Abstract
Background
Partner notification is an important component of public health test and treat interventions. To enhance this essential function, we assessed the potential for molecular methods to supplement routine partner notification and corroborate HIV networks.
Methods
All persons diagnosed with HIV infection in Wake County, NC during 2012–2013 and their disclosed sexual partners were included in a sexual network. A dataset containing HIV-1 pol sequences collected in NC during 1997–2014 from 15,246 persons was matched to HIV-positive persons in the network and used to identify putative transmission clusters. Both networks were compared.
Results
The partner notification network comprised 280 index cases and 383 sexual partners and high-risk social contacts (n=131 HIV-positive). Of the 411 HIV-positive persons in the partner notification network, 181 (44%) did not match to a HIV sequence, 61 (15%) had sequences but were not identified in a transmission cluster, and 169 (41%) were identified in a transmission cluster. More than half (59%) of transmission clusters bridged sexual network partnerships that were not recognized in the partner notification; most of these clusters were dominated by men who have sex with men.
Conclusions
Partner notification and HIV sequence analysis provide complementary representations of the existent partnerships underlying the HIV transmission network. The partner notification network components were bridged by transmission clusters, particularly among components dominated by men who have sex with men. Supplementing the partner notification network with phylogenetic data highlighted avenues for intervention.
Keywords: HIV-1, molecular epidemiology, partner notification, sexual networks, surveillance, North Carolina
Introduction
Across the Southern United States (US), including North Carolina (NC), the HIV epidemic has persisted in large connected sexual networks, particularly among men who have sex with men (MSM).1–5 The South is the epicenter of the US epidemic, accounting for a disproportionate number of HIV infections.1 HIV incidence continues to rise among Black and Hispanic/Latino MSM,6 despite widespread prevention efforts. Entry into a sexual network composed largely of Black MSM increases the likelihood of contracting HIV,3 highlighting the importance of enumerating sexual networks. An improved understanding of sexual networks will aid in the development of enhanced interventions to reach Black and Hispanic/Latino MSM. Time-intensive efforts to reach members of densely-connected sexual networks often result in analysis of incomplete networks, due in part to anonymous partners, persons who cannot be located, and interview refusal.7
Phylogenetic analysis of HIV sequences is an excellent adjunct to enumerating networks and allows tracking of local transmission patterns. HIV phylogenies based on sequence similarity and inference of common ancestors can identify putative transmission clusters.8,9 While these methods are increasingly used to understand HIV transmission dynamics within sub-populations,10–12 use of sequence data to complement sexual networks as understood by contacts elicited during partner notification services (PNS) is understudied.13 Sequence data has potential to add structure to the sexual network through genetic linkage of network components that erroneously appear disjointed due to inability to locate network members.14–16 In San Diego, for example, HIV genetic clusters combined with PNS data from recently-infected MSM increased membership in putative transmission networks.15 In an investigation of spatiotemporally-clustered acute HIV infections in NC, phylogenetics revealed multiple transmission chains rather than a single outbreak.17 Such analyses demonstrate that sequence data can enhance our knowledge of sexual networks. Analysis of phylogenetic transmission cluster growth can also point to groups in which HIV transmission continues to occur,18 signaling the need for immediate intervention.19,20
We investigated the sexual network constructed from PNS data in Wake County, NC, and compared this with HIV transmission clusters using pol sequences routinely collected statewide. Our objective was to assess the overlap between networks derived through PNS and sequence analysis to identify areas where interventions could be intensified.
Methods
Study Setting and Design
Wake County is a metropolitan county in central NC that accounts for approximately 10% of statewide annual new HIV diagnoses.21 In 2012, Wake County had a population of approximately 963,000 persons, including >2,800 persons living with HIV and an incidence of 16.3 cases per 100,000 person-years.21
We conducted a cross-sectional analysis of Wake County residents ≥18 years of age who were newly diagnosed with HIV-1 during 2012–2013 and their social and sexual contacts reported during routine PNS. These data were compared with 15,246 HIV genetic sequences collected among HIV cases in NC 1997–2014. The University of North Carolina Biomedical Institutional Review Board approved the study.
Study Population
Disease intervention specialists (DIS), employed by NC Department of Health and Human Services (DHHS) or Wake County DHHS, attempt to interview all newly diagnosed persons (referred to as index cases) and collect information about their partners for tracing and testing. In NC, high risk social contacts are elicited at the discretion of each DIS when perceived to increase case finding without overly burdening investigations.22,23 Using standardized data abstraction, we collected demographics, HIV testing history, and HIV-related laboratory results for index cases, and sexual and social contact data.
Acute HIV infection (AHI) was identified through the NC Screening and Tracing Active Transmission (STAT) Program,24 and defined by a positive HIV RNA test and negative or indeterminate HIV antibody, or a positive HIV antibody within 30 days of confirmed negative testing. Cases who did not meet the AHI definition but were reported to STAT with a positive antibody test with seronegative documentation and/or symptoms compatible with AHI within 3 months of first positive HIV test were classified as recent HIV infection (RHI). For persons diagnosed with AHI or RHI, DIS interviews focus on partnerships within 2 or 6 months prior to diagnosis, respectively.
Sexual Network Construction
We constructed the sexual network using name-based partnership data collected during PNS interviews with index cases. All network members were de-identified after network construction to preserve patient confidentiality. A socio-sexual network comprises discrete components (at least two people directly or indirectly connected) and singletons (isolated persons if no partners are disclosed or located). The network was created using the igraph25 package in R.26
HIV-1 Sequences and Transmission Cluster Identification
HIV-1 pol sequences (full length protease and partial reverse transcriptase) were extracted from genotypes performed by LabCorp®, the largest reference laboratory in NC, and sampled between 1997 and mid-2014 from patients accessing clinical care. Demographic variables available included birth date, gender, and sampling site. Geographic location of sampling site was categorized by NC-DHHS HIV Field Service Region (Figure, Supplemental Digital Content (SDC) 1).
Index and HIV-positive partners were probabilistically matched to the statewide sequence dataset by birth date, gender, and laboratory test dates. We considered nonmatching sequences as background references for cluster construction. All analyses used the earliest sequence per individual. The final dataset included 15,246 sequences. A random subset of 100 sequences is available in GenBank, accession numbers KY579388-KY579812.
Sequences were aligned using MUSCLE27 and edited manually in BioEdit,28 with a final sequence alignment length of 1,497 bases. Maximum-likelihood (ML) phylogenies were constructed in FastTree29 with the generalized time-reversible model.30 Statistical support of clades was assessed with local support values using the Shimodaira-Hasegawa-like test (SH-test).31 Putative transmission clusters were identified using ClusterPicker v1.332 and defined as clades with 1) high branch support (≥0.90 SH-test), 2) maximum pairwise genetic distance <3.5% between all sequences, and 3) inclusion of a sequence from at least one index or partner case.
Putative clusters were confirmed with the Bayesian Markov Chain Monte Carlo (MCMC) approach in BEAST v1.8.2.33 Analyses were conducted using the SRD06 nucleotide substitution model, a lognormal relaxed molecular clock model, and the Bayesian Skyline model as coalescent tree prior. The MCMC chain was run for 50–100 million generations, sampling every 10,000 generations. Convergence of the estimates was considered satisfactory when the effective sample size calculated in Tracer v1.6.034 was >200 in all parameters; 10% of generations were discarded as burn-in. The maximum clade credibility tree was summarized using TreeAnnotator v1.8.2,33 keeping the median height over the posterior distribution of trees. Clades with posterior probability ≥0.95 were considered highly supported and analyzed further.
Statistical Analyses
We compared membership in transmission clusters and sexual network components. Clusters involving ≥2 cases (index or partners) were characterized by demographic features and compared to case location within and across network components. Time of most recent common ancestor (MRCA) and cluster age were estimated based upon timing of branching in the phylogeny.
Results
Study Population
In total, 280 persons newly diagnosed with HIV were reported in Wake County from 2012–2013; 83% (n=232) were male, 65% (n=183) were Black, and 40% (n=112) were younger than 30 years. Many (27%, n=75) were concurrently diagnosed with AIDS and 4% (n=11) were diagnosed during AHI. Among 235 index cases with CD4 count data, the median first CD4 count was 338 cells/mm3 (IQR 130–525 cells/mm3); 31% had CD4 count <200 cells/mm3. Among 147 cases with viral load results within 3 months of diagnosis, the median was 4.9 log copies/mL (IQR 4.3–5.3 log copies/mL) (Table 1).
Table 1.
Index cases diagnosed 2012–2013 in Wake County, NC and their partners in the sociosexual network (N=663).
| Index (n=280) | Partner (n=383) | ||||
|---|---|---|---|---|---|
| n | (%) | n | (%) | ||
| Gender | |||||
| Male | 232 | (83) | 327 | (85) | |
| Female | 44 | (16) | 53 | (14) | |
| Transgender (M to F) | 4 | (1) | 3 | (1) | |
| Race/ethnicity | |||||
| non-Hispanic White | 69 | (25) | 120 | (31) | |
| non-Hispanic Black | 183 | (65) | 238 | (62) | |
| Hispanic or Latino | 23 | (8) | 12 | (3) | |
| Other | 5 | (2) | 8 | (2) | |
| unknown | 0 | 5 | (1) | ||
| Age at index case's HIV diagnosis (years)* | |||||
| ≤ 19 | 5 | (2) | 28 | (7) | |
| 20–29 | 107 | (38) | 178 | (46) | |
| 30–39 | 54 | (19) | 87 | (23) | |
| 40–49 | 67 | (24) | 49 | (13) | |
| ≥ 50 | 47 | (17) | 26 | (7) | |
| unknown | 0 | 15 | (4) | ||
| median (IQR) | 34 | (25–45) | 28 | (23–37) | |
| HIV status | |||||
| Positive, with HIV sequence | 148 | (53) | 82 | (21) | |
| Positive, no HIV sequence | 132 | (47) | 49 | (13) | |
| Negative | --- | 148 | (39) | ||
| unknown | --- | 104 | (27) | ||
| Year of HIV diagnosis | n=131 | ||||
| < 2006 | --- | 31 | (24) | ||
| 2006–2010 | --- | 59 | (45) | ||
| 2011 | --- | 16 | (12) | ||
| 2012 | 131 | (47) | 11 | (8) | |
| 2013 | 149 | (53) | 9 | (7) | |
| 2014 | --- | 5 | (4) | ||
| HIV stage at diagnosis | |||||
| Acute / Recent | 23 | (8) | --- | ||
| Chronic, non-AIDS | 182 | (65) | --- | ||
| Chronic, AIDS | 75 | (27) | --- | ||
| CD4 count closest to diagnosis (cells/mm3) | n=235 | ||||
| < 200 | 74 | (31) | --- | ||
| ≥ 200 | 161 | (69) | --- | ||
| Viral load (log copies/mL)† | n=147 | n=60 | |||
| ≤ 3 | 8 | (5) | 29 | (48) | |
| > 3–5 | 78 | (53) | 10 | (17) | |
| > 5–5.7 | 44 | (30) | 1 | (2) | |
| > 5.7 | 17 | (12) | 20 | (33) | |
| median (IQR) | 4.9 | (4.3–5.3) | 3.3 | (2.9–7.7) | |
| Number of sexual and social partners reported‡ | n=225 | ||||
| 0 | 15 | (7) | --- | ||
| 1 | 78 | (35) | --- | ||
| 2 | 42 | (19) | --- | ||
| 3–5 | 61 | (27) | --- | ||
| ≥ 6 | 29 | (13) | --- | ||
Among partners, for earliest record associated with an index case
Within 3 months of diagnosis for index patients and within 12 months prior to index case diagnosis for partners
Among those reached for interview; includes located and anonymous partners
Partner Notification Network
DIS interviewed 225/280 index cases (80%), who reported 854 sex partners and 34 social contacts (average 4 contacts per person; number of sex partners ranged 0–50). Approximately half (50%; 446/888) of contacts (414 sexual and 32 social contacts) had enough locating information for DIS to begin investigation. The 446 partnerships investigated (Table 2) resulted in 383 unique non-index case partners (Table 1): 36 were index cases themselves, 19 were named by ≥2 index cases, and 3 were index cases who were also named as partners more than once. Although 48/383 (13%) partners were not located during investigation, we included them in the network. Of 383 partners, 39% were HIV-negative, 34% (n=131) were HIV-positive, and 27% HIV status was unknown. Most HIV-positive non-index partners (81%; 106/131) were diagnosed before 2012. Thirty-six percent (138/383) of partners resided outside of Wake County, including 22 (6%) residing out of state and 6 (2%) with unknown location of residence.
Table 2.
Partnerships reported by index cases with located members of the sociosexual network (N=446).
| Sociosexual Network Partnerships (N=446)* |
|||
|---|---|---|---|
|
| |||
| n | (%) | ||
| Partnership type | |||
| Sexual | 414 | (93) | |
| Social only | 32 | (7) | |
| Pair gender | |||
| Male – Male | 355 | (80) | |
| Male – Transgender | 5 | (1) | |
| Male – Female | 85 | (19) | |
| Female – Female | 1 | (0.2) | |
| Index case | |||
| Index – Index | 42 | (9) | |
| Index – Partner | 404 | (91) | |
| HIV serostatus | |||
| Positive – Positive (concordant) | 181 | (41) | |
| Positive – Negative (discordant) | 159 | (36) | |
| Positive – unknown | 106 | (24) | |
| Pair race | |||
| Black – Black | 261 | (59) | |
| White – White | 98 | (22) | |
| Hispanic/Latino – Hispanic/Latino | 8 | (2) | |
| Black – White | 40 | (9) | |
| White – Hispanic/Latino | 17 | (4) | |
| Black – Hispanic/Latino | 7 | (2) | |
| Other | 15 | (3) | |
104 singletons in the network are not represented in this table
The PNS network included 663 persons (Table 1), with 280 index cases and 383 partners. Most network members were Black (63% vs. 29% White and 5% Hispanic/Latino), MSM or men who have sex with transgender women (MST) (61%), and young (median age 30 years, IQR 24–42). Persons of color were more likely to be HIV-positive (74% Latino and 66% Black) compared to White persons (53%). MSM index cases were more likely to have partners who could not be located than men only reporting female partners (37% vs. 29%).
Overall, 176/280 index cases were connected to at least one other person in the network. The remaining 104 singletons represented 37% of index cases; 55 (53%) reported zero partners and 49 provided information for 1–50 partners, though none could be located. The sexual network was sparsely connected, comprising 104 singletons and 137 network components (≥2 persons). Component sizes ranged from 2–65 persons; the three largest included 20, 26, and 65 people (Figure 1a). Most (62%, n=85) components only included MSM and MST.
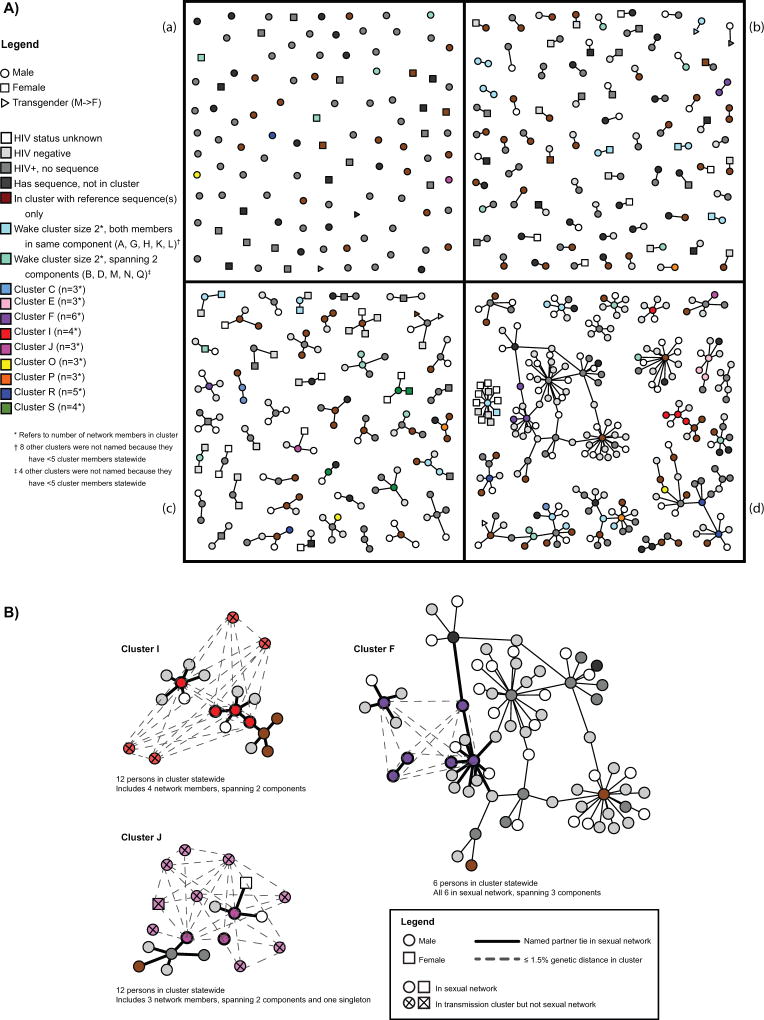
Figure 1. Sexual network showing phylogenetic cluster membership and gender (A), and selected sexual network components showing cluster members and genetic distance statewide (B).
1A) Sexual and social network compiled from contract tracing depicting HIV status and phylogenetic transmission cluster, Wake County, NC during 2012–2013. Graph shows gender (node shape), cluster membership with respect to gene sequence availability and cluster membership of other persons represented in this sexual network (node color), and partnerships disclosed by index cases (lines connecting nodes). The graph is split into quadrants by number of persons in each component: (a) singletons (n=104 persons), (b) dyads (n=75 components), (c) components size 3 (n=22), 4 (n=10), or 5 (n=12), and (d) components size 6 or larger (n=18 components comprising 243 persons).
1B) Selected phylogenetic transmission clusters (F, I, and J) show sexual network components spanned and additional cluster members statewide who were not part of the Wake County-based sexual network. Graph shows gender (node shape), appearance in sexual network or only transmission cluster (diagonal cross in node shape), transmission cluster status (node color), and connections between nodes. Having a named partner tie (i.e., connection in the sociosexual network) is represented by a solid line and being ≤1.5% pairwise genetic distance in the transmission cluster is represented by a dashed line. Component orientation matches Figure 1a.
NC, North Carolina
We assessed characteristics of the 446 partnerships (93% sexual and 7% social), which included 559 persons across 137 network components (excluding 104 singletons) [Table 2]. Most partnerships involved either MSM or MST (81%), were among people of the same race (82%), and included at least one Black person (71%). Nearly 25% (n=106) of partnerships were between an index case and a person with unknown HIV status. Among 340 partnerships where HIV status was documented for both people, 53% involved two HIV-infected persons (n=181). Most (80%) of the 131 HIV-infected partners received their diagnoses before the index cases (median 2.5 years, IQR 1 month-5.5 years).
Transmission Clusters
Over half of HIV-positive cases (56%; 230/411) matched to a pol sequence, including 53% (148/280) index cases and 63% (82/131) HIV-positive partners. Cases who had sequences were similar to those without sequences with respect to gender and age. Among index cases, Whites were more likely than persons of color to have sequences (64% vs. 49%, p=0.04), as well as those diagnosed in 2012 compared to 2013 (63% vs. 44%, p=0.002).
We identified 116 clusters involving ≥1 person from the network, with a total of 800 persons including 103 index cases (70% those with sequences), 66 partners (80% those with sequences), and 631 background sequences (Figure 2). In the initial ML analysis, 117 clusters were identified but two sequences failed to cluster in the confirmatory BEAST analyses. The 116 confirmed clusters had median size two members (range 2–36 persons); only three clusters were non-B subtypes (A1, CRF02_AG, CRF06_cpx).
Figure 2. Phylogenetic tree of HIV pol gene sequences showing transmission clusters.
Maximum-likelihood tree constructed for display purposes using sequences (n=800) identified in confirmed phylogenetic transmission clusters among 15,246 HIV-1 positive persons sampled in North Carolina 1997–2014. Confirmed clusters had posterior probability >0.98 in the Bayesian analysis and include at least one index or partner case identified during partner notification of new HIV diagnoses in Wake County, 2012–2013. Index cases (new diagnoses in 2012–2013) are indicated by red circles and partner cases are indicated with blue circles at the tips of the tree. Clusters in grey boxes involve ≥2 cases from the partner notification network. Clusters with letters (A–S) are the Wake clusters that meet these criteria and also include ≥5 persons statewide. Branch support, using the Shimodaira-Hasegawa-like test values, is included for the Wake clusters.
Among 230 index cases (n=148) and partners (n=82) with sequences, we evaluated associations with cluster membership. Cluster members were more likely to be male (77% vs. 52% female, p=0.006), men reporting male contacts (83% vs. 67% heterosexual and 57% no partners reported, p=0.001), Black (80% vs. 69% White and 33% Latino, p=0.001), and younger (mean age 35 vs. 38 years, p=0.04), compared to cases with sequences who were not in a cluster. Cluster members had more connections in the network than did cases with sequences who did not cluster (2 vs. 1 mean partners, p=0.001).
Most clusters included only one index case or partner from the network; 34 (29%) including ≥2 index cases were denoted “Wake” clusters for further analysis (Table 3 shows Wake clusters with ≥5 total cluster members). Wake clusters included 287 persons (56 index cases, 31 partners, and 200 background sequences) [Figure 2]; two (6%) comprised only two partners with no index cases. All Wake clusters were subtype B and most were male-dominated; seven (21%) included ≥50% women. More than half (59%; n=20) of Wake clusters only included persons sampled from the same eleven-county geographic region (Figure 2). Most (74%; 61/82) clusters with only one person from PNS were clusters with ≥50% members sampled in the same region, including 22 clusters with 100% members sampled in the same region.
Table 3.
Transmission clusters that included 5 or more persons statewide and at least two members of the Wake County-based sexual network of adults diagnosed with HIV during 2012–2013 and their contacts (n=235).
| Statewide | Wake Network | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||
| Cluster ID |
Cluster Size |
Max Genetic Distance (%) |
Sampling Year (median (IQR)) |
Estimated Cluster Age (years) |
Most Recent Common Ancestor |
# Male: # Female |
# of Network Persons |
# Index: # Partner |
Max Genetic Distance (%) |
# Components Spanned* |
| A | 5 | 0.95 | 2013 (2013-2013) | 4.7 | 2009 | 4:1 | 2 | 1:1 | 0.00 | 1 |
| B | 5 | 1.65 | 2012 (2012–2013) | 7.6 | 2005 | 5:0 | 2 | 2:0 | 1.02 | 2 |
| C | 5 | 2.58 | 2011 (2009–2012) | 11.7 | 2002 | 4:0† | 3 | 1:2 | 2.58 | 2 |
| D | 6 | 2.05 | 2012 (2008–2014) | 8.1 | 2006 | 6:0 | 2 | 1:1 | 0.96 | 2 |
| E | 6 | 0.95 | 2012 (2010–2013) | 6.5 | 2007 | 6:0 | 3 | 2:1 | 0.68 | 1 |
| F | 6 | 1.56 | 2012 (2012-2012) | 5.5 | 2007 | 6:0 | 6 | 3:3 | 1.56 | 3 |
| G | 7 | 2.92 | 2010 (2005–2012) | 18.5 | 1993 | 1:6 | 2 | 2:0 | 1.40 | 1 |
| H | 8 | 2.94 | 2007 (2003–2012) | 18.6 | 1995 | 8:0 | 2 | 2:0 | 0.61 | 1 |
| I | 8 | 1.56 | 2013 (2012–2014) | 7.6 | 2006 | 8:0 | 4 | 3:1 | 1.15 | 2 |
| J | 12 | 3.42 | 2011 (2009–2012) | 12.2 | 2001 | 11:1 | 3 | 2:1 | 1.24 | 3 |
| K | 14 | 3.33 | 2007 (2007–2010) | 18.3 | 1995 | 6:7† | 2 | 1:1 | 1.27 | 1 |
| L | 14 | 3.22 | 2011 (2008–2013) | 18.3 | 1995 | 14:0 | 2 | 1:1 | 0.07 | 1 |
| M | 15 | 3.59 | 2010 (2008–2011) | 14.7 | 1999 | 15:0 | 2 | 1:1 | 0.94 | 2 |
| N | 16 | 2.33 | 2009 (2008–2013) | 12.5 | 2001 | 16:0 | 2 | 2:0 | 0.47 | 2 |
| O | 16 | 2.11 | 2010 (2009–2012) | 8.8 | 2004 | 15:1 | 3 | 1:2 | 1.24 | 3 |
| P | 20 | 3.26 | 2010 (2008–2012) | 13.8 | 2000 | 20:0 | 3 | 3:0 | 3.22 | 3 |
| Q | 23 | 3.24 | 2008 (2007–2011) | 17.3 | 1997 | 10:13 | 2 | 2:0 | 0.07 | 2 |
| R | 23 | 2.95 | 2012 (2012–2013) | 12.0 | 2002 | 23:0 | 5 | 4:1 | 1.83 | 4 |
| S | 36 | 3.26 | 2012 (2011–2013) | 12.4 | 2002 | 34:2 | 4 | 4:0 | 2.54 | 3 |
Includes number of network singletons and components that included at least one person from the Wake County sexual network
Gender unknown for one person in this cluster
Wake cluster maximum genetic distance was 1.67% (IQR: 1.04–2.93%) statewide and 0.95% (IQR: 0.32–1.28%) when restricted to network members (Table 3). Median estimated cluster age prior to the index case diagnosis was 8.5 years (IQR: 5.1–12.9 years) with median MRCA estimated to occur in 2005 (range 2000–2007).
Partner Notification Network and Transmission Cluster Overlap
The PNS network included 663 persons: 280 index cases and 383 contacts who formed 104 singletons plus 559 persons in 446 partnerships (Figure 1a). Among 230 network members with sequences, including 45 singletons, 169/230 (73%) were in one of 116 statewide transmission clusters that included at least one network member. The 169 persons spanned 82 network components and 23 singletons; the remaining 61 persons who were not in a cluster spanned 36 network components and 22 singletons. Among the 23 singletons in a cluster (51% singletons with sequences), 8 (35%) did not name any partners and the remainder disclosed at least one partner, though none could be located. The median cluster size among singletons was 4 persons (range 2–23).
Among 446 partnerships, 70 (16%) included two HIV-positive persons with sequences; of these, 83% (58/70) were sexual connections. All male-female pairs were in the same cluster, whereas only 34% of male-male pairs were in the same transmission cluster (χ2 p<0.001). Of the 383 contacts, 27 (7%) were only identified as social contacts of an index case; 11 had a sequence, of which 9 were in a statewide cluster with no one else from the PNS network and 2 were in a Wake cluster; one clustered with another PNS social contact (statewide cluster size 2) and the other clustered with the index case who disclosed the contact as a social connection (pairwise genetic distance 1.3%, statewide cluster size 14).
Eighty-seven persons were in 34 Wake clusters (defined as ≥2 persons from PNS network), which included 2–6 network members and spanned 56 PNS network components plus 12 singletons. Overall, 41% (14/34) Wake clusters covered only one network component; 1 included three network members and the rest included two. The Wake clusters that covered only one component were more likely to include ≥50% women (36% [5/14] vs. 10% [2/20] spanning multiple components).
Among 19 Wake clusters with ≥5 persons statewide (Table 3), 6 (32%) covered only one component, where all network persons in the cluster were also linked by named partner ties. The remaining 13 spanned multiple components, where the phylogenetic relationships bridged located partnerships: 7 (37%) spanned two components, 5 (26%) spanned three, and 1 (5%) spanned four components. For example, the three network members in Cluster J spanned two components and one singleton (Figure 1a, quadrants a, c, and d), although there were 12 people in the cluster statewide (Figure 1b). The maximum genetic distance between any pair of network members in Cluster J was 1.24%, despite each of the 3 network members being in different components (Table 3, Cluster J). Of 13 clusters with ≥5 members statewide (Table 3) that spanned multiple components, 9 (69%) included only men.
There was no significant difference by sampling year, cluster age, or statewide genetic distance between Wake clusters that covered single or spanned multiple components. However, the mean genetic distance among persons in the Wake cluster was significantly smaller when the cluster covered only one component (0.66% vs. 1.23%, p=0.03).
Discussion
This study sought to explore the benefits of combining molecular data with sociosexual network data obtained during routine partner notification services from persons newly diagnosed with HIV in a single large county in NC. The study drew on a statewide dataset of over 15,000 HIV-1 sequences from persons sampled between 1997 and mid-2014. We overlaid the genetic data and sociosexual network constructed from partner notification records to obtain a more comprehensive picture of the epidemic and identify gaps in PNS, particularly among male-dominated sexual network components.
More than half of local transmission clusters bridged sexual network components that appeared disconnected, demonstrating that molecular data can detect unobserved links in the sexual network. Furthermore, despite not having any partners identified in the network, over half of singletons with sequences were in a statewide cluster. For each set of disconnected network components or singletons in the same transmission cluster, at least one connection is not represented in the PNS network. Some of the disagreement may be explained by differing collection periods, as sequence sampling time for the clusters was not limited by time period. Many index cases were likely infected for years, so partners reported at diagnosis may not reflect the network at the time of infection. Additionally, some persons in the network were only social contacts, so their inclusion increased PNS network component size and may have increased the effect of bridging by the transmission clusters if they were in a different cluster than the index case. However, they represented only 2 of 87 network members in a Wake cluster.
Partner notification is limited by missing data due to persons not being diagnosed or located and partnerships not being disclosed or not occurring during the DIS interview time period. Stigma and discrimination faced by MSM contribute to interview bias and may reduce willingness to disclose partners to health authorities. Previous HIV sexual network studies in NC found that a high proportion of partners cannot be located3,35 and MSM tend to have more undisclosed partners,36 causing components to appear disjointed and impacting PNS network completeness. However, this completeness is precisely what we wanted to investigate and adding sequences offered some correction to the observed network.
Accordingly, local transmission clusters, particularly those that spanned multiple components, were more likely to be male-dominated. This reflects the current epidemic in NC, where the overall rate of new diagnoses remains elevated with ongoing transmission among young men37 and demonstrates the value of supplementing partner notification with another method that portrays transmission networks differently. By overlaying phylogenetic data onto the sexual network, we were able to identify components with ongoing transmission. Persons in either network may benefit from interventions such as offering pre-exposure prophylaxis to HIV-negative partners or linkage to care support to HIV-positive persons who are not virally suppressed.38 A substantial proportion of incident HIV cases in NC are attributed to persons who are diagnosed and aware of their status at the time of transmission;35 determining which network components have unidentified partners and which clusters have unsuppressed members may help guide intervention targets. Additionally, the smaller genetic distance amongst persons in the sexual network compared to other cases in NC indicates that applying these interventions locally could have an immediate local benefit.
We combined methodologies previously used to describe HIV transmission networks. While several studies have used sequence data to construct transmission networks,2,39–46 few have compared these to PNS networks.16,20,47 To our knowledge, none compared PNS networks constructed from surveillance data using all known incident HIV diagnoses made in a large, defined administrative area. We used all incident diagnoses in our area of interest and matched to all available sequences from one laboratory that serves most patients in this area. We included partnership and demographic data, allowing us to compare groups. We found that male-male PNS pairs were less likely to be in the same transmission cluster, and that male-dominated clusters are more likely to bridge PNS components. The percentage of named partners with genetically similar virus in this largely-MSM population was similar to what was found among MSM in New York City (NYC). Similar to NYC, heterosexual pairs in this population were more likely to cluster than MSM pairs.16
Combining PNS and molecular data can lead to an improved representation above what is possible with either alone,10,12 as both methods have limitations. Sequence analysis is limited by inability to infer directionality and missing data for persons who have not been diagnosed or who do not have sequences available.48 In NC, genotyping is routinely performed at entry to clinical care, so failure to receive a diagnosis or link to care will impact phylogenetic network completeness. Black persons with HIV infection are less likely to link to care,49–51 which is reflected in the lower proportion of Black persons in our study with sequences. Additionally, sequences stemmed from only one laboratory and some of the cases without sequences may seek care from providers who use other labs, affecting cluster comprehensiveness. Still, characteristics associated with cluster membership in our study, including younger age,2,52,53 Black race,52 being male,52 and being MSM,36,45,52 agree with previous studies in the US.
While there is no accepted genetic distance criteria to define transmission clusters,8 traditional cut-offs of <1.5% genetic distance difference allow a focus on only on recent transmissions. We used a higher cluster threshold within the range of multiple other studies8 to permit the characterization of transmission dynamics over longer time periods in the region. Our focus is not on source attribution or using the sequences to confirm transmissions between known partners, but to identify ongoing, local transmission networks using available sequence and routinely collected PNS data. Additionally, most sequences were from chronically-infected persons, so genetic distances between connected persons are expected to be larger due to greater time since infection and we did not want to restrict our analysis to recent partnerships.
Both HIV phylogenetic and PNS data portray networks differently and care must be taken not to misinterpret results. Although the combination of these data provide new insights into network structure, potential ethical and privacy concerns must be considered. HIV genetic clustering does not imply direct person-to-person transmission or direction of transmission;48 thus these data should not be used for identification of first-degree partnerships or confirming transmission from one person to another.
The HIV sequence analysis recognized ongoing transmission chains among high-risk persons, notably MSM, which was not detected through routine partner notification. Persons who experience the most stigma and those at highest risk, MSM or not, such as those who engage in transactional sex or have anonymous partnerships, are more difficult to reach and may therefore be absent from the PNS network. Molecular approaches provide clues to gaps in PNS and direction for case finding and partner elicitation efforts.54 By adding HIV sequences to the PNS network, we were able to successfully identify localized areas where infected persons were missing from the network, demonstrating the value of integrating molecular data into routine partner tracing and testing.
Supplementary Material
Map showing the North Carolina field service regions for HIV and STD control. Wake County, marked with a star, is one of eleven counties in Region 4.
NC, North Carolina; STD, Sexually Transmitted Disease
Acknowledgments
We would like to thank the NC Department of Health and Human Services and the disease intervention specialists serving Wake County.
sources of support: The project described was supported by the National Center for Advancing Translational Sciences (NCATS), NIH, through Grant Award Number UL1TR001111 (AMD and IAD) and the National Institute of Allergy and Infectious Diseases (NIAID), NIH, through Grant Award Numbers K08AI112432-02 (AMD). DKP was supported by the NIH T32 Grant Award 5T32AI070114-10 to UNC-Chapel Hill. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Conflicts of interest: For the remaining authors none were declared.
Presented in part at CROI 2016 (Boston, MA, USA, February 2016)
“HIV Transmission Hotspot Detection Combining Sexual and Phylogenetic Network Analysis”
Accession Numbers for Sequences in GenBank (n=100): KY579388-KY579812
References
- 1.National Center for HIV/AIDS VH, STD, and TB Preventio. HIV in the Southern United States. Atlanta, GA: Centers for Disease Control and Prevention; May, 2016. 2016. [Google Scholar]
- 2.Dennis AM, Hue S, Hurt CB, et al. Phylogenetic insights into regional HIV transmission. AIDS. 2012;26(14):1813–1822. doi: 10.1097/QAD.0b013e3283573244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hurt CB, Beagle S, Leone PA, et al. Investigating a sexual network of black men who have sex with men: implications for transmission and prevention of HIV infection in the United States. J Acquir Immune Defic Syndr. 2012;61(4):515–521. doi: 10.1097/QAI.0b013e31827076a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McKellar MS, Cope AB, Gay CL, et al. Acute HIV-1 infection in the Southeastern United States: a cohort study. AIDS research and human retroviruses. 2013;29(1):121–128. doi: 10.1089/aid.2012.0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oster AM, Wejnert C, Mena LA, et al. Network analysis among HIV-infected young black men who have sex with men demonstrates high connectedness around few venues. Sexually transmitted diseases. 2013;40(3):206–212. doi: 10.1097/OLQ.0b013e3182840373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. HIV in the United States: At a glance. Atlanta, GA: Jun, 2017. 2017. [Google Scholar]
- 7.Hogben M, McNally T, McPheeters M, et al. The effectiveness of HIV partner counseling and referral services in increasing identification of HIV-positive individuals a systematic review. American journal of preventive medicine. 2007;33(2 Suppl):S89–100. doi: 10.1016/j.amepre.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 8.Hassan AS, Pybus OG, Sanders EJ, et al. Defining HIV-1 transmission clusters based on sequence data. AIDS. 2017;31(9):1211–1222. doi: 10.1097/QAD.0000000000001470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aldous JL, Pond SK, Poon A, et al. Characterizing HIV transmission networks across the United States. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2012;55(8):1135–1143. doi: 10.1093/cid/cis612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grabowski MK, Redd AD. Molecular tools for studying HIV transmission in sexual networks. Current opinion in HIV and AIDS. 2014;9(2):126–133. doi: 10.1097/COH.0000000000000040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oster AM, Pieniazek D, Zhang X, et al. Demographic but not geographic insularity in HIV transmission among young black MSM. AIDS. 2011;25(17):2157–2165. doi: 10.1097/QAD.0b013e32834bfde9. [DOI] [PubMed] [Google Scholar]
- 12.Vasylyeva TI, Friedman SR, Paraskevis D, et al. Integrating molecular epidemiology and social network analysis to study infectious diseases: Towards a socio-molecular era for public health. Infection, genetics and evolution : journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2016 doi: 10.1016/j.meegid.2016.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delva W, Leventhal GE, Helleringer S. Connecting the dots: network data and models in HIV epidemiology. AIDS. 2016;30(13):2009–2020. doi: 10.1097/QAD.0000000000001184. [DOI] [PubMed] [Google Scholar]
- 14.Peters PJ, Gay CL, Beagle S, et al. HIV Infection Among Partners of HIV-Infected Black Men Who Have Sex with Men — North Carolina, 2011–2013. MMWR. 2014;63(5):90–94. [PMC free article] [PubMed] [Google Scholar]
- 15.Smith DM, May SJ, Tweeten S, et al. A public health model for the molecular surveillance of HIV transmission in San Diego, California. AIDS. 2009;23(2):225–232. doi: 10.1097/QAD.0b013e32831d2a81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wertheim JO, Kosakovsky Pond SL, Forgione LA, et al. Social and Genetic Networks of HIV-1 Transmission in New York City. PLoS pathogens. 2017;13(1):e1006000. doi: 10.1371/journal.ppat.1006000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dennis AM, Pasquale DK, Billock R, et al. Integration of Contact Tracing and Phylogenetics in an Investigation of Acute HIV Infection. Sexually transmitted diseases. 2017 doi: 10.1097/OLQ.0000000000000726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lewis F, Hughes GJ, Rambaut A, et al. Episodic sexual transmission of HIV revealed by molecular phylodynamics. PLoS Med. 2008;5(3):e50. doi: 10.1371/journal.pmed.0050050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brenner BG, Roger M, Routy JP, et al. High rates of forward transmission events after acute/early HIV-1 infection. The Journal of infectious diseases. 2007;195(7):951–959. doi: 10.1086/512088. [DOI] [PubMed] [Google Scholar]
- 20.Brenner BG, Roger M, Stephens D, et al. Transmission clustering drives the onward spread of the HIV epidemic among men who have sex with men in Quebec. The Journal of infectious diseases. 2011;204(7):1115–1119. doi: 10.1093/infdis/jir468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.NC Department of Health and Human Services. North Carolina 2012 HIV/STD Surveillance Report. Raleigh, NC: NC Division of Public Health; Jul, 2013. 2013. [Google Scholar]
- 22.Centers for Disease Control and Prevention. Recommendations for Partner Services Programs for HIV Infection, Syphilis, Gonorrhea, and Chlamydial infection. Vol. 7. Atlanta, GA: Nov, 2008. 2008. [PubMed] [Google Scholar]
- 23.Dailey Garnes NJ, Moore ZS, Cadwell BL, et al. Previously undiagnosed HIV infections identified through cluster investigation, North Carolina, 2002–2007. AIDS and behavior. 2015;19(4):723–731. doi: 10.1007/s10461-014-0913-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuruc JD, Cope AB, Sampson LA, et al. Ten Years of Screening and Testing for Acute HIV Infection in North Carolina. J Acquir Immune Defic Syndr. 2016;71(1):111–119. doi: 10.1097/QAI.0000000000000818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Csardi G, Nepusz T. The igraph software package for complex network research. InterJournal. 2006 Complex Systems:1695. [Google Scholar]
- 26.R: A Language and Environment for Statistical Computing [computer program] Vienna, Austria: R Foundation for Statistical Computing; 2015. [Google Scholar]
- 27.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic acids research. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hall T. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 98/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 29.Price MN, Dehal PS, Arkin AP. FastTree: Computing Large Minimum Evolution Trees with Profiles instead of a Distance Matrix. Molecular biology and evolution. 2009;26(7):1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tavaré S. Some Probabilistic and Statistical Problems in the Analysis of DNA Sequences. American Mathematical Society: Lectures on Mathematics in the Life Sciences. 1986;17:57–86. [Google Scholar]
- 31.Shimodaira H, Hasegawa M. Multiple Comparisons of Log-Likelihoods with Applications to Phylogenetic Inference. Molecular biology and evolution. 1999;16(8):1114–1116. [Google Scholar]
- 32.Struck D, Lawyer G, Ternes AM, et al. COMET: adaptive context-based modeling for ultrafast HIV-1 subtype identification. Nucleic acids research. 2014;42(18):e144. doi: 10.1093/nar/gku739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Drummond AJ, Rambaut A. BEAST: Bayesian Evolutionary Analysis by Sampling Trees. BMC Evolutionary Biology. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tracer v1.6 [computer program] 2014 [Google Scholar]
- 35.Cope AB, Powers KA, Kuruc JD, et al. Ongoing HIV Transmission and the HIV Care Continuum in North Carolina. PloS one. 2015;10(6):e0127950. doi: 10.1371/journal.pone.0127950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan PA, Hogan JW, Huang A, et al. Phylogenetic Investigation of a Statewide HIV-1 Epidemic Reveals Ongoing and Active Transmission Networks Among Men Who Have Sex With Men. J Acquir Immune Defic Syndr. 2015;70(4):428–435. doi: 10.1097/QAI.0000000000000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.NC HIV/STD Surveillance Unit. 2015 North Carolina HIV/STD Surveillance Report. Raleigh, NC: NC Division of Public Health; Aug, 2016. 2016. [Google Scholar]
- 38.Amirkhanian YA. Social networks, sexual networks and HIV risk in men who have sex with men. Curr HIV/AIDS Rep. 2014;11(1):81–92. doi: 10.1007/s11904-013-0194-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Callegaro A, Svicher V, Alteri C, et al. Epidemiological network analysis in HIV-1 B infected patients diagnosed in Italy between 2000 and 2008. Infection, genetics and evolution : journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2011;11(3):624–632. doi: 10.1016/j.meegid.2011.01.019. [DOI] [PubMed] [Google Scholar]
- 40.Wertheim JO, Leigh Brown AJ, Hepler NL, et al. The global transmission network of HIV-1. The Journal of infectious diseases. 2014;209(2):304–313. doi: 10.1093/infdis/jit524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Skoura L, Metallidis S, Buckton AJ, et al. Molecular and epidemiological characterization of HIV-1 infection networks involving transmitted drug resistance mutations in Northern Greece. The Journal of antimicrobial chemotherapy. 2011;66(12):2831–2837. doi: 10.1093/jac/dkr386. [DOI] [PubMed] [Google Scholar]
- 42.Ross LL, Horton J, Hasan S, et al. HIV-1 Transmission Patterns in Antiretroviral Therapy-Naive, HIV-Infected North Americans Based on Phylogenetic Analysis by Population Level and Ultra-Deep DNA Sequencing. PloS one. 2014;9(2):e89611. doi: 10.1371/journal.pone.0089611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ng KT, Ong LY, Lim SH, et al. Evolutionary history of HIV-1 subtype B and CRF01_AE transmission clusters among men who have sex with men (MSM) in Kuala Lumpur, Malaysia. PloS one. 2013;8(6):e67286. doi: 10.1371/journal.pone.0067286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hue S, Clewley JP, Cane PA, et al. HIV-1 pol gene variation is sufficient for reconstruction of transmissions in the era of antiretroviral therapy. AIDS. 2004;18(5):719–728. doi: 10.1097/00002030-200403260-00002. [DOI] [PubMed] [Google Scholar]
- 45.Antoniadou ZA, Kousiappa I, Skoura L, et al. Short Communication: Molecular Epidemiology of HIV Type 1 Infection in Northern Greece (2009–2010): Evidence of a Transmission Cluster of HIV Type 1 Subtype A1 Drug-Resistant Strains Among Men Who Have Sex with Men. AIDS research and human retroviruses. 2014;30(3):225–232. doi: 10.1089/aid.2013.0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buskin SE, Ellis GM, Pepper GG, et al. Transmission Cluster of Multiclass Highly Drug-Resistant HIV-1 Among 9 Men Who Have Sex With Men in Seattle/King County, WA, 2005–2007. J Acquir Immune Defic Syndr. 2008;49(2):205–211. doi: 10.1097/QAI.0b013e318185727e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Little SJ, Kosakovsky Pond SL, Anderson CM, et al. Using HIV networks to inform real time prevention interventions. PloS one. 2014;9(6):e98443. doi: 10.1371/journal.pone.0098443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dennis AM, Herbeck JT, Brown AL, et al. Phylogenetic studies of transmission dynamics in generalized HIV epidemics: an essential tool where the burden is greatest? J Acquir Immune Defic Syndr. 2014;67(2):181–195. doi: 10.1097/QAI.0000000000000271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hall HI, Frazier EL, Rhodes P, et al. Paper presented at: AIDS. Washington, D.C.: 2012. FRLBX05 - Oral Abstract: Continuum of HIV care: differences in care and treatment by sex and race/ethnicity in the United States. 2012. [Google Scholar]
- 50.Bradley H, Hall HI, Wolitski RJ, et al. Vital Signs: HIV Diagnosis, Care, and Treatment Among Persons Living with HIV — United States, 2011. MMWR. 2014;63(47):1113–1117. [PMC free article] [PubMed] [Google Scholar]
- 51.Singh S, Bradley H, Hu X, et al. Men Living with Diagnosed HIV Who Have Sex with Men: Progress Along the Continuum of HIV Care — United States, 2010. MMWR. 2014;63(38):829–833. [PMC free article] [PubMed] [Google Scholar]
- 52.Lubelchek RJ, Hoehnen SC, Hotton AL, et al. Transmission clustering among newly diagnosed HIV patients in Chicago, 2008 to 2011: using phylogenetics to expand knowledge of regional HIV transmission patterns. J Acquir Immune Defic Syndr. 2015;68(1):46–54. doi: 10.1097/QAI.0000000000000404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Castor D, Low A, Evering T, et al. Transmitted drug resistance and phylogenetic relationships among acute and early HIV-1-infected individuals in New York City. J Acquir Immune Defic Syndr. 2012;61(1):1–8. doi: 10.1097/QAI.0b013e31825a289b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brenner B, Wainberg MA, Roger M. Phylogenetic inferences on HIV-1 transmission: implications for the design of prevention and treatment interventions. AIDS. 2013;27(7):1045–1057. doi: 10.1097/QAD.0b013e32835cffd9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Map showing the North Carolina field service regions for HIV and STD control. Wake County, marked with a star, is one of eleven counties in Region 4.
NC, North Carolina; STD, Sexually Transmitted Disease