Abstract
Melatonin, the primary indoleamine hormone of the mammalian pineal gland, is known to have a plethora of neuroregulatory, neuroprotective and other properties. Melatonergic signalling is mediated by its two GPCRs, MT1 and MT2, which are widely expressed in the mammalian CNS. Melatonin levels and receptor expression often show a decrease during normal ageing, and this reduction may be accelerated in some disease states. Depleted melatonergic signalling has been associated with neuropsychiatric dysfunction and impairments in cognition, memory, neurogenesis and neurorestorative processes. The anticonvulsant and mood stabilizer, valproic acid (VPA), up‐regulates melatonin MT1 and/or MT2 receptor expression in cultured cells and in the rat brain. VPA is known to affect gene expression through several mechanisms, including the modulation of intracellular kinase pathways and transcription factors, as well as the inhibition of histone deacetylase (HDAC) activity. Interestingly, other HDAC inhibitors, such as trichostatin A, which are structurally distinct from VPA, can also up‐regulate melatonin receptor expression, unlike a VPA analogue, valpromide, which lacks HDAC inhibitory activity. Moreover, VPA increases histone H3 acetylation along the length of the MT1 gene promoter in rat C6 cells. These findings indicate that an epigenetic mechanism, linked to histone hyperacetylation/chromatin remodelling and associated changes in gene transcription, is involved in the up‐regulation of melatonin receptors by VPA. Epigenetic induction of MT1 and/or MT2 receptor expression, in areas where these receptors are lost because of ageing, injury or disease, may be a promising therapeutic avenue for the management of CNS dysfunction and other disorders.
Linked Articles
This article is part of a themed section on Recent Developments in Research of Melatonin and its Potential Therapeutic Applications. To view the other articles in this section visit http://onlinelibrary.wiley.com/doi/10.1111/bph.v175.16/issuetoc
Abbreviations
- ALS
amyotrophic lateral sclerosis
- BDNF
brain‐derived neurotrophic factor
- CpG
5′‐C‐phosphate‐G‐3′
- CREB
cAMP response element binding protein
- DNMT
DNA methyltransferase
- GDNF
glial cell line‐derived neurotrophic factor
- GSK3β
glycogen synthase kinase 3β
- HAT
histone acetyltransferase
- HDAC
histone deacetylase
- HP1
heterochromatin protein 1
- Nrf2‐ARE
NF‐E2‐related factor 2‐antioxidant responsive element
- SCN
suprachiasmatic nucleus
- SMC
structural maintenance of chromosomes
- VPA
valproic acid
Introduction
Melatonin receptors
The mammalian http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=224 receptor subtypes, http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=287 and http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=288, are localized on neuronal dendrites and somata in areas that include the cerebral cortex, basal forebrain, hippocampus, basal ganglia, diencephalon and mesencephalon (Lacoste et al., 2015). The anatomical distribution patterns of MT1 and MT2 receptors in the adult rat brain, which are detailed in recent reports (Lacoste et al., 2015; Ng et al., 2017), occur in a complementary fashion, in which relative increases in MT1 receptor expression correspond with relative decreases in MT2 receptors, or vice versa. For example, in the hypothalamus, the expression of MT1 receptors is relatively high in the suprachiasmatic nucleus (SCN), whereas that of MT2 receptors is relatively high in the supraoptic nucleus. This property may be indicative of their unique functional specializations (Lacoste et al., 2015).
Functional studies have revealed that both melatonin receptor subtypes are coupled to various pertussis‐sensitive and insensitive G proteins. A cornerstone of melatonergic signalling involves the inhibition of adenylate cyclase activity and attenuation of cAMP accumulation, PKA activity and cAMP response element binding protein (CREB) phosphorylation (Brydon et al., 1999); however, the involvement of several other signal transduction pathways has been reported (Dubocovich et al., 2010). Studies of melatonin receptor regulation in the ovine pars tuberalis demonstrated that stimulation of cAMP production, by forskolin or cholera toxin, increases MT1 receptor expression (Barrett et al., 1996). Since cAMP levels regulate the transcription of many genes through the cAMP/PKA/CREB pathway, the transcription factor CREB has been implicated in the regulation of melatonin receptors (Barrett et al., 1996). Cloning of the proximal 1.5 kb region of the rat MT1 promoter (GenBank AY228510) allowed for a more precise analysis of the regulation of MT1 receptor transcription. MT1 promoter activity assays have identified paired‐like homeodomain transcription factor 1, a transcription factor widely distributed throughout the rodent and ovine pituitary, as an important regulator of MT1 receptor transcription (Johnston et al., 2003).
Protective effects of melatonin
Antioxidant effects
The CNS is highly vulnerable to damage from oxidative stress because of its large composition of lipids, high demands for oxidative metabolism and, relatedly, production of toxic metabolites (Wang and Michaelis, 2010). Melatonin exerts many protective effects in the mammalian CNS. The NF‐E2‐related factor 2‐antioxidant responsive element (Nrf2‐ARE) signalling cascade is one of the intracellular pathways that mediates the regulation of antioxidant gene expression by melatonin. Melatonin induces Nrf2 expression, as well as the downstream targets of Nrf2‐ARE signalling in multiple models of peripheral (Tripathi and Jena, 2010) and central oxidative stress (Wang et al., 2012). Recent studies in Klotho mutant mice show that the Nrf2‐related antioxidant actions of melatonin involve MT2 receptor‐mediated activation of the ERK pathway (Shin et al., 2015).
In addition to regulating intracellular antioxidant pathways, melatonin, as well as its metabolites, has direct free radical scavenging capabilities. As such, a single melatonin molecule can forage and neutralize multiple highly toxic reactive oxygen, nitrogen and hydroxyl radicals at any one time (Rosen et al., 2006). Although free radical scavenging and avoidance are widely beneficial properties, they are especially protective of mitochondrial integrity because of the increased susceptibility of this organelle to oxidative damage. Melatonin readily enters the mitochondrial matrix, where it limits electron leakage from the electron transport chain and free radical production (Reiter et al., 2003). While the direct free radical scavenging effects of melatonin do not involve its receptors, other antioxidant effects, such as maintaining glutathione levels and stimulating the activities of catalase and superoxide dismutase, are mediated by melatonin receptors (Rezzani et al., 2006).
Neurotrophic factor modulation
Melatonin is thought to promote neuronal development, differentiation and survival in the nervous system by modulating neurotrophic factor expression. Melatonin, as well as its analogues, has been shown to up‐regulate neurotrophic factors such as glial cell line‐derived neurotrophic factor (GDNF; Armstrong and Niles, 2002; Kong et al., 2008b), nerve growth factor (Pongsa‐Asawapaiboon et al., 1998) and brain‐derived neurotrophic factor (BDNF; Molteni et al., 2010; Boulle et al., 2014). Reduced expression of these targets has been implicated in the pathogenic mechanisms driving several brain disorders. Therefore, controlling the expression of these and other neurotrophic factors is thought to have therapeutic potential for both neurodegenerative (Allen et al., 2013) and psychiatric disorders (Castrén, 2014).
There is limited information about the mechanisms involved in the modulation of neurotrophic factors by melatonin. The melatonin‐induced increase in GDNF mRNA expression in C6 cells, which express both MT1 and MT2 receptors (Armstrong and Niles, 2002), may be mediated by the MT1 subtype, as this effect is not blocked by the MT2 antagonist, http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=1358) (Niles and Armstrong, 2002). The increase in GDNF protein levels induced by melatonin in primary astrocytes is blocked by the PI3K/Akt antagonist, wortmannin and the non‐selective melatonin receptor antagonist, http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=1363, indicating the involvement of this pathway and melatonin receptors in this effect (Kong et al., 2008a). Recently, melatonin treatment in the presence of the pro‐inflammatory cytokine, IL‐18, was found to increase the mRNA and protein expression of GDNF and BDNF in neural stem cells, whereas luzindole blocked these effects, indicating that this effect is mediated by stimulation of melatonin receptors (Li et al., 2017). As discussed later, epigenetic regulation of gene transcription, including chromatin remodelling via histone modifications, has been linked to synaptic plasticity, neurotrophic factor induction, neuroprotection and related effects (Schmidt et al., 2013; Harrison et al., 2015). Evidence that melatonin can acetylate histone proteins in cultured cells and in vivo (Sharma et al., 2008; Niles et al., 2013) suggests that epigenetic induction of gene expression may underlie its effects on neurotrophic factors and other neuroprotective targets. Moreover, blockade of melatonin‐induced histone acetylation by luzindole, in human SH‐SY5Y neuroblastoma cells, indicates the involvement of melatonin receptors in these effects (Pan and Niles, 2015).
Melatonergic signalling in psychiatric and neurodegenerative disease
Several in vitro and in vivo studies have revealed aberrations in the expression patterns of MT1 and/or MT2 receptors in various models of CNS disorders. Post‐mortem analyses of human brain tissue from related disorders have also revealed changes in melatonin receptor expression patterns, but these findings are complicated by the fact that the affected patients may have received prolonged treatment with therapeutic drugs, which may induce changes in receptor expression independent of the disease (Hirsch‐Rodriguez et al., 2007).
Aberrant melatonergic function has been implicated in various neuropsychiatric conditions. The potential therapeutic use of melatonin in neuropsychiatric disorders has been reviewed recently with a note on the need for selective targeting of receptor subtypes (Mahmood et al., 2016), which exhibit distinct functional specialization. For example, a study of sleep regulation, using receptor knockout mice, revealed that MT1 and MT2 receptor signalling regulate rapid eye movement sleep and non‐rapid eye movement sleep respectively (Comai et al., 2013). The sleep‐promoting role of MT2 receptors is further supported by the observation that MT2 receptor ligands produce a hypnotic effect, which is eliminated in mice lacking MT2 receptors (Ochoa‐Sanchez et al., 2011). MT2 ligands have also been observed to have an antinociceptive effect in rodents, which can be blocked by MT2 receptor antagonists, implying a role for this receptor in pain regulation (Lopez‐Canul et al., 2015). Increased MT1 receptor expression, but not MT2, has been observed in the hypothalamic tissue of depressed patients post‐mortem (Wu et al., 2013). Mice lacking the expression of MT1 receptors exhibit neurobiological variations characteristic of depression, including hyperstress responses, psychomotor disturbances and increased anhedonic and depressive‐like behaviours in comparison to their wild‐type counterparts (Comai et al., 2015), suggesting a role for the MT1 subtype in the aetiology of depression.
Abnormal melatonin receptor expression profiles have also been noted in various neurodegenerative conditions. For example, rodent models revealed that MT1 receptor expression was reduced in the spinal cords of transgenic amyotrophic lateral sclerosis (ALS) mice (Zhang et al., 2013). Analysis of post‐mortem brain tissue revealed increased MT1 (Savaskan et al., 2002) and decreased MT2 receptor expression (Savaskan et al., 2005) in the hippocampus of Alzheimer's disease patients. The reason for the different expression of these receptor subtypes in Alzheimer's disease awaits clarification but could involve discrete differences in their localization or other neurodegenerative changes associated with this disorder. Other studies have reported regional decreases in melatonin receptor expression in Alzheimer's disease, including reduced hypothalamic, pineal and cortical MT1 expression (Brunner et al., 2006; Wu et al., 2007), as well as pineal, cortical and retinal MT2 expression (Brunner et al., 2006; Savaskan et al., 2007), relative to healthy controls. Similarly, post‐mortem analysis of brains from patients with Parkinson's disease revealed decreases in both MT1 and MT2 receptor expression in the substantia nigra as well as the amygdala in comparison to healthy controls (Adi et al., 2010). Consequently, impairments in melatonergic signalling related to aberrant MT1 and/or MT2 expression profiles could contribute to the overall deterioration of the nervous system in these disorders. Evidence that the neuroprotective effect of melatonin in a transgenic mouse model of ALS involves inhibition of receptor interacting protein‐2‐induced activation of the caspase‐1 pathway via the MT1 receptor (Zhang et al., 2013) supports the importance of melatonin receptors in brain preservation. Drug‐induced up‐regulation of melatonin receptors, as proposed in this review, is a potential strategy for restoring depleted melatonin receptor expression in areas where it is lost as a result of normal ageing, injury or disease.
Valproic acid and melatonin receptors
http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=7009 (VPA) or 2‐propylpentanoic acid is a branched short‐chain fatty acid with well‐known anticonvulsive and mood‐stabilizing effects. A recent study has shown that VPA may be an effective adjunct to antidepressant therapies in treatment‐resistant depression (Ghabrash et al., 2016). In addition to the multiple targets and pathways influenced by VPA (Monti et al., 2009), increasing evidence indicates an interaction between this psychotropic agent and the melatonergic system. Treatment with VPA enhances the expression of tryptophan hydroxylase, which contributes to melatonin biosynthesis (Qiu et al., 2015) and reduces the sensitivity of melatonin to light (Hallam et al., 2005). Initial studies on the effects of VPA on melatonin receptor expression were completed in rat C6 glioma cells (Castro et al., 2005; Kim et al., 2008) and in human MCF‐7 breast cancer cells (Jawed et al., 2007). This work revealed a dose‐dependent relationship in which VPA caused a robust increase in MT1 receptor mRNA and/or protein (Castro et al., 2005; Jawed et al., 2007; Kim et al., 2008). These findings set the groundwork for in vivo studies, which showed that chronic VPA administration causes a robust increase in MT1 and MT2 receptor mRNA levels in the rat hippocampus (Niles et al., 2012). Moreover, an in situ hybridization study revealed significant increases in MT2 receptor expression in the CA1‐3 and dentate gyrus regions of the rat hippocampus, following a similar treatment with VPA (Bahna et al., 2014). As discussed below, this up‐regulation of melatonin receptors, especially the MT1 subtype, is thought to involve an epigenetic mechanism.
Epigenetic mechanisms
The term epigenetics refers to heritable changes in gene expression, which do not involve an alteration in DNA sequence. Three major mechanisms including DNA methylation, regulation of transcription by non‐coding RNAs and histone modification are involved in the epigeneticregulation of gene expression. Dysfunction of any of these interacting mechanisms can result in the abnormal expression or silencing of genes and the possible onset of ‘epigenetic diseases’ (Egger et al., 2004; Hwang et al., 2017)).
DNA methylation
The methylation of 5‐cytosine residues to 5‐methylcytosine, in cytosine‐guanine (CpG) dinucleotides, is an established epigenetic mechanism for gene silencing. The methylation of CpG sites within the mammalian genome is maintained by a number of DNA methyltransferases (DNMTs). DNMT3A and DNMT3B are involved in the de novo methylation of DNA, while DNMT1 performs a maintenance function by methylating the complementary strand in hemimethylated DNA before its replication (Bayraktar and Kreutz, 2017). It is now apparent that DNA demethylation can occur via deaminases, which catalyse the conversion of 5‐methylcytosine to thymidine. A second mechanism involves ten‐eleven translocation enzymes that reverse the methylation status of DNA by the successive oxidation of 5‐methylcytosine to 5‐carboxylcytosine, which is subjected to a base‐excison‐repair process to regenerate cytosine (Nabel and Kohli, 2011).
Regulation of transcription by non‐coding RNAs
Another category of epigenetic regulators includes non‐coding RNAs such as microRNAs (miRNAs), small interfering RNAs and long non‐coding RNAs. To date, the most studied are miRNAs, which are thought to target more than 60% of human genes (Friedman et al., 2009) and to regulate gene transcription primarily by suppression of target translation or induction of mRNA decay (Huntzinger and Izaurralde, 2011). Their ability to regulate gene expression in pathways linked to major physiological processes, including neurodevelopment, neurogenesis and neuroprotection, has implicated miRNAs in various central pathological states such as neurodegeneration and psychiatric dysfunction (Kim et al., 2016; Boone et al., 2017).
Histone modification
Chromatin consists of nucleosomes, each composed of about 146 base pairs of genomic DNA, which is wrapped around an octamer of core histone proteins consisting of two copies each of H2A, H2B, H3 and H4. Histone proteins have long N‐terminal tails that are key targets in multiple post‐translational modifications including acetylation, methylation, phosphorylation, sumoylation, ubiquitination and ADP‐ribosylation. Acetylation and methylation are the most widely studied of these covalent modifications, which cause conformational changes in chromatin structure and influence the access of transcription factors and other regulatory proteins to their DNA targets (Bannister and Kouzarides, 2011). Enzymes that alter the electrostatic interactions between the negatively charged DNA and the positively charged histone core, via histone acetylation or deacetylation, are involved in the epigenetic regulation of gene transcription, as illustrated in Figure 1. Histone acetylation, which is catalysed by histone acetyltransferase (http://www.guidetopharmacology.org/GRAC/FamilyDisplayForward?familyId=872) enzymes from various families (Roth et al., 2001), neutralizes the positive charge on histone proteins, causing a reduced affinity between the histone protein and the DNA strand. The associated chromatin assumes a more loosened structure, known as euchromatin, which exposes regulatory genetic sequences and permits the binding of transcription factors to activate transcription (Morse, 2007). The reverse process, deacetylation, is catalysed by histone deacetylase (http://www.guidetopharmacology.org/GRAC/FamilyDisplayForward?familyId=848) enzymes, which cause chromatin condensation leading to the termination of transcriptional activity (Bannister and Kouzarides, 2011).
Figure 1.
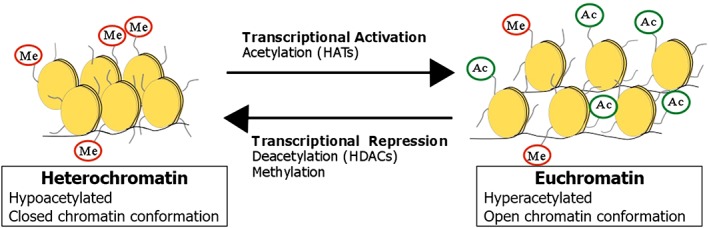
Regulation of chromatin conformation by histone acetylation and methylation. Histone acetylation, methylation or other covalent modifications of lysine residues on the N‐terminal tail of the histone core can alter gene transcription via changes in the accessibility of DNA to regulatory factors.
Based on gene sequence, subcellular localization or functional differences, 11 zinc‐dependent HDACs have been assigned to different classes as follows: Class I (HDAC 1, 2, 3, 8), Class IIa (HDAC 4, 5, 7, 9), Class IIb (HDAC 6, 10) and Class IV (HDAC 11). Class III HDACs are a distinct family of nicotinamide adenine dinucleotide‐dependent enzymes, termed sirtuins. Several HDAC inhibitors have been examined in studies aimed at identifying clinically relevant epigenetic regulators. Of particular relevance to this review, VPA inhibits the activity of multiple Class I and Class II HDACs (Gurvich et al., 2004), which are involved in critical aspects of CNS physiology, including neurodevelopment and cognition (Morris and Monteggia, 2013).
Epigenetic regulation of melatonin receptors by VPA
VPA can directly inhibit HDAC activity, which results in the conversion of chromatin to an acetylated and transcriptionally active conformation (Phiel et al., 2001). Several factors are involved in the maintenance of chromatin structure in the normal state, such as the structural maintenance of chromosomes (SMC) family of proteins, which are required for the general stabilization of chromosome conformation (Yokomori, 2003), and heterochromatin protein 1 (HP1), which maintains DNA in the heterochromatin state (Maison and Almouzni, 2004). VPA treatments stimulate a conformational change in chromatin structure, from a highly condensed state to a more loosened structure, which corresponds with a decreased expression of the proteins which promote heterochromatin stability (Marchion et al., 2005; Felisbino et al., 2014).
As noted earlier, VPA treatment up‐regulates melatonin receptors in cultured mammalian cells and the rat brain (Castro et al., 2005; Niles et al., 2012), but the mechanism(s) involved awaits clarification. Recent studies indicate that an epigenetic mechanism, specifically that of MT1 promoter histone acetylation due to HDAC inhibition, underlies the transcriptional activation of the MT1 receptor gene by VPA (Bahna and Niles, 2017). This is supported by the observation that structurally diverse drugs, such as the potent HDAC inhibitor, trichostatin A, also increase the expression of MT1 receptors (Kim et al., 2008), whereas valpromide, a VPA analogue without HDAC inhibitory properties, does not (Bahna and Niles, 2017).
VPA and other HDAC inhibitors can alter the demethylation of DNA, either globally or gene‐specfically, by enhancing DNA demethylase activity via histone acetylation or other mechanisms (Detich et al., 2003). This is important as several CpG islands are located within the promoter regions of many genes, which, when methylated, suppress transcription (Cedar and Bergman, 2009). As noted earlier, diverse epigenetic mechanisms interact in the regulation, amplification and fine‐tuning of gene expression (Egger et al., 2004). For example, the inhibition of HDAC activity and the attenuation of DNA methylation induced by VPA are not mutually exclusive events but rather are thought to be in a dynamic correlation and can impose compounded effects on gene transcription (Milutinovic et al., 2007). In view of the foregoing, it is possible that DNA demethylation contributes to the regulation of melatonin receptors by VPA.
Chromatin decondensation surrounding the MT1 promoter region allows transcription factors to access regulatory sequences along the MT1 promoter to initiate transcription of the gene. The ability of VPA to prevent heterochromatin formation following histone acetylation (Marchion et al., 2005), suggests that it can influence the transcriptional kinetics of MT1 receptors. It is interesting to note that histone acetylation is thought to precede the modulation of HP1 and SMC protein expression by VPA (Marchion et al., 2005), suggesting that the conformational changes in chromatin structure induced by VPA can cause prolonged induction of the melatonin receptor (Figure 2).
Figure 2.
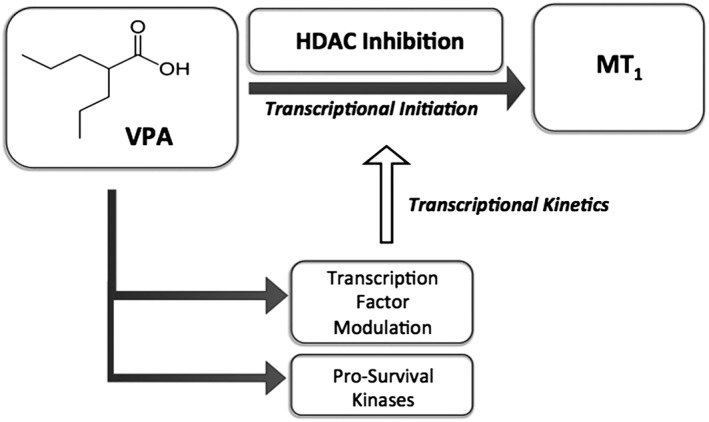
Proposed mechanism for the induction of MT1 receptors by VPA. VPA activates the transcription of MT1 receptors by causing chromatin remodelling, mediated by HDAC inhibition and MT1 promoter hyperacetylation. VPA also acts via intracellular kinases to regulate the expression of transcription factors that control the activity of the MT1 promoter, suggesting a role for this drug in the transcriptional kinetics of MT1 receptor expression.
Molecular regulation of melatonin receptors by VPA
VPA influences the expression and binding affinities of various transcription factors in cultured cells, as well as in the brain (Monti et al., 2009). These effects may involve its influence on the post‐translational modifications of various intracellular kinases, which regulate the activation and subsequent gene expression of transcription factors. VPA is a positive regulator of the AMP‐activated protein kinase (Avery and Bumpus, 2014), MAPK, PI3K and Akt (PKB) and PKC signalling pathways, and a negative regulator of glycogen synthase kinase 3β (GSK3β) signalling (Monti et al., 2009). We have reported that pharmacological inhibition of MAPK (Castro et al., 2005), PI3K/Akt, PKC or GSK3β does not affect melatonin MT1 receptor up‐regulation (Bahna and Niles, 2017), further supporting our epigenetic hypothesis.
Although the above findings suggest that these kinase pathways are not involved in the up‐regulation of MT1 receptors by VPA, caution should be exercised when interpreting the effects of kinase inhibitors, which exhibit limited selectivity (Bain et al., 2007). Moreover, given the multiple cellular and molecular effects of VPA, its administration could affect not just melatonin receptors but also other targets or pathways, which may in turn influence melatonergic function. One such target is tetrahydrobiopterin (BH4), an essential cofactor in the biosynthesis of neurotransmitters including 5‐HT and catecholamines, and the enzyme, nitric oxide synthase, which produces nitric oxide (Werner et al., 2011), a modulator of glutamate release (Neitz et al., 2011). Given the functional interaction between melatonin and neurotransmitters such as 5‐HT (Matheus et al., 2010), it is possible that melatonergic activity following VPA treatment could also be modulated by other pathways or in vivo systems activated by this pleiotropic drug.
Recently, VPA was reported to inhibit internalization of the MT1 receptor and its cAMP signalling by interfering with the association between the receptor and β‐arrestin‐2 (Hong et al., 2016). If this effect, which was observed in an in vitro expression system, translates to the in vivo mammalian CNS, it is possible that it could negatively affect the anticipated benefits of melatonin receptor up‐regulation. Nonetheless, the novel idea of using epigenetic drugs to restore deficient melatonin receptor expression is worth exploring in various in vivo models of neurological or psychiatric disorders, where potential benefits would depend on the net effect of VPA or other HDAC inhibitors on melatonin receptor expression and function. Given the neuroprotective, anti‐manic and other effects of VPA treatment, the up‐regulation of melatonin receptors by this drug in conjunction with the administration of agonists (e.g. melatonin, http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=198 and http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=1356), which are similarly beneficial, could produce enhanced therapeutic outcomes.
Clinical relevance of epigenetic up‐regulation of melatonin receptor expression
The protective effects of increasing melatonin receptor expression have been demonstrated using several in vitro models of glutamate toxicity. Melatonin treatments in cultured embryonic rat VSC4.1 motor neurons transfected with MT1 and/or MT2 receptors were found to have a large increase in cell survival markers and a concurrent suppression in apoptotic and inflammatory markers (Das et al., 2013). Melatonin treatments in untransfected cells did not have the same effect, emphasizing the importance of receptor density in the neuroprotective effects of this hormone (Das et al., 2013). The beneficial effects of melatonin receptor overexpression have also been explored in the periphery. In human MCF‐7 breast cancer cells, melatonin was found to have a greater antiproliferative effect in MT1‐transfected cells than in untransfected cells (Yuan et al., 2002).
A limitation of these studies is how melatonin receptor overexpression is achieved. Gene therapy is a challenging approach and may not be a clinically suitable treatment option for a large number of patients (Gonin et al., 2005). There is increasing evidence that dysregulated epigenetic processes underlie several neurological dysfunctions, including Parkinson's and Alzheimer's disease (Urdinguio et al., 2009; Masliah et al., 2013) and especially psychiatric disorders such as depression and schizophrenia (Houtepen et al., 2016; Pishva et al., 2017). Diverse psychopharmacological agents including antidepressants, antipsychotics and mood stabilizers can influence epigenomic profiles, as reflected by histone protein modifications and DNA methylation (Boks et al., 2012). While the mechanisms driving melatonin receptor depletion are not yet known, accruing evidence proposes a role for dysregulated epigenetic processes in the pathogenesis of many neurological and neurodegenerative diseases (Saha and Pahan, 2006; Coppedè, 2014). This implies that the reduction in melatonin receptor expression in the diseased state may be a maladaptive response caused by an imbalance in epigenetic activities associated with disease progression. Given the central role of GPCRs in mediating the effects of diverse therapeutics, evidence that GPCR expression can be regulated by epigenetic mechanisms suggests that this approach of receptor regulation may be beneficial for a wide range of CNS disorders (Dogra et al., 2016). As illustrated in Figure 3, epigenetic reprogramming of gene expression by VPA (or other HDAC inhibitors) to regain transcriptional control of the melatonin receptor gene can be a therapeutic strategy used to offset imbalances in receptor expression and, relatedly, melatonergic signalling. Some examples of neuropsychiatric disorders, which may benefit from the epigenetic manipulation of melatonin receptor expression, are discussed below.
Figure 3.
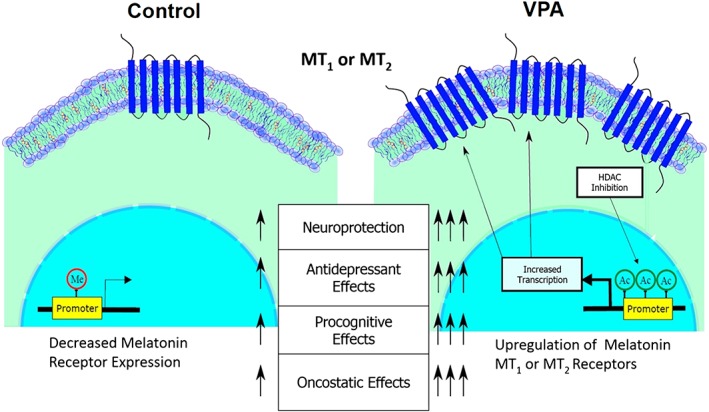
An overview of the potential therapeutic benefits of epigenetic up‐regulation of melatonin receptors. While VPA is shown in this illustration, a similar epigentic up‐regulation of melatonin receptors can be induced by other HDAC inhibitors.
Psychiatric disorders
It is well known that disruptions in the circadian rhythms of biological processes such as sleep and the secretion of hormones including melatonin are linked to mood disorders (Germain and Kupfer, 2008; Srinivasan et al., 2009). Melatonin has shown efficacy in alleviating insomnia and sleep abnormalities in depressed patients (Ferracioli‐Oda et al., 2013). These beneficial effects are thought to involve the modulation of circadian function via melatonin receptors in the SCN, where the MT1 subtype can inhibit neuronal activity while the MT2 receptor plays a role in phase‐shifting rhythms (Liu et al., 1997). Adjunctive administration of slow‐release melatonin decreased insomnia in patients with treatment‐resistant depression, but it did not augment the effect of antidepressant treatment (Dalton et al., 2000). Other studies suggest that melatonin may be beneficial in reversing symptoms associated with anxiety and depression (Hansen et al., 2014), among other psychiatric conditions (Mahmood et al., 2016). The melatonin analogue, agomelatine, which couples agonistic activity at MT1 and MT2 receptors with antagonistic action at the http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=8 receptor, is the first melatonergic agent approved for psychiatric treatment. Agomelatine has shown promise in the management of depression and other psychiatric conditions (Bourin and Prica, 2009; Spadoni et al., 2011; Laudon and Frydman‐Marom, 2014). Another melatonin MT1/MT2 agonist, ramelteon, improves sleep quality and duration across age groups including older insomniacs (Pandi‐Perumal et al., 2007; Schroeck et al., 2016). These findings suggest that the melatonergic system is a worthwhile target for future psychiatric treatment strategies (Catena‐Dell'Osso et al., 2012).
Increasing evidence indicates that epigenetic dysfunction may contribute to psychiatric conditions, such as major depressive disorder and schizophrenia (Dogra et al., 2016; Hoffmann et al., 2017). As noted earlier, melatonin itself can induce histone acetylation in cultured brain‐derived cells or in the mammalian brain by a mechanism that appears to involve MT1 receptors and HAT (Pan and Niles, 2015). This raises the interesting possibility that combination therapy with VPA (or other HDAC inhibitors) and melatonin agonists could normalize epigenetic function with associated stabilization of brain physiology and related therapeutic effects.
Ageing and neurodegenerative disease
It is well‐known that melatonin acts via its receptors to confer an assortment of neuroprotective and neurorestorative effects in the aged and neurodegenerative brain. Melatonin receptors are depleted during physiological ageing (Sánchez‐Hidalgo et al., 2009), as well as in various neurodegenerative states, as mentioned previously. This reduction may contribute to the overall deterioration of the CNS in neurodegenerative disorders. Melatonin treatments have been associated with delayed disease onset and progression, as well as decreased mortality. For example, melatonin attenuates dopaminergic cell loss in the nigrostriatal pathway in the Parkinson's disease brain of animal models and restores the associated motor function deficits (Capitelli et al., 2008; Ma et al., 2009; Carriere et al., 2015). Other studies show that melatonin administration reverses impairments in cognition by augmenting adult hippocampal neurogenesis (Chern et al., 2012; Liu et al., 2013). Additional support for this notion comes from the observation that chronic melatonin treatment causes enhanced dendritic maturation and arborization in the mouse hippocampus (Ramirez‐Rodriguez et al., 2011). This is important because increasing adult hippocampal neurogenesis has been found to be capable of improving cognition (Sahay et al., 2011).
In keeping with the above, low melatonin levels and disrupted circadian rhythmicity have been found in patients with Alzheimer's disease (Wu et al., 2003). Melatonin has been reported to protect against memory impairment, synaptic dysfunction and neurodegeneration, by activating the PI3K/Akt/GSK3β pathway, in a mouse model of Alzheimer's disease (Ali and Kim, 2015). The protective effects of melatonin on the aged and neurodegenerative brain further include the modulation of energy metabolism, circadian function (Jenwitheesuk et al., 2014) and suppression of the accumulation of toxic‐free radicals and other metabolic by‐products in the CNS (Reiter et al., 2003). The ability to optimize melatonergic function by restoring depleted melatonin receptor populations in the CNS holds clear therapeutic promise in ageing and neurodegeneration.
Conclusions
GPCRs act as molecular sensors that convert extracellular stimuli into intracellular responses. The expression of melatonin receptors, as well as other GPCRs, is often depleted in the aged and/or diseased state. Although the mechanisms driving these receptor abnormalities are not yet characterized, increasing evidence suggests that epigenetic processes influence the development of several CNS diseases, which may also involve aberrant GPCR expression. VPA, as well as other HDAC inhibitors, up‐regulates the expression of melatonin receptors via a mechanism that involves histone acetylation/chromatin remodelling with associated gene transcription. The epigenetic control of melatonin receptor expression provides a novel therapeutic strategy for offsetting the negative trajectory of melatonergic impairment associated with ageing and/or disease. However, the successful utilization of this strategy will require clarification of important issues, including which of the multiple HDAC isoforms inhibited by VPA or other HDAC inhibitors are specifically linked to the regulation of the MT1 or MT2 receptors, in order to permit selective targeting of melatonin receptor subtypes in future therapeutic approaches.
Nomenclature of targets and ligands
Key protein targets and ligands in this article are hyperlinked to corresponding entries in http://www.guidetopharmacology.org, the common portal for data from the IUPHAR/BPS Guide to PHARMACOLOGY (Southan et al., 2016), and are permanently archived in the Concise Guide to PHARMACOLOGY 2015/16 (Alexander et al., 2015a,b).
Conflict of interest
The authors declare no conflicts of interest.
Acknowledgements
This review is based on work supported by the Natural Sciences and Engineering Research Council of Canada. We thank Daniel Cadete for assistance with illustrations.
Bahna, S. G. , and Niles, L. P. (2018) Epigenetic regulation of melatonin receptors in neuropsychiatric disorders. British Journal of Pharmacology, 175: 3209–3219. 10.1111/bph.14058.
References
- Adi N, Mash DC, Ali Y, Singer C, Shehadeh L, Papapetropoulos S (2010). Melatonin MT1 and MT2 receptor expression in Parkinson's disease. Med Sci Monit 16: BR61–BR67. [PubMed] [Google Scholar]
- Alexander SPH, Davenport AP, Kelly E, Marrion N, Peters JA, Benson HE et al (2015a). The Concise Guide to PHARMACOLOGY 2015/16: G protein‐coupled receptors. Br J Pharmacol 172: 5744–5869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander SPH, Fabbro D, Kelly E, Marrion N, Peters JA, Benson HE et al (2015b). The Concise Guide to PHARMACOLOGY 2015/16: Enzymes. Br J Pharmacol 172: 6024–6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali T, Kim MO (2015). Melatonin ameliorates amyloid beta‐induced memory deficits, tau hyperphosphorylation and neurodegeneration via PI3/Akt/GSk3β pathway in the mouse hippocampus. J Pineal Res 59: 47–59. [DOI] [PubMed] [Google Scholar]
- Allen SJ, Watson JJ, Shoemark DK, Barua NU, Patel NK (2013). GDNF, NGF and BDNF as therapeutic options for neurodegeneration. Pharmacol Ther 138: 155–175. [DOI] [PubMed] [Google Scholar]
- Armstrong KJ, Niles LP (2002). Induction of GDNF mRNA expression by melatonin in rat C6 glioma cells. Neuroreport 13: 473–475. [DOI] [PubMed] [Google Scholar]
- Avery LB, Bumpus NN (2014). Valproic acid is a novel activator of AMP‐activated protein kinase and decreases liver mass, hepatic fat accumulation, and serum glucose in obese mice. Mol Pharmacol 85: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahna SG, Niles LP (2017). Epigenetic induction of melatonin MT 1 receptors by valproate: neurotherapeutic implications. Eur Neuropsychopharmacol 27: 828–832. [DOI] [PubMed] [Google Scholar]
- Bahna SG, Sathiyapalan A, Foster JA, Niles LP (2014). Regional upregulation of hippocampal melatonin MT receptors by valproic acid: therapeutic implications for Alzheimer's disease. Neurosci Lett 576C: 84–87. [DOI] [PubMed] [Google Scholar]
- Bain J, Plater L, Elliott M, Shpiro N, Hastie CJ, McLauchlan H et al (2007). The selectivity of protein kinase inhibitors: a further update. Biochem J 408: 297–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T (2011). Regulation of chromatin by histone modifications. Cell Res 21: 381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett P, MacLean A, Davidson G, Morgan PJ (1996). Regulation of the Mel 1a melatonin receptor mRNA and protein levels in the ovine pars tuberalis: evidence for a cyclic adenosine 3′,5′‐monophosphate‐independent Mel 1a receptor coupling and an autoregulatory mechanism of expression. Mol Endocrinol 10: 892–902. [DOI] [PubMed] [Google Scholar]
- Bayraktar G, Kreutz MR (2017). Neuronal DNA methyltransferases: epigenetic mediators between synaptic activity and gene expression? Neuroscientist. 10.1177/1073858417707457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boks MP, de Jong NM, Kas MJH, Vinkers CH, Fernandes C, Kahn RS et al (2012). Current status and future prospects for epigenetic psychopharmacology. Epigenetics 7: 20–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone DK, Weisz HA, Bi M, Falduto MT, Torres KEO, Willey HE et al (2017). Evidence linking microRNA suppression of essential prosurvival genes with hippocampal cell death after traumatic brain injury. Sci Rep 7: 6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulle F, Massart R, Stragier E, Païzanis E, Zaidan L, Marday S et al (2014). Hippocampal and behavioral dysfunctions in a mouse model of environmental stress: normalization by agomelatine. Transl Psychiatry 4: e485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourin M, Prica C (2009). Melatonin receptor agonist agomelatine: a new drug for treating unipolar depression. Curr Pharm Des 15: 1675–1682. [DOI] [PubMed] [Google Scholar]
- Brunner P, Sözer‐Topcular N, Jockers R, Ravid R, Angeloni D, Fraschini F et al (2006). Pineal and cortical melatonin receptors MT1 and MT2 are decreased in Alzheimer's disease. Eur J Histochem 50: 311–316. [PubMed] [Google Scholar]
- Brydon L, Roka F, Petit L, de Coppet P, Tissot M, Barrett P et al (1999). Dual signaling of human mel1a melatonin receptors via G i2, G i3, and G q/11 proteins. Mol Endocrinol 13: 2025–2038. [DOI] [PubMed] [Google Scholar]
- Capitelli C, Sereniki A, Lima MM, Reksidler AB, Tufik S, Vital MA (2008). Melatonin attenuates tyrosine hydroxylase loss and hypolocomotion in MPTP‐lesioned rats. Eur J Pharmacol 594: 101–108. [DOI] [PubMed] [Google Scholar]
- Carriere C, Kang N, Niles L (2015). Chronic low‐dose melatonin treatment maintains nigrostriatal integrity in an intrastriatal rotenone model of Parkinson's disease. Brain Res 1633: 115–125. [DOI] [PubMed] [Google Scholar]
- Castrén E (2014). Neurotrophins and psychiatric disorders In: Lewin G., Carter B. (eds) Neurotrophic Factors. Handbook of Experimental Pharmacology, vol 220 Springer: Berlin, Heidelberg, pp. 461–479. [DOI] [PubMed] [Google Scholar]
- Castro LMR, Gallant M, Niles LP (2005). Novel targets for valproic acid: up‐regulation of melatonin receptors and neurotrophic factors in C6 glioma cells. J Neurochem 95: 1227–1236. [DOI] [PubMed] [Google Scholar]
- Catena‐Dell'Osso M, Marazziti D, Rotella F, Bellantuono C (2012). Emerging targets for the pharmacological treatment of depression: focus on melatonergic system. Curr Med Chem 19: 428–437. [DOI] [PubMed] [Google Scholar]
- Cedar H, Bergman Y (2009). Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet 10: 295–304. [DOI] [PubMed] [Google Scholar]
- Chern C‐M, Liao J‐F, Wang Y‐H, Shen Y‐C (2012). Melatonin ameliorates neural function by promoting endogenous neurogenesis through the MT2 melatonin receptor in ischemic‐stroke mice. Free Radic Biol Med 52: 1634–1647. [DOI] [PubMed] [Google Scholar]
- Comai S, Ochoa‐Sanchez R, Gobbi G (2013). Sleep‐wake characterization of double MT₁/MT₂ receptor knockout mice and comparison with MT₁ and MT₂ receptor knockout mice. Behav Brain Res 243: 231–238. [DOI] [PubMed] [Google Scholar]
- Comai S, Ochoa‐Sanchez R, Dominguez‐Lopez S, Bambico FR, Gobbi G (2015). Melancholic‐Like behaviors and circadian neurobiological abnormalities in melatonin MT1 receptor knockout mice. Int J Neuropsychopharmacol 18: pyu075–pyu075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppedè F (2014). The potential of epigenetic therapies in neurodegenerative diseases. Front Genet 5: 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton EJ, Rotondi D, Levitan RD, Kennedy SH, Brown GM (2000). Use of slow‐release melatonin in treatment‐resistant depression. J Psychiatry Neurosci 25: 48–52. [PMC free article] [PubMed] [Google Scholar]
- Das A, Wallace G, Reiter RJ, Varma AK, Ray SK, Banik NL (2013). Overexpression of melatonin membrane receptors increases calcium‐binding proteins and protects VSC4.1 motoneurons from glutamate toxicity through multiple mechanisms. J Pineal Res 54: 58–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detich N, Bovenzi V, Szyf M (2003). Valproate induces replication‐independent active DNA demethylation. J Biol Chem 278: 27586–27592. [DOI] [PubMed] [Google Scholar]
- Dogra S, Sona C, Kumar A, Yadav PN (2016). Epigenetic regulation of G protein coupled receptor signaling and its implications in psychiatric disorders. Int J Biochem Cell Biol 77: 226–239. [DOI] [PubMed] [Google Scholar]
- Dubocovich ML, Delagrange P, Krause DN, Sugden D, Cardinali DP, Olcese J (2010). International Union of Basic and Clinical Pharmacology. LXXV. Nomenclature, classification, and pharmacology of G protein‐coupled melatonin receptors. Pharmacol Rev 62: 343–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger G, Liang G, Aparicio A, Jones PA (2004). Epigenetics in human disease and prospects for epigenetic therapy. Nature 429: 457–463. [DOI] [PubMed] [Google Scholar]
- Felisbino MB, Gatti MSV, Mello MLS (2014). Changes in chromatin structure in NIH 3T3 cells induced by valproic acid and trichostatin A. J Cell Biochem 115: 1937–1947. [DOI] [PubMed] [Google Scholar]
- Ferracioli‐Oda E, Qawasmi A, Bloch MH (2013). Meta‐analysis: melatonin for the treatment of primary sleep disorders. PLoS One 8: e63773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK‐H, Burge CB, Bartel DP (2009). Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19: 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain A, Kupfer DJ (2008). Circadian rhythm disturbances in depression. Hum Psychopharmacol 23: 571–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghabrash MF, Comai S, Tabaka J, Saint‐Laurent M, Booij L, Gobbi G (2016). Valproate augmentation in a subgroup of patients with treatment‐resistant unipolar depression. World J Biol Psychiatry 17: 165–170. [DOI] [PubMed] [Google Scholar]
- Gonin P, Buchholz CJ, Pallardy M, Mezzina M (2005). Gene therapy bio‐safety: scientific and regulatory issues. Gene Ther 12 (Suppl 1): S146–S152. [DOI] [PubMed] [Google Scholar]
- Gurvich N, Tsygankova OM, Meinkoth JL, Klein PS (2004). Histone deacetylase is a target of valproic acid‐mediated cellular differentiation. Cancer Res 64: 1079–1086. [DOI] [PubMed] [Google Scholar]
- Hallam KT, Olver JS, Norman TR (2005). Effect of sodium valproate on nocturnal melatonin sensitivity to light in healthy volunteers. Neuropsychopharmacology 30: 1400. [DOI] [PubMed] [Google Scholar]
- Hansen MV, Andersen LT, Madsen MT, Hageman I, Rasmussen LS, Bokmand S et al (2014). Effect of melatonin on depressive symptoms and anxiety in patients undergoing breast cancer surgery: a randomized, double‐blind, placebo‐controlled trial. Breast Cancer Res Treat 145: 683–695. [DOI] [PubMed] [Google Scholar]
- Harrison IF, Crum WR, Vernon AC, Dexter DT (2015). Neurorestoration induced by the HDAC inhibitor sodium valproate in the lactacystin model of Parkinson's is associated with histone acetylation and up‐regulation of neurotrophic factors. Br J Pharmacol 172: 4200–4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch‐Rodriguez E, Imbesi M, Manev R, Uz T, Manev H (2007). The pattern of melatonin receptor expression in the brain may influence antidepressant treatment. Med Hypotheses 69: 120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A, Sportelli V, Ziller M, Spengler D (2017). Epigenomics of major depressive disorders and schizophrenia: early life decides. Int J Mol Sci 18: E1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong LJ, Jiang Q, Long S, Wang H, Zhang LD, Tian Y et al (2016). Valproic acid influences mtnr1a intracellular trafficking and signaling in a beta‐arrestin 2‐dependent manner. Mol Neurobiol 53: 1237–1246. [DOI] [PubMed] [Google Scholar]
- Houtepen LC, van Bergen AH, Vinkers CH, Boks MP (2016). DNA methylation signatures of mood stabilizers and antipsychotics in bipolar disorder. Epigenomics 8: 208–216. [DOI] [PubMed] [Google Scholar]
- Huntzinger E, Izaurralde E (2011). Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet 12: 99–110. [DOI] [PubMed] [Google Scholar]
- Hwang J‐Y, Aromolaran KA, Zukin RS (2017). The emerging field of epigenetics in neurodegeneration and neuroprotection. Nat Rev Neurosci 18: 347–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jawed S, Kim B, Ottenhof T, Brown GM, Werstiuk ES, Niles LP (2007). Human melatonin MT1 receptor induction by valproic acid and its effects in combination with melatonin on MCF‐7 breast cancer cell proliferation. Eur J Pharmacol 560: 17–22. [DOI] [PubMed] [Google Scholar]
- Jenwitheesuk A, Nopparat C, Mukda S, Wongchitrat P, Govitrapong P (2014). Melatonin regulates aging and neurodegeneration through energy metabolism, epigenetics, autophagy and circadian rhythm pathways. Int J Mol Sci 15: 16848–16884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston JD, Messager S, Ebling FJ, Williams LM, Barrett P, Hazlerigg DG (2003). Gonadotrophin‐releasing hormone drives melatonin receptor down‐regulation in the developing pituitary gland. Proc Natl Acad Sci U S A 100: 2831–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B, Rincon Castro LM, Jawed S, Niles LP (2008). Clinically relevant concentrations of valproic acid modulate melatonin MT(1) receptor, HDAC and MeCP2 mRNA expression in C6 glioma cells. Eur J Pharmacol 589: 45–48. [DOI] [PubMed] [Google Scholar]
- Kim Y, Zhang Y, Pang K, Kang H, Park H, Lee Y et al (2016). Bipolar disorder associated microRNA, miR‐1908‐5p, regulates the expression of genes functioning in neuronal glutamatergic synapses. Exp Neurobiol 25: 296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong P‐J, Byun J‐S, Lim S‐Y, Lee J‐J, Hong S‐J, Kwon K‐J et al (2008a). Melatonin Induces Akt phosphorylation through melatonin receptor‐ and PI3K‐dependent pathways in primary astrocytes. Korean J Physiol Pharmacol 12: 37–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X, Li X, Cai Z, Yang N, Liu Y, Shu J et al (2008b). Melatonin regulates the viability and differentiation of rat midbrain neural stem cells. Cell Mol Neurobiol 28: 569–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacoste B, Angeloni D, Dominguez‐Lopez S, Calderoni S, Mauro A, Fraschini F et al (2015). Anatomical and cellular localization of melatonin MT and MT receptors in the adult rat brain. J Pineal Res 58: 397–417. [DOI] [PubMed] [Google Scholar]
- Laudon M, Frydman‐Marom A (2014). Therapeutic effects of melatonin receptor agonists on sleep and comorbid disorders. Int J Mol Sci 15: 15924–15950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Li X, Chan MTV, Wu WKK, Tan D, Shen J (2017). Melatonin antagonizes interleukin‐18‐mediated inhibition on neural stem cell proliferation and differentiation. J Cell Mol Med. 10.1111/jcmm.13140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Weaver DR, Jin X, Shearman LP, Pieschl RL, Gribkoff VK et al (1997). Molecular dissection of two distinct actions of melatonin on the suprachiasmatic circadian clock. Neuron 19: 91–102. [DOI] [PubMed] [Google Scholar]
- Liu Y, Ni C, Tang Y, Tian X, Zhou Y, Qian M et al (2013). Melatonin attenuates isoflurane‐induced acute memory impairments in aged rats. Basic Clin Pharmacol Toxicol 113: 215–220. [DOI] [PubMed] [Google Scholar]
- Lopez‐Canul M, Palazzo E, Dominguez‐Lopez S, Luongo L, Lacoste B, Comai S et al (2015). Selective melatonin MT2 receptor ligands relieve neuropathic pain through modulation of brainstem descending antinociceptive pathways. Pain 156: 305–317. [DOI] [PubMed] [Google Scholar]
- Ma J, Shaw VE, Mitrofanis J (2009). Does melatonin help save dopaminergic cells in MPTP‐treated mice? Parkinsonism Relat Disord 15: 307–314. [DOI] [PubMed] [Google Scholar]
- Mahmood D, Muhammad BY, Alghani M, Anwar J, el‐ Lebban N, Haider M (2016). Advancing role of melatonin in the treatment of neuropsychiatric disorders. Egypt J Basic Appl Sci 3: 203–218. [Google Scholar]
- Maison C, Almouzni G (2004). HP1 and the dynamics of heterochromatin maintenance. Nat Rev Mol Cell Biol 5: 296–304. [DOI] [PubMed] [Google Scholar]
- Marchion DC, Bicaku E, Daud AI, Sullivan DM, Munster PN (2005). Valproic acid alters chromatin structure by regulation of chromatin modulation proteins. Cancer Res 65: 3815–3822. [DOI] [PubMed] [Google Scholar]
- Masliah E, Dumaop W, Galasko D, Desplats P (2013). Distinctive patterns of DNA methylation associated with Parkinson disease: identification of concordant epigenetic changes in brain and peripheral blood leukocytes. Epigenetics 8: 1030–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheus N, Mendoza C, Iceta R, Mesonero JE, Alcalde AI (2010). Melatonin inhibits serotonin transporter activity in intestinal epithelial cells. J Pineal Res 48: 332–339. [DOI] [PubMed] [Google Scholar]
- Milutinovic S, D'Alessio AC, Detich N, Szyf M (2007). Valproate induces widespread epigenetic reprogramming which involves demethylation of specific genes. Carcinogenesis 28: 560–571. [DOI] [PubMed] [Google Scholar]
- Molteni R, Calabrese F, Pisoni S, Gabriel C, Mocaer E, Racagni G et al (2010). Synergistic mechanisms in the modulation of the neurotrophin BDNF in the rat prefrontal cortex following acute agomelatine administration. World J Biol Psychiatry 11: 148–153. [DOI] [PubMed] [Google Scholar]
- Monti B, Polazzi E, Contestabile A (2009). Biochemical, molecular and epigenetic mechanisms of valproic acid neuroprotection. Curr Mol Pharmacol 2: 95–109. [DOI] [PubMed] [Google Scholar]
- Morris MJ, Monteggia LM (2013). Unique functional roles for class I and class II histone deacetylases in central nervous system development and function. Int J Dev Neurosci 31: 370–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse RH (2007). Transcription factor access to promoter elements. J Cell Biochem 102: 560–570. [DOI] [PubMed] [Google Scholar]
- Nabel CS, Kohli RM (2011). Molecular biology. Demystifying DNA demethylation. Science 333: 1229–1230. [DOI] [PubMed] [Google Scholar]
- Neitz A, Mergia E, Eysel UT, Koesling D, Mittmann T (2011). Presynaptic nitric oxide/cGMP facilitates glutamate release via hyperpolarization‐activated cyclic nucleotide‐gated channels in the hippocampus. Eur J Neurosci 33: 1611–1621. [DOI] [PubMed] [Google Scholar]
- Ng KY, Leong MK, Liang H, Paxinos G (2017). Melatonin receptors: distribution in mammalian brain and their respective putative functions. Brain Struct Funct. 10.1007/s00429-017-1439-6. [DOI] [PubMed] [Google Scholar]
- Niles LP, Armstrong KJ (2002). Modulation of GDNF expression by melatonin. Program No. 691.2. 2002. Abstract Viewer/Itinerary Planner. Washington DC: Society for Neuroscience. Online.
- Niles LP, Sathiyapalan A, Bahna S, Kang NH, Pan Y (2012). Valproic acid up‐regulates melatonin MT1 and MT2 receptors and neurotrophic factors CDNF and MANF in the rat brain. Int J Neuropsychopharmacol 15: 1343–1350. [DOI] [PubMed] [Google Scholar]
- Niles LP, Pan Y, Kang S, Lacoul A (2013). Melatonin induces histone hyperacetylation in the rat brain. Neurosci Lett 541: 49–53. [DOI] [PubMed] [Google Scholar]
- Ochoa‐Sanchez R, Comai S, Lacoste B, Bambico FR, Dominguez‐Lopez S, Spadoni G et al (2011). Promotion of non‐rapid eye movement sleep and activation of reticular thalamic neurons by a novel MT2 melatonin receptor ligand. J Neurosci 31: 18439–18452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Niles LP (2015). Epigenetic mechanisms of melatonin action in human SH‐SY5Y neuroblastoma cells. Mol Cell Endocrinol 402: 57–63. [DOI] [PubMed] [Google Scholar]
- Pandi‐Perumal SR, Srinivasan V, Poeggeler B, Hardeland R, Cardinali DP (2007). Drug Insight: the use of melatonergic agonists for the treatment of insomnia‐focus on ramelteon. Nat Clin Pract 3: 221–228. [DOI] [PubMed] [Google Scholar]
- Phiel CJ, Zhang F, Huang EY, Guenther MG, Lazar MA, Klein PS (2001). Histone deacetylase is a direct target of valproic acid, a potent anticonvulsant, mood stabilizer, and teratogen. J Biol Chem 276: 36734–36741. [DOI] [PubMed] [Google Scholar]
- Pishva E, Rutten BPF, van den Hove D (2017). DNA methylation in major depressive disorder. Adv Exp Med Biol 978: 185–196. [DOI] [PubMed] [Google Scholar]
- Pongsa‐Asawapaiboon A, Asavaritikrai P, Withyachumnarnkul B, Sumridthong A (1998). Melatonin increases nerve growth factor in mouse submandibular gland. J Pineal Res 24: 73–77. [DOI] [PubMed] [Google Scholar]
- Qiu H‐M, Yang J‐X, Jiang X‐H, Hu X‐Y, Liu D, Zhou Q‐X (2015). Enhancing tyrosine hydroxylase and tryptophan hydroxylase expression and improving oxidative stress involved in the antidepressant effect of sodium valproate on rats undergoing chronic unpredicted stress. Neuroreport 26: 1145–1150. [DOI] [PubMed] [Google Scholar]
- Ramirez‐Rodriguez G, Ortiz‐Lopez L, Dominguez‐Alonso A, Benitez‐King GA, Kempermann G (2011). Chronic treatment with melatonin stimulates dendrite maturation and complexity in adult hippocampal neurogenesis of mice. J Pineal Res 50: 29–37. [DOI] [PubMed] [Google Scholar]
- Reiter RJ, Tan D‐X, Mayo JC, Sainz RM, Leon J, Czarnocki Z (2003). Melatonin as an antioxidant: biochemical mechanisms and pathophysiological implications in humans. Acta Biochim Pol 50: 1129–1146. [PubMed] [Google Scholar]
- Rezzani R, Rodella LF, Bonomini F, Tengattini S, Bianchi R, Reiter RJ (2006). Beneficial effects of melatonin in protecting against cyclosporine A‐induced cardiotoxicity are receptor mediated. J Pineal Res 41: 288–295. [DOI] [PubMed] [Google Scholar]
- Rosen J, Than NN, Koch D, Poeggeler B, Laatsch H, Hardeland R (2006). Interactions of melatonin and its metabolites with the ABTS cation radical: extension of the radical scavenger cascade and formation of a novel class of oxidation products, C2‐substituted 3‐indolinones. J Pineal Res 41: 374–381. [DOI] [PubMed] [Google Scholar]
- Roth SY, Denu JM, Allis CD (2001). Histone acetyltransferases. Annu Rev Biochem 70: 81–120. [DOI] [PubMed] [Google Scholar]
- Saha RN, Pahan K (2006). HATs and HDACs in neurodegeneration: a tale of disconcerted acetylation homeostasis. Cell Death Differ 13: 539–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahay A, Scobie KN, Hill AS, O'Carroll CM, Kheirbek MA, Burghardt NS et al (2011). Increasing adult hippocampal neurogenesis is sufficient to improve pattern separation. Nature 472: 466–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez‐Hidalgo M, Guerrero Montávez JM, Carrascosa‐Salmoral MDP, Naranjo Gutierrez MDC, Lardone PJ, de la Lastra Romero CA (2009). Decreased MT1 and MT2 melatonin receptor expression in extrapineal tissues of the rat during physiological aging. J Pineal Res 46: 29–35. [DOI] [PubMed] [Google Scholar]
- Savaskan E, Olivieri G, Meier F, Brydon L, Jockers R, Ravid R et al (2002). Increased melatonin 1a‐receptor immunoreactivity in the hippocampus of Alzheimer's disease patients. J Pineal Res 32: 59–62. [DOI] [PubMed] [Google Scholar]
- Savaskan E, Ayoub MA, Ravid R, Angeloni D, Fraschini F, Meier F et al (2005). Reduced hippocampal MT2 melatonin receptor expression in Alzheimer's disease. J Pineal Res 38: 10–16. [DOI] [PubMed] [Google Scholar]
- Savaskan E, Jockers R, Ayoub M, Angeloni D, Fraschini F, Flammer J et al (2007). The MT2 melatonin receptor subtype is present in human retina and decreases in Alzheimer's disease. Curr Alzheimer Res 4: 47–51. [DOI] [PubMed] [Google Scholar]
- Schmidt HD, McGinty JF, West AE, Sadri‐Vakili G (2013). Epigenetics and psychostimulant addiction. Cold Spring Harb Perspect Med 3: a012047–a012047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeck JL, Ford J, Conway EL, Kurtzhalts KE, Gee ME, Vollmer KA et al (2016). Review of safety and efficacy of sleep medicines in older adults. Clin Ther 38: 2340–2372. [DOI] [PubMed] [Google Scholar]
- Sharma R, Ottenhof T, Rzeczkowska PA, Niles LP (2008). Epigenetic targets for melatonin: induction of histone H3 hyperacetylation and gene expression in C17.2 neural stem cells. J Pineal Res 45: 277–284. [DOI] [PubMed] [Google Scholar]
- Shin E‐J, Chung YH, Le H‐LT, Jeong JH, Dang D‐K, Nam Y et al (2015). Melatonin attenuates memory impairment induced by Klotho gene deficiency via interactive signaling between MT2 receptor, ERK, and Nrf2‐related antioxidant potential. Int J Neuropsychopharmacol. 10.1093/ijnp/pyu105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southan C, Sharman JL, Benson HE, Faccenda E, Pawson AJ, Alexander SPH et al (2016). The IUPHAR/BPS guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. Nucl Acids Res 44: D1054–D1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spadoni G, Bedini A, Rivara S, Mor M (2011). Melatonin receptor agonists: new options for insomnia and depression treatment. CNS Neurosci Ther 17: 733–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan V, Pandi‐Perumal SR, Trakht I, Spence DW, Hardeland R, Poeggeler B et al (2009). Pathophysiology of depression: role of sleep and the melatonergic system. Psychiatry Res 165: 201–214. [DOI] [PubMed] [Google Scholar]
- Tripathi DN, Jena GB (2010). Effect of melatonin on the expression of Nrf2 and NF‐kappaB during cyclophosphamide‐induced urinary bladder injury in rat. J Pineal Res 48: 324–331. [DOI] [PubMed] [Google Scholar]
- Urdinguio RG, Sanchez‐Mut JV, Esteller M (2009). Epigenetic mechanisms in neurological diseases: genes, syndromes, and therapies. Lancet Neurol 8: 1056–1072. [DOI] [PubMed] [Google Scholar]
- Wang X, Michaelis EK (2010). Selective neuronal vulnerability to oxidative stress in the brain. Front Aging Neurosci 2: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Ma C, Meng C‐J, Zhu G‐Q, Sun X‐B, Huo L et al (2012). Melatonin activates the Nrf2‐ARE pathway when it protects against early brain injury in a subarachnoid hemorrhage model. J Pineal Res 53: 129–137. [DOI] [PubMed] [Google Scholar]
- Werner ER, Blau N, Thöny B (2011). Tetrahydrobiopterin: biochemistry and pathophysiology. Biochem J 438: 397–414. [DOI] [PubMed] [Google Scholar]
- Wu YH, Feenstra MG, Zhou JN, Liu RY, Torano JS, Kan HJV et al (2003). Molecular changes underlying reduced pineal melatonin levels in Alzheimer disease: alterations in preclinical and clinical stages. J Clin Endocrinol Metab 88: 5898–5906. [DOI] [PubMed] [Google Scholar]
- Wu YH, Ursinus J, Zhou JN, Scheer FA, Ai‐Min B, Jockers R et al (2013). Alterations of melatonin receptors MT1 and MT2 in the hypothalamic suprachiasmatic nucleus during depression. J Affect Disord 148: 357–367. [DOI] [PubMed] [Google Scholar]
- Wu Y‐H, Zhou J‐N, Heerikhuize JV, Jockers R, Swaab DF (2007). Decreased MT1 melatonin receptor expression in the suprachiasmatic nucleus in aging and Alzheimer's disease. Neurobiol Aging 28: 1239–1247. [DOI] [PubMed] [Google Scholar]
- Yokomori K (2003). SMC protein complexes and the maintenance of chromosome integrity. Curr Top Microbiol Immunol 274: 79–112. [DOI] [PubMed] [Google Scholar]
- Yuan L, Collins AR, Dai J, Dubocovich ML, Hill SM (2002). MT(1) melatonin receptor overexpression enhances the growth suppressive effect of melatonin in human breast cancer cells. Mol Cell Endocrinol 192: 147–156. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Cook A, Kim J, Baranov SV, Jiang J, Smith K et al (2013). Melatonin inhibits the caspase‐1/cytochrome c/caspase‐3 cell death pathway, inhibits MT1 receptor loss and delays disease progression in a mouse model of amyotrophic lateral sclerosis. Neurobiol Dis 55: 26–35. [DOI] [PMC free article] [PubMed] [Google Scholar]


