Abstract
We here describe a novel method for MYD88L265P mutation detection and minimal residual disease monitoring in Waldenström macroglobulinemia, by droplet digital polymerase chain reaction, in bone marrow and peripheral blood cells, as well as in circulating cell-free DNA. Our method shows a sensitivity of 5.00×10−5, which is far superior to the widely used allele-specific polymerase chain reaction (1.00×10−3). Overall, 291 unsorted samples from 148 patients (133 with Waldenström macroglobulinemia, 11 with IgG lymphoplasmacytic lymphoma and 4 with IgM monoclonal gammopathy of undetermined significance) were analyzed: 194 were baseline samples and 97 were followup samples. One hundred and twenty-two of 128 (95.3%) bone marrow and 47/66 (71.2%) baseline peripheral blood samples scored positive for MYD88L265P. To investigate whether MYD88L265P detection by droplet digital polymerase chain reaction could be used for minimal residual disease monitoring, mutation levels were compared with IGH-based minimal residual disease analysis in 10 patients, and was found to be as informative as the classical, standardized, but not yet validated in Waldenström macroglobulinemia, IGH-based minimal residual disease assay (r2=0.64). Finally, MYD88L265P detection by droplet digital polymerase chain reaction on plasma circulating tumor DNA from 60 patients showed a good correlation with bone marrow findings (bone marrow median mutational value 1.92×10−2, plasma circulating tumor DNA value: 1.4×10−2, peripheral blood value: 1.03×10−3). This study indicates that droplet digital polymerase chain reaction assay of MYD88L265P is a feasible and sensitive tool for mutation screening and minimal residual disease monitoring in Waldenström macroglobulinemia. Both unsorted bone marrow and peripheral blood samples can be reliably tested, as can circulating tumor DNA, which represents an attractive, less invasive alternative to bone marrow for MYD88L265P detection.
Introduction
In recent years, the MYD88L265P mutation has been recurrently identified in Waldenström macroglobulinemia (WM),1,2 an indolent lymphoplasmacytic lymphoma (LPL) characterized by the accumulation in the bone marrow (BM) of monoclonal lymphocytes, lymphoplasmacytic cells and plasma cells, responsible for monoclonal IgM protein secretion.3 Several studies using different techniques such as Sanger sequencing, polymerase chain reaction (PCR) and allele-specific quantitative PCR (ASqPCR), on CD19-sorted BM samples, found that about 90% of WM patients carry the MYD88L265P mutation, while it is present in 14–29% of “activated B-cell” diffuse large B-cell lymphomas,4,5 6–10% of marginal zone lymphomas, 3–5% of chronic lymphocytic leukemia and is absent in multiple myeloma and in non-IgM monoclonal gammopathies of uncertain significance (MGUS).6–8 Therefore, MYD88L265P is now considered a hallmark of WM and may be helpful in the differential diagnosis from other lymphoproliferative neoplasms with overlapping clinical features, such as multiple myeloma.9,10 Furthermore, it might represent an ideal marker for minimal residual disease (MRD) monitoring in a disease whose therapeutic scenario is rapidly changing, with many newly available and highly effective drugs.11–17 Moreover, MYD88L265P has been demonstrated in CD19-sorted BM samples from 50–80% of patients with IgM-MGUS, an asymptomatic phase which represents a pre-neoplastic condition, suggesting a potential role in disease progression.6,8,18–20 BM biopsy is mandatory for the differential diagnosis between WM and IgM-MGUS, but patients with an asymptomatic M component do not readily agree to undergo such an invasive procedure. The availability of accurate diagnostic tools based on the use of peripheral blood (PB), or even urine samples, would overcome this problem and avoid the risk of misclassification of patients. Additionally, the current MYD88L265P ASqPCR method lacks sensitivity (1.00×10−3) and is not, therefore, suitable for MRD.6,21 Indeed, ASqPCR is suboptimal for testing specimens such as unsorted BM or even PB, which contains low concentrations of circulating tumor cells (especially after immunochemotherapy), or for assessing cell-free tumor DNA (ctDNA), usually present in only very small amounts in plasma, including cerebrospinal fluid and pleural effusions.22,23 Recently, digital PCR has been shown to be a powerful technique that provides improved sensitivity, precision and reproducibility, overcoming some of the pitfalls of qPCR.24,25 We here describe a newly developed, highly sensitive, droplet digital PCR (ddPCR) assay for the identification of the MYD88L265P mutation in patients affected by WM or LPL, suitable for screening and MRD monitoring on BM and PB cells, as well as on cell-free DNA.
Methods
Patients and samples collection
BM, PB, plasma and urine samples were collected at baseline and during follow up from patients affected by WM, IgM-MGUS or IgG-secreting LPL (Online Supplementary Figure S1). Consecutive patients were included in this study and were classified based on the 2008 World Health Organization classification criteria.3 PB from 40 healthy subjects and BM from 20 patients with multiple myeloma were used as negative controls. Sample collection and storage as well as nucleic acid extraction procedures are described in the Online Supplementary Appendix.
All patients provided written informed consent to the use of their biological samples for research, in accordance with Institutional Review Board provisions and the Declaration of Helsinki. This study was approved by the local ethics committee.
Droplet digital polymerase chain reaction assays for detection of the MYD88L265P mutation
The mutation detection assay was designed as reported in Online Supplementary Figure S2A. ddPCR was performed using the QX100 Droplet Digital PCR system (Bio-Rad Laboratories) as detailed in the Online Supplementary Appendix. Samples were tested in triplicate and results are expressed as merge of wells. The cut-off for defining the presence of the mutation was set based on the highest MYD88L265P level detected within the control group and is indicated in figures as a red dashed line. Samples with a ratio value below the red dashed line are considered MYD88L265P wild-type (WT). Each experiment included a no template control, a known highly mutated positive control sample (mutation rate = 70%, mutated/WT ratio 7×10−1), previously tested by Sanger sequencing, as reported by Treon et al.,1 and a negative control (healthy donor or multiple myeloma gDNA). dMIQE guidelines (Minimum Information for Publication of Quantitative Digital PCR Experiments) for ddPCR experiments are listed in Online Supplementary Table S1.25
Allele-specific quantitative polymerase chain reaction assay for detection of the MYD88L265P mutation
The sensitivity of MYD88L265P ddPCR was compared to that of the ASqPCR assay previously described by Xu et al.6 on a standard curve of 10-fold serial dilutions. In parallel, 100 WM samples from the Torino series and 23 patients from the Salamanca series were analyzed, following the strategy described by Jiménez et al.21 Additionally, MYD88L265P mutation detection by ddPCR was compared to that of the qBiomarker™ Somatic Mutation Assay kit for MYD88_85940 (Qiagen) on 15 samples from the University of Pisa.
Allele-specific quantitative polymerase chain reaction assay for tumor-specific IGH-VDJ rearrangement
In order to perform a comparison to the worldwide standardized ASqPCR technique for immunoglobulin-VDJ rearrangement (IGH-VDJ) MRD analysis, patient-specific IGH-VDJ was amplified and directly sequenced26 (Online Supplementary Figure S2B). This IGH-based MRD analysis was performed according to the Euro-MRD guidelines.27
Statistical analysis
Associations between categorical variables were analyzed by the Fisher exact test, while Mann-Whitney and Kruskal-Wallis tests were used for the inference on continuous variables. Results of the analyses of continuous variables are expressed as the median (range); ddPCR and ASqPCR results are expressed as mutated:WT ratio. The interrater agreement on categorical data was estimated by computing the Fleiss kappa (k) index. All reported P-values were estimated by the two-sided exact method with the conventional 5% significance level. Data were analyzed as of July 2017 using R 3.4.1 (R Foundation for Statistical Computing, Vienna, Austria).28
Results
Patients and samples
Overall, 291 samples from 148 patients from the three series were analyzed (133 WM, 1 amyloid-associated IgM-LPL, 10 IgG-LPL, 4 IgM-MGUS): 194 samples were taken at baseline in active disease (128 BM, 66 PB) and 97 samples during follow up (43 BM and 54 PB). The baseline samples were defined either “treatment-naïve” (143/194, 73.7%, 99 BM and 44 PB) or “relapsed” (51/194, 26.2%, 29 BM and 22 PB), according to previous exposure to chemoimmunotherapy. All the follow-up samples were taken for the purpose of studying MRD at different time points after specific treatment. The distribution of the study population is summarized in Online Supplementary Figure S1, while the patients’ main clinical features are detailed in Table 1.
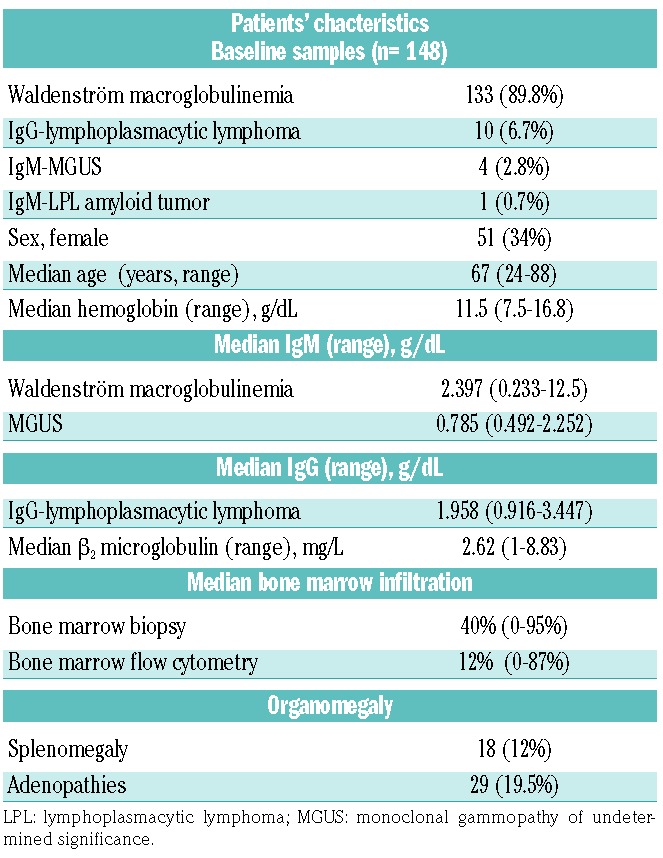
Table 1.
Patients’ clinical and biological features at baseline.
Detection limit of droplet digital polymerase chain reaction
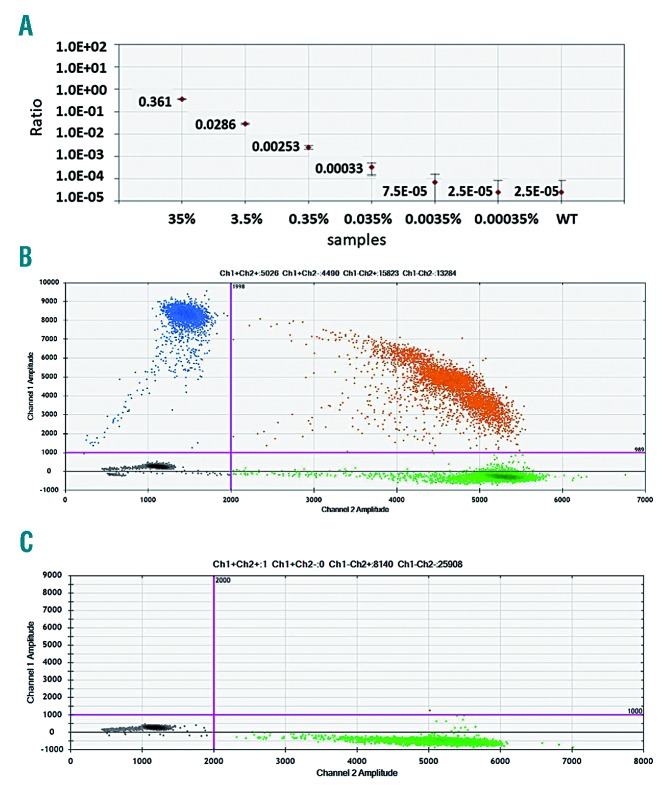
The detection limit of ddPCR was determined using a serial dilution of MYD88L265P mutated gDNA in WT DNA at levels of 35, 3.5, 0.35, 0.035 and 0.0035%, corresponding to 10500, 1050, 105, 10.5 and 1 mutated copy present in 100 ng of gDNA (30,000 copies) which is the overall quantity of gDNA loaded per well. We identified the limit of detection using a statistical method based on binomial distribution, as previously reported.29 This analysis indicated that we were able to detect the mutation, with a good degree of confidence, when the level of mutated copies was more than 0.035% (10 mutated copies in 30000 WT). This value mirrored the cut-off mutated:WT ratio we identified based on the control group (40 healthy subjects) (Figure 1A). Additional reproducibility tests confirmed that the above calculated limit of detection and the experimentally detected cut-off, emerging from the control group, are equally reliable (Online Supplementary Table S2).
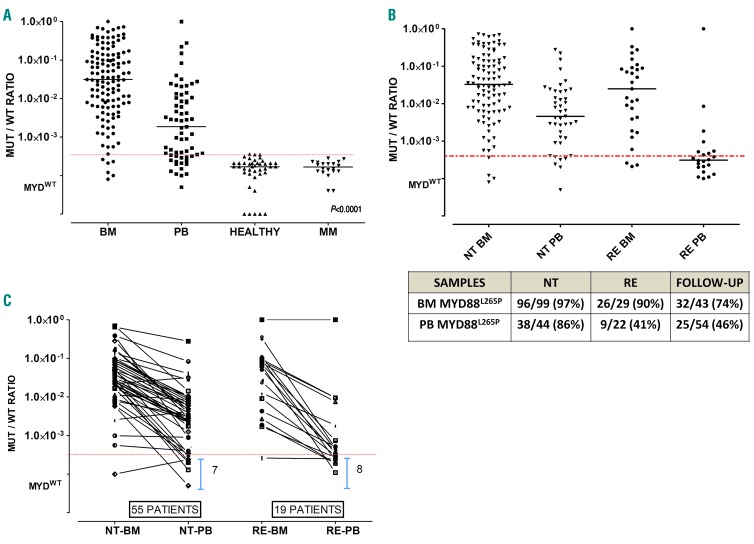
Figure 1.
MYD88L265P mutation at baseline is lower in peripheral blood (PB) than in bone marrow (BM). (A) Waldenström macroglobulinemia (WM) patients show statistically significant differences in mutated/wild-type ratio (MUT/WT) between BM and PB samples (P<0.0001). The control group of healthy subjects and multiple myeloma (MM) patients show MUT/WT ratios below the limit of detection. The dashed red line shows the cut-off for mutational status. Symbols below the red line represent MYD88WT samples. (B) MUT/WT ratio differences between relapsed (RE) and Naïve to Treatment (NT) patients, at baseline, is higher in PB than in BM samples. (C) MYD88L265P MUT/WT ratio at baseline in pared BM/PB samples from 55 NT and 19 RE patients, highlight the differences between biological specimens suggesting that for RE patients, PB is less reliable than BM.
MYD88L265P screening by droplet digital polymerase chain reaction in baseline samples
Overall 142/148 (96%) patients were identified as having the MYD88L265P mutation. We observed a 91.6% mutation rate (33/36) among relapsed patients and 97.3% rate (109/112) among treatment-naïve patients (P=0.15). Notably, no one of the 6 MYD88L265P WT patients (4 WM with histological BM invasion of 20%, 30%, 30% and 60%) or 2 patients with IgG LPL (with 20% and 30% histological BM invasion) showed either alternative MYD88 or CXCR4 mutations, as investigated by Sanger sequencing on unselected cells.10,12,30 All BM samples from 20 patients with multiple myeloma used as negative controls were below the limit of detection (defined as previously described) and below the mutation cut-off ratio established based on 40 PB samples from healthy individuals (mutated:WT ratio <3.4×10−4) (Figure 1A). Moreover, to confirm the specificity of our assay for mutational screening, 15 patients with mantle cell lymphoma, 10 with follicular lymphoma and 10 with chronic lymphocytic leukemia were tested for the MYD88L265P mutation. All samples from the patients with follicular lymphoma or mantle cell lymphoma were WT, whereas one of the 10 patients with chronic lymphocytic leukemia showed a mutation ratio of 4.4×10−4, as already described in this disease.6
Looking at the single tissues, 95.3% (122/128) of baseline BM and 71.2% (47/66) of PB samples scored positive for MYD88L265P (BM median mutation burden 3.60×10−2, range: 2.00×10−4 – 7.30 ×10−1; PB median 5.00×10−3, range: 1.00×10−4 – 2.80×10−1) (Figure 1A). Notably, among the PB samples there was a statistically significant difference in mutation rate between samples from relapsed patients (median burden 4.00×10−4, range: 1.00×10−4 – 1.00×10−3) and treatment-naïve patents (median burden 2.80×10−3, range: 2.00×10−4 – 1.00×10−2), supporting the possible negative impact on mutation detection of previous treatment on PB samples (P<0.0001) (Figure 1B).
Seventy-four patients in this series had paired baseline BM and PB samples. Overall, in this subgroup of patients, the rate of MYD88L265P mutation detection by ddPCR was 93% (69/74) on BM and 72% (53/74) on PB samples. Accordingly, the detection rates were higher among treatment-naïve patients than among relapsed patients: 95% (52/55) on BM and 82% (45/55) on PB versus 89% (17/19) on BM and 42% (8/19) on PB, respectively (P=0.014). In addition, 15 of these 74 patients (20%) showed BM-mutated/PB-WT discordance, such discordance being more frequent among relapsed cases than among treatment-naïve cases (8/19, 42% vs. 7/55, 13%; P=0.02) (Figure 1C).
Finally, a test of within-run reproducibility showed uniform results across the experiments, with an inter-assay Standard Deviation (SD) of 1.3% for the MYD88L265P positive control and 0.01% for WT gDNA samples.
Comparison of MYD88L265P droplet digital polymerase chain reaction versus allele-specific quantitative polymerase chain reaction assays
Once the ddPCR assay had been optimized, the sensitivity of the MYD88L265P ddPCR was compared to that of ASqPCR on a standard curve of 10-fold serial dilutions constructed with a highly MYD88L265P mutated WM sample (70%, mutated/WT ratio 7.00×10−1, diluted to 35%), previously identified by Sanger sequencing.1 Whereas the ASqPCR standard curve confirmed the reported sensitivity of 1.00×10−3 (0.25% out of 6000 WT),6 ddPCR reached a 1.5 log higher sensitivity (0.0035% out of 30000 WT) also because of the larger amount of gDNA used in ddPCR than in ASqPCR (100 ng vs. 20 ng) (Figure 2 and Online Supplementary Table S2). Overall, 100 WM samples (48 BM, 52 PB, 40 baseline and 60 follow up) from 48 patients, as well as 20 control samples (15 healthy subjects and 5 with multiple myeloma) were tested by both methods (Online Supplementary Figure S3). An example of mutation analysis performed by both methods on 2 patients is presented in Online Supplementary Figure S4. Of the 40 baseline samples tested by both methods, 35 (87.5%) scored positive and 4 (10%) scored negative for MYD88L265P by both ddPCR and ASqPCR (with there being only one discordant ddPCR+/ASqPCR− case), while higher number of discordances were observed among follow-up samples: 13/60 (21.7%) ddPCR+/ASqPCR−, and 11/60 (18.3%) ddPCR−/ASqPCR+ (Online Supplementary Table S3). Indeed, the strength of agreement was very good in the baseline cases (Cohen κ=0.8) but poor in follow-up samples (Cohen κ=0.2), resembling what had been previously shown for low burden infiltrated samples.31 All control samples scored negative by both methods (median: ddPCR 1.75×10−4 (range 3.10×10−4 – 2.70×10−5) and ASqPCR DCT=10 (range DCT=8.4–10.7, setting DCT=8 as the cut-off for negativity).
Figure 2.
Digital droplet polymerase chain reaction sensitivity test and example plots of controls (MUT vs. WT). (A) ddPCR assay measured on a 10-fold dilution standard curve, shows a sensitivity of 0.0035% MUT (1 mutated copy in 30000 WT). (B) Example of ddPCR result plot for the MYD88L265P control. (C) Example of ddPCR result plot for the MYD88WT control. MUT: mutated; WT: wild-type.
Comparison of MYD88L265P and IGH-based digital droplet polymerase chain reaction assays for the purposes of minimal residual disease detection
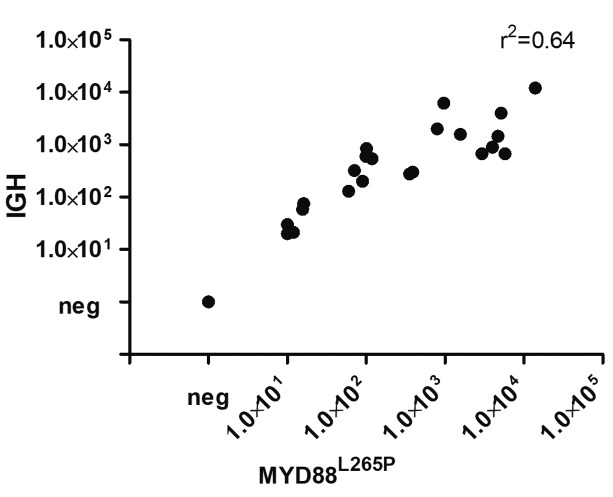
To investigate whether ddPCR of MYD88L265P could be used for MRD detection, we compared it to the highly sensitive IGH-based MRD ddPCR assay. Only patients with available follow-up samples were screened for IGH rearrangements. A clonal VDJ rearrangement was detected in 34/52 (65%) patients, as expected in WM.32 All these patients scored positive for MYD88L265P by ddPCR. We, therefore, tested 10 informative patients, at baseline and during clinical follow up, with both techniques. Overall, there was good concordance (r2=0.64) between the two methods (P<0.0001) in the 23 samples (18 BM, 5 PB) (Figure 3).
Figure 3.
MYD88L265P assay and IGH-based approaches are superimposable. Comparison of MYD88 and IGH-based minimal residual disease monitoring (expressed as copies of mutation in 1×105 cells) in 23 samples from 10 patients shows a good degree of correlation (r2=0.64).
MYD88L265P digital droplet polymerase chain reaction on circulating tumor DNA
In order to investigate the feasibility and the sensitivity advantages of ddPCR-based MYD88L265P mutation detection on plasma ctDNA vs. PB gDNA, paired samples from 60 patients were analyzed. Interestingly, a higher median MYD88L265P mutated / WT ratio was detected in plasma ctDNA (1.4×10−2) than in PB (1.03 ×10−3) (P<0.001) (Online Supplementary Figure S5), while no statistically significant difference was observed between ctDNA and BM samples from 32 patients (1.92×10−2 vs. 1.4×10−2; P=0.2). Figure 4 shows the matches among BM, plasma and PB MYD88L265P mutation quantification. Of note, 2 samples scored WT in all three compartments (Figure 4 and Online Supplementary Figure S6), while in 3 other patients MYD88L265P was detected in BM and PB, but scored WT in plasma ctDNA; these samples were excluded from the analysis since negativity, confirmed by RNAseP analysis, was ascribed to pre-analytical issues, related to sample collection and storage. Two cases were erroneously collected in lithium-heparin tubes instead of K3EDTA, while one case was collected correctly but processed more than 6 h after the blood had been drawn, confirming the importance of collection tubes and processing timing for good quality ctDNA, as already reported.33–35
Figure 4.
Cell tumor DNA from plasma mirrors the bone marrow mutation level. MYD88L265P mutated/wild-type (MUT/WT) ratio in bone marrow (BM), peripheral blood (PB) and plasma (PL) paired samples from 32 patients show no statistical differences between BM and PL. The dashed red line shows the cut-off for mutational status. Symbols below the red line represent MYD88WT samples. RE: relapsed; NT: Naïve-to-Treatment patients.
Minimal residual disease monitoring in Waldenström macroglobulinemia by MYD88L265P droplet digital polymerase chain reaction
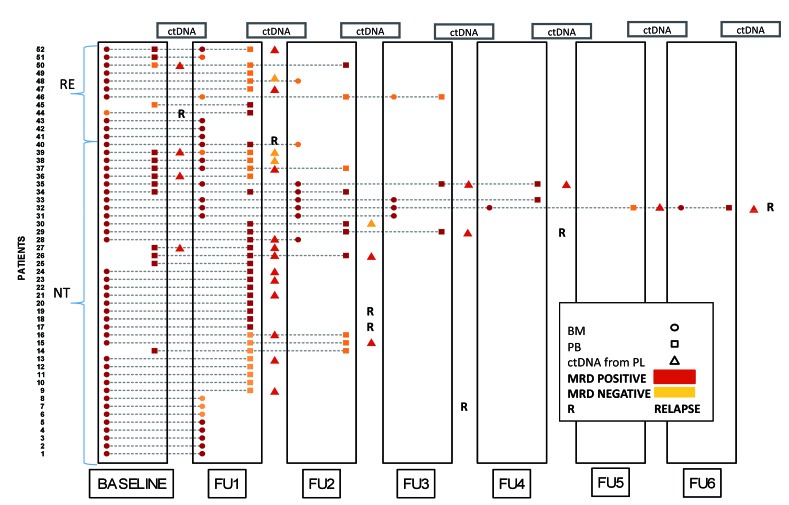
To explore the potential role of MYD88L265P mutation detection by ddPCR for MRD monitoring and therapy response evaluation, we focused our analysis on 52 patients who had at least one available follow-up sample. These patients’ features and the treatment they received Supplementary Table S4. MRD monitoring of this series is graphically depicted in Figure 5. All the treatment-naïve patients were MYD88L265P mutated at baseline (either on BM or PB or both), while 2 relapsed patients (one BM and one PB) scored WT (as false negative results, due to previous rituximab exposure, see below). After therapy, 6/22 informative cases showed MRD negativization on BM (27%) and 17/35 (49%) on PB, further suggesting a faster clearance of MRD from PB than from BM. At subsequent time points, the majority of patients remained MRD-positive (namely, patients 26, 28–31 and 33–35), some remained MRD-negative (patients 14–16, 37, 46 and 48), while a few show alternating results (patients 32, 40 and 50). Given the limited series of patients and the retrospective nature of the analysis, it is difficult to draw conclusions on the efficacy of single therapeutic schedules in clearing MRD in WM. However, the high potential of MRD shrinkage of regimens containing either fludarabine or bendamustine (each combined with rituximab) is highlighted in the Online Supplementary Methods (Online Supplementary Figure S7A–C).
Figure 5.
MYD88L265P can be used for disease monitoring. MYD88L265P monitored in 52 patients with at least one follow-up (FU) sample. ctDNA: cell tumor DNA; RE: relapsed at baseline; NT: Naïve to Treatment at baseline; MRD: minimal residual disease; PL: plasma; BM: bone marrow; PB: peripheral blood.
Of note, there were 2 false-negative cases among relapsed patients in this series. Patient 44, who had a baseline BM sample depicted as WT (meaning “MRD negative”), but with a borderline mutated:WT ratio, followed by PB positivity in the first follow-up sample (taken after 3 years). The other false-negative case was patient 45, whose PB relapsed sample was negative at baseline, but then reverted to positive during the subsequent follow up (“MRD reappearance”).
Additionally, 27 paired PB/plasma follow-up samples highlight the role that ctDNA might have for MYD88L265P mutation detection in pre-treated cases, as well as for MRD monitoring. Of note, 14/27 were concordantly positive and 3/27 concordantly negative, while 9/27 scored positive only in plasma and 1/27 positive on PB but not on plasma. Interestingly, the 9 plasma positive cases “rescued” by ctDNA analysis showed MYD88L265P levels in plasma comparable to those detected in paired BM samples (see Figure 5: patients 52, 50 and 37).
Discussion
ASqPCR has been widely used for MYD88L265P detection in WM, providing a higher level of sensitivity than Sanger sequencing.6,21 However, the recently developed ddPCR simplifies and improves the accuracy of MRD monitoring.24,31
This study describes a new ddPCR tool for MYD88L265P screening and MRD monitoring in WM. Our results can be summarized as follows:
ddPCR is a sensitive tool for MYD88L265P detection in WM and provides higher resolution compared to the canonical ASqPCR;
MYD88L265P ddPCR assay is particularly useful for reliable screening of low infiltrated WM specimens, such as unsorted BM or PB (routinely used in clinical practice);
both BM and PB are informative in treatment-naïve patients when tested for MYD88L265P by ddPCR; however, in relapsed patients (mainly exposed to rituximab), PB has a 1 log lower median mutated:WT ratio compared to BM, likely related to the efficacy of the anti-CD20 antibody in clearing the circulating tumor cells: therefore, BM analysis should be preferred for these patients;
the MYD88L265P ddPCR assay can be reliably used for MRD purposes, being as informative as the classical, standardized but less applicable, IGH-based MRD assay, not yet validated in WM;
the MYD88L265P ddPCR assay targeting ctDNA is very promising for identifying the mutation in less invasively collected tissues, such as plasma or urine, and in samples from pre-treated patients.
The here described MYD88L265P ddPCR assay showed an overall mutation detection rate on baseline unselected mononuclear cells samples of 95.3% in BM and 71.2% in PB. These data might help to solve the still open debate on whether to use sorted versus unsorted BM mononuclear cells to assess the MYD88L265P mutation36 and to extend the implementation of the ddPCR mutation assay to general diagnostic laboratories that do not routinely perform cell selection. At present, only 40% of MYD88L265P cases can be detected by ASqPCR using unselected PB cells, with an overall sensitivity of 39.5% and specificity of 100%.37
In our study we observed that the ddPCR assay on unselected cells greatly improved the rate of MYD88L265P detection compared to that achieved by ASqPCR. In fact, an analysis of 74 paired BM/PB baseline samples showed an overall detection rate of 93% (69/74) in BM and 72% (53/74) in PB. Among 69 patients with detectable mutations in BM samples, the sensitivity for MYD88L265P mutation detection by ddPCR on paired PB samples was 77% (53/69). The highly sensitive results of MYD88L265P ddPCR assay on unsorted samples makes this assay ideal for diagnostic use in clinical routine, avoiding the costs and technical requirements of cell sorting. In addition, our data confirmed, as also reported by Xu et al.,37 that PB samples are suboptimal for mutational screening in previously treated patients, evidence confirmed by the high rate of false-negative results. The sensitivity of our assay in PB samples dropped from 85% in treatment-naïve patients to 47% in relapsed patients. Thus, a BM sample is essential to accurately identify MYD88L265P WT status in pre-treated patients (strongly suggested, for example, before starting an expensive therapy with ibrutinib).
This study shows that, besides its potential diagnostic role, MYD88L265P can effectively and easily be used for MRD monitoring in WM, achieving similar results to the less-applicable IGH-based MRD assay. In fact, monitoring allele levels can also provide insights into treatment effectiveness in a disease whose therapeutic scenario is rapidly changing, with many new and highly effective drugs.13 Of note, a report claimed the achievement of the first “molecular remission” in WM treated with carfilzomib.38 However, the deepness of molecular response matters, as does the sensitivity of the assay used for MYD88L265P detection, which was quite modest in that case (1.00×10−3)6 compared to the newly developed flow cytometry assays39 and with the ddPCR strategy described in this manuscript.
We observed that the agreement between ddPCR and ASqPCR results was weaker in follow-up samples than in baseline ones. Further evaluations on large, prospective series of patients are needed to assess whether ddPCR could be useful for the identification of false-positive ASqPCR results, as was recently demonstrated in a next-generation sequencing/real-time quantitative PCR comparison performed in acute lymphoblastic leukemia.40 Moreover, a methodological validation against IGH-based MRD detection and multiparametric flow cytometry, as well as correlations with clinical impact, are eagerly awaited in this setting and are currently ongoing in series of external samples.
The most innovative aspect of our study is the impact that ctDNA might have on MYD88L265P mutation detection and MRD monitoring. We showed that ctDNA mirrors the BM mutational burden much better than gDNA from PB at baseline. Moreover, our data suggest that ctDNA might be able to reflect the dynamic changes in tumor burden in response to treatment, with higher sensitivity than PB, which is particularly promising in pretreated cases (Figures 3 and 4). Similar promising data have been obtained with ctDNA extracted from exosomes from plasma and urine in a pivotal series of patients (D Drandi et al., 2018, unpublished data), although further investigations, as well as technical optimization, are needed. Nevertheless, these data add to the previously published experiences on the promising role of ctDNA analysis in lymphoproliferative disorders other than WM.41–50 Indeed, ctDNA represents an alternative, less invasive and “patient-friendly” tissue source for mutational analysis, and eventually MRD, which is especially attractive for screening and monitoring asymptomatic patients, such as those with MGUS. (MYD88L265P ddPCR screening on a large group of IgM-MGUS patients is currently ongoing). However, plasma collection needs particular care since blood collection tubes and drawing procedures, as well known, can affect the stability of ctDNA.33,34 Pre-analytical standardized procedures are a prerequisite for clinical application and for consolidation of the promising potential of ctDNA analysis.
Finally, we described the impact of different therapies on MRD clearance in WM (i.e. chlorambucil, rituximab monotherapy), RCD (rituximab, cyclophosphamide, dexamethasone), FCR (fludarabine, cyclophosphamide, rituximab) and bendamustine-containing regimens (Online Supplementary Appendix). We acknowledge that the limitations of the present study include the composite and retrospective nature of the series of patients characterized, moreover, by little outcome information. One of the main goals of this manuscript was the technical description of the MYD88L265P ddPCR assay on unsorted cells. The illustration of the broad range of possible applications, including those on liquid biopsy, makes this approach potentially practical for implementation in routine diagnostic laboratories. We are aware that this assay represents exploratory research, whose real advantages and predictive values need to be further validated in the context of prospective clinical trials. For this purpose, we are currently investigating MRD assessment by ddPCR on gDNA and ctDNA in the context of a phase II prospective study of relapsed WM patients, sponsored by the Fondazione Italiana Linfomi (EudraCT number: 2013-005129-22). Moreover, further efforts to establish uniform guidelines regarding standardization procedures for sample collection and methodological validation are currently ongoing, both at national and European levels.
In conclusion, our study shows that ddPCR is a feasible and highly sensitive assay for MYD88L265P mutational screening and MRD monitoring in WM, particularly in samples harboring low concentrations of circulating tumor cells. For this reason, plasma ctDNA represents a promising tissue source, and might be an attractive, less invasive alternative to BM for MYD88L265P detection, also beyond the scenario of WM.
Supplementary Material
Acknowledgments
The authors would like to thank all the patients who participated in the study. We are grateful to Luca Arcaini, Alfredo Benso, Stefano Di Carlo, Angelo Fama, Paola Ghione, Idanna Innocenti, Luca Laurenti, Giacomo Loseto, Gianfranco Politano, Marzia Varettoni and Silvia Zibellini for their scientific advice and to Antonella Fiorillo and Sonia Perticone for administrative support.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/103/6/1029
This work was supported by: Progetto di Rilevante Interesse Nazionale (PRIN2009) from Ministero Italiano dell’Università e della Ricerca (MIUR), Roma, Italy [7.07.02.60 AE01]; Progetto di Ricerca Sanitaria Finalizzata 2009 [RF-2009-1469205] and 2010 [RF-2010-2307262 to S.C.], A.O.S. Maurizio, Bolzano/Bozen, Italy; Fondi di Ricerca Locale, Università degli Studi di Torino, Italy; Fondazione Neoplasie Del Sangue (Fo.Ne.Sa), Torino, Italy; Associazione Da Rosa, Torino, Italy; Comitato Regionale Piemontese (Gigi Ghirotti),Italy and Ricerca biomedica condotta da giovani ricercatori - Fondazione Cariplo (project codes: 2016-0476), Dr. Jiménez was supported by a grant from the Sociedad Espanola de Hematologia y Hemoterapia (SEHH) 2016.
References
- 1.Treon SP, Xu L, Yang G, et al. MYD88 L265P somatic mutation in Waldenström’s macroglobulinemia. N Engl J Med. 2012;367(9):826–833. [DOI] [PubMed] [Google Scholar]
- 2.Yang G, Zhou Y, Liu X, et al. A mutation in MYD88 (L265P) supports the survival of lymphoplasmacytic cells by activation of Bruton tyrosine kinase in Waldenström macroglobulinemia. Blood. 2013;122(7): 1222–1232. [DOI] [PubMed] [Google Scholar]
- 3.Campo E, Swerdlow SH, Harris NL, Pileri S, Stein H, Jaffe ES. The 2008 WHO classification of lymphoid neoplasms and beyond: evolving concepts and practical applications. Blood. 2011;117(19):5019–5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiménez C, Sebastián E, Chillón MC, et al. MYD88 L265P is a marker highly characteristic of, but not restricted to, Waldenström’s macroglobulinemia. Leukemia. 2013;27(8): 1722–1728. [DOI] [PubMed] [Google Scholar]
- 5.Poulain S, Roumier C, Decambron A, et al. MYD88 L265P mutation in Waldenstrom macroglobulinemia. Blood. 2013;121(22): 4504–4511. [DOI] [PubMed] [Google Scholar]
- 6.Xu L, Hunter ZR, Yang G, et al. MYD88 L265P in Waldenstrom macroglobulinemia, immunoglobulin M monoclonal gammopathy, and other B-cell lymphoproliferative disorders using conventional and quantitative allele-specific polymerase chain reaction. Blood. 2013;121(11):2051–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ngo VN, Young RM, Schmitz R, et al. Oncogenically active MYD88 mutations in human lymphoma. Nature. 2011;470(7332):115–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Varettoni M, Arcaini L, Zibellini S, et al. Prevalence and clinical significance of the MYD88 (L265P) somatic mutation in Waldenstrom’s macroglobulinemia and related lymphoid neoplasms. Blood. 2013;121(13):2522–2528. [DOI] [PubMed] [Google Scholar]
- 9.Swerdlow SH, Campo E, Pileri SA, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127(20):2375–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Treon SP, Cao Y, Xu L, Yang G, Liu X, Hunter ZR. Somatic mutations in MYD88 and CXCR4 are determinants of clinical presentation and overall survival in Waldenstrom macroglobulinemia. Blood. 2014;123(18):2791–2796. [DOI] [PubMed] [Google Scholar]
- 11.Treon SP. How I treat Waldenström macroglobulinemia. Blood. 2009;114(12): 2375–2385. [DOI] [PubMed] [Google Scholar]
- 12.Treon SP, Xu L, Hunter Z. MYD88 Mutations and response to ibrutinib in Waldenström’s macroglobulinemia. N Engl J Med. 2015;373(6):584–586. [DOI] [PubMed] [Google Scholar]
- 13.Leblond V, Kastritis E, Advani R, et al. Treatment recommendations from the Eighth International Workshop on Waldenström’s Macroglobulinemia. Blood. 2016;128(10):1321–1328. [DOI] [PubMed] [Google Scholar]
- 14.Castillo JJ, Hunter ZR, Yang G, Argyropoulos K, Palomba ML, Treon SP. Future therapeutic options for patients with Waldenström macroglobulinemia. Best Pract Res Clin Haematol. 2016;29(2):206–215. [DOI] [PubMed] [Google Scholar]
- 15.Abeykoon JP, Yanamandra U, Kapoor P. New developments in the management of Waldenström macroglobulinemia. Cancer Manag Res. 2017;973–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hunter ZR, Yang G, Xu L, Liu X, Castillo JJ, Treon SP. Genomics, signaling, and Treatment of Waldenström macroglobulinemia. J Clin Oncol. 2017;35(9): 994–1001. [DOI] [PubMed] [Google Scholar]
- 17.Tedeschi A, Picardi P, Ferrero S, et al. Bendamustine and rituximab combination is safe and effective as salvage regimen in Waldenström macroglobulinemia. Leuk Lymphoma. 2015;56(9):2637–2642. [DOI] [PubMed] [Google Scholar]
- 18.Kyle RA. Long-term follow-up of IgM monoclonal gammopathy of undetermined significance. Blood. 2003;102(10): 3759–3764. [DOI] [PubMed] [Google Scholar]
- 19.Landgren O, Staudt L. MYD88 L265P somatic mutation in IgM MGUS. N Engl J Med. 2012;367(23):2255–2257. [DOI] [PubMed] [Google Scholar]
- 20.Varettoni M, Zibellini S, Arcaini L, et al. MYD88 (L265P) mutation is an independent risk factor for progression in patients with IgM monoclonal gammopathy of undetermined significance. Blood. 2013; 122(13):2284–2285. [DOI] [PubMed] [Google Scholar]
- 21.Jiménez C, Chillón Mdel C, Balanzategui A, et al. Detection of MYD88 L265P mutation by real-time sllele-specific oligonucleotide polymerase chain reaction. Appl Immunohistochem Mol Morphol. 2014;22(10):768–773. [DOI] [PubMed] [Google Scholar]
- 22.Gustine JN, Meid K, Hunter ZR, Xu L, Treon SP, Castillo JJ. MYD88 mutations can be used to identify malignant pleural effusions in Waldenström macroglobulinaemia. Br J Haematol. 2018;180(4):578–581. [DOI] [PubMed] [Google Scholar]
- 23.Komatsubara KM, Sacher AG. Circulating tumor DNA as a liquid biopsy: current clinical applications and future directions. Oncology. 2017;31(8):618–627. [PubMed] [Google Scholar]
- 24.Hindson BJ, Ness KD, Masquelier DA, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011;83(22):8604–8610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huggett JF, Whale A. Digital PCR as a novel technology and its potential implications for molecular diagnostics. Clin Chem. 2013;59(12):1691–1693. [DOI] [PubMed] [Google Scholar]
- 26.Ladetto M, Donovan JW, Harig S, et al. Real-Time polymerase chain reaction of immunoglobulin rearrangements for quantitative evaluation of minimal residual disease in multiple myeloma. Biol Blood Marrow Transplant. 2000;6(3):241–253. [DOI] [PubMed] [Google Scholar]
- 27.van der Velden VHJ, Cazzaniga G, Schrauder A, et al. Analysis of minimal residual disease by Ig/TCR gene rearrangements: guidelines for interpretation of real-time quantitative PCR data. Leukemia. 2007;21(4):604–611. [DOI] [PubMed] [Google Scholar]
- 28.Fleiss JL, Levin B, Paik MC. Statistical Methods for Rates and Proportions. Hoboken, NJ, USA: John Wiley & Sons, Inc.; 2003. [Google Scholar]
- 29.Uchiyama Y, Nakashima M, Watanabe S, et al. Ultra–sensitive droplet digital PCR for detecting a low–prevalence somatic GNAQ mutation in Sturge–Weber syndrome. Sci Rep. 2016;6(1):22985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hunter ZR, Xu L, Yang G, et al. The genomic landscape of Waldenstrom macroglobulinemia is characterized by highly recurring MYD88 and WHIM-like CXCR4 mutations, and small somatic deletions associated with B-cell lymphomagenesis. Blood. 2014;123(11):1637–1646. [DOI] [PubMed] [Google Scholar]
- 31.Drandi D, Kubiczkova-Besse L, Ferrero S, et al. Minimal residual disease detection by droplet digital PCR in multiple myeloma, mantle cell lymphoma, and follicular lymphoma. J Mol Diagnostics. 2015;17(6):652–660. [DOI] [PubMed] [Google Scholar]
- 32.Petrikkos L, Kyrtsonis M-C, Roumelioti M, et al. Clonotypic analysis of immunoglobulin heavy chain sequences in patients with Waldenström’s macroglobulinemia: correlation with MYD88 L265P somatic mutation status, clinical features, and outcome. Biomed Res Int. 2014;2014:809103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kang Q, Henry NL, Paoletti C, et al. Comparative analysis of circulating tumor DNA stability In K3EDTA, Streck, and CellSave blood collection tubes. Clin Biochem. 2016;49(18):1354–1360. [DOI] [PubMed] [Google Scholar]
- 34.El Messaoudi S, Rolet F, Mouliere F, Thierry AR. Circulating cell-free DNA: Preanalytical considerations. Clin Chim Acta. 2013;424: 222–230. [DOI] [PubMed] [Google Scholar]
- 35.Devonshire AS, Whale AS, Gutteridge A, et al. Towards standardisation of cell-free DNA measurement in plasma: controls for extraction efficiency, fragment size bias and quantification. Anal Bioanal Chem. 2014;406(26):6499–6512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gustine J, Meid K, Xu L, Hunter ZR, Castillo JJ, Treon SP. To select or not to select? The role of B-cell selection in determining the MYD88 mutation status in Waldenström macroglobulinaemia. Br J Haematol. 2017;176(5):822–824. [DOI] [PubMed] [Google Scholar]
- 37.Xu L, Hunter ZR, Yang G, et al. Detection of MYD88 L265P in peripheral blood of patients with Waldenström’s Macroglobulinemia and IgM monoclonal gammopathy of undetermined significance. Leukemia. 2014;28(8):1698–1704. [DOI] [PubMed] [Google Scholar]
- 38.Treon SP, Tripsas CK, Meid K, et al. Carfilzomib, rituximab, and dexamethasone (CaRD) treatment offers a neuropathy-sparing approach for treating Waldenström’s macroglobulinemia. Blood. 2014;124(4):503–510. [DOI] [PubMed] [Google Scholar]
- 39.Paiva B, Montes MC, García-Sanz R, et al. Multiparameter flow cytometry for the identification of the Waldenström’s clone in IgM-MGUS and Waldenström’s Macroglobulinemia: new criteria for differential diagnosis and risk stratification. Leukemia. 2014;28(1):166–173. [DOI] [PubMed] [Google Scholar]
- 40.Kotrova M, van der Velden VHJ, van Dongen JJM, et al. Next-generation sequencing indicates false-positive MRD results and better predicts prognosis after SCT in patients with childhood ALL. Bone Marrow Transplant. 2017;52(7):962–968. [DOI] [PubMed] [Google Scholar]
- 41.Kurtz DM, Green MR, Bratman SV, et al. Noninvasive monitoring of diffuse large B-cell lymphoma by immunoglobulin high-throughput sequencing. Blood. 2015;125(24):3679–3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roschewski M, Dunleavy K, Pittaluga S, et al. Circulating tumour DNA and CT monitoring in patients with untreated diffuse large B-cell lymphoma: a correlative bio-marker study. Lancet Oncol. 2015;16(5): 541–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rossi D, Diop F, Spaccarotella E, et al. Diffuse large B-cell lymphoma genotyping on the liquid biopsy. Blood. 2017;129(14): 1947–1957. [DOI] [PubMed] [Google Scholar]
- 44.Rustad EH, Coward E, Skytøen ER, et al. Monitoring multiple myeloma by quantification of recurrent mutations in serum. Haematologica. 2017;102(7):1266–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sarkozy C, Huet S, Carlton VEH, et al. The prognostic value of clonal heterogeneity and quantitative assessment of plasma circulating clonal IG-VDJ sequences at diagnosis in patients with follicular lymphoma. Oncotarget. 2017;8(5):8765–8774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bruscaggin A, Spina V, Di Trani M, et al. Genotyping of classical Hodgkin lymphoma on the liquid biopsy. Hematol Oncol. 2017;3564–3565. [Google Scholar]
- 47.Camus V, Sarafan-Vasseur N, Bohers E, et al. Digital PCR for quantification of recurrent and potentially actionable somatic mutations in circulating free DNA from patients with diffuse large B-cell lymphoma. Leuk Lymphoma. 2016;57(9): 2171–2179. [DOI] [PubMed] [Google Scholar]
- 48.Alcaide M, Yu S, Bushell K, et al. Multiplex doplet digital PCR quantification of recurrent somatic mutations in diffuse large B-cell and follicular lymphoma. Clin Chem. 2016;62(9):1238–1247. [DOI] [PubMed] [Google Scholar]
- 49.Hattori K, Sakata-Yanagimoto M, Suehara Y, et al. Clinical significance of disease-specific MYD88 mutations in circulating DNA in primary central nervous system lymphoma. Cancer Sci. 2018;109(1):225–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hiemcke-Jiwa LS, Minnema MC, Radersma-van Loon JH, et al. The use of droplet digital PCR in liquid biopsies: a highly sensitive technique for MYD88 p.(L265P) detection in cerebrospinal fluid. Hematol Oncol. 6 December 2017. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.