Abstract
Purpose
Preeclampsia is known to be a leading cause of mortality and morbidity among mothers and their infants. Approximately 3–8% of all pregnancies in the US are complicated by preeclampsia and another 5–7% by hypertensive symptoms. However, less is known about its long-term influence on infant neurobehavioral development. The current review attempts to demonstrate new evidence for imprinting gene dysregulation caused by hypertension, which may explain the link between maternal preeclampsia and neurocognitive dysregulation in offspring.
Method
Pub Med and Web of Science databases were searched using the terms “preeclampsia,” “gestational hypertension,” “imprinting genes,” “imprinting dysregulation,” and “epigenetic modification,” in order to review the evidence demonstrating associations between preeclampsia and suboptimal child neurodevelopment, and suggest dysregulation of placental genomic imprinting as a potential underlying mechanism.
Results
The high mortality and morbidity among mothers and fetuses due to preeclampsia is well known, but there is little research on the long-term biological consequences of preeclampsia and resulting hypoxia on the fetal/child neurodevelopment. In the past decade, accumulating evidence from studies that transcend disciplinary boundaries have begun to show that imprinted genes expressed in the placenta might hold clues for a link between preeclampsia and impaired cognitive neurodevelopment. A sudden onset of maternal hypertension detected by the placenta may result in misguided biological programming of the fetus via changes in the epigenome, resulting in suboptimal infant development.
Conclusion
Furthering our understanding of the molecular and cellular mechanisms through which neurodevelopmental trajectories of the fetus/infant are affected by preeclampsia and hypertension will represent an important first step toward preventing adverse neurodevelopment in infants.
Keywords: Genomic imprinting, Placenta Epigenetics, Preeclampsia, Neurobehavioral development
Preeclampsia, genomic imprinting, and child neurobehavioral development
Preeclampsia, a hypertensive disorder unique to human pregnancy, is one of the most common and potentially modifiable metabolic problems, and affects approximately 3–8% of all pregnancies [1, 2]. In the United States among 2,748,302 births in 2008, there were 104,850 cases of preeclampsia [3]. It is defined by the onset of hypertension (140/90 mm HG) after 20 weeks’ gestation in the index pregnancy, accompanied by 300 mg of protein in a 24-h urine specimen, or persistent >30 mg/dL (1+) protein on a dipstick [2, 4]. While the etiology of preeclampsia is still elusive, it remains a leading cause of mortality and morbidity among mothers and their babies [4–6].
Recent research has demonstrated that preeclampsia is associated with an increased risk for impaired early language development, lower neurocognitive functioning, and ADHD [7–10]. And there is accumulating evidence to show that pregnancy-induced hypertensive disorders, especially preeclampsia, can have potentially long-term consequences for the offspring’s neurobehavioral development [11, 12] via changes in the epigenome. In recent animal studies, genomic imprinting perturbations have been linked to both hypertensive problems and neurodevelopmental syndromes [13]. Similarly, in humans, adverse pregnancy outcomes have been associated with genomic imprinting perturbations, leading to neurodevelopmental syndromes [14] and subsequent disorders [15, 16]. Delineating some of the underlying biological mechanisms by which preeclampsia influences the trajectory of optimal/suboptimal child development and functioning can help us in our aim to uncover the mechanisms of dysregulated neurobehavioral development and related mental disorders via epigenetic changes.
Severe preeclampsia may cause symptoms such as hypertension, proteinuria, eclampsia, cerebral edema, cerebral hemorrhage, long-term neurocognitive dysfunction, blindness, liver swelling, and other liver damage leading to elevated serum transaminase, oliguria, thrombocytopenia, pulmonary edema necrotizing pancreatitis. All of these symptoms can be fatal to mothers, and, importantly, to their child in utero, resulting in greater mortality in mothers and varying degrees of child morbidity after birth [2, 17]. The main impact of preeclampsia on the fetus is malnourishment, resulting from uteroplacental vascular insufficiency hypoxia, which restricts nutrient supplies and oxygen flow from the placenta to the fetus [18, 19], which affects approximately 30% of preeclampsia cases. This leads to various perinatal and neonatal problems, including intrauterine growth restriction (IUGR, defined as birth weight less than the 10th percentile) [6, 18, 20–25], emergency C-section [6], preterm delivery [24, 26, 27], reduced birth weight [6, 28, 29], lower APGAR scores [30], more frequent and prolonged neonatal intensive care unit (NICU) stays [6, 20, 31], and increased acute respiratory distress syndromes after birth [32]. In some cases, fetal damage is so severe that it results in fetal demise, such as stillbirth and neonatal death [32, 33]. Beyond birth, while the long-term health and developmental consequences of exposure to maternal preeclampsia for the surviving child are relatively unexplored, there is some evidence for suboptimal neurocognitive development among infants with IUGR [34, 35], which is one of the major fetal/child consequences of preeclampsia [6, 20–25]. Recently, with the leadership of the NICHD, growing efforts have been made to find associations between preeclampsia and health consequences in offspring, including IUGR [34–36], preterm birth [37–40], LBW [41–43], and child neurobehavioral development [44–46].
The extent of adverse neurobehavioral and other developmental consequences for surviving infants with perinatal problems has been investigated less frequently. Many and colleagues [47], for example, reported that IQ at age 3 years was significantly lower among IUGR children with maternal preeclampsia, compared to those without (85.5 versus 96.9, p = .03) [47]. Similarly, Cheng and colleagues documented that preterm infants of preeclamptic mothers, compared to those of non-preeclamptic mothers, had a compromised neurodevelopmental index [48], as measured by the Bayley Scales of Infant and Toddler Development—Second Edition (Bayley-II) [49]. Children born to mothers with preeclampsia had a lower Mental Developmental Index (MDI) than children born to mothers without at 2 years of age (p = .04), while there was no significant difference between the two groups on the Physical Developmental Index (PDI) (p = .56). Furthermore, they reported that preeclampsia was associated with an over tenfold increased risk of mildly delayed MDI (p = .007), after controlling for demographic and biomedical confounders. Temperament is thought of as an early biological characteristic similar to a personality trait. Although it is not typically considered to be a neuropsychological construct, there are many overlaps. In particular, the temperamental construct of effortful control is very similar to the neuropsychological construct of attention and executive functioning [50]. More recently, anger and negative emotionality have gained considerable attention as temperamental traits that are closely linked to the limbic system in the brain [51–53].
Thus, further investigation of the underlying epigenetic mechanisms that show how preeclampsia or pregnancy-induced hypertensive problems are associated with early emerging temperament profiles of the infant as well as child suboptimal neurodevelopment, using validated clinical instruments such as the Early Childhood Behavior Questionnaire (ECBQ) [54] and the Bayley Scales of Infant and Toddler Development—Third Edition (Bayley-III) [49], is warranted. In this paper, we are going to propose that imprinting dysregulation might be the intermediate underlying conditions that could link maternal preeclampsia and possible suboptimal neurodevelopment in offspring.
Method
PRISMA guidelines for conducting and reporting systematic reviews were followed during this analysis [55]. Studies were selected using PubMED and Web of Science. Three categories (A: preeclampsia, B: imprinting genes, and C: child neurodevelopment) were first searched separately. The following keywords were used: For category A, “preeclampsia” or “gestational hypertension”; for category B, “imprinting genes,” “imprinting dysregulation,” or “epigenetic modification”; and for category C, “neurodevelopment,” “neuro-behavior,” or “developmental problems.” Results were further limited to (1) English language articles, (2) human studies, and (3) participants aged 10 years or younger. The initial search returned 28,649 results in category A, 14,426 results in category B, and 72,667 results in category C. There were 138 articles that covered both categories B and C, and there were 9 that covered both categories A and B. The titles and abstracts of those 138 and 19 articles were examined against previous exclusion criteria. Among those, 75 articles were examined against previous exclusion criteria.
Placenta Epigenetics and neurodevelopment
The medical condition of the mother affects the nature of the in utero condition (i.e., environment) and perturbs gene expression (i.e., genes), which governs the developmental trajectories of offspring. When information from these two areas, environment and genes, are examined together, they can provide us with important clues as to how the mother’s medical complications influence the trajectories of child development.
Over the past decade, researchers have begun to consider how the placenta, which is usually discarded at birth, could hold important information about the relationship between the pre- and perinatal environment and cognitive neurobehavioral outcomes in the developing child, over and above the effects of postnatal environment. Many studies have linked poor placentation during pregnancy with a wide range of chronic neurodevelopmental disorders in children [36], including Autism Spectrum Disorder (ASD) [56–59], Attention Deficit Hyperactivity Disorder (ADHD) [11], and learning disabilities [60]. Moreover, recent data from the CDC showed a 23% increase in identified cases of ASD between 2009 and 2011 [61]. It is important to note that placenta tissue should not be viewed as a surrogate (to the brain) but as a target tissue in understanding the genes–environment interplay. The placenta develops from the extra-embryonic cell layer of the blastocyst, as opposed to the embryonic cell mass that will differentiate into the fetus. In an effort to understand the mechanisms of how the environment gets “under the skin,” attention has been focused on the potential importance of the link between the placenta and fetal/child brain development.
The placenta has been shown to produce an array of neuropeptide hormones that are analogues to those produced by the hypothalamus and the pituitary gland including GnRH, TRH, CRH, and oxytocin [62]. Rapid advancements in the discovery of integrated regulation of neuropeptide homeostasis within the brain and placenta [63, 64] have led to the concept that the placenta may be like a “third brain” linking the developed (maternal) and developing (fetal) brains [64, 65]. Maternal perturbations are conveyed to the fetus via the placenta, in the expression of transporters that regulate the flux of glucose, amino acids, and vitamins required for growth and development [66]. Thus, the placenta serves as the “master regulator” in utero and plays a highly functional role in shaping fetal development [65].
Imprinted genes expressed in the placenta and fetoplacental development
Fetoplacental development begins with a complex and highly coordinated set of epigenetic events that take place few hours after fertilization and before the implantation of the fertilized egg [66–68]. During this relatively short but very active window an almost complete reprogramming of the genome methylation takes place accompanied by a reorganization of the histone coding [68, 69]. Other, less characterized, epigenetic events also contribute to preparing the newly fused parental genomes for implantation and embryogenesis [68]. At this stage, specific genomic regions carrying a unique set of multilayer epigenetic signals inherited from the germline, known as Imprinting Control Regions (ICRs), are spared this epigenome reprogramming wave (Fig. 1) [66, 70]. ICRs control the allele-specific expression of clusters of over 100 genes (or about 1% of the protein-coding genes) distributed across the human genome. Genes which are monoallelically expressed through the action of these ICRs are known as imprinted genes (Table S1) with their expressed allele determined by the parent of origin. Imprinted genes are thus physically present as two copies but functionally haploid with about 50% of them maternally expressed and 50% paternally expressed [70, 71].
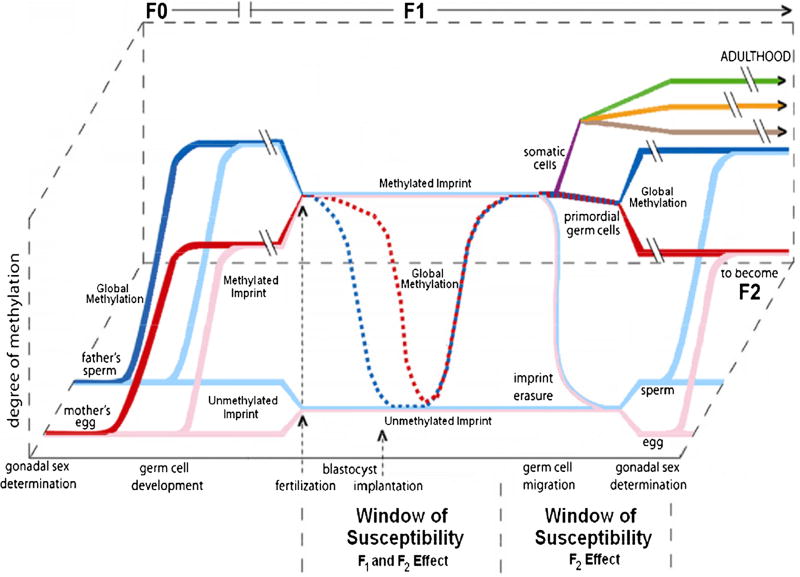
Fig. 1.
Fetal imprinting reprogramming of DNA methylation marks during the early zygote developmental phases. F0 sperm and egg carry global (red and blue lines) and imprinting specific methylation signals (pink and light blue). After fertilization, global methylation is reprogrammed at the blastocyst stadium. Imprinting signals are maintained unaltered to generate an embryo with distinct parental contributions. Imprinting reprogramming takes place only in the primordial germ cells later in development to generate gametes carrying imprinting marks according to the sex of the developing embryo. Perturbations of the imprinting profiles at the blastocyst stage can directly affect the embryo and also the gametes (F1 and F2 windows). Somatic cells separately develop from the embryo carrying the parental imprinting signals and the newly reprogrammed global methylation setting (purple line). They later rearrange their methylation status coherently with the adult tissue they will originate (green, orange, and brown lines) Adapted from Lambertini et al. [66]; Perera and Herbstman [69]
A number of imprinted genes play critical roles in regulating the fetoplacental growth and development and instructing postnatal development [13, 72]. Based on their functional roles, imprinted genes have been classified in four main categories: (1) genes that directly regulate fetal growth; (2) genes that indirectly regulate fetal growth by modifying the function of the placenta; (3) genes that modulate metabolic processes postnatally; and (4) genes that modify behavior postnatally [13, 72, 73].
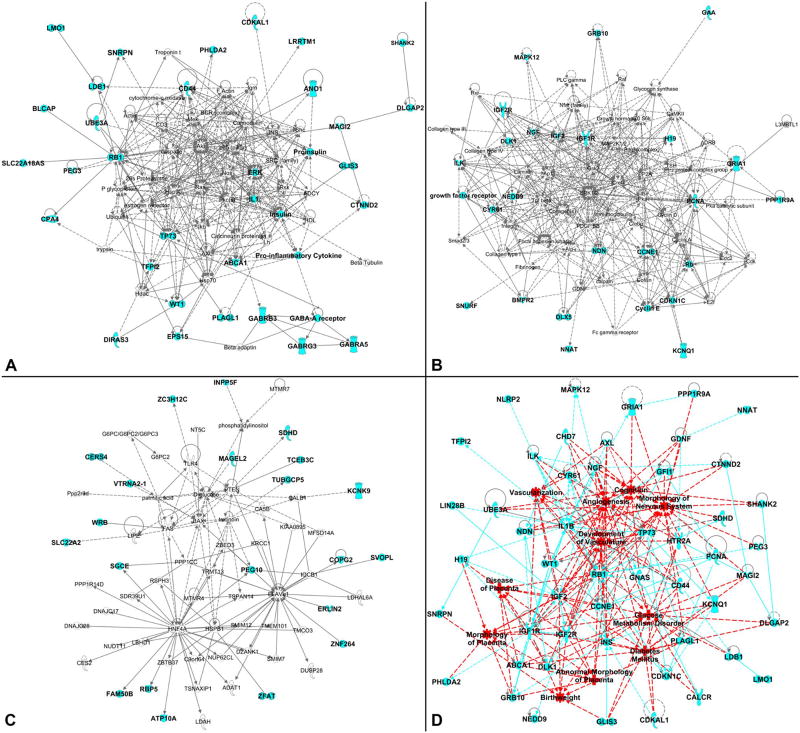
We confirmed the reported findings by feeding the full set of imprinted genes to the online Ingenuity Pathway Analysis (IPA) software. IPA identified 8 main gene networks (Table 1 and Table S2) tied to organ development (including the placenta), metabolic regulation, and the development and functioning of the brain and nervous system (i.e., neurodevelopment), heart and vascular system, and metabolic organs (e.g., pancreas). The first three networks, including the majority of the input genes, are presented in Fig. 2a–c, and the remaining five networks are available in Figure S1.
Table 1.
Network distribution of imprinted genes as determined by the Ingenuity Pathway Analysis (IPA) database
| Network | Network-associated diseases and functions | Genes in network |
Imprinted genes in network |
% Imprinted genes in network |
Figure |
|---|---|---|---|---|---|
| 1 | Gene expression, neurological disease, organismal injury and abnormalities | 70 | 31 | 44 | 1a |
| 2 | Gene expression, cellular development, cellular growth and proliferation | 70 | 25 | 36 | 1b |
| 3 | Post-translational modification, endocrine system disorders, metabolic disease | 70 | 21 | 30 | 1c |
| 4 | Cardiovascular system development and function, tissue morphology, cell-mediated immune response | 70 | 18 | 26 | S1A |
| 5 | Cell death and survival, organismal injury and abnormalities, cancer | 70 | 18 | 26 | S1B |
| 6 | Cardiovascular system development and function, carbohydrate metabolism, post-translational modification | 2 | 1 | 50 | S1C |
| 7 | Cardiovascular disease, cell cycle, cell death and survival | 2 | 1 | 50 | S1C |
| 8 | Connective tissue development and function, skeletal and muscular system development and function, nervous system development and function | 2 | 1 | 50 | S1C |
| Total imprinted genes included in the IPA networks | 116 | ||||
Network number, associated diseases and functions, genes included in the network and number and percentage of imprinted genes in the network are reported. Reference to the figure depicting the network structure is also reported
Fig. 2.
IPA network analysis for imprinted genes (see Table 1 and S2 for more details). A total of 131 genes were entered into IPA software (see Table S1 for the full list). a Depiction of Network 1 (IPA label is “gene expression, neurological disease, organismal injury and abnormalities”). This network includes 31 imprinted genes out of 70 network members. b Depiction of Network 2 (IPA label is “gene expression, cellular development, cellular growth and proliferation”). This network includes 25 imprinted genes out of 70 network members. c Depiction of Network 3 (IPA label is “post-translational modification, endocrine system disorders, metabolic disease”). This network includes 21 imprinted genes out of 70 network members. d Depiction of the imprinted super-pathway determined by scoring the imprinted genes associated with the development and functioning of the placenta, brain and nervous system, heart and vascular system, metabolic organs (mainly pancreas), and metabolism regulation. For all panels, the light blue shading identifies the input imprinted genes. In d, red shading identifies disease and function areas as labeled by the IPA, light blue lines (either dashed or continuous) identify connections between genes, red lines (either dashed or continuous) identify connections between disease and function areas as well as connections between disease and function areas and imprinted genes
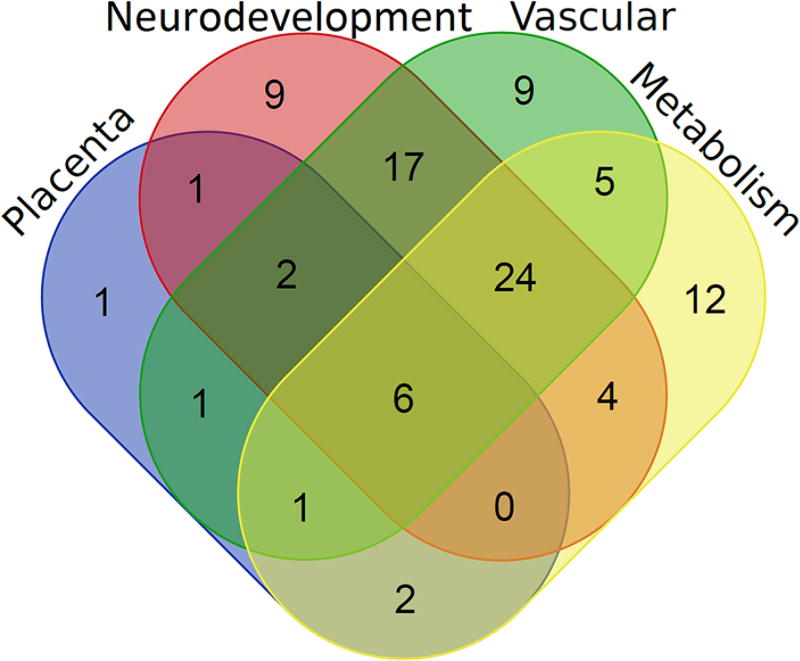
The IPA tool connects genes with known diseases and functions, and we can further generate an imprinted super-pathway which shows the gene network involved in the development and functioning of the placenta, brain, heart and vasculature, and pancreas, as well as those with important metabolic functions (Fig. 2d). Figure 3 shows the overlap among the genes between different organ systems, including placenta, neurodevelopmental, vascular and metabolic genes (see Tables S3 and S4 for details).
Fig. 3.
Venn diagram showing the overlap between the imprinted genes involved in the development and functioning of placenta, brain and nervous system, heart and vascular system, metabolic organs (mainly pancreas), and metabolism regulation (see Table S4 for additional details)
Furthermore, recent research suggests that a subset of imprinted genes expressed in the placenta may regulate maternal adaptations to pregnancy, i.e., controlling the function of the mother, by regulating the production of placental hormones [74]. The timely expression of imprinted genes has been shown to play an important role in fetoplacental development [13, 75, 76]. These findings are in agreement with the functional importance of imprinted genes which, as shown for other such genes [77, 78], once altered, can lead to serious phenotypic consequences [79, 80], some being lethal [81, 82], which may be further enhanced by the constitutional haplo-insufficiency of imprinted genes [83].
The endurance of ICRs in the face of extensive epigenetic reprogramming early in development and through the life course raises many interesting points. As the ICR epigenetic setup is not reprogrammed at fertilization, it represents one of the few known instances of epigenetic inheritance [84]. This in turn may explain the influence of parental environments prior to conception in determining fetal development. Environmentally driven alterations of the ICR epigenetic status in the parents’ gametes at different stages of life may be preserved. Similarly, environmental exposure may influence the process that protects ICRs from the epigenome reprogramming wave happening at fertilization. As both types of exposure occur very early, the consequences are likely to be wide-ranging impacting the whole embryo and potentially detectable in most tissues [85]. Environmental exposures occurring after implantation that alter the ICR epigenetic setup or function also carry the potential for modifying the fetoplacental development trajectory at different stages [69, 86–88]. Lastly, it must be mentioned that the effects of the alteration of the ICR epigenetic setup can extend beyond the F1 generation. Primordial germ cells (PGCs) are the only embryonic cells that actually undergo ICR reprogramming in order to generate gametes that carry the ICR epigenetic setup specific to the sex of the developing embryo [89]. Alterations that affect the process of erasure, reestablishment, or maintenance during maturation into gametes could be passed to the F2 generation. To summarize, the epigenetic setup of ICRs can be considered as a recording devices of past exposures [5] acting as lasting environmental biosensors of the intrauterine status as conveyed by the mother (e.g., changes in blood pressure). Of note, alterations of both the ICR epigenetic setup and imprinted genes have been linked to different pregnancy and newborn outcomes [86, 89–96].
Imprinted genes as a sensor of pregnancy-induced hypertensive disorder and predictor of child development
Imprinted genes have been found to respond to common environmental stimuli. For example, peri-conceptional and prenatal exposures to both insufficient and excessive maternal nutrient intake have been found to leave lasting signals on the methylation profile of several imprinted domains, including imprinted genes INS, IGF2, GNASAS, and MEG3 [65].
Previous studies have examined the expression of imprinted genes in the placenta from preeclamptic pregnancies, often with conflicting results. For example, the maternally expressed imprinted gene CYCLIN-DEPENDENT KINASE INHIBITOR 1C (CDKN1C) has variously been reported to be significantly decreased [97], increased [98, 99], or unaltered [100] in preeclamptic placentas. These conflicting results may be explained by a difference in mode of delivery, given recent evidence demonstrating significantly increased CDKN1C expression in laboring versus non-laboring placentas [101]. Some mothers carrying babies with loss-of-function of CDKN1C have a very severe form of preeclampsia called HELLP (Hemolysis, Elevated Liver enzymes, Low platelet count) syndrome which is a life-threatening complication [102]. There are data from an animal model to suggest loss-of-function of CDKN1C in the placenta may contribute to preeclampsia-like symptoms in the mother. Genetically unaltered female mice carrying Cdkn1c loss-of-function fetuses exhibit increased blood pressure and proteinuria during pregnancy [103]. However, in a separate study, maternal symptoms were less apparent initially suggesting an environmental component [104]. Loss-of-function of Cdkn1c in the mouse placenta was associated with increased trophoblast proliferation and a narrowed intervillous space in some studies [103, 105]. A narrowed intervillous space could impede uteroplacental blood flow, which, combined with the shallow trophoblast invasion observed, could contribute to the development of preeclampsia-like symptoms [103]. However, a more recent study revealed a very different placental phenotype with a severely disorganized placenta late in gestation, and with maternal blood hemorrhaging into the blood spaces suggesting that genetic background could influence the phenotypic consequences of loss-of-function of Cdkn1c [106]. Taken together, these studies highlight the importance of further research investigating the relationship between preeclampsia and CDKN1C.
A second imprinted gene, PLECKSTRIN HOMOLOGYLIKE DOMAIN, FAMILY A, MEMBER 2 (PHLDA2), has also been linked to preeclampsia. PHLDA2 was found to be highly overexpressed in placentas from preeclamptic pregnancies [107]. The phenotype of female mice carrying fetuses with loss-of-function of Phlda2 has not been reported, but overexpression of PHLDA2 in a human placental cell line resulted in impaired cell proliferation, migration, and invasion [107]. Placental PHLDA2 expression may also be important in preeclampsia.
Around 70% of the known imprinted genes are expressed in the placenta [91], and recent work has demonstrated that alterations in placental imprinted gene expression are associated with infant neurodevelopmental outcomes [108]. In two small pilot studies on 50 placenta samples from the Stress in Pregnancy (SIP) Study, we found both an alteration of the methylation profile of the ICR that regulates the parent-of-origin-specific expression of two key imprinted genes, IGF2 and H19 (imprinted in the opposite direction), in correlation with maternal stress in pregnancy and an alteration of the global DNA methylation in correlation with preeclampsia [72]. We also found that imprinted gene expression in the placenta correlates with fetal growth and development, as measured by head circumference and birth weight [87, 108]. Furthermore, the imprinting status of each imprinted locus, as defined by the ICR methylation status, has been shown to exert different effects on the reactivation of the silent allele of imprinted genes at that specific locus [91, 108].
Understanding the molecular mechanisms for fetal programming, through exposure to pregnancy-related medical problems, such as preeclampsia and pregnancy-induced hypertension, is a promising, but neglected, area of research, while as a whole the area of fetal programming represents an important first step toward prevention of lifelong negative developmental and mental health consequences for offspring. To achieve this, epigenetically informative longitudinal research that follows a birth cohort from a period in utero through childhood is the key to understanding how maternal preeclampsia in utero can influence an infant’s developmental trajectory by increasing vulnerability to cognitive and neurobehavioral impairment. If imprinting gene profiles are determined to be early biomarkers for impaired cognitive neurobehavioral development, they could be used as biomarkers to design more targeted preventive measures for childhood developmental problems by alleviating and reducing the risk for maternal preeclampsia during pregnancy.
Conclusion
In sum, gaining further understanding regarding the ways in which common pregnancy-induced problems such as preeclampsia may lead to suboptimal fetal/infant development, specifically impaired neurobehavioral development via dysregulation in genomic imprinting status in the placenta, and may yield effective clues for the prevention of neurodevelopmental disorders in early childhood. Prenatal influence caused by preeclampsia or hypertensive disorders will not fully explain the cause of the neurodevelopmental problems, as there are also many postnatal assaults that influence the risk for neurodevelopmental disorders in offspring, as well as other possible prenatal mechanism. However, if epigenetic pathways can be used as potential tools for identifying high-risk infants, it is the first step toward developing prevention plans for full-fledged disorders later in childhood, perhaps through educating pregnant mothers. To this end, it is important to encourage collaboration among obstetricians, pediatricians, child psychiatrists, as well as early childhood educators to encourage research around the peri- and prenatal periods to reduce the risk for impaired neurodevelopmental disorders in childhood. Knowledge gained from such studies could contribute to an enhanced capacity for early prevention. At the same time, it will help inform and educate pregnant mothers about the importance of prenatal monitoring of blood pressure, weight gain, and metabolic functioning during pregnancy for the health of their offspring.
Supplementary Material
Acknowledgments
This work was supported by the NIMH Grants K01 MH080062, ARRA supplement K01 MH080062S and R01MH102729 (to YN), and MRC Grant MR/MD013960/1 (to AJ).
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00404-017-4347-3) contains supplementary material, which is available to authorized users.
Author Contributions Y Nomura: Protocol/project development, Manuscript writing/editing, Study Design; R John: Manuscript writing/editing, Placenta Biology, and Epigenetics; AB Janssen: Manuscript writing/editing, Translational Epigenetics; C Davey: Manuscript writing/editing, Critical Review; J Finik: Manuscript writing/editing, Literature Review, Literature Search; J Buthmann: Manuscript writing/editing, Literature Review, Literature Search; V Glover: Manuscript writing/editing, Critical Review; L Lambertini: Manuscript writing/editing, Translational Epigenetics.
Compliance with ethical standards
Ethical approval This review article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent This article does not contain any studies with human participants that required Informed consents.
Conflict of interest There is no conflict of interest with any author in the study.
References
- 1.McCalla CO, Nacharaju VL, Muneyyirci-Delale O, Glasgow S, Feldman JG. Placental 11β-hydroxysteroid dehydrogenase activity in normotensive and pre-eclamptic pregnancies. Steroids. 1998;63:511–515. doi: 10.1016/s0039-128x(98)00056-7. [DOI] [PubMed] [Google Scholar]
- 2.Williams MA, Miller RS, Qiu C, Cripe SM, Gelaye B, Enquobahrie D. Associations of early pregnancy sleep duration with trimester-specific blood pressures and hypertensive disorders in pregnancy. Sleep. 2010;33:1363–1371. doi: 10.1093/sleep/33.10.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Osterman MJ, Martin JA, Mathews TJ, Hamilton BE. Expanded data from the new birth certificate, 2008. National vital statistics reports: from the Centers for Disease Control and Prevention, National Center for Health Statistics, National Vital Statistics System. 2011;59:1–28. [PubMed] [Google Scholar]
- 4.Podymow T, August P. Hypertension in pregnancy. Adv Chronic Kidney Dis. 2007;14:178–190. doi: 10.1053/j.ackd.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 5.Chang J, Elam-Evans LD, Berg CJ, Herndon J, Flowers L, Seed KA, Syverson CJ. Pregnancy-related mortality surveillance–united states, 1991–1999. Morbidity and mortality weekly report. Surveill Summ. 2003;52:1–8. [PubMed] [Google Scholar]
- 6.Saadat M, Nejad SM, Habibi G, Sheikhvatan M. Maternal and neonatal outcomes in women with preeclampsia. Taiwan J Obstetr Gynecol. 2007;46:255–259. doi: 10.1016/S1028-4559(08)60029-7. [DOI] [PubMed] [Google Scholar]
- 7.Getahun D, Rhoads GG, Demissie K, Lu SE, Quinn VP, Fassett MJ, Wing DA, Jacobsen SJ. In utero exposure to ischemic-hypoxic conditions and attention-deficit/hyperactivity disorder. Pediatrics. 2012;131:e53–e61. doi: 10.1542/peds.2012-1298. [DOI] [PubMed] [Google Scholar]
- 8.Mann JR, McDermott S. Are maternal genitourinary infection and pre-eclampsia associated with adhd in school-aged children? J Atten Disord. 2010;15:667–673. doi: 10.1177/1087054710370566. [DOI] [PubMed] [Google Scholar]
- 9.Silva D, Colvin L, Hagemann E, Bower C. Environmental risk factors by gender associated with attention-deficit/hyperactivity disorder. Pediatrics. 2013;133:e14–e22. doi: 10.1542/peds.2013-1434. [DOI] [PubMed] [Google Scholar]
- 10.Whitehouse AJO, Robinson M, Newnham JP, Pennell CE. Do hypertensive diseases of pregnancy disrupt neurocognitive development in offspring? Paediatr Perinat Epidemiol. 2012;26:101–108. doi: 10.1111/j.1365-3016.2011.01257.x. [DOI] [PubMed] [Google Scholar]
- 11.Nomura Y, Marks DJ, Grossman B, Yoon M, Loudon H, Stone J, Halperin JM. Exposure to gestational diabetes mellitus and low socioeconomic status. Arch Pediatr Adolesc Med. 2012;166:337. doi: 10.1001/archpediatrics.2011.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ornoy A, Ratzon N, Greenbaum C, Wolf A, Dulitzky M. School-age children born to diabetic mothers and to mothers with gestational diabetes exhibit a high rate of inattention and fine and gross motor impairment. J Pediatr Endocrinol Metabol. 2001;14:681–690. doi: 10.1515/jpem.2001.14.s1.681. [DOI] [PubMed] [Google Scholar]
- 13.Bressan FF, De Bem THC, Perecin F, Lopes FL, Ambrosio CE, Meirelles FV, Miglino MA. Unearthing the roles of imprinted genes in the placenta. Placenta. 2009;30:823–834. doi: 10.1016/j.placenta.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 14.Davies W, Isles AR, Wilkinson LS. Imprinted gene expression in the brain. Neurosci Biobehav Rev. 2005;29:421–430. doi: 10.1016/j.neubiorev.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 15.Roseboom T, de Rooij S, Painter R. The dutch famine and its long-term consequences for adult health. Early Hum Dev. 2006;82:485–491. doi: 10.1016/j.earlhumdev.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 16.Susser E, Hoek HW, Brown A. Neurodevelopmental disorders after prenatal famine: the story of the dutch famine study. Am J Epidemiol. 1998;147:213–216. doi: 10.1093/oxfordjournals.aje.a009439. [DOI] [PubMed] [Google Scholar]
- 17.Rasmussen S, Irgens LM. Fetal growth and body proportion in preeclampsia. Obstet Gynecol. 2003;101:575–583. doi: 10.1016/s0029-7844(02)03071-5. [DOI] [PubMed] [Google Scholar]
- 18.Henriksen T, Clausen T. The fetal origins hypothesis: placental insufficiency and inheritance versus maternal malnutrition in well-nourished populations. Acta Obstet Gynecol Scand. 2002;81(2):112–114. doi: 10.1034/j.1600-0412.2002.810204.x. [DOI] [PubMed] [Google Scholar]
- 19.Wu G, Imhoff-Kunsch B, Girard AW. Biological mechanisms for nutritional regulation of maternal health and fetal development. Paediatr Perinat Epidemiol. 2012;26(Suppl 1):4–26. doi: 10.1111/j.1365-3016.2012.01291.x. [DOI] [PubMed] [Google Scholar]
- 20.Bramham K, Briley AL, Seed P, Poston L, Shennan AH, Chappell LC. Adverse maternal and perinatal outcomes in women with previous preeclampsia: A prospective study. Am J Obstet Gynecol. 2011;204(512):e1–12.e9. doi: 10.1016/j.ajog.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Habli M, Levine RJ, Qian C, Sibai B. Neonatal outcomes in pregnancies with preeclampsia or gestational hypertension and in normotensive pregnancies that delivered at 35, 36, or 37 weeks of gestation. Am J Obstet Gynecol. 2007;197(406):e1–06.e7. doi: 10.1016/j.ajog.2007.06.059. [DOI] [PubMed] [Google Scholar]
- 22.Jelin AC, Cheng YW, Shaffer BL, Kaimal AJ, Little SE, Caughey AB. Early-onset preeclampsia and neonatal outcomes. J Matern-Fetal Neonat Med. 2010;23:389–392. doi: 10.1080/14767050903168416. [DOI] [PubMed] [Google Scholar]
- 23.Liggins GC, Howie RN. A controlled trial of antepartum glucocorticoid treatment for prevention of the respiratory distress syndrome in premature infants. Pediatrics. 1972;50:515–525. [PubMed] [Google Scholar]
- 24.Ødegard RA, Vatten LJ, Nilsen ST, Salvesen KÅ, Austgulen R. Preeclampsia and fetal growth. Obstetr Gynecol. 2000;96:950–955. [PubMed] [Google Scholar]
- 25.Swank M, Nageotte M, Hatfield T. Necrotizing pancreatitis associated with severe preeclampsia. Obstetr Gynecol. 2012;120:453–455. doi: 10.1097/AOG.0b013e31824fc617. [DOI] [PubMed] [Google Scholar]
- 26.Benediktsson R, Lindsay RS, Noble J, Seckl JR, Edwards CRW. Glucocorticoid exposure in utero: New model for adult hypertension. Lancet. 1993;341:339–341. doi: 10.1016/0140-6736(93)90138-7. [DOI] [PubMed] [Google Scholar]
- 27.Edwards CRW, Benediktsson R, Lindsay RS, Seckl JR. Dysfunction of placental glucocorticoid barrier: link between fetal environment and adult hypertension? Lancet. 1993;341:355–357. doi: 10.1016/0140-6736(93)90148-a. [DOI] [PubMed] [Google Scholar]
- 28.Kalder M, Ulrich S, Hitschold T, Berle P. [fetal development in mild and severe pre-eclampsia: Correlation with maternal laboratory parameters and doppler ultrasound] Zeitschrift fur Geburtshilfe und Neonatologie. 1995;199:13–17. [PubMed] [Google Scholar]
- 29.Liu C-M, Cheng P-J, Chang S-D. Maternal complications and perinatal outcomes associated with gestational hypertension and severe preeclampsia in taiwanese women. Taiwan yi zhi=J Formos Med Assoc. 2008;107:129–138. doi: 10.1016/S0929-6646(08)60126-6. [DOI] [PubMed] [Google Scholar]
- 30.Leuner B, Gould E. Structural plasticity and hippocampal function. Annu Rev Psychol. 2010;61:111–140. doi: 10.1146/annurev.psych.093008.100359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones BP, Bell EA, Maroof M. Epidural labor analgesia in parturient with von willebrand’s disease type iia and severe preeclampsia. Anesthesiology. 1999;90:1219–1220. doi: 10.1097/00000542-199904000-00043. [DOI] [PubMed] [Google Scholar]
- 32.Masoura S, Kalogiannidis I, Margioula-Siarkou C, Diamanti E, Papouli M, Drossou-Agakidou V, Prapas N, Agorastos T. Neonatal outcomes of late preterm deliveries with preeclampsia. Minerva Ginecol. 2012;64:109–115. [PubMed] [Google Scholar]
- 33.Nomura Y, Lambertini L, Rialdi A, Lee M, Mystal EY, Grabie M, Manaster I, Huynh N, Finik J, et al. Global methylation in the placenta and umbilical cord blood from pregnancies with maternal gestational diabetes, preeclampsia, and obesity. Reprod Sci. 2013;21:131–137. doi: 10.1177/1933719113492206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bos AF, Einspieler C, Prechtl HFR. Intrauterine growth retardation, general movements, and neurodevelopmental outcome: a review. Dev Med Child Neurol. 2001;43:61. doi: 10.1017/s001216220100010x. [DOI] [PubMed] [Google Scholar]
- 35.Tolsa CB, Zimine S, Warfield SK, Freschi M, Rossignol AS, Lazeyras F, Hanquinet S, Pfizenmaier M, Hüppi PS. Early alteration of structural and functional brain development in premature infants born with intrauterine growth restriction. Pediatr Res. 2004;56:132–138. doi: 10.1203/01.PDR.0000128983.54614.7E. [DOI] [PubMed] [Google Scholar]
- 36.Arcangeli T, Thilaganathan B, Hooper R, Khan KS, Bhide A. Neurodevelopmental delay in small babies at term: a systematic review. Ultrasound Obstet Gynecol. 2012;40(3):267–275. doi: 10.1002/uog.11112. [DOI] [PubMed] [Google Scholar]
- 37.Allin M, Matsumoto H, Santhouse AM, Nosarti C, AlAsady MHS, Stewart AL, Rifkin L, Murray RM. Cognitive and motor function and the size of the cerebellum in adolescents born very pre-term. Brain. 2001;124:60–66. doi: 10.1093/brain/124.1.60. [DOI] [PubMed] [Google Scholar]
- 38.Marlow N, Wolke D, Bracewell MA, Samara M. Neurologic and developmental disability at six years of age after extremely preterm birth. N Engl J Med. 2005;352:9–19. doi: 10.1056/NEJMoa041367. [DOI] [PubMed] [Google Scholar]
- 39.Moster D, Lie RT, Markestad T. Long-term medical and social consequences of preterm birth. N Engl J Med. 2008;359:262–273. doi: 10.1056/NEJMoa0706475. [DOI] [PubMed] [Google Scholar]
- 40.Straub H, Adams M, Kim JJ, Silver RK. Antenatal depressive symptoms increase the likelihood of preterm birth. Am J Obstet Gynecol. 2012;207(329):e1–29.e4. doi: 10.1016/j.ajog.2012.06.033. [DOI] [PubMed] [Google Scholar]
- 41.Hack M, Schluchter M, Cartar L, Rahman M, Cuttler L, Borawski E. Growth of very low birth weight infants to age 20 years. Pediatrics. 2003;112:e30–e38. doi: 10.1542/peds.112.1.e30. [DOI] [PubMed] [Google Scholar]
- 42.Mikkola K. Neurodevelopmental outcome at 5 years of age of a national cohort of extremely low birth weight infants who were born in 1996–1997. Pediatrics. 2005;116:1391–1400. doi: 10.1542/peds.2005-0171. [DOI] [PubMed] [Google Scholar]
- 43.Sung I-K, Vohr B, Oh W. Growth and neurodevelopmental outcome of very low birth weight infants with intrauterine growth retardation: comparison with control subjects matched by birth weight and gestational age. J Pediatr. 1993;123:618–624. doi: 10.1016/s0022-3476(05)80965-5. [DOI] [PubMed] [Google Scholar]
- 44.Davis EP, Glynn LM, Schetter CD, Hobel C, Chicz-Demet AS, Man CA. Prenatal exposure to maternal depression and cortisol influences infant temperament. J Am Acad Child Adolesc Psychiatry. 2007;46:737–746. doi: 10.1097/chi.0b013e318047b775. [DOI] [PubMed] [Google Scholar]
- 45.Huizink AC, Robles de Medina PG, Mulder EJH, Visser GHA, Buitelaar JK. Stress during pregnancy is associated with developmental outcome in infancy. J Child Psychol Psychiat. 2003;44:810–818. doi: 10.1111/1469-7610.00166. [DOI] [PubMed] [Google Scholar]
- 46.Marcus S, Lopez JF, McDonough S, MacKenzie MJ, Flynn H, Neal CR, Gahagan S, Volling B, Kaciroti N, et al. Depressive symptoms during pregnancy: Impact on neuroendocrine and neonatal outcomes. Infant Behav Dev. 2011;34:26–34. doi: 10.1016/j.infbeh.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Many A, Fattal A, Leitner Y, Kupferminc MJ, Harel S, Jaffa A. Neurodevelopmental and cognitive assessment of children born growth restricted to mothers with and without preeclampsia. Hypertens Pregnancy. 2003;22:25–29. doi: 10.1081/PRG-120016791. [DOI] [PubMed] [Google Scholar]
- 48.Cheng S-W, Chou H-C, Tsou K-I, Fang L-J, Tsao P-N. Delivery before 32 weeks of gestation for maternal preeclampsia: neonatal outcome and 2-year developmental outcome. Early Hum Dev. 2004;76:39–46. doi: 10.1016/j.earlhumdev.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 49.Bayley N. In: Bayley scales of infant and toddler development. third. Assessment H, editor. PsycTESTS Dataset. Psych. Corporation; San Antonio: 2006. [Google Scholar]
- 50.Posner MI, Rothbart MK, Sheese BE, Voelker P. Developing attention: behavioral and brain mechanisms. Adv Neurosci. 2014;2014:1–9. doi: 10.1155/2014/405094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dougherty LR, Klein DN, Olino TM, Dyson M, Rose S. Increased waking salivary cortisol and depression risk in preschoolers: the role of maternal history of melancholic depression and early child temperament. J Child Psychol Psychiatry. 2009;50:1495–1503. doi: 10.1111/j.1469-7610.2009.02116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Madsen KS, Jernigan TL, Iversen P, Frokjaer VG, Mortensen EL, Knudsen GM, Baaré WFC. Cortisol awakening response and negative emotionality linked to asymmetry in major limbic fibre bundle architecture. Psychiatry Res: Neuroimaging. 2012;201:63–72. doi: 10.1016/j.pscychresns.2011.07.015. [DOI] [PubMed] [Google Scholar]
- 53.Whittle S, Allen NB, Lubman DI, Yücel M. The neurobiological basis of temperament: Towards a better understanding of psychopathology. Neurosci Biobehav Rev. 2006;30:511–525. doi: 10.1016/j.neubiorev.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 54.Putnam SP, Gartstein MA, Rothbart MK. Measurement of fine-grained aspects of toddler temperament: the early childhood behavior questionnaire. Infant Behav Dev. 2006;29:386–401. doi: 10.1016/j.infbeh.2006.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moher D, Liberati A, Tetzlaff J, Altman DG PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med. 2009;15(4):264–269. doi: 10.7326/0003-4819-151-4-200908180-00135. [DOI] [PubMed] [Google Scholar]
- 56.Walker CK, Krakowiak P, Baker A, Hansen RL, Ozonoff S, Hertz-Picciotto I. Preeclampsia, placental insufficiency, and autism spectrum disorder or developmental delay. JAMA Pediatr. 2015;169(2):154–162. doi: 10.1001/jamapediatrics.2014.2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walker CK, Ashwood P, Hertz-Picciotto I. Preeclampsia, placental insufficiency, autism, and antiphospholipid anti-bodies-reply. JAMA Pediatr. 2015;169(6):606–607. doi: 10.1001/jamapediatrics.2015.0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schroeder DI, Schmidt RJ, Crary-Dooley FK, Walker CK, Ozonoff S, Tancredi DJ, Hertz-Picciotto I, LaSalle JM. Placental methylome analysis from a prospective autism study. Mol Autism. 2016;7:51. doi: 10.1186/s13229-016-0114-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boyle CA, Decoufle P, Yeargin-Allsopp M. Prevalence and health impact of developmental disabilities in us children. Pediatrics. 1994;93:399–403. [PubMed] [Google Scholar]
- 60.Pastor PN, Reuben CA. Vital and health statistics, series 10, number 206, PsycEXTRA Dataset. American Psychological Association (APA); 2002. Attention deficit disorder and learning disability: United states, 1997–98. [PubMed] [Google Scholar]
- 61.Baio J. Prevalence of autism spectrum disorders–autism and developmental disabilities monitoring network, 14 sites, united states, 2008. In: Prevention, C.f.D.C.a, editor. Morbidity and mortality weekly report. Surveillance summaries. 2012. pp. 1–19. [PubMed] [Google Scholar]
- 62.Liu JH. Endocrinology of pregnancy, creasy and resnik’s maternal-fetal medicine: principles and practice. Elsevier, BV: 2009. pp. 111–124. [Google Scholar]
- 63.Petraglia F, Coukos G, Volpe A, Genazzani AR, Vale W. Involvement of placental neurohormones in human parturition. Ann NY Acad Sci. 1991;622:331–340. doi: 10.1111/j.1749-6632.1991.tb37878.x. [DOI] [PubMed] [Google Scholar]
- 64.Yen SS. The placenta as the third brain. J Reprod Med. 1994;39:277–280. [PubMed] [Google Scholar]
- 65.Lambertini L, Lee TL, Chan WY, Lee MJ, Diplas A, Wetmur J, Chen J. Differential methylation of imprinted genes in growth-restricted placentas. Reprod Sci. 2011;18:1111–1117. doi: 10.1177/1933719111404611. [DOI] [PubMed] [Google Scholar]
- 66.Lambertini L, Lee M-J, Marsit JC, Che J. Genomic imprinting in human placenta. Recent Advances in Research on the Human Placenta. 2012 InTech. [Google Scholar]
- 67.Reik W. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 68.Santos F, Dean W. Epigenetic reprogramming during early development in mammals. Reproduction. 2004;127:643–651. doi: 10.1530/rep.1.00221. [DOI] [PubMed] [Google Scholar]
- 69.Perera F, Herbstman J. Prenatal environmental exposures, epigenetics, and disease. Reprod Toxicol. 2011;31:363–373. doi: 10.1016/j.reprotox.2010.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.DUJ The genomic imprinting website. Shortitle The genomic imprinting website. http://www.geneimprint.com/site/what-is-imprinting.
- 71.Garg P, Borel C, Sharp AJ. Detection of parent-of-origin specific expression quantitative trait loci by cis-association analysis of gene expression in trios. PLoS One. 2012;7:e41695. doi: 10.1371/journal.pone.0041695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Charalambous M, da Rocha ST, Ferguson-Smith AC. Genomic imprinting, growth control and the allocation of nutritional resources: consequences for postnatal life. Curr Opin Endocrinol Diabetes Obes. 2007;14:3–12. doi: 10.1097/MED.0b013e328013daa2. [DOI] [PubMed] [Google Scholar]
- 73.Shaikh MG. 2.4.1 hypothalamic dysfunction (hypothalamic syndromes), Oxford Textbook of Endocrinology and Diabetes. Oxford University Press (OUP); 2011. [Google Scholar]
- 74.John RM. Epigenetic regulation of placental endocrine lineages and complications of pregnancy. Biochm Soc Trans. 2013;41:701–709. doi: 10.1042/BST20130002. [DOI] [PubMed] [Google Scholar]
- 75.McMinn J, Wei M, Schupf N, Cusmai J, Johnson EB, Smith AC, Weksberg R, Thaker HM, Tycko B. Unbalanced placental expression of imprinted genes in human intrauterine growth restriction. Placenta. 2006;27:540–549. doi: 10.1016/j.placenta.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 76.Tycko B, Morison IM. Physiological functions of imprinted genes. J Cell Physiol. 2002;192:245–258. doi: 10.1002/jcp.10129. [DOI] [PubMed] [Google Scholar]
- 77.Newman JRS, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, DeRisi JL, Weissman JS. Single-cell proteomic analysis of s. Cerevisiae reveals the architecture of biological noise. Nature. 2006;441:840–846. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- 78.Zaitoun I, Downs KM, Rosa GJM, Khatib H. Upregulation of imprinted genes in mice: An insight into the intensity of gene expression and the evolution of genomic imprinting. Epigenetics. 2010;5:149–158. doi: 10.4161/epi.5.2.11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Elowitz MB. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 80.Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A. Regulation of noise in the expression of a single gene. Nat Genet. 2002;31:69–73. doi: 10.1038/ng869. [DOI] [PubMed] [Google Scholar]
- 81.Blake WJ, KÆrn M, Cantor CR, Collins JJ. Noise in eukaryotic gene expression. Nature. 2003;422:633–637. doi: 10.1038/nature01546. [DOI] [PubMed] [Google Scholar]
- 82.Fraser HB, Hirsh AE, Giaever G, Kumm J, Eisen MB. Noise minimization in eukaryotic gene expression. PLoS Biol. 2004;2:e137. doi: 10.1371/journal.pbio.0020137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Batada NN, Hurst LD. Evolution of chromosome organization driven by selection for reduced gene expression noise. Nat Genet. 2007;39:945–949. doi: 10.1038/ng2071. [DOI] [PubMed] [Google Scholar]
- 84.Guerrero-Bosagna C, Skinner MK. Environmentally induced epigenetic transgenerational inheritance of phenotype and disease. Mol Cell Endocrinol. 2012;354:3–8. doi: 10.1016/j.mce.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dilinoy DC, Weidman JR, Jirtle RL. Epigenetic gene regulation: linking early developmental environment to adult disease. Reprod Toxicol. 2007;23:297–307. doi: 10.1016/j.reprotox.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 86.Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet. 2007;8:253–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kappil M, Lambertini L, Chen J. Environmental influences on genomic imprinting. Curr Environ Health Rep. 2015;2:155–162. doi: 10.1007/s40572-015-0046-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lambertini L. Genomic imprinting. Curr Opin Pediatr. 2014;26:237–242. doi: 10.1097/MOP.0000000000000072. [DOI] [PubMed] [Google Scholar]
- 89.Lambertini L, Marsit CJ, Sharma P, Maccani M, Ma Y, Hu J, Chen J. Imprinted gene expression in fetal growth and development. Placenta. 2012;33:480–486. doi: 10.1016/j.placenta.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen J, Li Q, Rialdi, Mystal A, EY Ly, Finik J, Davey J, Lambertini T, Nomura LY. Influences of maternal stress during pregnancy on the epi/genome: comparison of placenta and umbilical cord blood. Depress Anxiety. 2014;3:1–6. doi: 10.4172/2167-1044.1000152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Diplas AI, Lambertini L, Lee M-J, Sperling R, Lee YL, Wetmur JG, Chen J. Differential expression of imprinted genes in normal and iugr human placentas. Epigenetics. 2009;4:235–240. doi: 10.4161/epi.9019. [DOI] [PubMed] [Google Scholar]
- 92.Green BB, Kappil M, Lambertini L, Armstrong DA, Guerin DJ, Sharp AJ, Lester BM, Chen J, Marsit CJ. Expression of imprinted genes in placenta is associated with infant neurobehavioral development. Epigenetics. 2015;10:834–841. doi: 10.1080/15592294.2015.1073880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lambertini L, Diplas AI, Lee M-J, Sperling R, Chen J, Wetmur JG. A sensitive functional assay reveals frequent loss of genomic imprinting in human placenta. Epigenetics. 2008;3:261–269. doi: 10.4161/epi.3.5.6755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lambertini L, Diplas AL, Wetmur J, Lee MJ, Chen J. Evaluation of genomic imprinting employing the analysis of loss of imprinting (loi) at the rna level: preliminary results. Eur J Oncol. 2009;14:161–169. [Google Scholar]
- 95.Marsit CJ, Lambertini L, Maccani MA, Koestler DC, Houseman EA, Padbury JF, Lester BM, Chen J. Placenta-imprinted gene expression association of infant neurobehavior. J Pediatr. 2012;160(854–60):e2. doi: 10.1016/j.jpeds.2011.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mathers JC. Early nutrition: Impact on epigenetics, nutrigenomics—opportunities in Asia. S. Karger AG; 2007. pp. 42–48. [DOI] [PubMed] [Google Scholar]
- 97.Kawasaki K, Kondoh E, Chigusa Y, Ujita M, Murakami R, Mogami H, Brown JB, Okuno Y, Konishi I. Reliable preeclampsia pathways based on multiple independent microarray data sets. Mol Hum Reprod. 2014;21:217–224. doi: 10.1093/molehr/gau096. [DOI] [PubMed] [Google Scholar]
- 98.Enquobahrie DA, Meller M, Rice K, Psaty BM, Siscovick DS, Williams MA. Differential placental gene expression in preeclampsia. Am J Obstet Gynecol. 2008;199(566):e1–66. e11. doi: 10.1016/j.ajog.2008.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Unek G, Ozmen A, Mendilcioglu I, Simsek M, Korgun ET. The expression of cell cycle related proteins pcna, ki67, p27 and p57 in normal and preeclamptic human placentas. Tissue Cell. 2014;46:198–205. doi: 10.1016/j.tice.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 100.Bourque DK, Avila L, Peñaherrera M, von Dadelszen P, Robinson WP. Decreased placental methylation at the h19/igf2 imprinting control region is associated with normotensive intrauterine growth restriction but not preeclampsia. Placenta. 2010;31:197–202. doi: 10.1016/j.placenta.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 101.Janssen AB, Tunster SJ, Savory N, Holmes A, Beasley J, Parveen SAR, Penketh RJA, John RM. Placental expression of imprinted genes varies with sampling site and mode of delivery. Placenta. 2015;36:790–795. doi: 10.1016/j.placenta.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Romanelli V, Belinchón A, Campos-Barros A, Heath KE, García-Miñaur S, Martínez-Glez V, Palomo R, Mercado G, Gracia R, et al. Cdkn1c mutations in hellp/preeclamptic mothers of beckwith–wiedemann syndrome (bws) patients. Placenta. 2009;30:551–554. doi: 10.1016/j.placenta.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 103.Kanayama N. Deficiency in p57kip2 expression induces preeclampsia-like symptoms in mice. Mol Hum Reprod. 2002;8:1129–1135. doi: 10.1093/molehr/8.12.1129. [DOI] [PubMed] [Google Scholar]
- 104.Knox KS, Baker JC. Genome-wide expression profiling of placentas in the p57kip2 model of pre-eclampsia. Mol Hum Reprod. 2007;13:251–263. doi: 10.1093/molehr/gal116. [DOI] [PubMed] [Google Scholar]
- 105.Takahashi K. P57kip2 regulates the proper development of labyrinthine and spongiotrophoblasts. Mol Hum Reprod. 2000;6:1019–1025. doi: 10.1093/molehr/6.11.1019. [DOI] [PubMed] [Google Scholar]
- 106.Tunster SJ, Van de Pette M, John RM. Fetal overgrowth in the cdkn1c mouse model of beckwith-wiedemann syndrome. Dis Models Mech. 2011;4:814–821. doi: 10.1242/dmm.007328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jin F, Qiao C, Luan N, Shang T. The expression of the imprinted gene pleckstrin homology-like domain family a member 2 in placental tissues of preeclampsia and its effects on the proliferation, migration and invasion of trophoblast cells jeg-3. Clin Exp Pharmacol Physiol. 2015;42:1142–1151. doi: 10.1111/1440-1681.12468. [DOI] [PubMed] [Google Scholar]
- 108.Weksberg R. Imprinted genes and human disease. Am J Med Genet. 2010;154C:317–320. doi: 10.1002/ajmg.c.30268. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.