Summary
Heart failure (HF) is the end-stage of all heart disease and arguably constitutes the greatest unmet therapeutic need in cardiovascular medicine today. Classic epidemiological studies have established clinical risk factors for HF, but the cause remains poorly understood in many cases. Biochemical analyses of small case-control series and animal models have described a plethora of molecular characteristics of HF, but a single unifying pathogenic theory is lacking. Heart failure appears to result not only from cardiac overload or injury but also from a complex interplay among genetic, neurohormonal, metabolic, inflammatory, and other biochemical factors acting on the heart. Recent development of robust, high-throughput tools in molecular biology provides opportunity for deep molecular characterization of population-representative cohorts and HF cases (molecular epidemiology), including genome sequencing, profiling of myocardial gene expression and chromatin modifications, plasma composition of proteins and metabolites, and microbiomes. The integration of such detailed information holds promise for improving understanding of HF pathophysiology in humans, identification of therapeutic targets, and definition of disease subgroups beyond the current classification based on ejection fraction which may benefit from improved individual tailoring of therapy. Challenges include: 1) the need for large cohorts with deep, uniform phenotyping; 2) access to the relevant tissues, ideally with repeated sampling to capture dynamic processes; and 3) analytical issues related to integration and analysis of complex datasets. International research consortia have formed to address these challenges and combine datasets, and cohorts with up to 1 million participants are being collected. This paper describes the molecular epidemiology of HF and provides an overview of methods and tissue types and examples of published and ongoing efforts to systematically evaluate molecular determinants of HF in human populations.
Key Words: genetics, heart failure, metabolomics, molecular epidemiology, proteomics
Abbreviations and Acronyms: ATP, adenosine triphosphate; CAD, coronary artery disease; DCM, dilated cardiomyopathy; GWAS, genome-wide association study; HCM, hypertrophic cardiomyopathy; HF, heart failure; LVEF, left ventricular ejection fraction; MS, mass spectrometry; nMR, nuclear magnetic resonance
Central Illustration
Heart failure (HF) is the end stage of all heart disease, characterized by inability of the heart to maintain sufficient output of blood for the demands of the body, resulting in frequent hospitalizations for breathing difficulties and fluid overload, risk of deadly arrhythmias, impaired quality of life, and high mortality (1) (Central Illustration). Medical therapy for HF has developed significantly over the past decades, with improved outcomes after the introduction of neurohormonal antagonists, but mortality remains high (2). The HF burden is substantial with an estimated prevalence of 2% in the United States and lifetime risk of 20% (3). Epidemiological studies show worrisome trends including globally increasing HF prevalence (4) and increasing incidence in young adults in Western countries (5). Improved preventive and therapeutic tools are thus urgently needed.
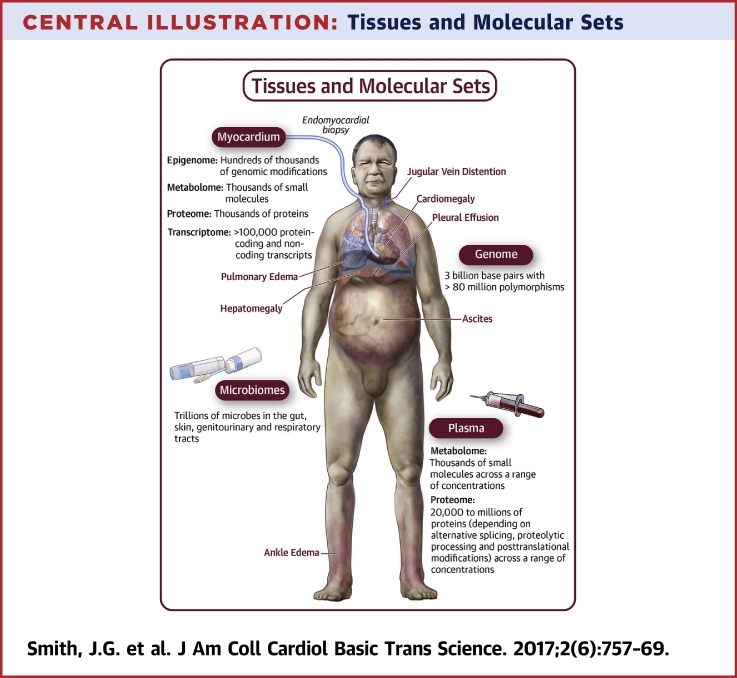
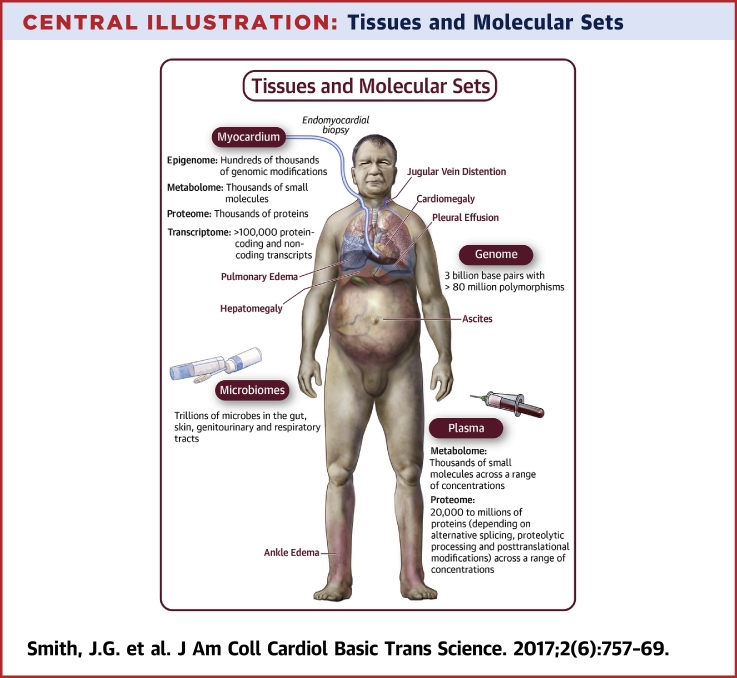
Central Illustration.
Tissues and Molecular Sets
Common signs of heart failure include enlargement of the heart (cardiomegaly) and fluid accumulation in the lungs (pulmonary edema), pleural cavity (pleural effusion), abdomen (ascites), legs (ankle edema), distended jugular veins, and liver (hepatomegaly). Locations for tissue sampling and sets of molecules for systematic studies of the molecular epidemiology of heart failure are indicated. Heart samples are obtained through endomyocardial biopsy (left section), from deceased donors, or donors undergoing heart transplantation. Plasma samples are obtained by centrifugation of blood from peripheral or central venipuncture. DNA for genome analysis can be isolated from any tissue, but most easily from leukocytes in blood samples. Microbial samples are obtained from urine, stool, genital, respiratory, skin, or oral swabs.
Epidemiology is the study of disease occurrence and determinants of disease in populations and the application of such knowledge to disease control. Classic prospective, population-based epidemiological cohorts were instrumental for identification of modifiable risk factors for coronary artery disease (CAD), including smoking, hyperlipidemia, hypertension, and diabetes mellitus (6). Essentially the same risk factors have been associated with HF, consistent with a large subset of HF cases caused by CAD 7, 8. However, the predictive accuracy of conventional risk factors for HF incidence is modest (8) due largely to causative heterogeneity of HF, improved outcomes in CAD, and overlap with comorbid conditions in other organ systems (3). In such studies, as well as in clinical practice, the cause of HF in an individual often remains poorly understood.
Information on molecular states may enhance understanding of causes and pathophysiological processes, improve risk prediction, identify new therapeutic targets, and improve subclassification for targeted therapy (precision medicine). Comprehensive analyses of molecular states in HF have mainly been restricted to small human case-control studies and animal models, which have resulted in a wide array of biochemical characteristics of HF. Thus, HF appears to result not only from cardiac overload or injury but also from a complex interplay among genetic, neurohormonal, metabolic, inflammatory, and other biochemical factors with individually modest effects acting on the heart. However, the pathophysiological role of many biochemical characteristics remains unclear, and a single unifying pathogenic theory is lacking (9). In order to better understand the relevance of a particular molecular state in patients with HF, the distribution of such states in the general population also needs to be characterized.
In recent years, the development of reproducible, high-throughput tools for molecular profiling has paved the way for the burgeoning field of molecular epidemiology: the coupling of information on molecular states to population-level disease data. Thus, thousands of human samples can now rapidly be characterized for genome sequence, myocardial gene expression profiles and chromatin modifications, plasma composition of proteins and metabolites, and microbiomes (Central Illustration, Table 1), comprehensive molecular profiling approaches that are agnostic to previous knowledge.
Table 1.
Glossary
| Epigenome | Chemical modifications of DNA or histones, such as methylation, that may influence gene expression and chromatin conformation |
| Genetic polymorphism | Genetic variant that is common in the general population, usually biallelic, limited to a single nucleotide, and defined as minor allele frequency ≥5% |
| Genome | Complete set of DNA in an organism, including all genes, that is present in all cells; the human genome consists of 3 billion base pairs |
| Genome-wide association study | Test for association of a genome-wide set of genetic variants with a phenotype, typically common variants (polymorphisms) |
| High-throughput sequencing | Methods to determine nucleotide sequence at high throughput based on multiple parallelized reactions; alternative to the classical, low-throughput Sanger sequencing |
| Linkage analysis | Method in genetics for discovery of chromosomal regions transmitted with disease in a family through genotyping of polymorphic sites distributed across the genome |
| lncRNA | Long, nonprotein-coding RNA transcripts with diverse and often unclear functions |
| Mendelian randomization | Observational method of using genetic variation to examine causal effect of a modifiable exposure on an outcome |
| Metabolome | A heterogeneous group of small molecules (metabolites), including amino acids, carbohydrates, lipids |
| Microarray | Technology for parallel testing of multiple analytes from mixture, based on a small glass or plastic slide to which multiple reagents are attached |
| Microbiome | Collection of all genomes in a microbial ecosystem |
| MicroRNA | Small non–protein coding RNA molecule (usually about 22 nucleotides) that functions in RNA silencing and post-transcriptional regulation of gene expression |
| Peripartum cardiomyopathy | A form of dilated cardiomyopathy with onset during the final month of pregnancy or in the 6 months after delivery |
| Proteome | Full set of proteins expressed in an organism |
| Sequencing depth | Number of times a given nucleotide has been read in a sequencing experiment, high depth facilitates distinguishing sequencing errors |
| Throughput | Number of samples and/or analytes undergoing analysis in a certain timeframe |
| Transcriptome | The full set of RNA transcripts in an organism |
This article describes the application of molecular epidemiology tools to HF. An overview of the molecular methods and tissue types used is provided, with particular emphasis on the genome, myocardium, plasma, and the microbiomes. Recent examples of such studies are described, along with expectations based on previous case-control studies.
Study Design in Molecular Epidemiology of HF
Prospective, population-based cohort study is the preferred study design for molecular epidemiology as previously described in detail (10), although most studies in the HF field published to date are case-control studies. Population-based cohort designs offer several advantages over other analytic study designs in observational epidemiology such as case-control studies, including avoidance of selection bias, recall bias, and differential misclassification bias, and allow study of trajectories and of multiple endpoints. Such designs offer opportunities for both pathophysiological insight into mechanisms involved in disease development and for disease outcomes (in the inception cohort), facilitating both primordial risk prediction and prediction of outcomes in distinct disease subsets (Table 2). With regard to HF, the ideal population-based cohort study should have several characteristics. First, given the heterogeneous nature of HF, very large cohorts are needed to capture sufficient numbers of incident events for robust analysis of relevant disease subsets. Several very large cohorts are currently being collected for study of the molecular epidemiology of a wide range of chronic diseases, as exemplified by the Precision Medicine Initiative in the United States, which aims to recruit 1 million participants, and the U.K. Biobank, which has recruited 500,000 participants. As costs are significant for several assays with high analyte throughput, a powerful and often more feasible approach than assaying the entire cohort is comparison of incident cases with a population-representative subcohort in a case-cohort or nested case-control design (11). Second, phenotypic characterization of participants should be uniform and detailed, including modern imaging modalities (echocardiography or cardiac magnetic resonance imaging) which is necessary for the HF diagnosis. Heart failure should be clearly defined, given the overlap with a multitude of comorbidities that present with similar symptoms (e.g., pulmonary disease, renal disease, thyroid disease, and anemia) and may modify the severity of HF (1). Information on established causes and mechanistic information, such as systolic and diastolic cardiac function, should also be obtained. Third, sampling of tissues for molecular profiling should be performed from as many of the relevant tissues as possible (Central Illustration). A major impediment is the difficulty of obtaining heart tissue. Fourth, serial examinations for repeated sampling are needed for analysis of processes that are dynamic over time, such as for transcriptomics, proteomics, and metabolomics, whereas the genome may be assayed at a single timepoint. Tissue sampling should ideally also be performed at different stages of HF development, including in subjects with early signs of heart injury, myocardial remodeling, and those with overt HF.
Table 2.
Study Populations and Selected Findings
| General Community | Myocardial Injury | Intermediate HF Phenotypes | Symptomatic HF | Outcomes in HF | |
|---|---|---|---|---|---|
| Context | Population-based, risk factors | Acute myocardial infarction or myocarditis | Myocardial remodeling | Symptom onset and diagnosis | Decompensation, arrhythmia, death, LVAD, or transplantation |
| Genetic variants | >300 independent SNPs for blood pressure, 113 for diabetes | 73 SNPs for CAD | >150 SNPs for systolic function, diastolic function, left ventricular mass, left ventricular diameter, QRS duration, QT duration and heart rate, 25 for AF, 7 for valvular disease | Sarcomere gene mutations (including MYBPC3 founder mutations, TTNtv), 8 SNPs for HF and cardiomyopathy | Mutations in LMNA, LAMP2, and MYH7; SNP at 5q22 (death), SNPs at 4q25 and 16q22 for AF, an SNP near hypocretin receptor-2 for myocardial recovery |
| Myocardial profile | — | — | — | Sarcomere changes (α- to β-myosin switch, titin isoform switch), reduced calcium channel expression, increased NP expression, metabolic changes (reduced mitochondrial fatty acid and glucose oxidation, reduced phosphocreatine, increased ketone oxidation) | Same as for HF (death) Increased glycolytic metabolites, lactate and amino acids (LVAD unloading) |
| Plasma profile | BCAAs for diabetes | TMAO for CAD | — | Increased catecholamines, angiotensin II, NPs, ST2, galectin-3, endothelin, CRP, TNFα, ketone bodies, lactate, reduced iron | Same as for HF, TMAO (death) |
| Microbiome composition | Strains that generate BCAAs for diabetes | Strains that generate TMAO for CAD | — | — | Strains that generate TMAO |
Overview of populations under study in the molecular epidemiology of heart failure, with a selection of key findings from different molecular sets. Some forms of HF are not preceded by myocardial injury or overload, such as monogenic cardiomyopathies, but likely influence intermediate phenotypes.
AF = atrial fibrillation; BCAAs = branched-chain amino acids; CAD = coronary artery disease; HF = heart failure; LVAD = left ventricular assist device; NPs = natriuretic peptides; SNP = single nucleotide polymorphism; TMAO = trimethylamine-N-oxide; TTNtv = titin-truncating variants.
The ideal cohort for HF based on all the requirements outlined above is not feasible to collect. Instead, international collaborative research consortia have formed to improve sample sizes by bringing together multiple population-based cohorts for molecular epidemiology research, as exemplified in Table 3. A major challenge for consortia is harmonization of HF phenotypes and molecular assays across cohorts. In genomics, imputation of genotypes from different array-based genotyping platforms to common reference panels has been a successful approach (12), whereas centralized analysis offers advantages for gene expression and plasma profiling assays. The analysis and integration of the large, multidimensional datasets generated by molecular assays constitutes another challenge. To date, data analysis has largely focused on robust detection of individual disease-associated markers with linear effects on disease risk, which can be separated from stochastic noise in data by using strict significance thresholds to control for multiple testing burden. With accumulating numbers of such molecules identified, analysts will need to embrace more complex models such as machine learning algorithms (13) to make use of the information content of datasets into predictive, diagnostic, and prognostic tools and complete understanding of HF pathophysiology across different causative contexts. The use of phenomapping, a form of cluster analysis to identify new disease subsets that may be subject to targeted therapies, was recently demonstrated by using clinical data in HF cases with preserved left ventricle ejection fraction LVEF (14) and would likely be even more powerful when unbiased biological parameters are included.
Table 3.
Design Considerations in Prospective Molecular Epidemiology Studies of HF
| Study Design Feature | HF Implications | Cohort Examples |
|---|---|---|
| Sample size | Very large cohorts or combined cohorts needed due to disease heterogeneity | PMI (n = 1 million), UK Biobank (n = 500,000), consortia for genomics (CHARGE-HF, GENIUS-CHD, HERMES, MGC, TOPMed) and plasma profiles (HOMAGE, inHForm) |
| Phenotypic characterization | Heterogeneous phenotype: need for standardized definitions and cardiac imaging | ESC diagnostic algorithm in several cohorts, Framingham criteria |
| Tissue sampling | Multiple tissues involved, heart most important but difficult to obtain | Multi-tissue: GTEx, heart tissue: MAGNet |
| Follow-up | Repeat sampling warranted | Biannual examinations in Framingham Heart Study, annual sampling in inHForm project |
CHARGE-HF = Heart Failure working group of the Cohorts for Heart and Aging Research consortium; GENIUS-CHD = GENetIcs of sUbSequent Coronary Heart Disease; GTEx = Genotype-Tissue Expression project; HERMES = Heart Failure Molecular Epidemiology for Therapeutic Targets; HOMAGE = Heart Omics in AGEing consortium; inHForm = INtegrative omics of Heart Failure to infORM discovery of novel drug targets and clinical biomarkers; MAGNet = Myocardial Applied Genomics Network; MGC = Myocardial Genetics Consortium; PMI = Precision Medicine Initiative; TOPMed = National Heart Lung Blood Institute Trans-Omics for Precision Medicine Program; other abbreviations as in Table 2.
Population Genetics of HF
Epidemiological studies have established a heritable component to all-cause HF beyond clinical risk factors 15, 16, but limited work has examined the genetic alleles that contribute to such heritability in populations. Many studies over the past 30 years have focused on individual families with monogenic forms of HF, where disease is transmitted across generations by rare mutations according to the laws of inheritance described by Gregor Mendel, such as hypertrophic cardiomyopathy (HCM) or dilated cardiomyopathy (DCM). Most cases of HF in the population, however, are not transmitted according to Mendel’s laws but represent the end result of 1 or multiple diseases that negatively influence the myocardium, after which the myocardium may progress to HF through adverse remodeling processes. Both the risk for myocardial injury and the myocardial response to damage likely result from complex interactions between environment and multiple rare and common genetic (polygenic) variants of individually modest effect. The mapping of the 3 billion base pairs of the human genome, followed by whole-genome resequencing in large populations have resulted in a catalog of >80 million genetic variants, many of which are common in human populations (polymorphisms) (17). Phenotypic consequences of such variants have been mapped over the past 10 years in genome-wide association studies (GWASs), with an increasing number of studies focusing on HF. In addition, an increasing number of studies of population cohorts are using high-throughput sequencing methods to generate exome-wide or even whole-genome sequence data (Table 4) to facilitate discovery of low-frequency and population-specific variants influencing susceptibility to HF and other diseases.
Table 4.
High-Throughput Molecular Assays in Molecular Epidemiology
| Molecular set | Method | Description |
|---|---|---|
| Genome | DNA microarrays | Genotyping of 100,000 to 1 million SNVs from which additional SNVs can be imputed for GWAS or exome-targeted analysis |
| Exome sequencing | Sequencing of the 30 million base pairs that encode proteins | |
| Whole-genome sequencing | Sequencing of the entire genome of an organism, typically at lower sequencing depth than exome sequencing | |
| Transcriptome | RNA microarrays | Gene expression profiling for all protein-coding genes and select noncoding transcripts |
| RNA sequencing | Sequencing of all RNA transcripts | |
| Proteome | Mass spectrometry | Method for unbiased detection of proteins in a mixture by mass and charge, usually coupled to liquid or gas chromatography or electrophoresis for sample separation |
| Affinity-based methods | Methods for protein detection based on protein isolation by an affinity reagent (e.g., antibody or aptamer) coupled to a reporter system for detection, can be robustly multiplexed for thousands of proteins with recent methods | |
| Metabolome | Mass spectrometry | Method for unbiased detection of metabolites in a mixture by mass and charge |
| nMR spectroscopy | Method for detection of soluble compounds in a mixture based on content of atoms of a certain spin, typically hydrogen | |
| Epigenome | ChIP | Method to identify histone modifications or transcription factor binding using antibodies, coupled to sequencing or microarrays to determine location |
| MeDIP | Method to identify DNA methylation using antibodies, coupled to sequencing or microarrays to determine location | |
| Bisulfite sequencing | Method to identify DNA methylation by bisulfite treatment and sequencing | |
| Hi-C | Method to determine chromatin conformation at high throughput | |
| Microbiome | 16S rRNA sequencing | Method to determine species of a prokaryotic organism by sequencing the rRNA gene |
ChIP = chromatin immunoprecipitation; MeDIP = methylation DNA immunoprecipitation; nMR = nuclear magnetic resonance; SNV = single-nucleotide variant.
Monogenic HF
Genes in which rare mutations cause HCM and DCM have been successfully characterized by using linkage analysis and targeted DNA sequencing. For HCM, mutations in the genes encoding 2 sarcomere proteins, cardiac myosin binding protein-C (MYBPC3) and β-myosin heavy chain (MYH7), explain approximately 50% of clinically recognized cases (18). For DCM, truncating mutations in the giant gene encoding the sarcomere protein titin (TTN) explain up to 25% of familial DCM cases, and many other genes have been implicated in certain families but explain only a minimal proportion of cases 19, 20. Collectively, such studies have highlighted the central role of cardiac sarcomere dysfunction in cardiomyopathy (18) and, although the biochemical and biophysical mechanisms by which sarcomere gene mutations cause disease remain incompletely understood, have recently resulted in development of promising sarcomere-targeted therapies to reduce the hypercontractility observed in HCM (21).
Polygenic contributions to HF
The importance of sarcomere mutations to overall HF in the population remains unclear. Sarcomere mutations have been described as relatively common, with reported estimates of 0.6% of the population carrying a likely pathogenic variant and 1.5% carrying a titin-truncating variant 22, 23. Founder mutations in MYBPC3, resulting in a mild HCM phenotype with late onset, have been reported to be prevalent in 4% of South Asians (mutation estimated to have arisen 30,000 years ago, conferring a 7-fold increased risk of HF) (24) and in Iceland where a mutation dating to the 15th century accounts for 60% of HCM (25).
GWAS studies have identified genetic polymorphisms conferring susceptibility to many conditions predisposing to HF, with 73 loci for CAD, 113 for diabetes, 25 for atrial fibrillation; 1 locus for aortic valvular stenosis, 6 for mitral valve prolapse, and >300 for blood pressure regulation 26, 27, 28, 29, 30, 31. Findings have confirmed a role for established pathways such as LDL cholesterol in CAD, the cardiac pacemaker channel of the sinoatrial node (HCN4) in atrial fibrillation, and vasoactive hormones such as natriuretic peptides and endothelin in blood pressure regulation but also implicated many previously unsuspected pathways. A total of 8 loci have also been associated with HF or cardiomyopathy in GWAS. Two intronic loci on chromosomes 1p36 and 3p14 have been associated with all-cause HF in case-control studies (32), of which the 1p36 locus has been reproducibly associated with both ischemic HF and DCM 32, 33, 34. Resequencing of the 1p36 locus and patch clamp experiments in recombinant cell lines suggested that the causal variant is a loss-of-function variant in the gene encoding a major renal chloride channel (CLCNKA), also linked to salt sensitivity (35). A GWAS in 4 combined population-based studies nominated additional polymorphisms at 2 loci on chromosomes 12q14 and 15q22, which have not yet been replicated (36). GWAS has also identified 2 loci for DCM, in BAG3 (34) and HCG22 (37), and a nonsynonymous polymorphism in the FHOD3 gene for HCM, encoding a protein implicated in sarcomere formation (38). Recently, an exome-centered study nominated 6 additional loci for DCM at a less strict significance threshold, including loci encoding the TTN gene and the phospholamban gene (PLN), a regulator of diastolic function through calcium ATPase in the cardiac sarcoplasmic reticulum, and the FHOD3 locus (39). An intergenic polymorphism near the PTHLH gene has been associated with peripartum cardiomyopathy, encoding a parathyroid hormone-related hormone involved in formation of the mammary glands and calcium metabolism. Motivated by these initial successes in small studies with <3,000 cases, international collaborative consortia for GWAS have been formed for more well-powered genomic analyses: for example, the HERMES (Heart Failure Molecular Epidemiology for Therapeutic Targets) consortium included >30,000 cases and >600,000 controls from >40 population-based cohorts, clinical trials, and clinical case collections to focus on both HF incidence and outcomes in HF (manuscript in preparation). Similarly, the GENIUS-CHD (GENetIcs of sUbSequent Coronary Heart Disease) consortium brings together data for nearly 250,000 subjects with CAD from >50 studies for HF and other outcomes (40).
Intermediate HF phenotypes
Important intermediate HF phenotypes reflecting precursor stages such as myocardial hypertrophy and dysfunction can be quantitatively determined from cardiac magnetic resonance imaging, echocardiography, or the electrocardiogram. Such markers are objective and, due to their quantitative nature spanning the full population distribution, variability can be more powerful to study in genetic studies than dichotomous phenotypes based on arbitrary cutoffs, such as the LVEF ≤40% used in clinical practice to indicate systolic dysfunction. In the EchoGen consortium, meta-analysis of GWAS in up to 32,212 subjects identified 18 loci for 5 characteristics of the LV (systolic function, diastolic function, end-diastolic diameter, mass, and aortic root size) including a locus on 6q22 for end-diastolic diameter, near the gene encoding phospholamban, also associated with DCM 41, 42. Four additional loci have been identified in African Americans (43) and 1 in subjects with hypertension (44). Several studies of electrocardiographic phenotypes have been conducted, including LV mass (45), QRS voltage duration (45), QT duration (46), and heart rate (47). In aggregate, these studies have identified >150 genetic loci, many of which are currently explored mechanistically. Although individual polymorphisms explain little of the population variability of each intermediate phenotype, the additive contribution of polymorphisms may explain as much as 20% to 50% of variability (48). Expanded analyses of these phenotypes are ongoing, as are integrative analyses of intermediate phenotypes within the MGC (Myocardial Genetics Consortium, unpublished data, December 2017).
Genetic contribution to HF outcomes
A heritable component has been described for adverse outcomes in HF (Lindgren M et al., unpublished data, December 2017). Certain forms of cardiomyopathy, e.g., with mutations in the LMNA, LAMP2, or MYH7 gene, are known to have particularly poor outcomes and may contribute to such heritability. A few studies have also explored the association of genetic polymorphisms with outcomes in HF. In a GWAS including 4,698 HF cases from 9 cohorts, an intergenic polymorphism on chromosome 5q22 was associated with all-cause mortality (49). Functional characterization indicated that the polymorphism influenced enhancer binding and expression of the cytokine thymic stromal lymphoprotein gene (TSLP) involved in T-lymphocyte signaling. In candidate gene studies, components of the β-adrenergic receptor signaling pathway have also been associated with HF survival 50, 51. In a study of LVEF recovery in patients with systolic HF, a genetic variant on chromosome 6p12 was reproducibly associated with LVEF improvement of ≥20% during follow-up. Experiments suggested that the polymorphism disrupts a transcription factor binding site for the downstream gene hypocretin receptor-2 and that the agonist of this receptor (hypocretin) was associated with improved systolic function in an HF mouse model (52). In another study, 2 polymorphisms on chromosomes 4q25 and 16q22 were strongly associated with atrial fibrillation in HF patients, present in up to 50% of HF cases and a determinant of adverse outcomes (53). Furthermore, a significant interaction of the 16q22 polymorphism was observed with HF for risk of atrial fibrillation, suggesting that the gene influenced by this variant may modulate the atrial substrate in the context of HF and increased filling pressures.
Mendelian randomization
One of the main advantages of genomics is the potential to infer causality for molecular states (54). Proteomic and metabolic markers—like clinical factors—provide a snapshot of a dynamic process and may thus be confounded by a multitude of factors, whereas genetic variants are inherited at conception and thus not subject to confounding factors. Genetics can therefore be leveraged to evaluate the cause of such factors by testing whether genetic variants associated with increased levels of a certain plasma protein or other characteristics also are associated with outcome, a concept termed “Mendelian randomization.” A published example for HF is the observation that genetically elevated body mass index (based on a polymorphism in the FTO gene) is associated with increased HF risk, suggesting that population weight control may reduce the burden of HF (55). With more well-powered cohorts such as HERMES, Mendelian randomization studies will become an increasingly powerful tool to nominate therapeutic and preventive strategies for HF.
Molecular Profiling of the Heart in HF
Population-based cohorts with heart tissue would be particularly valuable, as information on dynamic molecular states in the heart may reflect cause-specific mechanisms of heart injury, the cardiac response to damage, remodeling processes, response to therapy, and potential for recovery of function (Table 2). Some degree of recovery of systolic function is not uncommon with modern therapy, but the mechanisms are poorly understood 56, 57. In principle, heart tissue can be accessed postmortem, from explanted hearts of heart transplant patients, or from small endomyocardial biopsies obtained through cardiac catheterization. With the limitations inherent to such cohorts, resources with hundreds to thousands of patients are currently being established. The MAGNet (Myocardial Applied Genomics Network) consortium has collected explanted hearts and unused donor hearts from >500 patients with and without HF (58). The Genotype-Tissue Expression project (GTEx) collects postmortem samples and has performed whole-genome genotyping and RNA sequencing of >40 tissues from >400 subjects, with left ventricle and atrial appendage samples from >200 subjects (59). Detailed information on the myocardial transcriptome, proteome and metabolome is emerging from these datasets.
Myocardial transcriptome and proteome
Gene expression profiling in human heart tissue in GTEx, MAGNet, and several smaller case-control studies has provided an atlas of the transcriptome in failing and nonfailing myocardium 58, 60. In these datasets, gene expression in myocardial tissue is strongly dominated by sarcomere and mitochondrial components and other transcripts abundantly expressed in the voluminous cardiomyocytes. Enrichment protocols are therefore required to characterize gene expression in other cardiac cell types, such as fibroblasts and immune cells, which may be particularly important in myocardial remodeling and recovery processes. With this cardiomyocyte predominance in mind, well-described characteristics of gene expression in HF include widespread changes in splicing of sarcomere genes (61), a transition from α-myosin to β-myosin expression (62), a titin isoform switch (63), lower expression of key calcium channels including SERCA2a and RYR2 (64), an upregulation of natriuretic peptides (64), and a switch from expression of enzymes involved in fatty acid oxidation to glycolysis (65). A unified theory to explain these structural and metabolic characteristics of HF has not been clearly formulated, but many of them are considered manifestations of a physiological response to increasing strain resulting in hypertrophy with similarities to gene expression in the embryonic heart (9). Some reports have suggested cause-specific gene expression profiles, with pronounced differences in sarcomeric transcription in HCM, immune activation in DCM, and increased expression of enzymes involved in fatty acid oxidation in diabetes-related cardiomyopathy 66, 67.
Following the sequencing of the human genome, a surprising finding was pervasive transcription of many genomic regions previously considered “junk DNA.” Many such non–protein-encoding RNA species, including microRNAs and long noncoding RNAs (lncRNAs), are thought to regulate transcription of protein-encoding genes. Several microRNAs have been associated with HF, for example, mir-208a has been implicated as a strain-induced regulator of hypertrophy and the switch from α- to β-myosin transcription (68); mir-21 in fibrosis and hypertrophy (69); mir-425 in atrial natriuretic peptide expression (70); and mir-133 in cardiomyocyte hypertrophy (71). The interpretation of microRNA functions are complicated by widespread presence of target sites in the genome, and the fact that many miRNAs can be detected with remarkably stable concentrations in blood suggests wide-ranging effects across tissues. Several lncRNAs, apparently acting through diverse mechanisms, have also been associated with HF (72). In contrast to miRNAs, some reports suggest cell-type specific effects of many lncRNAs (73).
Expression of mRNA and proteins in a tissue is often modestly correlated, as protein abundance depends not only on gene transcriptional rate but also on mRNA stability, protein synthesis rate, and post-translational regulation and degradation (74). However, limited data are available for the myocardial proteome in humans. Two-dimensional gel electrophoresis and, more recently, mass spectrometry (MS), powerful technologies that may detect up to 10,000 proteins from a tissue sample, have been applied to DCM samples in a few studies (75). For MS, the dominance of sarcomeric and mitochondrial proteins necessitates depletion of these proteins. Catalogs of proteins expressed in the human heart, based on immunohistochemistry and MS, are available online through the Human Protein Atlas and MaxQB (see URLs provided in Useful Resources).
In important molecular epidemiology studies, data on genetic variation and transcriptional profiles have been integrated in the GTEx and MAGNet consortia, identifying genetic determinants for myocardial gene expression and, in Mendelian randomization designs, supporting evidence for a causal role of certain myocardial transcripts in HF (Morley M et al., unpublished data, December 2017) (59).
Myocardial epigenome
Like gene expression, patterns of chemical modifications to the DNA strand and histones as well as chromatin conformation are dynamic and differ among cell types. Such epigenetic factors may be influenced by environmental factors and contribute to disease susceptibility by influencing gene expression through modulation of transcription factor accessibility to cis-regulatory elements. Different chromatin states can now be assessed at high throughput using a range of methods (Table 4). Such tools are currently being coupled with gene expression profiles to understand how alterations in chromatin structure interact with transcription factors and noncoding RNAs to drive HF pathogenesis. Recent studies have identified loci in which CpG island and promoter methylation are associated with gene expression changes in DCM and HF, as reviewed in detail elsewhere (76). A comprehensive catalog of chromatin states across 111 human tissues and cell lines is available through the National Institutes of Health ROADMAP Epigenomics Project and will be expanded as part of the International Human Epigenome Consortium. Such data can be integrated with genomic and transcriptomic data to nominate functional motifs in the genome and prioritize causal variants, as exemplified by studies of myocardial gene expression (77) and QT and QRS interval durations (78).
Myocardial metabolome
Comprehensive analysis of the large and heterogeneous group of small molecules involved in human metabolism (lipids, amino acids, and carbohydrates) can be achieved in tissues using multiple approaches, including MS and nMR spectroscopy. The complex metabolome is influenced both by dietary intake and the regulatory effects of genes and proteins. Several myocardial differences in metabolite profile have been described in HF, but the mechanisms remain incompletely understood. The heart consumes more energy than any other organ, cycling approximately 6 kg of ATP every day to convert chemical energy stored in nutrients to mechanical energy used in the actin-myosin interaction of myofibrils. The heart is capable of metabolizing fatty acids, carbohydrates, lactate, ketone bodies, and certain amino acids as substrates for ATP generation, but under normal conditions more than two thirds of ATP in the myocardium is generated by oxidation of fatty acids, whereas, for example, the brain mainly relies on glucose (79). Classic studies found reduced reliance on fatty acid oxidation for energy generation in HF and increasing reliance on glycolysis (80). Furthermore, ATP generation from glucose seems to be disconnected from glucose oxidation pathways in the mitochondria and increasingly shuttled to lactate generation. Structural abnormalities in mitochondria have been described in HF, along with reduced activity of the respiratory carriers and oxidative phosphorylation (81). Levels of phosphocreatine, a small molecule phosphorylated by creatine kinase in the mitochondria that rapidly diffuses to myofibrils for energy transfer, are also reduced in HF (81). These observations have been reported mainly in hypertensive HF, ischemic HF, and DCM. In contrast, a different metabolite profile has been described for the restrictive form of cardiomyopathy associated with diabetes, including myocardial insulin resistance, reduced glucose and lactate metabolism, enhanced fatty acid oxidation, and intracellular fatty acid accumulation (67).
A recent metabolomics study based on MS in human heart tissue from DCM patients and nonfailing controls confirmed substantially lower levels of fatty acids in HF and of long-chain acylcarnitines, the primary substrates for mitochondrial fatty acid oxidation. Levels of acetyl-coenzyme A (CoA) species were increased in failing hearts, but Krebs cycle intermediates were decreased. Importantly, increased use of ketone bodies and increased myocardial expression of genes encoding key rate-limiting enzymes for myocardial oxidation of ketone bodies was observed, suggesting that increased oxidation of ketone bodies, produced by the liver from fatty acids, may be an important energy source in the failing heart (65).
Another recent study used MS to characterize the myocardial metabolome in serial myocardial biopsies from HF patients at the time of implantation of a ventricular assist device and after device explantation at the time of transplantation (82). The authors observed increased metabolites reflecting glycolysis after assist device unloading (pyruvate and lactate), without a corresponding increase in early tricarboxylic cycle intermediates. Markers of mitochondrial oxidative function and mitochondrial volume density were reduced compared to nonfailing hearts both before and after unloading, indicating irreversibility of glucose oxidation and mitochondrial dysfunction in HF. Increased levels of amino acids were also observed, implicating amino acids as another substrate of increasing importance in HF to fuel the tricarboxylic cycle and generate ATP.
The metabolic characteristics of the failing heart described above have sparked considerable interest in metabolic modulation as therapy for HF. Although neurohormonal antagonists used to treat HF lower metabolic demands in the myocardium, therapies directly targeting metabolism to further inhibit mitochondrial fatty acid oxidation (carnitine palmitoyl transferase inhibitor, long-chain 3-ketoacyl-CoA thiolase inhibitor), stimulate mitochondrial glucose oxidation (pyruvate dehydrogenase stimulator) or improve insulin sensitivity (GLP-1 agonist) have not been successful in smaller trials 83, 84. It remains unclear whether boosting ketone formation may improve cardiac function, which could help explain the decreased HF hospitalizations and cardiovascular mortality observed in patients with diabetes randomized to therapy with sodium-glucose cotransporter-2 inhibitors in the EMPA-REG OUTCOME (Empagliflozin Cardiovascular Outcome Event Trial in Type 2 Diabetes Mellitus Patients-Removing Excess Glucose) study or whether interventions to reduce reliance on ketone bodies would be desirable. Larger cohorts with comprehensive analysis of metabolomes and careful attention to HF subgroups and confounders are warranted to nominate other therapeutic targets. The integration of genetics, transcriptomics, and proteomics with metabolomics can also shed further light on the mechanisms of metabolic remodeling. For example, several monogenic metabolic diseases such as Danon disease, Pompe disease, and Fabry disease can result in cardiomyopathy.
Plasma Profiles in HF
Many tissues are influenced by reduced cardiac output and congestion in HF and may contribute to its progression, but like heart tissue, sampling of such tissues is difficult and associated with significant risks. However, blood samples can be easily obtained and represent the molecular states of all tissues, as molecules from tissue leakage and signaling can be monitored in blood. Profiling the circulating components of blood in populations at different stages of HF risk (Table 2), therefore, can identify pathophysiological pathways and may identify novel therapeutic targets and inform precision medicine. Indeed, established medical therapies for HF target circulating markers, such as catecholamines, angiotensin, and aldosterone, which have all been shown to be higher in HF patients, correlate with HF severity and decline after treatment with neurohormonal antagonists 85, 86, 87. More recently, therapies targeting natriuretic peptides (neprilysin inhibitors) and iron (intravenous iron substitution), shown to be increased and decreased in plasma samples of HF patients, respectively, have shown beneficial effects in clinical trials. Many other circulating molecules have been shown to be perturbed in HF, as summarized elsewhere (86), but the relationships between HF and most proteins and metabolites present in plasma remain unexplored.
Plasma proteome
The plasma proteome is a complex mixture of thousands of proteins, including carrier proteins such as albumin; immune system effectors including immunoglobins and complement factors; hemostatic factors; tissue messengers such as natriuretic peptides and interleukins; and tissue leakage products such as troponin and creatine kinase. This diversity in plasma protein function is accompanied by a diversity in protein abundance, with reference intervals for known plasma proteins in healthy subjects spanning more than 11 orders of magnitude (88). Application of proteome-wide, untargeted MS to plasma is therefore limited by the need for complex pre-analytical sample preparation stages, particularly for low-abundance proteins, resulting in substantial manual workflows and low sample throughput. Multiplexing of targeted immunoassays are therefore receiving increasing attention for plasma proteomics. Immunoassays are the workhorses for measurement of individual proteins but have been limited for proteomic applications by long development times, cross-reactivity preventing multiplexing, specificity issues, and incomplete sensitivity to detect proteins in the lower range of the abundance spectrum (below picograms per milliliter). Emerging technologies that address these issues include nucleotide-labeled immunoassays and aptamer reagents which can be automated for efficient multiplexing of thousands of proteins at high throughput, coupling of affinity capture methods to MS for improved specificity, and ultrasensitive detection systems to measure low-abundance proteins (88).
Aptamer proteomics has recently been applied to large cohorts to identify hundreds of novel protein biomarkers for CAD (89). No such comprehensive studies have yet been reported for HF, but many proteins in pathways such as inflammation, oxidative stress, extracellular matrix remodeling, neurohormones, and myocyte injury and stress have been associated with HF in targeted analyses (86). Antagonists for some markers strongly associated with HF in plasma, and supportive results from animal models have moved to clinical trials, such as endothelin and tumor necrosis factor α, but did not show beneficial effects in clinical trials 90, 91. The coupling of protein profiles to genetic data to evaluate causality will therefore be important steps to nominate therapeutic targets. Comprehensive proteomic profiling with affinity-based tools and integration with metabolomics and genetic information is currently ongoing in large population-cohorts with HF including the French HOMAGE (Heart Omics in AGEing) consortium (92) and the Swedish project INtegrative omics of Heart Failure to infORM discovery of novel drug targets and clinical biomarkers ([inHForm] Smith JG, in preparation).
Plasma metabolome
The plasma metabolome provides a complex snapshot of the global metabolic state of all tissues. It is influenced by the many causes of HF, systemic metabolic consequences of HF, comorbidities, medications, nutritional state and other environmental factors. One of the most robust signals from the application of MS and nMR spectroscopy to population-based studies has been the associations of higher branched-chain amino acids with incident diabetes and CAD (93). Although the association with HF remains unclear, animal models of HF have described branched-chain amino acid catabolic defects in the myocardium (94). Similarly, the metabolite trimethylamine-N-oxide (TMAO) has been robustly associated with CAD and has also been associated with outcome in HF (see discussion in the following text) (95). Studies of targeted metabolites have also reported robust associations of both increased plasma lactate and ketone bodies with HF, consistent with observations from myocardial metabolomic analyses 65, 96, 97. Recently, a few small case-control studies and a population-based cohort with nearly 2,000 subjects have evaluated comprehensive plasma metabolite profiles in HF with a combined total of 877 cases (98). These studies have reported a diverse set of metabolic profiles, with little overlap across studies. Studies have been limited by differences in case-control selection, assays used, statistical methodology, and small samples. Several ongoing projects aim to more comprehensively evaluate plasma metabolite profiles with centralized procedures in large population cohorts and various forms of HF including the HOMAGE consortium and the inHForm project.
Microbiomes in HF
The human body is estimated to harbor at least as many microbial cells as human cells (99). Recent years have seen the development of tools to study the microbiome at scale, such as high-throughput sequencing of the ribosomal subunit 16S rRNA, which is highly species-specific and subject to slow evolution rate. The Human Microbiome project was undertaken by characterization of the microbiomes in multiple locations from a large cohort, totaling 4,788 specimens from 242 adults. The overall composition of microbe communities were found to be remarkably stable over time within individuals and families but may be profoundly influenced by dietary and other environmental factors and differ greatly among individuals. Plasma abundance of 2 metabolites associated with CAD as discussed above have been linked to gut microbiomes, for example, metabolism of dietary phosphatidylcholine and l-carnitine by gut microbes results in production of TMAO, for which plasma levels are strongly associated with CAD and outcomes in HF and renal disease (95). Similarly, plasma branched-chain amino acid concentrations, consistently associated with diabetes and CAD (93), have been associated with 2 microbial species in the gut (Prevotella copri and Bacteroides vulgatus) (100). Sample collection for microbiomics is ongoing in many population cohorts, as such samples have not previously been biobanked.
Conclusions
A wide array of molecular characteristics of HF have been described. The integration of high-throughput molecular biology techniques (omics), agnostic to previous knowledge, in epidemiological cohorts offers opportunities to improve understanding of the causes of HF; mechanisms of HF development that are likely to include both cause-specific and shared components; and outcomes in HF. Challenges include the need for very large cohorts, uniform definitions, and detailed phenotyping, ideally with serial sampling of dynamic molecular profiles and heart tissue. Very large cohorts with hundreds of thousands of participants and deep phenotyping have been established, and international consortia have formed to combine cohorts. Over the next few years we shall learn whether these efforts will bear fruit into improved therapies and precision medicine for HF.
Useful Resources
CHARGE consortium: http://www.chargeconsortium.com/; ENGAGE consortium: http://www.euengage.org/; GENIUS-CHD consortium: http://www.genius-chd.com/; GTEx consortium: http://gtexportal.org/home/; HERMES consortium: http://www.hermesconsortium.org/; HOMAGE consortium: http://www.homage-hf.eu/; Human Metabolome Database: http://www.hmdb.ca/; Human Microbiome Project: http://hmpdacc.org/; Human Protein Atlas: http://www.proteinatlas.org; MAGNet consortium: http://www.med.upenn.edu/magnet/index.shtml; MaxQB database: http://maxqb.biochem.mpg.de/mxdb; National Heart, Lung, and Blood Institute TOPMed program: https://www.nhlbi.nih.gov/research/resources/nhlbi-precision-medicine-initiative/topmed; Precision Medicine Initiative: https://www.nih.gov/research-training/allofus-research-program; Roadmap Epigenomics Project: http://www.roadmapepigenomics.org/; UK Biobank: http://www.ukbiobank.ac.uk/.
Footnotes
This research was supported by the European Research Council, the Wallenberg Center for Molecular Medicine in Lund, the Swedish Heart-Lung Foundation, the Swedish Research Council, the Crafoord Foundation, governmental funding of clinical research within the Swedish National Health Service, and Skåne University Hospital in Lund. Dr. Smith has reported that he has no relationships relevant to the contents of this paper to disclose.
The author attests he is in compliance with human studies committees and animal welfare regulations of the authors' institutions and Food and Drug Administration guidelines, including patient consent where appropriate. For more information, visit the JACC: Basic to Translational Scienceauthor instructions page.
References
- 1.Jessup M., Brozena S. Heart failure. N Engl J Med. 2003;348:2007–2018. doi: 10.1056/NEJMra021498. [DOI] [PubMed] [Google Scholar]
- 2.Shafazand M., Schaufelberger M., Lappas G., Swedberg K., Rosengren A. Survival trends in men and women with heart failure of ischaemic and non-ischaemic origin: data for the period 1987-2003 from the Swedish Hospital Discharge Registry. Eur Heart J. 2009;30:671–678. doi: 10.1093/eurheartj/ehn541. [DOI] [PubMed] [Google Scholar]
- 3.Roger V.L. Epidemiology of heart failure. Circ Res. 2013;113:646–659. doi: 10.1161/CIRCRESAHA.113.300268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lozano R., Naghavi M., Foreman K. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christiansen M.N., Kober L., Weeke P. Age-specific trends in incidence, mortality, and comorbidities of heart failure in Denmark, 1995 to 2012. Circulation. 2017;135:1214–1223. doi: 10.1161/CIRCULATIONAHA.116.025941. [DOI] [PubMed] [Google Scholar]
- 6.Mahmood S.S., Levy D., Vasan R.S., Wang T.J. The Framingham heart study and the epidemiology of cardiovascular disease: a historical perspective. Lancet. 2014;383:999–1008. doi: 10.1016/S0140-6736(13)61752-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kannel W.B., D'Agostino R.B., Silbershatz H., Belanger A.J., Wilson P.W., Levy D. Profile for estimating risk of heart failure. Arch Intern Med. 1999;159:1197–1204. doi: 10.1001/archinte.159.11.1197. [DOI] [PubMed] [Google Scholar]
- 8.Smith J.G., Newton-Cheh C., Almgren P. Assessment of conventional cardiovascular risk factors and multiple biomarkers for the prediction of incident heart failure and atrial fibrillation. J Am Coll Cardiol. 2010;56:1712–1719. doi: 10.1016/j.jacc.2010.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braunwald E., Bristow M.R. Congestive heart failure: fifty years of progress. Circulation. 2000;102:IV14–IV23. doi: 10.1161/01.cir.102.suppl_4.iv-14. [DOI] [PubMed] [Google Scholar]
- 10.Psaty B.M., O'Donnell C.J., Gudnason V. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium: design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet. 2009;2:73–80. doi: 10.1161/CIRCGENETICS.108.829747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wacholder S. Practical considerations in choosing between the case-cohort and nested case-control designs. Epidemiology. 1991;2:155–158. doi: 10.1097/00001648-199103000-00013. [DOI] [PubMed] [Google Scholar]
- 12.de Bakker P.I., Ferreira M.A., Jia X., Neale B.M., Raychaudhuri S., Voight B.F. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum Mol Genet. 2008;17:R122–R128. doi: 10.1093/hmg/ddn288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deo R.C. Machine learning in medicine. Circulation. 2015;132:1920–1930. doi: 10.1161/CIRCULATIONAHA.115.001593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shah S.J., Katz D.H., Selvaraj S. Phenomapping for novel classification of heart failure with preserved ejection fraction. Circulation. 2015;131:269–279. doi: 10.1161/CIRCULATIONAHA.114.010637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee D.S., Pencina M.J., Benjamin E.J. Association of parental heart failure with risk of heart failure in offspring. N Engl J Med. 2006;355:138–147. doi: 10.1056/NEJMoa052948. [DOI] [PubMed] [Google Scholar]
- 16.Lindgren M.P., Smith J.G., Li X., Sundquist J., Sundquist K., Zoller B. Sibling risk of hospitalization for heart failure: a nationwide study. Int J Cardiol. 2016;223:379–384. doi: 10.1016/j.ijcard.2016.08.067. [DOI] [PubMed] [Google Scholar]
- 17.Genomes Project C., Auton A., Brooks L.D. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burke M.A., Cook S.A., Seidman J.G., Seidman C.E. Clinical and mechanistic insights into the genetics of cardiomyopathy. J Am Coll Cardiol. 2016;68:2871–2886. doi: 10.1016/j.jacc.2016.08.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herman D.S., Lam L., Taylor M.R. Truncations of titin causing dilated cardiomyopathy. N Engl J Med. 2012;366:619–628. doi: 10.1056/NEJMoa1110186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hershberger R.E., Hedges D.J., Morales A. Dilated cardiomyopathy: the complexity of a diverse genetic architecture. Nat Rev Cardiol. 2013;10:531–547. doi: 10.1038/nrcardio.2013.105. [DOI] [PubMed] [Google Scholar]
- 21.Green E.M., Wakimoto H., Anderson R.L. A small-molecule inhibitor of sarcomere contractility suppresses hypertrophic cardiomyopathy in mice. Science. 2016;351:617–621. doi: 10.1126/science.aad3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bick A.G., Flannick J., Ito K. Burden of rare sarcomere gene variants in the Framingham and Jackson heart study cohorts. Am J Hum Genet. 2012;91:513–519. doi: 10.1016/j.ajhg.2012.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roberts A.M., Ware J.S., Herman D.S. Integrated allelic, transcriptional, and phenomic dissection of the cardiac effects of titin truncations in health and disease. Sci Transl Med. 2015;7:270ra6. doi: 10.1126/scitranslmed.3010134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dhandapany P.S., Sadayappan S., Xue Y. A common MYBPC3 (cardiac myosin binding protein C) variant associated with cardiomyopathies in South Asia. Nat Genet. 2009;41:187–191. doi: 10.1038/ng.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adalsteinsdottir B., Teekakirikul P., Maron B.J. Nationwide study on hypertrophic cardiomyopathy in Iceland: evidence of a MYBPC3 founder mutation. Circulation. 2014;130:1158–1167. doi: 10.1161/CIRCULATIONAHA.114.011207. [DOI] [PubMed] [Google Scholar]
- 26.Thanassoulis G., Campbell C.Y., Owens D.S. Genetic associations with valvular calcification and aortic stenosis. N Engl J Med. 2013;368:503–512. doi: 10.1056/NEJMoa1109034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howson J.M.M., Zhao W., Barnes D.R. Fifteen new risk loci for coronary artery disease highlight arterial-wall-specific mechanisms. Nat Genet. 2017;49:1113–1119. doi: 10.1038/ng.3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christophersen I.E., Rienstra M., Roselli C. Large-scale analyses of common and rare variants identify 12 new loci associated with atrial fibrillation. Nat Genet. 2017;49:946–952. doi: 10.1038/ng.3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scott R.A., Scott L.J., Magi R. An expanded genome-wide association study of type 2 diabetes in Europeans. Diabetes. 2017;66:2888–2902. doi: 10.2337/db16-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoffmann T.J., Ehret G.B., Nandakumar P. Genome-wide association analyses using electronic health records identify new loci influencing blood pressure variation. Nat Genet. 2017;49:54–64. doi: 10.1038/ng.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dina C., Bouatia-Naji N., Tucker N. Genetic association analyses highlight biological pathways underlying mitral valve prolapse. Nat Genet. 2015;47:1206–1211. doi: 10.1038/ng.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cappola T.P., Li M., He J. Common variants in HSPB7 and FRMD4B associated with advanced heart failure. Circ Cardiovasc Genet. 2010;3:147–154. doi: 10.1161/CIRCGENETICS.109.898395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garnier S., Hengstenberg C., Lamblin N. Involvement of BAG3 and HSPB7 loci in various etiologies of systolic heart failure: Results of a European collaboration assembling more than 2000 patients. Int J Cardiol. 2015;189:105–107. doi: 10.1016/j.ijcard.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 34.Villard E., Perret C., Gary F. A genome-wide association study identifies two loci associated with heart failure due to dilated cardiomyopathy. Eur Heart J. 2011;32:1065–1076. doi: 10.1093/eurheartj/ehr105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cappola T.P., Matkovich S.J., Wang W. Loss-of-function DNA sequence variant in the CLCNKA chloride channel implicates the cardio-renal axis in interindividual heart failure risk variation. Proc Natl Acad Sci U S A. 2011;108:2456–2461. doi: 10.1073/pnas.1017494108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith N.L., Felix J.F., Morrison A.C. Association of genome-wide variation with the risk of incident heart failure in adults of European and African ancestry: a prospective meta-analysis from the cohorts for heart and aging research in genomic epidemiology (CHARGE) consortium. Circ Cardiovasc Genet. 2010;3:256–266. doi: 10.1161/CIRCGENETICS.109.895763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meder B., Ruhle F., Weis T. A genome-wide association study identifies 6p21 as novel risk locus for dilated cardiomyopathy. Eur Heart J. 2014;35:1069–1077. doi: 10.1093/eurheartj/eht251. [DOI] [PubMed] [Google Scholar]
- 38.Wooten E.C., Hebl V.B., Wolf M.J. Formin homology 2 domain containing 3 variants associated with hypertrophic cardiomyopathy. Circ Cardiovasc Genet. 2013;6:10–18. doi: 10.1161/CIRCGENETICS.112.965277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Esslinger U., Garnier S., Korniat A. Exome-wide association study reveals novel susceptibility genes to sporadic dilated cardiomyopathy. PLoS One. 2017;12 doi: 10.1371/journal.pone.0172995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patel R.S., Asselbergs F.W., The GENIUS-CHD consortium Eur Heart J. 2015;36:2674–2676. [PubMed] [Google Scholar]
- 41.Vasan R.S., Glazer N.L., Felix J.F. Genetic variants associated with cardiac structure and function: a meta-analysis and replication of genome-wide association data. JAMA. 2009;302:168–178. doi: 10.1001/jama.2009.978-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wild P.S., Felix J.F., Schillert A. Large-scale genome-wide analysis identifies genetic variants associated with cardiac structure and function. J Clin Invest. 2017;127:1798–1812. doi: 10.1172/JCI84840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fox E.R., Musani S.K., Barbalic M. Genome-wide association study of cardiac structure and systolic function in African Americans: the Candidate Gene Association Resource (CARe) study. Circ Cardiovasc Genet. 2013;6:37–46. doi: 10.1161/CIRCGENETICS.111.962365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arnett D.K., Meyers K.J., Devereux R.B. Genetic variation in NCAM1 contributes to left ventricular wall thickness in hypertensive families. Circ Res. 2011;108:279–283. doi: 10.1161/CIRCRESAHA.110.239210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van der Harst P., van Setten J., Verweij N. 52 Genetic loci influencing myocardial mass. J Am Coll Cardiol. 2016;68:1435–1448. doi: 10.1016/j.jacc.2016.07.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arking D.E., Pulit S.L., Crotti L. Genetic association study of QT interval highlights role for calcium signaling pathways in myocardial repolarization. Nat Genet. 2014;46:826–836. doi: 10.1038/ng.3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eppinga R.N., Hagemeijer Y., Burgess S. Identification of genomic loci associated with resting heart rate and shared genetic predictors with all-cause mortality. Nat Genet. 2016;48:1557–1563. doi: 10.1038/ng.3708. [DOI] [PubMed] [Google Scholar]
- 48.Yang J., Manolio T.A., Pasquale L.R. Genome partitioning of genetic variation for complex traits using common SNPs. Nat Genet. 2011;43:519–525. doi: 10.1038/ng.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith J.G., Felix J.F., Morrison A.C. Discovery of Genetic Variation on Chromosome 5q22 associated with mortality in heart failure. PLoS Genet. 2016;12 doi: 10.1371/journal.pgen.1006034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liggett S.B., Cresci S., Kelly R.J. A GRK5 polymorphism that inhibits beta-adrenergic receptor signaling is protective in heart failure. Nat Med. 2008;14:510–517. doi: 10.1038/nm1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cresci S., Kelly R.J., Cappola T.P. Clinical and genetic modifiers of long-term survival in heart failure. J Am Coll Cardiol. 2009;54:432–444. doi: 10.1016/j.jacc.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perez M.V., Pavlovic A., Shang C. Systems genomics identifies a key role for hypocretin/orexin receptor-2 in human heart failure. J Am Coll Cardiol. 2015;66:2522–2533. doi: 10.1016/j.jacc.2015.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith J.G., Melander O., Sjogren M. Genetic polymorphisms confer risk of atrial fibrillation in patients with heart failure: a population-based study. Eur J Heart Fail. 2013;15:250–257. doi: 10.1093/eurjhf/hfs176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Newton-Cheh C., Smith J.G. What can human genetics teach us about the causes of cardiovascular disease? J Am Coll Cardiol. 2010;55:2843–2845. doi: 10.1016/j.jacc.2009.11.097. [DOI] [PubMed] [Google Scholar]
- 55.Fall T., Hagg S., Magi R. The role of adiposity in cardiometabolic traits: a Mendelian randomization analysis. PLoS Med. 2013;10 doi: 10.1371/journal.pmed.1001474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Basuray A., French B., Ky B. Heart failure with recovered ejection fraction: clinical description, biomarkers, and outcomes. Circulation. 2014;129:2380–2387. doi: 10.1161/CIRCULATIONAHA.113.006855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Margulies K.B., Matiwala S., Cornejo C., Olsen H., Craven W.A., Bednarik D. Mixed messages: transcription patterns in failing and recovering human myocardium. Circ Res. 2005;96:592–599. doi: 10.1161/01.RES.0000159390.03503.c3. [DOI] [PubMed] [Google Scholar]
- 58.Liu Y., Morley M., Brandimarto J. RNA-Seq identifies novel myocardial gene expression signatures of heart failure. Genomics. 2015;105:83–89. doi: 10.1016/j.ygeno.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.GTEx Consortium Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mele M., Ferreira P.G., Reverter F. Human genomics. The human transcriptome across tissues and individuals. Science. 2015;348:660–665. doi: 10.1126/science.aaa0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kong S.W., Hu Y.W., Ho J.W. Heart failure-associated changes in RNA splicing of sarcomere genes. Circ Cardiovasc Genet. 2010;3:138–146. doi: 10.1161/CIRCGENETICS.109.904698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lowes B.D., Gilbert E.M., Abraham W.T. Myocardial gene expression in dilated cardiomyopathy treated with beta-blocking agents. N Engl J Med. 2002;346:1357–1365. doi: 10.1056/NEJMoa012630. [DOI] [PubMed] [Google Scholar]
- 63.Neagoe C., Kulke M., del Monte F. Titin isoform switch in ischemic human heart disease. Circulation. 2002;106:1333–1341. doi: 10.1161/01.cir.0000029803.93022.93. [DOI] [PubMed] [Google Scholar]
- 64.Kao D.P., Lowes B.D., Gilbert E.M. Therapeutic molecular phenotype of beta-blocker-associated reverse-remodeling in nonischemic dilated cardiomyopathy. Circ Cardiovasc Genet. 2015;8:270–283. doi: 10.1161/CIRCGENETICS.114.000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bedi K.C., Jr., Snyder N.W., Brandimarto J. Evidence for Intramyocardial disruption of lipid metabolism and increased myocardial ketone utilization in advanced human heart failure. Circulation. 2016;133:706–716. doi: 10.1161/CIRCULATIONAHA.115.017545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nanni L., Romualdi C., Maseri A., Lanfranchi G. Differential gene expression profiling in genetic and multifactorial cardiovascular diseases. J Mol Cell Cardiol. 2006;41:934–948. doi: 10.1016/j.yjmcc.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 67.Boudina S., Abel E.D. Diabetic cardiomyopathy revisited. Circulation. 2007;115:3213–3223. doi: 10.1161/CIRCULATIONAHA.106.679597. [DOI] [PubMed] [Google Scholar]
- 68.van Rooij E., Sutherland L.B., Qi X., Richardson J.A., Hill J., Olson E.N. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 69.Thum T., Gross C., Fiedler J. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980–984. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- 70.Arora P., Wu C., Khan A.M. Atrial natriuretic peptide is negatively regulated by microRNA-425. J Clin Invest. 2013;123:3378–3382. doi: 10.1172/JCI67383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Care A., Catalucci D., Felicetti F. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 72.Devaux Y., Zangrando J., Schroen B. Long noncoding RNAs in cardiac development and ageing. Nat Rev Cardiol. 2015;12:415–425. doi: 10.1038/nrcardio.2015.55. [DOI] [PubMed] [Google Scholar]
- 73.Liu S.J., Horlbeck M.A., Cho S.W. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science. 2017;355 doi: 10.1126/science.aah7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li J.J., Biggin M.D. Gene expression. Statistics requantitates the central dogma. Science. 2015;347:1066–1067. doi: 10.1126/science.aaa8332. [DOI] [PubMed] [Google Scholar]
- 75.Kooij V., Venkatraman V., Tra J. Sizing up models of heart failure: Proteomics from flies to humans. Proteomics Clin Appl. 2014;8:653–664. doi: 10.1002/prca.201300123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Di Salvo T.G., Haldar S.M. Epigenetic mechanisms in heart failure pathogenesis. Circ Heart Fail. 2014;7:850–863. doi: 10.1161/CIRCHEARTFAILURE.114.001193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Das A., Morley M., Moravec C.S. Bayesian integration of genetics and epigenetics detects causal regulatory SNPs underlying expression variability. Nat Commun. 2015;6:8555. doi: 10.1038/ncomms9555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang X., Tucker N.R., Rizki G. Discovery and validation of sub-threshold genome-wide association study loci using epigenomic signatures. eLife. 2016;5:e10557. doi: 10.7554/eLife.10557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Taegtmeyer H. Failing heart and starving brain: ketone bodies to the rescue. Circulation. 2016;134:265–266. doi: 10.1161/CIRCULATIONAHA.116.022141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bing R.J. Myocardial metabolism. Circulation. 1955;12:635–647. doi: 10.1161/01.cir.12.4.635. [DOI] [PubMed] [Google Scholar]
- 81.Neubauer S. The failing heart: an engine out of fuel. N Engl J Med. 2007;356:1140–1151. doi: 10.1056/NEJMra063052. [DOI] [PubMed] [Google Scholar]
- 82.Diakos N.A., Navankasattusas S., Abel E.D. Evidence of glycolysis up-regulation and pyruvate mitochondrial oxidation mismatch during mechanical unloading of the failing human heart. Implications for cardiac reloading and conditioning. J Am Coll Cardiol Basic Trans Science. 2016;1:432–444. doi: 10.1016/j.jacbts.2016.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Heggermont W.A., Papageorgiou A.P., Heymans S., van Bilsen M. Metabolic support for the heart: complementary therapy for heart failure? Eur J Heart Fail. 2016;18:1420–1429. doi: 10.1002/ejhf.678. [DOI] [PubMed] [Google Scholar]
- 84.Margulies K.B., Hernandez A.F., Redfield M.M. Effects of liraglutide on clinical stability among patients with advanced heart failure and reduced ejection fraction: a randomized clinical trial. JAMA. 2016;316:500–508. doi: 10.1001/jama.2016.10260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Swedberg K., Eneroth P., Kjekshus J., Wilhelmsen L. Hormones regulating cardiovascular function in patients with severe congestive heart failure and their relation to mortality. CONSENSUS Trial Study Group. Circulation. 1990;82:1730–1736. doi: 10.1161/01.cir.82.5.1730. [DOI] [PubMed] [Google Scholar]
- 86.Braunwald E. Biomarkers in heart failure. N Engl J Med. 2008;358:2148–2159. doi: 10.1056/NEJMra0800239. [DOI] [PubMed] [Google Scholar]
- 87.Cohn J.N., Levine T.B., Olivari M.T. Plasma norepinephrine as a guide to prognosis in patients with chronic congestive heart failure. N Engl J Med. 1984;311:819–823. doi: 10.1056/NEJM198409273111303. [DOI] [PubMed] [Google Scholar]
- 88.Smith J.G., Gerszten R.E. Emerging affinity-based proteomic technologies for large-scale plasma profiling in cardiovascular disease. Circulation. 2017;135:1651–1664. doi: 10.1161/CIRCULATIONAHA.116.025446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ngo D., Sinha S., Shen D. Aptamer-based proteomic profiling reveals novel candidate biomarkers and pathways in cardiovascular disease. Circulation. 2016;134:270–285. doi: 10.1161/CIRCULATIONAHA.116.021803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mann D.L. Inflammatory mediators and the failing heart: past, present, and the foreseeable future. Circ Res. 2002;91:988–998. doi: 10.1161/01.res.0000043825.01705.1b. [DOI] [PubMed] [Google Scholar]
- 91.Handoko M.L., de Man F.S., Vonk-Noordegraaf A. The rise and fall of endothelin receptor antagonists in congestive heart failure. Eur Respir J. 2011;37:484–485. doi: 10.1183/09031936.00145910. [DOI] [PubMed] [Google Scholar]
- 92.Jacobs L., Thijs L., Jin Y. Heart “omics” in AGEing (HOMAGE): design, research objectives and characteristics of the common database. J Biomed Res. 2014;28:349–359. doi: 10.7555/JBR.28.20140045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang T.J., Larson M.G., Vasan R.S. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17:448–453. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sun H., Olson K.C., Gao C. Catabolic defect of branched-chain amino acids promotes heart failure. Circulation. 2016;133:2038–2049. doi: 10.1161/CIRCULATIONAHA.115.020226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kitai T., Kirsop J., Tang W.H. Exploring the microbiome in heart failure. Curr Heart Fail Rep. 2016;13:103–109. doi: 10.1007/s11897-016-0285-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lommi J., Kupari M., Koskinen P. Blood ketone bodies in congestive heart failure. J Am Coll Cardiol. 1996;28:665–672. doi: 10.1016/0735-1097(96)00214-8. [DOI] [PubMed] [Google Scholar]
- 97.Matsushita K., Williams E.K., Mongraw-Chaffin M.L. The association of plasma lactate with incident cardiovascular outcomes: the ARIC study. Am J Epidemiol. 2013;178:401–409. doi: 10.1093/aje/kwt002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang T.J., Gupta D.K. Metabolite profiles in heart failure: looking for unique signatures in a heterogeneous syndrome. J Am Coll Cardiol. 2015;65:1521–1524. doi: 10.1016/j.jacc.2015.02.019. [DOI] [PubMed] [Google Scholar]
- 99.Sender R., Fuchs S., Milo R. Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell. 2016;164:337–340. doi: 10.1016/j.cell.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 100.Pedersen H.K., Gudmundsdottir V., Nielsen H.B. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature. 2016;535:376–381. doi: 10.1038/nature18646. [DOI] [PubMed] [Google Scholar]