Abstract
High-throughput sequencing efforts in molecular tumour diagnostics detect increasing numbers of novel variants, including variants predicted to affect splicing. In silico prediction tools can reliably predict the effect of variant disrupting canonical splice sites; however, experimental validation is required to confirm aberrant splicing. Here, we present RNA analysis performed for 13 canonical splice site variants predicted or known to result in splicing in the cancer predisposition genes MLH1, MSH2, MSH6, APC and BRCA1. Total nucleic acid was successfully isolated for 10 variants from eight formalin-fixed paraffin-embedded (FFPE) tumour tissues and two B-cell lines. Aberrant splicing was confirmed in all six variants known to result in splicing. Of one known variant in the B-cell line, aberrant splicing could only be detected after formalin fixation, which indicated that formalin fixation could possibly inhibit RNA degradation. Aberrant splicing was concluded in three of four predicted splice variants of uncertain significance, supporting their pathogenic effect. With this assay, somatic splice variants can be easily and rapidly analysed, enabling retrospective analysis to support the pathogenicity of variants predicted to result in splicing when only FFPE material is available.
Introduction
Most mammalian genes consist of multiple exons interspersed by long intronic sequences [1]. To create mature mRNA, the introns must be correctly identified and ‘spliced out’, and the exons joined together [1]. The spliceosome, the splicing machinery responsible for this process, recognises conserved motifs at or near the intron ends and a branch site within the intron [1]. Splice regulatory elements (SRE) near exon–intron boundaries, such as serine–arginine rich (SR) or heterogeneous nuclear ribonucleoparticle (hnRNP) proteins, are indispensable for correct splice site identification [2]. These elements can enhance or repress splicing and play an important role in alternative splicing [1, 2].
Of the variants that cause disease, 15–60% are proposed to disrupt splicing [3]. Included are variants in the canonical splice site, which directly alter the canonical splice site efficiency, but also intronic and exonic variants that alter the SREs or result in the creation of a new splice site or activation of a cryptic splice site [3, 4]. The latter could result in inclusion of a pseudoexon, an intronic sequence wrongly interpreted as an exon. Exon skipping or inclusion of a pseudoexon often results in a shift of the open reading frame, resulting in a premature stop codon or leading to a non- or less-functional protein. The mechanism of nonsense-mediated decay (NMD) in which mRNAs with a premature termination codon are (partially) degraded can remove aberrant mRNAs encoding for truncated proteins, ensuring mRNA quality [5].
Current assessment whether variants result in aberrant RNA transcripts often consists of in silico prediction with bioinformatics prediction tools, sometimes followed by reverse transcriptase polymerase chain reaction (RT-PCR) analysis of RNA extracted from blood [6–8] or functional splicing reporter minigene assays [4, 9]. Assessing alternative splicing using RNA-seq data has been described, but until now this has only been shown to be feasible with high-quality RNA [10, 11]. Additionally, splicing microarrays can be used for large-scale identification of splicing differences but are not always implemented in current diagnostics [3]. Although high-quality patient RNA analysis is usually preferred, this RNA is not always available, or the analysis is hampered because of degradation of aberrant transcripts through NMD [1].
With the current high-throughput sequencing methods applied in molecular tumour diagnostics, many variants are found, most of uncertain significance (VUS); hence, functional tests are required to classify these variants. Of these unclassified variants, a percentage is predicted to affect splicing. Specific kits are available to isolate RNA from formalin-fixed paraffin-embedded (FFPE) tissue blocks, and previous studies show that PCR, RT-PCR and even next-generation sequencing (NGS) are possible on these RNA samples [12, 13]. RNA analysis on RNA isolated from the FFPE tissue is currently not standardly performed but would enable the analysis of somatic splice site variants.
In the current study, the effect of splice site variants was examined in multiple cancer susceptibility genes, MLH1, MSH2, MSH6, APC and BRCA1. MLH1, MSH2 and MSH6 are part of the mismatch repair (MMR) pathway. Pathogenic heterozygous germ line variants in the MMR genes cause Lynch Syndrome, an autosomal dominant predisposition for colorectal, endometrial and other cancers [14]. Other known causes of MMR deficiency are somatic MLH1 promoter hypermethylation and the recently described biallelic somatic inactivation of the MMR genes caused by somatic variants [15–17]. Pathogenic germ line variants in the APC gene are known to result in familial adenomatous polyposis, a dominant disorder characterised by the occurrence of hundreds to even thousands of adenomas throughout the colon [18, 19]. In a small percentage of patients, the tumour phenotype can be explained by mosaic APC variants [20–22]. These variants can be easily detected by screening multiple adenomas, because the APC variant is present with a higher variant allele frequency in the tumour [20, 23]. Pathogenic variants in the BRCA1 gene, a key player in the nucleotide excision repair pathway, result in a high susceptibility to breast and ovarian cancers [24, 25]. Because BRCA-mutation status affects treatment strategies (PARP-inhibitors), the ability to detect and functionally assess both germ line and somatic mutations in BRCA1 and BRCA2 must increase [26, 27]. With the shift towards increased diagnostic screening of tumour tissue for all three syndromes, more somatic variants are found, which require functional tests to assess their pathogenicity.
The aim of our study was to investigate the possibility of analysing RNA isolated from the FFPE tissue to assess the effect of germ line and somatic variants predicted to affect splicing. We hypothesised that formalin fixation could inhibit RNA degradation, enabling the detection of aberrant RNA in FFPE tissues.
Materials and methods
Selection of variants
In total, 13 variants in the cancer susceptibility genes MLH1, MSH2, MSH6, APC and BRCA1 were tested for their effect on splicing (Supplemental Table 1). Of all variants, eight were somatic variants found between 2014 and 2017 with NGS in a previous study [28] or through molecular tumour diagnostic NGS, with all having a variant allele frequency of at least 12%. Five were germ line splice site variants, all previously demonstrated to result in aberrant RNA (Supplemental Table 1). The MLH1 c.(453+1_454-1)_(545+1_546-1)del, a germ line genomic exon 6 deletion, was added as a positive control.
Total nucleic acid isolation and cDNA synthesis
For 11 variants, total nucleic acid was obtained from tissue cores punched from FFPE blocks embedded between 2009 and 2016. Tumour areas were marked on a hematoxylin and eosin stained slide by a pathologist. Tissue cores from the corresponding area on the FFPE block were punched with a 0.6-mm biopsy needle. Total nucleic acid (undivided RNA/DNA) was isolated from the obtained punches and microdissected areas with a Tissue Preparation System with VERSANT Tissue Preparation Reagents (Siemens Healthcare Diagnostics, Tarrytown, NY, USA) as previously described [29]. For two variants (MLH1 c.791-1G>C and MSH2 c.1511-2A>G), no FFPE tissue was available, but Epstein-Barr virus (EBV)-immortalised B-cells were cultured. Additionally, RNA from the MSH2 c.1511-2A>G B-cell line was isolated after incubating the cells with 4% formalin for 5 h. RNA from the B-cell lines and three colorectal cancer cell lines (SW480, SW837 and LS180) was isolated using a Nucleospin RNA isolation kit (Macherey-Nagel-06/2015, Rev.17, Düren, Germany) according to the manufacturer’s protocol. Colorectal cancer cell lines were used as a positive control for RNA expression.
All cDNA was synthesised using OligoDT’s and random primers with the SuperScript VILO cDNA synthesis Kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol using total nucleic acid. To show that RNA was successfully isolated and cDNA was synthesised, HNRNPM and/or CPSF6 housekeeping gene expression was assessed. Also, gene expression of the tested genes (MLH1, MSH2, MSH6, APC and BRCA1) was assessed using qPCR primers.
Splice site prediction/variant nomenclature
For (canonical) splice site prediction, Alamut (Interactive Biosoftware, Rouen, France) was used. This software package includes the in silico splice site prediction algorithms SpliceSite Finder (SSF), MaxEntScan (MES) (http://genes.mit.edu/burgelab/maxent/Xmaxentscan_scoreseq.html), NNSPLICE (http://www.fruitfly.org/seq_tools/splice.html) and Human Splicing Finder (HSF, http://www.umd.be/HSF/). Variants were annotated according to the Human Genetics Variation Society (HGVS) guidelines. Recommendations of the HGVS were followed to use the term “variant” instead of “mutation” (http://www.hgvs.org/mutnomen/recs.html). The following Genbank reference sequences were used: NM_000249.2/NG_007109.2 for MLH1, NM_000251.2/NG_007110.2 for MSH2, NM_000179.2/NG_007111.1 for MSH6, NM_000038.5/NG_008481.4 for APC and NM_007294.3/NG_005905.2 for BRCA1. All variants have been submitted to the LOVD databases. The MMR and APC variants were submitted to www.insight-database.org (patient IDs 00033694-00033800 and 00033702-00033705), and the BRCA1 variants were added to https://databases.lovd.nl/shared (patient IDs 00144415 and 00144416).
Primer design and PCR
For all variants with an (predicted) exon skip, two primer pairs were created to amplify two exon–exon boundaries. Primers were used in three combinations: Forward1/Reverse1, Foward2/Reverse2 and Forward1/Reverse2. For all variants with a partial exon skip or pseudoexon insertion as (predicted) RNA effect, primers were designed to amplify the exon–exon boundary. All primer sequences are listed in Supplemental Table 2.
Real-time PCR was used to amplify the exon–exon boundaries and to assess the expression of the affected gene. All PCR reactions were performed on a CFX96 touch Realtime PCR machine (Bio-rad, Hercules, CA, USA) with the following PCR program: 95 °C for 5 min (1 cycle), 95 °C for 15 s, 60 °C for 30 s and 72 °C for 30 s (38 cycles), followed by a melt curve from 65 °C to 95 °C with a 0.5 °C increment for 5 s with plate read. When no PCR product was detected, PCR was repeated with high cDNA input and 44 instead of 38 cycles. Because of limited cDNA, only F1/R2 was repeated for variants with two primer pairs, when the first PCR failed. All PCR products were analysed on a Qiaxcel capillary electrophoresis system (Qiagen, Hilden, Germany) and sequenced with Sanger Sequencing.
Results
MMR variants
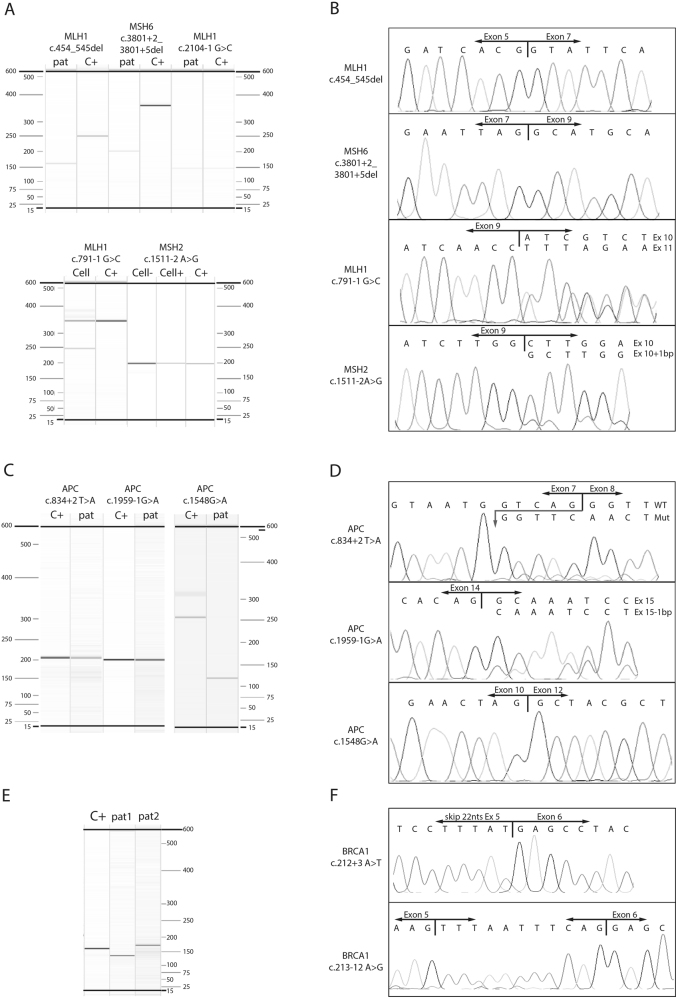
Five MMR splice variants and one MLH1 genomic exon deletion in RNA isolated from FFPE tissue were analysed for their effect on splicing (Table 1). Total nucleic acid was isolated from the FFPE blocks and converted to cDNA using OligoDT’s and random primers. The quality of cDNA was evaluated by detecting the expression of housekeeping genes (HKG) HNRNPM and/or CPSF6. From five FFPE tissue blocks, HKG expression was detected, and in three of the five, cDNA from the affected MMR gene could be amplified and analysed (MLH1 c.(453+1_454-1)_(545+1_546-1)del, MLH1 c.2104G>C and MSH6 c.3801+1_3801+5del). The amplified products of the three MMR cDNAs from FFPE tissues were measured with Qiaxcel (Fig. 1a) and sequenced (Fig. 1b). Size determination of the cDNAs carrying the MLH1 c.(453+1_454-1)_(545+1_546-1)del and MSH6 c.3801+1_3801+5del variants showed only a product size smaller than that of the wildtype (WT) control, whereas MLH1 c.2104-1G>C only showed a product comparable in size with that of the WT product. Sequencing showed an aberrant product in two of the three FFPE samples; the MLH1 genomic exon 6 deletion (r.454_545del) and an r.3647_3801 deletion in the MSH6 c.3801+1_3801+5del sample (Fig. 1b), whereas for MLH1 c.2104-1G>C, sequencing was normal, as was that for the WT control.
Table 1.
Results of splicing assay
| Gene | Variant | Source RNA | Variant frequency | (predicted) RNA effect | Result |
|---|---|---|---|---|---|
| A. Variants known to result in splicing | |||||
| MLH1 | c.(453+1_454-1)_(545+1_546-1)dela | FFPE, CRC45 | NAb | r.? | Genomic exon deletion shown in RNAc |
| MLH1 | c.791-1G>Ca | Cell line | 50% | r.791_884del | Aberrant splicing as reported |
| MLH1 | c.793C>Ta | FFPE, CRC63 | NA | r.791_884del (major), WT (minor) | Unable to isolate nucleic acid |
| MLH1 | c.1731G>A | FFPE, CRC51 | 35% | r.1668_1731del | No MMR RNA expression detected |
| MSH2 | c.1511-2A>Ga | Cell line | 80% | r.1510_1511ins1511-1 | Aberrant splicing as reportedd |
| MSH6 | c.3801+1_3801+5dela | FFPE, Pol67 | NA | r.3647_3801del | Aberrant splicing as reportedc |
| APC | c.1548G>A | FFPE, Pol68 | 28% | r.1409_1548del | Aberrant splicing as reportedc |
| BRCA1 | c.213-12A>G | FFPE, EC74 | 93% | r.212_213ins213-12_213-1 | Aberrant splicing as reportedc |
| B. Variants predicted to result in splicing | |||||
| MLH1 | c.885-2A>G | FFPE, CRC45 | 57% | r.(885_889del)e | No MMR RNA expression detected |
| MLH1 | c.2104-1G>C | FFPE, EC55 | 36% | r.(2104_2105del)e | Only WT product |
| APC | c.834+2T>A | FFPE, Pol60 | 12% | r.(830_834del)e | Aberrant splicing as predicted |
| APC | c.1959-1G>A | FFPE, Pol67 | 40% | r.(1959del)e | Aberrant splicing as predicted |
| BRCA1 | c.212+3A>T | FFPE, OC46 | 54% | r.(191_212del)e | Aberrant splicing as predictedc |
Source RNA shows cell line or formalin-fixed paraffin-embedded (FFPE) tissue block, with type of tumour and age of onset
CRC colorectal cancer, EC endometrial cancer, OC ovarian cancer, Pol polyps, WT wildtype, nt nucleotides, NA not assessed
aGermline variants. Variants were detected with Sanger sequencingPlease check if the term "r.?" is ok in Table 1.yes, this is correct
bLOH was detected in the tumour
cNo WT product detected
dAberrant product only detected after formalin fixation of the cells. Genbank reference sequences NM_000249.2 (MLH1), NM_000251.2 (MSH2), NM_000179.2 (MSH6), NM_000038.5 (APC) and NM_007294.3 (BRCA1) were used
ePredicted RNA effect is based on nearest cryptic splice site, which could possibly act as a new 3′ or 5′ splice site
Fig. 1.
Size determination and sequencing results. Qiaxcel results showing the size of the MMR variants (a), the APC variants (c) and BRCA1 variants (e) of patient material (pat), cell lines (Cell) and control RNA isolated from colorectal cell lines (C+). The MSH2 cell line was analysed with (Cell+) and without (Cell−) formalin fixation of the cells. The BRCA1 variants were analysed with the same primers, pat1 shows the BRCA1 c.212+3A>T and pat2 shows the BRCA1 c.213-12A>G. b, d and f Sanger sequencing results of variants showing aberrant products
Additionally, RNA isolated from the two EBV-immortalised B-cell lines carrying an MSH2 c.1511-2A>G and an MLH1 c.791-1G>C splice site variant was tested. Size determination of the cDNA from the two cell lines showed a product of approximately 350 bp (comparable with the WT control) and a product of 250 bp for the MLH1 c.791-1G>C sample and a product comparable in size with that of the WT for the MSH2 c.1511-2A>G sample. Sequencing detected an aberrant product only in the MLH1 c.791-1G>C sample, which showed an r.791_884 deletion. To mimic FFPE conditions, 4% formalin was added to fixate EBV- immortalised B-cells carrying the MSH2 variant. After fixation, RNA was isolated, and cDNA was synthesised following the same protocol as that for the non-formalin fixed cells. cDNA from the formalin-fixed cells was tested and showed a size comparable with that of the WT (Fig. 1a), and an aberrant product was detected with Sanger Sequencing (Fig. 1b).
APC variants
Three APC variants were analysed for their effect on splicing (Table 1). RNA was successfully isolated from all three FFPE blocks, shown by the detection of HKG expression. From all three samples, cDNA from APC could be amplified (Fig. 1c) and sequenced (Fig. 1d). Compared with the control without the variant, the difference in size of APC c.1548G>A was almost 125 bp. Sequencing showed an aberrant product for all three variants: an APC r.830_834 deletion in the APC c.834+2T>A sample; a one-nucleotide deletion (r.1959del) in the APC c.1959-1G>A sample; and an r.1409_1548 deletion in the APC c.1548G>A sample (Fig. 1d).
BRCA1 variants
Two BRCA1 splice site variants were analysed for their effect on splicing (Table 1). RNA was successfully isolated from both FFPE blocks (shown by HKG detection), and BRCA1 cDNA was amplified and analysed. PCR was performed with the same primers for both variants. Size determination indicated a smaller (BRCA1 c.212+3A>T) and a slightly larger (BRCA1 c.213-12A>G) product compared with that of the WT control (C+, Fig. 1e). Sequencing showed a BRCA1 r.191_212 deletion (BRCA1 c.212+3A>T) and an inclusion of the last 11 nucleotides of intron 5 corresponding to the BRCA1 c.213-11_c.213-1 sequence (BRCA1 c.213-12A>G, Fig. 1f).
Discussion
We performed RNA analysis for six splice site variants known to result in aberrant splicing and five variants predicted to result in aberrant splicing using RNA isolated from FFPE tissues. For the six variants shown previously to result in aberrant splicing, the reported splice effect was confirmed for four, and for the other two variants, RNA analysis was not possible because either RNA isolation from FFPE tissue failed (no expression of HKG) or the affected gene (i.e., MLH1) did not show expression in the presence of positive HKG expression. In all four confirmed splice effects, no WT product was identified. For two variants (MLH1 c.(453+1_454-1)_(545+1_546-1)del and BRCA1 c.213-12A>G), this result was expected because of the high variant allele frequencies (approximately 100%, as detected with Sanger Sequencing and 93%, respectively). The APC c.1548G>A and MSH6 c.3801+1_3801+5del had variant allele frequencies of 28% and 50%, respectively, but only aberrant product was detected, which could be due to preferential amplification of the smaller (aberrant) product or possible FFPE-induced RNA degradation. For the APC variant predicted to result in an exon 11 skip, the F2/R2 primers that amplified the boundary of exon 11–exon 12 produced a product, which showed that WT cDNA product with exon 11 was present.
For two variants, MLH1 c.791-1G>C and MSH2 c.1511-2A>G, RNA was isolated from EBV-immortalised B-cell lines because the FFPE tissue was not available. Both previously resulted in an (partial) exon skip [4, 30–32], which changed the reading frame and led to a premature stop codon; however, in the current study, aberrant splicing was only confirmed for one of the two variants (MLH1 c.791-1G>C), whereas only WT transcript was detected for the MSH2 c.1511-2A>G. The B-cell lines were cultured without NMD inhibitors, and we hypothesised that the aberrant MSH2 RNA was possibly degraded through NMD. Detection of MSH2 c.1511-2A>G RNA and no detection of the MLH1 c.791-1G>C RNA is consistent with previous studies in which NMD inhibitors were omitted [33, 34]. Notably, the aberrant RNA was detected after formalin fixation of the B-cells carrying the MSH2 c.1511-2A>G, which is consistent with our hypothesis that aberrant RNA in the FFPE tissue can be detected because formalin fixation prevents the degradation of RNA.
For the five variants predicted to affect the canonical splice site, three resulted in aberrant splicing, showing the predicted splice effect. For the other two, in one (MLH1 c.2104-1G>C), only the WT transcript was detected, and in the other, no expression of the affected gene (MLH1) was detected in the presence of normal HKG expression. The APC c.834+2T>A and APC c.1959-1G>A showed WT and aberrant product, which can be explained by the variant allele frequencies (12% and 40%, respectively). The BRCA1 c.212+3A>T had a variant allele frequency of 54%, although only the aberrant product was detected, which was possibly due to preferential amplification of the smaller (aberrant) product or because no RNA expression of the WT allele occurred. However, FFPE-related RNA degradation could not be excluded.
Two of the five variants predicted to result in splicing (MLH1 c.885-2A>G and APC c.1959-1G>A) have previously been described to be (likely) pathogenic, even though the functional effect was unknown [35, 36]. The remaining three variants (MLH1 c.2104-1G>C, APC c.834+2T>A and BRCA1 c.212+3A>T) are novel, although the BRCA1 c.212+3A>G variant has been described to result in aberrant splicing. Aberrant RNA was detected in three variants predicted to affect splicing, while RNA could not be assessed for the pathogenic MLH1 c.885-2A>G variant. The RNA effect seen was conform to the predicted effect, confirming that in silico splice prediction tools are reliable in their predictions, particularly for variants disrupting the canonical splice site, although experimental analysis is required [4, 7, 37]. For two novel variants, this study supports their pathogenic effect.
Interestingly, no aberrant splicing was shown for the pathogenic MLH1 c.2104-1G>C variant, possibly because the splice effect falls outside the amplification window. This shows the limitations of the current study. First, RNA from FFPE tissue blocks is often degraded to fragments less than ~300 bases in length [38]. To analyse this RNA, small amplicons must be designed instead of large amplicons containing multiple exons, as is preferred in leucocyte RNA testing. The design of the primers is very specific for the variant and is based on splicing prediction software. Although these algorithms are described as accurate for variants in the canonical splice site [4, 7, 8], aberrant products that are not predicted and fall outside the amplification window of the assay will not be detected. Second, when an aberrant product is not detected, poor RNA quality, a wrongly predicted effect or no splice effect of the variant can be implicated, but no expression or complete RNA degradation of the mutant product by NMD can also occur. Therefore, the assay can confirm aberrant splicing, but from negative assay results, one cannot accurately conclude that no aberrant splicing occurs or that lack of aberrant product is due to other factors. Negative results should be confirmed with other methods, such as a minigene splicing assay [4, 9]. Further evidence of FFPE-based RNA analysis should be obtained from a larger study with more samples and variants. Targeted RNA-seq of the RNA isolated from the FFPE tissue might enable high-throughput analysis of somatic splice site variants. Finally, possibly because of a size-related amplification bias, the reaction seems to favour the aberrant splice product. Leaky splicing (a partial splice defect with also the ability to express normal RNA) might remain unnoticed as a result of this.
FFPE blocks were collected from the archives and were embedded between 2000 and 2016, with most (n = 9) embedded after 2009. Notably, from the two samples embedded before 2009, no RNA was successfully isolated from the FFPE block, indicating that isolating RNA from the FFPE tissue might not be possible in older blocks. From eight of the nine blocks embedded after 2009, RNA was successfully isolated, showing WT and/or aberrant RNA, and combined with the formalin fixation results from the cell lines, showed that formalin fixation possibly inhibited RNA degradation. With the analysis of the RNA from the FFPE tissues, the splicing effect of somatic variants that are only present in the tumour can be analysed. Furthermore, available material can be retrospectively analysed, without having to request RNA from leukocyte DNA, which is not always available.
RNA analysis on total nucleic acid isolated from the FFPE tissue blocks is a valuable tool for fast and easy detection of aberrant splicing, offering additional support for the pathogenicity of a (predicted) splice variant. While the current study focuses on MMR, APC and BRCA1 gene variants, this method could be applied on splice variants in other genes as well. With this assay, we correctly showed the splice effect of six known splice site variants and showed the splice effect of three variants predicted to affect splicing. This assay can be used to analyse somatic variants found in the FFPE tumour tissue, with formalin fixation possibly inhibiting RNA degradation, and can be easily implemented in current molecular tumour diagnostics to help classify the high number of variants of uncertain significance currently found with high-throughput sequencing.
Electronic supplementary material
Acknowledgements
The authors would like to acknowledge Marina Ventayol Garcia for technical assistance. This work was supported by the Dutch Cancer Society under study number UL2012-5542.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Electronic supplementary material
The online version of this article (10.1038/s41431-018-0153-z) contains supplementary material, which is available to authorised users.
References
- 1.Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet. 2002;3:285–98. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 2.Brillen AL, Schoneweis K, Walotka L, Hartmann L, Muller L, Ptok J, et al. Succession of splicing regulatory elements determines cryptic 5’ss functionality. Nucleic Acids Res. 2016;45:4202–16. doi: 10.1093/nar/gkw1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang GS, Cooper TA. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat Rev Genet. 2007;8:749–61. doi: 10.1038/nrg2164. [DOI] [PubMed] [Google Scholar]
- 4.van der Klift HM, Jansen AM, van der Steenstraten N, Bik EC, Tops CM, Devilee P, et al. Splicing analysis for exonic and intronic mismatch repair gene variants associated with Lynch syndrome confirms high concordance between minigene assays and patient RNA analyses. Mol Genet Genom Med. 2015;3:327–45. doi: 10.1002/mgg3.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hentze MW, Kulozik AE. A perfect message: RNA surveillance and nonsense-mediated decay. Cell. 1999;96:307–10. doi: 10.1016/S0092-8674(00)80542-5. [DOI] [PubMed] [Google Scholar]
- 6.Whiley PJ, de la Hoya M, Thomassen M, Becker A, Brandao R, Pedersen IS, et al. Comparison of mRNA splicing assay protocols across multiple laboratories: recommendations for best practice in standardized clinical testing. Clin Chem. 2014;60:341–52. doi: 10.1373/clinchem.2013.210658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vreeswijk MP, Kraan JN, van der Klift HM, Vink GR, Cornelisse CJ, Wijnen JT, et al. Intronic variants in BRCA1 and BRCA2 that affect RNA splicing can be reliably selected by splice-site prediction programs. Hum Mutat. 2009;30:107–14. doi: 10.1002/humu.20811. [DOI] [PubMed] [Google Scholar]
- 8.Spurdle AB, Couch FJ, Hogervorst FB, Radice P, Sinilnikova OM, Group IUGVW. Prediction and assessment of splicing alterations: implications for clinical testing. Hum Mutat. 2008;29:1304–13. doi: 10.1002/humu.20901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooper TA. Use of minigene systems to dissect alternative splicing elements. Methods. 2005;37:331–40. doi: 10.1016/j.ymeth.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 10.Sultan M, Schulz MH, Richard H, Magen A, Klingenhoff A, Scherf M, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–60. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- 11.Bisognin A, Pizzini S, Perilli L, Esposito G, Mocellin S, Nitti D, et al. An integrative framework identifies alternative splicing events in colorectal cancer development. Mol Oncol. 2014;8:129–41. doi: 10.1016/j.molonc.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hedegaard J, Thorsen K, Lund MK, Hein AM, Hamilton-Dutoit SJ, Vang S, et al. Next-generation sequencing of RNA and DNA isolated from paired fresh-frozen and formalin-fixed paraffin-embedded samples of human cancer and normal tissue. PLoS ONE. 2014;9:e98187. doi: 10.1371/journal.pone.0098187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gouveia GR, Ferreira SC, Ferreira JE, Siqueira SA, Pereira J. Comparison of two methods of RNA extraction from formalin-fixed paraffin-embedded tissue specimens. Biomed Res Int. 2014;2014:151724. doi: 10.1155/2014/151724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lynch HT, de la Chapelle A. Hereditary colorectal cancer. N Engl J Med. 2003;348:919–32. doi: 10.1056/NEJMra012242. [DOI] [PubMed] [Google Scholar]
- 15.Mensenkamp AR, Vogelaar IP, van Zelst-Stams WA, Goossens M, Ouchene H, Hendriks-Cornelissen SJ, et al. Somatic mutations in MLH1 and MSH2 are a frequent cause of mismatch-repair deficiency in Lynch syndrome-like tumors. Gastroenterology. 2014;146:643–6 e8. doi: 10.1053/j.gastro.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 16.Geurts-Giele WR, Leenen CH, Dubbink HJ, Meijssen IC, Post E, Sleddens HF, et al. Somatic aberrations of mismatch repair genes as a cause of microsatellite-unstable cancers. J Pathol. 2014;234:548–59. doi: 10.1002/path.4419. [DOI] [PubMed] [Google Scholar]
- 17.Kuismanen SA, Holmberg MT, Salovaara R, de la Chapelle A, Peltomäki P. Genetic and epigenetic modification of MLH1 accounts for a major share of microsatellite-unstable colorectal cancers. Am J Pathol. 2000;156:1773–9. doi: 10.1016/S0002-9440(10)65048-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Powell SM, Petersen GM, Krush AJ, Booker S, Jen J, Giardiello FM, et al. Molecular diagnosis of familial adenomatous polyposis. N Engl J Med. 1993;329:1982–7. doi: 10.1056/NEJM199312303292702. [DOI] [PubMed] [Google Scholar]
- 19.Groden J, Thliveris A, Samowitz W, Carlson M, Gelbert L, Albertsen H, et al. Identification and characterization of the familial adenomatous polyposis coli gene. Cell. 1991;66:589–600. doi: 10.1016/0092-8674(81)90021-0. [DOI] [PubMed] [Google Scholar]
- 20.Spier I, Drichel D, Kerick M, Kirfel J, Horpaopan S, Laner A, et al. Low-level APC mutational mosaicism is the underlying cause in a substantial fraction of unexplained colorectal adenomatous polyposis cases. J Med Genet. 2016;53:172–9. doi: 10.1136/jmedgenet-2015-103468. [DOI] [PubMed] [Google Scholar]
- 21.Yamaguchi K, Komura M, Yamaguchi R, Imoto S, Shimizu E, Kasuya S, et al. Detection of APC mosaicism by next-generation sequencing in an FAP patient. J Hum Genet. 2015;60:227–31. doi: 10.1038/jhg.2015.14. [DOI] [PubMed] [Google Scholar]
- 22.Out AA, van Minderhout IJ, van der Stoep N, van Bommel LS, Kluijt I, Aalfs C, et al. High-resolution melting (HRM) re-analysis of a polyposis patients cohort reveals previously undetected heterozygous and mosaic APC gene mutations. Fam Cancer. 2015;14:247–57. doi: 10.1007/s10689-015-9780-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jansen AM, Crobach S, Geurts-Giele WR, van den Akker BE, Garcia MV, Ruano D, et al. Distinct patterns of somatic mosaicism in the APC gene in neoplasms from patients with unexplained adenomatous polyposis. Gastroenterology. 2017;152:546–9 e3. doi: 10.1053/j.gastro.2016.10.040. [DOI] [PubMed] [Google Scholar]
- 24.Hall JM, Lee MK, Newman B, Morrow JE, Anderson LA, Huey B, et al. Linkage of early-onset familial breast cancer to chromosome 17q21. Science. 1990;250:1684–9. doi: 10.1126/science.2270482. [DOI] [PubMed] [Google Scholar]
- 25.Rahman N, Stratton MR. The genetics of breast cancer susceptibility. Annu Rev Genet. 1998;32:95–121. doi: 10.1146/annurev.genet.32.1.95. [DOI] [PubMed] [Google Scholar]
- 26.Mafficini A, Simbolo M, Parisi A, Rusev B, Luchini C, Cataldo I, et al. BRCA somatic and germline mutation detection in paraffin embedded ovarian cancers by next-generation sequencing. Oncotarget. 2016;7:1076–83. doi: 10.18632/oncotarget.6834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koczkowska M, Zuk M, Gorczynski A, Ratajska M, Lewandowska M, Biernat W, et al. Detection of somatic BRCA1/2 mutations in ovarian cancer - next-generation sequencing analysis of 100 cases. Cancer Med. 2016;5:1640–6. doi: 10.1002/cam4.748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jansen AM, van Wezel T, van den Akker BE, Ventayol Garcia M, Ruano D, Tops CM, et al. Combined mismatch repair and POLE/POLD1 defects explain unresolved suspected Lynch syndrome cancers. Eur J Hum Genet. 2016;24:1089–92. doi: 10.1038/ejhg.2015.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Eijk R, Stevens L, Morreau H, van Wezel T. Assessment of a fully automated high-throughput DNA extraction method from formalin-fixed, paraffin-embedded tissue for KRAS, and BRAF somatic mutation analysis. Exp Mol Pathol. 2013;94:121–5. doi: 10.1016/j.yexmp.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 30.Wijnen J, Khan PM, Vasen H, Menko F, vanderKlift H, vandenBroek M, et al. Majority of hMLH1 mutations responsible for hereditary nonpolyposis colorectal cancer cluster at the exonic region 15-16. Am J Hum Genet. 1996;58:300–7. [PMC free article] [PubMed] [Google Scholar]
- 31.Wijnen J, Khan PM, Vasen H, van der Klift H, Mulder A, van Leeuwen-Cornelisse I, et al. Hereditary nonpolyposis colorectal cancer families not complying with the Amsterdam criteria show extremely low frequency of mismatch-repair-gene mutations. Am J Hum Genet. 1997;61:329–35. doi: 10.1086/514847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Casey G, Lindor NM, Papadopoulos N, Thibodeau SN, Moskow J, Steelman S, et al. Conversion analysis for mutation detection in MLH1 and MSH2 in patients with colorectal cancer. JAMA. 2005;293:799–809. doi: 10.1001/jama.293.7.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bianchi F, Rosati S, Belvederesi L, Loretelli C, Catalani R, Mandolesi A, et al. MSH2 splice site mutation and endometrial cancer. Int J Gynecol Cancer. 2006;16:1419–23. doi: 10.1111/j.1525-1438.2006.00572.x. [DOI] [PubMed] [Google Scholar]
- 34.Thompson BA, Goldgar DE, Paterson C, Clendenning M, Walters R, Arnold S, et al. A multifactorial likelihood model for MMR gene variant classification incorporating probabilities based on sequence bioinformatics and tumor characteristics: a report from the Colon Cancer Family Registry. Hum Mutat. 2013;34:200–9. doi: 10.1002/humu.22213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Terdiman JP, Gum JR, Jr., Conrad PG, Miller GA, Weinberg V, Crawley SC, et al. Efficient detection of hereditary nonpolyposis colorectal cancer gene carriers by screening for tumor microsatellite instability before germline genetic testing. Gastroenterology. 2001;120:21–30. doi: 10.1053/gast.2001.20874. [DOI] [PubMed] [Google Scholar]
- 36.Lagarde A, Rouleau E, Ferrari A, Noguchi T, Qiu J, Briaux A, et al. Germline APC mutation spectrum derived from 863 genomic variations identified through a 15-year medical genetics service to French patients with FAP. J Med Genet. 2010;47:721–2. doi: 10.1136/jmg.2010.078964. [DOI] [PubMed] [Google Scholar]
- 37.Colombo M, De Vecchi G, Caleca L, Foglia C, Ripamonti CB, Ficarazzi F, et al. Comparative in vitro and in silico analyses of variants in splicing regions of BRCA1 and BRCA2 genes and characterization of novel pathogenic mutations. PLoS ONE. 2013;8:e57173. doi: 10.1371/journal.pone.0057173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cronin M, Pho M, Dutta D, Stephans JC, Shak S, Kiefer MC, et al. Measurement of gene expression in archival paraffin-embedded tissues: development and performance of a 92-gene reverse transcriptase-polymerase chain reaction assay. Am J Pathol. 2004;164:35–42. doi: 10.1016/S0002-9440(10)63093-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.