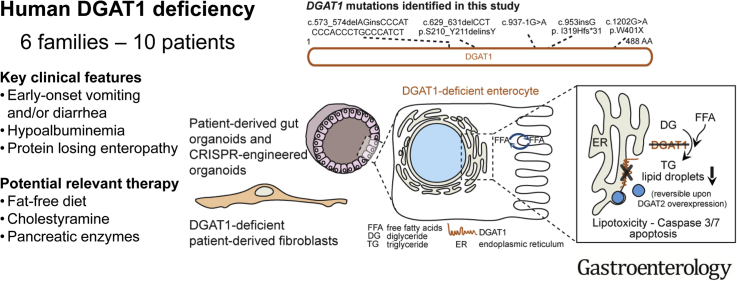
Abstract
Background & Aims
Congenital diarrheal disorders are rare inherited intestinal disorders characterized by intractable, sometimes life-threatening, diarrhea and nutrient malabsorption; some have been associated with mutations in diacylglycerol-acyltransferase 1 (DGAT1), which catalyzes formation of triacylglycerol from diacylglycerol and acyl-CoA. We investigated the mechanisms by which DGAT1 deficiency contributes to intestinal failure using patient-derived organoids.
Methods
We collected blood samples from 10 patients, from 6 unrelated pedigrees, who presented with early-onset severe diarrhea and/or vomiting, hypoalbuminemia, and/or (fatal) protein-losing enteropathy with intestinal failure; we performed next-generation sequencing analysis of DNA from 8 patients. Organoids were generated from duodenal biopsies from 3 patients and 3 healthy individuals (controls). Caco-2 cells and patient-derived dermal fibroblasts were transfected or transduced with vectors that express full-length or mutant forms of DGAT1 or full-length DGAT2. We performed CRISPR/Cas9-guided disruption of DGAT1 in control intestinal organoids. Cells and organoids were analyzed by immunoblot, immunofluorescence, flow cytometry, chromatography, quantitative real-time polymerase chain reaction, and for the activity of caspases 3 and 7.
Results
In the 10 patients, we identified 5 bi-allelic loss-of-function mutations in DGAT1. In patient-derived fibroblasts and organoids, the mutations reduced expression of DGAT1 protein and altered triacylglycerol metabolism, resulting in decreased lipid droplet formation after oleic acid addition. Expression of full-length DGAT2 in patient-derived fibroblasts restored formation of lipid droplets. Organoids derived from patients with DGAT1 mutations were more susceptible to lipid-induced cell death than control organoids.
Conclusions
We identified a large cohort of patients with congenital diarrheal disorders with mutations in DGAT1 that reduced expression of its product; dermal fibroblasts and intestinal organoids derived from these patients had altered lipid metabolism and were susceptible to lipid-induced cell death. Expression of full-length wildtype DGAT1 or DGAT2 restored normal lipid metabolism in these cells. These findings indicate the importance of DGAT1 in fat metabolism and lipotoxicity in the intestinal epithelium. A fat-free diet might serve as the first line of therapy for patients with reduced DGAT1 expression. It is important to identify genetic variants associated with congenital diarrheal disorders for proper diagnosis and selection of treatment strategies.
Keywords: CDD, Genomic, PLE, 3-D Culture Model
Abbreviations used in this paper: B-LCL, B lymphoblastoid cell line; BSA, bovine serum albumin; CDD, congenital diarrheal disorder; cDNA, complementary DNA; DG, diacylglycerol; DGAT1, diacylglycerol-acyltransferase 1; hSI-EM, human small intestine expansion medium; FFA, free fatty acid; GI, gastrointestinal; OA, oleic acid; PB, PiggyBac transposon; PBS, phosphate-buffered saline; PLE, protein-losing enteropathy; sgRNA, single-guide RNA; SSC, Side Scatter; TG, triacylglycerol; WT, wild-type
Graphical abstract
What You Need to Know.
Background and Context
Mutations in DGAT1 have recently been identified in patients with congenital diarrheal disorders (CDDs), but the underlying molecular pathomechanisms have remained largely elusive.
New Findings
The authors identified 10 patients with DGAT1 deficiency representing the largest cohort to date, linking gut epithelial lipid metabolism and lipotoxicity to CDD; and rescued aberrant lipid metabolism with isoenzyme DGAT2.
Limitations
Although the authors show exogenous DGAT1 or DGAT2 expression or proteasome inhibitors may overcome defects, future studies may need to address how that knowledge can be translated to targeted therapies.
Impact
The authors highlight the importance of identifying the genetic defect in patients with CDD, and showcase further use of gut organoid technology to study rare diseases of the gastrointestinal tract.
Congenital diarrheal disorders (CDDs) are a group of rare inherited intestinal disorders that are characterized by intractable, sometimes life-threatening, diarrhea and nutrient malabsorption. CDDs can be classified based on their aberrations in absorption and transport of nutrients and electrolytes, enterocyte differentiation and polarization, enteroendocrine cell differentiation, or dysregulation of the intestinal immune response.1 Congenital protein-losing enteropathy (PLE) is a type of CDD that is characterized by increased protein loss from the gastrointestinal (GI) system. Patients with PLE often suffer from hypoproteinemia, fat malabsorption, fat-soluble vitamin deficiencies, and malnutrition. Recently, we have identified germline loss-of-function mutations in CD55 as a major monogenic etiology for congenital PLE.2
Previously, mutations in the gene encoding diacylglycerol-acyltransferase 1 (DGAT1) were found to underlie a syndrome of diarrhea and congenital PLE.3, 4, 5, 6 DGAT1 and its isozyme DGAT2 (encoding for diacylglycerol-acyltransferase 2) are responsible for the conversion of diacylglycerol (DG) and fatty acyl-CoA to triacylglycerol (TG) in humans.7, 8 TG is the main energy substrate stored in human adipose tissue, is essential for milk production in the mammary gland, and is part of the very low-density lipoprotein–mediated transport of lipids to peripheral tissue.9, 10 In the human small intestine, DGAT1 is the only highly expressed enzyme, whereas DGAT2 is mainly expressed in the liver.3, 11 In enterocytes, TG is stored in lipid droplets or packaged into chylomicrons before transport into the lymphatic system.8, 12, 13
The pathomechanism responsible for intestinal failure and PLE in DGAT1 deficiency has remained unclear. Through next-generation sequencing, we identified 10 additional patients from 6 unrelated pedigrees with 5 different, novel bi-allelic mutations in DGAT1 leading to severe, sometimes fatal course of PLE and fat intolerance. We took this unique opportunity to further shed light on the fundamental pathomechanisms of human DGAT1 deficiency.
Materials and Methods
Study Approval
The study was approved by the responsible local ethics committees (Ethics Commission of the Medical University of Vienna and Institutional Review Board of the University Medical Center Utrecht). All participants provided written informed consent for the collection of samples and subsequent analysis.
DNA Sequencing
Whole-exome sequencing was performed on patients as previously described.14, 15 Targeted panel sequencing was performed as previously described.16 Conventional Sanger sequencing was performed for validation and segregation analysis of variants.
Cell Culture
Organoids were generated from duodenal biopsies that were obtained from 3 healthy controls and 3 patients during duodenoscopy for diagnostic purposes, as described in detail in the Supplementary Materials and Methods. The healthy controls were patients suspected of celiac disease or inflammatory bowel disease, who did not show abnormalities on endoscopic and histological examinations.
Caco-2 cells and patient-derived fibroblasts were cultured in Dulbecco’s modified Eagle’s medium with/without GlutaMax and high glucose (Life Technologies, Carlsbad, CA) supplemented with 10% heat-inactivated fetal bovine serum (GE Health Care, Little Chalfont, UK), 100 U/mL penicillin (Gibco, Waltham, MA), 100 μg/mL streptomycin (Gibco), and 1 mM sodiumpyruvate (Gibco) at 37°C and 5% CO2. Patient-derived Epstein-Barr virus B lymphoblastoid cell line (B-LCL) was maintained in RPMI 1640 with 10% heat-inactivated fetal bovine serum, 100 U/mL penicillin (Gibco), and 100 μg/mL streptomycin (Gibco).
CRISPR/Cas9 Knockout of DGAT1
Plasmid constructs for the expression of DGAT1 single-guide RNAs (sgRNA) and Cas9 nuclease were generated as previously described and outlined in Supplementary Material and Methods.17 Three DGAT1 sgRNAs targeting exon 7 and 1 sgRNA targeting intron 6-7 were designed. Two different sgRNA (mix)-plasmids were used for transfection: sgRNA#2 and a mix of sgRNA#6, sgRNA#7, and sgRNA#8 (sgRNA#678). Hygromycin-resistance was achieved by co-transfection with the PiggyBac (PB) Transposon System (plasmids PB-Hygromycin and PB-Transposase were kindly provided by Bon-Kyoung Koo).
Transfection of healthy intestinal organoids was performed by electroporation, as described previously18 and extensively in Supplementary Materials and Methods.
Lipid Droplet Assays
Oleic acid (OA) was conjugated to bovine serum albumin (BSA) as described in Supplementary Materials and Methods. Organoids were grown in expansion medium (EM) on black clear-bottom 96-well imaging plates (Corning Life Sciences, Corning, NY). On day 6, the organoids were incubated with 1 mM OA/BSA for 17 hours in presence or absence of 0.1 μM DGAT1 inhibitor (AZD 3988; Tocris, Bristol, UK). Organoids were then fixed in 4% formaldehyde for 30 minutes at room temperature. Cells were washed in phosphate-buffered saline (PBS) and stained with 0.025 mg/mL LD54019 and 4′,6-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich, St Louis, MO) in PBS for 15 minutes at room temperature in the dark. Cells were washed and stored in PBS. Imaging of the organoids was performed using a Leica (Wetzlar, Germany) SP8X laser-scanning confocal microscope outfitted with a white light laser. The acquired stacks were processed and analyzed with Fiji/ImageJ (National Institutes of Health, Bethesda, MD)20, 21; shown are maximum projections of approximately 15-μm stacks.
For flow cytometry analysis, organoids were grown and treated with OA/BSA in the same manner as for the confocal analysis. After overnight incubation with 1 mM OA/BSA, the cells were harvested by pipetting and dissociated using TrypLE Express (ThermoFisher, Waltham, MA) until single cells were acquired. The cells were then fixed and stained in the same manner as for the confocal analysis and assayed using a BD FACS Canto II (BD Biosciences, San Jose, CA).
Fibroblasts were seeded at 6 × 104 cells in a 6-well plate and allowed to adhere overnight. Cells were then treated with OA positive control from the Lipid Droplet Fluorescence Assay (#500001; Cayman Chemical, Ann Arbor, MI) at 1:4000 dilution for 24 hours. Cells were fixed and stained according to the manufacturer’s protocol and assayed using BD FACS Fortessa (BD Biosciences, Franklin Lakes, NJ).
Cloning and Retrovirus Production
DGAT1 (ENST00000528718.5) and DGAT2 (ENST00000228027.11) complementary DNAs (cDNAs) were amplified by polymerase chain reaction from HEK293 cDNA library and cloned into pDONR221 using BP reaction according to the manufacturer’s protocol (Thermo Fisher). LR reaction was performed into pFMIG for wild-type (WT) DGAT1 and DGAT2 with N-terminal Streptavidin-HA tag.
Quantitative Real-Time Polymerase Chain Reaction
RNA was isolated from Caco-2 cells or organoids grown in either EM or differentiation medium (DM) for 5 days, used to synthesize cDNA by using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA) and amplified with SYBR green supermix (BIO-Rad) in a Light Cycler96 (BIO-Rad) according to the manufacturer’s protocol. Details on analysis and primers are given in Supplementary Materials and Methods.
Western Blotting
Cell lysates were generated and Western blotting was performed as described in the Supplementary Materials and Methods.
Thin Layer Chromatography
Organoids were grown in EM for 7 days and then prepared for Folch extraction as described previously22 and in the Supplementary Materials and Methods. Thin layer chromatography was performed by spotting the isolated lipid phase on aluminium-backed silica plates (Merck Millipore, Burlington, MA). As reference samples 1,3-dipentadecanoin (DG, C15:0/-/C15:0), tripentadecanoin (TG, C15:0/C15:0/C15:0) and triheptadecanoin (TG, C17:0/C17:0/C17:0) were included. The plates were then developed in a mobile phase of hexane:diethylether:acetic acid (60:15:2). The lipid bands were visualized by spraying the plates with a solution of 10% CuSO4 (wt/vol) in 10% H2SO4 (vol/vol) and subsequently heating the plates to 120°C for 30 minutes, as described previously23 to calculate DG/TG ratios, band intensities were quantified by Fiji/ImageJ.20, 21
Lipotoxicity Assays
For the propidium iodide staining, organoids were grown and incubated with varying concentrations of OA as described for the lipid droplet confocal assay. The organoids were then washed with Hank’s balanced salt solution (HBSS) (Gibco), and stained with Hoechst 1 μg/mL (Sigma-Aldrich) and propidium iodide 0.1 mg/mL (ThermoFisher) in HBSS at room temperature for 15 minutes. Organoids were imaged by an inverted Olympus IX53 epifluorescence microscope (Tokyo, Japan).
For the Caspase-Glo 3/7 assay (Promega, Madison, WI), organoids were grown on TC-treated 96-well plates (Greiner, Kremsmunster, Austria) and incubated with OA as was done for the propidium iodide staining. Organoids were washed and resuspended in HBSS and transferred to a white-walled 96-well plate (Greiner). The assay was performed according to the manufacturer’s protocol and luminescence was measured on a Tristar 2 luminometer (Berthold Technologies, Oak Ridge, TN).
Statistical Analysis
Data are presented as mean ± SD. Experiments were performed with a minimum of 3 replications. Statistical significance was determined at P ≤ .05 using 2-way analysis of variance with Tukey’s multiple comparison test, a Mann-Whitney U test, or a Student t test where appropriate. Significance is indicated as P ≤ .05 (*), P ≤ .01 (**), or P ≤ .001 (***)or P < .0001 (∗∗∗∗).
Results
Clinical Phenotype
We investigated 10 patients from 5 consanguineous families and 1 family of unknown consaguinity with unaffected parents. All of the patients had a history of intestinal failure due to congenital diarrhea and/or vomiting, resulting in failure to thrive. (Figure 1A, Table 1, Supplementary Table 1, see Supplementary Materials and Methods for clinical details). These 10 patients come from 4 Turkish families originating from Turkey and 2 Caucasian families from The Netherlands. In summary, 8 of 10 cases showed early-onset PLE characterized by hypoalbuminemia, hypogammaglobulinemia, and intractable diarrhea, with 1 patient developing marked steatorrhea. Patients 7 and 8 presented with severe vomiting only, which resulted in failure to thrive in the older sibling. Food containing fat induced abdominal pain and vomiting soon after ingestion and serum lipid profiles of the 10 patients were variable (Supplementary Table 2). Some patients had normal levels of serum TG, whereas 1 patient exhibited hypertriglyceridemia. Most showed a reduced level of high-density lipoprotein, with normal levels of low-density lipoprotein, very low-density lipoprotein, and cholesterol.
Figure 1.
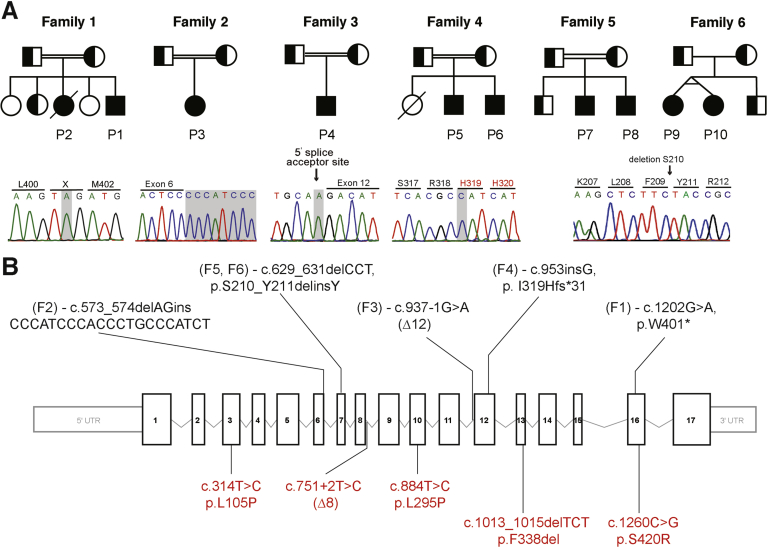
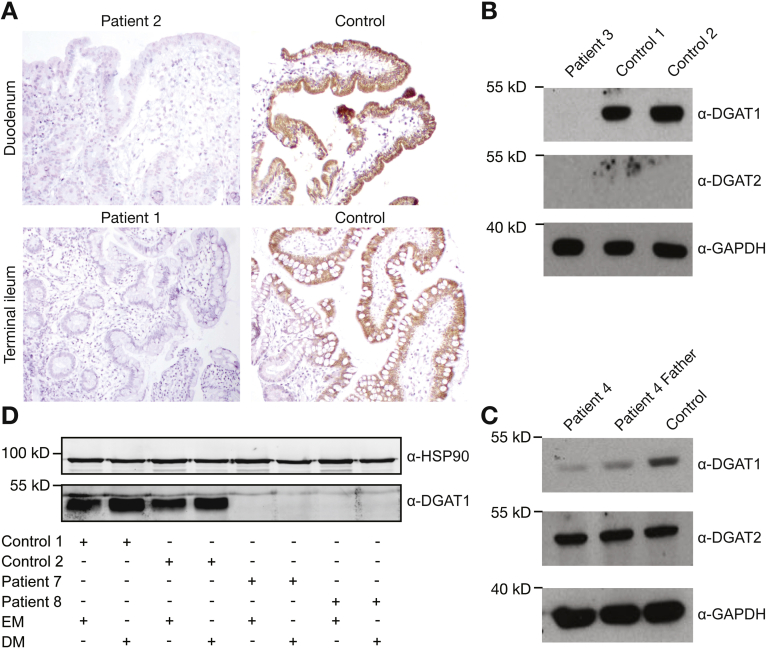
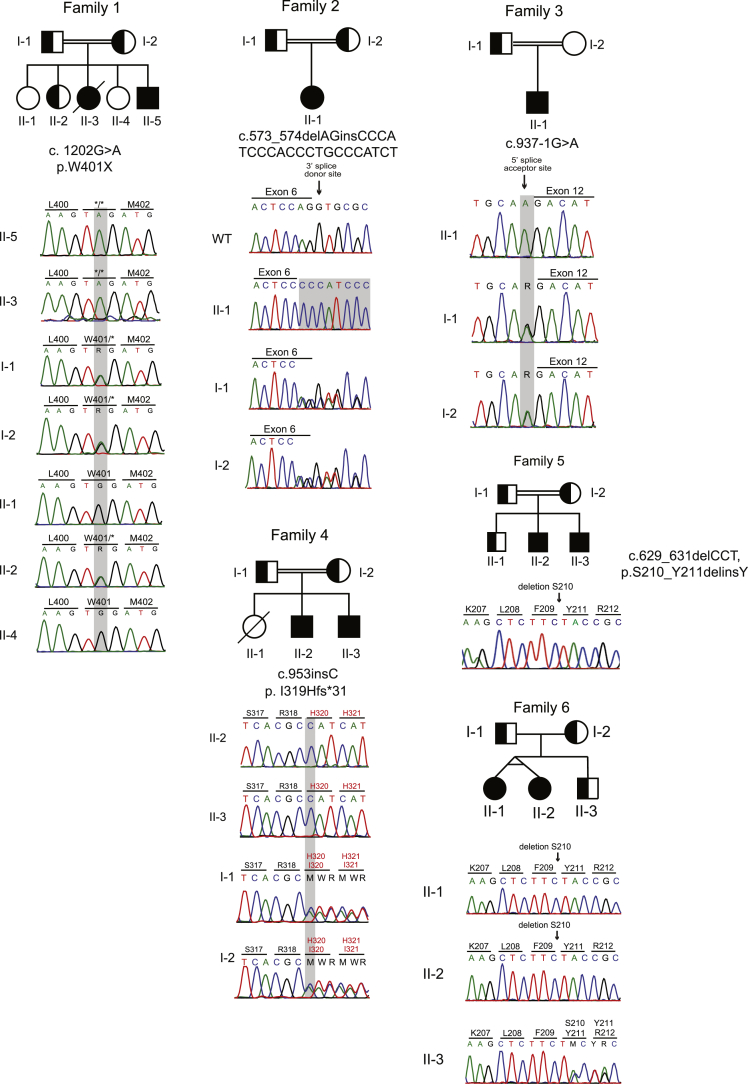
Pedigrees, mutations, and genetic location of 6 families with DGAT1 deficiency. (A) Pedigrees of families with DGAT1 deficiency and chromatograms showing mutation in affected patients. Filled shapes indicate affected individuals, half-filled are heterozygous for mutation indicated, and empty shapes indicate WT. (B) Exonic scheme of DGAT1 showing mutations identified in this study in black and previously identified mutations in red.3, 4, 5, 6
Table 1.
Patient Characteristics
| Patient ID | P1 | P2 | P3 | P4 | P5 | P6 | P7 | P8 | P9 | P10 |
|---|---|---|---|---|---|---|---|---|---|---|
| Demographics | ||||||||||
| Current age and gender | 4.5 y, M | Deceased, F | 2 y, F | 8 y, M | 2 y, M | 6 y, M | 14 y, M | 17 y, M | 10 y, F | 10 y, F |
| Country of origin and ethnicity | Turkey, Turkish | Turkey, Turkish | Turkey, Turkish | Turkey, Turkish | Turkey, Turkish | Turkey, Turkish | The Netherlands, Caucasian | The Netherlands, Caucasian | The Netherlands, Caucasian | The Netherlands, Caucasian |
| Age of clinical onset | Birth | Birth | 3 wk | 2 mo | 40 d | 2.5 mo | First month | First month | Birth | Birth |
| Disease manifestations | ||||||||||
| Failure to thrive | Yes | Yes | Yes | Yes | Yes | Yes | No | Yes | Yes | Yes |
| Vomiting | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Diarrhea | Yes, fatty | Yes | Yes | Yes | Yes, bloody and watery | Yes, watery | No | No | Yes | Yes |
| Hypoalbuminemia | Yes | Yes | Yes | Yes | Yes | Yes | No | No | Yes | Yes |
| Hypogammaglobulinemia | Yes | Yes | Yes | Yes | Yes | Yes | ND | ND | Yes | Yes |
| Edema | Yes | Yes | Yes | No | No | No | No | No | No | No |
| GI examinations (endoscopy and imaging) | Normal | Normal | Normal | Normal | ND | Normal | Normal | Normal | Normal | Normal |
| Hematoxylin-eosin and electron microscopy | Normal | Normal | Duodenal enterocytic lipid accumulation, microvilli are shortened and rarefied | Normal | ND | Focal vacuolization at one area and partially blunted villi | Normal histology (on fat-free diet) | Normal histology (on fat-free diet) | Misdiagnosis of atypical MVID based on CD10 positive globules on LM and laterally located microvilli on EM | Misdiagnosis of atypical MVID based on CD10 positive globules on LM and laterally located microvilli on EM |
| Extra-GI manifestations | Recurrent infections, otitis media | Recurrent infections | Corneal cystine crystal accumulation and intermittent metabolic acidosis | ND | Hepatomegaly and jaundice | Recurrent infections | Gilles de la Tourette syndrome | Gilles de la Tourette syndrome | Recurrent infections | Recurrent infections |
| Clinical course | ||||||||||
| Treatments | Fat-free formula and MCT, albumin infusions | Albumin infusions | Albumin, TPN | Cholestyramine | Creon pancreatic lipase, hydrolyzed formula | Creon pancreatic lipase | Monthly infusion of Intralipid and Omegaven suppletion of lipid-soluble vitamins | Monthly infusion of Intralipid and Omegaven suppletion of lipid-soluble vitamins | TPN and small bowel transplantation | TPN, recently started with fat-free formula |
| Outcome | Asymptomatic and normal growth with low-fat diet + MCT oil + fat-free formula | Patient passed away at 6 mo due to sepsis | Stool frequency once a day with Basic F formula feeding | Reduced stool volume and frequency on cholestyramine treatment | Stool frequency reduced on Creon treatment; weight and height are still below 3rd percentile | Stool frequency reduced on Creon treatment; after 2 y, symptoms resolved spontaneously | Enteral feeding without fat | Enteral feeding without fat | Tolerates enteral feeding, but still stunted | Tolerates enteral feeding, but still stunted |
EM, electron microscopy; F, female; GI, gastrointestinal; LM, light microscopy; M, male; MCT, medium-chain triglyceride; MVID, microvillus inclusion disease; ND, not determined; TPN, total parenteral nutrition.
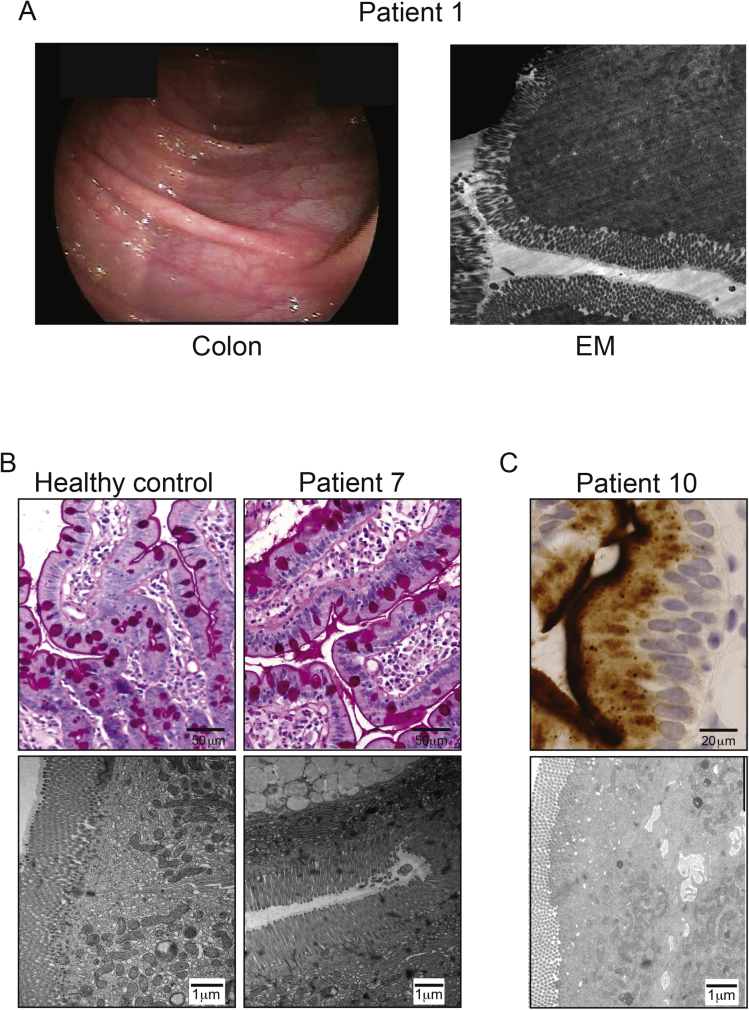
Endoscopy was performed on patients 1 to 3 and 7 to 10, which showed no macroscopic abnormalities of the duodenum and colon (Supplementary Figures 1 and 2). Histopathology of a duodenal biopsy from patient 1 showed marked flattening of the villi (Figure 2 and Supplementary Figure 2) and patient 3 showed marked shortening of the villi (Supplementary Figure 1A), Electron microscopy of a duodenal biopsy of patient 3 showed lack of microvilli (Supplementary Figure 1A). Patients 7 and 8 showed normal pathology, although the biopsies were taken under fat-restricted diet (Supplementary Figure 1B). Duodenal biopsies from patient 10 showed lateral microvilli and cytoplasmic CD10 staining (Supplementary Figure 1C), which led to the misdiagnosis of atypical microvillus inclusion disease.22
Supplementary Figure 1.
Histologic and endoscopic characteristics of intestines of DGAT1-deficient patients. (A) Colonoscopy (left) and electron microscopy (EM, right) revealed normal intestinal structures in patient 1. (B) Periodic acid-Schiff and EM of patient 7 were obtained during fat-free diet and do not show abnormalities. (C) Patient 10 shows cytosolic CD10 staining and lateral microvilli on EM.
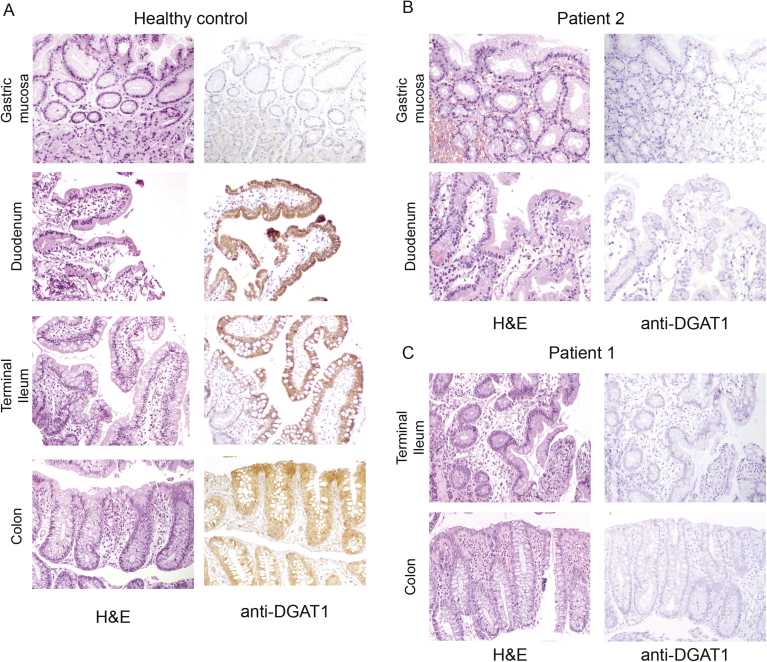
Supplementary Figure 2.
Hematoxylin-eosin (H&E) and immunohistochemical staining for DGAT1 in (A) healthy controls, (B) patient 2, and (C) patient 1.
Figure 2.
DGAT1 protein expression in patient-derived material. (A) Immunohistochemistry of DGAT1 in control and patient 1 and patient 2 ileal and duodenal biopsy, respectively. (B) Western blot showing lack of DGAT1 and DGAT2 in patient 3 fibroblast lysate, but normal expression of DGAT1 in healthy control fibroblasts. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control. (C) Western blot showing lower expression of potentially nonfunctional DGAT1 protein and normal DGAT2 protein level from Epstein-Barr virus–derived B lymphoblastoid cell line of patient 4, a parent, and a healthy control. GAPDH was used as a loading control. (D) Western blot for DGAT1 protein expression in undifferentiated (EM) and differentiated (DM) organoids from 2 healthy controls and patients 7 and 8. HSP90 was used as loading control. Results are representative of 3 independent experiments.
Eight patients received various treatments that led to resolution or significant improvement of the GI symptoms. Most patients were placed on a fat-restricted diet, which alleviated their GI symptoms. Addition of medium-chain TG was tolerated, and some patients were infused with intravenous essential fatty acid supplements Intralipid and Omegaven and fat-soluble vitamins. Patient 4 received cholestyramine and patients 5 and 6 received pancreatic lipase due to low fecal elastase level, both of which reduced daily stool frequency. Patients 9 and 10 are twins and patient 9 received an intestinal transplant. The various treatments and outcomes are summarized in Supplementary Table 3.
Some patients developed extra-GI manifestations. In patients 1, 2, and 6, recurrent episodes of unspecific infections were recorded. Patients 9 and 10 had recurrent episodes of septicemia due to catheter-related blood stream infections. None of our patients showed any pattern of unusual or opportunistic infections that might be overtly associated with fecal loss of immunoglobulins or lymphopenia. Treatment of infections was successful with antibiotics, antifungal drugs, or prophylactic intravenous immunoglobulin, as outlined in Supplementary Table 3. Patient 5 had hepatomegaly and jaundice, and liver biopsy revealed fibrosis and cholestasis. Patients 7 and 8 were diagnosed with Gilles de la Tourette syndrome and were treated with dexamphetamine.
Identification of Novel DGAT1 Mutations
Whole-exome sequence analysis was performed in patients 1, 3, 7, 8, 9, and 10, while targeted panel sequencing was performed for patients 4 and 5. Variants in affected siblings (patients 2 and 6) were identified using conventional Sanger sequencing. Collectively, we identified 5 novel homozygous variants in the gene DGAT1 (Mendelian Inheritance in Man: 604900, GenBank: NM_012079.5). These variants segregated with the disease under the assumption of autosomal recessive inheritance, with heterozygous carriers being unaffected (Figure 1A). The variants are neither present nor reported as rare heterozygous variants in the gnomAD database,24 and were predicted to be deleterious using Combined Annotation Dependent Depletion25 prediction tool (Supplementary Table 1).
In family 1, we identified a homozygous nonsense mutation at amino acid 401 leading to an early stop codon (c.1202G>A, p.W401X). In family 2, we identified a homozygous insertion deletion (c.573_574delAGinsCCCATCCCACCCTGCCCATCT) in exon 6 of DGAT1. In family 3, we identified a homozygous splice site acceptor mutation (c. 937-1G>A) preceding exon 12. In family 4, we identified a homozygous single base-pair insertion, leading to a frameshift and early stop codon (c.953insG, p.I319Hfs*31) in exon 12. In families 5 and 6, we identified a homozygous 3 base-pair deletion (c.629_631delCCT, p.S210_Y211delinsY) in exon 7 (Figure 1, Supplementary Figure 3). Altogether, we identified 5 novel disease-causing homozygous mutations in DGAT1 in 6 patients of Turkish origin and 4 of Dutch origin.
Supplementary Figure 3.
Family pedigrees and Sanger sequencing histograms of DGAT1 deficiency. Filled shapes indicate affected individuals, half-filled are heterozygous for mutation indicated, and empty shapes indicate wild type.
Consequences of DGAT1 Mutations
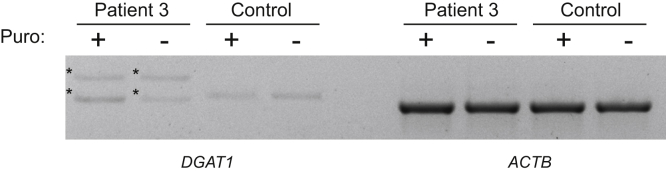
We proceeded to study the consequences of DGAT1 mutation on available material. Immunohistochemistry on GI biopsies from patient 1 and patient 2 showed a lack of DGAT1 protein expression specifically in the epithelium of the duodenum, ileum, and colon (Figure 2A and Supplementary Figure 2), whereas DGAT1 protein was not detected in the gastric mucosa of control or patient material (Supplementary Figure 2). In patient 3, reverse-transcription polymerase chain reaction of cDNA from patient-derived fibroblast showed aberrant splicing (Supplementary Figure 4), which ultimately led to undetectable protein expression on Western blot (Figure 2B). In patient 4, Epstein-Barr virus–derived B-lymphoblastic cell line showed a highly reduced expression of DGAT1 (Figure 2C). Intestinal organoids derived from patients 7 and 8 showed normal mRNA levels on differentiation (data not shown), but protein was absent (Figure 2D). Similar data were obtained from patient 9 (data not shown). Together, we show that all the novel DGAT1 mutations identified led to aberrant protein expression.
Supplementary Figure 4.
Defective mRNA splicing in patient 3. (A) Gel showing aberrant DGAT1 transcripts in mRNA isolated from patient 3 fibroblasts with and without a 4-hour puromycin treatment. * denotes aberrant transcript. ACTB was used as control.
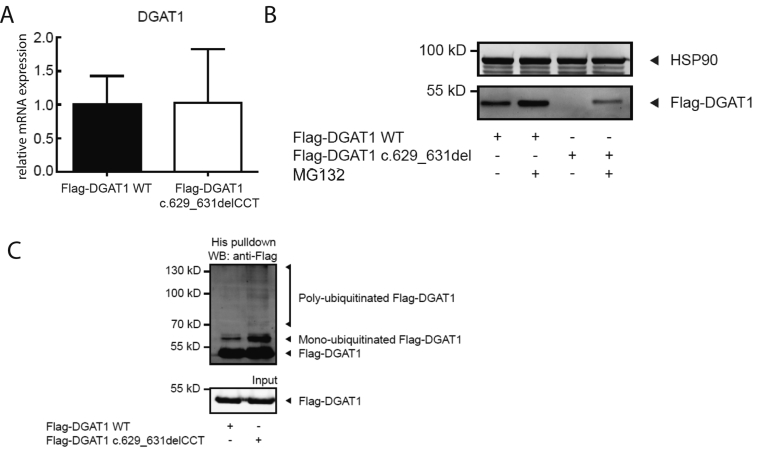
To confirm that the DGAT1 c.629_631delCCT mutation specifically leads to reduced DGAT1 protein levels without affecting mRNA levels, Caco-2 cells were stably transfected with Flag-DGAT1 WT or Flag-DGAT1 c.629_631delCCT. Indeed, DGAT1 mRNA expression was similar in both cell lines (Supplementary Figure 5A), but DGAT1 protein was not detectable when DGAT1 was mutated (Supplementary Figure 5B). Incubation with the proteasome inhibitor MG132 shows that the loss of protein is at least partially due to proteasomal degradation of the mutant DGAT1 (Supplementary Figure 5B). To further investigate the increased proteasomal degradation of DGAT1 c.629_631delCCT, we determined the level of ubiquitination of the mutant protein. Therefore, Caco-2 cells were co-transfected with His-ubiquitin and Flag-DGAT1 WT or Flag-DGAT1 c.629_631delCCT and treated with MG132. A ubiquitin pulldown assay showed that ubiquitination of Flag-DGAT1 c.629_631delCCT was increased compared with ubiquitination of Flag-DGAT1 WT (Supplementary Figure 5C).
Supplementary Figure 5.
DGAT1 c.629_631delCCT results in increased proteasomal degradation of DGAT1. Caco-2 cells were stably transfected with indicated constructs. (A) qRT-PCR analysis of DGAT1 mRNA expression, relative to HPRT1, in Caco-2 cells stably transfected with indicated constructs. Average and standard deviation of 3 independent experiments are shown. (B) Transfected Caco-2 cells were untreated or treated with 2 μM MG132 for 16 hours. Protein expression was determined by Western blot analysis using anti-Flag and anti-HSP90 antibodies. Results are representative of 3 independent experiments. (C) Caco-2 cells were transiently transfected with His-ubiquitin and indicated constructs and treated with 2 μM MG132 for 16 hours. Ubiquitination of DGAT1 was determined by a His pulldown and subsequent Western blot analysis using anti-Flag antibodies. Results are representative of 3 independent experiments.
Loss of DGAT1 Leads to Aberrant Lipid Metabolism
Free fatty acids (FFAs) can be processed for energy production through beta oxidation, or stored in the form of lipid droplets on its incorporation into TG. In enterocytes, this lipid droplet formation is required for the transport of long-chain FFAs into chylomicrons, before being excreted across the basolateral membrane of enterocytes. Recently, it was shown that DGAT1 mutant fibroblasts accumulated less lipid droplets when incubated with OA, an 18-carbon FFA.5 We hypothesized that DGAT1 deficiency will lead to aberrant lipid droplet formation in patient-derived cells.
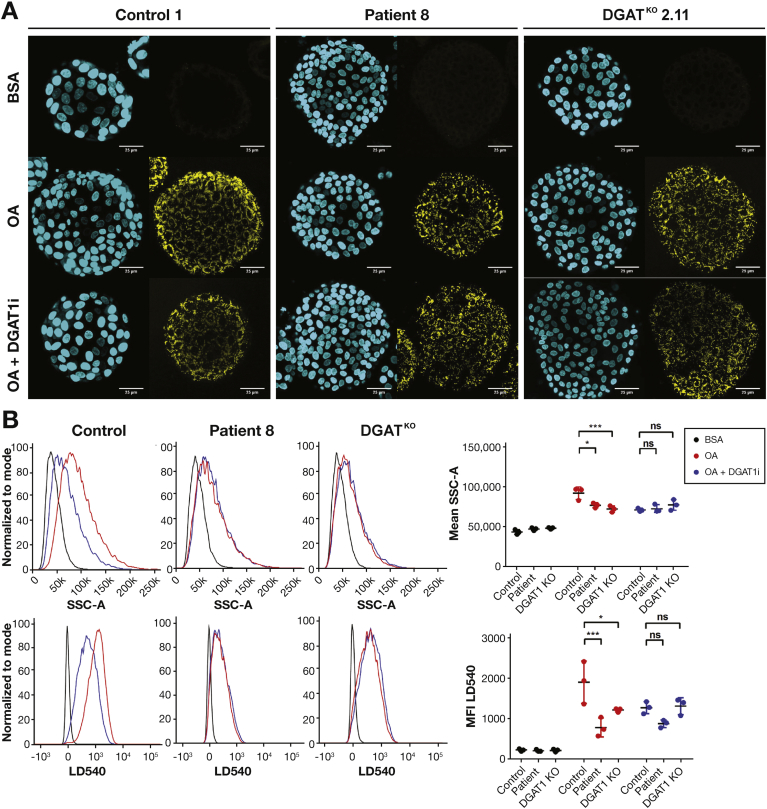
We performed a staining for LD540, which binds neutral lipids,19, 26 on intestinal organoids from patients 7, 8, and 9 after 16 hours incubation with BSA-coupled OA. Using fluorescence imaging, we observed an increase in lipid droplet formation in healthy control organoids, which was significantly reduced in DGAT1 mutant patient-derived organoids from patients 7, 8, and 9 and DGAT1 knockout (DGAT1KO) organoids that were generated using CRISPR/Cas9 genome editing for DGAT1 in healthy control organoids (Figure 3A and Supplementary Figure 6 and Supplementary Table 4). Formation of lipid droplets was at least partially dependent on DGAT1, as the use of a selective DGAT1 inhibitor (DGAT1i) AZD 3988 on healthy control organoids resulted in a reduction of lipid droplet accumulation as shown by decreased LD540 staining (Figure 3A). This was further confirmed and quantified using a flow cytometry–based assay, in which healthy control organoids showed an increased granularity due to the accumulation of lipid droplets (side scatter area, SSC-A) and increased LD540 staining on incubation with OA. Absence of DGAT1 (patients or KO) or DGAT1 inhibition caused a significant reduction in granularity and LD540 staining compared with healthy controls (Figure 3B).
Figure 3.
Loss of DGAT1 results in decreased lipid droplet formation in organoids. (A) Immunofluorescent images of 4′,6-diamidino-2-phenylindole (DAPI) (blue) and LD540 (yellow) staining of organoids from healthy control, DGAT1 mutant patient 8 (P8) and DGAT1KO organoids after 17-hour incubation with vehicle control (BSA), 1 μM OA, or 1 μM OA + 0.1 μM DGAT1 inhibitor (OA+DGAT1i). Representative images of 3 healthy controls, 3 patients (patients 7–9), and 3 CRISPR/Cas9 genome-edited DGAT1-knock-out (DGAT1KO) organoids. (B) Representative histograms of SSC and LD540 staining in organoids from controls, patients, and DGAT1KO organoids as described in (A). Upon OA stimulation, control organoids accumulate lipid droplets and show increased SSC and LD540, which was severely reduced in patient-derived and DGAT1KO cells. Mean fluorescence intensity (MFI) of SSC and LD540 was plotted for n = 3 per group. Statistical analysis was done using a 2-way analysis of variance with Tukey’s multiple comparison test. Mean ± SD is indicated; * P ≤.05, *** P ≤.001.
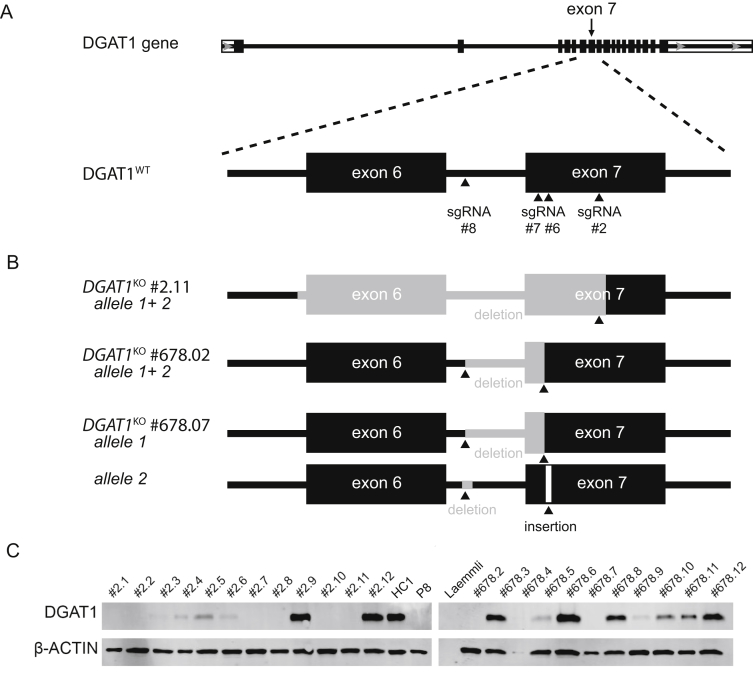
Supplementary Figure 6.
Generation of DGAT1 knockouts in healthy human intestinal organoids. (A) Targeting strategy for the generation of DGAT1 knockout organoids using CRISPR/Cas9 genome editing. (B) Schematic view of insertion-deletion mutations of 3 DGAT1KO clones. Gray areas represent deleted sequences from the WT DGAT1 gene. (C) Western blot analysis of DGAT1 expression in DGAT1KO organoids compared with healthy control 1 (HC1) and patient 8 (P8). Actin, loading control.
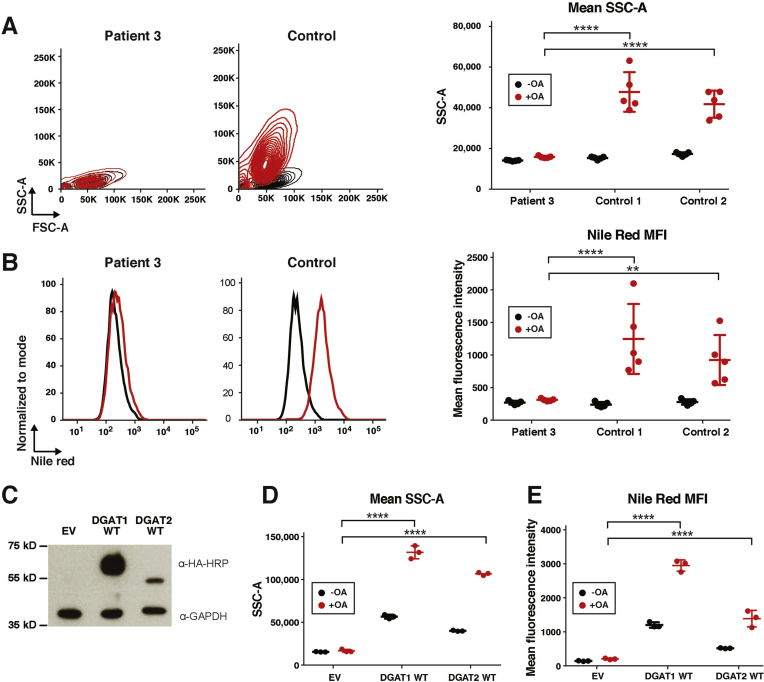
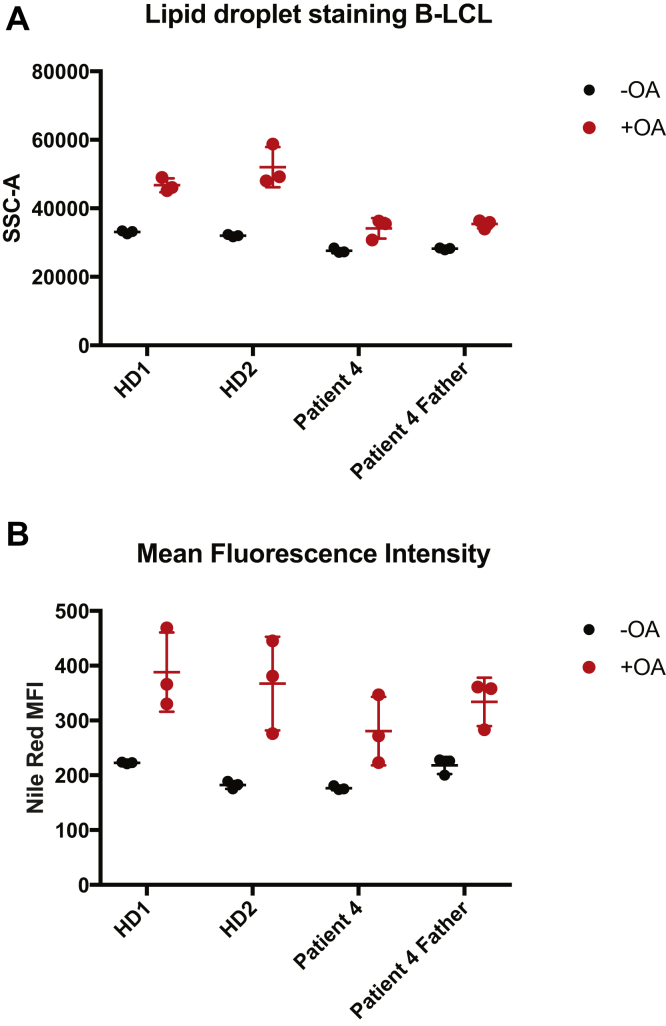
In addition, we incubated normal donor and patient 3–derived fibroblasts with OA and quantified lipid droplet formation by flow cytometry. Normal donor fibroblasts accumulated lipid droplets on incubation with OA, which was shown by increased granularity (SSC-A) and Nile Red staining, a dye that binds to neutral lipids such as TG.27 In contrast, patient-derived dermal fibroblast from patient 3 failed to accumulate lipid droplets, as they showed a lack of granularity (Figure 4A) and Nile Red staining (Figure 4B). In addition, we show that this lack of lipid droplet formation was not detected in B-LCL derived from patient 4 (Supplementary Figure 7), presumably due to the expression of DGAT2 in this cell type (Figure 2).
Figure 4.
Loss of DGAT1 results in decreased lipid droplet formation in fibroblasts. (A) Left: Representative contour plots for forward (FSC-A) and SSC-A of patient 3 and 2 control fibroblasts with and without OA addition. Right: Mean SSC-A of 5 technical replicates. (B) Left: Representative histogram of Nile Red mean fluorescence intensity (MFI) of patients 3 and 2 control fibroblasts with and without OA addition. Right: MFI of 5 technical replicates. (C) Western blot showing retroviral-mediated delivery of exogenous DGAT1 and DGAT2 on patient 3 fibroblasts. (D) Mean SSC-A and (E) MFI of Nile Red staining on patient 3 fibroblasts reconstituted with empty vector (EV), WT DGAT1, or WT DGAT2. Statistical analysis was done using 2-way analysis of variance with Tukey’s multiple comparison test. Mean ± SD is indicated; **P< .01 or ****P < .0001.
Supplementary Figure 7.
Lipid droplet staining in Epstein-Barr virus–derived B-lymphoblastic cell line (EBV B-LCL) of patient 4 with and without OA. (A) Mean SSC-A and (B) Nile Red mean fluorescence intensity (MFI) staining of lipid droplet stained EBV B-LCLs.
WT DGAT1 and DGAT2 Rescues Lipid Droplet Formation in DGAT1 Deficiency
We reconstituted fibroblasts from patient 3 with WT DGAT1 and DGAT2 protein using a retroviral delivery system. We successfully reconstituted DGAT1 protein expression, and overexpressed DGAT2 in these cells (Figure 4C). The DGAT1-reconstituted fibroblasts were able to incorporate OA into lipid droplets as seen by the increased granularity and increased Nile Red fluorescence (Figure 4D and E). The phenotype was also rescued by overexpression of DGAT2, resulting in restored granularity and Nile Red staining on addition of OA (Figure 4D and E). We concluded that reconstitution of DGAT1 rescues the altered lipid metabolism phenotype, and that DGAT2 can partially rescue altered lipid droplet formation in DGAT1-deficient fibroblasts.
Loss of DGAT1 Specifically Inhibits TG Formation
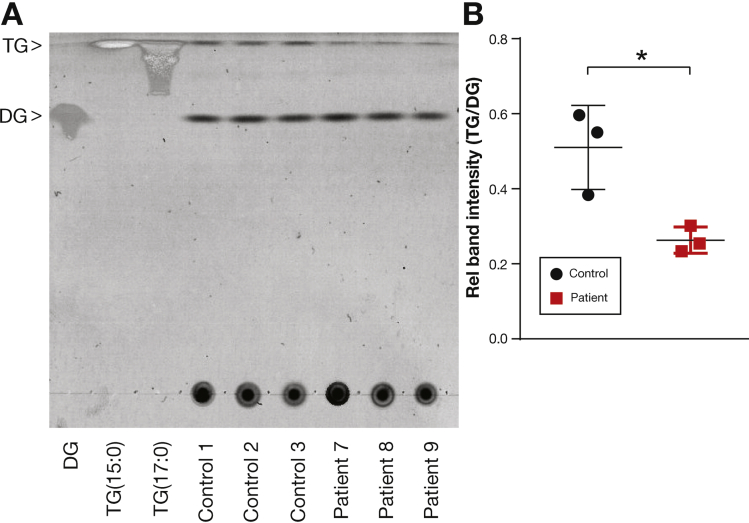
To substantiate the specificity of DGAT1 deficiency for causing aberrant TG formation, we performed thin layer chromatography to measure the levels of TG and DG in organoids derived from 3 healthy controls and patients 7, 8, and 9 (Figure 5A). By quantification of respective DG and TG band intensities, we determined that the TG/DG ratio in healthy control organoids was significantly higher compared with patient-derived organoids (Figure 5B). These results indicate a DGAT1-dependent loss of TG synthesis in patient-derived intestinal organoids, whereas levels of DG were comparable.
Figure 5.
Loss of DGAT1 results in decreased triglyceride (TG) formation. (A) Organoids from 3 healthy controls and patients 7, 8, and 9 were grown in EM for 7 days. Lipid extracts were isolated and run by thin layer chromatography to determine levels of diacylglycerol (DG) and triglyceride (TG). Relative positions were indicated by reference samples for DAG and TG. (B) Ratio of intensity of TG over DAG for each sample. Statistical analysis was done using a Mann-Whitney U test. Mean ± SD is indicated, ∗P < .05.
Loss of DGAT1 Results in Increased Sensitivity to Lipid-induced Toxicity
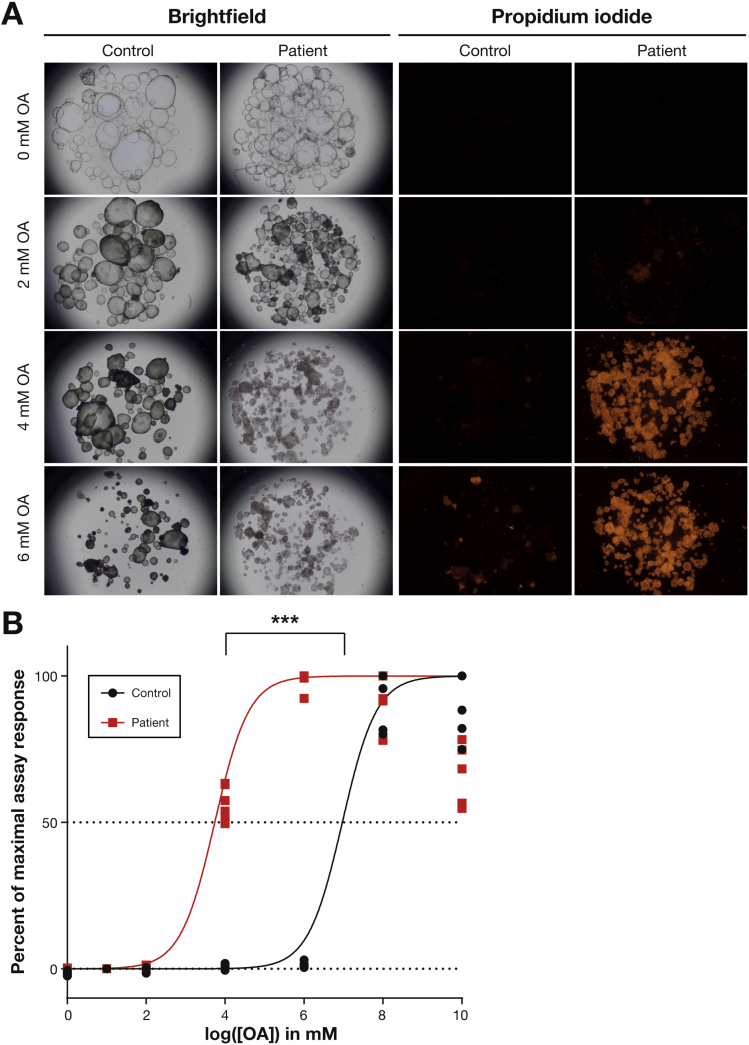
Most DGAT1-deficient patients reported in this study suffered from PLE after ingestion of dietary lipids, which can be the result of either intestinal mucosal injury or lymphatic abnormalities.28 To assess whether exposure to lipids leads to mucosal injury in DGAT1-deficient patients, we performed lipotoxicity assays in healthy control and DGAT1-deficient organoids. We incubated the organoids with varying concentrations of BSA-coupled OA in EM and assessed cell death by brightfield microscopy and propidium iodide staining (Figure 6A). Remarkably, we observed 100% cell death at 4 mM OA in patient-derived organoids, whereas cell death was still almost absent at 6 mM OA in control organoids. To further quantify these findings, we determined lipid-induced caspase-mediated cell death using a Caspase-Glo 3/7 assay. We show that DGAT1-deficient cells are more sensitive to lipotoxic stress compared with healthy control organoids and undergo programmed cell death on treatment with OA (Figure 6B). By nonlinear regression analysis, the median lethal dose was determined to be approximately 7 mM OA for healthy control organoids and 4 mM OA for patient-derived organoids (P < 0.001). Overall, these results indicate that DGAT1-deficient organoids are more susceptible to lipid-induced toxicity, which may reflect the clinical feature of PLE in DGAT1-deficient patients that occurs on ingestion of fat.
Figure 6.
Loss of DGAT1 results in increased sensitivity to lipotoxic stress. Organoids of 3 healthy controls and 3 patients (7–9) were grown in EM and incubated overnight with a range of oleic acid (OA) concentrations. (A) Representative images showing brightfield and propidium iodide staining for cell death of the organoids after incubation with OA. (B) Caspase-Glo 3/7 assay for apoptotic cells after incubating organoids with OA. Samples were normalized for vehicle control values and the maximum value for each sample was set to 100% assay response. Median lethal dose (LD50) was calculated by regression analysis. Statistical analysis on LD50 values was done using a Student’s t test, ***P ≤ .001.
Discussion
Lipid metabolism is an important physiological function within the human body that includes the digestion and absorption of lipid products from food. Inborn errors of lipid metabolism can result in a wide range of symptoms, from neurological impairment to hypertriglyceridemia. Recently, lipid metabolism disorders have been linked to CDD, such as in the case of Niemann-Pick disease type C with inflammatory bowel disease due to impaired autophagy.29 In the case of cytoplasmic TG metabolism disorders, none of the other deficiencies have been reported to develop any GI phenotype.30
Previous studies on DGAT1 deficiency have identified a total of 3 distinct homozygous and 2 compound heterozygous mutations in DGAT1.3, 4, 5, 6 These patients suffered from severe congenital diarrhea and PLE, clinical features that are shared with most of our patient cohort. Although this shared phenotype further confirms the involvement of DGAT1 in intestinal failure, limited functional data or potential therapeutic options have been reported thus far.
Currently, there is no genotype-phenotype correlation in DGAT1 deficiency, as patients develop varied clinical history ranging from complete resolution of GI symptoms to a lethal course of disease. As previously described and also observed in our study, discordant phenotypes associated with identical mutations in siblings and in different, unrelated families are of interest. Whether this phenomenon, as well as the novel clinical features, such as normal serum TG and hepatic involvement, observed in our patients indeed extend the spectrum of the clinical phenotype of DGAT1 deficiency, or are unspecific secondary effects of treatment, cannot be inferred from our study. By analogy, the unsatisfactory clinical outcome described for patient 9 after the small bowel transplantation may be attributed to known posttransplant complications rather than unsuccessful correction of the DGAT1 deficiency in the intestine.
To provide further insight into the pathomechanism of DGAT1 deficiency, we have produced cell models that recapitulate an altered lipid metabolism in vitro. Here, we show that patient-derived organoids can recapitulate the molecular pathomechanism of DGAT1 deficiency. We have demonstrated aberrant lipid metabolism as evidenced by reduced lipid droplet and TG formation on incubation with OA in both patient-derived organoids and fibroblasts. In addition, we show for the first time that DGAT1-deficient cells are more susceptible to lipid-induced toxicity, which provides a plausible explanation for the clinical PLE symptoms in DGAT1 deficiency and possibly other forms of PLE, such as primary intestinal lymphangiectasia.31 The fact that dietary fat restriction can even restore normal fecal protein clearance in PLE further supports the concept that cellular lipotoxicity may be one of the driving forces for ongoing fecal protein loss in untreated PLE. Whether this lipotoxicity-induced enterocyte dysfunction is caused by endoplasmic reticulum stress or induced autophagy, which has been implicated in lipid metabolism disorders such as Niemann-Pick disease type C with inflammatory bowel disease,29 remains to be determined. Moreover, elucidation of the precise role of DGAT1 in common lipotoxicity-related diseases, such as type 2 diabetes, nonalcoholic fatty liver disease, and metabolic cardiomyopathy, may be the first step toward new therapy strategies for obesity-related disorders.
Of note, the novel technology of patient-derived intestinal organoids provided an unprecedented look into pathobiology of the cells in the GI tract. In the future, a more systematic use and collection of organoids from potential DGAT1 deficiency and other patients with intestinal failure will allow for better dissection of the disease mechanism involving the gut epithelium.
As described in this study and in previous literature, DGAT1-deficient patients usually do well on a fat-restricted diet. An early introduction of such a diet might even have prevented the development of a full-blown PLE in patients 7 and 8. In addition, the administration of short chain fatty acid proves to be a good supplement to the diet for some patients. Intravenous administration of essential fatty acid was also well-tolerated, presumably as it bypasses absorption through the gut epithelium as well. Whether patients benefit from novel ways of treatment with available drugs, such as cholestyramine and pancreas enzymes, as empirically applied for patient 4 and patients 5 and 6, respectively, would have to be attempted and evaluated in additional patients and in carefully designed trials. Before treatment, neither diagnosis of bile acid diarrhea nor exocrine pancreas insufficiency was formally tested or firmly established, because the observation of slightly decreased fecal elastase levels may be unspecific due to fecal dilution in severe chronic diarrhea. Although the course of disease evidently can be mild, early diagnosis of DGAT1 remains vital, as all patients were severely ill during the first few months of life and thus far one patient suffered from a lethal course of disease. Additionally, the impact on the quality of life during chronic treatment (monthly infusions or through a central line that includes risks of infections) and the variable genotype-phenotype relationships of DGAT1 patients should not deter the search for new therapeutic options for these patients.
In pursuit of possible therapeutic strategies, we determined that proteasome inhibitors could provide a potential therapeutic option in case of proteasome-mediated protein degradation, as shown for the mutation in families 5 and 6 (Supplementary Figure 4). In addition, we determined that DGAT2 might be able to compensate for the lack of DGAT1 function. DGAT2 is an isozyme of DGAT1 that does not share any homology in protein sequence or domains.11 However, DGAT2 shares functional characteristics with DGAT1, catalyzing the formation of TG from DG and fatty acyl-CoA in the liver. In this study, we show that DGAT2 might be able to compensate for this phenotype, as shown by OA addition on DGAT2-expressing B-LCL and exogenous expression of DGAT2 in DGAT1-deficient fibroblasts. A single heterozygous, autosomal dominant DGAT2 mutation has been described in a family with Charcot-Marie-Tooth syndrome,32 but no GI involvement was reported, possibly because DGAT2 is not highly expressed in the human intestine.3 Therapeutic strategies to induce DGAT2 expression might potentially provide an additional, viable treatment strategy in DGAT1 deficiency.
Previous studies performed with Dgat1−/− mice implicated a beneficial role of DGAT1 inhibition in obesity through changing metabolic landscapes.33, 34, 35 These studies resulted in the development of DGAT1 inhibitors for use in human obesity.36 However, participants in a clinical trial of DGAT1 inhibitor Pradigastat developed side effects of diarrhea and nausea,37, 38 similar to the phenotype of DGAT1 deficiency. Dgat1−/− mice did not develop a GI phenotype, presumably due to the intestinal expression of DGAT2 and diacylglycerol transacylase, which might compensate for the lack of DGAT1.39 Our data suggest that in humans, lack of TG formation and packaging is detrimental in the context of GI epithelium, and that a serious note of caution on the clinical use of DGAT1 inhibitors should be provided. This finding accentuates the lack of available knowledge of intestinal lipid uptake and metabolism, and emphasized how the use of clinically relevant in vitro systems can help to fill this gap.
In summary, we here described a large cohort of DGAT1-deficient patients, and extended our knowledge of the pathomechanism of DGAT1 deficiency. Our findings expand the differential diagnosis of vomiting and congenital diarrhea in neonates. Clinicians should maintain a high suspicion for DGAT1 deficiency in cases of unexplained vomiting, especially when associated with failure to thrive and PLE, and could consider a fat-free diet with supplementation of essential fatty acids and fat-soluble vitamins as a first line of therapy. In conclusion, the findings described in this article show that DGAT1 mutations not only cause congenital diarrhea and PLE, but are also linked to fat intolerance.
Acknowledgments
We thank Theresa Waidacher, Imre Schene, Nicola Fenderico, and Bon-Kyoung Koo for technical and material assistance during the project. We thank Tatjana Hirschmugl for the graphical abstract of this manuscript.
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This work was supported by a DOC Fellowship of the Austrian Academy of Sciences (24486) to Rico Chandra Ardy, OeNB Jubiläumsfonds (16678) to Thomas Müller, The Netherlands Organisation for Scientific Research (NWO-ZonMW; VIDI 016.146.353) to Sabine Middendorp, and the European Research Council (ERC StG 310857) to Kaan Boztug.
Author names in bold indicate shared first authorship.
Note: To access the supplementary material accompanying this article, visit the online version of Gastroenterology at www.gastrojournal.org, and at https://doi.org/10.1053/j.gastro.2018.03.040.
Contributor Information
Sabine Middendorp, Email: s.middendorp@umcutrecht.nl.
Kaan Boztug, Email: kaan.boztug@rud.lbg.ac.at.
Supplementary Clinical Description of the 10 Patients
Patient 1 presented with watery, nonbloody, nonmucoid diarrhea that commenced immediately after birth. He was severely dehydrated; breast milk was discontinued and changed to lactose-free formula, but diarrhea persisted. A diagnosis of microvillus inclusion disease (MVID) was considered after endoscopic biopsy. At 25 days of age, patient 1 was admitted to another hospital, where he required recurrent albumin infusions (AIs) to correct the hypoalbuminemia. Immunological tests were normal except for slight decrease in immunoglobulin (Ig)G and IgM. Fat-soluble vitamins A, D, and E levels were low. Stool steatocrit was positive. Infection, food allergy, cystic fibrosis, and prohormone convertase deficiency had been excluded by appropriate laboratory tests. A second endoscopy and colonoscopy were performed, and the previous endoscopic biopsy was reanalyzed. Congenital enteropathies (Tufting enteropathy, MVID, inflammatory bowel disease, autoimmune enteropathy, and food-protein–induced enteropathy) were excluded. Treatment with amino acid–based formula was not effective, but a slight clinical improvement was observed with basic casein hydrolysate and galactomin 19 formula. He was discharged at 4.5 months of age with some weight gain. Diarrhea and vomiting persisted after his discharge at home, and 2 weeks later, he was referred to the current attending physician for dehydration, weight loss (470 g over 15 days), severe hypoalbuminemia (1.8 g/dL), hyponatremia (130 mEq/L), and hypokalemia (3 mEq/L). On admission, he was malnourished (weight 3680 g [<3%], height 58.5 cm [3%]) and severely dehydrated. Fluid replacement therapy, AI, and total parenteral nutrition (TPN) were started. Laboratory tests showed low serum IgG 1.53 g/L (3.04–12.31), total cholesterol 63 mg/dL, high-density lipoprotein (HDL) 25 mg/dL, low-density lipoprotein (LDL) 12 mg/dL, very low-density lipoprotein (VLDL) 27 mg/dL, triglyceride 135 mg/dL, and vitamin B12 101 ng/mL and steatorrhea was observed. Workup included extensive infectious, immunologic, and hormonal studies, all of which were negative or normal. Upper gastrointestinal endoscopy and ileocolonoscopy, biopsies, and electron microscopy were normal. During his hospitalization, bulky, watery, and greasy diarrhea was observed, and Sudan stain of stool continuously revealed massive droplets of fat. Basic-F (fat-free formula [FFF]) was started for fat malabsorption, and within 3 weeks, a dramatic improvement and subsequent complete resolution of his symptoms was observed and further TPN was not needed. Extensively hydrolyzed formula rich in medium-chain triglyceride (MCT) was gradually added to the FFF. He was discharged at 7 months of age with a combination of these formulas. During the follow-up, solid food with daily MCT oil supplementation was commenced gradually. The child exhibited elevated fasting triglyceride level (207 mg/dL) at 8 months of age, which was decreased with adding omega-3 fatty acids. Due to recurrent episodes of vomiting, cough, and otitis media during follow-up, long-term prophylactic antibiotic with sulfamethoxazole-trimethoprim was added at 12 months of age. He is currently 4 years and 4 months old, with normal growth and development with the combination of low-fat–age-appropriate diet, MCT supplementation, and FFF.
Patient 2, a sibling of patient 1, also presented with nonbilious vomiting and watery, nonbloody, nonmucoid diarrhea 2 to 3 times a day and required repeated hospitalizations for dehydration due to diarrhea. At 5 months, she was referred to a tertiary hospital due to failure to thrive, peripheral edema, and severe hypoalbuminemia (1.8 g/dL, reference 3.2–5.0 g/dL). The stool was negative for leukocytes. Standard laboratory tests, including urinalysis; acute-phase reactants; serum IgA, IgM, and IgE; serum and urine amino acids; artery blood gases; tandem mass spectroscopy; and thyroid function tests were normal. Serum IgG level was slightly reduced (204 mg/dL, reference 304–1230 mg/dL). Serum lipid profile was normal. Triglyceride level was 84 mg/dL (reference <150 mg/dL), total cholesterol 62 mg/dL (reference <170 mg/dL), LDL 28 mg/dL (reference <110 mg/dL), HDL 18 mg/dL (reference 40–60 mg/dL), VLDL 16 mg/dL (reference <30 mg/dL). Upper gastrointestinal endoscopy and biopsy were normal. She required recurrent AIs to correct the hypoalbuminemia and died at 6 months of age due to sepsis.
Patient 3, a female infant with lack of dysmorphic features, was born via vaginal delivery at 39 weeks of gestation following an uneventful prenatal history, weighing 3500 g, measuring 48 cm body length and 35 cm head circumference at birth. The healthy Turkish parents were not aware of a consanguineous relationship but share geographical origin. This small village was founded by the children of 3 siblings, collectively indicating parental consanguinity. A sister of the patient’s father died at the age of 11 months from vomiting and diarrhea. Apart from that, family history was inconspicuous. Because of watery diarrhea (7 times a day, 150 g/kg per day) starting at the age of 3 weeks, weight loss, and severe dystrophy, she was admitted to a pediatric clinic weighing 3400 g. Parenteral nutrition was immediately started and the patient was also continuously fed through a nasogastric catheter. Stool analyses showed normal values for α1 antitrypsin (13 mg/dL, reference 90–200 mg/dL), fecal elastase (>500 μg/dL, reference >200 μg/g), stool sodium (66 mmol/L, reference 20–25 mEq/L) and potassium (31.7 mmol/L, reference 50–75 mEq/L), excluding congenital sodium and chloride diarrhea. Serum bile acids were mildly increased (38,9 μmol/L, reference <10 μmol/L), amylase 7 U/L (reference <30 U/L) and lipase 13 U/L (reference 13–60 U/L) were normal. Unfortunately, no information on fecal bile acid levels is available. The patient exhibited hypoproteinemia (3.5–4.7 g/dL, reference 5.7–8.7 g/dL) and hypoalbuminemia (2.2–2.7 g/dL, reference 3.2–4.7 g/dL). Metabolic screening was without pathological findings. Stool cultures as well as polymerase chain reaction (PCR) for adenovirus, rotavirus, and enterovirus in the stool were negative. Serum immunoglobulin levels were normal. Food mix allergen-specific IgE in serum was negative. A gastroduodenoscopy performed at the age of 3 months showed duodenal microvillus atrophy. Light microscopy analysis of the biopsy of the duodenum excluded enteric anendocrinosis. Electron microscopy revealed a deteriorated integrity of the microvilli, and no microvillus inclusions (Supplementary Figure 1). The patient continuously failed to thrive, and at the age of 11 months, kidney stones of several millimeters in diameter were found, along with lower-extremity edema, corneal cystine crystal accumulation, and metabolic acidosis. Renal function tests and urine analyses were repeatedly normal, and leukocyte cysteine levels were normal with 0.047 nmol/mg protein (0–0.3 nmol/mg). The patient received 1 g/kg albumin every other day due to protein-losing enteropathy and decreasing albumin values. She was fed via a nasogastric tube on elemental and semi-elemental formula. Additionally, parenteral nutrition (protein 2 g/kg per day and lipid 1 g/kg per day) was administered. A second endoscopy of the upper gastrointestinal tract was performed due to persisting diarrhea. Because milky deposits (lipid accumulation) were seen in the duodenum, chylomicron retention disease and abetalipoproteinemia were considered. The patient's triglyceride levels were increased (up to 370 mg/dL [0–150 mg/dL]), whereas total cholesterol was in the normal range (96 mg/dL [0–200 mg/dL]) and HDL and LDL were decreased (HDL 9.5 up to 22 mg/dL [40–60 mg/dL], LDL 23 mg/dL [60–130 mg/dL]). The patient’s parents’ lipid profiles were normal.
At this point, whole-exome sequencing established the diagnosis of a congenital protein-losing enteropathy due to DGAT1 mutation. Subsequently, oral MCT administration was started. Parenteral nutrition support continued. Subsequent to observing hypocalcemia, secondary hyperparathyroidism was diagnosed (parathyroid hormone: 312 pg/mL, reference 15–65 pg/mL, 25-hydroxyvitamin D 18 ng/mL, reference 25–80 ng/mL, calcium 6.5 mg/dL, reference 8.7–10.4 mg/dL), and vitamin D and calcium supplementation was initiated. The patient is now 13 months old. The stool amount declined from 150 g/kg per day during the first application to 100 g/kg per day in the past 3 months. The stool frequency decreased from 7 times a day to 3 to 4 times a day. She weighs 3850 g (<3 percentile), is 57 cm (<3 percentile) long, and has a head circumference of 38 cm (<3 percentile). She vomits 3 to 4 times every day, has extreme flatulence and abdominal distention. Parenteral nutrition support continues. For this purpose, the patient is hospitalized for 3 to 4 weeks and then she has a maximum break of 1 week without parenteral nutrition. Her psychomotor development is compatible with 3 to 4 months.
Targeted gene sequencing had excluded infantile nephropathic cystinosis, and whole-exome sequencing excluded microvillus inclusion disease and variants in other known genes causing isolated and syndromic forms of congenital diarrheas.
Patient 4 was referred to the attending physician due to intractable diarrhea, which started when he was 2 months old. Intestinal and colonic endoscopic and histopathological investigations were normal. Infectious, metabolic disorders, malignancies, cystic fibrosis, and congenital glycosylation defect were excluded. Longitudinal measurement of lipid profile showed normal levels of total cholesterol, LDL, HDL, VLDL, and triglyceride. Serum immunoglobulin levels were normal. He had marked clinical improvement on treatment with cholestyramine, which resulted in the reduction of stool volume and frequency. At the latest follow-up, the patient weight is at 9.5% and height is at 4.5% of Turkish boys. He does not have any vomiting or diarrhea and is still on cholestyramine treatment.
Patient 5 was born at term to consanguineous parents. He was hospitalized at 40 days of age because of vomiting, bloody diarrhea, and failure to thrive. He was started on amino acid–based formula due to a suspected cow milk allergy. At the age of 5 months, abdominal and cranial ultrasonography were normal and upper gastrointestinal endoscopy and biopsy revealed nonspecific results. He had low IgA, IgM, and IgG levels. At a follow-up, the patient was found to have low serum albumin (2.2g/dL, normal 3.2 g/dL) and total protein. At 1 year of age, he was referred to the current attending physician because of hepatomegaly and jaundice for a liver biopsy. He is below the third percentile in height and weight. Physical examination revealed abdominal distention, cutis marmoratus, 8 cm length of liver below the right costal margin. Laboratory assessment of liver function and immunoglobulin levels were as follows: aspartate aminotransferase 240 U/L, alanine aminotransferase 111 U/L, γ-glutamyltransferase 335 U/L, total/direct bilirubin levels 2.95/2.05 mg/dL, serum IgA 1.59 g/L, IgM 1,14 g/L, IgG 9.14 g/L. He has watery stools 4 to 5 times per day. He also has milk protein intolerance (bloody diarrhea occurs after formula feeding). Liver biopsy revealed hepatocanalicular and ductular cholestasis, paucity of bile ducts, mixed type hepatosteatosis (10%), porto-portal fibrosis, and fibrotic activity of 3/6. His daily stool frequency had reduced with the administrations of pancreatic enzymes (Creon), but his weight and height are below the third percentile.
Patient 6, a sibling of patient 5, was admitted to hospital when he was 2.5 months of age because of watery diarrhea (at least 10 times a day). He presented hyponatremia, hypochloremia, hypoalbuminemia, hypocalcemia. Sweat chloride test was normal. His daily stool frequency had reduced with the administrations of pancreatic enzymes (Creon), but he had recurrent infections and was hospitalized. He was also placed on amino acid–based formula. Endoscopy and colonoscopy was normal at the age of 2 years. Duodenum biopsy revealed focal vacuolization at one area and partially blunted villi. Colon biopsy was normal. Liver profile: aspartate aminotransferase 149 U/L, alanine aminotransferase 56 U/L γ-glutamyltransferase 70 U/L. Total serum protein was normal (4.38 g/dL), but low albumin level (2.59 g/dL) was observed. He has low IgA serum level but normal IgG and IgM levels. Free thyroxine–thyroid stimulating hormone levels were normal. His lipase and amylase levels were within normal range (13/46 U/L respectively). Lipid profile revealed low HDL 21 mg/dL (40–60 mg/dL), LDL 14 mg/dL (<130 mg/dL), VLDL 36 mg/dL (<40 mg/dL), triglyceride 184 mg/dL (<200 mg/dL), and normal total cholesterol 59 mg/dL (<200). Vitamin E level was deficient, but vitamin A and D levels were normal. He had bilateral nephrocalcinosis on abdominal ultrasonography. Urine organic acid analysis and serum amino acid chromatography were normal. After 2 years of age, his symptoms improved spontaneously, and he was lost to follow-up afterward. His weight is now 16 kg (<third percentile), height 104 cm (<third percentile).
Family 5 is a consanguineous Dutch family, with the parents sharing ancestors 7 generations ago, and consists of 3 brothers of which 2 were affected (patients 7 and 8). The oldest boy (patient 8) presented in the first month of life with vomiting and failure to thrive. Physical examination showed a dystrophic boy without further abnormalities, and a normal serum albumin (4.35 g/dL). A scintigraphy was performed, which showed delayed gastric emptying. Extensive additional research did not reveal a diagnosis. Initially the patient was treated with TPN, which reversed the symptoms. By experiment, a fat-restricted diet was introduced at the age of 7 months, which enabled enteral feeding again. Incidental ingestion of low amounts of fat causes abdominal pain and vomiting within 1 hour after ingestion. The boy currently receives bimonthly infusions of Intralipid and Omegaven and supplementation of fat-soluble vitamins, leading to normal plasma levels of fat-soluble vitamins and fatty acids, except for linoleic acid, which is just below normal. He was diagnosed with Gilles de la Tourette syndrome at the age of 7 years, which was treated with dexamphetamine. The younger brother (patient 7) presented with vomiting in the first days of life, which was reversed after immediate introduction of a fat-restricted diet. Like his brother, he is supplemented with fat-soluble vitamins and monthly infusions of Intralipid and Omegaven, leading to normal plasma levels of fat-soluble vitamins and fatty acids, except for marginally decreased levels of linoleic acid. He was also diagnosed with Gilles de la Tourette syndrome.
Family 6 is a Dutch family of unknown consanguinity, which consists of a monozygotic female twin pair and a male sibling of which the twin pair was affected, patients 9 and 10. They were born after a pregnancy of 34 6/7 weeks. Patient 9 had a birth weight of 2540 g and a bilateral clubfoot. During the first week of life, she developed severe watery diarrhea on bottle feeding. This subsided after a short period with TPN after which she tolerated full enteral feeding on Neocate for some months. However, the diarrhea returned and was accompanied by severe vomiting, also after small amounts of oral feeding. Therefore, enteral feeding was stopped and she was totally dependent on TPN. Protein-losing enteropathy (PLE) was present with a high fecal alpha-1-antitrypsin (9.1 mg/g feces, reference 0.0–1.1 mg/g feces) and a low serum albumin (23.8 g/L, reference 36–42 g/L). Microscopy of the small intestinal mucosa showed minimal villus atrophy, with generally a normal brush border on periodic acid-Schiff staining, but with unspecific CD10-positive globules in the cytoplasm of the enterocyte. As numerous microvilli were seen on the lateral membranes of the enterocyte, a misdiagnosis of variant microvillus inclusion disease was made. Given this diagnosis, and the dependence on TPN, she received a small bowel transplantation at the age of 3.5 years. After the small bowel transplantation, TPN was stopped and polymeric enteral nutrition was tolerated. Her clinical course after small bowel transplantation was complicated by post transplantation lymphoproliferative disease, autoimmune problems, and recurrent (pulmonary) infections. Currently, she is malnourished and still stunted despite a high caloric diet with a polymeric formula for which we started additional parenteral nutrition. After small bowel transplantation, it is sometimes difficult to unequivocally define the origin of failure to thrive, which could be related to at least partially ongoing disease, side effects of immunosuppression, or difficulties in reestablishment of sufficient enteral nutrition necessary to promote catch-up growth. Clinically, we do not have hints for persisting PLE following partial correction of intestinal DGAT1 deficiency due to isolated small bowel transplantation, and intestinal biopsies did not show signs of chronic rejection.
The identical twin sister, patient 10, had a birth weight of 1745 g without any congenital abnormalities. She had the same clinical course as her sister. She also developed severe watery diarrhea on bottle feeding in the first week of life, which subsided on TPN. Subsequently she was fed with Neocate for some months. As her twin sister, she presented with watery diarrhea and vomiting at the age of 5 months, for which enteral feeding was stopped and she was totally dependent of TPN. Results of laboratory investigations were very similar to her sister, with a PLE and an intestinal histology that suggested the diagnosis of atypical MVID. She was also put on the waiting list for small bowel transplantation. However, during the waiting list period (and after the transplantation of her sister) enteral feeding was successfully reintroduced with carbohydrates (first) and proteins (secondly), whereas the introduction of fats was complicated by motility problems (vomiting). At first, she did not tolerate MCT ingestion at all (vomiting directly after ingestion), but MCT was tolerated within months. Long-chain triglycerides were tolerated only in small amounts and oral supplementation of docosahexaenoic acid and amino acids were tolerated. After 4 months, she was not dependent on TPN anymore and she was taken off the waiting list for small bowel transplantation. Her diet consisted of a mixture of Hydrolyzed Whey Protein/Maltodextrin mix, Fantomalt, Liquigen, Calogen, Phlexy-vits, and Key Omega. We tried to switch to a standard oligomeric feeding formula after 2 years, but she did not tolerate this. Although she tolerated enteral feeding, she was still stunted and PLE was still present. For this reason, we recently started a fat-free diet with enteral supplementation of fat-soluble vitamins and monthly intravenous infusions of SMOF lipids.
Supplementary Materials and Methods
Cell Culture
Crypts were isolated from biopsies as described previously1 and resuspended in medium without growth factors (GF), consisting of Advanced Dulbecco’s modified Eagle’s medium/F12 (Gibco, Waltham, MA), 100 U mL−1 penicillin-streptomycin (Gibco), 10 mM HEPES (Gibco), and Glutamax (Gibco). Matrigel (Corning, Tewksbury, MA) was added to a final concentration of 70% and plated on prewarmed cell culture 24-well plates (Corning). After matrigel polymerization, organoid culture medium (human small intestine expansion medium [hSI-EM]) was added consisting of GF-medium, 50% Wnt-conditioned medium, 20% Rspondin-1-conditioned medium, 10% Noggin-conditioned medium, 50 ng/mL murine epidermal growth factor (PeproTech, Rocky Hill, NJ), 10 mM nicotinamide (Sigma), 1.25 mM N-acetyl (Sigma), B27 (Gibco), 500 nM TGF-α inhibitor A83–01 (Tocris, Bristol, UK), 10 μM P38 inhibitor SB202190 (Sigma), and 100 μg/mL Primocin (InvivoGen, San Diego, CA). During experiments, Primocin was left out. Organoids were cultured in 37°C and 5% CO2 and medium was refreshed every 2 to 3 days. The organoids were passaged 1:3 to 1:6 every week after mechanical disruption or 1:10 to 1:20 as single cells (Trypsin/EDTA or TrypLE Express). For single cells, 10 μM ROCK inhibitor Y-27632 (Abcam, Cambridge, UK) was added for the first 2 to 3 days of the culture. To induce differentiation, organoids were cultured in differentiation medium (hSI-DM, which is hSI-EM lacking WNT3A, nicotinamide, and SB202190) for 5 days, as described previously.1
Immunohistochemistry
Biopsies obtained from patients and siblings were either freshly or previously mounted onto glass slides. Staining was done using a standard immunohistochemistry protocol with the Lab Vision UltraVision LP Detection System: HRP (horseradish peroxidase) Polymer (Ready-To-Use) (TL-060-HL; Thermo Scientific, Waltham, MA) DGAT1 protein was detected using a rabbit polyclonal IgG (H-255; Santa Cruz Biotechnology, Inc., Dallas, TX) at 1:400 dilution.
CRISPR/Cas9 Knockout of DGAT1
Corresponding DNA-oligos were ligated into the plasmid vector pSpCas9(BB) (Addgene PX459). Before transfection, the organoids were primed by removing WNT3A and Rspondin-1 and addition of 5 μM CHIR99021, 1.25% dimethyl sulfoxide, and 10 μM Y-27632 (ROCK inhibitor) to the culture medium as described previously.2 For transfection, 1 × 106 single cells in 80 μL BTXpress buffer (BTX Genetronics, Holliston, MA) were mixed with 10 μL DGAT1 sgRNA construct plasmid (1 μg/μL), 7.2 μL PB-Hygromycin plasmid (1 μg/μL), and 2.8 μL PB-Transposase plasmid (1 μg/μL). Transfection was performed in a NEPA21 Electroporator with settings as described previously.2 After transfection, priming medium was used for 5 days before replacement by normal hSI-EM and selection with Hygromcyin B (100 μg/mL; Invitrogen, Carlsbad, CA). The organoids were passaged 1:4 by mechanical disruption at day 9. Single organoids were picked 16 days after transfection, disrupted into single cells by trypsinization with TripLE, and scaled up as clonal organoid cell lines. Several clones per transfection were screened for DGAT1 protein expression by Western blot (Supplementary Figure 6C). Organoids transfected with sgRNA#2 achieved 10/12 DGAT1KO clones, whereas organoids transfected with sgRNA#678 achieved 3/11 DGAT1KO clones. Finally, 3 DGAT1KO clones were further characterized by genotyping (Data not shown). Clone DGAT1KO #2–11 showed a deletion of 280 base pairs (bp) on both alleles (NG_034192.1; 13347–13627). Clone DGAT1KO #678–02 showed a deletion of 83 bp on both alleles (NG_034192.1; 13487–13570). Clone DGAT1KO #678–07 showed a deletion of 83 bp on one allele (NG_034192.1; 13487–13570); the other allele has a deletion of 14 bp (NG_034192.1; 13485–13499) and an insertion of 1 bp (NG_034192.1; T between 13486–13487) (Supplementary Figure 6B).
Reverse-Transcriptase Polymerase Chain Reaction
Protein synthesis inhibitor puromycin in a concentration of 20 mg/12 mL was added to patient 3 and control fibroblast culture 4 hours before harvesting for total RNA extraction. Total RNA was extracted using the Qiagen RNAeasy mini kit (Qiagen, Hilden, Germany). RNA was reverse transcribed into complementary DNA (cDNA) by a standard protocol using M-MLV reverse transcriptase (Promega, Madison, WI). cDNA was PCR-amplified using GoTaq polymerase and buffer (Promega) in 35 cycles of 20 seconds at 95°C, 30 seconds at 60°C, and 30 seconds at 72°C in addition to a 7-minute final extension. The amplicons were cleaned using Microcon centrifugal filter for agarose gel–excised PCR products, and subsequently sequenced using forward (gtaaaacgacggccagtGTGGACCCCATCCAGGTGG) and reverse (caggaaacagctatgacTGAGCCAGATGAGGTGATTGG) primers and run on an agarose gel.
Quantitative Real-Time Polymerase Chain Reaction
RNA was isolated from Caco-2 cells or organoids grown in either EM or DM for 5 days using the RNeasy Mini kit (Qiagen) or TRIzol LS Reagent (Invitrogen), respectively, according to the manufacturer’s protocol. cDNA was synthesized using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA) and amplified using SYBR green supermix (Bio-Rad) in a Light Cycler96 (Roche, Basel, Switzerland) according to the manufacturer’s protocol. The comparative Ct method was used to quantify the data. The relative quantity was defined as 2−ΔΔCt. In organoids and Caco-2 cells, HP1 and HPRT1 were used as housekeeper genes, respectively. Sucrase-isomaltase (SI) and LGR5 were used to determine the stem cell vs differentiation status of organoids. Primers used were DGAT1-forward (FW): cgacgtgggagccgc, DGAT1-reverse (RV): gctcaagatcagcatcacca, HP1-FW: cccacgtcccaagatggat, HP1-RV: ctgatgcaccactcttctggaa, SI-FW: ggacactggcttggagacaac, SI-RV: tccagcgggtacagagatgat, LGR5-FW: gaatcccctgcccagtctc, LGR5-RV: attgaaggcttcgcaaattct, HPRT1-FW: tgacactggcaaaacaatgca, HPRT1-RV: ggtccttttcaccagcaagct.
Western Blotting
Caco-2 cells or organoids were lysed in Laemmli buffer (0.12 mol/L Tris-HCl pH 6.8, 4% sodium dodecyl sulfate [SDS], 20% glycerol, 0.05 μg/μL bromophenol blue, 35 mmol/L β-mercaptoethanol) and incubated at 100°C for 5 minutes. Fibroblasts were lysed in RIPA buffer (50 mM HEPES, 150 mM NaCl, 10% glycerol, 1% Triton-X100, 2 mM EDTA, 1% sodium deoxycholate, 50 mM NaF, 10 mM NaO3V4) supplemented with protease inhibitor cocktail (1:200, P8340; Sigma-Aldrich, St Louis, MO) then supplemented with Laemmli buffer. The protein concentration was measured by performing a Lowry or Bradford protein assay. Equal amounts of proteins were separated by SDS–polyacrylamide gel electrophoresis, transferred to a polyvinylidene difluoride membrane (Merck Millipore, Billerica, MA), blocked with 5% milk protein in TBST (0.3% Tween, 10 mM Tris-HCl pH 8, and 150 mM NaCl in H2O) and probed with primary antibodies. The membranes were washed with TBST and incubated with appropriate secondary antibodies. Immunocomplexes were detected using the LI-COR (Lincoln, NE) Odyssey. Caco-2 cells transfected with Flag-DGAT1 c.629_631delCCT were left untreated or treated with 2 μM MG132 (Cayman Chemicals, Ann Arbor, MI) for 16 hours.
The primary antibodies used were mouse anti-Flag M2 (1:3000; F3165; Sigma-Aldrich), rabbit anti-DGAT1 (1:1000; ab181180; Abcam), rabbit anti-DGAT1 (1:1000, sc-32861; Santa Cruz Biotechnology), rabbit anti-DGAT2 (1:1000, bs-12998R; Bioss Antibodies, Woburn, MA), mouse anti-GAPDH (1:3000, sc-32233; Santa Cruz Biotechnology), anti-HA-HRP conjugate (1:3000, H6533; Sigma-Aldrich), and rabbit anti-HSP90 (1:10000; kindly provided by Professor L.J. Braakman). The secondary antibodies used were donkey anti-mouse IgG IRDye 680 (1:10000; 926-32222; LI-COR) and donkey anti-rabbit IgG IRDye 680RD (1:10000; 926-68073; LI-COR), goat anti-mouse IgG-HRP (1:15000, 554002; BD Biosciences), and goat anti-rabbit IgG-HRP (1:15000, 172-1019; Bio-Rad).
Lipid Droplet Assays
Oleic acid (OA) was conjugated to bovine serum albumin (BSA) by heating the OA in phosphate-buffered saline (PBS) to 70°C for 1 hour. The OA suspension was then thoroughly vortexed and slowly added to a solution of fatty acid–free BSA in PBS kept at 37°C in a molar ratio of OA:BSA (5:1) to a final stock concentration of 10 mM OA and 2 mM BSA.
Thin Layer Chromatography
Folch extraction was performed by harvesting organoids, washed in PBS and spun down, and the cell pellets were frozen in ice-cold methanol on dry ice to stop cell metabolism. Last, the cell pellets were homogenized using a bullet blender, using glass bullets of 0.1-mm diameter for 2 minutes. Folch extraction was performed similar to the original protocol.3 In short, chloroform and dH2O were added to the cell lysate in methanol in a ratio of methanol:chloroform:water (1:2:1). The mixture was vortexed thoroughly, and incubated for 10 minutes in a shaking heat block at 37˚C. Afterward, the lysates were spun down at 20,000 relative centrifugal force for 5 minutes to achieve a phase separation. The bottom phase (chloroform/methanol) was carefully collected and evaporated at 40˚C under flow of nitrogen. The residual fatty precipitate was dissolved in chloroform:methanol (2:1) and collected in glass vials.
Ubiquitin Pulldown Assay
Caco-2 cells were grown in 10-cm dishes and co-transfected with 3 μg pcDNA3, Flag-DGAT1 WT, or Flag-DGAT1 c.629_631delCCT and 3 μg His-ubiquitin. MG132 (2 μM) was added 32 hours posttransfection and the cells were incubated at 37°C for 16 hours. After incubation with His-ubiquitin, transfected cells were once washed with PBS and lysed in lysis buffer pH 8.0 containing 8 M urea (GE Healthcare, Little Chalfont, UK), 7 mM NaH2PO4 (Merck, Whitehouse Station, NJ), 100 mM Na2HPO4 (Merck), 10 mM Tris-HCl pH 8.0 (Roche), 0.2% Triton-X100 (Sigma-Aldrich), 10 mM imidazole (Sigma-Aldrich), and 5 mM N-ethylmaleimide (Sigma-Aldrich); 10 mM β-mercaptoethanol (Sigma-Aldrich) and Ni-NTA agarose beads (Qiagen) were added and the cells were tumbled for 2 hours at room temperature. The beads were washed twice in buffer pH 8.0, 2 times in buffer pH 6.3 (12 mM urea, 100 mM NaH2PO4, 30 mM Na2HPO4, 15 mM Tris-HCl pH 6.3, 0.3% Triton X-100, 30 mM imidazole), and once in wash buffer (100 mM NaCl [Sigma-Aldrich], 20% glycerol [Sigma-Aldrich], 20 mM Tris-HCl pH 8.0, 2 mM dithiothreitol [GE Healthcare] and 10 mM imidazole). Sample buffer (0.12 M Tris-HCl pH 6.8, 4% SDS, 20% glycerol, 0.05 μg/μL bromophenol blue and 35 mM β-mercaptoethanol) was added. The samples were incubated at 100°C for 5 minutes and subjected to Western blot.
Supplementary Table 1.
Genetic Description of DGAT1-deficient Patients in This Study and in Previous Reports
| Patient | Position Chr. 8 | Reference | Alternate | DGAT1 mutation | DGAT1 mutated protein | CADD score |
|---|---|---|---|---|---|---|
| 1 | 145540731 | C | T | c.1202G>A | p.W401X | 38 |
| 2 | 145540731 | C | T | c.1202G>A | p.W401X | 38 |
| 3 | 145542123 | CCT | CAGATGGGCAGGGTGGGATGGG | c.573_574delAGinsCCCATCCCACCCTGCCCATCT | - | 25.7 |
| 4 | 145541252 | C | T | c. 937-1G>A | - | 24.9 |
| 5 | 145541233 | T | TG | c.953insC | p. I319Hfs *31 | 35 |
| 6 | 145541233 | T | TG | c.953insC | p. I319Hfs *31 | 35 |
| 7 | 145541968 | TAGG | T | c.629_631delCCT | p.S210_Y211delinsY | 18.9 |
| 8 | 145541968 | TAGG | T | c.629_631delCCT | p.S210_Y211delinsY | 18.9 |
| 9 | 145541968 | TAGG | T | c.629_631delCCT | p.S210_Y211delinsY | 18.9 |
| 10 | 145541968 | TAGG | T | c.629_631delCCT | p.S210_Y211delinsY | 18.9 |
| Haas et al. | 145541756 | A | G | c.751+2T>C | p.A226_R250del | 22.9 |
| Stephen et al. (Fam 1) | 145541756 | A | G | c.751+2T>C | p.A226_R250del | 22.9 |
| Stephen et al. (Fam 2) | 145541457 | A | G | c.884T>C | p.L295P | 31 |
| Gluchowski et al. | 145542706 | A | G | c.314T>C | p.L105P | 29.3 |
| Ratchford et al. | 145541074 | TAGA | T | c.1013_1015delTCT | p.F338del | 22.3 |
| 145540567 | C | G | c.1260C>G | p.S420R | 31 |
CADD, combined annotation dependent depletion; Chr, chromosome.
Supplementary Table 2.
Serum Lipid Values of DGAT1-deficient Patients in This Study
| Patient | Age on test date | Triglyceride (mg/dL) | HDL (mg/dL) | LDL (mg/dL) | VLDL (mg/dL) | Cholesterol (mg/dL) |
|---|---|---|---|---|---|---|
| 1 | 5 mo | 135 (<150) | 25a (40–60) | 12 (<110) | 27 (<30) | 63 (<170) |
| 2 | 5 mo | 84 (<150) | 16a (40–60) | 28 (<110) | 16 (<30) | 62 (<170) |
| 3 | 11 mo | 370a (<200) | 9.5a (40–60) | 23 (<110) | ND | 96 (<170) |
| 4 | 8 y | 123 (<180) | Normal | 110a (<110) | ND | 181a (<170) |
| 5 | ND | ND | ND | ND | ND | ND |
| 6 | 2 y | 184 (<200) | 21a (40–60) | 13 (<110) | 36a (<30) | 59 (<170) |
| 7 | 9 y | 142 (<180) | 32a (40–60) | ND | ND | 139 (<170) |
| 8 | 12 y | 88 (<180) | 30a (40–60) | ND | ND | 112 (<170) |
| 9 | 22 mo | 124 (<180) | 24a (40–60) | ND | Normal | 139 (<170) |
| 10 | 20 mo | 165 (<180) | ND | ND | ND | 128 (<170) |
NOTE. Parentheses indicate normal values.
HDL, high-density lipoprotein; LDL, low-density lipoprotein; ND, not determined; VLDL, very low-density lipoprotein.
Indicates abnormal value
Supplementary Table 3.
Compilation of Clinical Characteristics and Specific Treatment of DGAT1-deficient Patients in This Study
| Patient ID | P1 | P2 | P3 | P4 | P5 | P6 | P7 | P8 | P9 | P10 |
|---|---|---|---|---|---|---|---|---|---|---|
| Recurrent infections | Yes | Yes | No | No | No | Yes | No | No | Yes | Yes |
| Type of infections | Upper respiratory tract infections | NA | Central line–related fungal infection | NA | NA | Upper respiratory infections, recurrent otitis media | NA | NA |
|
|
| Treatment for infectious episodes | Antibiotics IV or PO | NA | Amphotericin B IV | NA | Prophylactic immunoglobulins IV | Antibiotics PO | NA | NA | Antibiotics IV | Antibiotics IV |
| Response to treatment for recurrent infection | Well responded | NA | NA | NA | Well responded | Well responded | NA | NA | Well responded | Well responded |
| Signs of liver disease | NA | NA | NA | Elevated liver enzymes | Jaundice and hepatomegaly. Elevated liver enzymes. Liver biopsy: hepatocanalicular and ductular cholestasis, paucity of bile ducts, mixed type; hepatosteatosis (10%), porto-portal fibrosis and fibrotic activity of 3/6. |
Elevated liver enzymes | NA | NA | Elevated liver enzymes and jaundice. Toxic liver injury due to effects of long-term treatment with antifungal drugs (confirmed by liver biopsy). |
Elevated liver enzymes and jaundice. Liver biopsy normal. |
| Fecal elastase 1 levels (normal range: > 500 μg/g) |
ND | NA | ND | NA | 130 μg/g | ND | NA | NA | >500 μg/g | >500 μg/g |
| Serum albumin level before treatment (normal range: >3.2 g/dL) | 1.8 g/dL | 1.8 g/dL | 2.2 g/dL | 2.8 g/dL | 2.2 g/dL | 2.59 g/dL | NA | 4.35 g/dL | 2.38 g/dL | 3.2 g/dL |
| Fecal-alpha-1-antitrypsin level (normal range: <5 mg/g) |
NA | ND | Low | ND | NA | NA | NA | NA | 9.1 mg/g | 7.6 mg/g |
| Specific treatment | Fat-free formula and medium chain triglyceride Albumin IV |
Albumin IV | Albumin IV, TPN Fat-free formula and supplementation of medium chain triglyceride |
Cholestyramine | Creon hydrolyzed formula | Creon | Monthly infusion of Intralipid and Omegaven supplementation of lipid-soluble vitamins | Monthly infusion of Intralipid and Omegaven supplementation of lipid-soluble vitamins | TPN and small bowel transplantation | TPN and fat-free formula |
| Clinical course after treatment | Significant improvement | Deceased | Significant improvement of diarrhea | Significant improvement; diarrhea stopped after treatment |
Mild improvement. His daily stool frequency had reduced with the administrations Creon, but his weight and height are below 3rd percentile. | No improvement of failure to thrive, but significant improvement of diarrhea. After 2 years of age, his symptoms improved spontaneously. His weight is now 16 kg (3rd–10th percentile), height 104 cm (3rd percentile) |
Significant improvement | Significant improvement | Completely resolved PLE. Tolerates polymeric enteral feeding. Still stunted and underweight by causes secondary to intestinal transplantation and several autoimmune problems. |
NA |
IV, intravenous; NA, not available; ND, not done; PLE, protein-losing enteropathy PO, per os; TPN, total parenteral nutrition.
Supplementary Table 4.
sgRNAs Used to Target DGAT1 for CRISPR/Cas9 Gene Editing
| sgRNA ID | Core sequence DNA | Position DGAT1 gene | Predicted cleavagea |
|---|---|---|---|
| sgRNA#2 | GGCACCATGAGTTGACGTCG | Exon 7 | 13621-13622 (AC) |
| sgRNA#6 | TGTGCGCCATCAGCGCCAGC | Exon 7 | 13570-13571 (TG) |
| sgRNA#7 | GTGCGCCATCAGCGCCAGCA | Exon 7 | 13569-13570 (CT) |
| sgRNA#8 | GTGGCTGGCCCGAGACAGAT | Intron 6–7 | 13486-13487 (CT) |
sgRNA, single-guide RNA.
According NCBI Reference Sequence: NG_034192.1; Homo sapiens diacylglycerol O-acyltransferase 1 (DGAT1).
Author names in bold designate shared co-first authorship.
References
- 1.Canani R.B., Castaldo G., Bacchetta R. Congenital diarrhoeal disorders: advances in this evolving web of inherited enteropathies. Nat Rev Gastroenterol Hepatol. 2015;12:293–302. doi: 10.1038/nrgastro.2015.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ozen A., Comrie W.A., Ardy R.C. CD55 deficiency, early-onset protein-losing enteropathy, and thrombosis. N Engl J Med. 2017;377:52–61. doi: 10.1056/NEJMoa1615887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haas J.T., Winter H.S., Lim E. DGAT1 mutation is linked to a congenital diarrheal disorder. J Clin Invest. 2012;122:4680–4684. doi: 10.1172/JCI64873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stephen J., Vilboux T., Haberman Y. Congenital protein losing enteropathy: an inborn error of lipid metabolism due to DGAT1 mutations. Eur J Hum Genet. 2016;24:1268–1273. doi: 10.1038/ejhg.2016.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gluchowski N.L., Chitraju C., Picoraro J.A. Identification and characterization of a novel DGAT1 missense mutation associated with congenital diarrhea. J Lipid Res. 2017;58:1230–1237. doi: 10.1194/jlr.P075119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ratchford T.L., Kirby A.J., Pinz H. Congenital diarrhea from DGAT1 mutation leading to electrolyte derangements, protein-losing enteropathy, and rickets. J Pediatr Gastroenterol Nutr. 2018;66:e82–e83. doi: 10.1097/MPG.0000000000001750. [DOI] [PubMed] [Google Scholar]
- 7.Yen C.L., Stone S.J., Koliwad S. DGAT enzymes and triacylglycerol biosynthesis. J Lipid Res. 2008;49:2283–2301. doi: 10.1194/jlr.R800018-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iqbal J., Hussain M.M. Intestinal lipid absorption. Am J Physiol Metab. 2009;296:E1183–E1194. doi: 10.1152/ajpendo.90899.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coleman R.A., Mashek D.G. Mammalian triacylglycerol metabolism: synthesis, lipolysis, and signaling. Chem Rev. 2011;111:6359–6386. doi: 10.1021/cr100404w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yen C.-L.E., Nelson D.W., Yen M.-I. Intestinal triacylglycerol synthesis in fat absorption and systemic energy metabolism. J Lipid Res. 2015;56:489–501. doi: 10.1194/jlr.R052902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cases S., Stone S.J., Zhou P. Cloning of DGAT2, a second mammalian diacylglycerol acyltransferase, and related family members. J Biol Chem. 2001;276:38870–38876. doi: 10.1074/jbc.M106219200. [DOI] [PubMed] [Google Scholar]
- 12.Abumrad N.A., Davidson N.O. Role of the gut in lipid homeostasis. Physiol Rev. 2012;92:1061–1085. doi: 10.1152/physrev.00019.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hussain M.M. Intestinal lipid absorption and lipoprotein formation. Curr Opin Lipidol. 2014;25:200–206. doi: 10.1097/MOL.0000000000000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Massink M.P.G., Créton M.A., Spanevello F. Loss-of-function mutations in the WNT co-receptor LRP6 cause autosomal-dominant oligodontia. Am J Hum Genet. 2015;97:621–626. doi: 10.1016/j.ajhg.2015.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salzer E., Daschkey S., Choo S. Combined immunodeficiency with life-threatening EBV-associated lymphoproliferative disorder in patients lacking functional CD27. Haematologica. 2013;98:473–478. doi: 10.3324/haematol.2012.068791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erman B., Bilic I., Hirschmugl T. Combined immunodeficiency with CD4 lymphopenia and sclerosing cholangitis caused by a novel loss-of-function mutation affecting IL21R. Haematologica. 2015;100:e216–e219. doi: 10.3324/haematol.2014.120980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ran F.A., Hsu P.D., Wright J. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujii M., Matano M., Nanki K. Efficient genetic engineering of human intestinal organoids using electroporation. Nat Protoc. 2015;10:1474–1485. doi: 10.1038/nprot.2015.088. [DOI] [PubMed] [Google Scholar]
- 19.Kruitwagen H.S., Oosterhoff L.A., Vernooij I.G.W.H. Long-term adult feline liver organoid cultures for disease modeling of hepatic steatosis. Stem Cell Reports. 2017;8:822–830. doi: 10.1016/j.stemcr.2017.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rueden C.T., Schindelin J., Hiner M.C. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics. 2017;18:529. doi: 10.1186/s12859-017-1934-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schindelin J., Arganda-Carreras I., Frise E. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wiegerinck C.L., Janecke A.R., Schneeberger K. Loss of syntaxin 3 causes variant microvillus inclusion disease. Gastroenterology. 2014;147:65–68.e10. doi: 10.1053/j.gastro.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 23.Grall A., Guaguère E., Planchais S. PNPLA1 mutations cause autosomal recessive congenital ichthyosis in golden retriever dogs and humans. Nat Genet. 2012;44:140–147. doi: 10.1038/ng.1056. [DOI] [PubMed] [Google Scholar]
- 24.Lek M., Karczewski K.J., Minikel E.V. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kircher M., Witten D.M., Jain P. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spandl J., White D.J., Peychl J. Live cell multicolor imaging of lipid droplets with a new dye, LD540. Traffic. 2009;10:1579–1584. doi: 10.1111/j.1600-0854.2009.00980.x. [DOI] [PubMed] [Google Scholar]
- 27.Greenspan P., Mayer E.P., Fowler S.D. Nile red: a selective fluorescent stain for intracellular lipid droplets. J Cell Biol. 1985;100:965–973. doi: 10.1083/jcb.100.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Braamskamp M.J.A.M., Dolman K.M., Tabbers M.M. Clinical practice: Protein-losing enteropathy in children. Eur J Pediatr. 2010;169:1179–1185. doi: 10.1007/s00431-010-1235-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwerd T., Pandey S., Yang H.T. Impaired antibacterial autophagy links granulomatous intestinal inflammation in Niemann-Pick disease type C1 and XIAP deficiency with NOD2 variants in Crohn’s disease. Gut. 2017;66:1060–1073. doi: 10.1136/gutjnl-2015-310382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu J.W., Yang H., Wang S.P. Inborn errors of cytoplasmic triglyceride metabolism. J Inherit Metab Dis. 2014;38:85–98. doi: 10.1007/s10545-014-9767-7. [DOI] [PubMed] [Google Scholar]
- 31.Wen J., Tang Q., Wu J. Primary intestinal lymphangiectasia: four case reports and a review of the literature. Dig Dis Sci. 2010;55:3466–3472. doi: 10.1007/s10620-010-1161-1. [DOI] [PubMed] [Google Scholar]
- 32.Hong Y Bin, Kang J., Kim J.H. DGAT2 mutation in a family with autosomal-dominant early-onset axonal Charcot-Marie-Tooth disease. Hum Mutat. 2016;37:473–480. doi: 10.1002/humu.22959. [DOI] [PubMed] [Google Scholar]
- 33.Smith S.J., Cases S., Jensen D.R. Obesity resistance and multiple mechanisms of triglyceride synthesis in mice lacking Dgat. Nat Genet. 2000;25:87–90. doi: 10.1038/75651. [DOI] [PubMed] [Google Scholar]
- 34.Chen H., Smith S., Ladha Z. Increased insulin and leptin sensitivity in mice lacking acyl CoA: diacylglycerol acyltransferase 1. J Clin Invest. 2002;109:1049–1055. doi: 10.1172/JCI14672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen H.C., Jensen D.R., Myers H.M. Obesity resistance and enhanced glucose metabolism in mice transplanted with white adipose tissue lacking acyl CoA:diacylglycerol acyltransferase 1. J Clin Invest. 2003;111:1715–1722. doi: 10.1172/JCI15859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DeVita R.J., Pinto S. Current status of the research and development of diacylglycerol O-acyltransferase 1 (DGAT1) inhibitors. J Med Chem. 2013;56:9820–9825. doi: 10.1021/jm4007033. [DOI] [PubMed] [Google Scholar]
- 37.Meyers C.D., Tremblay K., Amer A. Effect of the DGAT1 inhibitor pradigastat on triglyceride and apoB48 levels in patients with familial chylomicronemia syndrome. Lipids Health Dis. 2015;14:8. doi: 10.1186/s12944-015-0006-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meyers C.D., Amer A., Majumdar T. Pharmacokinetics, pharmacodynamics, safety, and tolerability of pradigastat, a novel diacylglycerol acyltransferase 1 inhibitor in overweight or obese, but otherwise healthy human subjects. J Clin Pharmacol. 2015;55:1031–1041. doi: 10.1002/jcph.509. [DOI] [PubMed] [Google Scholar]
- 39.Buhman K.K., Smith S.J., Stone S.J. DGAT1 is not essential for intestinal triacylglycerol absorption or chylomicron synthesis. J Biol Chem. 2002;277:25474–25479. doi: 10.1074/jbc.M202013200. [DOI] [PubMed] [Google Scholar]
Supplementary References
- 1.Wiegerinck C.L., Janecke A.R., Schneeberger K. Loss of syntaxin 3 causes variant microvillus inclusion disease. Gastroenterology. 2014;147:65–68.e10. doi: 10.1053/j.gastro.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 2.Fujii M., Matano M., Nanki K. Efficient genetic engineering of human intestinal organoids using electroporation. Nat Protoc. 2015;10:1474–1485. doi: 10.1038/nprot.2015.088. [DOI] [PubMed] [Google Scholar]
- 3.Folch J., Lees M., Sloane Stanley G.H. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]