Abstract
The dynamic posttranslational modification O-linked β-N-acetylglucosamine glycosylation (O-GlcNAcylation) is present on thousands of intracellular proteins in the brain. Like phosphorylation, O-GlcNAcylation is inducible and plays important functional roles in both physiology and disease. Recent advances in mass spectrometry (MS) and bioconjugation methods are now enabling the mapping of O-GlcNAcylation events to individual sites in proteins. However, our understanding of which glycosylation events are necessary for regulating protein function and controlling specific processes, phenotypes, or diseases remains in its infancy. Given the sheer number of O-GlcNAc sites, methods are greatly needed to identify promising sites and prioritize them for time- and resource-intensive functional studies. Revealing sites that are dynamically altered by different stimuli or disease states will likely go a long way in this regard. Here, we describe advanced methods for identifying O-GlcNAc sites on individual proteins and across the proteome, and for determining their stoichiometry in vivo. We also highlight emerging technologies for quantitative, site-specific MS-based O-GlcNAc proteomics (O-GlcNAcomics), which allow proteome-wide tracking of O-GlcNAcylation dynamics at individual sites. These cutting-edge technologies are beginning to bridge the gap between the high-throughput cataloging of O-GlcNAcylated proteins and the relatively low-throughput study of individual proteins. By uncovering the O-GlcNAcylation events that change in specific physiological and disease contexts, these new approaches are providing key insights into the regulatory functions of O-GlcNAc in the brain, including their roles in neuroprotection, neuronal signaling, learning and memory, and neurodegenerative diseases.
For Table of Contents Use Only

The addition of β-N-acetylglucosamine to serine or threonine residues of proteins (O-GlcNAcylation) is a dynamic, inducible posttranslational modification (PTM) that is highly abundant in the brain. Unlike other forms of glycosylation, O-GlcNAcylation is found primarily on intracellular proteins and is not elaborated beyond the single GlcNAc residue. Two enzymes are responsible for O-GlcNAc cycling in higher mammals: the glycosyltransferase O-GlcNAc transferase (OGT) and the glycosidase O-GlcNAcase (OGA).1 To date, thousands of O-GlcNAc-modified proteins have been identified in the mammalian brain,2–4 and O-GlcNAc has been shown to regulate many important neuronal functions, such as activity-dependent gene expression, α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor (AMPAR) trafficking, calcium signaling, and metabolism.5 Notably, the modification may also contribute to higher-order brain functions. For instance, manipulating global O-GlcNAcylation levels profoundly alters synaptic plasticity, as well as learning and memory in vivo.6–12 Moreover, O-GlcNAc regulates the function of numerous proteins critical for learning and memory, including AMPAR subunit 2,7 protein kinase A,8 and cAMP response element-binding protein (CREB).13 Remarkably, mutation of only a single O-GlcNAcylation site on CREB increased its transcriptional activity and accelerated long-term memory consolidation in mice.13
In addition to its physiological roles in the brain, aberrant O-GlcNAcylation and OGT/OGA expression have been linked to several neurodegenerative disorders such as Alzheimer’s, Huntington’s, and Parkinson’s disease.11, 14–16 For instance, global levels of O-GlcNAcylation and neuronal OGT are significantly decreased in Alzheimer’s disease (AD) brains.11, 17, 18 Moreover, conditional genetic deletion of OGT in excitatory neurons of the mouse forebrain recapitulates many of the pathological and behavioral hallmarks of AD, including progressive neuronal loss, increased production of hyperphosphorylated tau and amyloidogenic amyloid β peptides, macroscopic plaque and tangle formation, and memory loss.11 Globally increasing O-GlcNAcylation has also been demonstrated to limit cognitive decline and neuronal pathology in mouse models of neurodegeneration.14, 19–22 Overall, it is becoming increasingly clear that O-GlcNAcylation plays a pivotal role in neuronal health and function, and modulating O-GlcNAc signaling may represent a promising neuroprotective strategy for neurodegenerative diseases.14, 15
Over the past decade, growing interest in O-GlcNAcylation has led to the discovery of many new functions for the modification and revealed its central importance for both brain physiology and pathology. However, the huge diversity of functions and substrates has created a major challenge for the field – namely, the prioritization of O-GlcNAcylation events for follow-up studies. Given the sheer number of O-GlcNAc modification sites, methods are greatly needed to identify promising sites and prioritize them for time- and resource-intensive functional studies. To address this challenge, it will be critical to move from a static picture of whether a protein is O-GlcNAcylated to a more dynamic view and identify context-specific changes in O-GlcNAcylation, such as sites that are induced by specific stimuli or altered during development or disease progression. Identifying these key sites and developing methods to selectively modulate their glycosylation status may also prove critical for therapeutic intervention in neurodegenerative disease, as globally altering O-GlcNAcylation levels could have deleterious, compensatory, or potentially unforeseen off-target effects.22–26
The importance of monitoring the dynamics of O-GlcNAcylation at specific sites is further underscored by the observation that OGT and OGA exhibit exquisite selectivity for particular substrates, depending on the cellular context.26–30 For instance, early quantitative proteomics studies by our laboratory showed that robust excitatory stimulation of the brain in vivo induces O-GlcNAcylation on only a subset of OGT substrates, while pharmacological inhibition of OGA affects almost an entirely different subset.27 This suggests that OGT can discriminate among its various substrates in response to specific cellular signals. OGT can also exhibit exquisite selectivity for different sites within the same protein. For example, neuronal depolarization induces CREB glycosylation at only a single site, Ser40, while other glycosylation sites in CREB remain unchanged.13 Thus, different sites within the same protein can show distinct temporal dynamics, which may enable O-GlcNAc to regulate various aspects of a protein’s function. Finally, several studies have observed that changes in O-GlcNAcylation at the protein level often do not correlate with changes at individual sites. This is exemplified by the transcriptional repressor methyl CpG binding protein 2 (MeCP2), whose loss of function underlies Rett syndrome.31 Although the O-GlcNAcylation level on the total MeCP2 population decreases upon neuronal depolarization, the O-GlcNAcylation level on the Ser80-phosphorylated subpopulation of MeCP2 significantly increases.32 These examples clearly demonstrate that O-GlcNAcylation undergoes complex regulation and site-specific dynamics in the brain (Figure 1).
Figure 1.
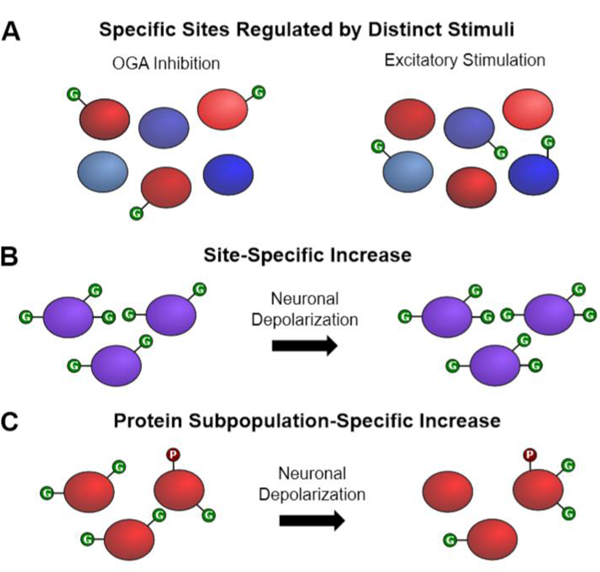
O-GlcNAcylation undergoes complex regulation and site-specific dynamics in the brain. (A) Inhibition of OGA increases O-GlcNAcylation on a subset of OGT substrates (in red), whereas excitatory neuronal stimulation with kainic acid increases O-GlcNAcylation on a different subset (in blue). (B) Neuronal stimulation increases O-GlcNAcylation of CREB at S40, while the other two sites remain unchanged. (C) Overall O-GlcNAcylation of MeCP2 decreases upon neuronal depolarization; however, for a subset of phosphorylated MeCP2, O-GlcNAcylation increases.
In this perspective, we will highlight state-of-the art technologies for identifying O-GlcNAc sites and tracking their context-specific dynamics. We will first focus on methods for mapping glycosylation sites within proteins and for quantifying site occupancy or stoichiometry. We will then describe emerging technologies for quantitative, mass spectrometry (MS)-based O-GlcNAc proteomics (O-GlcNAcomics) and discuss how such approaches are providing new, systems-level insights into the site-specific dynamics of O-GlcNAc in response to different cellular or disease states. Our intention is to provide an overview of these technologies with an emphasis on their potential impact on the field of neurobiology. Overall, the development and implementation of quantitative O-GlcNAcomics methods should greatly accelerate the prioritization of O-GlcNAcylation sites for further study and are a critical next step toward advancing our understanding of the functions of O-GlcNAc in the brain.
O-GLCNAC SITE IDENTIFICATION ON INDIVIDUAL PROTEINS
The identification of O-GlcNAc sites is often crucial for elucidating the function of the modification in the context of specific proteins. However, this task can be challenging as there is no known consensus sequence for OGT, and the O-GlcNAc modification occurs substoichiometrically on many proteins. Moreover, the O-GlcNAc moiety is frequently found within a stretch of serine or threonine residues and is readily lost upon collision-induced dissociation (CID) MS due to the lability of the O-glycosidic linkage.33, 34 As a result, O-GlcNAcylation sites are often ambiguous, difficult to localize to a single serine or threonine residue, or require confirmation with time-consuming site-directed mutagenesis studies.35, 36
In response to these challenges, Hart and coworkers developed the BEMAD (β-elimination followed by Michael addition with dithiothreitol) approach,37 which chemically converts O-GlcNAcylated serine and threonine residues to thiol-containing derivatives that are stable during CID. As other O-linked modifications such as phosphorylation are also derivatized in the process, this technique is not well suited for the selective mapping of O-GlcNAc sites unless it is combined with some form of enrichment for O-GlcNAcylated peptides.35, 38, 39 Our laboratory has developed a chemoenzymatic labeling strategy that accomplishes this task.36, 40–43 The approach allows for selective, quantitative labeling of O-GlcNAcylated peptides or proteins with an unnatural azido- or ketone-containing galactose sugar (N-azidoacetylgalactosamine (GalNAz)42 or 2-acetonyl-2-deoxy-α-D-galactose (ketogal)40) using an engineered β−1,4-galactosyltransferase (Y289L GalT).44 The azide or ketone functionality enables the attachment of different reporter groups (e.g., biotin, fluorescent dyes) using bioorthogonal chemistry.36, 40, 42, 43 Thus, following labeling with biotin, O-GlcNAcylated proteins can be captured using streptavidin resin and then simultaneously derivatized and eluted using BEMAD (Figure 2A). This dual chemoenzymatic labeling/BEMAD strategy has proven effective for mapping O-GlcNAcylation sites on individual proteins35, 38, 39, 45, 46 and has been applied on a proteome-wide level,38, 46–48 albeit with less success in site identification compared to the methods discussed below. Using this approach, several residues near the active site of calcium/calmodulin-dependent protein kinase type IV (CaMKIV), an important kinase whose signaling functions have been linked to learning, memory, and neurodegeneration,39, 49–51 were identified.39 Site-directed mutagenesis studies demonstrated that O-GlcNAcylation of these sites modulates the phosphorylation and subsequent activation of CaMKIV.39, 52 Overall, although BEMAD has some limitations (such as being prone to false positives and causing peptide degradation47, 53), it remains a useful method for mapping O-GlcNAc sites and is amenable to traditional MS workflows.
Figure 2.

Strategies for mapping O-GlcNAc sites. (A) Chemoenzymatic labeling/BEMAD strategy. O-GlcNAcylated peptides are chemoenzymatically labeled with a biotin moiety, enriched by streptavidin capture, and then eluted using BEMAD prior to LC-MS/MS analysis. (B) Cleavable biotin-Dde-alkyne strategy. O-GlcNAcylated peptides are chemoenzymatically labeled with a biotin-Dde-alkyne derivative and enriched by streptavidin capture. Quantitative release of O-GlcNAcylated peptides is then achieved using hydroxylamine or hydrazine, which imparts an additional positive charge on the peptide and thereby facilitates LC-MS/MS analysis.
With advances in MS instrumentation, the direct mapping of O-GlcNAc sites, without chemical conversion of the labile O-GlcNAc moiety, has also become possible in some cases. Electron-transfer dissociation (ETD) enables the observation of O-GlcNAcylated serine and threonine fragment ions in MS/MS spectra without O-glycosidic bond cleavage,27, 54 thereby localizing the exact modification site. Typically, O-GlcNAcylated proteins of interest are overexpressed, immunoprecipitated, and analyzed directly by liquid chromatography (LC)-MS/MS following in-gel digestion.13, 36 We and others have successfully employed this approach to identify crucial, regulatory O-GlcNAcylation sites on neuronal proteins involved in learning and memory, signal transduction, and metabolism, including CREB and the mitochondrial motor-adaptor protein Milton.13, 55 Given the limitations of the BEMAD approach discussed above, the use of ETD-MS/MS to map exact O-GlcNAc sites is generally preferred if appropriate instrumentation is available.
Difficult-to-detect or low-abundance O-GlcNAc sites on proteins require techniques for the highly efficient enrichment and release of O-GlcNAcylated peptides. While labeling O-GlcNAcylated peptides with a biotin tag provides an effective enrichment strategy,27, 38 the strength of the biotin-streptavidin interaction can hinder efficient release of the peptides from the resin. To overcome this problem, we recently developed a biotin tag containing a chemically cleavable 1-(4,4-dimethyl-2,6-dioxocyclohex-1-ylidene)ethyl (Dde) functionality to selectively capture O-GlcNAcylated peptides after chemoenzymatic labeling.56 The O-GlcNAcylated peptides are then quantitatively released from streptavidin resin using hydrazine or hydroxylamine (Figure 2B).36 Importantly, this cleavage event leaves O-GlcNAcylated peptides with an additional positive charge that significantly improves their ETD fragmentation efficiency. Ultimately, this approach greatly increases the likelihood of successful identification and sequencing of O-GlcNAcylated peptides.36, 56–59 It is worth noting that an additional advantage of the biotin-Dde tag is that produces three signature ions upon fragmentation by higher-energy collisional dissociation (HCD), which result from cleavage of the glycosidic bond between GlcNAc and GalNAz (300.1 m/z), the water adduct of the previous fragment (318.1 m/z), and cleavage between GlcNAc and the serine/threonine residue (503.2 m/z).36 These signature ions enable unambiguous assignment of the modification at the peptide level, and the increased ETD fragmentation efficiency (due to the additional positive charge) increases the likelihood that an O-GlcNAcylation event can be definitively localized to a single residue. In fact, using this strategy, we identified four new O-GlcNAc sites on OGT that were previously undetectable by other methods, all of which were on tryptic peptides spanning multiple sites of O-GlcNAcylation.56 We believe that this improved enrichment/release workflow will be widely applicable for discovering low-abundance sites or sites that were previously ambiguous due to the lability of O-GlcNAc or poor glycopeptide ionization/fragmentation. Thus, the development of new chemoenzymatic labeling tags coupled with cutting-edge MS technologies has significantly reduced the barriers to the unambiguous assignment of O-GlcNAc sites, and in the future, the complete mapping of sites on nearly any protein of interest should become increasingly routine.
O-GLCNAC SITE MAPPING ACROSS THE PROTEOME
The identification of O-GlcNAc sites across the proteome requires effective methods for the enrichment of O-GlcNAcylated proteins or peptides from complex biological samples. One powerful, widely used approach involves the chemoenzymatic tagging strategy described above. In early studies, we applied this strategy to enrich O-GlcNAcylated proteins from the mammalian brain and identified 34 O-GlcNAcylated peptides corresponding to 25 proteins. These proteins had a wide range of important neuronal functions, including the regulation of gene expression, neurotransmission, and synaptic plasticity.38 Similar to the biotin-Dde tag described above, the biotin tag in this case also provided a unique fragmentation pattern upon MS/MS analysis that enabled us to conclusively identify the modification. This approach was subsequently improved through the development of a photocleavable biotin tag (biotin-PC)60 and the chemically cleavable biotin-Dde tag described above (Figure 2B).56 Using the biotin-PC tag, Hart and colleagues identified 458 O-GlcNAc sites on 195 proteins in the brains of WT and 3xTg (a common mouse model of AD61) mice.3 Impressively, 168 of the identified O-GlcNAcylated proteins had not been previously described and included multiple proteins involved in synaptic plasticity and neuronal signaling, cytoskeletal organization, and neuronal gene expression.3 More recently, the chemically cleavable biotin-Dde tag has significantly improved our ability to comprehensively map O-GlcNAc sites across the proteome. In a direct side-by-side comparison, the biotin-Dde tag outperformed the biotin-PC tag, enabling the identification of 414 unique O-GlcNAcylated peptides from human embryonic kidney 293T cells, as compared to the 227 unique O-GlcNAcylated peptides identified using the biotin-PC tag (unpublished). Building further on this approach, we have designed a next-generation, chemically cleavable tag that has greater thermal stability compared to biotin-Dde and has enabled the mapping of over 1,300 O-GlcNAc sites across ~600 proteins in 293T cells (manuscript in preparation).
An alternative to chemoenzymatic labeling strategies involves the use of lectin weak affinity chromatography with wheat germ agglutinin to enrich O-GlcNAcylated peptides. Using this approach combined with high-pH reversed-phase fractionation and ETD-MS/MS, Burlingame and coworkers identified 1,750 unique sites of O-GlcNAcylation from fractionated mouse synaptosomes, the most sites described thus far in a single experiment.2 This study also interrogated the phosphoproteome on these same samples given the precedent for a high degree of crosstalk between these two modifications.1, 26 Interestingly, all but one of the extensively O-GlcNAcylated proteins were extensively phosphorylated. Moreover, protein kinases as a class were found to be more extensively O-GlcNAcylated in synaptosomes, suggesting an intriguing potential for crosstalk at the level of enzyme activity. However, at the site level, the number of O-GlcNAc sites that were also phosphorylation sites did not exceed what would be expected by chance, and there was no propensity for O-GlcNAc and phosphorylation sites to cluster in primary sequence or three-dimensional space.
Collectively, studies of the O-GlcNAcylated proteome have provided critical insights into the physiological functions of O-GlcNAc in the brain. For instance, these analyses have revealed a large number of pre- and postsynaptic scaffolding proteins, suggesting important roles for the modification in synaptic communication and function.62 Moreover, the unbiased identification of O-GlcNAc sites has facilitated follow-up studies to understand the function of O-GlcNAc in the context of specific proteins, as demonstrated by a recent study by Pratt and coworkers on the effects of site-specific O-GlcNAcylation on α-synuclein aggregation.63, 64 However, despite the power of O-GlcNAcomic approaches, these technologies provide only a static snapshot of the O-GlcNAcome, without regard for the stoichiometry or dynamics of specific sites. As described below, knowledge of the site occupancy and inducibility of specific sites can provide insights into the importance of specific glycosylation events and the cellular contexts in which they function.
METHODS FOR MEASURING DYNAMIC O-GLCNAC STOICHIOMETRY
Determining the stoichiometry of glycosylation on a protein, or at a specific site, is critical for assessing whether that glycosylation event is likely important. The modification is more likely to be functionally relevant and to operate in a regulatory capacity if a significant population of the protein is modified and the modification is dynamically cycled. Glycosylation stoichiometries can be challenging to quantify and thus are known for only a small fraction of the O-GlcNAcome. To quantify O-GlcNAc stoichiometries, large amounts of purified protein (e.g., for high-pH anion-exchange chromatography with pulsed amperometric detection)65–67 or synthetic O-GlcNAcylated peptide standards encompassing the sites of interest are typically required. In order to overcome these challenges, we have developed two methods to detect and quantify the O-GlcNAc-modified subpopulation. The first approach employs the chemoenzymatic biotin/streptavidin enrichment strategy described above, in combination with immunoblotting to detect the protein of interest. The amount of a captured (i.e. O-GlcNAcylated) protein is quantified relative to the total amount of that protein to give an approximate measure of stoichiometry.36, 41, 68 The second approach allows for direct visualization and quantification of the O-GlcNAc-modified subpopulation via chemoenzymatic labeling of the proteins with resolvable polyethylene glycol (PEG) tags (Figure 3A). Specifically, O-GlcNAcylated proteins from cell lysates are labeled with a keto-galactose or azido-galactose sugar as described above and then reacted with an aminooxy-, alkyne-, or dibenzocyclooctyne-functionalized PEG tag of defined molecular mass (typically 2-kDa or 5-kDa). Importantly, the PEG tag shifts the molecular weight of the glycosylated species and allows them to be resolved from the non-glycosylated species by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).4, 32, 36, 43 After immunoblotting to detect the protein(s) of interest, simple inspection of the mass-shifted bands can rapidly establish whether a protein is singly, doubly or multiply glycosylated. Moreover, in vivo glycosylation stoichiometries can be readily calculated by quantifying the relative intensities of each band. One caveat with chemoenzymatic labeling, however, is that GalT can also label complex glycans that contain a terminal GlcNAc sugar. For known membrane proteins, complex N-linked glycans can be removed using Peptide:N-glycosidase F.36 In addition, it is possible that chemoenzymatic labeling may differ in efficiency for different sites, although the use of excess reagents ensures highly efficient labeling, and both of these methods have been consistently demonstrated to provide accurate measurements of the O-GlcNAcylation stoichiometry on a range of peptide and protein substrates.32, 36, 43, 56, 69
Figure 3.

Chemoenzymatic labeling with PEG tags to determine O-GlcNAcylation stoichiometry. (A) A PEG tag of defined mass (2-kDa or 5-kDa) is installed on O-GlcNAcylated proteins in cell lysates using Y289L GalT and copper (I)-catalyzed azide-alkyne cycloaddition (CuAAC) chemistry. (B) Neurons expressing either WT or S40A CREB fused to a FLAG tag were depolarized with KCl for the times indicated before lysis and chemoenzymatic labeling with PEG. Labeled proteins were then subjected to SDS-PAGE and Western blotting for CREB or FLAG. Note the presence of two major CREB glycoforms, mono- and di-glycosylated, and the loss of KCl-induced O-GlcNAcylation upon mutation of Ser40.
When used in combination with site-directed mutagenesis, this approach allows for rapid interrogation of O-GlcNAc stoichiometry at specific sites and across multiple conditions. For instance, we found that approximately 30% of CREB was mono-glycosylated in resting neurons. Upon KCl-mediated depolarization, glycosylation was induced specifically at a single site, Ser40 (Figure 3B).13, 32 Strikingly, mutation of Ser40 to alanine resulted in increased CREB-dependent gene transcription, and significantly accelerated long-term memory formation in mice.13 Overall, this powerful method enables the measurement of O-GlcNAcylation stoichiometries on endogenous proteins of interest without the need for protein purification, advanced instrumentation, or expensive radiolabels.
QUANTITATIVE O-GLCNACOMICS
Systems-level approaches to quantify site-specific changes in O-GlcNAcylation across the proteome can help find the ‘needle in the haystack’ – namely, the glycosylation events that are functionally important in a specific cellular or disease context. We will describe here three promising technologies for quantitative site-specific O-GlcNAcomics: QUIC-Tag, quantitative CEPC, and quantitative IsoTaG.
The quantitative isotopic and chemoenzymatic tag (QUIC-Tag) method, developed in 2007, was the first quantitative O-GlcNAcomics approach.27 This approach combines chemoenzymatic biotin tagging for O-GlcNAcylated peptide enrichment with stable isotope dimethyl labeling for quantitative proteomics (Figure 4A). Using QUIC-Tag, we observed changes in O-GlcNAcylation at specific sites following stimulation of rats with kainic acid, a kainate-type glutamate receptor agonist that produces widespread excitatory stimulation of the brain.27 These studies provided the first evidence that O-GlcNAcylation is dynamically modulated in vivo by neuronal activity. Moreover, we found that different subsets of O-GlcNAc sites respond to different neuronal stimuli, with kainic acid affecting a distinct set of sites compared to the OGA inhibitor O-(2-acetamido-2deoxy-D-glucopyranosylidene)amino-N-phenylcarbamate (PUGNAc). Overall, our findings highlight key differences in the occupancy and temporal dynamics of specific sites in response to different stimuli and demonstrate the complex regulation of O-GlcNAcylation in neurons. With recent advances in MS instrumentation and the new chemoenzymatic tags described above, this strategy can now be adapted to enable more comprehensive, quantitative comparisons across many different functional states.
Figure 4.

Strategies for quantitative O-GlcNAcomics. (A) QUIC-Tag strategy. O-GlcNAcylated proteins are chemoenzymatically labeled with biotin, digested, captured, and subjected to isotopic dimethyl labeling before LC-MS/MS analysis. (B) Quantitative CEPC. O-GlcNAcylated proteins are digested, TMT labeled, and fractionated before chemoenzymatic labeling with a biotin-PC tag. The labeled peptides are then enriched, eluted from the beads by UV cleavage, and subjected to LC-MS/MS analysis. (C) Quantitative IsoTaG. Metabolically labeled O-GlcNAzylated proteins are functionalized with an acid-cleavable biotin linker containing an isotopic label (IsoTaG Probe) using CuAAC. Labeled proteins are then enriched, digested, eluted from the beads, and analyzed by LC-MS/MS. Importantly, glycopeptides can then be readily identified by their unique isotopic signature in MS1. In all cases, the relative O-GlcNAcylation levels on peptides derived from two or more samples can be calculated from the ratio between light and heavy isotopically-labeled peptides (QUIC-Tag and CEPC) or the ratio of intensities after label-free quantification (IsoTaG).
The quantitative chemical/enzymatic photochemical cleavage (CEPC) strategy relies on isotopic labeling (e.g., stable isotope labeling using amino acids in cell culture (SILAC)) or isobaric labeling (e.g., tandem mass tags (TMTs)) of peptides derived from cell lysates, followed by chemoenzymatic labeling using Y289L GalT and biotin-PC to enrich O-GlcNAcylated peptides (Figure 4B).70 Recently, Liu and coworkers exploited this technique to quantify site-specific differences in O-GlcNAcylation between AD and non-diseased brain tissue.4 After TMT labeling followed by chemoenzymatic labeling, enrichment, and photochemical cleavage, they determined 623 ratios at the O-GlcNAcylated peptide level and mapped 1,094 sites. Twenty-four of these sites showed significant differences in O-GlcNAcylation between AD and control tissues that were not due to changes in protein expression. Most of the sites were found on proteins involved in pre- and post-synaptic organization and structure, such as synaptopodin, ankyrin-3, integrin beta-4, and neurofascin.4 Overall, these quantitative O-GlcNAcomics studies show that dysregulated O-GlcNAcylation occurs on multiple, select neuronal proteins during the development of sporadic AD and identify several novel candidates for in-depth investigations into the mechanisms underlying AD pathogenesis.
Finally, Pitteri and colleagues have recently reported combining the isotope-targeted glycoproteomics (IsoTaG)71, 72 strategy with label-free quantification to profile changes in O-GlcNAcylation (Figure 4C).73 This approach relies on metabolic incorporation of N-azidoacetylglucosamine (GlcNAz) into O-GlcNAcylated proteins.74 Briefly, cells are incubated with peracetylated GalNAz, which is converted to uridine diphosphate (UDP)-GlcNAz by the UDP-galactose 4-epimerase pathway.75 As OGT tolerates UDP-GlcNAz as a substrate, O-GlcNAcylated proteins can be labeled with GlcNAz and then conjugated to an acid-cleavable biotin-alkyne linker containing an isotopic label (IsoTaG Probe) using CuAAC. It is important to note, however, that other GlcNAc- and N-acetylgalactosamine-containing complex glycans will also be metabolically labeled,72, 74 and the incorporation of GlcNAz is substoichiometric, reducing the labeling efficiency compared to chemoenzymatic strategies. The labeled glycoproteins are enriched by streptavidin capture, proteolytically digested, and the resulting glycopeptides are released by cleavage of the linker and analyzed by LC-MS/MS. Importantly, the cleaved IsoTaG Probe imparts a readily detectable isotope pattern to the glycopeptides which allows them to be definitively identified and targeted in subsequent LC-MS/MS runs.72, 73 Using this strategy, Woo et al. mapped 851 O-GlcNAc sites in primary human T cells across 77 MS runs.73 They also quantified changes in O-GlcNAcylation in response to T cell activation using label-free quantification and identified differences in the levels of 518 O-linked glycopeptides, corresponding to 227 proteins, between resting and activated T cells. One caveat with such comparisons is that the non-natural GlcNAz sugar may or may not be incorporated into proteins with the same relative ratios as the endogenous O-GlcNAc modification and thus the differences in O-GlcNAcylation at specific sites may not reflect physiological conditions. Nonetheless, diverse site-specific dynamics were observed on several highly O-GlcNAcylated proteins, including CREB, nuclear pore complex protein Nup98, transcription factor jun-B, and protein phosphatase 1 regulatory subunit 12A. Given the interesting parallels between T cell and neuronal activation,76 these results may have important implications for neurons and shed light on common pathways and mechanisms shared by both cell types.
CONCLUSIONS AND FUTURE DIRECTIONS
Significant advances in O-GlcNAc detection, site mapping, and quantitative MS-based O-GlcNAcomics are uncovering key regulatory roles for O-GlcNAcylation in neuronal maintenance and health, learning and memory, and neurodegeneration. As these advanced methods develop and become widely available, the number of important discoveries will continue to grow and impact our understanding of many aspects of brain function. We have focused here on the emergence of new MS-based O-GlcNAcomics technologies for profiling and monitoring the dynamics of O-GlcNAc at thousands of sites simultaneously. These technologies have the potential to revolutionize our understanding of the modification by providing unprecedented insight into the specificity and kinetics of glycosylation at particular sites, by facilitating the much-needed prioritization of O-GlcNAcylation sites for functional studies, and by revealing those sites that are important in specific physiological and disease contexts.
Chemoenzymatic labeling strategies are particularly well-suited for quantitative, site-specific O-GlcNAcomics because they provide highly specific, nearly quantitative labeling and enrichment of low-abundance O-GlcNAcylated proteins or peptides. Bringing other advanced techniques for PTM profiling and quantitative proteomics to bear on the O-GlcNAc field would also open up even more exciting experimental possibilities. Moreover, with additional improvements in the design of new chemical tags and MS analysis workflows specially tuned for glycopeptides, we may soon find that the long-standing challenge of quantitative ‘shotgun O-GlcNAcomics’ has become a thing of the past.
The continued development of quantitative, site-specific O-GlcNAcomics methods will also help to overcome key gaps in our understanding of O-GlcNAcylation. For instance, the ability to compare O-GlcNAcylation levels across different cell populations or regions of the brain should provide new insights into cell-specific roles for the modification and how aberrant O-GlcNAcylation at the cellular level may give rise to macroscopic, tissue-level pathology. Quantitative O-GlcNAcomics should also enable specific O-GlcNAcylation events to be monitored with unprecedented temporal resolution. As such, they could be used to track dynamic O-GlcNAc cycling at specific sites under a limitless set of important physiological and disease-relevant conditions. For example, while global changes in O-GlcNAcylation and expression of OGT/OGA are observed during development, aging,77–80 and disease progression,11, 14, 17, 18 the site-specific changes in O-GlcNAcylation associated with these processes are largely uncharacterized.
In the future, the integration of quantitative O-GlcNAcomics data with other state-of-the-art ‘-omics’ techniques (e.g., interactomics, phosphoproteomics, transcriptomics, proteomics) will help uncover important correlations across biological systems and crosstalk between O-GlcNAcylation and other networks. We expect that such multimodal approaches will prove particularly powerful for addressing questions such as how O-GlcNAcylation regulates protein-protein interactions, kinase signaling, and gene expression. In addition to revealing new regulatory roles for O-GlcNAc, these efforts may shed light on long-standing questions regarding the complex regulation of OGT and OGA. Together, we believe that multimodal, systems-level approaches, coupled with rapidly emerging technologies for quantitative O-GlcNAcomics, will usher in a new era in our understanding of the functional roles of O-GlcNAc in the brain.
Acknowledgments
Funding
This research was supported by the National Institutes of Health (R01GM084724 to L.C.H.-W., T32GM007616 to A.W.S and T32GM008042, T32GM007616, and F30AG055314 to J.W.T), the UCLA-Caltech Medical Scientist Training Program (J.W.T), and the National Science Foundation Graduate Research Fellowship under Grant No. DGE‐1745301 (A.W.S).
Footnotes
Notes
The authors declare no competing financial interest.
REFERENCES
- [1].Hart GW, Slawson C, Ramirez-Correa G, and Lagerlof O (2011) Cross talk between O-GlcNAcylation and phosphorylation: Roles in signaling, transcription, and chronic disease, Annual Review of Biochemistry 80, 825–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Trinidad JC, Barkan DT, Gulledge BF, Thalhammer A, Sali A, Schoepfer R, and Burlingame AL (2012) Global identification and characterization of both O-GlcNAcylation and phosphorylation at the murine synapse, Molecular & Cellular Proteomics 11, 215–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Alfaro JF, Gong CX, Monroe ME, Aldrich JT, Clauss TR, Purvine SO, Wang Z, Camp DG 2nd, Shabanowitz J., Stanley P., Hart GW., Hunt DF., Yang F., and Smith RD. (2012) Tandem mass spectrometry identifies many mouse brain O-GlcNAcylated proteins including EGF domain-specific O-GlcNAc transferase targets, Proceedings of the National Academy of Sciences of the United States of America 109, 7280–7285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Wang S, Yang F, Petyuk VA, Shukla AK, Monroe ME, Gritsenko MA, Rodland KD, Smith RD, Qian W-J, Gong C-X, and Liu T (2017) Quantitative proteomics identifies altered O-GlcNAcylation of structural, synaptic and memory-associated proteins in Alzheimer’s disease, The Journal of Pathology 243, 78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hwang H, and Rhim H (2017) Functional significance of O-GlcNAc modification in regulating neuronal properties, Pharmacological Research. [DOI] [PubMed] [Google Scholar]
- [6].Tallent MK, Varghis N, Skorobogatko Y, Hernandez-Cuebas L, Whelan K, Vocadlo DJ, and Vosseller K (2009) In vivo modulation of O-GlcNAc levels regulates hippocampal synaptic plasticity through interplay with phosphorylation, Journal of Biological Chemistry 284, 174–181. [DOI] [PubMed] [Google Scholar]
- [7].Taylor EW, Wang K, Nelson AR, Bredemann TM, Fraser KB, Clinton SM, Puckett R, Marchase RB, Chatham JC, and McMahon LL (2014) O-GlcNAcylation of AMPA receptor GluA2 is associated with a novel form of long-term depression at hippocampal synapses, Journal of Neuroscience 34, 10–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Xie S, Jin N, Gu J, Shi J, Sun J, Chu D, Zhang L, Dai C.-l., Gu J.-h., Gong C-X, Iqbal K, and Liu F (2016) O-GlcNAcylation of protein kinase A catalytic subunits enhances its activity: A mechanism linked to learning and memory deficits in Alzheimer’s disease, Aging Cell 15, 455–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lagerlof O, Slocomb JE, Hong I, Aponte Y, Blackshaw S, Hart GW, and Huganir RL (2016) The nutrient sensor OGT in PVN neurons regulates feeding, Science 351, 1293–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Lagerlöf O, Hart GW, and Huganir RL (2017) O-GlcNAc transferase regulates excitatory synapse maturity, Proceedings of the National Academy of Sciences of the United States of America 114, 1684–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wang AC, Jensen EH, Rexach JE, Vinters HV, and Hsieh-Wilson LC (2016) Loss of O-GlcNAc glycosylation in forebrain excitatory neurons induces neurodegeneration, Proceedings of the National Academy of Sciences of the United States of America 113, 15120–15125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ma X, Liu H, Li J, Wang Y, Ding Y-H, Shen H, Yang Y, Sun C, Huang M, Tu Y, Liu Y, Zhao Y, Dong M-Q, Xu P, Tang T-S, and Guo C (2017) Polη O-GlcNAcylation governs genome integrity during translesion DNA synthesis, Nature Communications 8, 1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Rexach JE, Clark PM, Mason DE, Neve RL, Peters EC, and Hsieh-Wilson LC (2012) Dynamic O-GlcNAc modification regulates CREB-mediated gene expression and memory formation, Nature Chemical Biology 8, 253–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Yuzwa SA, and Vocadlo DJ (2014) O-GlcNAc and neurodegeneration: Biochemical mechanisms and potential roles in Alzheimer’s disease and beyond, Chemical Society Reviews 43, 6839–6858. [DOI] [PubMed] [Google Scholar]
- [15].Akan I, Olivier-Van Stichelen S, Bond MR, and Hanover JA (2017) Nutrient-driven O-GlcNAc in proteostasis and neurodegeneration, Journal of Neurochemistry 144, 7–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wani WY, Ouyang X, Benavides GA, Redmann M, Cofield SS, Shacka JJ, Chatham JC, Darley-Usmar V, and Zhang J (2017) O-GlcNAc regulation of autophagy and α-synuclein homeostasis; implications for Parkinson’s disease, Molecular Brain 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Liu F, Iqbal K, Grundke-Iqbal I, Hart GW, and Gong CX (2004) O-GlcNAcylation regulates phosphorylation of tau: a mechanism involved in Alzheimer’s disease, Proceedings of the National Academy of Sciences of the United States of America 101, 10804–10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Liu Y, Liu F, Grundke-Iqbal I, Iqbal K, and Gong CX (2009) Brain glucose transporters, O-GlcNAcylation and phosphorylation of tau in diabetes and Alzheimer’s disease, Journal of Neurochemistry 111, 242–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yuzwa SA, Shan X, Jones BA, Zhao G, Woodward ML, Li X, Zhu Y, McEachern EJ, Silverman MA, Watson NV, Gong C-X, and Vocadlo DJ (2014) Pharmacological inhibition of O-GlcNAcase (OGA) prevents cognitive decline and amyloid plaque formation in bigenic tau/APP mutant mice, Molecular Neurodegeneration 9, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Yuzwa SA, Shan X, Macauley MS, Clark T, Skorobogatko Y, Vosseller K, and Vocadlo DJ (2012) Increasing O-GlcNAc slows neurodegeneration and stabilizes tau against aggregation, Nature Chemical Biology 8, 393–399. [DOI] [PubMed] [Google Scholar]
- [21].Kim C, Nam DW, Park SY, Song H, Hong HS, Boo JH, Jung ES, Kim Y, Baek JY, Kim KS, Cho JW, and Mook-Jung I (2013) O-linked β-N-acetylglucosaminidase inhibitor attenuates β-amyloid plaque and rescues memory impairment, Neurobiology of Aging 34, 275–285. [DOI] [PubMed] [Google Scholar]
- [22].Olivier-Van Stichelen S, Wang P, Comly M, Love DC, and Hanover JA (2017) Nutrient-driven O-GlcNAc cycling impacts neurodevelopmental timing and metabolism, Journal of Biological Chemistry 292, 6076–6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Vosseller K, Wells L, Lane MD, and Hart GW (2002) Elevated nucleocytoplasmic glycosylation by O-GlcNAc results in insulin resistance associated with defects in Akt activation in 3T3-L1 adipocytes, Proceedings of the National Academy of Sciences 99, 5313–5318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Yang X, Ongusaha PP, Miles PD, Havstad JC, Zhang F, So WV, Kudlow JE, Michell RH, Olefsky JM, Field SJ, and Evans RM (2008) Phosphoinositide signalling links O-GlcNAc transferase to insulin resistance, Nature 451, 964–969. [DOI] [PubMed] [Google Scholar]
- [25].Tan EP, McGreal SR, Graw S, Tessman R, Koppel SJ, Dhakal P, Zhang Z, Machacek M, Zachara NE, Koestler DC, Peterson KR, Thyfault JP, Swerdlow RH, Krishnamurthy P, DiTacchio L, Apte U, and Slawson C (2017) Sustained O-GlcNAcylation reprograms mitochondrial function to regulate energy metabolism, Journal of Biological Chemistry 292, 14940–14962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Yang X, and Qian K (2017) Protein O-GlcNAcylation: emerging mechanisms and functions, Nature Reviews. Molecular Cell Biology 18, 452–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Khidekel N, Ficarro SB, Clark PM, Bryan MC, Swaney DL, Rexach JE, Sun YE, Coon JJ, Peters EC, and Hsieh-Wilson LC (2007) Probing the dynamics of O-GlcNAc glycosylation in the brain using quantitative proteomics, Nature Chemical Biology 3, 339–348. [DOI] [PubMed] [Google Scholar]
- [28].Levine ZG, and Walker S (2016) The biochemistry of O-GlcNAc transferase: which functions make It essential in mammalian cells?, Annual Review of Biochemistry 85, 631–657. [DOI] [PubMed] [Google Scholar]
- [29].Alonso J, Schimpl M, and van Aalten DMF (2014) O-GlcNAcase: promiscuous hexosaminidase or key regulator of O-GlcNAc signaling?, Journal of Biological Chemistry 289, 34433–34439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Groves JA, Maduka AO, O’Meally RN, Cole RN, and Zachara NE (2017) Fatty acid synthase inhibits the O-GlcNAcase during oxidative stress, Journal of Biological Chemistry 292, 6493–6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, and Zoghbi HY (1999) Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2, Nature Genetics 23, 185–188. [DOI] [PubMed] [Google Scholar]
- [32].Rexach JE, Rogers CJ, Yu SH, Tao J, Sun YE, and Hsieh-Wilson LC (2010) Quantification of O-glycosylation stoichiometry and dynamics using resolvable mass tags, Nature Chemical Biology 6, 645–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Greis KD, Hayes BK, Comer FI, Kirk M, Barnes S, Lowary TL, and Hart GW (1996) Selective detection and site-analysis of O-GlcNAc-modified glycopeptides by β-Elimination and tandem electrospray mass spectrometry, Analytical Biochemistry 234, 38–49. [DOI] [PubMed] [Google Scholar]
- [34].Wells L, Vosseller K, and Hart GW (2003) A role for N -acetylglucosamine as a nutrient sensor and mediator of insulin resistance, Cellular and Molecular Life Sciences 60, 222–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Whelan SA, and Hart GW (2006) Identification of O‐GlcNAc sites on proteins, Methods in Enzymology 415, 113–133. [DOI] [PubMed] [Google Scholar]
- [36].Thompson JW, Griffin ME, and Hsieh-Wilson LC (2017) Methods for the detection, study, and dynamic profiling of O-GlcNAc glycosylation, Methods in Enzymology 598, 101–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Wells L, Vosseller K, Cole RN, Cronshaw JM, Matunis MJ, and Hart GW (2002) Mapping sites of O-GlcNAc modification using affinity tags for serine and threonine post-translational modifications, Molecular & Cellular Proteomics 1, 791–804. [DOI] [PubMed] [Google Scholar]
- [38].Khidekel N, Ficarro SB, Peters EC, and Hsieh-Wilson LC (2004) Exploring the O-GlcNAc proteome: direct identification of O-GlcNAc-modified proteins from the brain, Proceedings of the National Academy of Sciences of the United States of America 101, 13132–13137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Dias WB, Cheung WD, Wang Z, and Hart GW (2009) Regulation of calcium/calmodulin-dependent kinase IV by O-GlcNAc modification, J Biol Chem 284, 21327–21337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Khidekel N, Arndt S, Lamarre-Vincent N, Lippert A, Poulin-Kerstien KG, Ramakrishnan B, Qasba PK, and Hsieh-Wilson LC (2003) A chemoenzymatic approach toward the rapid and sensitive detection of O-GlcNAc posttranslational modifications, Journal of the American Chemical Society 125, 16162–16163. [DOI] [PubMed] [Google Scholar]
- [41].Tai HC, Khidekel N, Ficarro SB, Peters EC, and Hsieh-Wilson LC (2004) Parallel identification of O-GlcNAc-modified proteins from cell lysates, Journal of the American Chemical Society 126, 10500–10501. [DOI] [PubMed] [Google Scholar]
- [42].Clark PM, Dweck JF, Mason DE, Hart CR, Buck SB, Peters EC, Agnew BJ, and Hsieh-Wilson LC (2008) Direct in-gel fluorescence detection and cellular imaging of O-GlcNAc-modified proteins, Journal of the American Chemical Society 130, 11576–11577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Clark PM, Rexach JE, and Hsieh-Wilson LC (2013) Visualization of O-GlcNAc glycosylation stoichiometry and dynamics using resolvable poly(ethylene glycol) mass tags, Current Protocols in Chemical Biology 5, 281–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Ramakrishnan B, and Qasba PK (2002) Structure-based design of β1,4-Galactosyltransferase I (β4Gal-T1) with equally efficient N-acetylgalactosaminyltransferase activity, Journal of Biological Chemistry 277, 20833–20839. [DOI] [PubMed] [Google Scholar]
- [45].Wang Z, Pandey A, and Hart GW (2007) Dynamic interplay between O-linked N-acetylglucosaminylation and glycogen synthase kinase-3-dependent phosphorylation, Mol Cell Proteomics 6, 1365–1379. [DOI] [PubMed] [Google Scholar]
- [46].Wang Z, Park K, Comer F, Hsieh-Wilson LC, Saudek CD, and Hart GW (2009) Site-specific GlcNAcylation of human erythrocyte proteins: potential biomarker(s) for diabetes, Diabetes 58, 309–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Vosseller K, Hansen KC, Chalkley RJ, Trinidad JC, Wells L, Hart GW, and Burlingame AL (2005) Quantitative analysis of both protein expression and serine/threonine post-translational modifications through stable isotope labeling with dithiothreitol, Proteomics 5, 388–398. [DOI] [PubMed] [Google Scholar]
- [48].Lund PJ, Elias JE, and Davis MM (2016) Global analysis of O-GlcNAc glycoproteins in activated human T cells, The Journal of Immunology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Giese KP, and Mizuno K (2013) The roles of protein kinases in learning and memory, Learn Mem 20, 540–552. [DOI] [PubMed] [Google Scholar]
- [50].Kang H, Sun LD, Atkins CM, Soderling TR, Wilson MA, and Tonegawa S (2001) An important role of neural activity-dependent CaMKIV signaling in the consolidation of long-term memory, Cell 106, 771–783. [DOI] [PubMed] [Google Scholar]
- [51].Fukushima H, Maeda R, Suzuki R, Suzuki A, Nomoto M, Toyoda H, Wu LJ, Xu H, Zhao MG, Ueda K, Kitamoto A, Mamiya N, Yoshida T, Homma S, Masushige S, Zhuo M, and Kida S (2008) Upregulation of calcium/calmodulin-dependent protein kinase IV improves memory formation and rescues memory loss with aging, Journal of Neuroscience 28, 9910–9919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Soderling TR (1999) The Ca2+–calmodulin-dependent protein kinase cascade, Trends in Biochemical Sciences 24, 232–236. [DOI] [PubMed] [Google Scholar]
- [53].Whelan SA, and Hart GW (2003) Proteomic approaches to analyze the dynamic relationships between nucleocytoplasmic protein glycosylation and phosphorylation, Circ Res 93, 1047–1058. [DOI] [PubMed] [Google Scholar]
- [54].Chalkley RJ, Thalhammer A, Schoepfer R, and Burlingame AL (2009) Identification of protein O-GlcNAcylation sites using electron transfer dissociation mass spectrometry on native peptides, Proceedings of the National Academy of Sciences of the United States of America 106, 8894–8899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Pekkurnaz G, Trinidad JC, Wang X, Kong D, and Schwarz TL (2014) Glucose regulates mitochondrial motility via Milton modification by O-GlcNAc transferase, Cell 158, 54–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Griffin ME, Jensen EH, Mason DE, Jenkins CL, Stone SE, Peters EC, and Hsieh-Wilson LC (2016) Comprehensive mapping of O-GlcNAc modification sites using a chemically cleavable tag, Molecular BioSystems 12, 1756–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Syka JEP, Coon JJ, Schroeder MJ, Shabanowitz J, and Hunt DF (2004) Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry, Proceedings of the National Academy of Sciences of the United States of America 101, 9528–9533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Swaney DL, McAlister GC, and Coon JJ (2008) Decision tree–driven tandem mass spectrometry for shotgun proteomics, Nature Methods 5, 959–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Xie L-Q, Shen C-P, Liu M-B, Chen Z-D, Du R-Y, Yan G-Q, Lu H-J, and Yang P-Y (2012) Improved proteomic analysis pipeline for LC-ETD-MS/MS using charge enhancing methods, Molecular BioSystems 8, 2692. [DOI] [PubMed] [Google Scholar]
- [60].Wang Z, Udeshi ND, O’Malley M, Shabanowitz J, Hunt DF, and Hart GW (2010) Enrichment and site mapping of O-linked N-acetylglucosamine by a combination of chemical/enzymatic tagging, photochemical cleavage, and electron transfer dissociation mass spectrometry, Molecular & Cellular Proteomics 9, 153–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Oddo S, Caccamo A, Shepherd JD, Murphy MP, Golde TE, Kayed R, Metherate R, Mattson MP, Akbari Y, and LaFerla FM (2003) Triple-transgenic model of Alzheimer’s disease with plaques and tangles, Neuron 39, 409–421. [DOI] [PubMed] [Google Scholar]
- [62].Feng W, and Zhang M (2009) Organization and dynamics of PDZ-domain-related supramodules in the postsynaptic density, Nature Reviews. Neuroscience 10, 87–99. [DOI] [PubMed] [Google Scholar]
- [63].Marotta NP, Lin YH, Lewis YE, Ambroso MR, Zaro BW, Roth MT, Arnold DB, Langen R, and Pratt MR (2015) O-GlcNAc modification blocks the aggregation and toxicity of the protein α-synuclein associated with Parkinson’s disease, Nature Chemistry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Levine PM, De Leon CA, Galesic A, Balana A, Marotta NP, Lewis YE, and Pratt MR (2017) O-GlcNAc modification inhibits the calpain-mediated cleavage of α-synuclein, Bioorganic & Medicinal Chemistry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Roquemore EP, Dell A, Morris HR, Panico M, Reason AJ, Savoy LA, Wistow GJ, Zigler JS, Earles BJ, and Hart GW (1992) Vertebrate lens alpha-crystallins are modified by O-linked N-acetylglucosamine, Journal of Biological Chemistry 267, 555–563. [PubMed] [Google Scholar]
- [66].Dong DLY, Xu Z-S, Hart GW, and Cleveland DW (1996) Cytoplasmic O-GlcNAc modification of the head domain and the KSP repeat motif of the neurofilament protein neurofilament-H, Journal of Biological Chemistry 271, 20845–20852. [DOI] [PubMed] [Google Scholar]
- [67].Arnold CS, Johnson GVW, Cole RN, Dong DLY, Lee M, and Hart GW (1996) The microtubule-associated protein tau Is extensively modified with O-linked N-acetylglucosamine, Journal of Biological Chemistry 271, 28741–28744. [DOI] [PubMed] [Google Scholar]
- [68].Yi W, Clark PM, Mason DE, Keenan MC, Hill C, Goddard WA 3rd, Peters EC, Driggers EM, and Hsieh-Wilson LC (2012) Phosphofructokinase 1 glycosylation regulates cell growth and metabolism, Science 337, 975–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Darabedian N, Thompson JW, Chuh KN, Hsieh-Wilson LC, and Pratt MR (2018) Optimization of chemoenzymatic mass-tagging by strain-promoted cycloaddition (SPAAC) for the determination of O-GlcNAc stoichiometry by Western blotting., Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Ma J, and Hart GW (2016) Mass Spectrometry-Based Quantitative O-GlcNAcomic Analysis, In Quantitative Proteomics by Mass Spectrometry (Sechi S., Ed.), pp 91–103, Humana Press, New York, NY. [DOI] [PubMed] [Google Scholar]
- [71].Woo CM, Iavarone AT, Spiciarich DR, Palaniappan KK, and Bertozzi CR (2015) Isotope-targeted glycoproteomics (IsoTaG): a mass-independent platform for intact N- and O-glycopeptide discovery and analysis, Nature Methods 12, 561–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Woo CM, Felix A, Byrd WE, Zuegel DK, Ishihara M, Azadi P, Iavarone AT, Pitteri SJ, and Bertozzi CR (2017) Development of IsoTaG, a chemical glycoproteomics technique for profiling intact N- and O-glycopeptides from whole cell proteomes, Journal of Proteome Research. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Woo CM, Lund PJ, Huang AC, Davis MM, Bertozzi CR, and Pitteri S (2018) Mapping and quantification of over 2,000 O-linked glycopeptides in activated human T cells with isotope-targeted glycoproteomics (IsoTaG), Molecular & Cellular Proteomics 17, 764–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Palaniappan KK, and Bertozzi CR (2016) Chemical glycoproteomics, Chemical Reviews 116, 14277–14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Boyce M, Carrico IS, Ganguli AS, Yu SH, Hangauer MJ, Hubbard SC, Kohler JJ, and Bertozzi CR (2011) Metabolic cross-talk allows labeling of O-linked-N-acetylglucosamine-modified proteins via the N-acetylgalactosamine salvage pathway, Proceedings of the National Academy of Sciences of the United States of America 108, 3141–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Dustin ML (2012) Signaling at neuro/immune synapses, Journal of Clinical Investigation 122, 1149–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Rex-Mathes M, Werner S, Strutas D, Griffith LS, Viebahn C, Thelen K, and Schmitz B (2001) O-GlcNAc expression in developing and ageing mouse brain, Biochimie 83, 583–590. [DOI] [PubMed] [Google Scholar]
- [78].Liu Y, Li X, Yu Y, Shi J, Liang Z, Run X, Li Y, Dai C. l., Grundke-Iqbal I, Iqbal K, Liu F, and Gong C-X (2012) Developmental regulation of protein O-GlcNAcylation, O-GlcNAc transferase, and O-GlcNAcase in mammalian brain, PLoS ONE 7, e43724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Fülöp N, Feng W, Xing D, He K, Nőt LG, Brocks CA, Marchase RB, Miller AP, and Chatham JC (2008) Aging leads to increased levels of protein O-linked N-acetylglucosamine in heart, aorta, brain and skeletal muscle in Brown-Norway rats, Biogerontology 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Yang YR, Song M, Lee H, Jeon Y, Choi E-J, Jang H-J, Moon HY, Byun H-Y, Kim E-K, Kim DH, Lee MN, Koh A, Ghim J, Choi JH, Lee-Kwon W, Kim KT, Ryu SH, and Suh P-G (2012) O-GlcNAcase is essential for embryonic development and maintenance of genomic stability, Aging Cell 11, 439–448. [DOI] [PubMed] [Google Scholar]


