Abstract
Diamond-Blackfan anemia (DBA) is a rare inherited bone marrow failure disorder linked predominantly to ribosomal protein gene mutations. Here the European DBA consortium reports novel mutations identified in the RPL15 gene in 6 unrelated individuals diagnosed with DBA. Although point mutations have not been previously reported for RPL15, we identified 4 individuals with truncating mutations p.Tyr81* (in 3 of 4) and p.Gln29*, and 2 with missense variants p.Leu10Pro and p.Lys153Thr. Notably, 75% (3 of 4) of truncating mutation carriers manifested with severe hydrops fetalis and required intrauterine transfusions. Even more remarkable is the observation that the 3 carriers of p.Tyr81* mutation became treatment-independent between four and 16 months of life and maintained normal blood counts until their last follow up. Genetic reversion at the DNA level as a potential mechanism of remission was not observed in our patients. In vitro studies revealed that cells carrying RPL15 mutations have pre-rRNA processing defects, reduced 60S ribosomal subunit formation, and severe proliferation defects. Red cell culture assays of RPL15-mutated primary erythroblast cells also showed a severe reduction in cell proliferation, delayed erythroid differentiation, elevated TP53 activity, and increased apoptosis. This study identifies a novel subgroup of DBA with mutations in the RPL15 gene with an unexpected high rate of hydrops fetalis and spontaneous, long-lasting remission.
Introduction
Diamond-Blackfan anemia (DBA) (OMIM# 105650) is an inherited bone marrow failure disorder that typically manifests in children under the age of one year. While the central phenotype is pure red cell aplasia, developmental delay and a number of physical malformations are also linked to DBA.1 These include (but are not limited to) craniofacial malformations, growth retardation, abnormalities in the extremities, heart defects, as well as urogenital defects.2,3 While most patients (>90%) are diagnosed within the first year of life, a minority present with anemia at birth. Hydrops fetalis due to severe intrauterine anemia is considered a very rare manifestation of DBA.4 To date, a total of 10 cases of DBA-associated hydrops have been reported in single cases.5–13 The clinical outcome of these patients was poor as compared to typical DBA (3 patients died perinatally, 4 patients were steroid unresponsive, 2 patients required steroid therapy, and 1 had unknown outcome).
Almost all of the mutations linked to DBA have been found in genes coding for ribosomal proteins (RPs).14 These RPs include: eS7 (RPS7), uS8 (RPS15A), eS10 (RPS10), eS17 (RPS17), eS19 (RPS19), eS24 (RPS24), eS26 (RPS26), eS27 (RPS27), eS28 (RPS28), uS14 (RPS29), uL18 (RPL5), uL5 (RPL11), eL15 (RPL15), eL18 (RPL18), uL24 (RPL26), eL27 (RPL27), eL31 (RPL31), uL29 (RPL35) and eL33 (RPL35A).15–28 The RPL15 gene has so far been reported in one patient who carried a large monoallelic microdeletion involving this gene.23 Non-RP genes linked to DBA, albeit very rarely involved, are TSR2 and GATA1.26,29 RP gene aberrations lead to haploinsufficiency, and in many cases result in reduction in the respective RP which impairs ribosome biogenesis, affecting both the processing pathways of pre-rRNA maturation and the assembly of the large or small ribosomal subunit.30,31 TP53 is a tumor suppressor protein and transcription factor that stabilizes in response to cell stress, such as DNA damage or nucleolar stress induced by ribosome biogenesis defects.32,33 Depending on the level of stress, stabilized TP53 will induce cell arrest, DNA repair, senescence and/or apoptosis. This pathway has been prominently implicated in the pathogenesis of DBA, with a number of studies suggesting that TP53 stabilization lies at the heart of the loss of erythroid progenitor cells in DBA bone marrow.34–36 In addition to activating apoptosis, DBA-linked RP gene mutations can impair cellular differentiation, alter the landscape of mRNAs on ribosomes, and induce autophagy.36–38
Therapy in DBA depends on the severity of anemia and on the response to oral glucocorticosteroids (GCS). GCS-non-responders receive chronic blood transfusions or can undergo hematopoietic stem cell transplantation (HSCT) which is often delayed because patients can achieve treatment independence.39 For unknown reasons, 20% of all DBA cases can become independent of steroid treatment and/or blood transfusions by the age of 25 years.39,40 To date, however, there are no predictive genetic markers that could improve decision making for timely HSCT. Given the significant risks that are associated with HSCT,41 especially if a matched sibling donor is not available, such predictive markers would be very valuable.
Here we describe 6 DBA cases associated with mutations in ribosomal protein gene RPL15. These patients display an unexpectedly high rate of hydrops fetalis and treatment independence, pointing to a new genotype-phenotype correlation in DBA which could be important for risk stratification.
Methods
Patients
Diagnosis of DBA was made based on typical features including aregenerative anemia with erythroid hypoplasia.1 Written informed consent was obtained from patients and/or parents prior to inclusion in this study, which was performed in accordance with the ethical standards of the Declaration of Helsinki. The study was approved by the institutional ethics committee (University of Freiburg, IRB n. 205/99 and n. 351/17).
Cell culture
Lymphoblastoid cell lines (LCLs) were derived from Epstein-Barr virus (EBV)-immortalization of peripheral mononuclear cells isolated from whole blood using Ficoll (GE Life Sciences) and grown in RPMI (Gibco) containing 10% fetal calf serum (FCS), 1% L-glutamine, and 1% penicillin/streptomycin, as previously described.42
DNA sequencing and bioinformatics
Targeted Sanger sequencing was performed as previously reported.43 Primer sequences are available upon request. Pathogenicity of mutations, evolutionary conservation across species, and the physicochemical difference between amino acids were evaluated using standard prediction tools as outlined in Online Supplementary Table S1.
Details of pre-rRNA processing analysis, Northern blots, polysome profiling, measurement of growth rate and de novo protein synthesis are available in the Online Supplementary Methods.
Erythroid red cell culture assays
Erythroid red cell culture assays were performed as previously described.36,44 Antibodies and stains for FACS analysis were PC7 or PE conjugated CD34 (Beckman coulter, Brea, CA, USA), APC conjugated CD36 (BD Biosciences San Jose, CA, USA), PE conjugated α4 integrin (Miltenyi, Paris, France), APC conjugated Band 3 (kindly provided by Mohandas Narla’s lab, NYBC, New York, USA), PE/Cy7 conjugated IL-3R (Miltennyi, Paris, France), PE/Cy7 (PE)-coupled GPA (Life Technologies Carlsbad, CA, USA), and DAPI (Sigma). FACS analysis was conducted on a BD Biosciences Influx flow cytometer (BD Biosciences San Jose, CA, USA). Data were analyzed using Kaluza software (Beckman Coulter, Brea, CA, USA). Antibodies for western blotting were TP53 (Sigma #5816), phospho-TP53 (Ser15, Cell Signaling), p21 (Cell Signaling #2947), actin (Sigma #Ac-15), and eL15 (Abcam #ab130992). Primers for real-time PCR used are as follows: TaqMhupRPL15F: CAGCCATCAGGTAAGCCAAGA; TaqMhuRPL15R: CAGCGGACCCTCAGAAGAAA; TaqMhupp21F: TTGCTGCCGCATGGG, TaqMhup21R: CCTTGTGGAGCCGGAGCT; TaqMhupactinF: CTGGAACGGTGAAGGTGACA; TaqMhupactinR: AAGGGACTTCCTGTAACAACGCA; TGTAGTGGATGGTGGTACAGTCAGA; TaqMB2MhF: GCGGCATCTTCAAACCTCC; TaqMB2MhR: TGACTTTGTCACAGCCCAAGATA. Real-time PCR was performed using the ABI 7900 Real Time PCR system and Taqman PCR mastermix (Life Technologies Carlsbad, CA, USA). Quantification of gene amplification was performed in duplicate using ΔΔCt method. The expression level of each gene was normalized using the housekeeping genes: actin, and β2 microglobulin.
Results
Identification of novel RPL15 mutations in 6 patients within EuroDBA registries
Approximately 30% of all registered DBA patients who have been tested for mutations in the most common DBA-linked genes (RPS19, RPL5, RPL11, RPS10, RPS26, RPS7, RPS17, RPS24, and RPL35a) still do not have an established genotype. Because a whole gene deletion of RPL15 has been reported before in one DBA patient but not in our cohorts,23 we used targeted Sanger sequencing of RPL15 to determine if mutations in this gene could be driving disease in patients without an established genotype. The national patient registries from EuroDBA partners in Germany, France, Italy and Israel were included in this study. As of November 2017, these cohorts represent a total of 985 patients. A complete description of the history and composition of the EuroDBA consortium has been recently published.45 Study outline and screening strategy are illustrated in Online Supplementary Figure S1.
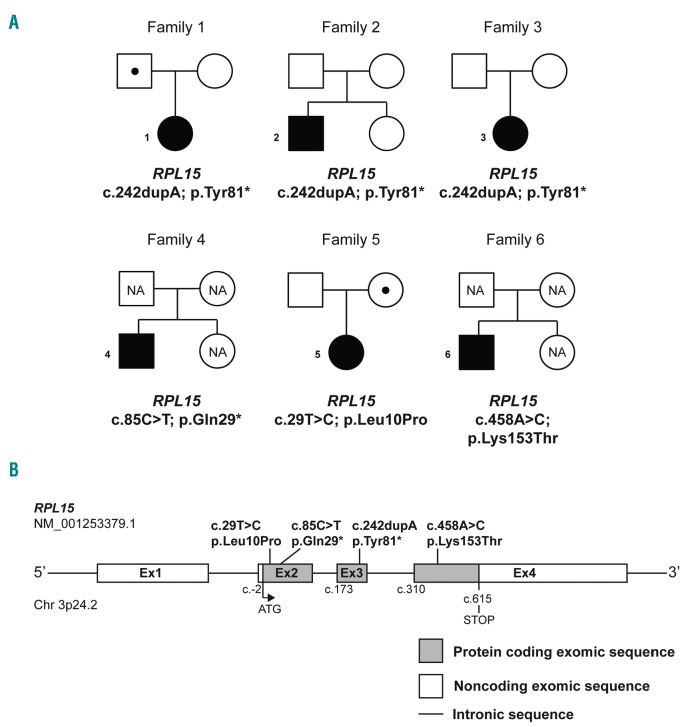
Four unrelated DBA patients from the German, and one each from the French and Israeli registries were identified carrying mutations in the RPL15 gene (NM_001253379.1), which encodes ribosomal protein eL15/RPL15 (Figure 1A and B). Out of the 6 RPL15-mutated patients, 3 unrelated patients (P1-3) carried the same novel indel mutation c.242dupA altering the tyrosine at position 81 to a stop codon: p.Tyr81* (Table 1 and Online Supplementary Table S1). Patient 4 (P4) carried another stop-gain mutation in RPL15 resulting in an even earlier protein truncation: c.85C>T; p.Gln29*. The Exome Aggregation Consortium (ExAC) reports that RPL15 is very intolerant to loss-of-function mutations with no reported cases present in over 60,000 individuals (pLI score =0.96), strongly suggesting the novel mutations p.Gln29* and p.Tyr81* found in our patients are highly deleterious (Online Supplementary Table S1). In addition, the analysis of family trios revealed that hotspot mutations arose de novo in two families (Table 1 and Figure 1A), while one patient (P1/DE071) inherited the mutation from her father who had been categorized as a DBA silent carrier due to very high erythrocyte adenosine deaminase (eADA) levels. The remaining two point mutations c.29T>C; p.Leu10Pro (P5) and c.458A>C; p.Lys153Thr (P6) affect highly conserved residues and, based on results from in silico prediction, are probably deleterious (Online Supplementary Table S1). Biomuta, DMDM, the Exac/GnomAD databases, and NCBI do not report any variants in the codon for Leu10. The GnomAD population database reports one variant in Lys153 (Lys153Arg; rs370700905) identified in 33 out of 232840 total alleles.
Figure 1.
Mutations in RPL15 are identified in patients with Diamond-Blackfan anemia (DBA). (A) Six unrelated pedigrees of individuals affected by DBA associated with RPL15 mutations. All families have one DBA-affected individual who is also a mutation carrier, as indicated with filled squares (male) or circles (female). Unaffected individuals are indicated by unfilled symbols. Unaffected mutation carriers are denoted by a dot symbol (●). NA: unaffected family members who were not investigated for the presence of mutations. Families 1–4 harbor heterozygous stopgain mutations in RPL15; families 5–6 carry heterozygous missense RPL15 mutations. (B) Schematic representation of human RPL15 depicting localization of the mutations identified in families 1–6.
Table 1.
Clinical characteristics of patients with RPL15 mutations.
Genotype-phenotype association for truncating RPL15 mutations: severe hematologic phenotype and rapid acquisition of treatment independence
All of the individuals with mutations in RPL15 presented with typical bone marrow erythroid hypoplasia, elevated eADA, and most of them presented with increased fetal hemoglobin (HbF) levels (Table 1). Notably, hydrops fetalis (considered the most severe hematologic phenotype of DBA) was associated only with truncating RPL15 mutations. The affected fetuses P2-4 required between four and nine intrauterine transfusions (Table 1). One individual (P2) with the p.Tyr81* hotspot mutation presented with various physical malformations, while the dysmorphic features in other patients were less severe (Table 1). Unexpectedly, all 3 patients carrying p.Tyr81* substitution (P1-3) attained a rapid treatment independence both with and without steroid treatment (Figure 2A), while the fourth patient with the RPL15 mutation c.85C>T; p.Gln29* responded to steroids; however, the therapy was discontinued due to overt toxicity.
Figure 2.
Longitudinal mutations in RPL15 are identified in individuals diagnosed with Diamond-Blackfan anemia (DBA). (A) Clinical evolution of P1-P3 carrying truncating hotspot mutation RPL15 p.Tyr81*. Patient 1 manifested with DBA after birth and after one transfusion achieved spontaneous remission at the age of six months. A relapse occurred five years later and after a short course of steroids the patient attained treatment independence. Patients 2 and 3 had a similar clinical course with hydrops fetalis and prenatal intrauterine transfusions, and achieved treatment independence either spontaneously or after one course of steroids. (B) Sanger sequencing results of the recurring mutation in RPL15 from initial diagnoses as well as post remission. Arrows indicate the inserted A nucleotide. FUP: follow up; BM: bone marrow; PB: peripheral blood; LCL: lymphoblastoid cell lines.
Based on published observations of genetic revertant mosaicism as a “repair mechanism” in other bone marrow failure syndromes,46 we speculated that the hematologic remission in our patients was attained via genetic reversion (e.g. uniparental disomy). Sequencing of DNA in P1-3 either at multiple time points or after the acquisition of treatment independence did not reveal any changes in mutation load (Figure 2B).
Mutations in RPL15 recapitulate specific pre-rRNA processing defects found in RP depleted cells
Mutations in DBA-linked RP genes result in haploinsufficiency of the encoded RP. Since the majority of RPs incorporate into pre-ribosomal particles in a progressive manner concurrently with pre-rRNA maturation, a lack of one RP impairs pre-rRNA processing in an explicit way that is reproducible in most cell types.47 Online Supplementary Figure S2 illustrates the normal pre-rRNA processing pathway that begins with a single transcribed strand of 47S pre-rRNA that then undergoes a complex series of cleavages and trimmings to ultimately result in the mature 18S, 28S, and 5.8S strands of rRNA found in ribosomes. To investigate the functional consequences of the newly identified RP gene mutations reported here, EBV-immortalized lymphoblast cell lines (LCLs) were generated and the pre-rRNA processing of these LCLs was compared to HeLa cells depleted of eL15.
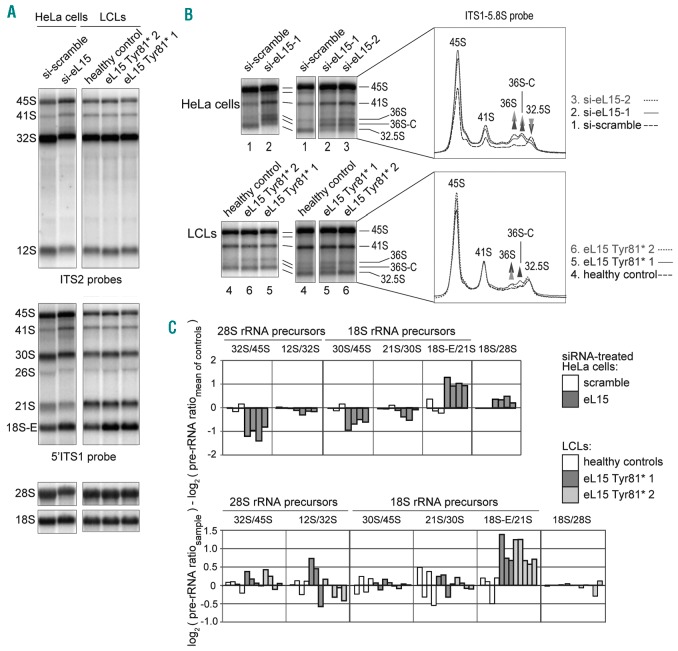
As previously reported,23 depletion of eL15 in HeLa cells resulted in a decrease in 32S and 12S pre-rRNAs (Figure 3A and C). This was accompanied by the accumulation of the 36S and 36S-C precursors, which are inconspicuous in normal cells, and a concomitant drop in 32.5S pre-rRNA (Figure 3B). In addition, these cells displayed lower levels of 30S pre-rRNA and higher levels of 41S and 18S-E pre-rRNAs (Figure 3A and C). This phenotype indicates that depletion of eL15 affects cleavage of the ITS1 at site 2, which promotes direct cleavage of early precursors at site E and formation of 18S-E and 36S pre-rRNAs. The 36S precursor is then trimmed by the 5’-3’ exoRNase XRN2 to produce the 36S-C and 32.5S pre-rRNAs.48 Sucrose gradient analyses confirmed the efficiency of the eL15 siRNAs in reducing the free 60S subunit peak and inducing the formation of half-mers, as expected from the deficiency of an RPL protein (Online Supplementary Figure S3). Consistent with eL15 partial loss-of-function, an increase of 36S and 36S-C precursors was observed in the patient LCLs carrying the eL15 p.Tyr81* variant (Figure 3B). In addition, both cell lines displayed a marked increase of the amount of 18S-E precursors translating into a higher 18S-E/21S ratio, similar to eL15-depleted cells (Figure 3A and C). This phenotype is similar to that previously observed in LCLs harboring a large heterozygous deletion in RPL15.23
Figure 3.
Mutations in RPL15 recapitulate specific pre-rRNA processing defects found in eL15 depleted cells. (A) Northern blot analysis of siRNA-treated HeLa cells or lymphoblastoid cell lines (LCLs) derived from individuals with Diamond-Blackfan anemia (DBA). Radiolabeled probes against ITS2 (top panel), ITS1 (middle panel), 18S or 28S (lower panel) rRNA sequences were used to blot 3μg total RNA isolated from cells. (B) Longer exposures of upper molecular weight pre-rRNA species observed in the northern blots from (A). Intensity profiles of the lanes is shown in the right-hand boxes. (C) Quantification of rRNA precursors in siRNA-treated HeLa cells (top) or LCLs (bottom) derived from individuals with DBA. The results of single experiments performed for each sample are displayed as multiple bars. Pre-rRNA ratios are normalized by dividing by the mean of the control samples.
RPL15 mutations impair 60S ribosomal subunit formation, cell proliferation, and de novo protein synthesis
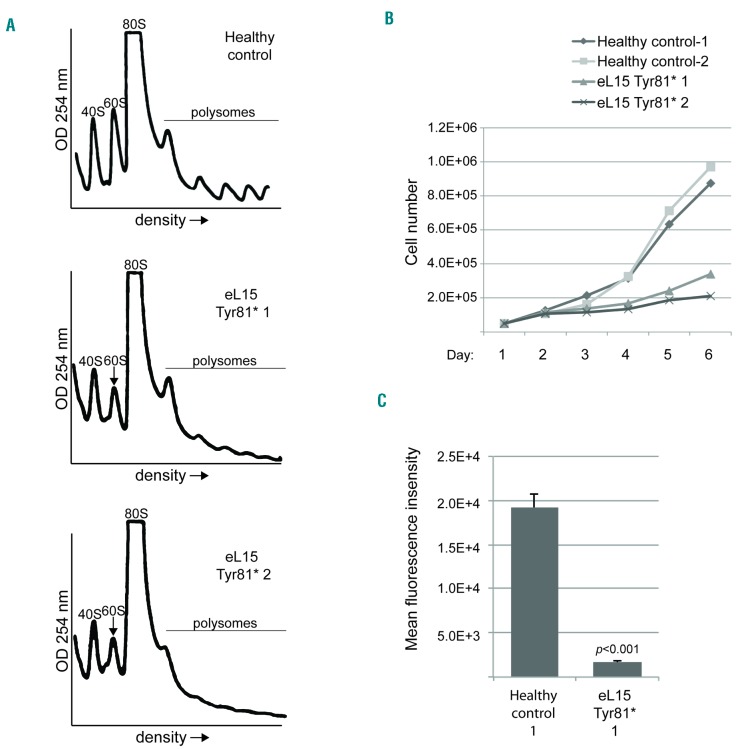
Defective processing of pre-rRNA in cells often results in cells that are unable to fully form ribosomal subunits. As such, pathogenic variants of RPs linked to DBA very often impair biogenesis of ribosomal subunits in LCLs.37 Polysome profiling was performed to determine if the observed pre-rRNA biogenesis defects resulted in impaired biogenesis of large ribosomal subunits. Figure 4A shows polysome profiles of LCL extracts derived from a healthy individual or patients carrying the p.Tyr81* variant in eL15. As expected, the LCLs from the healthy individual reveal an equivalent ratio of 40S to 60S peaks, while the LCLs derived from patients reveal a substantial reduction of 60S peaks compared to 40S peaks. These results suggest the pre-rRNA defects depicted in Figure 3 go on to impair biogenesis of large 60S ribosomal subunits.
Figure 4.
RPL15 mutations impair cell proliferation, de novo protein synthesis, and 60S ribosomal subunit formation. (A) Representative (n=3) polysome profiles of lymphoblastoid cell line (LCL) extracts derived from healthy individuals or individuals with Diamond-Blackfan anemia (DBA) carrying the RPL15 c.242dupA mutation. The 40S small subunit, 60S subunit, 80S monosome, and polysomes are labeled. Arrows point to the reduced 60S peaks in cells with RPL15 mutations. (B) Growth curve of LCLs derived from individuals with DBA or healthy controls over six days. Standard Deviations for healthy control-1 cells on days 1-5 are 2.6e4, 1.5e4, 3.8e4, 6.9e4, 1.0e5; healthy control-2 cells are 8.8e3, 3.0e4, 2.9e4, 7.5e4, 8.5e4; RPL15 c.242dupA-1 are 2.1e4, 1.9e4, 1.6e4, 2.3e4, 2.2e4; and RPL15 c.242dupA-2 are 9.5e3, 1.7e4, 1.7e4, 2.2e4, 2.5e4. (C) Measurement of the amount of de novo protein synthesis in 30 minutes in LCLs derived from healthy individuals or a DBA patient carrying the eL15 Tyr81* variant using Click-iT® analysis.
The impairment of ribosome biogenesis in cells can slow the rate of protein synthesis and cell proliferation. RP gene mutations linked to DBA are reported to slow the rate of cell proliferation and increase apoptosis.36 In agreement, we found that LCLs carrying the eL15 p.Tyr81* variant proliferated far more slowly than healthy control LCLs (Figure 4B). Moreover, measurement of de novo protein synthesis by Click-iT® labeling analysis revealed a severe reduction in the synthesis rate of LCLs carrying the eL15 variant (Figure 4C and Online Supplementary Figure S4).
Erythroid cell culture assays of primary RPL15 c.242dupA cells reveal severe proliferation defects, differentiation delays, and TP53-related apoptosis
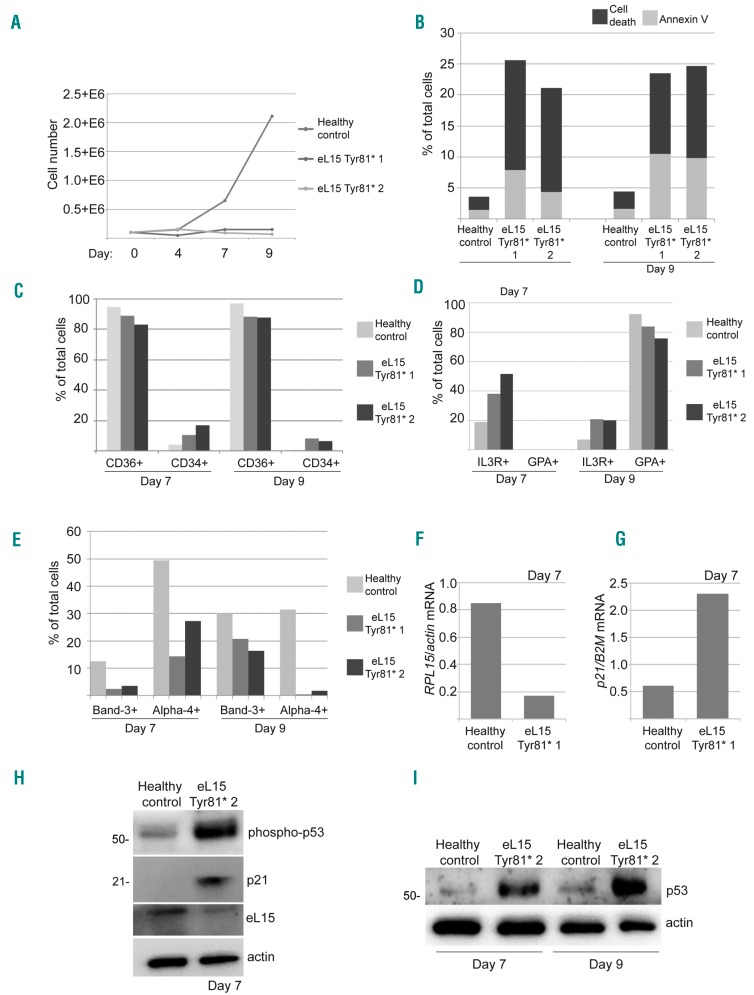
Erythroid cell culture assays show that hematopoietic progenitor cells can reveal reduced proliferation rates, delayed differentiation, and increased TP53-induced apoptosis in a manner that is largely dependent on which RP gene is haploinsufficient.36 We performed these assays in CD34+ cells isolated from bone marrow mononuclear cells (BM-MNC) of 2 patients with truncating RPL15 mutations p.Tyr81*. Compared to healthy control, patients BM-MNC showed a higher rate of cell death and apoptosis (Figures 5B and Online Supplementary Figure S5). FACS analysis revealed CD36 downregulation and CD34 upregulation on both day 7 and day 9 in the Tyr81* mutant cells as compared to healthy controls (Figures 5C and Online Supplementary Figure S5). In addition, Tyr81* mutant cells retain a higher expression of the IL3 receptor that is normally down-regulated in the erythroid lineage, and express less erythroid-specific glycophorin A (GPA), Band-3, or alpha-4 integrin compared to healthy control cells (Figure 5D and E and Online Supplementary Figure S5). Additionally, expression of RPL15 mRNA was lower (Figure 5F) and of p21 mRNA higher in Tyr81* mutant cells (Figure 5G). The observed p21 overexpression is likely TP53-mediated, as supported by western blot analysis confirming an increase in TP53 phosphorylation and increased p21 protein expression (Figure 5H). Finally, TP53 protein stabilization was observed in Tyr81* mutant patient cells (Figure 5I).
Figure 5.
Erythroid cell culture assays of primary RPL15 c.242dupA erythroid progenitor and precursor cells reveal severe erythroid proliferation defects, differentiation delays, and TP53-related apoptosis. (A) Proliferation curve of erythroid cells isolated from CD34+ cells from peripheral blood of 2 individuals with Diamond-Blackfan anemia (DBA) and an RPL15 mutation or a healthy control over nine days in liquid culture medium. (B) FACS analysis results of the percent of dead cells or apoptotic cells staining positive for Annexin V on days 7, 10, and 13 after plating in red cell culture medium. (C) FACS analysis results on days 7 and 9 of the percent of cells staining positive for CD36 and CD34. (D) FACS analysis results on days 7 and 9 of cells staining positive for IL3R and GPA. (E) FACS analysis results on days 7 and 9 of cells staining positive for Alpha-4 and Band-3. (F) Real-time PCR results of the ratio between RPL15 mRNA and actin mRNA in cells. (G) Real-time PCR results of the ratio between p21 mRNA and actin mRNA in cells. (H) Western blot analysis of cells from a healthy control or an individual with DBA and a mutation in RPL15 using antibodies against phosphorylated TP53, p21, eL15, or actin on day 7. (I) Western blot analysis of cells from a healthy control or an RPL15 with DBA and a mutation in RPL15 using antibodies against TP53 or actin on days 7 and 9.
Discussion
At present there are 19 RP genes associated with DBA, representing almost one-quarter of the 80 cytoplasmic RPs in human cells. Here, we describe a novel genetic subgroup of DBA due to mutations in the RPL15 gene and identify the truncating mutation c.242dupA; p.Tyr81* as a recurrent genetic cause in 3 unrelated patients. The experimental results show that LCLs carrying the p.Tyr81* variant reveal an array of molecular defects typically associated with a ribosomopathy phenotype. These include impaired pre-rRNA processing, reduced 60S ribosomal subunit formation, a reduction in de novo protein synthesis, and impaired cell proliferation. Further, mutant LCLs phenocopied biological changes observed in HeLa cells depleted of eL15. These results, similar to previous reports that DBA-linked RP mutations impair pre-rRNA processing, indicate that the RPL15 mutations reported here are likely pathogenic and result in haploinsufficiency.
The altered ribosome biogenesis may explain the observed reduction in global protein synthesis, substantiating previous reports discussing altered translation of specific mRNAs in DBA-mutant cells as a key component of disease pathogenesis.49,50 In support of this pathogenesis, we also found that hematopoietic stem cells with the eL15 Tyr81* variant that were induced to differentiate into erythrocytes revealed a substantial decrease in the number of erythroid colonies and delays in differentiation. These findings, in addition to increased apoptosis, TP53 stabilization, and p21 overexpression in RPL15-mutant hematopoiesis, suggest that eL15 plays a critical role in ribosome biogenesis and that the reduction of the protein by genetic haploinsufficiency drives severe stress in the nucleolus.
Hydrops fetalis arising from severe intrauterine anemia is an uncommon manifestation of DBA with 10 cases reported so far. The clinical outcome of these patients was poor and, unexpectedly, no spontaneous remission was observed. In contrast, 50% (3 of 6) of all patients with RPL15 mutations reported in our study showed prenatal manifestation with the necessity of intrauterine transfusions. Remarkably, the hydrops cases were observed only in the subgroup of patients carrying protein truncating mutations p.Tyr81* and p.Gln29*, with prevalence of hydrops reaching 75% (3 of 4).
A substantial percentage of individuals with DBA spontaneously achieve treatment independence at some point; however, the reasons for this remain elusive and no predictive biomarkers exist.39 In line with this, another unexpected finding was the rapid and sustained treatment independence achieved within four to 16 months after birth in all 3 unrelated DBA patients with the recurrent p.Tyr81* mutation. Although Patient 1 relapsed five years after achieving spontaneous remission, a short steroid course was very effective and resulted in treatment independence. These observations suggest that p.Tyr81* mutation carriers can achieve treatment independence with or without a previous course of steroids, and that a genotype itself may be an important biomarker for the expected clinical course. Our findings might help prospective clinical stratification to determine which individuals are more likely to become treatment independent without the need for HSCT. The reasons why the specific mutation p.Tyr81* drives treatment independence remain unknown and cannot be explained by a potential genetic reversion, which was not observed in our patients. One tempting speculation is that the remaining wild-type allele compensates for the haploinsufficiency (e.g. by epigenetic mechanisms) in the adult hematopoiesis, but the compensation might not be present or sufficient in the fetus. These findings warrant further studies on the future availability of more patient material.
In conclusion, our study establishes germline point mutations in the RPL15 gene as novel genetic etiology of DBA. Half of the individuals carrying these mutations manifest with hydrops fetalis, a phenotype that is very rarely observed in DBA patients. Finally, we establish that a recurrent DBA genotype, eL15 p.Tyr81*, is linked to treatment independence.
Supplementary Material
Acknowledgments
Very special thanks go to all the individuals who consented to participate in this study and their families. We thank Alexandra Fischer (Freiburg) for data management, Sandra Zolles, Sophia Hollander, Dirk Lebrecht, and Gunda Ruzaike (Freiburg) for technical assistance, Pritam Kumar Panda (Freiburg) for bioinformatic analysis, and Dr. Marije Bartels (UMC Utrecht) for critical reading of the manuscript.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/103/6/949
Funding
AM, ML, MW, LDC, NK, HT, PEG, and MFOD are supported under the framework of E-Rare-2, ERA-Net for Research on Rare Diseases (ZonMW #113301205 and #40-44000-98-1008 in the Netherlands; #BMBF 01GM1301 and 01GM1609 in Germany; Chief Scientist Office, Israeli Ministry of Health # 3-12844 in Israel; #ANR-15-RAR3-0007-04 and #ANR-12-RARE-0007-02 in France). MW and CN are supported by DKTK German Cancer Consortium, fot molecular diagnostics of pediatric malignancies. LDC was supported by the Laboratory of Excellence for Red Cells [(LABEX GR-Ex)-ANR Avenir-11-LABX-0005-02], the French National PHRC OFABD (DBA registry and molecular biology), and the “Fondation ARC pour la recherche contre le cancer”. LDC, MFOD and PEG are funded by the French National Research Agency [ANR-DBA-Multigenes-ANR-2015-AAP générique-CE12]. AA was supported by DBA Telethon grant GGP13177 and Fondazione Europea per la DBA and Gruppo di Sostegno DBA Italia.
References
- 1.Vlachos A, Ball S, Dahl N, et al. Diagnosing and treating Diamond Blackfan anaemia: results of an international clinical consensus conference. Br J Haematol. 2008; 142(6):859–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ball SE, McGuckin CP, Jenkins G, Gordon-Smith EC. Diamond-Blackfan anaemia in the U.K.: analysis of 80 cases from a 20-year birth cohort. Br J Haematol. 1996;94(4):645–653. [DOI] [PubMed] [Google Scholar]
- 3.Lipton JM, Atsidaftos E, Zyskind I, Vlachos A. Improving clinical care and elucidating the pathophysiology of Diamond Blackfan anemia: an update from the Diamond Blackfan Anemia Registry. Pediatr Blood Cancer. 2006;46(5):558–564. [DOI] [PubMed] [Google Scholar]
- 4.Vlachos A, Klein GW, Lipton JM. The Diamond Blackfan Anemia Registry: tool for investigating the epidemiology and biology of Diamond-Blackfan anemia. J Pediatr Hematol Oncol. 2001;23(6):377–382. [DOI] [PubMed] [Google Scholar]
- 5.Semmekrot BA, Haraldsson A, Weemaes CM, Smeets DF, Geven WB, Brunner HG. Absent thumb, immune disorder, and congenital anemia presenting with hydrops fetalis. Am J Med Genet. 1992;42(5):736–740. [DOI] [PubMed] [Google Scholar]
- 6.Van Hook JW, Gill P, Cyr D, Kapur RP. Diamond-Blackfan anemia as an unusual cause of nonimmune hydrops fetalis: a case report. J Reprod Med. 1995;40(12):850–854. [PubMed] [Google Scholar]
- 7.McLennan AC, Chitty LS, Rissik J, Maxwell DJ. Prenatal diagnosis of Blackfan-Diamond syndrome: case report and review of the literature. Prenat Diagn. 1996;16(4):349–353. [DOI] [PubMed] [Google Scholar]
- 8.Rogers BB, Bloom SL, Buchanan GR. Autosomal dominantly inherited Diamond-Blackfan anemia resulting in nonimmune hydrops. Obstet Gynecol. 1997;89(5 Pt 2):805–807. [DOI] [PubMed] [Google Scholar]
- 9.Scimeca PG, Weinblatt ME, Slepowitz G, Harper RG, Kochen JA. Diamond-Blackfan syndrome: an unusual cause of hydrops fetalis. Am J Pediatr Hematol Oncol. 1988. Fall;10(3):241–243. [PubMed] [Google Scholar]
- 10.Dunbar AE, 3rd, Moore SL, Hinson RM. Fetal Diamond-Blackfan anemia associated with hydrops fetalis. Am J Perinatol. 2003;20(7):391–394. [DOI] [PubMed] [Google Scholar]
- 11.Saladi SM, Chattopadhyay T, Adiotomre PN. Nomimmune hydrops fetalis due to Diamond-Blackfan anemia. Indian Pediatr. 2004;41(2):187–188. [PubMed] [Google Scholar]
- 12.Valsky DV, Daum H, Yagel S. Reversal of mirror syndrome after prenatal treatment of Diamond-Blackfan anemia. Prenat Diagn. 2007;27(12):1161–1164. [DOI] [PubMed] [Google Scholar]
- 13.Da Costa L, Chanoz-Poulard G, Simansour M, et al. First de novo mutation in RPS19 gene as the cause of hydrops fetalis in Diamond-Blackfan anemia. Am J Hematol. 2013;88(4):340–341. [DOI] [PubMed] [Google Scholar]
- 14.Boria I, Garelli E, Gazda HT, et al. The ribosomal basis of Diamond-Blackfan Anemia: mutation and database update. Hum Mutat. 2010;31(12):1269–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ban N, Beckmann R, Cate JH, et al. A new system for naming ribosomal proteins. Curr Opin Struct Biol. 2014;24:165–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cmejla R, Cmejlova J, Handrkova H, Petrak J, Pospisilova D. Ribosomal protein S17 gene (RPS17) is mutated in Diamond-Blackfan anemia. Hum Mutat. 2007; 28(12):1178–1182. [DOI] [PubMed] [Google Scholar]
- 17.Doherty L, Sheen MR, Vlachos A, et al. Ribosomal protein genes RPS10 and RPS26 are commonly mutated in Diamond-Blackfan anemia. Am J Hum Genet. 2010; 86(2):222–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Draptchinskaia N, Gustavsson P, Andersson B, et al. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat Genet. 1999;21(2):169–175. [DOI] [PubMed] [Google Scholar]
- 19.Farrar JE, Nater M, Caywood E, et al. Abnormalities of the large ribosomal subunit protein, Rpl35a, in Diamond-Blackfan anemia. Blood. 2008;112(5):1582–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gazda HT, Grabowska A, Merida-Long LB, et al. Ribosomal protein S24 gene is mutated in Diamond-Blackfan anemia. Am J Hum Genet. 2006;79(6):1110–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gazda HT, Preti M, Sheen MR, et al. Frameshift mutation in p53 regulator RPL26 is associated with multiple physical abnormalities and a specific pre-ribosomal RNA processing defect in diamond-black-fan anemia. Hum Mutat. 2012;33(7):1037–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gazda HT, Sheen MR, Vlachos A, et al. Ribosomal protein L5 and L11 mutations are associated with cleft palate and abnormal thumbs in Diamond-Blackfan anemia patients. Am J Hum Genet. 2008;83(6):769–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Landowski M, O’Donohue MF, Buros C, et al. Novel deletion of RPL15 identified by array-comparative genomic hybridization in Diamond-Blackfan anemia. Hum Genet. 2013;132(11):1265–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang R, Yoshida K, Toki T, et al. Loss of function mutations in RPL27 and RPS27 identified by whole-exome sequencing in Diamond-Blackfan anaemia. Br J Haematol. 2015;168(6):854–864. [DOI] [PubMed] [Google Scholar]
- 25.Mirabello L, Macari ER, Jessop L, et al. Whole-exome sequencing and functional studies identify RPS29 as a novel gene mutated in multicase Diamond-Blackfan anemia families. Blood. 2014;124(1):24–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gripp KW, Curry C, Olney AH, et al. Diamond-Blackfan anemia with mandibulofacial dystostosis is heterogeneous, including the novel DBA genes TSR2 and RPS28. Am J Med Genet A. 2014;164A(9):2240–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ikeda F, Yoshida K, Toki T, et al. Exome sequencing identified RPS15A as a novel causative gene for Diamond-Blackfan anemia. Haematologica. 2017;102(3):e93–e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mirabello L, Khincha PP, Ellis SR, et al. Novel and known ribosomal causes of Diamond-Blackfan anaemia identified through comprehensive genomic characterisation. J Med Genet. 2017;54(6):417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sankaran VG, Ghazvinian R, Do R, et al. Exome sequencing identifies GATA1 mutations resulting in Diamond-Blackfan anemia. J Clin Invest. 2012;122(7):2439–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choesmel V, Bacqueville D, Rouquette J, et al. Impaired ribosome biogenesis in Diamond-Blackfan anemia. Blood. 2007;109(3):1275–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferreira-Cerca S, Poll G, Gleizes PE, Tschochner H, Milkereit P. Roles of eukaryotic ribosomal proteins in maturation and transport of pre-18S rRNA and ribosome function. Mol Cell. 2005;20(2):263–275. [DOI] [PubMed] [Google Scholar]
- 32.Golomb L, Volarevic S, Oren M. p53 and ribosome biogenesis stress: the essentials. FEBS Lett. 2014;588(16):2571–2579. [DOI] [PubMed] [Google Scholar]
- 33.Kastan MB, Onyekwere O, Sidransky D, Vogelstein B, Craig RW. Participation of p53 protein in the cellular response to DNA damage. Cancer Res. 1991;51(23 Pt 1):6304–6311. [PubMed] [Google Scholar]
- 34.Dutt S, Narla A, Lin K, et al. Haploinsufficiency for ribosomal protein genes causes selective activation of p53 in human erythroid progenitor cells. Blood. 2011;117(9):2567–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jaako P, Flygare J, Olsson K, et al. Mice with ribosomal protein S19 deficiency develop bone marrow failure and symptoms like patients with Diamond-Blackfan anemia. Blood. 2011;118(23):6087–6096. [DOI] [PubMed] [Google Scholar]
- 36.Moniz H, Gastou M, Leblanc T, et al. Primary hematopoietic cells from DBA patients with mutations in RPL11 and RPS19 genes exhibit distinct erythroid phenotype in vitro. Cell Death Dis. 2012; 3:e356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pereboom TC, Bondt A, Pallaki P, et al. Translation of branched-chain aminotrans-ferase-1 transcripts is impaired in cells haploinsufficient for ribosomal protein genes. Exp Hematol. 2014;42(5):394–403.e4. [DOI] [PubMed] [Google Scholar]
- 38.Heijnen HF, van Wijk R, Pereboom TC, et al. Ribosomal protein mutations induce autophagy through S6 kinase inhibition of the insulin pathway. PLoS Genet. 2014; 10(5):e1004371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vlachos A, Muir E. How I treat Diamond-Blackfan anemia. Blood. 2010;116(19): 3715–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Narla A, Vlachos A, Nathan DG. Diamond Blackfan anemia treatment: past, present, and future. Semin Hematol. 2011; 48(2):117–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peffault de Latour R, Peters C, Gibson B, et al. Recommendations on hematopoietic stem cell transplantation for inherited bone marrow failure syndromes. Bone Marrow Transplant. 2015;50(9):1168–1172. [DOI] [PubMed] [Google Scholar]
- 42.Hui-Yuen J, McAllister S, Koganti S, Hill E, Bhaduri-McIntosh S. Establishment of Epstein-Barr virus growth-transformed lymphoblastoid cell lines. J Vis Exp. 2011;(57). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hirabayashi S, Flotho C, Moetter J, et al. Spliceosomal gene aberrations are rare, coexist with oncogenic mutations, and are unlikely to exert a driver effect in childhood MDS and JMML. Blood. 2012; 119(11):e96–99. [DOI] [PubMed] [Google Scholar]
- 44.Hu J, Liu J, Xue F, et al. Isolation and functional characterization of human erythroblasts at distinct stages: implications for understanding of normal and disordered erythropoiesis in vivo. Blood. 2013; 121(16):3246–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Da Costa L, O’Donohue MF, van Dooijeweert B, et al. Molecular approaches to diagnose Diamond-Blackfan anemia: The EuroDBA experience. Eur J Med Genet. 2017. October 26 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 46.Lo Ten Foe JR, Kwee ML, Rooimans MA, et al. Somatic mosaicism in Fanconi anemia: molecular basis and clinical significance. Eur J Hum Genet. 1997;5(3):137–148. [PubMed] [Google Scholar]
- 47.Henras AK, Plisson-Chastang C, O’Donohue MF, Chakraborty A, Gleizes PE. An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip Rev RNA. 2015;6(2):225–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Preti M, O’Donohue MF, Montel-Lehry N, Bortolin-Cavaille ML, Choesmel V, Gleizes PE. Gradual processing of the ITS1 from the nucleolus to the cytoplasm during synthesis of the human 18S rRNA. Nucleic Acids Res. 2013;41(8):4709–4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ludwig LS, Gazda HT, Eng JC, et al. Altered translation of GATA1 in Diamond-Blackfan anemia. Nat Med. 2014;20(7):748–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Horos R, Ijspeert H, Pospisilova D, et al. Ribosomal deficiencies in Diamond-Blackfan anemia impair translation of transcripts essential for differentiation of murine and human erythroblasts. Blood. 2012;119(1):262–272. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.