Abstract
Venomous organisms used in research were historically chosen based on size and availability. This opportunity-driven strategy created a species bias in which snakes, scorpions, and spiders became the primary subjects of venom research. Increasing technological advancements have enabled interdisciplinary studies using genomics, transcriptomics, and proteomics to expand venom investigation to animals that produce small amounts of venom or lack traditional venom producing organs. One group of non-traditional venomous organisms that have benefitted from the rise of -omic technologies is the Conoideans. The Conoidean superfamily of venomous marine snails includes, the Terebridae, Turridae (s.l), and Conidae. Conoidea venom is used for both predation and defense, and therefore under strong selection pressures. The need for conoidean venom peptides to be potent and specific to their molecular targets has made them important tools for investigating cellular physiology and bioactive compounds that are beneficial to improving human health. A convincing case for the potential of Conoidean venom is made with the first commercially available conoidean venom peptide drug Ziconotide (Prialt®), an analgesic derived from Conus magus venom that is used to treat chronic pain in HIV and cancer patients. Investigation of conoidean venom using -omics technology provides significant insights into predator-driven diversification in biodiversity and identifies novel compounds for manipulating cellular communication, especially as it pertains to disease and disorders.
Introduction
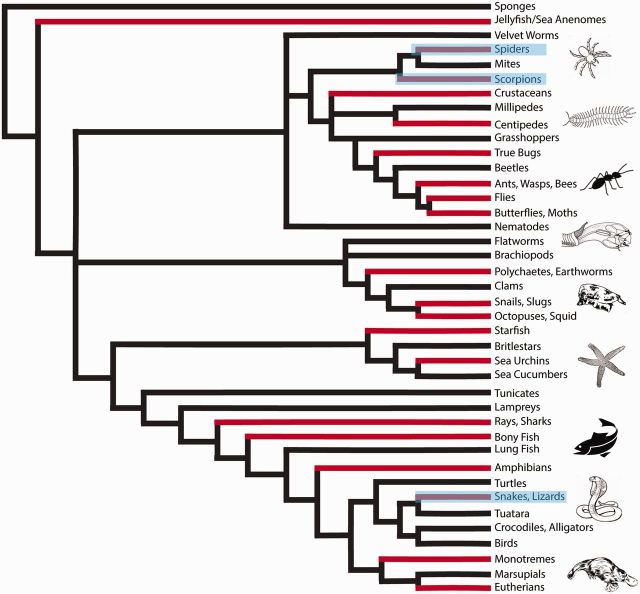
Venom is defined as any exogenous substance that is used to elicit an adverse effect in its target, and as a result a wide range of organisms from notorious snakes to lesser known leeches and bees are considered venomous (Fig. 1; Escoubas and King 2009; Casewell et al. 2013; King 2015; Petras et al. 2015). Historically, organisms used in venom research were chosen opportunistically, based on size and ease of collection, which largely focused on vertebrates, specifically snakes. Two genera of snakes account for almost 40% of all published venom toxin sequences in elapid snake venom research (Fry et al. 2008). Remarkably, one easy to collect genus (Naja) has been used to identify 40% of all three-finger snake venom toxins (3FTxs) sequenced. Only three studies have used harder to milk and less studied, non-front-fanged snakes to investigate 3FTx bioactivity (Fry et al. 2003; Pawlak et al. 2006, 2009; King 2015). The venom research strategy of size and accessibility can neglect the ecology, morphology, or evolutionary relatedness between organisms, resulting in a diversity of venomous animals, such as invertebrates, being effectively ignored (Modica and Holford 2010; Puillandre and Holford 2010; von Reumont et al. 2014b).
Fig. 1.
Biodiversity of venomous taxa. Phylogenetic reconstruction of the tree of life highlighting venomous organisms. Grey bars represent the clades that include venomous organisms. Highlighted clades represent the traditionally studied venomous taxa (scorpions, spiders, snakes, and lizards).
Invertebrates are underrepresented in venom research. Spiders, which are the most diverse group of venomous animals, with about 45,000 species, make up less than 5% of all venom research studies (Nentwig 2013). Analogous to the within taxa bias seen in snakes, of the 17,000 species of scorpions described, only ∼50 species have had their venom investigated (King 2015). It can be argued that the venom research bias existed largely due to lack of technological methods for effectively collecting and characterizing small quantities of venom. The deficiency of invertebrates has led to a dearth of information that has hindered venom research. However, recent technological advancements in the field of molecular biology and proteomics has increased the representation of marine cone snails, sea anemones, bees, and ants in venom studies (Norton and Olivera 2006; Moran et al. 2008; Barlow et al. 2009; Casewell et al. 2013; Sanggaard et al. 2014; Zhang et al. 2015). Extensive research on a broad range of organisms is imperative in order to effectively derive and test hypotheses about venom as it relates to species diversification, predator–prey interactions, and to describe the immense biodiversity of animals found on Earth (Fig. 1).
Rise of -omics
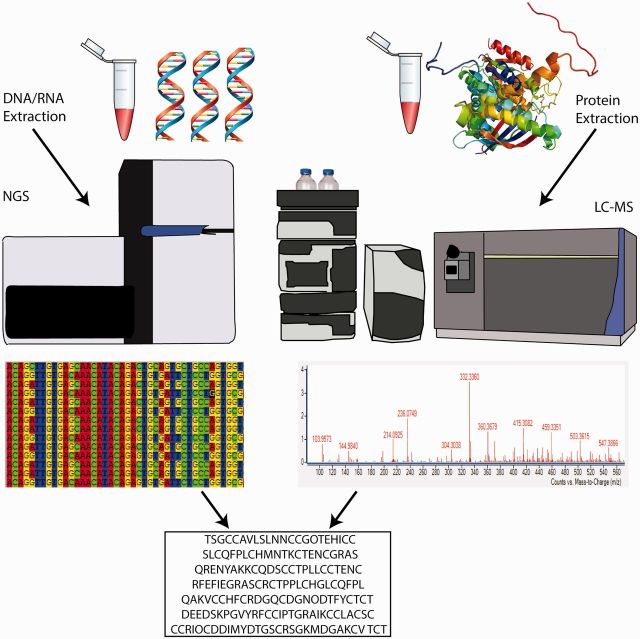
Early research on venom relied heavily on identifying proteins using Edman degradation and mass spectrometry (MS; Perkins et al. 1993a, 1993b). In conjunction with fractionation, MS allowed for the separation and identification of individual venom components. Development of soft ionization methods in the late 1980s, such as electrospray ionization (ESI; Fenn 2003), and matrix-assisted laser desorption (Karas and Hillenkamp 1988; Tanaka 2003) have revolutionized biological research. In particular, the ability to identify proteins directly from MS data is a powerful capability that soon demonstrated to be crucial for analyzing venoms. Quickly thereafter, research groups started to implement MS techniques to characterize snake venom (Perkins et al. 1993a, 1993b). MS approaches also enable the identification of isoforms of venom peptides, slight variations in sequences, and post translational modifications (Craig et al. 1999; Escoubas et al. 2008; Safavi-Hemami et al. 2014; Petras et al. 2015). Recent advancements in MS protocols have produced what are referred to as top-down methods, in which whole intact venom components can be identified (Breuker et al. 2008; Ueberheide et al. 2009; Anand et al. 2014; Sunagar et al. 2016). Although groundbreaking, in some organisms a proteome-only MS approach can be problematic. MS requires extraction of venom, which is impractical for organisms that do not readily store venom for delivery and other organisms that have hard-to-access venom delivery systems that prevent stimulation or extraction (von Reumont et al. 2014a). Additionally, MS methods for determining primary venom peptide sequences are largely dependent on downstream data analyses of source databases, such as Mascot or Genbank (Perkins et al. 1999; Bhatia et al. 2012). For model organisms with a rich complement of sequence databases, such as humans, mice, or drosophila, this is not an issue. In the case of non-model organisms, the application of MS methods for primary sequencing is severely limited by the database used. Non-model systems generally require de novo sequence assembly and source databases that are either missing or deficient. As a result, an integrated strategy, termed venomics (Calvete et al. 2007; Calvete 2014; Eichberg et al. 2015), in which MS proteomics is combined with next generation transcriptomic or genomic sequencing and bioinformatic methods is necessary to validate characterization of de novo venom peptides found in non-model organisms and to paint the full canvas of venom evolution and variation (Fig. 2; Fry et al. 2013; Sunagar et al. 2016). Using the multi-omic integrated venomic strategy, venom research has become more accessible to smaller, harder to collect, and understudied venomous taxa. The integrated venomic strategy has also broadened the scientific community engaged in venom research from traditional chemists and pharmacologists looking for bioactive compounds for drug discovery and development, to evolutionary biologists looking for anatomical and molecular characters to understand venom evolution through various taxa over time (Duda and Palumbi 1999; Moran et al. 2008; Favreau and Stöcklin 2009; Elmer et al. 2010; Koh and Kini 2012; Otvos et al. 2013; Gorson et al. 2015; Jouiaei et al. 2015; Zhang et al. 2015).
Fig. 2.
Venomics: an integrated NGS and proteomic strategy. An integrated multi -omics approach using genomic, transcriptomic, bioinformatic, and proteomic protocols to identify venom proteins and peptides. Application of a combined -omics strategy validates de novo venom peptide/protein identification and provides robust data to test hypotheses related to venom evolution and ecology. The sequences shown at the bottom are an example of a validated peptide database obtained from NGS and proteomics.
The honey bee, Apis mellifera, was the first venomous organism to have a fully sequenced genome using Sanger sequencing (Weinstock et al. 2006). Since then, the development of next generation sequencing (NGS) high throughput techniques has allowed rapid sequencing of other venomous organisms. The genomes of tarantula Acanthoscurria geniculate (Sanggaard et al. 2014), scorpion Mesobuthus martensii (Cao et al. 2013), velvet spider Stegodyphus mimosarum (Sanggaard et al. 2014), fire ant Soenopsis invicta (Wurm et al. 2011), and king cobra Ophiophagus hannah (Vonk et al. 2013) have all been sequenced using NGS technologies. With multiple platforms available, such as Illumina (Illumina, Inc., San Diego, California), 454 (Roche Applied Science, Penzberg, Germany), SOLiD (ThermoFisher Scientific, Waltham, Massachusetts), and Ion Torrent (ThermoFisher Scientific, Waltham, Massachusetts), genome sequencing of venomous organisms is becoming both accessible and affordable. However, genomics alone does not provide enough information for determining the exact mode and tempo of gene expression and does not give significant insight into differential gene expression within various tissue types (Sunagar et al. 2016).
While genomics is the study of the complete DNA composition of an organism, venom gland transcriptomics is the sequencing of mRNA specific to the venom gland or secretory tissue of a venomous organism and therefore a glimpse at the specific venom cocktail being used at the time by the animal (Durban et al. 2011; Dutertre et al. 2014; Gorson et al. 2015; Sunagar et al. 2016). Both transcriptomics and genomics enable the identification of certain domains of a venom protein, such as the signal and pre-pro regions that are rarely identified on the proteomic level as they are cleaved off after translation (Duda and Palumbi 1999; Espiritu et al. 2001; Kaas et al. 2010; Robinson and Norton 2014; Sunagar et al. 2016). Employing an integrated venomic strategy has enabled researchers to resolve previously unanswerable questions, such as identifying a correlation between varying venom compositions and differences in ecological and environmental factors. Several studies employing a combined genomics and transcriptomics approach have looked at venom variation between different developmental stages in snakes (Durban et al. 2011; Durban et al. 2013; Gibbs et al. 2013). Specifically, proteomics and transcriptomics were used to show venom variation in various populations of the Southern Pacific Rattlesnake, Crotalus oreganus helleri in the United States (Sunagar et al. 2014). Lectin β-chains, which are generally undergoing positive selection, were found to be evolving under negative selection in the C. oreganus helleri rattlesnake population found on Catalina Island (in the Pacific Ocean; Sunagar et al. 2014). The integrated venomics approach used in this study revealed that there can be different evolutionary selection pressures acting on different venom classes depending on the population site. In another study that used an integrative venomics approach, it was found that there were significant differences in the mature peptides being produced in different samples of venom from Conus consors (Biass et al. 2015). Proteomics and transcriptomics were used to analyze C. consors venom at three different stages: venom milked from the snail, venom extracted from the venom gland, and venom expressed in the transcriptome, effectively tracing the venom production and delivery process from the venom glad tissue to the point of venom envenomation of the prey. The surprising result was that the cocktail of venom peptides identified in the transcriptome, in the venom produced within the venom gland, and in the venom injected into the prey were not heavily correlated. Each venom compartment was distinctive in terms of peptide and protein content. This study emphasizes the complexity of the venom mechanism of Conodiean snails and indicates what is being secreted in the venom is not necessarily the same as what is being produced in the gland (Biass et al. 2015).
Advances in -omic technologies have increased the breadth of research being done on all organisms and have advanced research of non-model organisms.
Characterizing Conoidean Venom Evolution and Variation
The technological -omics advancements removed the barrier requiring large amounts of crude venom extracts and smaller-sized taxa such as centipedes, certain sea anemones, ants, and small scorpions have become the focal point of an ever increasing number of venom studies (Putnam et al. 2007; Moran et al. 2008; Calvete et al. 2009; Cao et al. 2013; Sanggaard et al. 2014; Xu et al. 2014). One non-model organism that has received a lot of attention using venomic technologies is the venomous marine snails in the Conoidean superfamily.
Conoidean snails are slow moving predators and therefore rely heavily on the efficacy of their venom (Azam et al. 2005; Yao et al. 2008; Kendel et al. 2013; King 2015). The dependence on venom for prey capture has led conoidean venom peptides to achieve incredible molecular specificity (Olivera 2002; Olivera et al. 2014). Conoideans subdue their prey using a venom apparatus made up of a proboscis, radular tooth, a radular sac, venom gland, and venom bulb (Fig. 3(A); Taylor 1990; Kantor et al. 2000; Modica and Holford 2010; Kantor and Puillandre 2012). Cone snails (Conus) are the most studied in the Conoidea (Puillandre et al. 2014; Puillandre et al. 2015); however, Conus comprises only ∼5% of the biodiverse group of venomous marine snails (Olivera et al. 1999; Holford et al. 2009; King 2015). Other non-Conus Conoideans, such as the Turridae (s.l.) family, which has more recently been divided into seven family groups (Tucker and Tenorio 2009; Bouchet et al. 2011), and the Terebridae family, also produce venom (Heralde et al. 2008; Aguilar et al. 2009; Gonzales and Saloma 2014; Gorson et al. 2015; Moon et al. 2016). Cone snails and terebrids dwell in shallow-water tropical marine habitats, while the majority of turrids can be found at greater depths (>200m; Taylor 1977). Terebrids and turrids (some less than 3 mm in length) have incredibly small venom ducts, producing limited amounts of venom, which initially inhibited their characterization. Using an integrated venomics strategy, venom research of terebrids and turrids has become more feasible (Castelin et al. 2012; Kendel et al. 2013; Gonzales and Saloma 2014; Gorson et al. 2015; Moon et al. 2016).
Fig. 3.
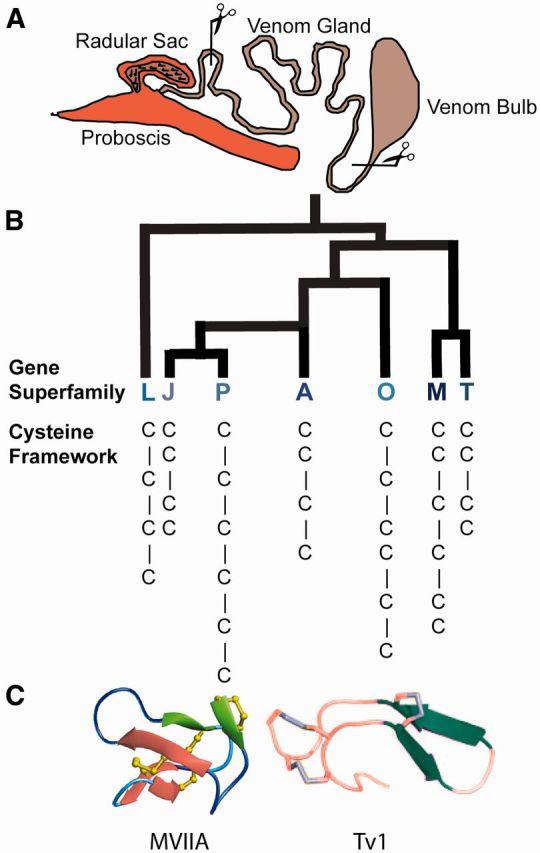
Conoidean venom characterization. (A) A generic representation of the Conoidean venom apparatus, which includes: a venom bulb that is contracted to push the venom through the venom gland, where the venom is being produced, a radular sac that contains hollowed teeth (harpoons) that are used to inject venom into the prey, and a proboscis that extends several times beyond the snails body size to deliver venom-filled radula to a prey target. Scissors shown represent the dissection of the venom duct for downstream analysis by transcriptomic, genomic or proteomic methods. (B) Identification of Conus venom peptide superfamilies and cysteine frameworks. (C) Conoidean venom peptides selected for bioactivity characterization. MVIIA is a peptide from Conus magus venom that produced the ziconotide (Prialt®) drug that is commercially available. MVIIA is in the O1 conotoxin gene superfamily and has a VI/VII cysteine framework. Tv1 is a peptide from Terebra variagata that has a cysteine pattern similar to the M-superfamily in cone snails and has a III cysteine framework, but has a peptide fold of antiparallel beta hairpins that is unique to known venom peptides. Tv1 and MVIIA are very distinct peptides illustrating the disparate complexity of Conoidean venom peptides.
Conoidean venoms are a complex mixture of small molecules, peptides and proteins (Norton and Olivera 2006; Gonzales and Saloma 2014; Gorson et al. 2015; Neves et al. 2015). Each Conoidean venom consists of >100 different peptides which contain a signal sequence, followed by a pro region, and a mature peptide at the C-terminus (Olivera et al. 1999; Lavergne et al. 2015). As cone snails are well studied, 3000 different peptides (conotoxins) have been identified from Conus venoms since the 1970s (Conoserver.org). The majority of conotoxins have been classified into venom gene superfamilies by examining the sequence identity of the signal sequence (Jacob and McDougal 2010; Robinson et al. 2014; Robinson and Norton 2014). A similar process is being used to characterize turrid and terebrid venom peptide superfamilies (Fig. 3(B); Heralde et al. 2008; Gonzales and Saloma 2014; Gorson et al. 2015). Different venom peptide superfamilies generally have distinct physiological targets and high specificity for those targets (Olivera et al. 1999). While there are similarities between the venoms of Conus, inter- and intraspecific variation exists, such as differences in the proportions of cysteine frameworks or venom gene superfamilies (Duda and Palumbi 1999; Olivera et al. 1999; Jakubowski et al. 2005; Romeo et al. 2008; Abdel-Rahman et al. 2011). Due to the high rates of non-synonymous mutations and the early divergence of Conoidean families, the venoms of terebrids and turrids (s.l.) vary significantly from the venoms of cone snails (Powell 1966; Duda and Palumbi 1999; Puillandre and Holford 2010; Gorson et al. 2015). The disparity in Conus and terebrid venom was recently revealed by looking at the variation in venom peptide superfamilies between Triplostephanus anilis and Terebra subulata (Gorson et al. 2015). Fourteen terebrid venom gene superfamilies were identified in the two terebrid species (TA, TB, TC, TD, TE, TF, TG, TH, TI, TJ, TK, TL, and TM). Of the fourteen terebrid superfamilies described, only one, TM, is homologous to a superfamily found in Conus marmoreus (H superfamily;Robinson and Norton 2014). The divergence of Conus and terebrid venom gene superfamilies suggests that terebrid venom peptides will have different structural features and physiological targets from Conus, thereby increasing the pool of bioactive compounds that can be explored for discovery of novel therapeutic drugs. Conoidean venoms are exceedingly effective candidates for drug discovery as they are: (1) rapid acting, (2) highly selective, and (3) very potent (Fig. 3(C)).
Potential of Conoidean Venom to Increase Drug Discovery and Development
Many bioactive peptides have evolved as a means of predation or defense, especially in venomous animals (Olivera et al. 1985; Gray et al. 1988; Olivera et al. 1999; Dutertre et al. 2014). The wide variety of biologically active venom peptides are a promising resource for drug discovery (Lewis and Garcia 2003; Favreau and Stöcklin 2009; Twede et al. 2009; Koh and Kini 2012; King 2015; Ortiz et al. 2015). The constant selective pressures acting on venom, due to the effects of the predator–prey arms race (Van Valen 1973; Dawkins and Krebs 1979; Casewell et al. 2013; Holding et al. 2016), enabled venom peptides to develop features to increase stability and prey molecular target affinity. Venom peptides tend to interfere with transmissions of ions in and out of cells, suggesting they would be effective tools for manipulating ion channel driven cell disorders such as pain or cancer (Miljanich 2004; Vetter and Lewis 2012; Lang et al. 2014). These properties make venom peptides more appealing than artificially or chemically conceived peptide-like compounds for which bioactivity is not guaranteed (Uhlig et al. 2014).
The past two and a half decades have seen an increase in the number of projects that are taking advantage of the Earth’s amazingly biodiverse group of venomous organisms to develop novel drugs, to create tools for diagnosing human diseases, and to create probes to help advance the study of molecular receptors and physiological pathways (Nisani et al. 2007; King 2011; Diochot et al. 2012; Casewell et al. 2013). As reptiles were the most accessible venomous organisms for quite some time, the majority of approved venom drugs were discovered from snake venom. Specifically, snake venom proteins targeting thrombin, integrin, and fibrinogen receptors were discovered (King 2011; Koh and Kini 2012). Captopril®, an angiotensin-converting enzyme (ACE) inhibitor synthesized to mimic a venom peptide from Brazilian lancehead snakes, is a breakthrough drug that validates venom-based drug discovery research (Cushman and Ondetti 1991). The venomic strategy has made it more affordable and practical to examine non-model venomous organisms for peptide or protein components that can lead to new therapeutics. Investigating more venomous organisms will greatly increase the amount of compounds available for drug discovery and development (Puillandre and Holford 2010; Casewell et al. 2013; King 2015).
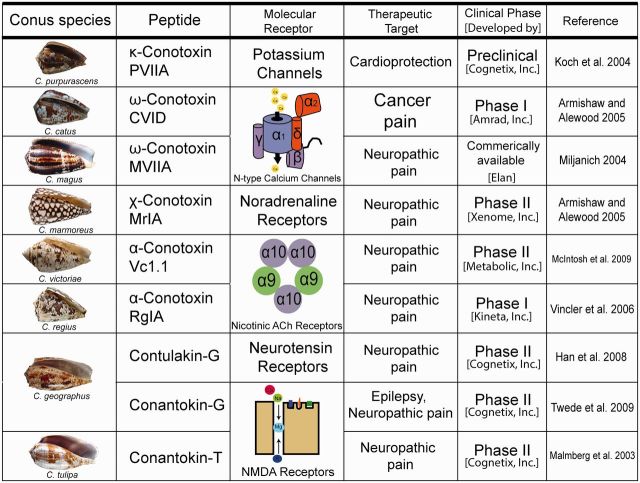
The >1 million estimated venom peptides expressed in conoidean venom are an immense resource for discovering novel compounds for therapeutic drug development. The majority of conoidean venom peptides are disulfide-rich molecules that have been shown to manipulate voltage and ligand gated potassium (K+), calcium (Ca2+), and sodium (Na+) channels, as well as nicotinic acetylcholine receptors, and noradrenaline transporters (Olivera 2002; Terlau and Olivera 2004; Becker and Terlau 2008; Fig. 4). As mentioned previously, ion channels and receptors are the molecular targets for cancer, pain, and other debilitating human disorders (Wood et al. 2004; Veiseh et al. 2007; Gkika and Prevarskaya 2011; Dave and Lahiry 2012; Lang et al. 2014). Ziconotide (Prialt®), a peptide from the venom of Conus magus, was approved for commercial use by the Food and Drug Administration in 2004 and is the first non-opiod analgesic (Olivera 2000; Miljanich 2004; Schmidtko et al. 2010). Similar to MVIIA, CVID, MrIA, Vc1.1, RgIA, Contulakin-G, and Conantokin-T are peptides synthesized from the natural secretions of Conus that are currently undergoing clinical trials to determine of their potential as pain therapeutics (Fig. 4; Malmberg et al. 2003; Miljanich 2004; Armishaw and Alewood 2005; Vincler et al. 2006; Han et al. 2008; McIntosh et al. 2009). Although most Conus peptides in pharmaceutical development are being used as analgesics, PVIIA shows promise as a therapy for myocardial infarction, and Conantokin-G for epilepsy (Fig. 4; Koch et al. 2004; Armishaw and Alewood 2005; Twede et al. 2009).
Fig. 4.
Snail venom peptides to drugs. Conoidean venom peptides under drug development.
Conclusion
The progress of -omics technologies triggered a domino effect in venom research. An integrated venomics strategy has enabled a broader range of venomous organisms, many of which are non-model organisms, difficult to acquire, and contain limited amounts of venom, to be examined and ultimately contribute to the understanding of venom evolution in biodiversity. Without the vast technological molecular improvements in the last decade, studies would most likely still revolve around snakes, spiders, and scorpions. The increasing amount of venom peptides identified from Conoideans that are now in clinical trials, demonstrates the importance of expanding the diversity of venomous species examined (Fig. 4). As -omics technology continues to improve, it will be easier and cheaper to add more species to the pool of venomous organisms under investigation, enabling researchers to resolve questions about venom convergence across the animal kingdom and increase the quantity of peptides available for drug discovery and development for the benefit of human health.
Acknowledgments
The authors thank all current and past members of the Holford group, especially the undergraduate students who have contributed to the research highlighted in this review: Aida Verdes, Stephen Jannetti, Prachi Anand, Danny Simpson, Abba Leffler, Patrick Kelly Peter Filipenko, Marouf Hossain, Yasmine Karma, Manjeet Kaur, Samer Khawaja, Emily Lau, Michael Lyudmer, Sujoy Manir, John Moon, Elena Pires, Carolina Santamaria, Chhime Sherpa, Hye Shin, Nicolette Somogyi, Alex Uvaydov, Laurel Yee, Henry Yelkin, and Musunri Michelle Yun.
Funding
M.H. acknowledges funding from Camille and Henry Dreyfus Teacher-Scholar Award, from Weill Cornell CTSC Award 5 UL1 TR000457-09 and from The National Science Foundation (NSF) awards CHE- 1247550 and CHE-1228921. J.G. support was provided by The Graduate Center of the City University of New York Science Scholarship.
References
- Abdel-Rahman MA, Abdel-Nabi IM, El-Naggar MS, Abbas OA, Strong PN. 2011. Intraspecific variation in the venom of the vermivorous cone snail Conus vexillum. Comp Biochem Physiol C Toxicol Pharmacol 154:318–25. [DOI] [PubMed] [Google Scholar]
- Aguilar MB, de la Rosa RA, Falcón A, Olivera BM, Heimer de la Cotera EP. 2009. Peptide pal9a from the venom of the turrid snail Polystira albida from the Gulf of Mexico: purification, characterization, and comparison with P-conotoxin-like (framework IX) conoidean peptides. Peptides 30:467–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand P, Grigoryan A, Bhuiyan MH, Ueberheide B, Russell V, Quinoñez J, Moy P, Chait BT, Poget SF, Holford M. 2014. Sample limited characterization of a novel disulfide-rich venom peptide toxin from terebrid marine snail Terebra variegata. PLoS One 9:e94122.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armishaw CJ, Alewood PF. 2005. Conotoxins as research tools and drug leads. Curr Prote Pept Sci 6:221–40. [DOI] [PubMed] [Google Scholar]
- Azam L, Dowell C, Watkins M, Stitzel JA, Olivera BM, McIntosh JM. 2005. Alpha-conotoxin BuIA, a novel peptide from Conus bullatus, distinguishes among neuronal nicotinic acetylcholine receptors. J Biol Chem 280:80–7. [DOI] [PubMed] [Google Scholar]
- Barlow A, Pook CE, Harrison RA, Wüster W. 2009. Coevolution of diet and prey-specific venom activity supports the role of selection in snake venom evolution. Proc Biol Sci 276:2443–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker S, Terlau H. 2008. Toxins from cone snails: properties, applications and biotechnological production. Appl Microbiol Biotechnol 79:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia S, Kil YJ, Ueberheide B, Chait BT, Tayo L, Cruz L, Lu B, Yates JR, Bern M. 2012. Constrained de novo sequencing of conotoxins. J Prot Res 11:4191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biass D, Violette A, Hulo N, Lisacek F, Favreau P, Stöcklin R. 2015. Uncovering intense protein diversification in a cone snail venom gland using an integrative venomics approach. J Prot Res 14:628–38. [DOI] [PubMed] [Google Scholar]
- Bouchet P, Kantor YI, Sysoev A, Puillandre N. 2011. A new operational classification of the Conoidea (Gastropoda). J Moll Stud 77:273–308. [Google Scholar]
- Breuker K, Jin M, Han X, Jiang H, McLafferty FW. 2008. Top-down identification and characterization of biomolecules by mass spectrometry. J Am Soc Mass Spec 19:1045–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvete JJ. 2014. Next-generation snake venomics: protein-locus resolution through venom proteome decomplexation. Exp Rev Prot 11:315–29. [DOI] [PubMed] [Google Scholar]
- Calvete JJ, Juárez P, Sanz L. 2007. Snake venomics. Strategy and applications. J Mass Spect 42:1405–14. [DOI] [PubMed] [Google Scholar]
- Calvete JJ, Sanz L, Angulo Y, Lomonte B, Gutiérrez JM. 2009. Venoms, venomics, antivenomics. FEBS Lett 583:1736–43. [DOI] [PubMed] [Google Scholar]
- Cao Z, Yu Y, Wu Y, Hao P, Di Z, He Y, Chen Z, Yang W, Shen Z, He X. et al. 2013. The genome of Mesobuthus martensii reveals a unique adaptation model of arthropods. Nat Commun 4:2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casewell NR, Wüster W, Vonk FJ, Harrison RA, Fry BG. 2013. Complex cocktails: The evolutionary novelty of venoms. Tren Ecol E 28:219–29. [DOI] [PubMed] [Google Scholar]
- Castelin M, Puillandre N, Kantor Yu I, Terryn Y, Cruaud C, Bouchet P, Holford MM. 2012. Macroevolution of venom apparatus innovations in auger snails (Gastropoda; Conoidea; Terebridae). Mol Phylogenet E 64:21–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig AG, Bandyopadhyay P, Olivera BM. 1999. Post-translationally modified neuropeptides from Conus venoms. Eur J Biochem 264:271–5. [DOI] [PubMed] [Google Scholar]
- Cushman D, Ondetti M. 1991. History of the design of captopril and related inhibitors of angiotensin converting enzyme. Hypertension 17:589–92. [DOI] [PubMed] [Google Scholar]
- Dave K, Lahiry A. 2012. Conotoxins: review and docking studies to determine potentials of conotoxin as an anticancer drug molecule. Curr Top Med Chem 12:845–51. [DOI] [PubMed] [Google Scholar]
- Dawkins R, Krebs JR. 1979. Arms races between and within species. Proc R Soc Lond B Biol Sci 205:489–511. [DOI] [PubMed] [Google Scholar]
- Diochot S, Baron A, Salinas M, Douguet D, Scarzello S, Dabert-Gay A-S, Debayle D, Friend V, Alloui A, Lazdunski M. et al. 2012. Black mamba venom peptides target acid-sensing ion channels to abolish pain. Nature 490:552–5. [DOI] [PubMed] [Google Scholar]
- Duda TF, Palumbi SR. 1999. Molecular genetics of ecological diversification: duplication and rapid evolution of toxin genes of the venomous gastropod Conus. Proc Natl Acad Sci USA 96:6820–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durban J, Juárez P, Angulo Y, Lomonte B, Flores-Diaz M, Alape-Girón A, Sasa M, Sanz L, Gutiérrez JM, Dopazo J. et al. 2011. Profiling the venom gland transcriptomes of Costa Rican snakes by 454 pyrosequencing. BMC Gen 12:259.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durban J, Pérez A, Sanz L, Gómez A, Bonilla F, Rodríguez S, Chacón D, Sasa M, Angulo Y, Gutiérrez JM. et al. 2013. Integrated “omics” profiling indicates that miRNAs are modulators of the ontogenetic venom composition shift in the Central American rattlesnake, Crotalus simus simus. BMC Gen 14:234.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutertre S, Jin A-H, Vetter I, Hamilton B, Sunagar K, Lavergne V, Dutertre V, Fry BG, Antunes A, Venter DJ. et al. 2014. Evolution of separate predation- and defence-evoked venoms in carnivorous cone snails. Nat Commun 5:3521.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichberg S, Sanz L, Calvete JJ, Pla D. 2015. Constructing comprehensive venom proteome reference maps for integrative venomics. Exp Rev Prot 12:557–73. [DOI] [PubMed] [Google Scholar]
- Elmer KR, Fan S, Gunter HM, Jones JC, Boekhoff S, Kuraku S, Meyer A. 2010. Rapid evolution and selection inferred from the transcriptomes of sympatric Crater Lake cichlid fishes. Mol Ecol 19(Suppl 1):197–211. [DOI] [PubMed] [Google Scholar]
- Escoubas P, King G. 2009. Venomics as a drug discovery platform. Exp Rev Prot 6:221–4. [DOI] [PubMed] [Google Scholar]
- Escoubas P, Quinton L, Nicholson GM. 2008. Venomics: Unravelling the complexity of animal venoms with mass spectrometry. J Mass Spec 43:279–95. [DOI] [PubMed] [Google Scholar]
- Espiritu DJD, Watkins M, Dia-Monje V, Cartier GE, Cruz LJ, Olivera BM. 2001. Venomous cone snails: Molecular phylogeny and the generation of toxin diversity. Toxicon 39:1899–916. [DOI] [PubMed] [Google Scholar]
- Favreau P, Stöcklin R. 2009. Marine snail venoms: use and trends in receptor and channel neuropharmacology. Curr Opin Pharmacol 9:594–601. [DOI] [PubMed] [Google Scholar]
- Fenn JB. 2003. Electrospray wings for molecular elephants (Nobel lecture). Angewandte Chemie - International Edition 42:3871–94. [DOI] [PubMed] [Google Scholar]
- Fry BG, Koludarov I, Jackson TNW, Holford M, Terrat Y, Casewell NR, Undheim EAB, Vetter I, Ali SA, Low DHW. et al. 2013. Seeing the woods for the trees: understanding venom evolution as a guide for biodiscovery In: King GF, editor. Venoms to drugs: venom as a source for the development of human therapeutics. London, UK: Royal Society of Chemistry. [Google Scholar]
- Fry BG, Lumsden NG, Wüster W, Wickramaratna JC, Hodgson WC, Manjunatha Kini R. 2003. Isolation of a neurotoxin (α-colubritoxin) from a nonvenomous colubrid: evidence for early origin of venom in snakes. J Mol E 57:446–52. [DOI] [PubMed] [Google Scholar]
- Fry BG, Scheib H, Van der Weerd L, Young B, McNaughtan J, Ramjan SFR, Vidal N, Poelmann RE, Norman J. 2008. Evolution of an arsenal structural and functional diversification of the venom system in the advanced snakes (Caenophidia). Mol Cell Prot 7:215–46. [DOI] [PubMed] [Google Scholar]
- Gibbs HL, Sanz L, Sovic MG, Calvete JJ. 2013. Phylogeny-based comparative analysis of venom proteome variation in a clade of rattlesnakes (Sistrurus sp.). PLoS One 8:e67220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkika D, Prevarskaya N. 2011. TRP channels in prostate cancer: the good, the bad and the ugly? Asi J Androl 13:673–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales DTT, Saloma CP. 2014. A bioinformatics survey for conotoxin-like sequences in three turrid snail venom duct transcriptomes. Toxicon 92:66–74. [DOI] [PubMed] [Google Scholar]
- Gorson J, Ramrattan G, Verdes A, Wright M, Kantor YI, Srinivasan R, Musunuri R, Packer D, Albano G, Qiu W. et al. 2015. Molecular diversity and gene evolution of the venom arsenal of terebridae predatory marine snails. Gen Biol E 7:1761–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WR, Olivera BM, Cruz LJ. 1988. Peptide toxins from venomous Conus snails. Ann Rev Biochem 57:665–700. [DOI] [PubMed] [Google Scholar]
- Han TS, Teichert RW, Olivera BM, Bulaj G. 2008. Conus venoms - a rich source of peptide-based therapeutics. Curr Pharm Des 14:2462–79. [DOI] [PubMed] [Google Scholar]
- Heralde FM, Imperial J, Bandyopadhyay PK, Olivera BM, Concepcion GP, Santos AD, Heralde FM, 3rd, Imperial J, Bandyopadhyay PK, Olivera BM. et al. 2008. A rapidly diverging superfamily of peptide toxins in venomous Gemmula species. Toxicon 51:890–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holding ML, Biardi JE, Gibbs HL. 2016. Coevolution of venom function and venom resistance in a rattlesnake predator and its squirrel prey. Proc R Soc Lond B Biol Sci 283No. 1829 p. 20152841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holford M, Puillandre N, Terryn Y, Cruaud C, Olivera B, Bouchet P. 2009. Evolution of the toxoglossa venom apparatus as inferred by molecular phylogeny of the Terebridae. Mol Biol E 26:15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob RB, McDougal OM. 2010. The M-superfamily of conotoxins: a review. Cell Mol Life Sci 67:17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubowski JA, Kelley WP, Sweedler JV, Gilly WF, Schulz JR. 2005. Intraspecific variation of venom injected by fish-hunting Conus snails. J Exp Biol 208:2873–83. [DOI] [PubMed] [Google Scholar]
- Jouiaei M, Sunagar K, Federman Gross A, Scheib H, Alewood PF, Moran Y, Fry BG. 2015. Evolution of an ancient venom: Recognition of a novel family of cnidarian toxins and the common evolutionary origin of sodium and potassium neurotoxins in sea anemone. Mol Biol E 32:1598–610. [DOI] [PubMed] [Google Scholar]
- Kaas Q, Westermann JC, Craik DJ. 2010. Conopeptide characterization and classifications: An analysis using ConoServer. Toxicon 55:1491–509. [DOI] [PubMed] [Google Scholar]
- Kantor Y, Taylor JD, Kantor Taylor JDYI. 2000. Formation of marginal radular teeth in Conoidea (Neogastropoda) and the evolution of the hypodermic envenomation mechanism. J Zool Lond 252:251–62. [Google Scholar]
- Kantor YI, Puillandre N. 2012. Evolution of the Radular Apparatus in Conoidea (Gastropoda: Neogastropoda) as Inferred from a Molecular Phylogeny. Malacologia 55:55–90. [Google Scholar]
- Karas M, Hillenkamp F. 1988. Laser desorption ionization of proteins with molecular masses exceeding 10,000 daltons. Anal Chem 60:2299–301. [DOI] [PubMed] [Google Scholar]
- Kendel Y, Melaun C, Kurz A, Nicke A, Peigneur S, Tytgat J, Wunder C, Mebs D, Kauferstein S. 2013. Venomous secretions from marine snails of the Terebridae family target acetylcholine receptors. Toxins (Basel) 5:1043–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King GF. 2011. Venoms as a platform for human drugs: translating toxins into therapeutics. Exp Opin Biol Ther 11:1469–84. [DOI] [PubMed] [Google Scholar]
- King GF. 2015. Venoms to drugs: venom as a source for the development of human therapeutics. The Royal Society of Chemistry (RSC Drug Discovery; ). London, UK. [Google Scholar]
- Koch ED, Olivera BM, Terlau H, Conti F. 2004. The binding of kappa-Conotoxin PVIIA and fast C-type inactivation of Shaker K+ channels are mutually exclusive. Biophys J 86:191–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh CY, Kini RM. 2012. From snake venom toxins to therapeutics–cardiovascular examples. Toxicon 59:497–506. [DOI] [PubMed] [Google Scholar]
- Lang F, Stournaras C, PTRS B. 2014. Ion channels in cancer : future perspectives and clinical potential Ion channels in cancer : future perspectives and clinical potential author for correspondence . Phil Trans Roy Soc B 369:20130108.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavergne V, Harliwong I, Jones A, Miller D, Taft RJ, Alewood PF. 2015. Optimized deep-targeted proteotranscriptomic profiling reveals unexplored Conus toxin diversity and novel cysteine frameworks. Proc. Natl. Acad. Sci. USA 112:E3782–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis RJ, Garcia ML. 2003. Therapeutic potential of venom peptides. Nat Rev Drug Discov 2:790–802. [DOI] [PubMed] [Google Scholar]
- Malmberg AB, Gilbert H, McCabe RT, Basbaum AI. 2003. Powerful antinociceptive effects of the cone snail venom-derived subtype-selective NMDA receptor antagonists conantokins G and T. Pain 101:109–16. [DOI] [PubMed] [Google Scholar]
- McIntosh JM, Absalom N, Chebib M, Elgoyhen AB, Vincler M. 2009. Alpha9 nicotinic acetylcholine receptors and the treatment of pain. Biochem Pharmacol 78:693–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miljanich GP. 2004. Ziconotide: neuronal calcium channel blocker for treating severe chronic pain. Curr Med Chem 11:3029–40. [DOI] [PubMed] [Google Scholar]
- Modica MV, Holford M. 2010. The neogastropoda: evolutionary innovations of predatory marine snails with remarkable pharmacological potential In: Evolutionary biology – concepts, molecular and morphological evolution. Berlin Heidelberg: Springer Editor: Pierre Pontarotti. [Google Scholar]
- Moon J, Gorson J, Wright ME, Yee L, Khawaja S, Shin H, Karma Y, Lochan R, Michelle Yun M, Holford M. 2016. Characterization and recombinant expression of terebrid venom peptide from Terebra gutttata. Toxins (Basel). 8:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran Y, Weinberger H, Sullivan JC, Reitzel AM, Finnerty JR, Gurevitz M. 2008. Concerted evolution of sea anemone neurotoxin genes is revealed through analysis of the Nematostella vectensis genome. Mol Biol E 25:737–47. [DOI] [PubMed] [Google Scholar]
- Nentwig W. 2013. Spider ecophysiology. Springer-Verlag Berlin Heidelberg. [Google Scholar]
- Neves JLB, Lin Z, Imperial JS, Antunes A, Vasconcelos V, Olivera BM, Schmidt EW. 2015. Small molecules in the cone snail arsenal. Org Lett 17:4933–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisani Z, Dunbar SG, Hayes WK. 2007. Cost of venom regeneration in Parabuthus transvaalicus (Arachnida: Buthidae). Comp Biochem Physiol A Mol Integr Physiol 147:509–13. [DOI] [PubMed] [Google Scholar]
- Norton RS, Olivera BM. 2006. Conotoxins down under. Toxicon 48:780–98. [DOI] [PubMed] [Google Scholar]
- Olivera BM. 2000. w-Conotoxin MVIIA: from marine snail venom to analgesic drug In: Fusetani N, editor. Drugs from the sea. Basel: Karger; p. 75–85. [Google Scholar]
- Olivera BM. 2002. Conus venom peptides: reflections from the biology of clades and species. Ann Rev Ecol Syst 33:25–47. [Google Scholar]
- Olivera BM, Gray WR, Zeikus R, McIntosh JM, Varga J, Rivier J, de Santos V, Cruz LJ. 1985. Peptide neurotoxins from fish-hunting cone snails. Science 230:1338–43. [DOI] [PubMed] [Google Scholar]
- Olivera BM, Showers Corneli P, Watkins M, Fedosov A. 2014. Biodiversity of cone snails and other venomous marine gastropods: evolutionary success through neuropharmacology. Ann Rev Anim Biosci 2:487–513. [DOI] [PubMed] [Google Scholar]
- Olivera BM, Walker C, Cartier GE, Hooper D, Santos AD, Schoenfeld R, Shetty R, Watkins M, Bandyopadhyay P, Hillyard DR. 1999. Speciation of cone snails and interspecific hyperdivergence of their venom peptides. Potential evolutionary significance of introns. Ann N Y Acad Sci 870:223–37. [DOI] [PubMed] [Google Scholar]
- Ortiz E, Gurrola GB, Schwartz EF, Possani LD. 2015. Scorpion venom components as potential candidates for drug development. Toxicon 93:125–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otvos RA, Heus F, Vonk FJ, Halff J, Bruyneel B, Paliukhovich I, Smit AB, Niessen WMA, Kool J. 2013. Analytical workflow for rapid screening and purification of bioactives from venom proteomes. Toxicon 76:270–81. [DOI] [PubMed] [Google Scholar]
- Pawlak J, Mackessy SP, Fry BG, Bhatia M, Mourier G, Fruchart-Gaillard C, Servent D, Ménez R, Stura E, Ménez A. et al. 2006. Denmotoxin, a three-finger toxin from the colubrid snake Boiga dendrophila (mangrove catsnake) with bird-specific activity. J Biol Chem 281:29030–41. [DOI] [PubMed] [Google Scholar]
- Pawlak J, Mackessy SP, Sixberry NM, Stura EA, Le Du MH, Menez R, Foo CS, Menez A, Nirthanan S, Kini RM. 2009. Irditoxin, a novel covalently linked heterodimeric three-finger toxin with high taxon-specific neurotoxicity. FASEB J 23:534–45. [DOI] [PubMed] [Google Scholar]
- Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data - Mascot. Electrophoresis 20:3551–67. [DOI] [PubMed] [Google Scholar]
- Perkins J, Smith B, Gallagher RT, Jones DS, Davis SC, Hoffman AD, Tomer KB. 1993a. Application of electrospray mass spectrometry and matrix-assisted laser desorption ionization time-of-flight mass spectrometry for molecular weight assignment of peptides in complex mixtures. J Am Soc Mass Spec 4:670–84. [DOI] [PubMed] [Google Scholar]
- Perkins JR, Parker CE, Tomer KB. 1993b. The characterization of snake venoms using capillary electrophoresis in conjunction with electrospray mass spectrometry: Black Mambas. Electrophoresis 14:458–68. [DOI] [PubMed] [Google Scholar]
- Petras D, Heiss P, Süssmuth RD, Calvete JJ. 2015. Venom proteomics of Indonesian king cobra, Ophiophagus hannah : integrating top-down and bottom-up approaches. J Prot Res 14:2539–56. [DOI] [PubMed] [Google Scholar]
- Powell A. 1966. The molluscan families Speightiidae and Turridae. An evaluation of the valid taxa, both recent and fossil, with lists of characteristics species. Bull Auckl Inst Mus 5:5–184. [Google Scholar]
- Puillandre N, Bouchet P, Duda TF, Kauferstein S, Kohn AJ, Olivera BM, Watkins M, Meyer C. 2014. Molecular phylogeny and evolution of the cone snails (Gastropoda, Conoidea). Mol Phylogenet E 78:290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puillandre N, Duda TF, Meyer C, Olivera BM, Bouchet P. 2015. One, four or 100 genera? A new classification of the cone snails. J Moll Stud 81:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puillandre N, Holford M. 2010. The Terebridae and teretoxins: Combining phylogeny and anatomy for concerted discovery of bioactive compounds. BMC Chem. Biol 10:7.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, Terry A, Shapiro H, Lindquist E, Kapitonov VV. et al. 2007. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 317:86–94. [DOI] [PubMed] [Google Scholar]
- Von Reumont BM, Blanke A, Richter S, Alvarez F, Bleidorn C, Jenner RA. 2014a. The first venomous crustacean revealed by transcriptomics and functional morphology: remipede venom glands express a unique toxin cocktail dominated by enzymes and a neurotoxin. Mol Biol E 31:48–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Reumont BM, Campbell LI, Jenner RA. 2014b. Quo Vadis venomics? A roadmap to neglected venomous invertebrates. Toxins (Basel) 6:3488–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson SD, Norton RS. 2014. Conotoxin gene superfamilies. Mar Drugs 12:6058–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson SD, Safavi-Hemami H, McIntosh LD, Purcell AW, Norton RS, Papenfuss AT. 2014. Diversity of conotoxin gene superfamilies in the venomous snail, Conus victoriae. PLoS One 9:e87648.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo C, Di Francesco L, Oliverio M, Palazzo P, Massilia GR, Ascenzi P, Polticelli F, Schininà ME. 2008. Conus ventricosus venom peptides profiling by HPLC-MS: a new insight in the intraspecific variation. J Sep Sci 31:488–98. [DOI] [PubMed] [Google Scholar]
- Safavi-Hemami H, Gajewiak J, Karanth S, Robinson SD, Ueberheide B, Douglass AD, Schlegel A, Imperial JS, Watkins M, Bandyopadhyay PK. et al. 2014. Specialized insulin is used for chemical warfare by fish-hunting cone snails. Proc Natl Acad Sci 11:1743–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanggaard KW, Bechsgaard JS, Fang X, Duan J, Dyrlund TF, Gupta V, Jiang X, Cheng L, Fan D, Feng Y. et al. 2014. Spider genomes provide insight into composition and evolution of venom and silk. Nat Commun 5:3765.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidtko A, Lötsch J, Freynhagen R, Geisslinger G. 2010. Ziconotide for treatment of severe chronic pain. Lancet 375:1569–77. [DOI] [PubMed] [Google Scholar]
- Sunagar K, Morgenstern D, Reitzel AM, Moran Y. 2016. Ecological venomics: How genomics, transcriptomics and proteomics can shed new light on the ecology and evolution of venom. J Prot 135:62–72. [DOI] [PubMed] [Google Scholar]
- Sunagar K, Undheim E. a B, Scheib H, Gren ECK, Cochran C, Person CE, Koludarov I, Kelln W, Hayes WK, King GF. et al. 2014. Intraspecific venom variation in the medically significant Southern Pacific Rattlesnake (Crotalus oreganus helleri): Biodiscovery, clinical and evolutionary implications. J Prot 99:68–83. [DOI] [PubMed] [Google Scholar]
- Tanaka K. 2003. The origin of macromolecule ionization by laser irradiation (Nobel lecture). Angew Chem Int Ed Engl 42:3860–70. [DOI] [PubMed] [Google Scholar]
- Taylor JD. 1977. Food and habitats of predatory gastropods on coral reefs. Rep Underw Assoc 2:17–34. [Google Scholar]
- Taylor JD. 1990. The anatomy of the foregut and relationships in the Terebridae. Malacologia 32:19–34. [Google Scholar]
- Terlau H, Olivera BM. 2004. Conus venoms: a rich source of novel ion channel-targeted peptides. Physiol Rev 84:41–68. [DOI] [PubMed] [Google Scholar]
- Tucker JK, Tenorio MJ. 2009. Systematic classification of recent and fossil conoidean gastropods : with keys to the genera of cone shells. Hackenheim: ConchBooks. [Google Scholar]
- Twede VD, Miljanich G, Olivera BM, Bulaj G. 2009. Neuroprotective and cardioprotective conopeptides: an emerging class of drug leads. Curr Opin Drug Discov Devel 12:231–9. [PMC free article] [PubMed] [Google Scholar]
- Ueberheide BM, Fenyö D, Alewood PF, Chait BT. 2009. Rapid sensitive analysis of cysteine rich peptide venom components. Proc Natl Acad Sci USA 106:6910–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlig T, Kyprianou T, Martinelli FG, Oppici CA, Heiligers D, Hills D, Calvo XR, Verhaert P. 2014. The emergence of peptides in the pharmaceutical business: From exploration to exploitation. EuPA Open Prot 4:58–69. [Google Scholar]
- Van Valen L. 1973. A new evolutionary theory. Evol The 1:1–30. [Google Scholar]
- Veiseh M, Gabikian P, Bahrami S-BB, Veiseh O, Zhang M, Hackman RC, Ravanpay AC, Stroud MR, Kusuma Y, Hansen SJ. et al. 2007. Tumor paint: a chlorotoxin:Cy5.5 bioconjugate for intraoperative visualization of cancer foci. Can Res 67:6882–8. [DOI] [PubMed] [Google Scholar]
- Vetter I, Lewis RJ. 2012. Therapeutic potential of cone snail venom peptides. Curr Top Med Chem 12:1546–52. [DOI] [PubMed] [Google Scholar]
- Vincler M, Wittenauer S, Parker R, Ellison M, Olivera BM, McIntosh JM. 2006. Molecular mechanism for analgesia involving specific antagonism of alpha9alpha10 nicotinic acetylcholine receptors. Proc Natl Acad Sci USA 103:17880–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonk FJ, Casewell NR, Henkel CV, Heimberg AM, Jansen HJ, McCleary RJR, Kerkkamp HME, Vos RA, Guerreiro I, Calvete JJ. et al. 2013. The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system. Proc Natl Acad Sci USA 110:20651–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock GM, Robinson GE, Gibbs RA, Weinstock GM, Weinstock GM, Robinson GE, Worley KC, Evans JD, Maleszka R, Robertson HM. et al. 2006. Insights into social insects from the genome of the honeybee Apis mellifera. Nature 443:931–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood JN, Boorman JP, Okuse K, Bake MD. 2004. Voltage-gated sodium channels and pain pathways. J Neurobiol 61:55–71. [DOI] [PubMed] [Google Scholar]
- Wurm Y, Wang J, Riba-Grognuz O, Corona M, Nygaard S, Hunt BG, Ingram KK, Falquet L, Nipitwattanaphon M, Gotzek D. et al. 2011. The genome of the fire ant Solenopsis invicta. Proc Natl Acad Sci USA 108:5679–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Duan Z, Di Z, He Y, Li J, Li Z, Xie C, Zeng X, Cao Z, Wu Y. et al. 2014. Proteomic analysis of the venom from the scorpion Mesobuthus martensii. J Prot 106:162–80. [DOI] [PubMed] [Google Scholar]
- Yao S, Zhang M-M, Yoshikami D, Azam L, Olivera BM, Bulaj G, Norton RS. 2008. Structure, dynamics, and selectivity of the sodium channel blocker mu-conotoxin SIIIA. Biochemistry 47:10940–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Gao B, Zhu S. 2015. Target-driven evolution of scorpion toxins. Sci Rep 5:14973.. [DOI] [PMC free article] [PubMed] [Google Scholar]