Abstract
The pathogenesis of systemic lupus erythematosus (SLE) is influenced by both genetic factors and epigenetic modifications; the latter is a result of exposure to various environmental factors. Epigenetic modifications affect gene expression and alter cellular functions without modifying the genomic sequences. CpG-DNA methylation, histone modifications, and miRNAs are the main epigenetic factors of gene regulation. In SLE, global and gene-specific DNA methylation changes have been demonstrated to occur in CD4+ T-cells. Moreover, histone acetylation and deacetylation inhibitors reverse the expression of multiple genes involved in SLE, indicating histone modification in SLE. Autoreactive T-cells and B-cells have been shown to alter the patterns of epigenetic changes in SLE patients. Understanding the molecular mechanisms involved in the pathogenesis of SLE is critical for the introduction of effective, target-directed and tolerated therapies. In this review, we summarize the recent findings that highlight the importance of epigenetic modifications and their mechanisms in SLE.
Keywords: DNA methylation, Epigenesis, Histones, microRNAs, Systemic lupus erythematosus
INTRODUCTION
Systemic lupus erythematosus (SLE) is a severe, chronic autoimmune disease that is characterized by the involvement of multiple organs including kidney, joints, and skin[1,2]. SLE is more prevalent in women (female:male ratio is 9:1); 70-90% of SLE patients are female. The increased frequency of SLE among women have been attributed to the effects of sex hormones on interferon (IFN)-α and toll-like receptor (TLR) as well as aberrant X chromosome inactivation or X chromosome dosage effects[3-7]. The main cause of this disease has not been determined yet, but it is thought to be multifactorial etiology, including the interaction of many genes, epigenetic factors viz DNA CpG methylation, histone tail modification, non-coding RNA (miRNAs, lncRNA, and siRNA), and environmental factors (sunlight, drugs, and infectious elements, especially Epstein-Barr virus)[4, 8-11].
The initial approaches, linkage analysis and candidate gene association studies, have identified 40 SLE-associated loci. The genome-wide association study could screen hundreds of thousands of single nucleotide polymorphisms (SNPs) and eight chromosome regions across genome in SLE patients[12-16]. The majority of SLE susceptibility genes encode the products involved in innate and adaptive immunity[17,18]. Among these varieties of elements for SLE etiology, today epigenetic factors are in the center of attention. Actually, epigenetics means beyond the genetics and includes some special changes in genome. It consist of three main modifications: DNA CpG methylation, histones modification (i.e. the addition of acetyl, methyl, and other chemical groups to some especial residue of histones), and lncRNA such as miRNA, in order to regulate mRNA expression[19]. Methylation modifications can occur through ultraviolet (UV) radiation, dietary contributions, and aging. Meanwhile, decrease in methylation level of several immune-related genes, e.g. TGAL (integrin alpha L chain, CD11a), CD40LG, TNFSF7 (CD70), KIR2DL4, and PRF1, can influence their expression in lupus T-cells. In addition, the increased H4 acetylation levels in monocytes, as one of the histone modifications, is frequently seen in SLE patients[20]. Several miRNAs, especially miR-21, miR-148a, and miR-126, can control the transcription of DNMT1 (DNA methyltransferase 1), a key component of DNA methylation[21].
A wide variety of studies have indicated epigenetic roles in SLE etiology. Epigenome-wide studies coupled with functional analysis of the epigenomic changes are able to determine novel important pathways and their mechanisms in the pathogenesis of some diseases like SLE. Given the importance of epigenetic factors, it can be expected that epigenetic therapy would be used and could be possible for SLE patients in the future, particularly when it is designed for target-specific regions within the genome. Therefore, in this review, we focus on the most important epigenetic factors and their mechanisms in SLE pathogenesis.
DNA methylation in SLE patients
Methylation of DNA is one of the most important epigenetic modifications that can change gene expression by adding methyl group to the deoxycytosine base in CpG dinucleotide, to form deoxymethylcytosine. DNA methylation modifications can influence gene expression and play an important role in SLE (Fig. 1). MECP2 (methyl-CpG-binding protein 2), MBD2 (methyl-CpG-binding domain), and DNMT1 are the main parts of DNA methylation processes in different cells. Increased expressions of both MBD2 and DNMT1 in SLE patients could cause DNA hypermethylation and gene dysregulation[22,23]. According to different analyses of CpG methylation, including CD4+ T-cells, CD19+ B-cells, and CD14+ monocytes, done in various immune cell types of several SLE patients, it can be assumed that lupus patients exhibit more global DNA hypomethylation in CD4+ T-cell[24]. DNA demethylation and over-expression also occur in several genes as well as in TNFSF7 (CD70) that are normally methylated. TNFSF7 encodes CD70 on B-cell that contributes to antibody production. CD70 hypo-methylation and overexpression in T-cells of SLE patients cause IgG overexpression and production[25].
Fig. 1.
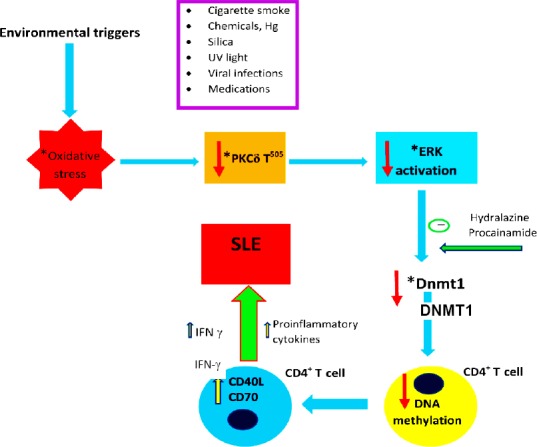
Schematic presentation of the proposed pathways in association with environmental factors that play a critical role in development of systemic lupus erythematosus (SLE). These processes take place inside the CD4+ T-cell. Hg, mercury; PKCδ T505, phosphorylated protein kinase C δ; ERK, extracellular signal-regulated kinase; DNMT1, DNA methyltransferase 1.
Environmental elements may affect epigenetic factors such as DNA methylation modifications. Exposure to UV light is associated with exacerbation of SLE. In addition, 73% of SLE patients have photosensitivity, which can be used as a diagnostic factor for this disease. Most cutaneous lesions occur in the light-exposed areas and can be triggered by sunlight exposure. UV light, especially UVB (290-320 nm), can reduce the expression of DNMT1 and that may cause T-cell auto-reactivity, accordingly inducing SLE[26,27]. Lymphocyte function-associated antigen-1 (LFA1) incorporates ITGAL and ITGB2 subunits, which is expressed on all leukocytes. Demethylation of the ITGAL gene promoter region requires an underlying mechanism for overexpression of LFA-1 on an auto-response set of T-cells in SLE patients. Indeed, LFA-1 overexpression, successively induces antibody production in B-cells, is believed to be concerned in T lymphocyte auto-reactivity in SLE[28,29].
DNA hypomethylation in CD4+ T-cells of SLE patients may happen under the influence of some chemical medications, together with 5-azacytidine, procainamide, and hydralazine, which could subsequently have impact on the expression of critical genes[30-32]. The 5-azacytidine is a cytosine analogue integrated into DNA during DNA replication and prevents DNA de novo methylation. Other DNA methylation inhibitors, like procainamide and hydralazine, are involved in CD4+ T-cells autoreactivity, in which autoreactive T-cell responds to MHC class II without the existence of exogenous antigen[33-35].
Patients with active lupus show lower methylated cytosine content (deoxymethylcytosine, about 4%) in their many genes of T-cells, as well as in ITGAL and TNFSF7. CD11a, perforin, and the KIR genes are also demethylated and overexpressed in patients with active, but not inactive, lupus. As a result, they can be used as a marker for distinguishing the disease activity[36].
DNA methylation can be one of the possible reasons for the SLE prevalence in females (∼90% of cases) through X chromosome inactivation. Female lupus patients display impaired DNA methylation on the inactive X chromosome. There are several X-linked genes that assist in SLE pathogenesis. DNA hypomethylation of CD40L on X chromosome plays an important role in the female predominance of SLE. Furthermore, the prevalence of SLE in women with Turner Syndrome (45, X0) is lower, but individuals with Klinefelter syndrome (47, XXY) have more potential for the progression of SLE[37-39]. It has been also shown that the hypermethylation of anti-inflammatory genes has a function in SLE pathogenesis[40]. HLA-DR alpha gene is hypermethylated in B-cells of SLE patients that may express a few amounts of HLA-DR antigen. The decreased expression of HLA-DR antigens and the HLA-DR alpha gene are associated with high anti-DNA antibody titers in patient’s serum[41].
Effects of cell signaling on DNA methylation in SLE patients
DNA methylation can be regulated through several signaling pathways viz extracellular signal-regulated kinase (ERK) pathway (PKC [protein kinase c]→ Ras→ Raf→ MEK→ ERK). PKC is a member of protein kinase enzymes family and has catalytic domain in its c-terminal that involves in regulating a number of proteins by phosphorylating their serine and threonine residues in the ERK pathway with its c-terminal domain as a catalytic domain. PKC is located on 3p21.31 and contributes significantly to many cellular processes, including regulation of cell growth and programmed cell death, additionally in B-cell negative selection[42-44]. PKCδ is one of the members of PKC family. PKCδ phosphorylation is diminished in lupus patients. Strong evidence has shown that defective PKCδ phosphorylation is associated with ERK pathway deficiency and lower DNMT1 gene expression, thereby influencing DNA demethylation and up-regulation of the several genes including CD11A, CD70, CD40L, the pro-inflammatory cytokine IL-17A as well as several interferon-regulated genes[45]. PKCδ is also phosphorylated in response to other stimuli and activates ERK pathway. However, in patient with active lupus, PKCδis not properly phosphorylated in response to some chemical compounds, such as phorbol myristate acetate.. This phenomenon can be linked to the disturbance of ERK pathway and the low level of DNMT1 in CD4+ T-cells[45,46]. Another reason could be the increased level of reactive oxygen species and reactive nitrogen intermediates in lupus, which can induce the inappropriate phosphorylation of PKCδ through post-translational modifications[47,48]. A recent study has shown homozygous missense mutation in PKCδ (c. 1294G>T; p. Gly432Trp) of juvenile SLE patients. This mutation affects catalytic domain of PKCδ, which causes the loss of PKCδ function and the early onset of juvenile SLE. Missense mutations of PKCδ can also lead to Mendelian juvenile-onset SLE through increased B-cell proliferation with the resistance of B-cell to B cell receptor and Ca2+-dependent apoptosis[49].
Methylation modifications have recently been considered as a diagnostic and a prognostic marker for detecting response to therapy, and also the level of DNA methylation can be a potential biomarker for disease activity[50]. There are several major methylation changes in SLE patients that are shown in the Table 1.
Table 1.
Some important genes and their methylation changes in SLE
| Gene | Cell type | Methylation level | Effect in SLE | Ref. |
|---|---|---|---|---|
| CD6 | CD3+ T-cells | + | Enhanced T-cell activation | [77,78] |
| CREM | CD3+ T-cells, CD4+ T-cells, Effector CD4d+ T-cells | + | Involved in the generation of DN T-cells and regulation of IL-2 and IL-17 in CD4+ T-cells | [77,78] |
| FOXP3 | Treg | + | Reduced number and altered function of regulatory T-cells | [77] |
| IL-2 | CD3+ T-cells, CD4+ T-cells, Effector CD4+ T-cells | ++ | Impaired production of regulatory T-cells, impaired function of cytotoxic CD8+ T-cells, effector CD4+ T-cell differentiation, and cytokine expression | [77,80] |
| CDKN1A | PBMCs | ++ | Potential effects on apoptosis and DNA repair | [77] |
| SOCS1 | CD4+xs T | ++ | Immune dysregulation | [20,77] |
| TREX1 | PBMCs | ++ | Impaired exonuclease function and cytosolic DNA accumulation and the survival of autoreactive cells | [77,81] |
Reduced DNA methylation;
increased DNA methylation; CD6, cluster of differentiation 6; FOXP3, forkhead-box-protein P3; CDKN1A, cyclin-dependent kinase inhibitor 1A; SOCS1, suppressor of cytokine signaling 1; PBMC, peripheral blood mononuclear cell; TREX1, three prime repair exonuclease 1
miRNAs in SLE
miRNAs have an important role in SLE pathogenesis and progression through their functions in humoral and cellular immune system and immune cell development[21]. Recent studies have shown a different expression pattern of miRNAs in peripheral blood mononucleated cells (PBMCs) of SLE patients that indicate their contribution in SLE[51,52]. The miR-146a is located on the susceptible and predisposing locus 5q33.3 in SLE pathogenesis, which regulates IFN pathway and is underexpressed in the PBMCs of lupus patients. The miR-146a down-regulation can induce IFN pathway activity by targeting key proteins such as IRAK1 and STAT1. Moreover, an A/G SNP (rs5095329) within the promoter of miR-146a reduces promoter activity and its expression, which is correlated with SLE[53,54]. The miR-125a is another down-regulated miRNA in PBMCs of SLE patients that can indirectly controls RANTES expression through binding to KLF13 mRNA in activated T-cells[55]. miR-3148 is predicted to bind 3’-UTR region of TLR7 to decrease the expression of TLR7, as a main component of the innate immune system, that finally leads to a high inflammatory response in SLE patients. However, SNP G allele rs385839 in the 3’-UTR of the TLR7 gene can inhibit its binding to miR-3148 to increase the expression of TLR7 at mRNA and protein levels through reducing mRNA degradation. However, individuals who carry C allele of this SNP in the 3’-UTR of TLR7 show a decreased amount of TLR7 level, resulting in mRNA degradation[56]. On the other hand, miR-1246 expression decreases in the B-cell of SLE patients and is attached to the 3’-UTR of early B-cell factor 1 (EBF1) mRNA. Therefore, miR-1246 overexpression causes EBF1 mRNA degradation. EBF1 is an important player in activation, development, and division of B-cell by triggering the AKT signaling pathway[57]. miR-155 is another miRNA that is associated with SLE. It inhibits MYD88 and TAB2 to block inflammatory response. In contrast, it can increase the inflammatory response in macrophage and induces type 1 interferon signaling by targeting suppressor of cytokine signaling 1 (SOCS1)[58,59]. miR-let-7a (let-7a) stimulates immune system responses and an inflammatory component production, which contributes to SLE pathogenesis. Its overexpression may result in hyperplasia and a pro-inflammatory response. IL-6 contains a potential binding site for let-7a in its 3’-UTR and can lead to its production[60].
Effects of miRNAs on DNA methylation in SLE patients
A previous study has shown that several miRNAs are regulated by epigenetic mechanisms, e.g. some miRNAs on the X chromosome can be influenced by DNA methylation during X chromosome inactiv-ation[61]. Some other studies have demonstrated that DNA demethylation occurs on the inactive X chromosome in female patients. Therefore, miRNAs such as miR-98, miR-188-3p, and let-7f-2, which are located on this chromosome, are highly expressed, which is likely due to the higher prevalence of SLE in females than males[62,63]. The overexpression of miR-148a and miR-126 (regulators of DNMT1) in CD4+ T-cells of SLE patints leads to global DNA hypomethylation. Each of these miRNAs directly inhibit DNMT1 through binding to its target 3’-UTR[61,64]. There are some miRNAs that affect DNMT1 indirectly. For instance, miR-29b reduces the expression of sp1, a DNMT1 transactivator, and miRNA-21 decreases the activity of Ras-MAPK-DNMT1 signaling pathway in T-cells of SLE patients[65]. Table 2 lists the miRNAs with their role in SLE (Table 2).
Table 2.
miRNAs in the pathogenesis of SLE
| miRNA | Target | Regulated process | Expression level in SLE | Ref. |
|---|---|---|---|---|
| miR-let-7a | IL-6 | IL-6 induction | UP | [60] |
| miR-let-7c | Blimp1, SOCS1 | Activation of DCs | UP | [82] |
| miR-125a | KLF13 | CCL5 induction in T-cells | DOWN | [55] |
| miR-146a | TRAF6, IRAK1, TRAF6, IRAK1, IRAK2, IRF5, STAT1 | NFκB mediated inflammatory response RIG-I-dependent anti-viral pathway Type I IFN induction and signaling | DOWN | [53,54,83] |
| miR-150 | SOCS1 | Renal fibrosis | DOWN | [84] |
| miR-155 | MyD88, TAB2 SOCS1, PP2Ac | TLR/IL-1 inflammatory pathway, Type I IFN signaling, IL-2 induction | UP | [59,85-87] |
| miR-17~92 | PTEN, BimRora, PHLPP2 | The proliferation of lymphocytes, Differentiation, and function of Tfh cells | UP | [65,88] |
| miR-23b | TAB2, TAB3, IKKα | IL-17, TNF-α, IL-1β signaling | DOWN | [89] |
| miR-30a | Lyn | Activation of B-cells | UP | [90] |
| miR-31 | RhoA | IL-2 induction in T-cells | DOWN | [91] |
UP, up-regulated; DOWN, down-regulated
Histones modifications in SLE patients
Acetylation, phosphorylation, and methylation of histones tails are the most important changes among histones modifications. A variety of enzymes and complexes of proteins, including lysine acetyl-transferases, histone deacelytase, lysine methyltransferases, and lysine demethylases, are responsible for creating specific epigenetic codes (histone methylation, histone acetylation, etc.). Epigenetic codes are extremely conserved and determine the phenotype and function of cells and tissues. Any destruction of histone codes contributes to the etiology of many diseases, especially autoimmune disorders like SLE. One of the histone code modifications occurs on lys9 and lys27 of H3 that can trigger chromatin compacting and gene silencing.
SLE patients have indicated decreased acetylation of whole histones and H3K9 methylation in their CD4+ T-cells. Promoter region of hematopoietic progenitor kinase 1 (HPK1) in CD4+ T-cells has trimethylated lys27 H3 that leads to the inhibition of HPK1 expression and assist in auto-immune response in SLE[66]. Down-regulation of HPK1 also results in the enhancement of T-cell level and the production of INF-γ and [54,67].
Evidence has shown that di-methylation of lys4 H3 increases the promoter of TNFSF7 (CD70) gene in SLE CD4+ cells, which is correlated with disease activity[68]. H3K4me3 level has also been demonstrated to elevate in some candidate genes, including PTPN22 and LRP1B, in PBMC of SLE patients[69].
Histone changes usually have relationship with DNA methylation; a methylated region of DNA has deacetylated histone that helps gene silencing and chromatin compacting. As an example, down- regulation of IL-2 expression with mediating cAMP-responsive element modulator in T-cells of lupus patients happens through both histone deacetylation and DNA hypermethylation[70]. Unusual histone acetylations have been observed near to IL-17 gene locus in T-cell of lupus patients[71].
Neutrophil extracellular traps (NETs) are chromatin structures that release from apoptotic blebs during apoptosis in many diseases like SLE[72]. Histones, which are secreted in NETosis process, are two to three times more acetylated on H2B-K12 and H4-K8, K12, and K16 and methylate on H3-K27 in SLE patients in contrast to healthy individuals, determining the association of the histones acetylation and methylation with apoptosis, NETs, and SLE[73,74]. Another histone modification in lupus patients is H3 and H4 acetylation, which is correlated with TNF-α locus and causes TNF-α hyper-expression in monocytes of SLE patients[75].
A genome-wide analysis and a global H4 acetylation analysis by ChIP-chip methodology show that the level of H4ac enhances in monocytes of SLE patients[71]. It has been suggested that 63% of genes having an abundance level of H4ac are associated with interferon regulatory factor 1 and the SLE pathogenesis[76]. Several major histone modifications in SLE are presented in Table 3.
Table 3.
Most important histone modifications within some genes in SLE
| Gene | Modification | Cell type | Effect in SLE | Ref. |
|---|---|---|---|---|
| CD8A, CD8B | H3K18 deacetylation, H3K27 trimethylation in DN T-cells | CD4+ T-cells, CD8+ T-cells, DN T-cells | Generation of CD3+ CD4− CD8− DN T-cells | [79] |
| ITGAL | Reduced H3K27 tri-methylation through histone demethylase | CD4+ T-cells | Increased T-cell-mediated inflammation | [92] |
| TNF | H3 acetylation | Monocytes | Increased monocyte maturation and pro-inflammatory cytokine expression | [75] |
| IL-2 | H3K18 deacetylation, H3K27 trimethylation | CD3+ T-cells, CD4+ T-cells, effector CD4+ T-cells | Impaired production of regulatory T-cells, reduced activation-induced cell death, and longer survival of autoreactive T-cells; impaired function of cytotoxic CD8+ T-cells, effector CD4+ T-cell differentiation, and cytokine expression | [93,94] |
Immune cells from SLE patients are characterized by a dysregulated gene expression profile. A significant proportion is caused by epigenetic alterations. A vast number of studies have identified several epigenetics factors, including miRNA, DNA methylation, and histone modification that are involved in SLE pathogenesis. This review tried to summarize some important areas of molecular pathogenesis of SLE. The presented data may facilitate the identification of new markers with possible application in diagnosis, prognosis, monitoring, or treatment of the disease.
Footnotes
CONFLICT OF INTEREST. None declared.
REFERENCES
- 1.Fairhurst AM, Wandstrat AE, Wakeland EK. Systemic lupus erythematosus: multiple immunological phenotypes in a complex genetic disease. Advances in immunology. 2006;92:1–69. doi: 10.1016/S0065-2776(06)92001-X. [DOI] [PubMed] [Google Scholar]
- 2.Ghodke-Puranik Y, Niewold TB. Immunogenetics of systemic lupus erythematosus: a comprehensive review. Journal of autoimmunity. 2015;64:125–136. doi: 10.1016/j.jaut.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shiari R, Yegane MH, farivar S, Parvaneh VJ, Mirjavadi SA. Neuropsychiatric Symptoms as The first manifestation of juvenile systemic lupus erythematosus: a complicated case with Klinefelter's syndrome. Iranian journal of child neurology. 2014;8(1):62–65. [PMC free article] [PubMed] [Google Scholar]
- 4.Tiffin N, Adeyemo A, Okpechi I. A diverse array of genetic factors contribute to the pathogenesis of systemic lupus erythematosus. Orphanet journal of rare diseases. 2013;8:2. doi: 10.1186/1750-1172-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chighizola C, Meroni PL. The role of environmental estrogens and autoimmunity. Autoimmunity reviews. 2012;11(6-7):A493–A501. doi: 10.1016/j.autrev.2011.11.027. [DOI] [PubMed] [Google Scholar]
- 6.Cooney CM, Bruner GR, Aberle T, Namjou-Khales B, Myers LK, Feo L, Li S, D'Souza A, Ramirez A, Harley JB, Scofield RH. 46,X,del (X)(q13) Turner's syndrome women with systemic lupus erythematosus in a pedigree multiplex for SLE. Genes and immunity. 2009;10(5):478–481. doi: 10.1038/gene.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shiari R, Farivar S. Juvenile systemic lupus erythematosus associated with Klinefelter's syndrome: a case report. Reumatologia clinica. 2010;6(4):212–213. doi: 10.1016/j.reuma.2009.09.016. [DOI] [PubMed] [Google Scholar]
- 8.Molokhia M, McKeigue P. Systemic lupus erythematosus: genes versus environment in high risk populations. Lupus. 2006;15(11):827–832. doi: 10.1177/0961203306070007. [DOI] [PubMed] [Google Scholar]
- 9.Criswell LA. The genetic contribution to systemic lupus erythematosus. Bulletin of the NYU hospital for joint diseases. 2008;66(3):176–183. [PubMed] [Google Scholar]
- 10.Kelly JA, Moser KL, Harley JB. The genetics of systemic lupus erythematosus: putting the pieces together. Genes and immunity. 2002;3:S71–S85. doi: 10.1038/sj.gene.6363885. [DOI] [PubMed] [Google Scholar]
- 11.Cervera R, Khamashta MA, Font J, Sebastiani GD, Gil A, Lavilla P, Doménech I, Aydintug AO, Jedryka-Góral A, de Ramón E. Systemic lupus erythematosus: clinical and immunologic patterns of disease expression in a cohort of 1,000 patients. Medicine (Baltimore) 1993;72(2):113–124. [PubMed] [Google Scholar]
- 12.Graham DSC. Genome-wide association studies in systemic lupus erythematosus: a perspective. Arthritis research and therapy. 2009;11(4):119. doi: 10.1186/ar2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsao BP. The genetics of human systemic lupus erythematosus. Trends in immunology. 2003;24(11):595–602. doi: 10.1016/j.it.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 14.Cui Y, Sheng Y, Zhang X. Genetic susceptibility to SLE: recent progress from GWAS. Journal of autoimmunity. 2013;41:25–33. doi: 10.1016/j.jaut.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 15.Farivar S, Tezerjani MD, Parvini N, Shiari R. Association of 1661A/G Cytotoxic T lymphocyte Antigen-4 (CTLA-4) Gene Polymorphism With a Clinical Subset of Iranian Children With Systemic Lupus Erythematosus. Thrita. 2014;3(1) 10.5812/thrita.16020. [Google Scholar]
- 16.Parvaneh VJ, Shiari R, Mahbobi L, Babaei D. Chronic granulomatous disease associated with systemic lupus erythematosus and systemic onset juvenile idiopathic arthritis. Pediatric Rheumatology online journal. 2014;12(Suppl 1):P169. [Google Scholar]
- 17.Beccastrini E, D'Elios MM, Emmi G, Silvestri E, Squatrito D, Prisco D, Emmi L. Systemic lupus erythematosus: immunopathogenesis and novel therapeutic targets. Internationakl journal of immunopathology and pharmacolofy. 2013;26(3):585–596. doi: 10.1177/039463201302600302. [DOI] [PubMed] [Google Scholar]
- 18.Farivar S, Hassani M, Shiari R. Interleukin-1 as a key factor in the development of inflammatory diseases. Archives of pediatric infectious diseases. 2014;2(4):e18177. [Google Scholar]
- 19.Zhao M, Liu S, Luo S, Wu H, Tang M, Cheng W, Zhang Q, Zhang P, Yu X, Xia Y, Yi N, Gao F, Wang L, Yung S, Chan TM, Sawalha AH, Richardson B, Gershwin ME, Li N, Lu Q. DNA methylation and mRNA and microRNA expression of SLE CD4+T cells correlate with disease phenotype. Journal of autoimmunity. 2014;54:127–136. doi: 10.1016/j.jaut.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 20.Zhao M, Wang J, Liao W, Li D, Li M, Wu H, Zhang Y, Gershwin ME, Lu Q. Increased 5-hydroxymethyl-cytosine in CD4+T cells in systemic lupus erythematosus. Journal of autoimmunity. 2016;69:64–73. doi: 10.1016/j.jaut.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 21.Yan S, Yim LY, Lu L, Lau CS, Chan VSF. MicroRNA regulation in systemic lupus erythematosus pathogenesis. Immune network. 2014;14(3):138–148. doi: 10.4110/in.2014.14.3.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu CC, Ou T, Wu CC, Li RN, Lin YC, Lin CH, Tsai WC, Liu HW, Yen JH. Global DNA methylation, DNMT1, and MBD2 in patients with systemic lupus erythematosus. Lupus. 2011;20(2):131–136. doi: 10.1177/0961203310381517. [DOI] [PubMed] [Google Scholar]
- 23.Sawalha AH, Webb R, Han S, Kelly JA, Kaufman KM, Kimberly RP, Alarcón-Riquelme ME, James JA, Vyse TJ, Gilkeson GS, Choi CB, Scofield RH, Bae SC, Nath SK, Harley JB. Common variants within MECP2 confer risk of systemic lupus erythematosus. PloS one. 2008;3(3):e1727. doi: 10.1371/journal.pone.0001727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Absher DM, Li X, Waite LL, Gibson A, Roberts K, Edberg J, Chatham WW, Kimberly RP. Genome-wide DNA methylation analysis of systemic lupus erythematosus reveals persistent hypomethylation of interferon genes and compositional changes to CD4+T-cell populations. PLoS genetics. 2013;9(8):e1003678. doi: 10.1371/journal.pgen.1003678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patel DR, Richardson BC. Epigenetic mechanisms in lupus. Current opinion in rheumatology. 2010;22(5):478–482. doi: 10.1097/BOR.0b013e32833ae915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang G, Zhang M, Li X, Zhang H, Chen W, Kan M, QWang YM. Ultraviolet B exposure of peripheral blood mononuclear cells of patients with systemic lupus erythematosus inhibits DNA methylation. Lupus. 2009;18(12):1037–1044. doi: 10.1177/0961203309106181. [DOI] [PubMed] [Google Scholar]
- 27.Wu Z, Li X, Qin H, Zhu X, Xu J, Shi W. Ultraviolet B enhances DNA hypomethylation of CD4+T cells in systemic lupus erythematosus via inhibiting DNMT1 catalytic activity. Journal of dermatological science. 2013;71(3):167–173. doi: 10.1016/j.jdermsci.2013.04.022. [DOI] [PubMed] [Google Scholar]
- 28.Yung RL, Richardson BC. Role of T cell DNA methylation in lupus syndromes. Lupus. 1994;3(6):487–491. doi: 10.1177/096120339400300611. [DOI] [PubMed] [Google Scholar]
- 29.Lu Q, Kaplan M, Ray D, Ray D, Zacharek S, Gutsch D, Richardson B. Demethylation of ITGAL (CD11a) regulatory sequences in systemic lupus erythematosus. Arthritis and rheumatology. 2002;46(5):1282–1291. doi: 10.1002/art.10234. [DOI] [PubMed] [Google Scholar]
- 30.Zhou Y, Lu Q. DNA methylation in T cells from idiopathic lupus and drug-induced lupus patients. Autoimmunity reviews. 2008;7(5):376–383. doi: 10.1016/j.autrev.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 31.Batchelor J, Welsh K, Tinoco RM, Dollery C, Hughes GR, Bernstein R, Ryna P, Naish PF, Aber GM, Bing RF, Russell GI. Hydralazine-induced systemic lupus erythematosus: influence of HLA-DR and sex on susceptibility. The lancet. 1980;1(8178):1107–1109. doi: 10.1016/s0140-6736(80)91554-8. [DOI] [PubMed] [Google Scholar]
- 32.Zhu X, Li F, Yang B, Liang J, Qin H, Xu J. Effects of ultraviolet B exposure on DNA methylation in patients with systemic lupus erythematosus. Experimental and therapeutic medicine. 2013;5(4):1219–1225. doi: 10.3892/etm.2013.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Richardson BC. Role of DNA methylation in the regulation of cell function: autoimmunity, aging and cancer. The Journal of nutrition. 2002;132(8 Suppl):2401S–2405S. doi: 10.1093/jn/132.8.2401S. [DOI] [PubMed] [Google Scholar]
- 34.Richardson B. Impact of aging on DNA methylation. Ageing research reviews. 2003;2(3):245–261. doi: 10.1016/s1568-1637(03)00010-2. [DOI] [PubMed] [Google Scholar]
- 35.Januchowski Ra, Prokop J, Jagodzinski PP. Role of epigenetic DNA alterations in the pathogenesis of systemic lupus erythematosus. Journal of applied genetics. 2004;45(2):237–248. [PubMed] [Google Scholar]
- 36.Pan Y, Sawalha AH. Epigenetic regulation and the pathogenesis of systemic lupus erythematosus. Translational research. 2009;153(1):4–10. doi: 10.1016/j.trsl.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 37.Stewart JJ. The female X-inactivation mosaic in systemic lupus erythematosus. Immunology today. 1998;19(8):352–357. doi: 10.1016/s0167-5699(98)01298-5. [DOI] [PubMed] [Google Scholar]
- 38.McDonald G, Cabal N, Vannier A, Umiker B, Yin RH, Orjalo AV, Jr, Johansson HE, Han JH, Imanish-Kari T. Female bias in systemic lupus erythematosus is associated with the differential expression of X-linked toll-like receptor 8. Frontiers in immunology. 2015;6:457. doi: 10.3389/fimmu.2015.00457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu Q, Wu A, Tesmer L, Ray D, Yousif N, Richardson B. Demethylation of CD40LG on the inactive X in T cells from women with lupus. The journal of immunology. 2007;179(9):6352–6358. doi: 10.4049/jimmunol.179.9.6352. [DOI] [PubMed] [Google Scholar]
- 40.Shirakawa F, Yamashita U, Suzuki H. Decrease in HLA-DR-positive monocytes in patients with systemic lupus erythematosus (SLE) The journal of immunology. 1985;134(6):3560–3562. [PubMed] [Google Scholar]
- 41.Sano H, Compton LJ, Shiomi N, Steinberg AD, Jackson RA, Sasaki T. Low expression of human histocompatibility leukocyte antigen-DR is associated with hypermethylation of human histocompatibility leukocyte antigen-DR alpha gene regions in B cells from patients with systemic lupus erythematosus. Journal of clinical investigation. 1985;76(4):1314–1322. doi: 10.1172/JCI112105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mellor H, Parker PJ. The extended protein kinase C superfamily. Biochemical Journal. 1998;332(Pt 2):281–292. doi: 10.1042/bj3320281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Newton AC. Protein kinase C: structure, function, and regulation. Journal of biological chemistry. 1995;270(480):28495–28498. doi: 10.1074/jbc.270.48.28495. [DOI] [PubMed] [Google Scholar]
- 44.Edwards AS, Newton AC. Phosphorylation at conserved carboxyl-terminal hydrophobic motif regulates the catalytic and regulatory domains of protein kinase C. Journal of biological chemistry. 1997;272(29):18382–18390. doi: 10.1074/jbc.272.29.18382. [DOI] [PubMed] [Google Scholar]
- 45.Gorelik G, Fang JY, Wu A, Sawalha AH, Richardson B. Impaired T cell protein kinase Cδactivation decreases ERK pathway signaling in idiopathic and hydralazine-induced lupus. The journal of immunology. 2007;179:5553–5563. doi: 10.4049/jimmunol.179.8.5553. [DOI] [PubMed] [Google Scholar]
- 46.Sawalha AH, Jeffries M, Webb R, Lu Q, Gorelik G, Ray D, Osban J, Knowlton N, Johnson K, Richardson B. Defective T-cell ERK signaling induces interferon-regulated gene expression and overexpression of methylation-sensitive genes similar to lupus patients. Genes and immunity. 2008;9(4):368–378. doi: 10.1038/gene.2008.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oelke K, Richardson B. Decreased T cell ERK pathway signaling may contribute to the development of lupus through effects on DNA methylation and gene expression. International reviews of immunology. 2004;23(3-4):315–331. doi: 10.1080/08830180490452567. [DOI] [PubMed] [Google Scholar]
- 48.Gorelik GJ, Yarlagadda S, Patel DR, Richardson BC. Protein kinase Cδoxidation contributes to ERK inactivation in lupus T cells. Arthritis and rheumatology. 2012;64(9):2964–2974. doi: 10.1002/art.34503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nanthapisal S, Omoyinmi E, Murphy C, Standing A, Eisenhut M, Eleftheriou D, Brogan PA. Early-onset juvenile SLE associated with a novel mutation in protein kinase C δ. Pediatrics. 2017;139(1):e20160781. doi: 10.1542/peds.2016-0781. [DOI] [PubMed] [Google Scholar]
- 50.Zhao S, Long H, Lu Q. Epigenetic perspectives in systemic lupus erythematosus: pathogenesis, biomarkers, and therapeutic potentials. Clinical reviews in allergy and immunology. 2010;39(1):3–9. doi: 10.1007/s12016-009-8165-7. [DOI] [PubMed] [Google Scholar]
- 51.Singh RP, Massachi I, Manickavel S, Singh S, Rao NP, Hasan S, Mc Curdy DK, Sharma S, Wong D, Hahn BH, Rehimi H. The role of miRNA in inflammation and autoimmunity. Autoimmunity reviews. 2013;12(12):1160–1165. doi: 10.1016/j.autrev.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 52.Dai R, Zhang Y, Khan D, Heid B, Caudell D, Crasta O, Ahmed SA. Identification of a common lupus disease-associated microRNA expression pattern in three different murine models of lupus. PloS one. 2010;5(12):e14302. doi: 10.1371/journal.pone.0014302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tang Y, Luo X, Cui H, Ni X, Yuan M, Guo Y, Huang X, Zhou H, de Vries N, Tak PP, Chen S, Shen N. MicroRNA-146a contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis and rheumatology. 2009;60(4):1065–1075. doi: 10.1002/art.24436. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Q, Long H, Liao J, Zhao M, Liang G, Wu X, Zhang P, Ding S, Luo S, Lu Q. Inhibited expression of hematopoietic progenitor kinase 1 associated with loss of jumonji domain containing 3 promoter binding contributes to autoimmunity in systemic lupus erythematosus. Journal of autoimmunity. 2011;37(3):180–189. doi: 10.1016/j.jaut.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 55.Zhao X, Tang Y, Qu B, Cui H, Wang S, Wang L, Luo X, Huang X, Li J, Chen S, Shen N. MicroRNA-125a contributes to elevated inflammatory chemokine RANTES levels via targeting KLF13 in systemic lupus erythematosus. Arthritis and rheumatology. 2010;62(11):3425–3435. doi: 10.1002/art.27632. [DOI] [PubMed] [Google Scholar]
- 56.Deng Y, Zhao J, Sakurai D, Kaufman KM, Edberg JC, Kimberly RP, Kamen DL, Gilkeson GS, Jacob CO, Scofield RH, Langefeld CD, Kelly JA, Ramsey-Goldman R, Petri MA, Reveille JD, Vilá LM, Alarcón GS, Vyse TJ, Pons-Estel BA, Argentine Collaborative Group. Freedman BI, Gaffney PM, Sivils KM, James JA, Gregersen PK, Anaya JM, Niewold TB, Merrill JT, Criswell LA, Stevens AM, Boackle SA, Cantor RM, Chen W, Grossman JM, Hahn BH, Harley JB, Alarcόn-Riquelme ME, BIOLUPUS and GENLES networks. Brown EE, Tsao BP. MicroRNA-3148 modulates allelic expression of toll-like receptor 7 variant associated with systemic lupus erythematosus. PLoS genetics. 2013;9(2):e1003336. doi: 10.1371/journal.pgen.1003336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Luo S, Liu Y, Liang G, Zhao M, Wu H, Liang Y, Qiu X, Dai Y, Yung S, Chan TM, Lu Q. The role of microRNA-1246 in the regulation of B cell activation and the pathogenesis of systemic lupus erythematosus. Clinical epigenetics. 2015;7:24. doi: 10.1186/s13148-015-0063-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang G, Tam LS, Li EK, Kwan BC, Chow KM, Luk CC, Li PK, Szeto CC. Serum and urinary cell–free MiR-146a and MiR-155 in patients with systemic lupus erythematosus. The Journal of rheumatology. 2010;37(12):2516–2522. doi: 10.3899/jrheum.100308. [DOI] [PubMed] [Google Scholar]
- 59.Tang B, Xiao B, Liu Z, Li N, Zhu ED, Li BS, Xie QH, Zhuang Y, Zou QM, Mao XH. Identification of MyD88 as a novel target of miR-155, involved in negative regulation of Helicobacter pylori-induced inflammation. FEBS letters. 2010;584(8):1481–1486. doi: 10.1016/j.febslet.2010.02.063. [DOI] [PubMed] [Google Scholar]
- 60.Chafin CB, Regna NL, Dai R, Caudell DL, Reilly CM. MicroRNA-let-7a expression is increased in the mesangial cells of NZB/W mice and increases IL-6 production in vitro. Autoimmunity. 2013;46(6):351–362. doi: 10.3109/08916934.2013.773976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pan W, Zhu S, Yuan M, Cui H, Wang L, Luo X, Li J, Zhou H, Tang Y, Shen N. MicroRNA-21 and microRNA-148a contribute to DNA hypomethylation in lupus CD4+T cells by directly and indirectly targeting DNA methyltransferase 1. The journal of immunology. 2010;184(12):6773–6781. doi: 10.4049/jimmunol.0904060. [DOI] [PubMed] [Google Scholar]
- 62.Hewagama A. Role of X-chromosome encoded miRNAs in autoimmunity: suppressing the suppressor and female predisposition. Rheumatology: current research. 2013;3:118. [Google Scholar]
- 63.Amarilyo G, La Cava A. miRNA in systemic lupus erythematosus. Clinical immunology. 2012;144(1):26–31. doi: 10.1016/j.clim.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 64.Zhao S, Wang Y, Liang Y, Zhao M, Long H, Ding S, Yin H, Lu Q. MicroRNA-126 regulates DNA methylation in CD4+T cells and contributes to systemic lupus erythematosus by targeting DNA methyl-transferase 1. Arthritis and rheumatology. 2011;63(5):1376–1386. doi: 10.1002/art.30196. [DOI] [PubMed] [Google Scholar]
- 65.Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, Henderson JM, Kutok JL, Rajewsky K. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nature immunology. 2008;9(4):405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Altorok N, Sawalha AH. Epigenetics in the pathogenesis of systemic lupus erythematosus. Current opinion in rheumatology. 2013;25(5):569–576. doi: 10.1097/BOR.0b013e328364206f. [DOI] [PubMed] [Google Scholar]
- 67.Sawasdikosol S, Alzabin S, Burakoff S. Hematopoietic progenitor kinase 1 for modulation of an immune response. Retrieved from: https://patents.google.com/ patent/US20070087988A1/lt .
- 68.Zhou Y, Qiu X, Luo Y, Yuan J, Li Y, Zhong Q, Zhao M, Lu Q. Histone modifications and methyl-CpG-binding domain protein levels at the TNFSF7 (CD70) promoter in SLE CD4+T cells. Lupus. 2011;20(13):1365–1371. doi: 10.1177/0961203311413412. [DOI] [PubMed] [Google Scholar]
- 69.Dai Y, Zhang L, Hu C, Zhang Y. Genome-wide analysis of histone H3 lysine 4 trimethylation by ChIP-chip in peripheral blood mononuclear cells of systemic lupus erythematosus patients. Clinical and experimental rheumatology. 2010;28(2):158–168. [PubMed] [Google Scholar]
- 70.Rauen T, Hedrich CM, Juang YT, Tenbrock K, Tsokos GC. cAMP-responsive element modulator (CREM) αprotein induces interleukin 17A expression and mediates epigenetic alterations at the interleukin-17A gene locus in patients with systemic lupus erythematosus. Journal of biological chemistry. 2011;286(50):43437–43446. doi: 10.1074/jbc.M111.299313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang Z, Song L, Maurer K, Petri MA, Sullivan KE. Global H4 acetylation analysis by ChIP-chip in systemic lupus erythematosus monocytes. Genes and immunity. 2010;11(2):124–133. doi: 10.1038/gene.2009.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Knight JS, Kaplan MJ. Lupus neutrophils:'NET'gain in understanding lupus pathogenesis. Current opinion in rheumatology. 2012;24(5):441–450. doi: 10.1097/BOR.0b013e3283546703. [DOI] [PubMed] [Google Scholar]
- 73.Liu CL, Tangsombatvisit S, Rosenberg JM, Mandelbaum G, Gillespie EC, Gozani OP, Alizadeh AA, Utz PJ. Specific post-translational histone modifications of neutrophil extracellular traps as immunogens and potential targets of lupus autoantibodies. Arthritis research and therapy. 2012;14(1):R25. doi: 10.1186/ar3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pieterse E, Hofstra J, Berden J, Herrmann M, Dieker J, Vlag J. Acetylated histones contribute to the immunostimulatory potential of neutrophil extracellular traps in systemic lupus erythematosus. Clinical and experimental immunology. 2015;179(1):68–74. doi: 10.1111/cei.12359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sullivan KE, Suriano A, Dietzmann K, Lin J, Goldman D, Petri MA. The TNFαlocus is altered in monocytes from patients with systemic lupus erythematosus. Clinical immunology. 2007;123(1):74–81. doi: 10.1016/j.clim.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Leung YT, Shi L, Maurer K, Song L, Zhang Z, Petri M, Sullivan KE. Interferon regulatory factor 1 and histone H4 acetylation in systemic lupus erythematosus. Epigenetics. 2015;10(3):191–199. doi: 10.1080/15592294.2015.1009764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sui W, Tan Q, Yang M, Yan Q, Lin H, Ou M, Xue W, Chen J, Zou T, Jing H, Guo L, Cao C, Sun Y, Cui Z, Dai Y. Genome-wide analysis of 5-hmC in the peripheral blood of systemic lupus erythematosus patients using an hMeDIP-chip. International journal of molecular medicine. 2015;35(5):1467–1479. doi: 10.3892/ijmm.2015.2149. [DOI] [PubMed] [Google Scholar]
- 78.Singer N, Richardson B, Powers D, Hooper F, Lialios F, Endres J, Bott CM, Fox DA. Role of the CD6 glycoprotein in antigen-specific and autoreactive responses of cloned human T lymphocytes. Immunology. 1996;88(4):537–543. [PMC free article] [PubMed] [Google Scholar]
- 79.Hedrich CM, Crispín JC, Rauen T, Ioannidis C, Koga T, Rodriguez NR, Apostolidis SA, Kyttaris VC, Tsokos GC. cAMP responsive element modulator (CREM) αmediates chromatin remodeling of CD8 during the Generation of CD3+CD4- CD8- T cells. Journal of biological chemistry. 2014;289(4):2361–2370. doi: 10.1074/jbc.M113.523605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Apostolidis SA, Lieberman LA, Kis-Toth K, Crispín JC, Tsokos GC. The dysregulation of cytokine networks in systemic lupus erythematosus. Journal of interferon and cytokine research. 2011;31(10):769–779. doi: 10.1089/jir.2011.0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Crispín JC, Hedrich CM, Tsokos GC. Gene-function studies in systemic lupus erythematosus. Nature reviews rheumatology. 2013;9(8):476–484. doi: 10.1038/nrrheum.2013.78. [DOI] [PubMed] [Google Scholar]
- 82.Kim SJ, Gregersen PK, Diamond B. Regulation of dendritic cell activation by microRNA let-7c and BLIMP1. The Journal of clinical investigation. 2013;123(2):823–833. doi: 10.1172/JCI64712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hou J, Wang P, Lin L, Liu X, Ma F, An H, Wang Z, Cao X. MicroRNA-146a feedback inhibits RIG-I-dependent Type I IFN production in macrophages by targeting TRAF6, IRAK1, and IRAK2. The journal of immunology. 2009;183(3):2150–2158. doi: 10.4049/jimmunol.0900707. [DOI] [PubMed] [Google Scholar]
- 84.Zhou H, Hasni SA, Perez P, Tandon M, Jang SI, Zheng C, Kopp JB, Austin H, 3rd, Balow JE, Alevizos I, Illei GG. miR-150 promotes renal fibrosis in lupus nephritis by downregulating SOCS1. Journal of the American society of nephrology. 2013;24(7):1073–1087. doi: 10.1681/ASN.2012080849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang P, Hou J, Lin L, Wang C, Liu X, Li D, Ma F, Wang Z, Cao X. Inducible microRNA-155 feedback promotes type I IFN signaling in antiviral innate immunity by targeting suppressor of cytokine signaling 1. The journal of immunology. 2010;185(10):6226–6233. doi: 10.4049/jimmunol.1000491. [DOI] [PubMed] [Google Scholar]
- 86.Lashine YA, Salah S, Aboelenein HR, Abdelaziz AI. Correcting the expression of miRNA-155 represses PP2Ac and enhances the release of IL-2 in PBMCs of juvenile SLE patients. Lupus. 2015;24(3):240–247. doi: 10.1177/0961203314552117. [DOI] [PubMed] [Google Scholar]
- 87.Ceppi M, Pereira PM, Dunand-Sauthier I, Barras E, Reith W, Santos MA, Pierre P. MicroRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells. Proceedings of the national academy of sciences. 2009;106(8):2735–2740. doi: 10.1073/pnas.0811073106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kang SG, Liu W-H, Lu P, Jin HY, Lim HW, Shepherd J, et al. MicroRNAs of the miR-17 [sim] 92 family are critical regulators of TFH differentiation. Nature immunology. 2013;14:849–857. doi: 10.1038/ni.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhu S, Pan W, Song X, Liu Y, Shao X, Tang Y, Liang D, He D, Wang W, Shi Y, Harely JB, Shen N, Qian Y. The microRNA miR-23b suppresses IL-17-associated autoimmune inflammation by targeting TAB2, TAB3 and IKK-α. Nature medicine. 2012;18(7):1077–1086. doi: 10.1038/nm.2815. [DOI] [PubMed] [Google Scholar]
- 90.Liu Y, Dong J, Mu R, Gao Y, Tan X, Li Y, Li Z, Yang G. MicroRNA-30a Promotes B Cell Hyperactivity in Patients With Systemic Lupus Erythematosus by Direct Interaction With Lyn. Arthritis and rheumatology. 2013;65(6):1603–1611. doi: 10.1002/art.37912. [DOI] [PubMed] [Google Scholar]
- 91.Fan W, Liang D, Tang Y, Qu B, Cui H, Luo X, Huang X, Chen S, Higgs BW, Jallal B, Yao Y, Harely JB, Shen N. Identification of microRNA-31 as a novel regulator contributing to impaired interleukin-2 production in T cells from patients with systemic lupus erythematosus. Arthritis and rheumatology. 2012;64(11):3715–3725. doi: 10.1002/art.34596. [DOI] [PubMed] [Google Scholar]
- 92.Yin H, Wu H, Zhao M, Zhang Q, Long H, Fu S, Lu Q. Histone demethylase JMJD3 regulates CD11a expression through changes in histone H3K27 tri-methylation levels in CD4+T cells of patients with systemic lupus erythematosus. Oncotarget. 2017;8(30):48938–48947. doi: 10.18632/oncotarget.16894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Crispín JC, Oukka M, Bayliss G, Cohen RA, Van Beek CA, Stillman IE, Kyttaris VC, Juang YT, Tsokos GC. Expanded double negative T cells in patients with systemic lupus erythematosus produce IL-17 and infiltrate the kidneys. The journal of immunology. 2008;181(12):8761–8766. doi: 10.4049/jimmunol.181.12.8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hedrich CM, Rauen T, Tsokos GC. cAMP-responsive element modulator (CREM) αprotein signaling mediates epigenetic remodeling of the human interleukin-2 gene implications in systemic lupus erythematosus. Journal of biological chemistry. 2011;286(50):43429–43436. doi: 10.1074/jbc.M111.299339. [DOI] [PMC free article] [PubMed] [Google Scholar]


