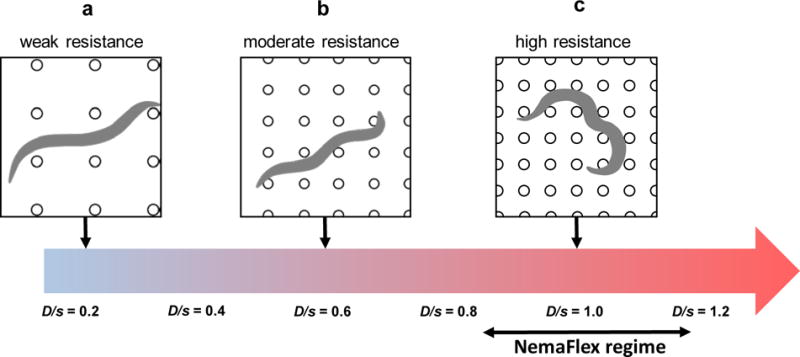
The nematode experiences increasing confinement (D/s) from left to right due to increasing density of pillars. (a) Weak resistance, D/s = 0.2, (b) moderate resistance, D/s = 0.6, and (c) strong resistance, D/s = 1.0. Large forces are expected under strong confinement due to enhanced mechanical resistance and constrained body shapes. The operating confinement regime of our NemaFlex system is highlighted.
In the moderately resistive regime, frequent contacts with more than two pillars occur (Fig. 7b) and the body wave conveniently fits into the space between pillars; the velocity is high because pillars provide transverse resistance enabling efficient forward locomotion. Forces are moderate because the nematode can fit in the available space with only minor adjustment of body posture. This picture is consistent with our results in A1 arena and further supported by the work of Ryu and coworkers36 who showed that when D/s ≈ 0.35 – 0.5 C. elegans swimming speed is enhanced.
In the highly-resistive regime which corresponds to our arenas A2 and A3, frequent contacts with multiple pillars occur (Fig. 7c). Animal velocity is low and forces are high because of the incompatibility between nematode gait and pillar configuration. Our observations are in agreement with the work by Johari et al. and Khare et al., who used arenas with D/s = 0.86 -1.6 and showed that animals exert strong forces23, 24.
The existence of these regimes indicates that NemaFlex device needs to operate in the highly resistive regime to score MEF. However, even in this highly resistive regime, individual pillar forces fluctuate from low to high as evidenced by our cumulative force distribution curves, thus requiring robust measurement and data analysis protocols for standardized strength measurement. Previous works have been unable to establish behavior- and gait-independent measures of strength that are indicative of MVF in C. elegans.
Maximum exertable force f95 is a reliable measure to score muscle strength in C. elegans
In this section, we show that MEF, our proposed metric of C. elegans strength, yields consistent values in highly resistive pillar arenas and that it is independent of gait and behavior. By using a drug that contracts muscles we confirm that MEF due to induced muscle contractions is the same as that calculated from force distribution due to voluntary muscle action. Finally, we demonstrate that MEF can be used to reliably score neuromuscular weakness in mutants.
Optimized pillar arena produces consistent readout of MEF
Given that even in the highly resistive pillar environments forces fluctuate, we asked whether MEF for the same individual changes due to different degrees of confinement in A1, A2, and A3 arenas. Our hypothesis was that if MEF is truly indicative of the maximum voluntary force then in the highly resistive arenas A2 and A3 we should obtain the same values for MEF. In Fig. 8a, we show the results for individual wild-type animals and find that the MEF in arenas A2 and A3 show a near-perfect correlation ( , calculated slope = 1.01 ± 0.05) while MEF is lower in A1 arena with a slope = 0.46 ± 0.07 shown in Fig. 8b. Thus f95 is consistent between the highly resistive A2 and A3 arenas.
Figure 8. Highly resistive pillar arenas produce consistent maximum exertable force.
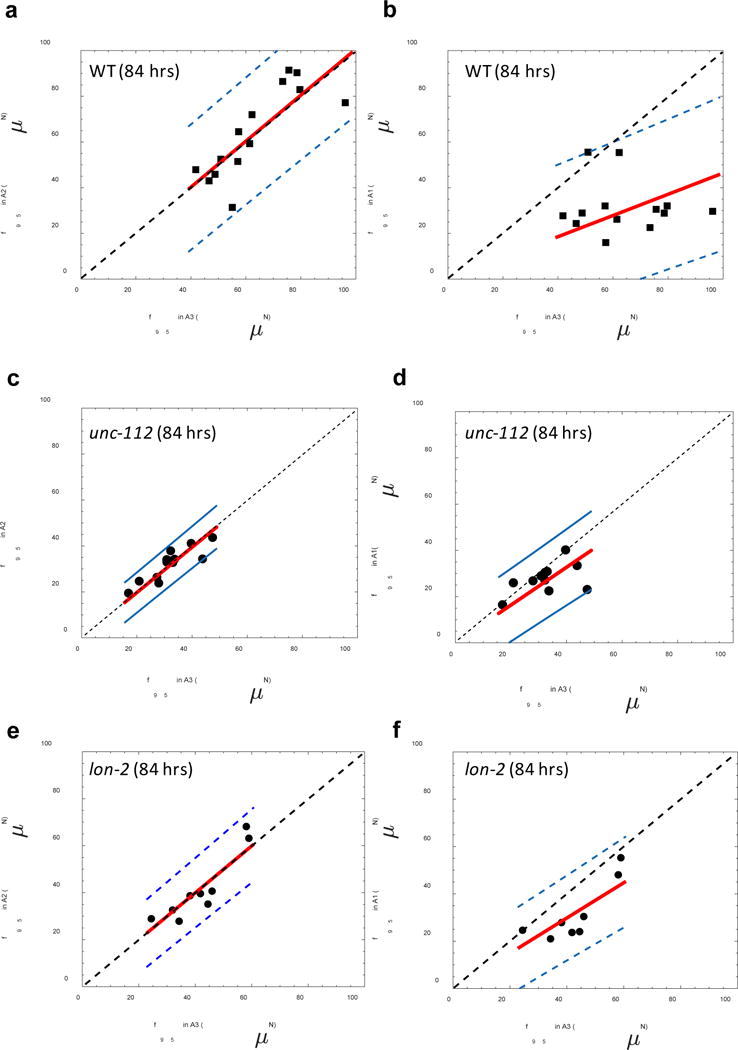
A comparison between maximum exertable force f95 measured for WT individuals in (a) section A2 and section A3 (n = 14, slope = 1.01 ± 0.05, r2 = 0.65) and (b) in section A1 and section A3 (n=13, slope = 0.46 ± 0.07, r2 = -0.55). A similar comparison is shown for unc-112 animals (n=13 individuals) in (c) A2 and A3 (n = 12, slope 0.99 ± 0.04, r2 = 0.67) and (d) A1 and A3 (n= 10, slope = 0.81 ± 0.07, r2 = -0.25). Comparison for lon-2 animals (n=10 individuals) in (e) A2 and A3 (n=9, slope=0.98± 0.05, r2 = 0.72) and (f) A1 and A3 (n= 8, slope = 0.75 ± 0.07, r2 = 0.67). The red line is the best-fit curve to the data, and the dashed black line has a slope of unity and passes through origin. The blue lines demarcate the 95% confidence interval region.
We also tested unc-112, a mutant that is weaker due to muscle defects26 and lon-2, a mutant that grows approximately 1.5 times longer than wild type. For unc-112, we find that the correlation of MEF values in arenas A1 and A3 is off from unity (slope = 0.81 ± 0.07), but in A2 and A3 we again observe a near-perfect correlation with slope 0.99 ± 0.04 (Fig. 8c, d). We find similar results for lon-2 which shows a correlation with slope 0.98 ± 0.05 in arenas A2 and A3 (Fig. 8e, f). Thus, in WT, unc-112 and lon-2 we obtain consistent f95 in the highly resistive A2 and A3 arenas. Previously Johari et al.23 and Khare et al.24 computed the time-average of the pillar forces favg to characterize C. elegans muscle force. We checked to see how well this measure correlates from the force data of individuals in A1, A2 and A3 arenas. We find that favg from different arenas are inconsistent in WT animals and unc-112 mutant (see Supplementary Note 4), suggesting that f95 is a much more reliable metric of strength.
Thus, our results from the composite arena provide compelling evidence that both the highly resistive arenas A2 and A3 maximize force exertion, and our strength metric f95 is invariant for a given individual as long as animals are confined to 0.85 < D/s < 1.16. We settled on using A3 arena for the NemaFlex device since maximal forces are more frequent in this arena than A2 arena (Fig. 5d,e) and smaller body-sized animals fit better in A3. The actual dimensions of the NemaFlex device are: a = 38.3 μm ± 0.4 μm, s = 61.7 μm ± 2.9 μm, h =71.8 ± 2.9 μm, with the pillars arranged in a square lattice within a ≈ 1 cm2 oval shaped arena (Fig. 1b).
It is possible that the limits of validity of confinement that we have established here to obtain consistent MEF readout may vary, for example for C. elegans of different sizes or mutants. In such cases, the composite-arena approach with individuals that we have demonstrated here is a useful means to identify the optimal confinement range for maximum force exertion.
MEF is independent of locomotory gait
To address whether the locomotory gait impacts MEF in the NemaFlex device we isolated episodes from our videos where the animal (WT, age = 60 hrs) was making a forward crawl or omega turn or reversal (Fig. 9a-d). We note that the forward crawl involved either a long-wave or a short-wave body posture with the short-wave being less frequent (Fig. 9a,b). The cumulative probability force distributions corresponding to each of these locomotory modes is shown in Fig. 9e. We find that the omega turns and reversals produce larger average force; however, the MEF estimates are very similar: f95 (long wave) = 20.98 μN, f95 (short wave) = 21.04 μN, f95 (reversal) = 22.38 μN, and f95 (omega) = 21.32 μN. Thus, the strength measure MEF provided by NemaFlex is not strongly dependent on animal gait.
Figure 9. NemaFlex quantitates maximum exertable force independent of C. elegans gait.
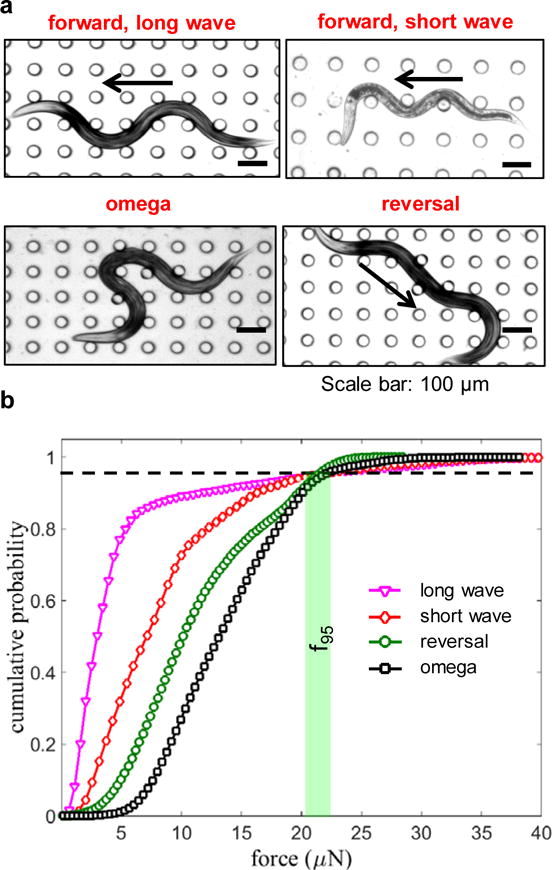
(a) Images showing the different gaits exhibited by crawling WT C. elegans in the pillar arena of the NemaFlex device. The arrows show direction of the animal motion. (b) The cumulative force distribution for the different gaits shown in (a). The horizontal dashed line indicates 95% probability, and the vertical bar highlights that the f95 values for each gait are very similar. Animal age = 60 hrs and D/s = 0.87 – 0.98.
MEF is insensitive to time-variation of behavior
Next, we evaluated whether MEF is sensitive to time variation of behavior by recording behavioral episodes of individuals. In a 30-second behavioral episode we typically observed forward crawls, omega turns, and reversals (c.f. Fig. 6). We analyzed contiguous images of 30-second duration with a random starting point in the video. In complementary sampling, we scored non-repeating, discrete images randomly (totaling 30-second duration) from the full behavioral episode. The f95 of individuals calculated either from contiguous or random sampling of the behavior is shown in Fig. 10a,b. We find that in both contiguous and discrete sampling f95 typically varied by less than 10% of the mean value. This less than 10% variation is commensurate with the error in force measurement shown in Fig. 3 indicating therefore that MEF is insensitive to short-time variation in behavior.
Figure 10. Maximum exertable force is independent of C. elegans behavior in the NemaFlex pillar arena.
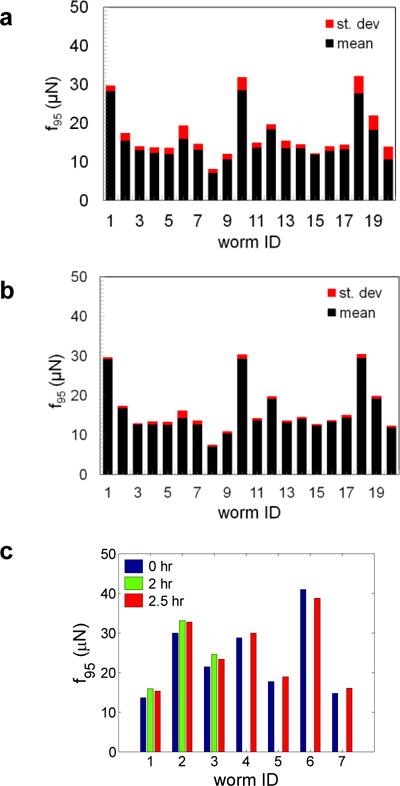
(a) MEF of WT individuals obtained from analyzing contiguous frames of 30-second duration with a randomly sampled starting point in the movie (movie length is 85 – 120 seconds). Data is shown as mean ± SD from N = 5 sampling trials. 17 individuals showed SD < 10%, while three showed SD between 10 - 16%. (b) MEF values obtained from non-repeating randomly sampled discrete frames. The movie sets are the same as in (a). Data is shown as mean ± SD from N = 5 trials. In this case all 20 individuals showed SD < 10%. (c) MEF of individuals evaluated at three time points: 0, 2, and 2.5 hours. Here a 2-minute episode was captured for each worm and a contiguous 30-second episode was analyzed to obtain MEF. Animal age = 60 hrs and D/s = 0.85 – 0.95.
We also sampled the behavior of an individual after a long time interval (0, 2, 2.5 hrs), and in this case the f95 values change by less than 10% between the three time points (Fig. 10c). Thus, the MEF is also unaffected by long-time variation in behavior. Together, these results suggest that sampling a 30-second behavioral episode is sufficient to determine the MEF of C. elegans and does not rely strongly on the details of behavior. We note that for some mutants it is possible that longer behavioral episodes may need to be tracked to achieve a statistically invariant MEF.
MEF before and after drug-induced muscle contractions is the same
As an important test to assess whether MEF measure corresponds to the maximum muscular strength, we induced muscle contractions by exposing each animal to the cholinergic agonist levamisole and comparing its f95 scores before and after drug treatment (Fig. 11a). Levamisole is known to bind to acetylcholine receptors in the body wall muscle of C. elegans, causing prolonged excitation of the muscles, shortening of body length, and eventually paralysis39. As shown in Fig. 11b, we indeed observe appreciable reduction in body length due to levamisole exposure (Supplementary Movie 4a & b).
Figure 11. NemaFlex quantitates maximum muscular strength in C. elegans.
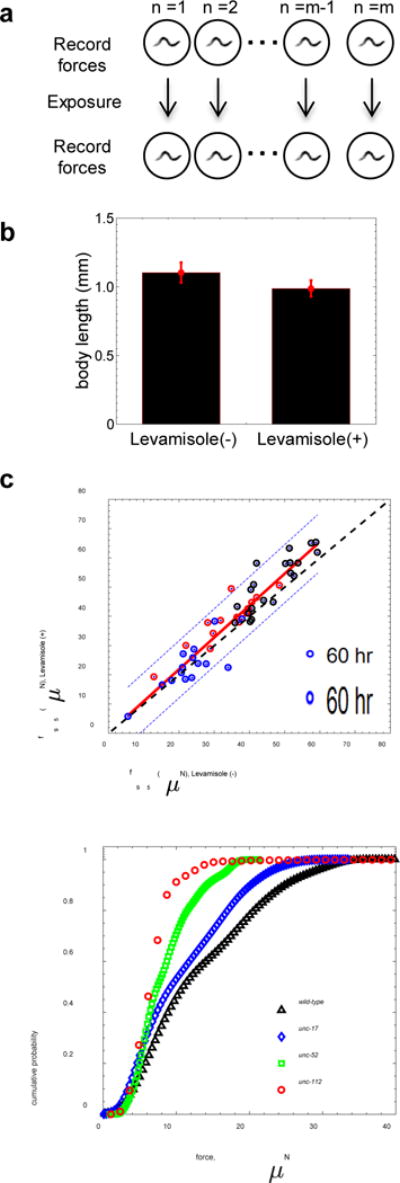
(a) A brief protocol for imaging and inducing muscle contraction on individual wild-type C. elegans with 1 mM levamisole. A 60-second episode is captured for each animal before levamisole treatment, and capturing continues for 60 to 200 seconds after the induction. (b) Levamisole treatment-induced muscle contraction causes the body length to decrease by 10.4 ± 3.2%. (age = 60 – 84 hrs, n=51, p < 0.0005). (c) Maximum strength of individual animals before and after the levamisole treatment for three different age groups – 60 hrs (n = 15), 76 hrs (n = 14) and 84 hrs (n = 20). The red line is a linear best fit of the pooled data: slope =1.09 +/− 0.06, intercept = -0.006, and r2 =0.89. Dashed blue lines show the 95% level confidence interval (n =49). The dashed black line represents f95lev+ = f95lev- (slope of 1 and intercept at origin). A two sample t-test confirms that NemaFlex is measuring the maximum muscular strength of the animal (p =0.24). For this data set, D/s = 0.95 - 1.02. (d) Comparison of population-level force distribution for wild type (n=20, N=3,475 data points) and three C. elegans muscular or neuromuscular mutants unc-52 (n=12, N=14,883 data points), unc-112 (n=5, N=2,992 data points), and unc-17 (n=20, N=6,997 data points). Wilcoxon rank-sum test confirms that NemaFlex is measuring neuromuscular weakness (p > 0.005). For this data set, D/s = 0.85 - 0.92.
In Fig. 11c, we show the force data for individual animals of three different ages (60, 76, and 84 hours) and find that the f95 value after levamisole treatment shows an excellent linear correlation with the f95 score of untreated crawling worms, i.e. f95lev+ f95lev- (slope for 60 hrs = 0.89 ± 0.19, slope for 76 hrs = 0.85 ± 0.13, and slope for 84 hrs = 1.08 ± 0.16), supporting that MEF quantitates the maximum muscular strength of C. elegans. It is remarkable that even though some animals are weak or strong to begin with due to differences in age or individual variability, the f95 after drug exposure correlates well for both the weak and strong animals, highlighting the robustness of our strength measure.
MEF detects muscle strength weakness in mutants
Given that NemaFlex measures the maximum strength of C. elegans, we tested its capability to detect strength changes due to genetic defects in body wall muscles and neuromuscular signaling. We measured strength in the muscle mutants unc-112 and unc-52, which have impaired production of the sarcomere proteins kindlin and perlecan respectively9, 40, 41. We also tested the mutant unc-17, which has decreased levels of acetylcholine, one of the major neurotransmitters responsible for muscle contractions39, 42. In all three mutants, we found that force production decreased compared to wild-type animals (f95 =11.92, 16.61, 22.74 μN, and 28.43 μN for unc-112, unc-52, unc-17, and wild-type respectively; Fig. 11d), confirming that the NemaFlex method can reveal neuromuscular weakness.
Influence of body size on muscular strength in C. elegans
Harnessing the full capability of NemaFlex for applications ranging from dissecting developmental biology to understanding neuromuscular disorders and aging requires evaluation of how C. elegans’ body size affects its muscular strength. For example, individuals from a synchronized culture of WT animals have differences in body size (Supplementary Figure 5), raising the question of whether some animals appear stronger simply because of their larger size. A similar question arises during mutant screenings in which some mutants may be smaller than WT or might develop more slowly than WT and therefore might be smaller at the time of comparison. In aging studies, it is also essential to decouple the strength changes due to frailty from those that might be attributed to animal size, which can change during adult life43.
Muscular strength depends strongly on body diameter but not length
To address the influence of body size, we measured the mid-body diameter, length, and strength of at least 80 age-matched individuals of WT and lon-2 mutants, which are longer than wild type (Fig. 12a-d). We find that even though the mean length of lon-2 mutants is ≈1.3 times that of wild-type animals (Fig. 12a, b), the muscular strength of these two populations is not statistically different (Fig. 12c), suggesting that the worm body length does not significantly influence the muscle strength recorded by NemaFlex. In striking contrast, we observe a strong increase of muscle strength with the worm body diameter (Fig. 12d). The data fit to the scaling relationship f95 ~ Dm, with m = 3.32 ± 0.48 and 3.04 ± 0.30 for WT and lon-2 animals respectively.
Figure 12. Influence of body size on C. elegans muscle strength.
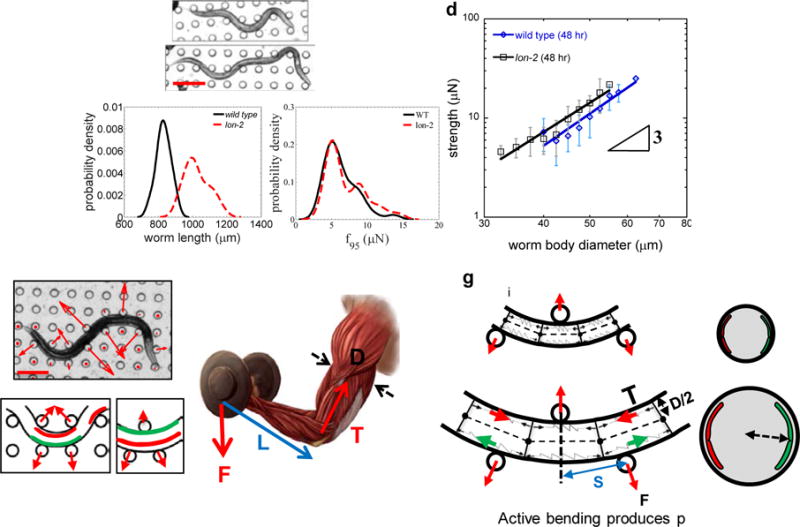
(a) Influence of the body length was evaluated by comparing the force production of wild type (top image) and a lon-2 mutant (bottom image). Scale bar, 200 μm. (b) Distribution of the body lengths of wild type (n = 89) and lon-2 (n = 37) at a single time point (48 hours) for similar diameter worms. The lon-2 worms are ≈ 1.3 times longer than wild type. (c) The strength distributions of wild-type and lon-2 worms are statistically similar. Animal populations are the same as in (b). (d) Both wild type (n = 94) and lon-2 (n = 84) show approximately a cubic dependency of strength on body diameter. MEF data for each population was binned using bin widths of 2.5 μm. Data shown is mean ± SD. (e) Active bending of worm body curvature produces pillar forces. The vector sum of the pillar forces (red arrows) is zero in (i). The nematode pushes pillars when trying to (ii) increase or (iii) decrease its curvature. Increase of the curvature is induced by tension from contracting muscles (red in (ii), green shows relaxing muscle). Similarly, decrease of the curvature is initiated by transferring the tension to the other pair of muscles (red in (iii)) by initiating contraction in the relaxed muscle section. Scale bar, 200 μm. (f) Bending moment analysis in the human muscle arm that is lifting a weight. See main text for description of the symbols. (g) A schematic of the worm body segment under active bending where muscles are shown as (i) springs resembling the contraction and relaxation of muscles. T is the muscle tension in the worm, D is the body diameter, F is the pillar force and s is the pillar spacing. (ii) Animals with larger diameters have more muscle cross-sectional area and therefore produce more force.
To understand the relationship between worm body diameter and muscular strength, we consider active bending of the worm body that produces pillar forces whose vector sum is nearly zero (Fig. 12e, i). These pillar forces are produced by C. elegans due to changes in body curvature induced by contraction or relaxation of its muscles. For the example shown in Fig. 12e, the curvature increase is induced by tension in the dorsal contracting muscle (Fig. 12e, ii), and it is decreased during dorsal relaxation (Fig. 12e, iii). The reason why thick animals exert higher forces is analogous to why thick elastic rods require more force to bend than thin rods. This qualitative argument may explain the influence of body diameter on C. elegans’ maximal muscle strength measures.
A biomechanics model explains D3 scaling
To develop a simple model that incorporates the effect of body diameter, we consider the bending moment (M) induced in the muscles of a human arm that is lifting a weight (F), i.e. M = F×L, where L is the lever arm (Fig. 12f). Applying this analogy to the bending moment introduced in the worm muscles while pushing against pillars, we get M = F×L ~ T×D, where T is the muscle tension in the worm body (Fig. 12g). Assuming the muscle tension is proportional to the cross-sectional area of the muscle (Am) and the lever arm is equivalent to pillar spacing, we have F ~ Am×D/s. Since Am is proportional to the worm body cross-section, i.e. Am ~ D2, we finally have
| (2) |
Eqn. (2) suggests that f95 should scale as the cube of worm body diameter and is inversely proportional to the pillar spacing.
We tested the relation F ~ D3, by considering wild-type animals of different age and three mutants (Table 2). We find that the data agrees reasonably well with the predicted cubic dependence on worm body diameter in all the tested cases. Interestingly, we observe that even in animals (WT, age = 48, 55 hrs) that have confinement less than our optimal range of 0.85 – 1.16, the exponent is close to 3. Furthermore, the fact that the f95 values of individuals were lower in arena A1, which has larger pillar spacing than arenas A2 and A3 is consistent with Eqn. (2). Indeed, we find that the ratios of f95 in A3 and A1 are approximately inversely proportional to the pillar spacing in the respective arena sections, supporting our simple scaling analysis.
Table 2.
Assessment of scaling of strength with body diameter in wild-type and mutant animals.
| Genotype | Age, hr | Pillar diameter a, μm | Pillar spacing s, μm | Confinement, D/s | Exponent, m | Sample size, n | Length of the animal, μm |
|---|---|---|---|---|---|---|---|
| wild type | 48 | 38.3 ± 0.5 | 61.7 ± 2.9 | 0.75 ± 0.05 | 3.12 ± 0.65 | 51 | 870.3 ± 64.8 |
| wild type | 55 | 0.77 ± 0.05 | 3.15 ± 0.64 | 53 | 974.7 ± 54.8 | ||
| wild type | 60 | 0.92 ± 0.07 | 3.32 ± 0.48 | 94 | 1062.8 ± 65.7 | ||
| wild type | 72 | 0.95 ± 0.06 | 3.17 ± 0.34 | 58 | 1188.7 ± 33.6 | ||
| lon-2 | 48 | 0.67 ± 0.09 | 3.04 ± 0.30 | 84 | 970.8 ± 125.5 | ||
| unc-52 | 60 | 0.89 ± 0.05 | 2.79 ± 0.32 | 95 | 915.9 ± 80.8 | ||
| unc-112 | 60 | 0.87 ± 0.07 | 3.30 ± 0.35 | 69 | 807.1 ± 34.6 |
Our data set shows that f95 ~ D3 and it is possible that deviations from the D3 scaling may occur in some mutants or older animals. In such cases deviations from D3 scaling can be potentially corrected by knowing the detailed anatomy of the muscle cross-sectional area. If deviations still exist then it might indicate intrinsic differences in the tension of the muscle fibers in the nematode. Investigation of such differences resulting from genetic defects or age is one of the main motivations for the development of the NemaFlex system.
IV. Conclusions and Outlook
C. elegans is a powerful genetic model with conserved muscle biology, and over the last few decades new insights have been gained into the assembly, maintenance, and regulation of striated muscle that are usually applicable to all animals. Kinematic measures, for example swim frequency or crawling speed, derived from locomotory assays have been typically used to assess neuromuscular function in C. elegans44. More recently, propulsive forces have been determined by tracking velocity fields and body kinematics during swimming45 by challenging worms to swim up inclined surfaces46 and by measuring drag coefficients of crawling animals47.
In a parallel effort, building on advances in microfluidics48, important attempts for measuring forces generated by moving C. elegans have been made using piezoresistance sensors20 and deformable pillars22–26. However, these prior efforts reported forces that vary depending on pillar arena geometry, worm velocity, and body posture;thus, none of these yields a maximum force that is independent of animal behavior and accounts for variations in animal size. Without a behavior- and size-independent strength measurement, it is difficult to standardize muscle force investigations in C. elegans.
Here we have optimized the micropillar forest design, defined the measurement conditions, and fine-tuned the workflow analysis so that NemaFlex can reliably score the maximum exertable force in C. elegans. To confirm that NemaFlex indeed reports behavior-independent strength, we evaluated different pillar spacings and duration of behavioral episodes and showed that animals push as hard as they can once they reach a certain level of confinement; a behavioral episode as low as 30 seconds is sufficient to capture a statistically invariant MEF for young adults. The force measures before and after levamisole-induced full muscle contractions were found to be similar, which supports our metric of maximum force. Indeed, the levamisole experiment shows that C. elegans MEF is similar to human MVF - a critical advance for the ability to compare experimental data across muscle studies.
Recognizing that animals from the same culture might have different body sizes or that mutations may cause variation in body size, we investigated the influence of body size on MEF. We showed that MEF is independent of the body length, but it strongly depends on body diameter. Importantly, we uncovered a relationship of strength with C. elegans diameter that is consistent with a scaling analysis in which bending moments due to maximum external force and internal muscle tension balance each other. Thus, we have produced a compensation factor for body size that enables animals of different age, genetic makeup, or experience to be fairly compared.
In humans, relative strength is a compelling indicator of muscle homeostasis, physical training, and disease. As we model these conditions in invertebrates, there is a critical need to establish standardized measures of strength that have relevance in human muscle physiology. The NemaFlex technology addresses this gap by providing a robust analog of MVF in vertebrates. With current throughput of strength evaluation being ~10 min/worm (loading, imaging, image analysis, and validation), the NemaFlex system is well equipped for targeted investigations on disease conditions that can be modeled in C. elegans. High throughput genetic and drug screens are also possible with NemaFlex, although this would require further microfluidic automation and parallelized analysis. At a more fundamental level, NemaFlex may help in defining molecular and cellular circuits of neuromuscular function49.
Supplementary Material
Acknowledgments
This work was partially supported by funding from NASA (Grant No. NNX15AL16G), NIH (Grant Nos. R21 AG050503, R01 AG051995-01A1), NSF CAREER (Grant No.1150836), CPRIT (Grant No. RP160806), and BBSRC (grant number N015894). We are grateful to Deepak Solomon for initial assistance in this project, Rebecca Gabrilska and Kendra Rumbaugh for assistance with bacterial cultures, Anna Hewitt for graphic rendition of figures, Ricardo Laranjeiro for useful discussions on exercise training of C. elegans and critical reading of the manuscript, and Alex Trindade for help with data analysis and statistics. We thank the Caenorhabditis Genetics Center (CGC) for providing some of the strains, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). We also thank members of the Driscoll and Blawzdziewicz lab for useful discussions.
Footnotes
Author contributions
MR designed and fabricated the devices, FB and MR wrote the image analysis code, MR and JEH performed the experiments and analyzed data, and HE maintained worm cultures and assisted with experiments. SAV, MD, NJS, JB and MR designed the experiments and interpreted the data. SAV, MD and MR wrote the paper. SAV supervised the study. All authors read and commented on the manuscript.
Conflicts of interest
There are no conflicts to declare.
References
- 1.Biewener A. Journal of Experimental Biology. 2016;219:285–294. doi: 10.1242/jeb.123935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frontera W, Ochala J. Calcified Tissue International. 2015;96:183–195. doi: 10.1007/s00223-014-9915-y. [DOI] [PubMed] [Google Scholar]
- 3.Ansved T. Acta Physiologica Scandinavica. 2001;171:359–366. doi: 10.1046/j.1365-201x.2001.00839.x. [DOI] [PubMed] [Google Scholar]
- 4.Lerario A, Bonfiglio S, Sormani M, Tettamanti A, Marktel S, Napolitano S, Previtali S, Scarlato M, Natali-Sora M, Mercuri E, Bresolin N, Mongini T, Comi G, Gatti R, Ciceri F, Cossu G, Torrente Y. Bmc Neurology. 2012;12 doi: 10.1186/1471-2377-12-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clark B, Manini T. Journals of Gerontology Series a-Biological Sciences and Medical Sciences. 2008;63:829–834. doi: 10.1093/gerona/63.8.829. [DOI] [PubMed] [Google Scholar]
- 6.Cheng S, Yang Y, Cheng F, Chen I, Wang R. International Journal of Gerontology. 2014;8:197–202. [Google Scholar]
- 7.Adams G, Caiozzo V, Baldwin K. Journal of Applied Physiology. 2003;95:2185–2201. doi: 10.1152/japplphysiol.00346.2003. [DOI] [PubMed] [Google Scholar]
- 8.Piazza N, Gosangi B, Devilla S, Arking R, Wessells R. Plos One. 2009;4 doi: 10.1371/journal.pone.0005886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gieseler K, Qadota H, Benian GM. Development, structure, and maintenance of C elegans body wall muscle (August 23 2016) WormBook, editor. The C elegans Research Community, WormBook. doi: 10.1895/wormbook.1.81.2. http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- 10.Moerman D, Fire A. 16 Muscle: Structure, Function, and Development. Cold Spring Harbor Monograph Archive; North America: 33, jan 1997 Available at: https://cshmonographs.org/index.php/monographs/article/view/5051%2C. [DOI] [PubMed] [Google Scholar]
- 11.Herndon L, Schmeissner P, Dudaronek J, Brown P, Listner K, Sakano Y, Paupard M, Hall D, Driscoll M. Nature. 2002;419:808–814. doi: 10.1038/nature01135. [DOI] [PubMed] [Google Scholar]
- 12.Chamberlain J, Benian G. Current Biology. 2000;10:R795–R797. doi: 10.1016/s0960-9822(00)00768-5. [DOI] [PubMed] [Google Scholar]
- 13.Higashibata A, Hashizume T, Nemoto K, Higashitani N, Etheridge T, Mori C, Harada S, Sugimoto T, Szewczyk NJ, Baba SA, Mogami Y, Fukui K, Higashitani A. 2016;2:15022. doi: 10.1038/npjmgrav.2015.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zengel J, Epstein H. Cell Motility and the Cytoskeleton. 1980;1:73–97. doi: 10.1002/cm.970010107. [DOI] [PubMed] [Google Scholar]
- 15.Benian G, Kiff J, Neckelmann N, Moermann D, Waterston R. Nature. 1989;342:45–50. doi: 10.1038/342045a0. [DOI] [PubMed] [Google Scholar]
- 16.Mercer K, Miller R, Tinley T, Sheth S, Qadota H, Benian G. Molecular Biology of the Cell. 2006;17:3832–3847. doi: 10.1091/mbc.E06-02-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hwang H, Barnes D, Matsunaga Y, Benian G, Ono S, Lu H. Scientific Reports. 2016;6 doi: 10.1038/srep19900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nahabedian J, Qadota H, Stirman J, Lu H, Benian G. Methods. 2012;56:95–102. doi: 10.1016/j.ymeth.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsunaga Y, Qadota H, Furukawa M, Choe H, Benian G. Molecular Biology of the Cell. 2015;26:2096–2111. doi: 10.1091/mbc.E14-05-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doll J, Harjee N, Klejwa N, Kwon R, Coulthard S, Petzold B, Goodman M, Pruitt B. Lab on a Chip. 2009;9:1449–1454. doi: 10.1039/b818622g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu P, Mao D, Martin R, Dong L. Lab on a Chip. 2012;12:3458–3466. doi: 10.1039/c2lc40459a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghanbari A, Nock V, Johari S, Blaikie R, Chen X, Wang W. Journal of Micromechanics and Microengineering. 2012;22:095009. [Google Scholar]
- 23.Johari S, Nock V, Alkaisi M, Wang W. Lab on a Chip. 2013;13:1699–1707. doi: 10.1039/c3lc41403e. [DOI] [PubMed] [Google Scholar]
- 24.Khare S, Awasthi A, Venkataraman V, Koushika S. Biomicrofluidics. 2015;9:014111. doi: 10.1063/1.4906905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiu Z, Tu L, Huang L, Zhu T, Nock V, Yu E, Liu X, Wang W. Biomicrofluidics. 2015;9 doi: 10.1063/1.4908595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Etheridge T, Rahman M, Gaffney C, Shaw D, Shephard F, Magudia J, Solomon D, Milne T, Blawzdziewicz J, Constantin-Teodosiu D, Greenhaff P, Vanapalli S, Szewczyk N. Faseb Journal. 2015;29:1235–1246. doi: 10.1096/fj.14-259119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edwards R, Young A, Hosking G, Jones D. Clinical Science and Molecular Medicine. 1977;52:283–290. doi: 10.1042/cs0520283. [DOI] [PubMed] [Google Scholar]
- 28.Sahaly R, Vandewalle H, Driss T, Monod H. European Journal of Applied Physiology. 2001;85:345–350. doi: 10.1007/s004210100451. [DOI] [PubMed] [Google Scholar]
- 29.Madsen O. European Journal of Applied Physiology and Occupational Physiology. 1996;74:206–210. doi: 10.1007/BF00377442. [DOI] [PubMed] [Google Scholar]
- 30.Richens B, Cleather D. Biology of Sport. 2014;31:157–161. doi: 10.5604/20831862.1099047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brenner S. Genetics. 1973;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McDonald JC, Duffy DC, Anderson JR, Chiu DT, Wu H, Schueller OJA, Whitesides GM. Electrophoresis. 2000;21:27–40. doi: 10.1002/(SICI)1522-2683(20000101)21:1<27::AID-ELPS27>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 33.Xian B, Shen J, Chen W, Sun N, Qiao N, Jiang D, Yu T, Men Y, Han Z, Pang Y, Kaeberlein M, Huang Y, Han J. Aging Cell. 2013;12:398–409. doi: 10.1111/acel.12063. [DOI] [PubMed] [Google Scholar]
- 34.Timoshenko S, Gere JM. Mechanics of Materials. Van Nostrand Reinhold Co.; New York, New York: 1972. [Google Scholar]
- 35.Lockery S, Lawton K, Doll J, Faumont S, Coulthard S, Thiele T, Chronis N, McCormick K, Goodman M, Pruitt B. Journal of Neurophysiology. 2008;99:3136–3143. doi: 10.1152/jn.91327.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park S, Hwang H, Nam S, Martinez F, Austin R, Ryu W. Plos One. 2008;3 doi: 10.1371/journal.pone.0002550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Albrecht D, Bargmann C. Nature Methods. 2011;8:599–605. doi: 10.1038/nmeth.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Majmudar T, Keaveny E, Zhang J, Shelley M. Journal of the Royal Society Interface. 2012;9:1809–1823. doi: 10.1098/rsif.2011.0856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rand JB. Acetylcholine. WormBook, editor. The C elegans Research Community, WormBook. 2007 Jan 30; http://www.wormbook.org. 10.1895/wormbook.
- 40.Rogalski TM, Williams BD, Mullen GP, Moerman DG. Genes & Development. 1993;7:1471–1484. doi: 10.1101/gad.7.8.1471. [DOI] [PubMed] [Google Scholar]
- 41.Rogalski T, Mullen G, Gilbert M, Williams B, Moerman D. Journal of Cell Biology. 2000;150:253–264. doi: 10.1083/jcb.150.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alfonso A, Grundahl K, Duerr J, Han H, Rand J. Science. 1993;261:617–619. doi: 10.1126/science.8342028. [DOI] [PubMed] [Google Scholar]
- 43.Zhang WB, Sinha DB, Pittman WE, Hvatum E, Stroustrup N, Pincus Z. Cell Systems. 2016;3:333–345. doi: 10.1016/j.cels.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buckingham SD, Sattelle DB. Invertebrate Neuroscience. 2008;8:121–131. doi: 10.1007/s10158-008-0077-3. [DOI] [PubMed] [Google Scholar]
- 45.Sznitman J, Shen X, Sznitman R, Arratia P. Physics of Fluids. 2010;22 [Google Scholar]
- 46.Yuan J, Ko H, Raizen D, Bau H. Journal of the Royal Society Interface. 2016;13 doi: 10.1098/rsif.2016.0612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rabets Y, Backholm M, Dalnoki-Veress K, Ryu W. Biophysical Journal. 2014;107:1980–1987. doi: 10.1016/j.bpj.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.San-Miguel A, Lu H. Microfluidics as a tool for C elegans research. WormBook, editor. The C elegans Research Community, WormBook. 2013 Sep 24; doi: 10.1895/wormbook.1.162.1. http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- 49.Butler V, Branicky R, Yemini E, Liewald J, Gottschalk A, Kerr R, Chklovskii D, Schafer W. Journal of the Royal Society Interface. 2015;12 doi: 10.1098/rsif.2014.0963. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


