Abstract
Knowledge of the number of DNA sequences targeted by the taxon-specific reference assays is essential for correct GM quantification and is key to the harmonisation of measurement results. In the present study droplet digital PCR (ddPCR) was used to determine the number of DNA target copies of taxon-specific assays validated for real-time PCR for the four main genetically modified (GM) crops. The transferability of experimental conditions from real-time PCR to ddPCR was also explored, as well as the effect of DNA digestion. The results of this study indicate that for each crop at least one taxon-specific assay can be identified as having a single DNA target. A short list of taxon-specific reference assays is proposed as best candidates for the relative quantification of GM events for soybean, maize, cotton and oilseed rape. The investigated assays could be in most cases transferred to ddPCR without further optimisation. The use of DNA digestion did not improve ddPCR characteristics such as rain and resolution at the conditions tested.
Keywords: Taxon-specific reference assay, Digital PCR (dPCR), Food, GMO, Maize, Soybean, Cotton, Oilseed rape, MIQE
Highlights
-
•
DdPCR was used to test number of targets of validated reference assays for 4 GM crops.
-
•
At least 1 taxon-specific assay for a single DNA target was identified per crop.
-
•
Most qPCR based assays tested can be transferred to ddPCR without optimisation.
-
•
DNA digestion does not improve ddPCR characteristics at the conditions tested.
1. Introduction
Key to the development of a harmonised monitoring system for genetically modified organisms (GMOs) in food and feed is that results expressing the GM content are reliable, comparable across laboratories and import/export regions, and that they comply with the regulations. A labelling threshold of 0.9% for adventitious or technically unavoidable presence of authorised GMOs is in force in the European Union for which the GMO content is measured relatively to the ingredient (species) (European Parliament and the Council of the European Union, 2003, 2004a, 2004b, 2014; European Commission, 2011; European and Parliament the Council of the European Union, 2013). The European Union Reference Laboratory for Genetically Modified Food and Feed (EURL GMFF), managed by the European Commission's Joint Research Centre, has validated so far in collaborative trials more than sixty event-specific methods using real-time PCR for the relative quantification of GMOs. These methods are used in routine monitoring for official controls. Appropriate quantification depends on the correct amplification and counting of two different targets: the GM event-specific DNA sequence and the taxon-specific sequence. The event-specific assay (an assay is herewith defined as the set of primers and probe, validated with known reaction mix and reaction conditions) is designed to target the insert-to-host genome sequence, which has been selected to be unique (single copy) in the plant genome, due to the random integration process of the DNA insert that occurs with the transformation technologies used so far. For the design of a taxon-specific assay there is in principle a wide selection of host genome sequences that comply with the demands for a standard real-time PCR assay. However, strict requirements have been set for quantitative real-time PCR (qPCR) methods submitted in the context of the authorisation of a new GMO in the European Union: the taxon-specific assay has to be specific to the crop of interest, stable across varieties and should address a single copy in the plant genome, if possible (EURL GMFF, 2015b). Thus, the design of a taxon-specific assay requires sufficient information about the genetics of the plant and necessitates proper optimisation and validation of the whole analytical procedure (Debode et al., 2017; Jacchia et al., 2015b, 2015a). The exercise can be particularly demanding when it comes to allopolyploid species such as cotton and oilseed rape, in which multiple sets of chromosomes (sub-genomes) are present, deriving from distinct species. However, the knowledge of the number of DNA targets per genome for a taxon-specific assay is essential for the correct quantification of a GM event and for the conversion of results of a GM event quantification performed in copy numbers into mass fractions (ratio of the measured GM mass to the total mass of the ingredient). Such conversion is ultimately required when it comes to the measurement of the so called 'low level presence' of GMOs in feed consignments (European Commission, 2011).
The European Network of GMO Laboratories (ENGL) has recently elaborated an approach on how to achieve this conversion (Corbisier et al., 2017). The determination of the corresponding conversion factor would employ digital PCR (dPCR), another technology exploiting DNA amplification and detection by using PCR chemistry (Vogelstein & Kinzler, 1999), which has the potential to become the next gold standard in GMO quantification (Corbisier, Bhat, Partis, Rui Dan Xie, & Emslie, 2010; Deprez et al., 2016; Dobnik, Štebih, Blejec, Morisset, & Žel, 2016; Fraiture et al., 2015; Iwobi, Gerdes, Busch, & Pecoraro, 2016; Lievens, Jacchia, Kagkli, Savini, & Querci, 2016). Unlike real-time PCR, where quantification requires a calibration system with standards at known GM DNA concentrations, dPCR does not necessitate a calibration with DNA and estimates the number of targeted copies per reaction, under certain assumptions directly (Sykes et al., 1992; Vogelstein & Kinzler, 1999).
In the present study this feature of dPCR was used to determine the number of DNA targets addressed by the taxon-specific assays for soybean, cotton, maize and oilseed rape listed below. The experimental data were backed by detailed bioinformatics analyses. The purpose of this work was to compile a short list of best candidate taxon-specific reference assays for the relative quantification of GMOs in the most commonly transformed crops (Parisi, Tillie, & Rodríguez-Cerezo, 2016). The taxon-specific assays for the following DNA targets validated in the frame of Regulation (EC) No 1829/2003 (2003) were tested: for soybean (Glycine max): Le1 A (EURL GMFF, 2012b) and Le1 B (EURL GMFF, 2015a); for cotton (Gossypium hirsutum): AdhC (EURL GMFF, 2012a), SAH7 (EURL GMFF, 2006) and acp1 (EURL GMFF, 2009); for maize (Zea mays): hmg (EURL GMFF, 2005), ZmAdh1 (EURL GMFF, 2014) and aldolase (EURL GMFF, 2016); and for oilseed rape (Brassica napus): ccf (EURL GMFF, 2013b), cruA (EURL GMFF, 2007) and FatA(A) (EURL GMFF, 2013a), plus FatA (Monsanto Biotechnology Regulatory Sciences, 2004). In this context the transferability of these real-time PCR assays to the QX200 droplet dPCR platform was investigated. The effect of DNA digestion on measurement results was also explored.
2. Materials and methods
2.1. Plant materials
Certified reference materials (CRMs) were used for DNA extraction: maize MON810 level 2, nominal 2% GMO in mass fraction (ERM-BF413ek) and NK603 level 4, nominal 2% GMO in mass fraction (ERM-BF415e); soybean 356043 level 3, nominal 10% GMO in mass fraction (ERM-BF425d) and DAS-68416 blank, nominal 0% GMO (ERM-BF432a); cotton GHB119 level 2, nominal 10% GMO in mass fraction (ERM-BF428c) and T304-40 blank, nominal 0% GMO (ERM-BF429a); oilseed rape 73496, nominal 10% GMO in mass fraction (ERM-BF434e), non-modified canola whole seed (AOCS 0304-A). Except for AOCS 0304-A, produced by the American Oil Chemists' Society, all other CRMs are from the European Commission's Joint Research Centre (JRC). DNA from B. napus, Brassica rapa, Brassica juncea, Brassica nigra, Brassica oleracea, Brassica carinata was prepared by the EURL GMFF.
2.2. Genomic DNA extraction and quality check
The genomic DNA used in this study was extracted with a CTAB DNA extraction method modified from ISO 21571 (International Organization for Standardization, 2005) (for soybean, oilseed rape and with an additional phenol-chloroform purification step for cotton when necessary) or a NucleoSpin® Food kit (Macherey-Nagel, for maize). In cases of suboptimal recovery or inhibition, the NucleoSpin® Food kit (for oilseed rape) or the foodproof® Sample Preparation kit III (Biotecon Diagnostics, for cotton) were used. The integrity of the extracted genomic DNA was tested by electrophoresis on a 1% [w/v] agarose gel stained with ethidium bromide, while the absence of PCR inhibitors was assessed through a real-time PCR inhibition run as described by Zel et al. (Žel et al., 2008). Only pure, non-inhibited, high molecular weight DNA was used. The extractions were done at least in duplicate with CRMs containing two different GM events per each crop; DNA samples were maintained at 4 °C for the duration of the experiments, then stored at −20 °C.
2.3. DNA quantification
Extracted genomic DNA was quantified fluorometrically with the Qubit® Fluorometer (Thermo Fisher Scientific) and the Qubit dsDNA BR assay Kit (Molecular Probes, Life Technologies) prior to its quality assessment for calculating the amount of DNA to be used in restriction digestion reactions. Before diluting for the ddPCR runs all DNA samples (digested and non-digested) were re-quantified together by using a Biorad VersaFluor fluorometer and the Quant-iT PicoGreen dsDNA Assay Kit (Molecular Probes) with a five point standard curve ranging from 1 to 500 ng/mL.
2.4. DNA digestion
The required DNA extracted from each CRM was enzymatically digested. For maize, soybean and oilseed rape, EcoRI (New England Biolabs) was used, while for cotton DraI (New England Biolabs) was used instead. These two restriction enzymes were chosen because they do not cut inside tested amplicons. TspRI (New England Biolabs) was also used in a limited set of cotton experiments. Its restriction site is present in the secondary target (herewith defined as the DNA target of an assay that presents mismatches in the primers and/or probe annealing sites) of the assay for acp1. The enzymatic digestions were performed in a final volume of 250 μL according to the manufacturer's conditions and inactivated appropriately. After digestion, 5 μL of the digestion solution was loaded on a 1% agarose gel, stained with ethidium bromide and visualised under UV light. If partial or incomplete digestion occurred, the digestion was repeated. Subsequently, samples were precipitated with ethanol (except for T304-40 digested with DraI which was not precipitated because of the limited amount available) and re-quantified. Since it was not possible to achieve complete DNA digestion with TspRI, the presence of undigested DNA was verified by comparing the λ values (average number of targets per droplet) measured with ddPCR for the primary and secondary targets of the assay for acp1. The amplification yield of the secondary acp1 target relative to the primary target decreased to 5% in digested samples, compared to 35% in non-digested samples.
2.5. Sample preparation
The total DNA content in samples for in-house testing of all the taxon-specific assays was quantified by PicoGreen (see above) and subsequently diluted to the concentration of interest in 0.1 × TE. The final concentration of the samples to be used in ddPCR was determined by taking into consideration the haploid genome masses of the different species [1.13 pg for soybean; 2.33 pg for cotton; 2.73 pg for maize and 1.15 pg for oilseed rape (Bennett & Leitch, 2012)] and by calculating the concentration needed to theoretically have 1 target copy/droplet in ddPCR, with a droplet size of 0.85 nL, as used by the QuantaSoft software (version 1.6.6.0320) (Corbisier et al., 2015). The DNA amount added to each ddPCR reaction was: 26.6 ng for soybean, 27.1 ng for oilseed rape, 54.8 ng for cotton and 64.2 ng for maize.
2.6. Real-time PCR
The inhibition runs for all materials and the qPCR for GHB119 cotton were performed following the validated methods (EURL GMFF, 2005, 2007, 2009, 2012a, 2012b) with an ABI 7900 platform (Life Technologies) or an ABI 7500 platform (Applied Biosystems).
2.7. Droplet digital PCR
Measurements were performed with the Biorad QX200 digital droplet platform using Twin.Tec 96 well PCR plates (Eppendorf). Reactions were set up using the ddPCR Supermix for Probes, no UNG (Biorad) with primers and probes at the final concentrations listed in Table S1 (Supplementary Information). The initial volume of the reaction mixture was 20 μL which, together with 70 μL of oil, resulted in a final volume of droplets in oil of 40 μL. Probes were either FAM or HEX labelled with a TAMRA, BHQ or MGB quencher (Table S1). Thermal cycling was performed on a Biorad C1000 Touch thermal cycler using the following thermal cycling protocol: 10 min at 95 °C, 45 × (15 s at 95 °C, 1 min at 60 °C), 10 min at 98 °C. Data were analysed and exported using the QuantaSoft 1.6.6.0320 software. No template controls were included for every method in every run.
2.8. Temperature gradient
It was set up using the built-in function of the C1000 Touch thermal cycler (Biorad) where each row can be set at a different temperature. The gradient protocol was: 10 min at 95 °C, 45 × (15 s at 95 °C, 1 min at 62-56 °C), followed by 10 min at 98 °C. The rows where samples were loaded had individual temperatures of 62, 59.8, 58.4 and 56 °C (from row A to H); the other 4 rows were left empty.
2.9. Data analysis
All calculations and model fitting were done using the software R version 3.3.1. Data were exported from the droplet reader and imported into R. Analysis of the droplet PCR results for λ values, percentage of rain, and resolution was done as described in (Lievens et al., 2016). R-scripts are available at https://github.com/Gromgorgel/ddPCR and as supplementary material. The percentage of rain was not calculated for reactions with multiple bands of droplets. Statistical t-tests were used to compare between results on various levels. Digital MIQE guidelines (Huggett et al., 2013) were taken into consideration when applicable to the present work.
2.10. Bioinformatics analyses
In order to evaluate in silico the target copy numbers of each reference gene, the primer and probe sequences used in the taxon-specific assays were analysed against the currently available genome sequence information (August 2017). Predictions of PCR amplification were made using e-PCR (NCBI, version 2.3.12) (Rotmistrovsky, Jang, & Schuler, 2004) with the following parameters: maximum 3 mismatches, maximum 3 gaps, amplicon size between 0 and 2000 bp. In addition, sequence similarity searches with the whole amplicon sequences were performed using BLASTN (NCBI, version 2.2.15) (Altschul, Gish, Miller, Myers, & Lipman, 1990). Reference genome sequences were downloaded from Genbank (at ftp://ftp.ncbi.nlm.nih.gov/genbank/). The following assemblies were used: B. napus (GCA_000751015.1), B. rapa (GCF_000309985.1), B. oleracea (GCF_000695525.1), Z. mays (GCA_000005005.6), G. max (GCF_000004515.4) and G. hirsutum (GCA_000987745.1).
3. Results and discussion
3.1. Determination of target copy numbers
The target copy number of each taxon-specific assay was estimated by comparing the results of bioinformatics analyses with the λ value measured with ddPCR. The quantification results obtained for the different assays targeting the same plant species were also compared by calculating all possible ratios between them, in order to exclude any influence of incorrect DNA quantification between the different DNA samples used, and for identifying possible differences between the two CRMs used for each species.
3.1.1. Bioinformatics analyses
Bioinformatics analyses with the primers and probes for Le1 A and Le1 B with the G. max genome indicated the presence of a single annealing site, with perfect matches for both assays for Le1 A and Le1 B. The same was also observed for maize assays for hmg and ZmAdh1. On the other hand, two target sequences were identified for maize assay for aldolase, one with perfect annealing of the primers and probe, and one with one mismatch in the reverse primer and one in the probe. Despite these mismatches, this second target is also expected to produce an amplification signal. Cotton is allotetraploid and resulted from the hybridization of ancestral A- and D-genome diploid species (Wendel, 1989); its genome contains 2 highly similar sub-genomes (Li et al., 2015). For the assay for AdhC the bioinformatics analyses suggested the presence of a single target site, with perfect annealing of the primers and probe. A second site was also identified, but is not predicted to amplify, since the forward primer has four mismatches and two gaps in the annealing region. For SAH7, two target sites were identified, one on chromosome 8 (A sub-genome) and one on chromosome 20 (D sub-genome); the annealing of primers and probe is perfect for both, but the two sites are expected to produce amplicons of different sizes (115 and 123 bp, respectively). For the primers and probe for acp1, two target sites were identified, one with perfect annealing of primers and probe, and a second with two mismatches in the primer forward, one in the primer reverse, and one in the probe (see Fig. S1, Supplementary Information). This second site could possibly produce an amplification signal. Oilseed rape is also allotetraploid, originating from the hybridization of the ancestors of the diploid species B. rapa (A sub-genome) and B. oleracea (C sub-genome) (Nagaharu, 1935). The genome sequence of B. napus is available, but it has not been assembled in a physical map yet, so it is difficult to determine on which sub-genome each target is found. However it can be inferred in most cases from the genome sequences of B. rapa and B. oleracea, which are also available. Both assays for ccf and cruA target the same loci in the oilseed rape genome, one corresponding to the cruciferin gene and the other to uncharacterised proteins with sequences similar to cruciferin. Neither one can be considered to target single copy number DNA sequences. The assay for ccf shows only perfect annealing with the target locus in the C sub-genome, while the one for cruA only shows perfect annealing with the target in the A sub-genome. Due to the presence of different mismatches in the binding regions for primers and probe of both assays with their secondary targets, and due to the incomplete assembly level of the B. napus genome, it is difficult to predict the final number of amplified targets based on sequence information alone. The assays for FatA and FatA(A) both target a putative fatty acid thioesterase gene that is present in both the A and C sub-genomes of B. napus. FatA(A) shows perfect annealing only with the A sub-genome target, while FatA only shows perfect annealing with the C sub-genome target. However, FatA(A) seems less likely to give an amplification signal from both sub-genomes than FatA because of the many mismatches in the probe region with the C sub-genome target. In both cases, the analysis of the B. napus genome shows the respective target putative fatty acid thioesterase gene in each B. napus sub-genome.
3.1.2. Droplet digital PCR results
DdPCR results are based on measurements of three technical replicates for each certified reference material and two different DNA extractions per certified reference material (biological replicates), for a total of 6 replicates per material, except for the assay for oilseed rape FatA(A) for which only 2 replicates were used for the digested samples (4 in total per material).
For soybean, the lectin gene was quantified using two different assays, for Le1 A and Le1 B, in two different CRMs for the GM events 356043 and DAS-68416, as described in the Materials and Methods section. The measured λ value was the same for the two assays, namely 0.802 (see Table 1), and close to the theoretical λ of 1. No significant difference was observed between data for the two reference materials used based on the ratio between λ values measured for Le1 A and Le1 B (Table 2). Since for soybean both tested assays target the same gene, it was decided to further confirm the assumption that lectin is present as a single copy in the soybean genome by quantifying with ddPCR the GM content of the 356043 soybean CRM used in this study. The certified property value for this material is expressed in mass fraction as being 100 g/kg with an expanded uncertainty (coverage factor k = 2) of 7 g/kg (Institute for Reference Materials and Measurements, Joint Research Centre, 2011). This should correspond to a GM content of 10% expressed as copy number ratio considering that the GM soybean used for the production of the CRM was homozygous for the GM insert and assuming that the reference assay's target is present as a single copy in the soybean genome. The quantification result obtained with ddPCR was 9.9% ± 0.2% (expanded measurement uncertainty with k = 4.3), which is in line with the certified value and confirms that the targets of the soybean reference assays tested are present as a single copy in the soybean genome. As a consequence both can be used indifferently for GMO quantification.
Table 1.
Measured λ values for the different assays, their comparison (paired t-test over the different CRMs) and the ratios between the different assays per crop. Only the results obtained with non-digested samples are shown. The significance of the t-test is expressed as either 0 (no significant difference) or 1 (significant difference, p Value and Significance highlighted in bold). Lambda A: measured λ value for target A; Lambda B: measured λ value for target B; p value: probability value of the t-test comparison of the measured λ value for target A and target B; Ratio: ratio between the measured λ value for target A and for target B.
| Crop | Target A | Target B | Lambda A | Lambda B | p Value | Significance | Ratio |
|---|---|---|---|---|---|---|---|
| soybean |
Le1 A |
Le1 B |
0.802 |
0.802 |
0.973 |
0 |
1.00 |
| maize |
ZmAdh1 | aldolase | 1.012 | 2.183 | 0.000 | 1 | 0.46 |
| ZmAdh1 | hmg | 1.012 | 1.149 | 0.000 | 1 | 0.88 | |
| aldolase |
hmg |
2.183 |
1.149 |
0.000 |
1 |
1.90 |
|
| cotton |
acp1 | AdhC | 0.881 | 0.916 | 0.310 | 0 | 0.96 |
| acp1 | SAH7 | 0.881 | 1.856 | 0.000 | 1 | 0.47 | |
| AdhC |
SAH7 |
0.916 |
1.856 |
0.000 |
1 |
0.49 |
|
| oilseed rape | ccf | cruA | 1.705 | 1.604 | 0.302 | 0 | 1.06 |
| ccf | FatA | 1.705 | 1.806 | 0.249 | 0 | 0.94 | |
| ccf | FatA(A) | 1.705 | 0.769 | 0.000 | 1 | 2.22 | |
| cruA | FatA | 1.604 | 1.806 | 0.063 | 0 | 0.89 | |
| cruA | FatA(A) | 1.604 | 0.769 | 0.000 | 1 | 2.08 | |
| FatA | FatA(A) | 1.806 | 0.769 | 0.000 | 1 | 2.35 |
Table 2.
Ratios between the measured λ values obtained by the different assays per CRM and their comparison (paired t-test over the different CRMs) using only the results obtained with non-digested samples. The last column lists the significance of a difference between the ratio of the λ values obtained with the two assay listed under ‘Assays’ for CRM A and for CRM B, which is calculated as follows: 1 (significant difference, highlighted in bold) is assigned if the p value is significant (highlighted in bold) and the difference between the two ratios is at least 20% of the lowest value. Ratio A: ratio between the λ values obtained with the two assays described in ‘Assays’ with the CRM A; ratio B: ratio between the λ values obtained with the two assays described in ‘Assays’ with CRM B; p value: probability value of the t-test comparison of the ratio calculated for CRM A and for CRM B.
| Crop | Assays | CRM A | CRM B | Ratio A | Ratio B | p Value | Significance |
|---|---|---|---|---|---|---|---|
| soybean |
Le1 A/Le1 B |
356043 |
DAS-68416 |
1.01 |
1.02 |
0.532 |
0 |
| maize |
ZmAdh1/aldolase | MON810 | NK603 | 0.47 | 0.45 | 0.017 | 0 |
| ZmAdh1/hmg | MON810 | NK603 | 0.87 | 0.89 | 0.322 | 0 | |
| aldolase/hmg |
MON810 |
NK603 |
1.84 |
2.00 |
0.001 |
0 |
|
| cotton |
acp1/AdhC | GHB119 | T304-40 | 0.96 | 1.00 | 0.515 | 0 |
| acp1/SAH7 | GHB119 | T304-40 | 0.49 | 0.48 | 0.733 | 0 | |
| AdhC/SAH7 |
GHB119 |
T304-40 |
0.51 |
0.48 |
0.041 |
0 |
|
| oilseed rape | ccf/cruA | 73496 | non-modified canola | 1.01 | 1.10 | 0.068 | 0 |
| ccf/FatA | 73496 | non-modified canola | 1.00 | 0.89 | 0.016 | 0 | |
| ccf/FatA(A) | 73496 | non-modified canola | 2.45 | 1.98 | 0.125 | 0 | |
| cruA/FatA | 73496 | non-modified canola | 1.00 | 0.81 | 0.000 | 1 | |
| cruA/FatA(A) | 73496 | non-modified canola | 2.33 | 1.83 | 0.028 | 1 | |
| FatA/FatA(A) | 73496 | non-modified canola | 2.46 | 2.30 | 0.583 | 0 |
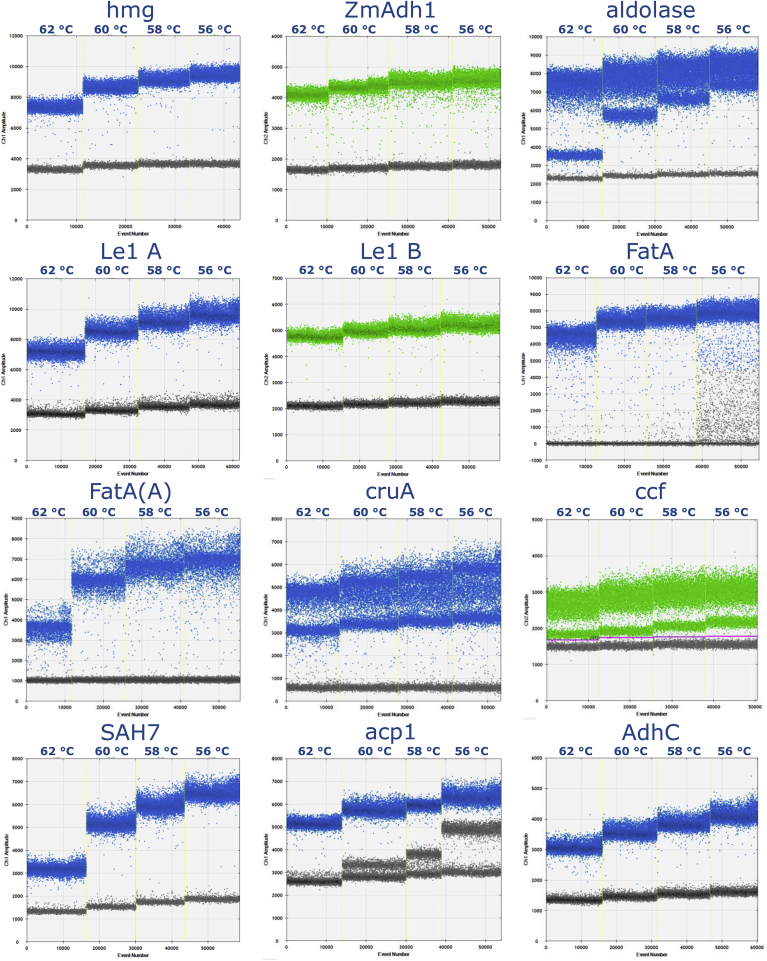
For cotton the ddPCR results were compared for three taxon-specific assays, for acp1, AdhC and SAH7, in two different CRMs. The measured λ values for acp1 and AdhC were 0.881 and 0.916, respectively, with no significant difference (Table 1). A λ of 1.856 was measured for SAH7, which is significantly different from the ones measured for acp1 and AdhC, respectively. The calculated λ ratios, close to 0.5 between acp1 and SAH7 and between AdhC and SAH7, and close to 1 between acp1 and AdhC (see Table 1) indicate the presence of the same number of targets for acp1 and AdhC, namely one, and two for SAH7. These results are in accordance with the bioinformatics analyses for AdhC and SAH7, while for acp1 the analyses identified a second binding site with mismatches in the primers and probe annealing site which could possibly be amplified and produce a signal (Fig. S1). In fact, on the droplet fluorescence plot of acp1 a second droplet population was identified next to the background, in addition to the population containing the positive droplets (see Fig. 1). This population of droplets seems to correspond to the second acp1 target, and is located closer to the negative or positive droplet population depending on the annealing/amplification temperature, as shown in Fig. 1. The presence of mismatches in the primers and probe region could be at the origin of this behaviour. At 60 °C the droplet population corresponding to the second acp1 target is automatically classified by the instrument's software as negative. When comparing the λ ratios obtained with the two reference materials, no biologically relevant differences were identified (Table 2). BioRad ddPCR is designed to work with Supermix as a reaction buffer, which is likely different in some respect from the reaction buffers that were used during the validation of the real-time PCR methods, although its composition is not released by the producer. Parameters (e.g. pH, MgCl2 concentration) influencing the heteroduplex formation during primer pairing could also be a source of variation. Thus, the effective amplification in real-time PCR of the second acp1 target was investigated, thanks to the presence of a restriction enzyme target site in the probe annealing site of the second acp1 target (see Fig. S1). Cotton genomic DNA extracted from GHB119 CRM (ERM-BF428c) was either digested with TspRI or not. Subsequently the GHB119 event was quantified relative to acp1, and to AdhC as control, in the digested and undigested samples by means of the validated qPCR protocols. If the second acp1 target would not amplify, there would be no quantification difference between the digested and the undigested sample. If the second acp1 target would amplify, an overestimation of the GM % of up to 100% would be expected for the digested material. The degree of overestimation depends on the fraction of the second target getting amplified, because the standard curve is prepared with 10% GHB119 undigested DNA, thus containing two acp1 putatively amplifiable target copies, like the 10% undigested sample, while the digested sample contains only one. The quantification results, available in Table S2 (Supplementary Information), indicate that the rate of amplification of the second acp1 target in real-time PCR is none, or very limited. In fact, the TspRI digested samples were measured to contain 10.77% GM, compared to the non-digested samples, with 10.43%. For cotton, only one among the validated assays has a single target, the one for AdhC. The one for SAH7 addresses two targets, both with a perfect match for the primers and probe, but with different amplicon length. The assay for acp1 also addresses two targets, but the rate of amplification of the non-intended target in real-time PCR seems to be very low. For quantification purposes, the assay for AdhC seems to be the best candidate because it addresses a single target. However, if this assay was not available to the testing laboratory, the one for SAH7 should be preferred over the assay for acp1, provided that it is taken into account that it amplifies two DNA targets. In fact SAH7 primers and probe anneal without mismatches to both target DNAs, which should thus be amplified and detected at the same rate regardless of the real-time PCR conditions.
Fig. 1.
Temperature gradient ddPCR results for the different taxon-specific assays. Results were all automatically analysed except for ccf, for which automatic analysis does not resolve positive from negative droplets.
Two maize CRMs were tested with three different taxon-specific assays for hmg, ZmAdh1 and aldolase. The λ values measured with the three assays are reported in Table 1. The difference between the measured λ value was significant for all compared assays, although the ratio between the λ values measured with ZmAdh1 and hmg is approaching 1 (0.88). The significant difference identified between the λ values measured by these two assays may be explained by the failed amplification of some hmg targets, or other effects which should be further investigated. This fact has to be taken into consideration when comparing ddPCR data of GM quantification relative to these two maize taxon-specific reference assays. The ratio between λ values is close to 0.5 when ZmAdh1 is compared to aldolase and to 2 when aldolase is compared to hmg (Table 1). These results indicate that the targets of hmg and ZmAhd1 are single, while aldolase has two targets in the maize genome, therefore confirming the observations of the bioinformatics analyses. The presence of two populations of positive droplets for the assay for aldolase further supports this finding (Fig. 1). The ratios between the different taxon-specific assays tested were also compared individually for each reference material. Despite the large overall data set, due to the combination of conditions and targets tested, the number of data per condition only reaches the bare minimum for some of the statistical analyses. As a consequence, caution has to be taken when interpreting p values. Not all what is flagged as statistically significant can also be considered as biologically relevant. Therefore results were considered only truly significant if p value was <0.05 and the difference between both λ values was at least 20% of the lowest value (Table 2). These results indicate that the presence of a single target site for hmg and ZmAdh1 and of two targets for aldolase is not limited to only one reference material, but is confirmed for both CRMs used in the study. Both the assay for hmg and for ZmAdh1could be used in GMO quantification, although one particular reaction mix recipe for hmg was reported to be more stable in terms of quantification performance according to Paternò et al. (2009). The assay for aldolase instead targets two sequences that amplify with different efficiencies due to the presence of mismatches in the reverse primer and in the probe (Fig. 1) and is consequently not recommended for use. Based on the position of the two populations of droplets, which are close to each other and distant from the background droplet population, it might be inferred that both targets are amplified in real-time PCR. This is only a speculation which should be further verified experimentally. Due to the absence of restriction enzyme target sites specific for one of the two aldolase DNA targets this aspect was not tested further in our laboratory.
For oilseed rape the number of target copies was estimated for four assays: for cruA, ccf, FatA and FatA(A). The measured λ values for the different assays and their comparison are listed in Table 1 where it can be observed that the λ values for ccf and FatA(A), FatA and FatA(A) and cruA and FatA(A) are significantly different. The ratios between these λ values are close to 2 for cruA/FatA(A), at 2.22 for ccf/FatA(A,) at 2.35 for FatA/FatA(A) and close to 1 for the ratios ccf/cruA, ccf/FatA and cruA/FatA (Table 1). This indicates that two targets are amplified for cruA, ccf and FatA, and one for FatA(A), considering the variability potentially deriving from the presence of mismatches in the primers and probe annealing regions of the secondary targets of the assays for ccf and for FatA. These calculations and considerations do not take into account an additional band of droplets that is present in ccf amplification plots, which can be distinguished from the background by visual inspection of the plot, but which is not recognised automatically by the analysis software used. When this additional band of droplets is manually included in the positive droplet count, ccf measured λ values approximately double, indicating that the targets amplified in ddPCR for ccf are in reality 4. For oilseed rape the complexity and the incomplete B. napus genome assembly level did not allow identifying reliably all the targets of the assays for cruA, FatA and ccf. Bioinformatics analyses could only predict that the assays for cruA and ccf would have multiple targets and that FatA was more likely to amplify two targets than FatA(A) on the basis of the number and type of mismatches identified. The experimental results confirm these findings. It is, however, difficult to predict which results are produced by these assays in real-time PCR, especially in the case of ccf, because the band of droplets in ddPCR belonging to the third and fourth copies is almost undistinguishable from the band of the background droplets, which might indicate that the third and fourth targets are not amplified in real-time PCR. This assumption was not further verified. Significant and biologically relevant differences between the two certified reference materials used for the analyses were observed only for cruA/FatA and cruA/FatA(A) ratios between the two oilseed rape CRMs (Table 2). A possible explanation is a difference in the genetic background of the materials used, leading to a variability for all three assays. In fact, the λ value measured for cruA is lower in one material (ERM-BF434e, GM event 73496), while λ values for FatA and FatA(A) are higher in the other one (non-modified canola). This accounts for a greater difference between the ratios rather than the absolute values. FatA(A) was developed to be specific for the A sub-genome of all three canola species B. napus (A and C sub-genomes), B. rapa (A sub-genome) and B. juncea (A and B sub-genomes) (Henderson, Harmon, & Zhong, 2016). When tested on 6 different Brassica species which contain the A, B and C sub-genomes (B. napus, B. rapa, B. juncea, B. nigra - B sub-genome, B. carinata - B and C sub-genomes, and B. oleracea - C sub-genome), all assays except for FatA(A) showed amplification in all species. For FatA(A) λ values compatible with the presence of one target were obtained for B. rapa and B. napus, very few positive droplets for B. juncea and no positives for B. nigra, B. carinata and B. olearecea. All the other assays showed amplification with all the species tested, with the λ values reported in Table S3 (Supplementary Information). Thus, the assay for FatA(A) showed amplification results only with the three canola species B. rapa, B. napus and B. juncea, for which the assay was specifically designed (Henderson et al., 2016). Even though FatA(A) measurements are not able to distinguish which of the above mentioned species is present, they can detect canola species in single copy and discriminate against impurities. The surprisingly low number of positive droplets resulting from FatA(A) in the B. juncea sample, an A sub-genome species, should be related to the variety that was used for the test. In fact, there is evidence showing that this assay can efficiently amplify B. juncea DNA in different varieties (Henderson et al., 2016). Based on these findings, the use of FatA(A) is recommended.
3.2. Transferability of real-time PCR assays to the QX200 ddPCR platform
The ddPCR reaction mix composition and reaction conditions were based on the validated methods (published at http://gmo-crl.jrc.ec.europa.eu/StatusOfDossiers.aspx). Among all the validated variations of the same assay, the highest concentration of primers and/or probe validated was chosen (see Table S1) with the modifications described below. This choice was based on the need to be as close as possible to the primers/probe range suggested for the Biorad ddPCR Supermix for Probes, which is 900 nM for the primers and 250 nM for the probe (Bio-Rad Laboratories, Inc, a). The modifications introduced concerned: the probe concentration for the assays for acp1 and FatA, raised from 50 to 150 nM; the probe concentration for the assay for ccf, lowered from 250 to 150 nM in order to increase the resolution; the number of PCR cycles performed, 45 for all assays (different from the validated methods for ZmAdh1, aldolase, FatA(A), Le1 B, which employ 40 cycles). Assays for Le1 A, Le1 B, hmg, ZmAdh1, SAH7, AdhC. FatA and FatA(A) displayed a good peak resolution (a measure of the separation between positive and negative droplet clouds, optimal above 2.5) and limited rain, measured as relative number of droplets classified as rain in relation to the total number of compartments generated (below 2.5%). These values are in agreement with the acceptance criteria indicated in Lievens et al. (2016). Measurements for aldolase and acp1 showed instead limited rain but poor resolution due to the presence of a second, lower band of positive droplets most likely belonging to the second amplified target; signals for cruA had suboptimal resolution and ccf measurements had very poor resolution despite the optimisation effort described above (Fig. 2, with detailed results available in Table S4, Supplementary Information). The use of a temperature gradient allowed the clear identification of a second target with mismatches in the primers/probe for those assays that had one: in particular for acp1 and aldolase, but also to some extent for cruA and ccf. In some cases changing the annealing temperature improved the specificity of the assay, for instance for acp1, for which the second target droplet population merged with the negative droplet population at 62 °C, and for aldolase, for which at 62 °C the second target droplet population could be more easily distinguished from the specific droplet population. However, it did not dramatically improve rain or resolution for those assays (for cruA and ccf) that did not satisfy the performance criteria for these parameters, as also described by (Lievens et al., 2016), see Fig. 1.
Fig. 2.
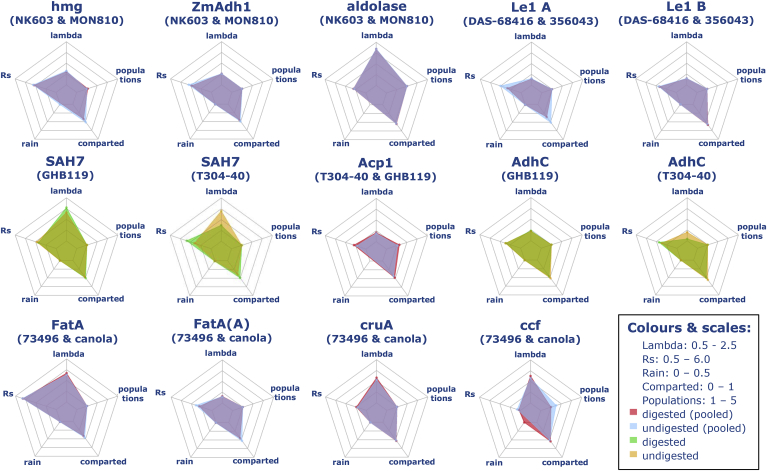
Comparison between digested and undigested template material per endogene. The mean ddPCR performance parameters are plotted as overlapping radar plots between reactions with digested and undigested template DNA. Where no significant difference was found between data for the two CRMs tested for each species results were pooled. The scale of each performance parameter's axis was adjusted to yield easily comparable plots (see legend). A different colour scheme was used depending on whether the data for the template materials were pooled or not. Lambda = measured λ values; Rs = resolution; Rain = ratio between partitions that were categorized as rain to the total number of partitions per reaction; Comparted = ratio of sample comparted to total sample volume; Population = number of populations identified (automatic analysis). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The study of the transferability of taxon-specific real-time PCR assays to the QX200 ddPCR platform was embedded in the main scope of this study. Except for the assay for ccf all others could be transferred from real-time PCR to ddPCR without problems. The use of ddPCR combined with a temperature gradient can reveal the presence of secondary targets, if there is a difference in amplification efficiency between them and the primary target. This was the case for the assays for aldolase, acp1, cruA and ccf. The temperature gradient can help identifying an annealing temperature at which the secondary target is not amplified. However, if this new annealing temperature is lower than the one used for the validation of the method, it is recommended to re-test the specificity of the method, as cross-reactivity might occur with other GMOs or potentially commingled species in food or feed. In some cases, ddPCR allows the use of a suboptimal reference assay for quantification, as in the case of acp1: the band of droplets of the secondary target can be excluded from the positive counts, thus allowing a correct GM quantification. This would not be possible with real-time PCR. For ccf the third and fourth target band of droplets could be confused with the background when the λ value is larger than 1 and the resolution is very low. The use of this assay is consequently not recommended in ddPCR without further optimisation.
3.3. Effects of DNA restriction enzyme digestion
To study the effects of DNA digestion on the performance of the taxon-specific assays tested in this study, all quantification experiments were performed in parallel with non-digested and digested aliquots of the same DNA. The DNA amount added per reaction was depending on the plant's genome size, and ranged between 26.6 ng for soybean and 64.2 ng for maize (calculated as described in 2.5). Digested versus non-digested DNAs were compared regarding the obtained λ values, resolution and percentage of rain. Digestion was performed with EcoRI for all crops except cotton, where DraI was used. The overall comparison between digested and non-digested DNAs evidenced p values close to 0.05 for resolution only, while all other parameters were not significantly different between digested and non-digested samples (see Table 3). The resolution was slightly lowered by DNA digestion, as can be seen from Table S4, summarising all results compared for λ values, resolution and rain, and observed in Fig. 2. The only exception are T304-40 cotton samples, whose DNA was not purified after DraI digestion. These samples had lower λ values, presumably an effect of incorrect quantification in the presence of the digestion buffer and enzyme. Fig. 2 also compares the results for sample compartmentalisation, which is the total volume of accepted droplets divided by the total loaded volume. The quality criterion defined in Lievens et al. (2016) for compartmentalisation is at least 30% for quantifications down to 1% GM. Since the taxon-specific assay targets are nominally 100% present in the materials used, the amount of compartmentalisation obtained is within this requirement. Another important practical consideration is that the process of DNA digestion and purification caused the loss of at least half of the initial amount of DNA.
Table 3.
Comparison of the overall effect of EcoRI and DraI digestion on λ values, resolution and rain. The p values are the probability value of the paired t-test comparison for each parameter. Within each sample and condition the results were averaged (i.e. average before compared to average after digestion).
| Restriction enzyme | Lambda p Value | Resolution p Value | Rain p Value |
|---|---|---|---|
| EcoRI | 0.223 | 0.052 | 0.126 |
| DraI | 0.256 | 0.068 | 0.513 |
There are conflicting statements in the scientific community about the necessity of digesting genomic DNA before the conduction of dPCR measurements. Digestion of genomic DNA is indispensable to obtain a random target distribution when the targets are on adjacent loci. DNA digestion or fragmentation is also considered useful to facilitate DNA distribution in the partitions by reducing large molecules to small fragments. This practice is usually recommended for both chamber and droplet dPCR, also by instrument manufacturers (Bio-Rad Laboratories, Inc, b; Bhat, Herrmann, Armishaw, Corbisier, & Emslie, 2009). However, the lack of DNA fragmentation did not induce a quantification bias in some cases (Morisset, Štebih, Milavec, Gruden, & Žel, 2013). With the samples and experimental conditions used in this study, DNA digestion prior to amplification did not have any positive effect on resolution, amplification, percentage of rain, and variability of results. Since for some samples more than 50% of DNA was lost in digestion and subsequent purification procedures, omitting the DNA digestion step may be convenient in terms of time and reagent consumption when the following conditions are met: the reference and the GMO target are not in linkage; the DNA amount used is at or below 26.6 ng for soybean, 27.1 ng for oilseed rape, 54.8 ng for cotton and 64.2 ng for maize; and the material from which DNA is extracted has a simple matrix similar to the unprocessed seed material used in this study.
4. Conclusions
In this study the number of targets for the most frequently used taxon-specific assays validated by the EURL GMFF in conjunction with different GM event-specific detection methods was assessed. The results confirm that not all reference genes are present in single copy. This fact has an influence on the relative quantification of GM events, as results are expressed in relation to a taxon-specific reference gene. In fact, knowledge of the exact copy number of a genomic sequence is crucial when it is used as a reference for relative quantification on a copy number basis and for the proper conversion between copy number ratios and mass fractions. Even more importantly, when mismatches are present in the primer/probe region of an assay's secondary target, its amplifiability in a real-time PCR experiment cannot be taken for granted and may vary with small changes in the reaction conditions, such as small differences in the annealing temperature between different instruments, or slight variations in the buffer or reaction mix composition or pH. Consequently, the choice of a reference assay that amplifies a single target is essential for correct relative GM quantification, as well as in other domains of nucleic acid quantification. Current work to characterise reference materials for their amount of target nucleic acid in copy number witnessed the relevance of the efforts and the trend of this development (Bhat & Emslie, 2016).
The absolute quantification provided by ddPCR has allowed us to elucidate the number of targets amplified in the genome of four crops (maize, soybean, cotton, oilseed rape) by the taxon-specific assays tested. At least one taxon-specific assay can be considered in single copy for each crop, and a short list of taxon-specific reference assays is proposed for the most commonly transformed crops that are best candidates for the relative quantification of GM events, with the considerations previously discussed: lec A and lec B for soybean; hmg and ZmAdh1 for maize; FatA(A) for oilseed rape; and AdhC or SAH7 for cotton (see Table 4). Moreover, the results of this study indicate that, with the exception of the assay for ccf, the taxon-specific validated assays investigated can be transferred to ddPCR without further optimisation, when the probe concentration is scaled up to a minimum of 150 nM. The use of DNA digestion has been evaluated and does not seem to improve the quality metrics (percentage of rain, resolution) of ddPCR tests, when DNA is extracted from simple matrixes (unprocessed seed material) and less than 64 ng DNA per reaction are used.
Table 4.
Short list of the taxon-specific reference assays for the most commonly transformed crops that are proposed as best candidates for the relative quantification of GM events. The number of estimated targets for each assay, the presence of mismatches in the primers and probe annealing regions of the secondary targets, and the recommendation for GMO analysis are presented in the table.
| Crop | Assay name | Estimated number of DNA targets | Presence of mismatches in secondary primers and probe annealing regions | Recommended for GMO analysis |
|---|---|---|---|---|
| soybean |
Le1 A | 1 | – | Yes |
| Le1 B |
1 |
– |
Yes |
|
| maize |
hmg | 1 | – | Yes |
| ZmAdh1 | 1 | – | Yes, although one reaction mix for hmg was reportedly more stable than the assay for ZmAdh1 in terms of quantification performance (Paternò et al., 2009) | |
| aldolase |
2 |
Yes, one mismatch in the reverse primer and one in the probe |
No, secondary target with mismatches in the primers and probe annealing region |
|
| cotton |
acp1 | 2 | Yes, two mismatches in the primer forward, one in the primer reverse, and one in the probe | No, secondary target with mismatches in the primers and probe annealing region |
| AdhC | 1 | – | Yes | |
| SAH7 |
2 |
No |
The assay for AdhC should be preferred, but SAH7 can also be used because primers and probe anneal without mismatches to both target DNAs |
|
| oilseed rape | cruA | 2 | Yes, position and number of mismatches not clear | No, secondary target with mismatches in the primers and probe annealing region |
| ccf | 4 | Yes, position and number of mismatches not clear | No, secondary target with mismatches in the primers and probe annealing region | |
| FatA(A) | 1 | – | Yes | |
| FatA | 2 | Yes, position and number of mismatches not clear | No, secondary target with mismatches in the primers and probe annealing region |
Acknowledgements
This work was financially supported by the European Commission.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.foodcont.2018.06.013.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
Table Making script.R
References
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bennett M.D., Leitch I.J. 2012. Plant DNA C-values database.http://data.kew.org/cvalues/CvalServlet?querytype=1 Release 6.0, December, 2012. Retrieved March 14, 2018, from. [DOI] [PubMed] [Google Scholar]
- Bhat S., Emslie K.R. Digital polymerase chain reaction for characterisation of DNA reference materials. Biomolecular Detection and Quantification. 2016;10:47–49. doi: 10.1016/j.bdq.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat S., Herrmann J., Armishaw P., Corbisier P., Emslie K.R. Single molecule detection in nanofluidic digital array enables accurate measurement of DNA copy number. Analytical and Bioanalytical Chemistry. 2009;394:457–467. doi: 10.1007/s00216-009-2729-5. [DOI] [PubMed] [Google Scholar]
- Bio-Rad Laboratories (a). ddPCR Supermix for Probes. Retrieved March 14, 2018, from http://www.bio-rad.com/webroot/web/pdf/lsr/literature/10026235.pdf.
- Bio-Rad Laboratories (b). Droplet Digital PCR Applications Guide. Retrieved March 14, 2018, from https://info.bio-rad.com/rs/bioradlaboratoriesinc/images/Bulletin_N6407.pdf.
- Corbisier P., Barbante A., Berben G., Broothaerts W., De Loose M., Emons H.…Trapmann S. Publications Office of the European Union; 2017. Recommendation for the unit of measurement and the measuring system to report traceable and comparable results expressing GM content in accordance with EU legislation.http://gmo-crl.jrc.ec.europa.eu/ENGL/docs/WG-UoM-Final-Report.pdf Retrieved from. [Google Scholar]
- Corbisier P., Bhat S., Partis L., Rui Dan Xie V., Emslie K.R. Absolute quantification of genetically modified MON810 maize (Zea mays L.) by digital polymerase chain reaction. Analytical and Bioanalytical Chemistry. 2010;396:2143–2150. doi: 10.1007/s00216-009-3200-3. [DOI] [PubMed] [Google Scholar]
- Corbisier P., Pinheiro L., Mazoua S., Kortekaas A.-M., Chung P.Y.J., Gerganova T.…Emslie K. DNA copy number concentration measured by digital and droplet digital quantitative PCR using certified reference materials. Analytical and Bioanalytical Chemistry. 2015;407:1831–1840. doi: 10.1007/s00216-015-8458-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debode F., Huber I., Macarthur R., Rischitor P.E., Mazzara M., Herau V.…Zel J. Inter-laboratory studies for the validation of two singleplex (tE9 and pea lectin) and one duplex (pat/bar) real-time PCR methods for GMO detection. Food Control. 2017;73:452–461. [Google Scholar]
- Deprez L., Corbisier P., Kortekaas A.-M., Mazoua S., Beaz Hidalgo R., Trapmann S. Validation of a digital PCR method for quantification of DNA copy number concentrations by using a certified reference material. Biomolecular Detection and Quantification. 2016;9:29–39. doi: 10.1016/j.bdq.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobnik D., Štebih D., Blejec A., Morisset D., Žel J. Multiplex quantification of four DNA targets in one reaction with Bio-Rad droplet digital PCR system for GMO detection. Scientific Reports. 2016;6(1) doi: 10.1038/srep35451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EURL GMFF . 2005. Event-specific method for the quantitation of maize line TC1507 using real-time PCR - validation report and protocol.http://gmo-crl.jrc.ec.europa.eu/StatusOfDossiers.aspx Retrieved March 14, 2018, from. [Google Scholar]
- EURL GMFF . 2006. Event-specific methods for the quantitation of the hybrid cotton line 281-24-236/3006-210-23 using real-time PCR - validation Report and Protocol.http://gmo-crl.jrc.ec.europa.eu/StatusOfDossiers.aspx Retrieved March 14, 2018, from. [Google Scholar]
- EURL GMFF . 2007. Event-specific method for the quantification of oilseed rape line Ms8 using real-time PCR - validation Report and Protocol.http://gmo-crl.jrc.ec.europa.eu/StatusOfDossiers.aspx Retrieved March 14, 2018, from. [Google Scholar]
- EURL GMFF . 2009. Event-specific methods for the quantification of cotton MON 88913 using real-time PCR - validation Report and Protocol.http://gmo-crl.jrc.ec.europa.eu/StatusOfDossiers.aspx Retrieved March 14, 2018, from. [Google Scholar]
- EURL GMFF . 2012. Event-specific method for the quantification of cotton T304-40 using real-time PCR - validation report and protocol.http://gmo-crl.jrc.ec.europa.eu/StatusOfDossiers.aspx Retrieved March 14, 2018, from. [Google Scholar]
- EURL GMFF . 2012. Event-specific method for the quantification of soybean FG72-validation report and protocol.http://gmo-crl.jrc.ec.europa.eu/StatusOfDossiers.aspx Retrieved March 14, 2018, from. [Google Scholar]
- EURL GMFF . 2013. Event-specific method for the quantification of oilseed rape DP-073496-4 using real-time PCR - validation report and protocol.http://gmo-crl.jrc.ec.europa.eu/StatusOfDossiers.aspx Retrieved March 14, 2018, from. [Google Scholar]
- EURL GMFF . 2013. Event-specific method for the quantification of oilseed rape MON88302 by real-time PCR - validation report and protocol.http://gmo-crl.jrc.ec.europa.eu/StatusOfDossiers.aspx Retrieved March 14, 2018, from. [Google Scholar]
- EURL GMFF . 2014. Event-specific method for the quantification of maize 5307 by real-time PCR - validation report and protocol.http://gmo-crl.jrc.ec.europa.eu/StatusOfDossiers.aspx Retrieved March 14, 2018, from. [Google Scholar]
- EURL GMFF . 2015. Event-specific method for the quantification of soybean DAS-44406-6 by real-time PCR - validation report and protocol.http://gmo-crl.jrc.ec.europa.eu/StatusOfDossiers.aspx Retrieved March 14, 2018, from. [Google Scholar]
- EURL GMFF . 2015. Definition of minimum performance requirements for analytical methods of GMO testing.http://gmo-crl.jrc.ec.europa.eu/doc/MPR%20Report%20Application%2020_10_2015.pdf April 20, Retrieved from. [Google Scholar]
- EURL GMFF . 2016. Event-specific method for the quantification of maize VCO-01981-5 using real-time PCR - validation report and protocol.http://gmo-crl.jrc.ec.europa.eu/StatusOfDossiers.aspx Retrieved March 14, 2018, from. [Google Scholar]
- European Commission Commission Regulation (EU) No 619/2011 of 24 June 2011 laying down the methods of sampling and analysis for the official control of feed as regards presence of genetically modified material for which an authorisation procedure is pending or the authorisation of which has expired Text with EEA relevance. Off J Eur Union. 2011;L 166:9–15. [Google Scholar]
- European Parliament and the Council of the European Union Regulation (EC) No 1829/2003 of the european parliament and of the Council of 22 september 2003 on genetically modified food and feed. Off J Eur Union. 2003;L 268:0001–0023. [Google Scholar]
- European Parliament and the Council of the European Union Regulation (EC) No 641/2004 on detailed rules for the implementation of Regulation (EC) No 1829/2003. Off J Eur Union. 2004;L 102:14–25. [Google Scholar]
- European Parliament and the Council of the European Union Regulation (EC) No 882/2004 on official controls performed to ensure the verification of compliance with feed and food law, animal health and animal welfare rules. Off J Eur Union. 2004;L165:1–141. [Google Scholar]
- European Parliament and the Council of the European Union Commission implementing Regulation (EU) No 120/2014 of 7 february 2014 amending Regulation (EC) No 1981/2006 on detailed rules for the implementation of article 32 of Regulation (EC) No 1829/2003 of the european parliament and the Council as regards the community reference laboratory for genetically modified organisms text with EEA relevance. Off J Eur Union. 2014;L 39:46–52. [Google Scholar]
- European and Parliament the Council of the European Union Commission implementing Regulation (EU) No 503/2013 of 3 april 2013 on applications for authorisation of genetically modified food and feed in accordance with Regulation (EC) No 1829/2003 of the european parliament and of the Council and amending commission Regulations (EC) No 641/2004 and (EC) No 1981/2006 text with EEA relevance. Off J Eur Union. 2013;L 157:1–48. [Google Scholar]
- Fraiture M.-A., Herman P., Taverniers I., De Loose M., Deforce D., Roosens N.H. Current and new approaches in GMO Detection: Challenges and solutions. BioMed Research International. 2015;2015:1–22. doi: 10.1155/2015/392872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson N., Harmon M., Zhong C.X. PCR-based detection and quantification of a transgenic glyphosate-tolerant canola using a novel Reference gene system. Food Analytical Methods. 2016;9:353–361. [Google Scholar]
- Huggett J.F., Foy C.A., Benes V., Emslie K., Garson J.A., Haynes R.…Bustin S.A. The digital MIQE Guidelines: Minimum information for publication of quantitative digital PCR experiments. Clinical Chemistry. 2013;59:892–902. doi: 10.1373/clinchem.2013.206375. [DOI] [PubMed] [Google Scholar]
- Institute for Reference Materials and Measurements, Joint Research Centre . 2011. Certificate of analysis ERM- BF425d.https://crm.jrc.ec.europa.eu/p/40455/40460/By-material-matrix/Plant-materials/ERM-BF425d-SOYA-356043-level-3-nominal-10-GMO/ERM-BF425d Retrieved March 14, 2018, from. [Google Scholar]
- International Organization for Standardization . 2005. ISO 21571:2005 Foodstuffs - methods of analysis for the detection of genetically modified organisms and derived products - nucleic acid extraction. Geneva. [Google Scholar]
- Iwobi A., Gerdes L., Busch U., Pecoraro S. Droplet digital PCR for routine analysis of genetically modified foods (GMO) – a comparison with real-time quantitative PCR. Food Control. 2016;69:205–213. [Google Scholar]
- Jacchia S., Nardini E., Bassani N., Savini C., Shim J.-H., Trijatmiko K.…Mazzara M. International Ring trial for the validation of an event-specific golden rice 2 quantitative Real-time polymerase chain Reaction method. Journal of Agricultural and Food Chemistry. 2015;63:4954–4965. doi: 10.1021/acs.jafc.5b00951. [DOI] [PubMed] [Google Scholar]
- Jacchia S., Nardini E., Savini C., Petrillo M., Angers-Loustau A., Shim J.-H.…Mazzara M. Development, optimization, and single laboratory validation of an event-specific Real-time PCR method for the detection and quantification of golden rice 2 using a novel taxon-specific assay. Journal of Agricultural and Food Chemistry. 2015;63:1711–1721. doi: 10.1021/jf505516y. [DOI] [PubMed] [Google Scholar]
- Lievens A., Jacchia S., Kagkli D., Savini C., Querci M. Measuring digital PCR Quality: Performance parameters and their optimization. PLoS One. 2016;11(5) doi: 10.1371/journal.pone.0153317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Fan G., Lu C., Xiao G., Zou C., Kohel R.J.…Yu S. Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nature Biotechnology. 2015;33:524–530. doi: 10.1038/nbt.3208. [DOI] [PubMed] [Google Scholar]
- Monsanto Biotechnology Regulatory Sciences . 2004. A recommended produce for real-time quantitative TaqMan® PCR for roundup Ready® canola RT73.http://gmo-crl.jrc.ec.europa.eu/detectionmethods/MON-Art47-pcrGT73rapeseed.pdf Retrieved March 14, 2018, from. [Google Scholar]
- Morisset D., Štebih D., Milavec M., Gruden K., Žel J. Quantitative analysis of food and feed samples with droplet digital PCR. PLoS One. 2013;8(5) doi: 10.1371/journal.pone.0062583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaharu U. Genome analysis in Brassica with special Reference to the experimental formation of B. Napus and peculiar mode of fertilization. Japanese Journal of Botany. 1935;7:389–452. [Google Scholar]
- Parisi C., Tillie P., Rodríguez-Cerezo E. The global pipeline of GM crops out to 2020. Nature Biotechnology. 2016;34(1):31–36. doi: 10.1038/nbt.3449. [DOI] [PubMed] [Google Scholar]
- Paternò A., Marchesi U., Gatto F., Verginelli D., Quarchioni C., Fusco C.…Ciabatti I. Finding the joker among the maize endogenous Reference genes for genetically modified organism (GMO) detection. Journal of Agricultural and Food Chemistry. 2009;57:11086–11091. doi: 10.1021/jf902560x. [DOI] [PubMed] [Google Scholar]
- Rotmistrovsky K., Jang W., Schuler G.D. A web server for performing electronic PCR. Nucleic Acids Research. 2004;32:W108–W112. doi: 10.1093/nar/gkh450. (Web Server) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykes P.J., Neoh S.H., Brisco M.J., Hughes E., Condon J., Morley A.A. Quantitation of targets for PCR by use of limiting dilution. BioTechniques. 1992;13:444–449. [PubMed] [Google Scholar]
- Vogelstein B., Kinzler K.W. Digital PCR. Proceedings of the National Academy of Sciences. 1999;96:9236–9241. doi: 10.1073/pnas.96.16.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel J.F. New World tetraploid cottons contain Old World cytoplasm. Proceedings of the National Academy of Sciences of the United States of America. 1989;86:4132–4136. doi: 10.1073/pnas.86.11.4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Žel J., Mazzara M., Savini C., Cordeil S., Camloh M., Štebih D.…Van den Eede G. Method validation and quality management in the flexible scope of Accreditation: An example of laboratories testing for genetically modified organisms. Food Analytical Methods. 2008;1(2):61–72. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table Making script.R