Abstract
Tissue Factor is a cell-surface glycoprotein expressed in various cells of the vasculature and is the principal regulator of the blood coagulation cascade and hemostasis. Notably, aberrant expression of Tissue Factor is associated with cardiovascular pathologies such as atherosclerosis and thrombosis. Here, we sought to identify factors that regulate Tissue Factor gene expression and activity. Tissue Factor gene expression is regulated by various transcription factors, including activating protein-1 and nuclear factor-κ B. The peptidyl-prolyl isomerase Pin1 is known to modulate the activity of these two transcription factors, and we now show that Pin1 augments Tissue Factor gene expression in both vascular smooth muscle cells and activated endothelial cells via activating protein-1 and nuclear factor-κ B signaling. Furthermore, the cytoplasmic domain of Tissue Factor contains a well-conserved phospho-Ser258-Pro259 amino-acid motif recognized by Pin1. Using co-immunoprecipitation and solution nuclear magnetic resonance spectroscopy, we show that the WW-domain of Pin1 directly binds the cytoplasmic domain of Tissue Factor. This interaction occurs via the phospho-Ser258-Pro259 sequence in the Tissue Factor cytoplasmic domain and results in increased protein half-life and pro-coagulant activity. Taken together, our results establish Pin1 as an upstream regulator of Tissue Factor-mediated coagulation, thereby opening up new avenues for research into the use of specific Pin1 inhibitors for the treatment of diseases characterized by pathological coagulation, such as thrombosis and atherosclerosis.
Introduction
Tissue Factor (TF), an integral cell-surface glycoprotein, is the initiator of the blood coagulation cascade and a key regulator of hemostasis.1,2 Aberrant expression of TF plays a crucial role in several coagulation-driven pathologies, such as thrombosis, atherosclerosis, and acute coronary syndromes,2–4 but also in endotoxemia, angiogenesis, and cancer.5–8 Many vascular cells express TF constitutively, including smooth muscle cells (SMCs), pericytes, and adventitial fibroblasts, while TF expression is undetectable in vascular endothelial cells (ECs).1 However, in both ECs and SMCs, the expression and activity of TF can be enhanced by pro-inflammatory signaling molecules such as tumor necrosis factor-α (TNF-α) and lipopolysaccharides (LPS), which induce TF expression via activating protein 1 (AP-1) and nuclear factor-kappa B (NF-κB) signaling.1,9,10 Consistently, inflammation-induced coagulation is completely abrogated by inhibition of TF activity in vivo.11
The TF protein contains two fibronectin type-III ectodomains, a single type-I transmembrane domain, and a twenty-amino acid cytoplasmic domain (TFCD), which plays an important role in several of the above-mentioned pathologies that involve TF.6–8 The TFCD can undergo multiple post-translational modifications, including palmitoylation at residue Cys245, phosphorylation at residues Ser253 and Ser258 by PKC-α and p38α-kinase, respectively, and ubiquitination at residue Lys255.12–15 These modifications act in concert to attenuate the interaction of the TFCD with the membrane lipid bilayer, as well as make it accessible to other interacting proteins that regulate TF activity.13,14,16
Peptidyl-prolyl cis-trans isomerase, NIMA-Interacting 1 (Pin1) is an enzyme that catalyzes cis-trans isomerization of proline residues that are preceded by a phosphorylated serine or threonine (a pSer/pThr-Pro motif) within its target proteins. The C-terminal isomerase domain of Pin1 binds the motif and catalyzes proline cis-trans isomerization, while the N-terminal WW-domain is responsible for mediating protein-protein interactions and target specificity.17–19 Conformational changes induced by Pin1-catalyzed proline isomerization have been shown to alter the phosphorylation, localization, stability, protein-protein interactions, and transcriptional activity of its target proteins, which include c-Jun, NF-κB, AP-1, p53, β-catenin, and the nuclear receptors PPARγ and Nur77.20–22
Here, we report that Pin1 enhances TF gene expression in activated vascular cells, and directly interacts with TF protein through a pSer258-Pro259 motif in the TFCD. We provide the solution structure of the TFCD in complex with the WW-domain of Pin1, which shows that this interaction requires both phosphorylation of Ser258 as well as trans-configuration of the pSer258-Pro259 peptide bond in the TFCD. Functionally, we demonstrate that Pin1 increases the protein half-life and pro-coagulant activity of TF in human vascular cells.
Methods
Mice
Mice in which the cytoplasmic domain of endogenous TF is deleted (TFΔCD mice)23 and wild-type litter mates were maintained under pathogen-free conditions at the Scripps Research Institute Animal Facility with approved protocols of the institutional Animal Care and Use Committee (protocols #08-0008 and #08-0009).
Cell culture
Human umbilical vein endothelial cells (HUVECs) and smooth muscle cells (SMCs) were isolated from umbilical cords anonymously collected at the Academic Medical Center, as previously described,22 in accordance with the institute’s ethical guidelines and with approval from the institute’s Medical Ethical Committee. Please see the Online Supplementary Methods for a detailed description of cell culture conditions.
Lentiviral transductions
Recombinant lentiviral particles encoding Pin1, shPin1, or backbone control constructs were produced as previously described.22 Cells were transduced at a multiplicity of infection of 100 for 24 hours (h), after which the medium was refreshed and cells were cultured for an additional 24 h before starting experiments.
Transfections and luciferase assays
HEK293T cells and SMCs were transfected using the CalPhos Mammalian Transfection Kit (Clontech) or Lipofectamine 3000 (Invitrogen), respectively. Cells were transfected according to the manufacturer’s instructions with wild-type, AP-1 binding site mutated, or NF-kB binding site mutated TF promoter luciferase reporter constructs (a gift from Nigel Mackman;24 Addgene #15442-15444) together with Pin1 or Pin1 mutants (described by van Tiel et al.22). The pRL-TK Renilla-reporter was co-transfected as internal control. After 24 h, cells were stimulated with 100 ng/mL 12-O-Tetradecanoylphorbol-13-acetate (PMA; Sigma) for 6 h. For NF-κB or AP-1 signaling inhibition, cells were treated with vehicle (DMSO), 10 μM BAY-117085 (Calbiochem), or 10 μM SP600125 (SelleckChem). Assays were performed using the dual-luciferase reporter assay and a Glomax Multi detection system (Promega).
Nuclear magnetic resonance spectroscopy
Nuclear magentic resonance (NMR) experiments were performed on Bruker DRX 600 MHz and 800 MHz spectrometers equipped with triple resonance 1H/13C/15N optimized for proton detection at 300K. Acquired data were converted from Bruker XWINNMR format to NMRVIEW v.5.3 format using NMRpipe software.25,26 All spectra were analyzed using NMRVIEW. All experiments were performed at the UH NMR Facility at the University of Houston. Please see the Online Supplementary Methods for a detailed description of NMR spectroscopy procedures.
TF protein half-life assays
Smooth muscle cells (SMCs) or HEK293T cells were transfected with Pin1 or Pin1 mutants and either TF or TFΔCD constructs. After 24 h, transfected cells were treated with 50 μg/mL cycloheximide (Sigma) for times indicated. TF protein levels were quantified by western blotting.
TF activity assays
Tissue factor activity was determined in SMCs, HUVECs, and EC-RF24 cells as previously described.27 Briefly, transduced cells were serum-starved overnight followed by stimulation with 50 ng/mL TNF-α (Peprotech) for 3 h. Cells were washed with PBS and incubated with 1 nM human Factor VIIa and 100 nM human Factor X (Kordia) at 37°C. Supernatant samples were collected in 100 mM EDTA, 50 mM Tris after 10, 20, and 30 minutes (min), incubated with 0.4 mM of FXa chromogenic substrate S-2222, and absorbance was measured at 405 nm.
Statistical analysis
Data are presented as mean±Standard Error of Mean. Significance was determined by unpaired two-tailed Student’s t-test or one- or two-way ANOVA with Bonferroni post-hoc correction as indicated in the figure legends. P<0.05 was considered statistically significant.
Results
Pin1 enhances TF gene expression via activation of NF-κB and AP-1
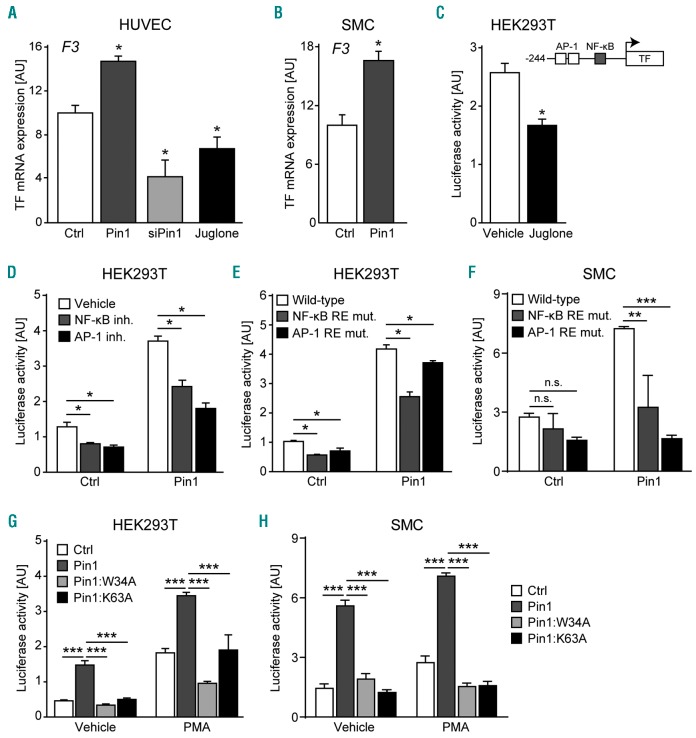
Pin1 modulates the activity of various transcription factors involved in TF gene expression.14,15 Therefore, we initiated our study by assessing the effect of Pin1 on TF gene expression. TF gene expression was measured in cultured ECs and SMCs after Pin1 gain and loss-of-function. Pin1 overexpression significantly increased TF mRNA levels in human SMCs and TNF-α-activated ECs, while knockdown of Pin1 by siRNA or Pin1 isomerase activity inhibition with Juglone28 resulted in significantly decreased TF mRNA expression in ECs (Figure 1A and B). Additionally, pharmacological inhibition of Pin1 isomerase activity with Juglone significantly decreased PMA-induced activation of the TF promoter in HEK293T cells (Figure 1C).
Figure 1.
Pin1 enhances Tissue Factor (TF) mRNA expression and TF promoter activation by NF-κB and AP-1. (A and B) Expression of the TF gene (F3) in human umbilical vein endothelial cells (HUVEC) and smooth muscle cells (SMC) after Pin1 overexpression, Pin1 knockdown (siPin1), or treatment with the Pin1 inhibitor Juglone. HUVECs were treated with TNF-α for 6 hours to induce TF gene expression. (C) Activity of the wild-type TF gene promoter luciferase reporter construct in PMA-stimulated HEK293T treated with Pin1 inhibitor Juglone or vehicle (DMSO) control. Inset shows schematic representation of the TF gene promoter luciferase reporter construct. (D) Activity of the wild-type TF gene promoter luciferase reporter construct after Pin1 overexpression in PMA-stimulated HEK293T and either vehicle (DMSO), NF-κB signaling inhibitor (BAY-117085), or AP-1 signaling inhibitor (SP600125) treatment. (E and F) Activity of luciferase reporter constructs containing either the wild-type TF gene promoter or the TF gene promoter in which the response element for NF-κB (NF-κB RE) or AP-1 (AP-1 RE) was mutated in PMA-stimulated HEK293T (E) or TNF-α-stimulated SMCs (F) transiently over-expressing Pin1. (G and H) Activity of the wild-type TF gene promoter luciferase reporter construct after transient overexpression of Pin1 or Pin1 mutants containing a disrupted WW-domain (Pin1:W34A) or lacking isomerase activity (Pin1:K63A) in HEK293T (G) or SMCs (H) left untreated or stimulated with PMA. Data are shown as mean±Standard Error of Mean. (A–C) P-values were calculated for comparisons between Pin1 overexpression, Pin1 knockdown (siPin1), or Juglone treatment versus control transduced (Ctrl) or vehicle-treated groups using the two-tailed Student’s t-test. (D–H) P-values were calculated using one-way ANOVA as indicated. *P<0.05, **P<0.01, ***P<0.001. AU: arbitrary units; mut: mutated.
The promoter of the TF gene contains binding motifs for NF-κB, AP-1, Sp1, and Egr-1, which are important transcription factors for both basal expression and stimulus-specific induction of TF expression.14,15 Consistent with our gene expression data, Pin1 overexpression markedly enhanced the activity of the wild-type TF promoter reporter construct in both HEK293T cells and SMCs (Figure 1D–F). However, this activation of the TF promoter by Pin1 was significantly diminished when treating HEK293T cells with inhibitors of NF-κB or AP-1 signaling (Figure 1D). Similarly, mutating either the NF-κB or AP-1 response element sequences to become non-functional, as previously described,24 greatly reduced the ability of Pin1 to activate the TF promoter in both HEK293T cells and SMCs (Figure 1E and F). Finally, neither a Pin1 mutant with a disrupted WW-domain (Pin1:W34A) nor a Pin1 mutant lacking isomerase activity (Pin1:K63A) could activate the TF promoter in HEK293T cells or SMCs (Figure 1G and H).
Taken together, these results show that Pin1 enhances TF gene expression and TF promoter activation via NF-κB and AP-1, and that this activation of the TF promoter is dependent on both Pin1 isomerase activity and a functional WW-domain.
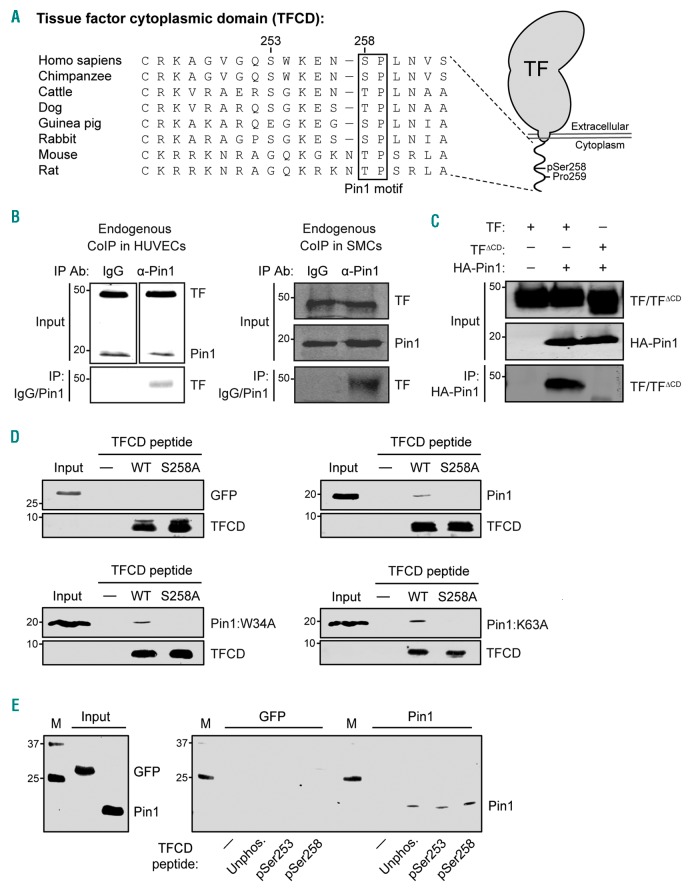
Pin1 interacts with the cytoplasmic domain of Tissue Factor
The TF cytoplasmic domain (TFCD) contains the well-conserved Ser258-Pro259 amino acid motif, which may function as a recognition site for Pin1 when Ser258 becomes phosphorylated (Figure 2A). We performed co-immunoprecipitation experiments in human SMCs and PMA-activated HUVECs, which showed that a pull-down with anti-Pin1 antibody resulted in co-precipitation of endogenous TF, while a pull-down with control IgG antibody did not, thereby indicating that TF and Pin1 interact in these two cell types (Figure 2B). Next, we performed co-immunoprecipitation experiments in HEK293T cells over-expressing HA-tagged Pin1 and either wild-type TF or truncated TF lacking the cytoplasmic domain (TFΔCD). These experiments showed that full-length TF interacts with Pin1, while the TFΔCD mutant does not, indicating that the TFCD is involved in the interaction with Pin1 (Figure 2C). Next, we performed pull-down assays with biotinylated peptides encoding the human TFCD, which confirmed our finding that Pin1 interacts with the TFCD, and showed that this interaction was completely disrupted by mutating the Ser258 residue in the TFCD (Figure 2D). This result is consistent with the observation that this residue is part of a Pin1 recognition motif. Furthermore, the interaction was not disturbed by mutations in either the WW-domain or isomerase domain of Pin1 Pin1:W34A and Pin1:K63A, respectively (Figure 2D). Finally, these interactions were specific, as control pull-downs without peptide did not precipitate Pin1, nor did TFCD peptides precipitate green fluorescent protein (GFP), a protein not predicted to bind the TFCD (Figure 2D).
Figure 2.
Pin1 interacts with Tissue Factor (TF) via the twenty-amino acid cytoplasmic domain (TFCD). (A) Amino acid sequence similarity of the TFCD in different species, showing strong conservation of the Pin1 recognition motif Ser/Thr-Pro (box). (B) Co-immunoprecipitation (CoIP) for TF and Pin1 in human PMA-stimulated HUVECs or smooth muscle cells (SMC) with control IgG or anti-Pin1 antibody (IP: IgG/Pin1) and analyzed by western blot with anti-TF antibody. Input TF and Pin1 levels are also shown. (C) CoIP for TF and Pin1 in HEK293T whole cell lysates over-expressing HA-tagged Pin1 and either full-length TF or TFΔCD using anti-HA antibody (IP: HA-Pin1) analyzed by western blot with anti-TF antibody. Input TF/TFΔCD and HA-Pin1 levels are also shown. (D) Pull-down assay with biotinylated peptides encoding wild-type human TFCD (WT) or TFCD with an S258A mutation. Full-length Pin1, Pin1 mutants with a disrupted WW-domain (Pin1:W34A) or lacking isomerase activity (Pin1:K63A), or GFP (as a negative control) were detected by western blot. (E) Pull-down assay with cysteine-linked TFCD-encoding peptides that were unphosphorylated or phosphorylated at either Ser253 or Ser258. Full-length Pin1 and GFP (as a negative control) were detected by western blot.
We next investigated the effect of phosphorylation on the interaction between Pin1 and TF using cysteine-linked peptides encoding the TFCD with varying states of phosphorylation. These experiments suggest that Pin1 has the highest affinity for the TFCD peptide phosphorylated at Ser258 (Figure 2E). However, it should be noted that these co-immunoprecipitation experiments are not quantitative. Furthermore, the observation that Pin1 seems to bind unphosphorylated and Ser253-phosphorylated TFCD peptides may also be due to TF-phosphorylating kinases present in the whole-cell lysates used for these pull-downs. The pull-down reactions were specific, as neither a pull-down without peptide or a pull-down for GFP resulted in precipitation of the target protein (Figure 2E).
Taken together, these results confirm that Pin1 interacts with TF and strongly suggest that this interaction involves the predicted pSer258-Pro259 Pin1 recognition motif in the TFCD.
Structural modalities mediating Pin1–TFCD complex formation
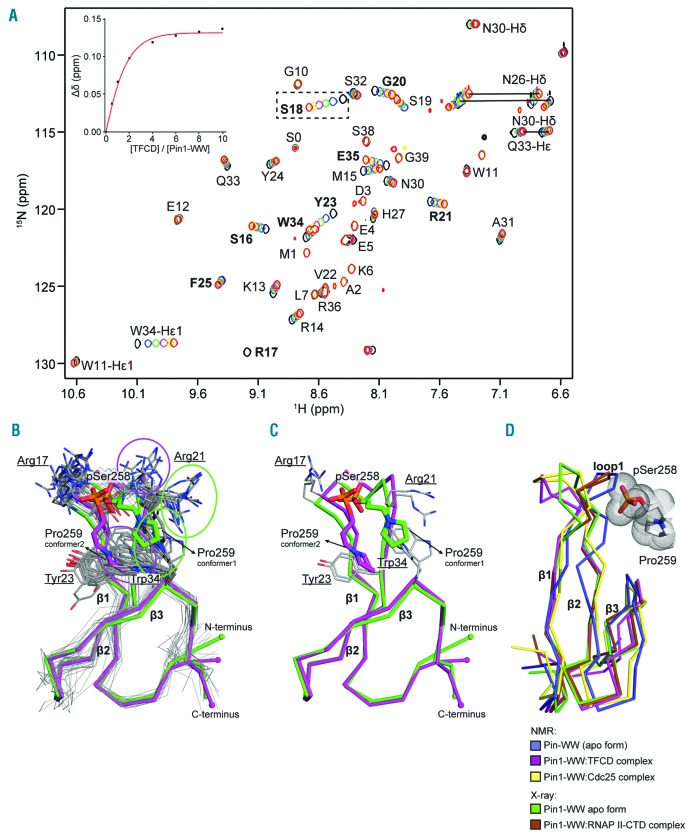
To study the interaction between the TFCD and Pin1 in more detail, we performed isothermal titration calorimetry (ITC) with Ser253 or Ser258-phosphorylated TFCD-encoding peptides and purified Pin1 WW-domain (Online Supplementary Figure S1). Consistent with our pull-down experiments, these assays showed that the Pin1 WW-domain interacts specifically with the TFCD peptide phosphorylated at Ser258 with a fitted Kd of approximately 137 μM (Online Supplementary Figure S1B and D).
To characterize the molecular basis of the WW-domain/TFCD interactions, we titrated the 15N-labeled WW-domain with an increasing concentration of pSer253/pSer258 TFCD peptide. Analysis of the 2D 1H/15N heteronuclear single-quantum correlation (HSQC) spectra showed that in all titrations, binding kinetics were in the fast-to-intermediate exchange regime, with at least 11 residues showing a large chemical shift (δγ > 0.1 ppm) (Figure 3A). Affected residues in the Pin1 WW-domain were located at the C-terminus of the β1-strand (S16), the β1-β2 loop (R17, S18, and G20), the β2-strand (R21, Y23, and F25), and the C-terminus of the β3-strand (W34 and E35) (Figure 3A; residues in bold). During our NMR titrations, the 1H/15N HSQC spectra showed line broadening for the Arg17 peak, resulting in its disappearance (Figure 3A). We attribute the broadening of the Arg17 line to its close proximity with the TFCD, with interactions occuring in the intermediate exchange regime in the NMR time scale (Figure 3B). A Kd of 133 ± 13 μM was determined from the δγ (ppm) for HN changes in Pin1 WW-domain residue Ser18 (Figure 3A; box around moving chemical shift and inset graph), which is similar to our calorimetric measurements showing a fitted Kd value of ~137 μM (Online Supplementary Figure S1).
Figure 3.
Nuclear magnetic resonance (NMR) spectroscopy shows interaction between twenty-amino acid cytoplasmic domain (TFCD) and Pin1 requires phosphorylation of Ser258 and trans-configuration of the pSer258-Pro259 peptide bond in the TFCD. (A) Superimposition of the assigned 1H/15N HSOC spectra of the Pin1 WW-domain with double phosphorylated TFCD (pSer253/pSer258) showing the chemical shift changes upon increasing amount of peptide to a 10× molar excess. A non-linear regression fit of the Ser18 (peak shift shown in black dotted box) was used to calculate the binding constant of the complex. (B) Bundle of 20 NMR conformers sampling into two major conformers. Sidechains of Pro259 (TFCD), Arg21 and Trp34 (Pin1 WW-domain) are indicated with circles. (C) Representative models of the two lowest-energy conformations from (B) are shown in green and magenta. The polypeptide backbones are shown as ribbons. Pin1 WW-domain residues are underlined. (D) Contact of the TFCD pSer258-Pro259 motif with the Pin1 WW-domain loop1 and comparison of NMR and X-ray structures.
We next prepared 13C/15N- isotopically-labeled Pin1 WW-domain in complex with double phosphorylated pSer253/pSer258 TFCD peptide for sequential backbone assignment and NMR structure determination. Analysis of the Cα and Cβ values based upon the weighted chemical shift index highlighted the three β-strands, which are characteristic of WW-domains (Online Supplementary Figure S2A). A total of 203 unique intra- and intermolecular distance constraints were derived from the analysis of the NOE spectroscopy (NOESY) experiments. We used these constraints together with the weighted chemical shift index (Online Supplementary Table S1 and Online Supplementary Figure S2A) to calculate the solution structure of the Pin1 WW-domain and TFCD complex (see Methods for more details). The 20 lowest energy conformers of the Pin1 WW-domain yielded a root-mean-square deviation (RMSD) of 1 Å (residues 6–39) (Figure 3B and Online Supplementary Figure S2B). The Pin1 WW-domain is a canonical WW-domain18 consisting of three twisted anti-parallel β-sheet strands with a conserved Trp-Trp motif located at the N-terminus of the first β-strand and C-terminus of the third β-strand, respectively.
Analysis of the Pin1 WW-domain–TFCD complex reveals that TFCD residues Asn257, pSer258 and Pro259 are involved in the binding interface of the Pin1 WW-domain, consistent with our pull-down assays. While the Pin1 WW-domain as a whole shows a well-defined, single conformation, the β1-β2 loop1 binding region of the WW-domain (residues 17 to 21) and the pSer258-Pro259 motif of TFCD show two distinct ensembles of conformers (Figure 3B and C), consistent with the fact that less NOEs were detected for the β1-β2 loop1. The features that principally drive the complex formation are the charge-charge interaction and the hydrophobic interaction between Trp34 (the second invariant Trp of the WW motif) with invariant Pro259. The ionic interactions between the phosphate of pSer258 with the positively charged guanidinium groups of Arg14, Arg17, and Arg21 also appeared to stabilize the complex. However, as mentioned earlier, the resonances for Arg17, located at position 1 of loop1, disappeared during the NMR titration due to intermediate exchange in the NMR timescale (Figure 3A), suggesting high flexibility around loop1. Arg21 connects loop1 to the β2-strand into two conformations and involves multiple polar contacts with the double phosphorylated TFCD at pSer258 (Figure 3B and C). Similarly, Trp34-driven hydrophobic packing of Pro259 induces two ensembles of Trp34 side chain rotamers and Pro259 configurations, and both ensemble states still adopt a trans-configuration of the pSer258-Pro259 peptide bond (Online Supplementary Table S1). For comparison, the previously determined interactions of Pin1 WW-domain with Cell division cycle 25 (Cdc25) and the C-terminal domain of the RNA polymerase II largest subunit (RNAP II-CTD) are also shown (Figure 3D).29,30
In order to further confirm the reliability of our structures, step-wise energy minimization was performed on all Pin1 WW-domain-TFCD complexes. The RMSD of heavy atoms/all atoms before and after energy minimization was less than 0.45 Å, while the RMSD of the backbone was less than 0.3 Å (Online Supplementary Figure S2B). Therefore, the interactions between the Pin1 WW-domain and the TFCD, such as the electrostatic interactions with pSer258 and the hydrophobic interactions of Pro259 with the WW-domain are consistent with the NMR structures calculated. The trans conformation of the pSer258-Pro259 peptide bond in the TFCD was stable during 10,000 steps of energy minimization, further demonstrating that our structures satisfy energy-of-motion (EOM) criteria with reasonable convergence parameters.
Taken together, our structural analyses show the interaction between TFCD and Pin1 involves phosphorylation of Ser258 and trans-configuration of the pSer258-Pro259 peptide bond in the TFCD.
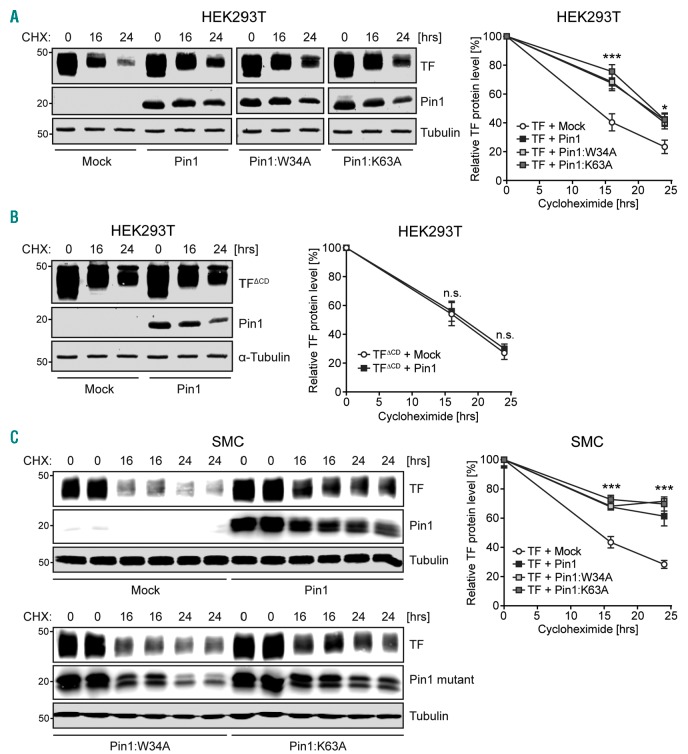
Pin1 extends TF protein half-life via the TFCD
Given our findings that Pin1 and TF interact and that Pin1 is known to enhance the protein half-life of several of its substrates,18 we sought to determine the effect of Pin1 on TF protein half-life using full-length TF and TF lacking the cytoplasmic domain in cycloheximide-treated HEK293T cells. Pin1 significantly enhanced the protein half-life of full-length TF (Figure 4A), but not that of TFΔCD (Figure 4B). Furthermore, Pin1 mutants with an inactive WW-domain (Pin1:W34A) or lacking isomerase activity (Pin1:K63A) were both still able to enhance the protein half-life of full-length TF (Figure 4A). Similar effects of Pin1 and Pin1 mutants on TF protein half-life were observed for endogenous TF protein in PMA-stimulated SMCs (Figure 4C). These results indicate that Pin1 can stabilize TF, and that this stabilization is dependent on interaction of Pin1 with the TFCD, but is not necessarily dependent on Pin1 isomerase activity.
Figure 4.
Pin1 increases Tissue Factor (TF) protein half-life via the twenty-amino acid cytoplasmic domain (TFCD). (A) HEK293T cells over-expressing full-length TF with or without Pin1 or Pin1 mutants were treated with cycloheximide (CHX) for times indicated in hours (hrs). TF protein levels were determined by western blot. The graph shows the amount of TF protein remaining after CHX treatment as a percentage of the starting TF protein level. (B) HEK293T cells over-expressing TFΔCD with or without Pin1 were treated with CHX for times indicated. TF protein levels were determined by western blot. The graph shows the amount of TF protein remaining after CHX treatment as a percentage of the starting TF protein level. (C) PMA-stimulated smooth muscle cells (SMC) over-expressing Pin1 or Pin1 mutants were treated with CHX for times indicated. Endogenous TF protein levels were determined by western blot. The graph shows the amount of TF protein remaining after CHX treatment as a percentage of the starting TF protein level. Data are shown as mean±Standard Error of Mean. P-values were calculated using two-way ANOVA. *P<0.05, **P<0.01, ***P<0.001 versus Mock-transfected controls.
Pin1 enhances TF pro-coagulant activity via the TFCD
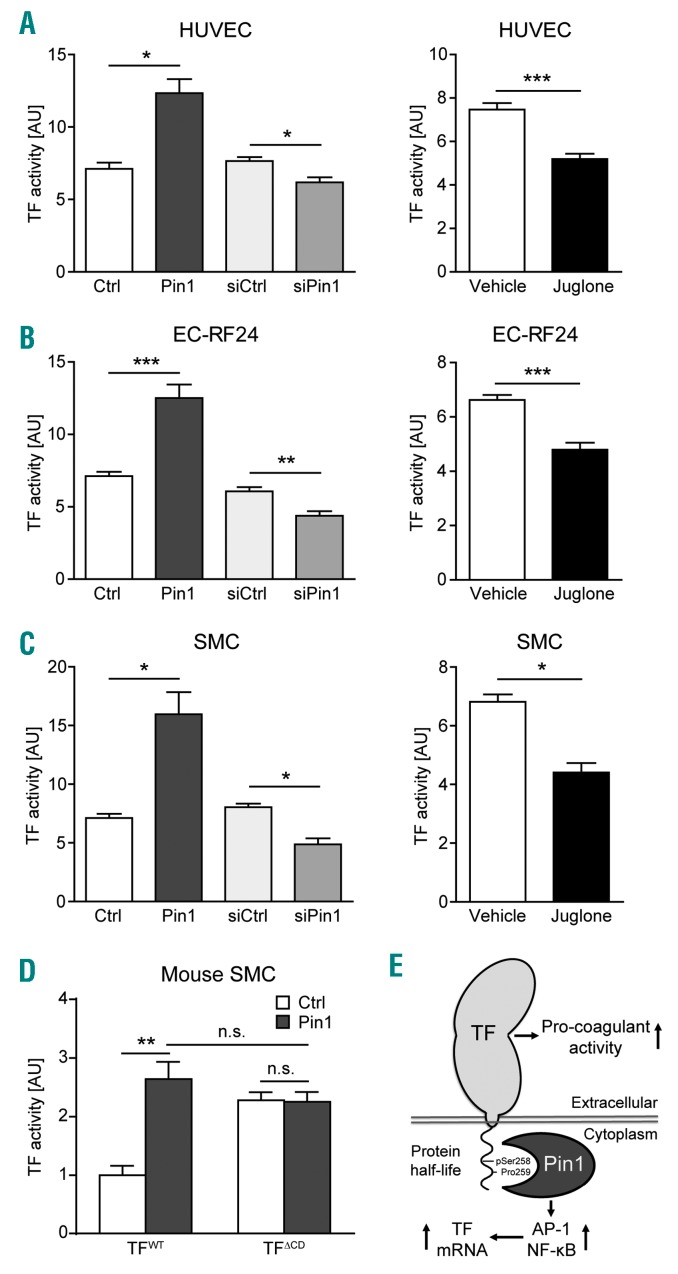
Having established that Pin1 enhances TF gene expression and TF protein half-life, we next examined the impact of Pin1 on the pro-coagulant activity of TF. We measured the Factor Xa-generating potential of the EC-RF24 cell line and primary human ECs and SMCs after Pin1 gain and loss-of-function. Pin1 overexpression markedly increased TF pro-coagulant activity in both ionomycin-treated SMCs and TNF-α treated EC-RF24 cells and ECs (Figure 5A–C). In contrast, knockdown of Pin1 by siRNA or inhibition of Pin1 isomerase activity by Juglone attenuated the TNF-α induced pro-coagulant activity of TF in these cells (Figure 5A–C). Furthermore, using SMCs derived from wild-type mice and mice expressing the TFΔCD truncated protein from the endogenous TF gene locus,23 we found that Pin1 can increase the activity of full-length TF, but not that of TF lacking the cytoplasmic domain (Figure 5D). Taken together, these results show that Pin1 interaction with TF via the TFCD strongly increases the pro-coagulation activity of TF.
Figure 5.
Pin1 enhances Tissue Factor (TF) pro-coagulant activity via the twenty-amino acid cytoplasmic domain (TFCD). (A–C) Factor Xa generation as a measure of TF activity in human human umbilical vein endothelial cells (HUVECs) (A), EC-RF24 cells (B), or smooth muscle cells (SMC) (C) after either overexpression or knockdown of Pin1 or treatment with the Pin1 inhibitor Juglone for 16 hours (hrs) followed by serum-starvation and treatment with ionomycin for 3 hrs (SMCs) or TNF-α for 16 hrs [EC-RF24 and endothelial cells (ECs)]. (D) Factor Xa generation as a measure of TF activity in mouse SMCs derived from wild-type mice (WT) or mice expressing TFΔCD from the endogenous TF gene locus after overexpression of Pin1 and stimulation with ionomycin for 3 hrs. (E) Summary of Pin1 effects on TF: Pin1 enhances the protein half-life and pro-coagulant activity of TF through interaction with the conserved pSer258-Pro259 motif in the TFCD and enhances the activity of the transcription factors AP-1 and NF-κB to increase TF gene expression. Data are shown as mean±Standard Error of Mean. P-values were calculated using two-tailed Student’s t-test (A–C) or two-way ANOVA (D). *P<0.05, **P<0.01, ***P<0.001. AU: arbitrary units.
Discussion
Tissue Factor plays a crucial role in initiating the extrinsic blood coagulation cascade. Therefore, detailed knowledge of the interacting proteins that modulate TF activity in vascular cells is essential to understand the regulation of thrombus formation under pathological conditions and may lead to novel intervention strategies for thrombotic disease. Here, we show that the peptidyl-prolyl isomerase Pin1 both enhances TF gene expression via activation of NF-κB and AP-1 signaling and directly interacts with TF through a well-conserved phosphorylated Ser258-Pro259 motif in its cytoplasmic domain. We elucidate the structural details of this interaction and show that Pin1 increases both the protein half-life and pro-coagulant activity of TF in vascular cells. Additional effects of Pin1 on TF activity may come from protease-activated receptor 2-induced release of TF on microvesicles, which was not studied in our experiments but were recently described.31
We demonstrate that Pin1 is a potent activator of TF gene expression in activated ECs and SMCs. The promoter of the human TF gene contains transcription factor binding sites for NF-κB, AP-1, Sp-1 and Egr-1, which are essential for inducing TF expression in many cell types.9,24 Interestingly, Pin1 has been shown to bind and regulate the activity of all four of these transcription factors in a cell type-dependent manner.15 The deletion of Egr-1 and Sp1 response elements in the TF promoter constructs revealed that there is a complete abrogation of TF promoter activity (data not shown). Therefore, the effect of Pin1 on activation of the TF gene promoter by Egr-1 and Sp1 cannot be determined.
Concerning our detailed structural analyses, we conclude that the TFCD-Pin1 WW-domain complex shows a larger contact area than what was observed in previous WW-domain/phosphopeptide NMR structures, but similar to the RNAP II-CTD:Pin1 crystal structure.32 Residues in the anchoring zone of the Pin1 WW-domain-TFCD complex showed apparent chemical shift perturbation (e.g. Tyr23) (Figure 3A), while these residues were stagnant in previous Pin1 NMR titrations.32 This discrepancy could potentially be the result of titration of the short pThr-Pro fragment of Cdc25 or tau in these experiments, rather than the full protein domain that was used here. Furthermore, the β1-β2 loop1 conformer shows structural similarity to the isolated ligand-free Pin1 WW-domain X-ray structures, as well as to the Cdc25 pThr peptide complex NMR structure, suggesting that loop1 undergoes a thermodynamic switch between ligand-bound and ligand-free states (Figure 3D).
Our calorimetric experiments revealed a weak interaction between the unphosphorylated TFCD and the Pin1 WW-domain (data not shown), with unphosphorylated TFCD having a lower affinity for Pin1 (Kd of 1.53 mM) than double phosphorylated TFCD (Kd of 137 μM). Similarly, our pull-down assays showed that unphosphorylated TF peptides can still pull-down Pin1 protein (Figure 2E). These two results suggest that Pin1 can interact with TF in both phosphorylation-specific and phosphorylation-independent manners. It has previously been shown that pre-attachment of Pin1 to an unphosphorylated “dynamic anchoring” region of c-Myc that is distant from its pSer-Pro motif is functionally important for the interaction between these two proteins.33 It could, therefore, be speculated that a similar distal, phosphorylation-independent interaction occurs between Pin1 and TF. Further studies are needed to confirm whether or not this is indeed the case.
It is also important to emphasize that intermolecular stacking of TFCD trans-Pro259 and Pin1 WW-domain Trp34 residues is highly orthologous to the intramolecular stacking between Trp8 and Pro37 of the Pin1 WW-domain. Given that mutational studies have shown that Trp8-Pro37 stacking can stabilize the WW-domain fold,32 we think that a similar, but intermolecular trans-Pro259 to Trp34 interaction-mechanism is utilized to promote the observed affinity between Pin1 and TF even when TFCD is unphosphorylated at Ser258. Indeed, complex formation due to the hydrophobic Pro-Trp stacking with unphosphorylated Ser-Pro or Ser-Thr motifs is consistent with previous studies in mammalian cell lines34 and in vitro studies of the WW-domain binding to unphosphorylated Ser-Pro motifs such as the cytoplasmic polyadenylation element binding (CPEB) protein.35,36
In our peptide pull-down and protein half-life experiments, we found that full-length Pin1 as well as Pin1 mutants with a disrupted WW-domain (Pin1:W34A) or lacking isomerase activity (Pin1:K63A) could bind the TFCD and stabilize TF protein. The discrepancy between these findings and our NMR data showing the importance of the Pin1 Trp34 residue in binding the TFCD can be explained by the fact that purified Pin1 WW-domain was used in our NMR studies, and that both domains of Pin1 can recognize and bind pSer-Pro motifs independently of each other.37 Therefore, it is possible that the Pin1 WW-domain mutant (Pin1:W34A) interacts with and stabilizes TF via its isomerase domain. However, whether the interaction between TF and Pin1 involves sequential and synergistic action of the Pin1 WW-domain and isomerase domain remains to be determined.
Several human and animal studies have shown that TF plays a crucial role in thrombosis development in vivo. We demonstrated that Pin1 has a regulatory role in FXa generation in both activated ECs and SMCs in vitro. Additional studies with Pin1-deficient mice may further delineate the role of Pin1 in TF gene expression, TF activity, and coagulation in vivo. However, Pin1-deficient mice already suffer from decreased body weight, testicular and retinal atrophies, and deficiencies in breast proliferative changes during pregnancy, which may interfere with in vivo coagulation experiments.38
In summary, our results demonstrate that Pin1 enhances TF gene expression, interacts with TF via the TFCD, and positively modulates TF half-life and pro-coagulant activity in vascular cells. Our findings suggest that Pin1 contributes to a hypercoagulable state in the local and systemic circulation. Specific Pin1 inhibitors could, therefore, be considered as potential novel therapeutic agents for the prevention of coagulation-driven pathologies, such as thrombosis and atherosclerosis.
Supplementary Material
Acknowledgments
We would like to thank Dr. Youlin Xia and the UH NMR facility-DOR for NMR data acquisition, CACDS for the structure calculations, and Amr Elnashai, Amy Sater, and Glen Legge for their support of this research.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/103/6/1073
Funding
This work was supported by the research program of the BioMedical Materials institute, co-funded by the Dutch Ministry of Economic Affairs as a part of Project P1.02 NEXTREAM. This work was also supported by the Rembrandt Institute for Cardiovascular Research (RICS-grant 2013), the Netherlands CardioVascular Research Initiative (CVON: 2012-08), and the National Institute of Health (grants HL-60742 and P01-HL16411).
References
- 1.Versteeg HH, Heemskerk JW, Levi M, Reitsma PH. New fundamentals in hemostasis. Physiol Rev. 2013;93(1):327–358. [DOI] [PubMed] [Google Scholar]
- 2.Furie B, Furie BC. Mechanisms of thrombus formation. N Engl J Med. 2008; 359(9):938–949. [DOI] [PubMed] [Google Scholar]
- 3.Taubman MB, Fallon JT, Schecter AD, et al. Tissue factor in the pathogenesis of atherosclerosis. Thromb Haemost. 1997; 78(1):200–204. [PubMed] [Google Scholar]
- 4.Moons AH, Levi M, Peters RJ. Tissue factor and coronary artery disease. Cardiovasc Res. 2002;53(2):313–325. [DOI] [PubMed] [Google Scholar]
- 5.Aras O, Shet A, Bach RR, et al. Induction of microparticle- and cell-associated intravascular tissue factor in human endotoxemia. Blood. 2004;103(12):4545–4553. [DOI] [PubMed] [Google Scholar]
- 6.Ahamed J, Niessen F, Kurokawa T, et al. Regulation of macrophage procoagulant responses by the tissue factor cytoplasmic domain in endotoxemia. Blood. 2007;109(12):5251–5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belting M, Dorrell M, Sandgren S, et al. Regulation of angiogenesis by tissue factor cytoplasmic domain signaling. Nat Med. 2004;10(5):502–509. [DOI] [PubMed] [Google Scholar]
- 8.Schaffner F, Versteeg HH, Schillert A, et al. Cooperation of tissue factor cytoplasmic domain and PAR2 signaling in breast cancer development. Blood. 2010; 23;116(26): 6106–6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mackman N. Regulation of the tissue factor gene. FASEB J. 1995;9(10):883–889. [DOI] [PubMed] [Google Scholar]
- 10.Schönbeck U, Mach F, Sukhova GK, et al. CD40 ligation induces tissue factor expression in human vascular smooth muscle cells. Am J Pathol. 2000;156(1):7–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levi M, ten Cate H, Bauer KA, et al. Inhibition of endotoxin-induced activation of coagulation and fibrinolysis by pentoxifylline or by a monoclonal anti-tissue factor antibody in chimpanzees. J Clin Invest. 1994;93(1):114–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ettelaie C, Collier ME, Featherby S, Greenman J, Maraveyas A. Oligoubiquitination of tissue factor on Lys255 promotes Ser253-dephosphorylation and terminates TF release. Biochim Biophys Acta. 2016;1863(11):2846–2857. [DOI] [PubMed] [Google Scholar]
- 13.Ettelaie C, Elkeeb AM, Maraveyas A, Collier ME. p38alpha phosphorylates serine 258 within the cytoplasmic domain of tissue factor and prevents its incorporation into cell-derived microparticles. Biochim Biophys Acta. 2013;1833(3):613–621. [DOI] [PubMed] [Google Scholar]
- 14.Sen M, Herzik M, Craft JW, et al. Spectroscopic Characterization of Successive Phosphorylation of the Tissue Factor Cytoplasmic Region. Open Spectrosc J. 2009;3:58–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dorfleutner A, Ruf W. Regulation of tissue factor cytoplasmic domain phosphorylation by palmitoylation. Blood. 2003; 102(12):3998–4005. [DOI] [PubMed] [Google Scholar]
- 16.Rothmeier AS, Liu E, Chakrabarty S, et al. Identification of the integrin-binding site on coagulation factor VIIa required for proangiogenic PAR2 signaling. Blood. 2018; 131(6):674–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ranganathan R, Lu KP, Hunter T, Noel JP. Structural and functional analysis of the mitotic rotamase Pin1 suggests substrate recognition is phosphorylation dependent. Cell. 1997;89(6):875–886. [DOI] [PubMed] [Google Scholar]
- 18.Joseph JD, Yeh ES, Swenson KI, Means AR. The peptidyl-prolyl isomerase Pin1. Prog Cell Cycle Res. 2003;5:477–487. [PubMed] [Google Scholar]
- 19.Wulf G, Finn G, Suizu F, Lu KP. Phosphorylation-specific prolyl isomerization: is there an underlying theme? Nat Cell Biol. 2005;7(5):435–441. [DOI] [PubMed] [Google Scholar]
- 20.Lu KP, Zhou XZ. The prolyl isomerase PIN1: a pivotal new twist in phosphorylation signalling and disease. Nat Rev Mol Cell Biol. 2007;8(11):904–916. [DOI] [PubMed] [Google Scholar]
- 21.Fujimoto Y, Shiraki T, Horiuchi Y, et al. Proline cis/trans-isomerase Pin1 regulates peroxisome proliferator-activated receptor gamma activity through the direct binding to the activation function-1 domain. J Biol Chem. 2010;285(5):3126–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Tiel CM, Kurakula K, Koenis DS, van der Wal E, de Vries CJ. Dual function of Pin1 in NR4A nuclear receptor activation: enhanced activity of NR4As and increased Nur77 protein stability. Biochim Biophys Acta. 2012;1823(10):1894–1904. [DOI] [PubMed] [Google Scholar]
- 23.Melis E, Moons L, De Mol M, et al. Targeted deletion of the cytosolic domain of tissue factor in mice does not affect development. Biochem Biophys Res Commun. 2001;286(3):580–586. [DOI] [PubMed] [Google Scholar]
- 24.Oeth P, Parry GC, Mackman N. Regulation of the tissue factor gene in human monocytic cells. Role of AP-1, NF-kappa B/Rel, and Sp1 proteins in uninduced and lipopolysaccharide-induced expression. Arterioscler Thromb Vasc Biol. 1997;17(2): 365–374. [DOI] [PubMed] [Google Scholar]
- 25.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol. 2004; 278:313–352. [DOI] [PubMed] [Google Scholar]
- 26.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6(3):277–293. [DOI] [PubMed] [Google Scholar]
- 27.van den Hengel LG, Osanto S, Reitsma PH, Versteeg HH. Murine tissue factor coagulant activity is critically dependent on the presence of an intact allosteric disulfide. Haematologica. 2013;98(1):153–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hennig L, Christner C, Kipping M, et al. Selective inactivation of parvulin-like peptidyl-prolyl cis/trans isomerases by juglone. Biochemistry. 1998;37(17):5953–5960. [DOI] [PubMed] [Google Scholar]
- 29.Zhou XZ, Kops O, Werner A, et al. Pin1-dependent prolyl isomerization regulates dephosphorylation of Cdc25C and tau proteins. Mol Cell. 2000;6(4):873–883. [DOI] [PubMed] [Google Scholar]
- 30.Verdecia MA, Bowman ME, Lu KP, Hunter T, Noel JP. Structural basis for phosphoserine-proline recognition by group IV WW-domains. Nat Struct Biol. 2000;7(8):639–643. [DOI] [PubMed] [Google Scholar]
- 31.Ettelaie C, Collier MEW, Featherby S, Greenman J, Maraveyas A. Peptidyl-prolyl isomerase 1 (Pin1) preserves the phosphorylation state of tissue factor and prolongs its release within microvesicles. Biochim Biophys Acta. 2018;1865(1):12–24. [DOI] [PubMed] [Google Scholar]
- 32.Wintjens R, Wieruszeski JM, Drobecq H, et al. 1H NMR study on the binding of Pin1 Trp-Trp domain with phosphothreonine peptides. J Biol Chem. 2001;276(27):25150–25156. [DOI] [PubMed] [Google Scholar]
- 33.Helander S, Montecchio M, Pilstål R, et al. Pre-Anchoring of Pin1 to Unphosphorylated c-Myc in a Fuzzy Complex Regulates c-Myc Activity. Structure. 2015;23(12):2267–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Macias MJ, Gervais V, Civera C, Oschkinat H. Structural analysis of WW-domains and design of a WW prototype. Nat Struct Biol. 2000;7(5):375–379. [DOI] [PubMed] [Google Scholar]
- 35.Nechama M, Lin CL, Richter JD. An unusual two-step control of CPEB destruction by Pin1. Mol Cell Biol. 2013;33(1):48–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schelhorn C, Martin-Malpartida P, Sunol D, Macias MJ. Structural Analysis of the Pin1-CPEB1 interaction and its potential role in CPEB1 degradation. Sci Rep. 2015;5:14990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Innes BT, Bailey ML, Brandl CJ, Shilton BH, Litchfield DW. Non-catalytic participation of the Pin1 peptidyl-prolyl isomerase domain in target binding. Front Physiol. 2013;4:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liou YC, Ryo A, Huang HK, et al. Loss of Pin1 function in the mouse causes phenotypes resembling cyclin D1-null phenotypes. Proc Natl Acad Sci USA. 2002; 99(3):1335–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.