Abstract
We analyzed, in an integrative fashion, the morphometry and structural integrity of the bilateral hippocampi and amygdalas in Alzheimer's disease (AD) using T1-weighted images and diffusion tensor images (DTIs). We detected significant hippocampal and amygdalar volumetric atrophies in AD relative to healthy controls (HCs). Shape analysis revealed significant region-specific atrophies with the hippocampal atrophy mainly being concentrated on the CA1 and CA2 while the amygdalar atrophy was concentrated on the basolateral and basomedial. In all structures, the structural integrity displayed a significantly decreased mean fractional anisotropy (FA) value and an increased mean trace value in AD. In addition to the inter-group comparisons, we systematically evaluated the discriminative power of our three types of features (volume, shape, and DTI), both individually and in their possible combinations, when differentiating between AD and HCs. We found the volume features to be redundant when the more sophisticated shape features were available. A combination of the shape and DTI features of the right hippocampus, with classification automatically performed by support vector machine, yielded the strongest classification result (overall accuracy, 94.6%; sensitivity, 95.5%; specificity, 93.3%).
Keywords: Hippocampus, Amygdala, Shape, Diffusion tensor imaging, Support vector machine, Linear discriminant analysis
1. Introduction
Alzheimer's disease (AD) is a progressive neurodegenerative disorder that is characterized by long-term memory loss, language impairment, and difficulty thinking. AD is the most common form of dementia, primarily affecting people over the age of 65 years. For research purposes, a probabilistic diagnosis of AD is usually based on neurological examinations and psychometric assessments such as the National Institute of Neurological and Communicative Disorders and Stroke and the Alzheimer's Disease and Related Disorders Association (NINCDS–ADRDA) criteria [1]. However, with the ever-growing development of brain imaging techniques, structural abnormalities of the human brain have been identified as effective biomarkers for AD, especially for prodromal cases, and have been included in the research criteria for the diagnosis of AD [2].
Among the anatomical structures of particular interest to the study of AD are the medial temporal lobe (MTL) structures, such as the hippocampus and the amygdala, primarily because of their active involvement in memory [3–6]. Morphometric abnormalities, in terms of both the global volume and the local shape, induced by the neuropathology of AD in the hippocampus and the amygdala have seen considerable investigation. Assessments for such abnormalities are usually conducted via the examination of structural magnetic resonance images (MRIs) such as the T1-weighted image. Generally, volumetric atrophy of those two MTL structures is a consistent observation in studies of AD, something that is particularly true for the hippocampus [7–14].
During the past decade, advancements in shape analysis methods have facilitated the detailed examination of hippocampal AD shape abnormalities, such as local surface atrophy [15–21]. However, studies of the impact of AD upon amygdalar shape have been relatively few in number. Only in recent years has the study of local morphometric properties (such as the localized surface area, as induced by a surface mesh representing the shape) of the amygdala in AD begun to gain momentum, with the detection of significant, region-specific atrophy being recorded using advanced computational algorithms [17,20,22–26].
With the advent of diffusion tensor imaging (DTI) technique comes new tools and new perspectives with which one can assess AD-related structural abnormality. DTI enables the detection of diffusional brain abnormalities at a stage where macrostructural abnormalities may not yet be visible. DTI measures the diffusion of water molecules in neural tissue and in so doing quantifies structural integrities, such as the white matter (WM) integrity. It is from such measurements that we know of significant decreases of WM integrity in subjects with mild cognitive impairment (MCI) and AD [27–30].
When quantifying structural integrity, there are two DTI indices which are most often looked to. The first, fractional anisotropy (FA), is a scalar value between zero and one that measures anisotropic water diffusion in the brain and reflects the degree to which directional diffusion occurs in the cellular structures of fiber tracts. FA provides useful information when examining fiber density, axonal diameter, and myelination in WM. A decrease in the FA value suggests a loss of fiber tract integrity and thus WM damage [31]. The second index, mean diffusivity (MD), measures the average diffusivity in the non-colinear directions of free diffusion [31]. An increase in MD indicates a loss of anisotropy and thus represents an increase in free water diffusion. Bringing these two DTI indices to bear in the analysis of the hippocampus and amygdala under AD is something yet to be fully attempted. There are two existing studies that consider the hippocampus [32,33] but none has approached the amygdala. This deficit may well be due to these structures lying outside the usual class that DTI is applied to; namely, WM structures as opposed to deep gray matter like the hippocampus and the amygdala. However, DTI may provide novel and unique angles for examining the hippocampal and amygdalar abnormalities induced by AD, especially in combination with the morphometric structural measures. Indeed, morphometric properties, in terms of volume and shape, and structural integrity, as measured by DTI indices, provide compatible and complementary information for understanding hippocampal and amygdalar abnormalities in the neuropathology of AD. With that in mind, this paper aims for an integrative investigation of both volume and shape alongside microstructural integrity with regard to the hippocampus and the amygdala of patients with AD as compared to healthy control (HC) subjects.
When examining shape, this study will make use of a well-established statistical shape analysis pipeline to explore the local morphometric properties of the hippocampus and the amygdala. This shape analysis pipeline lies in the framework of large deformation diffeomorphic metric mapping (LDDMM) [34], the key idea of which is to quantify local morphometric properties using a diffeomorphism that connects two coordinate systems of interest and is itself obtained as the endpoint of a geodesic. This pipeline has been successfully applied to various aspects of AD-related investigation, including the detection and quantification of subcortical surface area atrophies in MCI and AD [20] as well as preclinical AD [23,35], the quantification of rates of change in the localized surface areas of the hippocampus, the amygdala, and the ventricle in MCI and AD [24], the analysis of MTL shape neurodegeneration networks in preclinical AD [36], the interactive effects of the apolipoprotein E genotype and age upon hippocampal and amygdalar shapes in MCI and AD [37], and the estimation of the onset time for MTL morphometric abnormalities induced by AD [38]. In keeping with our previous studies, we will introduce high-field (7 Tesla) subregional divisions of the hippocampus (four subregions: CA1, CA2, CA3 combined with dentate gyrus, and subiculum) and the amygdala (four subregions: basolateral, basomedial, centromedial, and lateral nucleus) for a more detailed evaluation of the regionally specific shape abnormalities in AD.
Going beyond the quantification of AD-induced abnormalities of the hippocampus and the amygdala in terms of volume, shape, and DTI indices, for each structure, we will also explore the discriminative power of these features' corresponding biomarkers; its morphometric biomarkers, its DTI-derived biomarkers, as well as its combined multi-modality biomarkers will be systematically tested for the capability to differentiate between AD and HC. Points of investigation include whether combining different types of features will boost the classification accuracy and thus provide greater structure-specific discriminative abilities between AD and HC. Furthermore, we will explore the discriminative potential of introducing dimension reduction for the shape features. Guided by our previous work [20,37], we will consider principal component analysis (PCA) and Student's t-test as candidates for the dimension reduction techniques. For the discrimination algorithm, we will first examine the performance of linear discriminant analysis (LDA) before turning to support vector machine (SVM), the latter of which we include due to its superior performance in the context of high dimensional feature spaces and applicability to clinical disease categorization [39–43].
In summary, this study will present results from the following investigations: (1) quantitative comparisons of volumetric measurements of the bilateral hippocampi and amygdalas between HC and AD; (2) surface maps demonstrating the localized HC vs. AD surface area group differences in the bilateral hippocampi and amygdalas as well as their subdivisions into four compatible compartments; (3) comparisons between HC and AD in terms of the mean FA value and the mean trace value (the trace being three times the MD) within the bilateral hippocampi and amygdalas; and (4) structure-specific classification performance (overall accuracy, sensitivity, and specificity) in discriminating between AD and HC using various combinations of morphometric and microstructural features with two distinct machine learning techniques (LDA and SVM).
2. Materials and method
2.1. Participants and MRI dataset
A total of 29 patients with probable AD (M/F = 13/16, mean age = 67.5 ± 9.5 years), as assessed by the Alzheimer's criteria of the NINCDS–ADRDA [1], and 23 HC subjects (M/F = 11/12, mean age = 63.9 ± 7.5 years) were enrolled in this study at Tongji Hospital, Wuhan, China. Subjects with structural abnormalities that could produce dementia, such as cortical infarction, tumor, or subdural hematoma, along with those who had undergone treatment or had a concurrent illness (other than dementia) that interfered with cognitive function at the time of the MRI scan were not included in this study.
Of the total 52 subjects, 28 of the 29 AD subjects and all 23 of the HC subjects were scanned for structural data (T1-weighted image) acquisition using a 3D fast spoiled gradient-echo (FSPGR) sequence covering the whole brain of each subject with the following parameters: repetition time (TR) = 6.5 ms, echo time (TE) = 2.1 ms, inversion time (TI) = 400 ms, field of view (FOV) = 256 mm × 256 mm, phase FOV = 1, matrix = 256 × 256, slice thickness = 1.0 mm, slice gap = 0 mm, number of excitations (NEX) = 1, flip angle = 15°, and scan time = 4 min 8 s. For the acquisition of the DTI data, 23 AD and 15 HC subjects were scanned on a 3 Tesla (3 T) MR system (SignaHDxt, GE Healthcare, USA) with TR = 10,000 ms, TE = 83 ms, flip angle = 90°, matrix = 256 × 256, FOV = 240 mm × 240 mm, phase FOV = 1, slice thickness = 3.0 mm with no space, NEX = 1, total slice = 42, and b value = 1000 s/mm2 along 30 directions.
The reader should note that the total number of subjects scanned for both T1-weighted image and DTI data was 37 (22 AD and 15 HC subjects). Therefore, when evaluating the group differences in terms of morphometric properties (volume and shape), we used the 51 subjects who had T1 data; when evaluating the group differences in terms of the DTI indices, we used the 38 subjects having DTI data; and when evaluating the discriminative power of different combinations of features, we used the 37 subjects who had both T1 and DTI data. Details of each subject's demographics as well as the availability of imaging data are presented in Table 1.
Table 1.
Demographic information of each subject within the two groups — HC and AD.
| Gender | Age | T1 | DTI | MMSE | Gender | Age | T1 | DTI | MMSE | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| HC1 | F | 59 | Y | N | 28 | AD1 | F | 82 | Y | Y | 18 |
| HC2 | F | 65 | Y | Y | 28 | AD2 | F | 62 | Y | Y | 20 |
| HC3 | F | 63 | Y | Y | 29 | AD3 | M | 52 | Y | Y | 23 |
| HC4 | M | 55 | Y | Y | 28 | AD4 | F | 66 | Y | Y | 21 |
| HC5 | M | 55 | Y | N | - | AD5 | F | 59 | Y | N | 17 |
| HC6 | F | 59 | Y | Y | 29 | AD6 | M | 75 | Y | Y | 8 |
| HC7 | F | 61 | Y | Y | 28 | AD7 | F | 70 | Y | N | 21 |
| HC8 | M | 67 | Y | Y | 29 | AD8 | F | 61 | Y | Y | 15 |
| HC9 | M | 69 | Y | Y | 29 | AD9 | F | 76 | Y | N | 11 |
| HC10 | F | 56 | Y | N | - | AD10 | M | 60 | Y | Y | 16 |
| HC11 | M | 77 | Y | Y | 28 | AD11 | M | 78 | Y | Y | 17 |
| HC12 | F | 60 | Y | Y | 29 | AD12 | M | 65 | Y | N | - |
| HC13 | M | 65 | Y | Y | 29 | AD13 | M | 57 | Y | Y | 18 |
| HC14 | F | 64 | Y | Y | - | AD14 | F | 79 | Y | N | 23 |
| HC15 | M | 76 | Y | N | - | AD15 | F | 55 | Y | Y | 18 |
| HC16 | M | 56 | Y | N | - | AD16 | M | 59 | N | Y | 17 |
| HC17 | F | 56 | Y | N | - | AD17 | M | 60 | Y | Y | 15 |
| HC18 | F | 73 | Y | Y | 28 | AD18 | M | 73 | Y | Y | 25 |
| HC19 | F | 65 | Y | N | - | AD19 | F | 69 | Y | Y | - |
| HC20 | M | 58 | Y | Y | 29 | AD20 | F | 67 | Y | Y | 14 |
| HC21 | F | 82 | Y | Y | 27 | AD21 | F | 59 | Y | Y | 21 |
| HC22 | M | 63 | Y | Y | 28 | AD22 | M | 63 | Y | Y | - |
| HC23 | M | 65 | Y | N | - | AD23 | F | 84 | Y | Y | 15 |
| AD24 | F | 63 | Y | Y | - | ||||||
| AD25 | F | 56 | Y | Y | 12 | ||||||
| AD26 | M | 81 | Y | Y | 13 | ||||||
| AD27 | F | 65 | Y | Y | 17 | ||||||
| AD28 | M | 81 | Y | N | - | ||||||
| AD29 | M | 81 | Y | Y | 13 |
“F″ for Gender means female while “M” means male. “Y″ for T1 or DTI means the corresponding MRI modality is available whereas “N” means not available. MMSE — Mini-mental State Examination.
This study was approved by the internal institutional review board of Tongji Hospital and written informed consent was obtained from all participants; in the case of patients with dementia, consent was obtained from family members.
2.2. Volumetric segmentation from T1
For each of the 51 T1-weighted images, volumetric segmentations of the bilateral hippocampi and amygdalas were automatically obtained from a hierarchical segmentation pipeline [44] consisting of two steps; skull-stripping and brain structure segmentation. Built upon a two-level diffeomorphic multi-atlas likelihood-fusion algorithm in the framework of the random deformable template model [45], this segmentation algorithm has demonstrated superior performance when compared to other state-of-the-art methods, especially when applied to subcortical structures including the hippocampus and the amygdala. Detailed evaluations of the segmentation accuracy of this method can be found in our previous studies [44–46]. For the current study, we used 16 atlases (T1-weighted images) belonging to the same age range as the participants of this study. The four structures of interest (left and right hippocampus and amygdala) were manually delineated in each of the 16 atlases by a neuroanatomist with more than 15 years of experience in manual tracing. We visually examined the segmentation results and manual correction was employed when necessary to ensure a high segmentation quality. The volumetric measurement of each structure was calculated as the total number of voxels within that structure multiplied by the voxel resolutions (1 mm × 1 mm × 1 mm).
2.3. Shape processing
To extract the localized shape morphometrics, we created a 2-D triangulated surface contouring the boundary of each 3-D volumetric segmentation based on an approach similar to that which has been detailed and validated in an earlier study [20]. Briefly, for each 3-D volumetric segmentation corresponding to a structure of interest, its bounding surface was obtained by applying an optimized diffeomorphism to a template surface of that specific structure. These template surfaces for our structures of interest were created manually, ensuring sufficient smoothness and correct anatomical topology. Each optimized diffeomorphism was obtained through using LDDMM with appropriately selected parameters [47] to map the template segmentation to the scan-specific segmentation of the same structure. This surface-generation methodology was used to create the target shapes whose localized surface-based morphometrics, in terms of the surface areas associated to vertices of the triangulated mesh, were then extracted.
The localized surface-based morphometrics (the vertex-wise surface areas) of each target shape were quantified by a diffeomorphism that connected a common template shape to that target shape. The common template shape of each structure of interest was itself generated from the collection of 51 target surfaces using a Bayesian template estimation algorithm [48] to ensure “population averaging” by iteratively minimizing the overall metric distance from the common template surface to each target surface, in the setting of LDDMM, until convergence. Proceeding onwards, we used the LDDMM-surface mapping algorithm [49] to map the common template surface to each individual target surface, from which a scalar field was subsequently calculated as the log-determinant of the Jacobian of the diffeomorphism. This scalar field, indexed at each vertex of the common template surface, quantifies the factor by which the diffeomorphism expands or shrinks the localized surface area in the target relative to the template in a logarithmic scale; i.e. a positive value corresponds to a localized surface area expansion of the subject relative to the template while a negative value suggests a localized surface area contraction. This structure-specific and subject-specific scalar field, referred to as the deformation marker, is what we ultimately compared between the two groups and used as the set of shape features in our subsequent discriminant experiments.
2.4. DTI index extraction
All DTI datasets were processed using a fully-automated and web-based DTI data processing pipeline (a module implemented at www.mricloud.org) that includes: (1) DTI image quality control through the correction of mis-registration between diffusion weighted images caused by patient motion and eddy-current-induced distortion [50]; (2) DTI image corruption detection and rejection [51]; and (3) tensor calculation, which included providing the 3D FA and trace maps.
Instead of co-registering the DTI images and the T1-weighted images in subject-based pairs, we opted to perform the morphometric analysis based solely on the T1-weighted images while separately analyzing the microstructural qualities of the DTI images. To extract the mean FA and mean trace values within the bilateral hippocampi and amygdalas of each subject, we automatically segmented those four structures of interest from DTI images using a multi-modality extension of the aforementioned T1-based segmentation algorithm — the multi-modality diffeomorphic multi-atlas likelihood fusion, with multiple atlases created from DTI images. Details about this multi-atlas DTI segmentation algorithm and its performance, as well as creation of the multiple DTI atlases, can be found in our previous work [52].
2.5. Group comparison analysis
For inter-group comparisons of the localized shape morphometrics of the bilateral hippocampi and amygdalas, we employed the same linear regression model as described in [20], namely , where yk(s) is the deformation marker for subject s at vertex k of the structure-specific common template surface, g(s) is a binary group variable, Xcov(s) denotes the covariate information of subject s included in the analysis (in this study, we co-varied for age, gender, as well as the total intracranial volume (TIV) computed from the size of the brain's image after skull-stripping [44,53]), and εk(s) denotes a Gaussian noise structure. We tested the null hypothesis against the general hypothesis with the complete null hypothesis being simultaneously for all k. The statistical significance of a group difference is measured by a p-value obtained from Fisher's method of randomization; a non-parametric permutation test (a total of 40,000 permutations employed) was conducted by randomizing the model's residuals, more details on which can be found in Ref. [38]. To correct for multiple comparisons being performed simultaneously at all vertices of the template surface, we adjusted the p-values to control for familywise error rate (FWER) at a level of 0.05 based on the maximum statistic method [54]. For a direct comparison, we also present the statistical shape analysis results with the multiple comparison correction having been performed by controlling the false discovery rate (FDR) [55] at a level of 0.05. To examine the group differences between AD and HC in our global variables, the volumetric measurements and the DTI indices (mean FA and mean trace), we used the same linear regression model as that for shape, with the slight simplification of needing only one comparison at a time and being able to omit the multiple comparison correction. Please note that in the inter-group comparisons of DTI indices, we co-varied for age and gender but not TIV.
2.6. Template surface subdivision
With the help of accurate subsegmentations of the hippocampus and the amygdala based on high resolution MRI scans obtained from a high field scanner, we divided our common template surface for each of the four structures into multiple functionally-distinct anatomical subregions using the approach detailed in [20]. This subdivision was accomplished by manually sub-dividing the surfaces of high-field segmentations (obtained from a 7 T scanner with an image voxel resolution of 0.8 mm) and transferring the boundary definitions of those subregions to our population's common template surfaces using the geodesic positioning function provided by diffeomorphisms [56]. The geodesic positioning algorithm provides a diffeomorphic correspondence between the two atlas coordinate systems , with the corresponding diffeomorphism φ transporting the label maps defined on the 7 T high field atlas to the population template surfaces. Both the left and right hippocampus were subdivided into four subregions; CA1, CA2, CA3 combined with the dentate gyrus, and the subiculum. The bilateral amygdalas were also subdivided into four subregions; the basolateral, the basomedial, the centromedial, and the lateral nucleus.
2.7. Discriminant analysis
For each structure, there are three types of extracted features, the volumetric measurement (a scalar), the vertex-based shape deformation marker (a scalar field defined on the vertices of the template surface), and the DTI indices (two scalars). Since there are a total of 372 vertices on the left hippocampal template surface, 507 vertices on the right hippocampal template surface, 126 vertices on the left amygdalar template surface, and 200 vertices on the right amygdalar template surface, the shape features are of a rather high dimension when compared to the sample size (37 subjects). To reduce the dimension of the shape feature space for each structure, we performed PCA on the shape deformation markers of all the subjects to construct an orthonormal basis of principal components (PCs). This allowed our feature space of deformation markers to be linearly projected to the orthogonal directions that carry the greatest shape variance. We selected the first M PCs that account for 95% of the total variance and restricted our feature space to this M-dimensional subspace. In addition to PCA, we could further reduce the dimension based on the results of Student's t-tests; in this case, only those from that first M PCs whose projected coefficients differed significantly between the two groups at a level of 0.05 were retained.
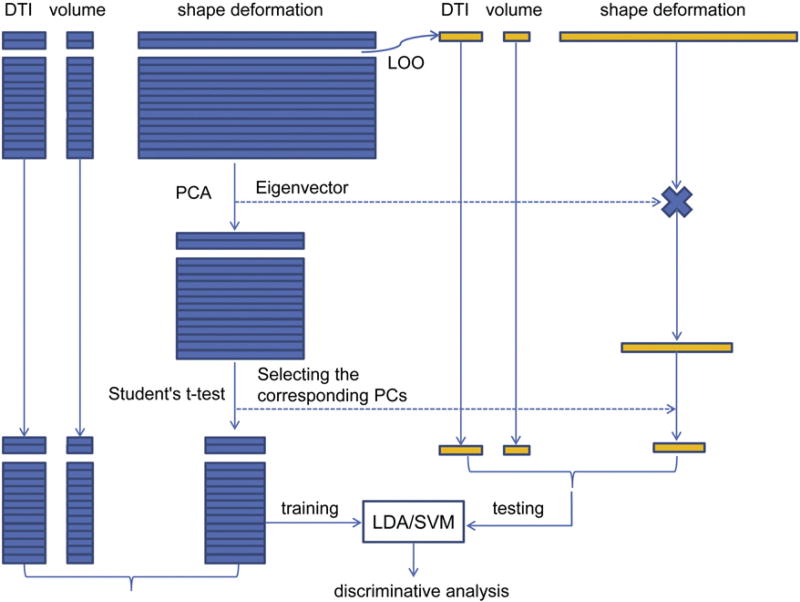
For the classification problem, LDA and SVM were both investigated as our machine learning algorithms to perform automated classification on our reduced feature spaces. To estimate the true classification rates, we used leave one out (LOO) as our cross-validation strategy; we set aside one subject at the very beginning of the discriminant analysis to be the testing subject and then proceeded to use the other 36 subjects as the training datasets. It is important to note that the PCA process occurred inside the LOO loop; that is, both the PCA and the Student's t-tests were performed upon the 36 training subjects and thus a total of 37 PCA and t-test based selection processes were performed per structure over the course of the experiment. The work flow of our discriminant analysis is illustrated in Fig. 1.
Fig. 1. Classification workflow.
Work flow of the classification experiments using various combinations of features in a leave-one-out fashion.
In this study, we first evaluated the discriminating capability of each of the three types of features (volume, shape, and DTI) of the left hippocampus, the right hippocampus, the left amygdala, and the right amygdala. Probing further, we then assessed the classification power yielded by combining different types of features, resulting in a total of four permutations: volume combined with shape, volume combined with DTI, shape combined with DTI, and shape combined with volume and DTI, for each structure of interest. It is worthy of note that in each case, given our initial feature space, we tested three stages of dimension reduction to evaluate the efficiency of our techniques; those being the original shape deformation markers, the shape deformation markers after PCA reduction, and the shape deformation markers after PCA and Student's t-test reductions. The classification accuracies collected from the range of settings were compared in terms of the overall classification accuracy (the percentage of all 37 subjects who were classified correctly), specificity (the percentage of all 22 AD subjects who were correctly identified as being AD), and sensitivity (the percentage of all 15 HC subjects who were correctly identified as being healthy).
3. Results
3.1. Group comparisons
3.1.1. Volume analysis
The mean and standard deviations of the volumetric measurements of the four structures of interest in both AD and HC are presented in Table 2, along with the p-values obtained from the inter-group comparisons of these measurements for each structure. In this acquired data, we found significant volumetric atrophies of the bilateral hippocampi and amygdalas in AD relative to those in HC (all p-values smaller than 0.05, see Table 2 for detailed values), with the atrophy of each structure in the right hemisphere being both stronger and more significant than its left hemisphere counterpart (as revealed by the group differences and the p-values). Fig. 2 gives structure-wise scatter plots of the volumetric measurements with separation by subject grouping, each having their group mean indicated. From these, the volumetric inter-group differences are apparent.
Table 2.
Top panel represents the mean and standard deviations of each structure-specific set of volumetric measurements, mean FA values, mean trace values for the bilateral hippocampi and amygdalas of the two groups (HC and AD).
| Left Amygdala | Right Amygdala | Left Hippocampus | Right Hippocampus | ||
|---|---|---|---|---|---|
| volume | HC | 1160.69 ± 161.62 | 1210.17 ± 159.75 | 3139.22 ± 296.43 | 3460.61 ± 312.25 |
| AD | 989.11 ± 272.42 | 976.64 ± 303.49 | 2695.25 ± 572.91 | 2844.32 ± 644.15 | |
| FA | HC | 0.1377 ± 0.0139 | 0.1277 ± 0.0127 | 0.1463 ± 0.0150 | 0.1456 ± 0.0171 |
| AD | 0.1283 ± 0.0224 | 0.1147 ± 0.0146 | 0.1289 ± 0.0178 | 0.1279 ± 0.0146 | |
| trace | HC | 0.0027 ± 0.0002 | 0.0028 ± 0.0002 | 0.0034 ± 0.0005 | 0.0034 ± 0.0005 |
| AD | 0.0032 ± 0.0005 | 0.0035 ± 0.0006 | 0.0046 ± 0.0006 | 0.0045 ± 0.0007 | |
| HC vs. AD p-values | Volume | 0.0294 | 0.0027 | 0.0032 | 0.0002 |
| Shape | 0.0249 | <2.5E-5 | 0.0022 | <2.5E-5 | |
| FA | 0.2071 | 0.0041 | 0.0047 | 0.0022 | |
| Trace | 0.0004 | 0.0002 | <2.5E-5 | <2.5E-5 |
Bottom panel lists the p-values in comparing each type of features (volume, shape, mean FA, mean trace) between HC and AD.
Fig. 2. Volumetric measurement of each subject for each structure.
Scatterplots of the volumetric measurements (in mm3) for the bilateral hippocampi and amygdalas in HC (blue diamonds) and AD (red outlined circles) subjects. Bars denote the mean volumetric measurements of each group.
3.1.2. Shape analysis
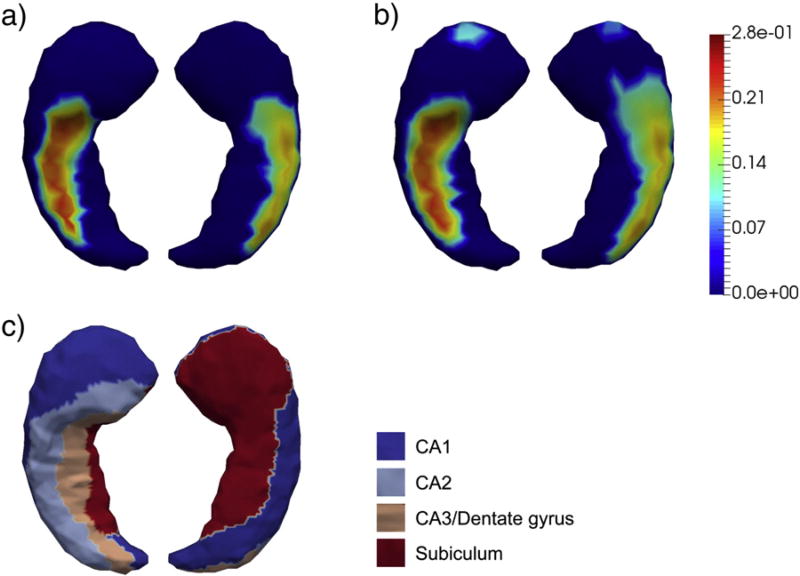
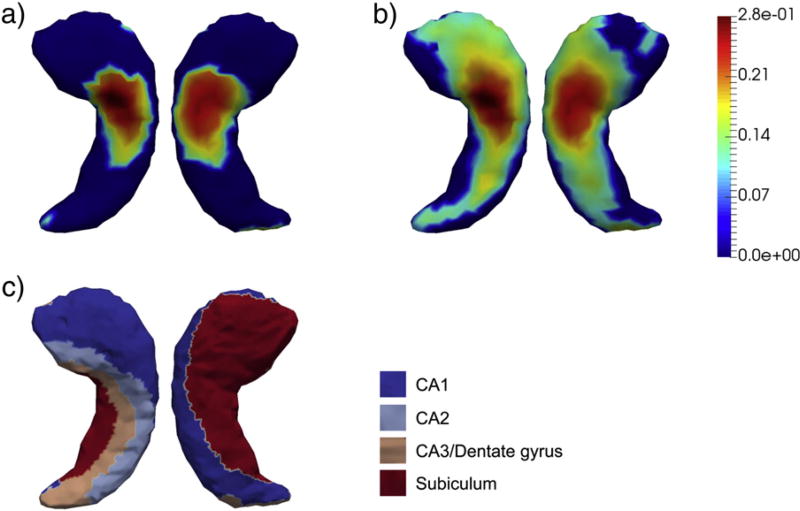
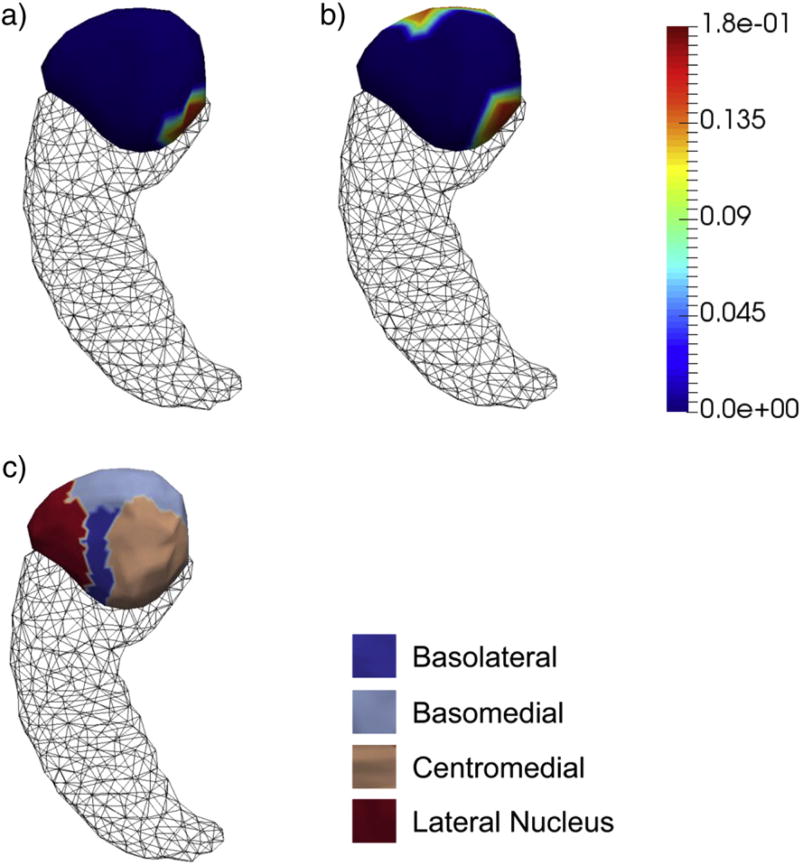
According to our vertex-based shape analyses, we detected significant group differences for each of the four structures upon rejecting the complete null hypothesis, the p-values of which are listed in Table 2. In Figs. 3–6, we present the shape analysis results at each vertex of the left hippocampus, the right hippocampus, the left amygdala, and the right amygdala respectively. In each of the four figures, panel (a) represents the shape difference maps after FWER-correction; that is, only vertices at which the significance of shape group differences survived the FWER multiple comparison correction are highlighted. Panel (b) represents the shape difference maps after FDR-correction, and panel (c) represents the subregional divisions of each template surface.
Fig. 3. Shape analysis results on the left hippocampus.
Panel (a) and panel (b) show the statistically significant vertex-wise surface area differences of the left hippocampus after FWER-correction and FDR-correction respectively. The color bar denotes the proportion of atrophy in AD relative to HC. Panel (c) denotes the four-compartment subdivisions of the study-specific template surface of the left hippocampus.
Fig. 6. Shape analysis results on the right amygdala.
Panel (a) and panel (b) show the statistically significant vertex-wise surface area differences of the right amygdala after FWER-correction and FDR-correction respectively. The color bar denotes the proportion of atrophy in AD relative to HC. Panel (c) denotes the four-compartment subdivisions of the study-specific template surface of the right amygdala.
As demonstrated in those four figures, significant regionally specific atrophies were detected on all four structures in AD relative to HC, with the magnitudes and the overall significance of the structures in the right hemisphere being much larger than those of the corresponding structures in the left hemisphere, which is consistent with our volumetric observations. Furthermore, the magnitude and significance of the hippocampal shape abnormalities are overall more substantial than those detected on the amygdala. The FWER-correction procedure was found to be more conservative than the FDR based correction, with the shape differences of many fewer vertices being determined to be statistically significant. As revealed by the template subdivisions of the left hippocampus in Fig. 3, the primarily affected regions of the left hippocampus belonged to CA1 and CA2. According to Fig. 4, the surface area atrophies on the right hippocampus were found on vertices belonging to parts of each subregion (CA1, CA2, CA3 combined with the dentate gyrus, and the subiculum), with the strongest atrophies occurring at vertices belonging to CA1, CA2, and the subiculum. The total number of vertices in the left amygdala that exhibited AD-related abnormalities is much smaller than those seen in the other three structures, being mainly concentrated on the centromedial region (see Fig. 5). For the right amygdala, as illustrated in Fig. 6, the significant group differences in terms of shape are strongly concentrated upon the basolateral and basomedial subregions.
Fig. 4. Shape analysis results on the right hippocampus.
Panel (a) and panel (b) show the statistically significant vertex-wise surface area differences of the right hippocampus after FWER-correction and FDR-correction respectively. The color bar denotes the proportion of atrophy in AD relative to HC. Panel (c) denotes the four-compartment subdivisions of the study-specific template surface of the right hippocampus.
Fig. 5. Shape analysis results on the left amygdala.
Panel (a) and panel (b) show the statistically significant vertex-wise surface area differences of the left amygdala after FWER-correction and FDR-correction respectively. The color bar denotes the proportion of atrophy in AD relative to HC. Panel (c) denotes the four-compartment subdivisions of the study-specific template surface of the left amygdala.
3.1.3. DTI analysis
The summarizing statistics (mean and standard deviation) and group-comparison p-values of the mean FA and mean trace values for the four structures of the two groups are listed in Table 2, from which we observed lower FA and higher trace on average in HC than in AD. According to the p-values listed in Table 2, all group differences in terms of the two DTI indices have reached statistical significance except the left amygdala FA. This again corresponds to our morphometric findings that the HC vs. AD group differences in both volume and shape of the left amygdala are the weakest to be found among the four structures. In Figs. 7 and 8, we give the scatter plots of the subject-specific mean FA and mean trace values respectively, within the bilateral hippocampi and amygdalas, as well as the mean values computed across each group.
Fig. 7. Mean FA value of each subject within each structure.
Scatterplots of the mean FA values within the left and right hippocampus and amygdala in HC (blue diamonds) and AD (red open circles) subjects. Bars denote the value of the mean FA averaged across each group.
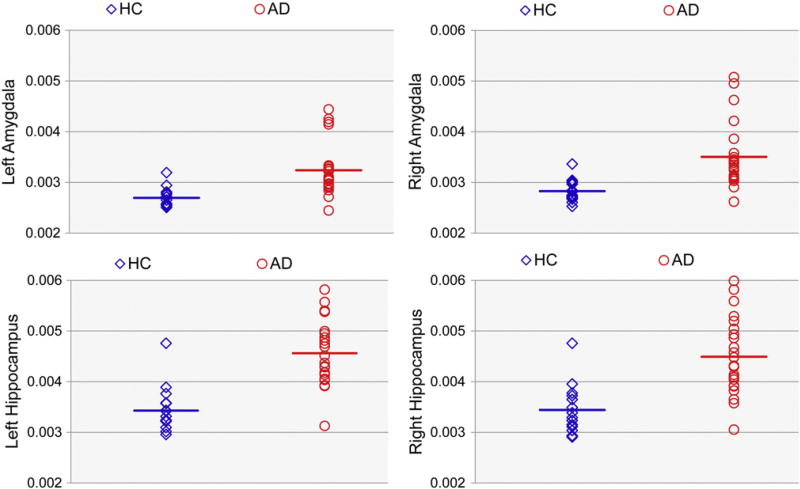
Fig. 8. Mean trace value of each subject within each structure.
Scatterplots of the mean trace values within the left and right hippocampus and amygdala in HC (blue diamonds) and AD (red open circles) subjects. Bars denote the value of the mean trace averaged across each group.
3.2. Discriminant analysis
The structural variability, as revealed by the shape deformation markers, was mainly concentrated on the first 12–13 PCs of the bilateral hippocampi and the first 6–7 PCs of the bilateral amygdalas. As there were a total of 37 instances of PCA for each structure, upon finding the first M PCs accounting for the 95% variability in our data, we saw that for the left hippocampus, M = 12 in 34 instances and M = 13 in the remainder; for the right hippocampus, M = 12 in 31 instances and M = 13 in the remainder; for the left amygdala, M = 6 in 33 instances and M = 7 in the remainder; and for the right amygdala, M = 6 in 17 instances and M = 7 in the remainder. The further reduction by Student's t-tests yielded a remaining 1–2 PCs for the left hippocampus (1 occurring in 16 cases and 2 in the other 21), 2–4 PCs for the right hippocampus (2 occurring in 6 cases, 3 in 30 of the others, and a single case of 4), 0–1 PC for the left amygdala (0 occurring twice and 1 occurring 35 times), and 0–1 PC for the right amygdala (0 occurring in 11 cases and 1 in the other 26). Given that it was a highly frequent occurrence for the Student's t-tests to exclude all of the amygdalar shape features, we concluded that the Student's t-test in addition to PCA was not a suitable dimension reduction procedure for the bilateral amygdalar features of this dataset. This issue may be resolved if there were more available subjects. Therefore, we will not report classification results involving amygdalar shape features after PCA and Student's t-tests.
3.2.1. Single feature analysis
In Table 3, we present the structure-wise classification rates, including the overall accuracy, the sensitivity, and the specificity, achieved when discriminating between AD and HC subjects using each one of the three feature types in isolation and when taking either LDA or SVM to be our classification algorithm. A careful examination of these results reveals that the hippocampus on average had a stronger AD-discriminating capability than the amygdala, regardless of the machine learning algorithm and the feature type used. Furthermore, for each of the four structures of interest, SVM yielded classification accuracies similar to those of LDA when using the morphometric features, either volume or shape. However, SVM was highly superior to LDA when using the DTI features to perform the automated classification. Another interesting observation is that, for each structure of interest, when using LDA, shape deformation features were able to deliver the highest classification accuracy among the feature types whereas DTI features yielded the best results when using SVM. In fact, when using only a single type of feature, the combination of SVM and the DTI features of the left hippocampus yielded the best performance (overall accuracy, 89.2%; sensitivity, 90.9%; specificity, 86.7%). Between the two types of morphometric features, volume and shape, we observed that, with appropriate dimension reduction technique, the shape features outperformed volumetrics irrespective of the classification technique.
Table 3.
Discrimination performances, as quantified by the overall accuracy (Acc), sensitivity (Sen), and Specificity (Spe), of using each structure-specific feature (volume, shape, and DTI) with two classification algorithms — LDA and SVM.
| LDA | SVM | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||||
| Volume | Shape | DTI | Volume | Shape | DTI | ||||||
|
|
|
||||||||||
| Original | PCA | PCA + ttest | Original | PCA | PCA + ttest | ||||||
| Left Hippocampus | Acc | 64.9% | 51.4% | 81.1% | 75.7% | 78.4% | 64.9% | 83.8% | 83.8% | 73.0% | 89.2% |
| Sen | 54.5% | 54.5% | 72.7% | 68.2% | 68.2% | 50.0% | 77.3% | 86.4% | 63.6% | 90.9% | |
| Spe | 80.0% | 46.7% | 93.3% | 86.7% | 93.3% | 86.7% | 93.3% | 80.0% | 86.7% | 86.7% | |
| Right Hippocampus | Acc | 73.0% | 62.2% | 73.0% | 86.5% | 81.1% | 78.4% | 78.4% | 70.3% | 86.5% | 83.8% |
| Sen | 68.2% | 63.6% | 59.1% | 81.8% | 72.7% | 63.6% | 72.7% | 63.6% | 81.8% | 86.4% | |
| Spe | 80.0% | 60.0% | 93.3% | 93.3% | 93.3% | 100.0% | 86.7% | 80.0% | 93.3% | 80.0% | |
| Left Amygdala | Acc | 59.5% | 40.5% | 73.0% | / | 59.5% | 59.5% | 62.2% | 70.3% | / | 83.8% |
| Sen | 59.1% | 36.4% | 63.6% | / | 36.4% | 45.5% | 59.1% | 68.2% | / | 81.8% | |
| Spe | 60.0% | 46.7% | 86.7% | / | 93.3% | 80.0% | 66.7% | 73.3% | / | 86.7% | |
| Right Amygdala | Acc | 62.2% | 40.5% | 59.5% | / | 78.4% | 62.2% | 56.8% | 56.8% | / | 86.5% |
| Sen | 59.1% | 36.4% | 45.5% | / | 68.2% | 63.6% | 59.1% | 40.9% | / | 86.4% | |
| Spe | 66.7% | 46.7% | 80.0% | / | 93.3% | 60.0% | 53.3% | 80.0% | / | 86.7% | |
Shape features were examined at three different dimensions (original — no dimension reduction; PCA — dimension reduced via PCA; PCA + ttest — dimension reduced via PCA followed by a Student's t-test).
Note: Student's t-tests were not applicable for the amygdalar shape features.
SVM was found to be generally superior to LDA in terms of discrimination based on high dimensional shape features. As shown in Table 3, the improvements brought about by the two dimension-reduction approaches, as applied to the shape features, have benefited LDA more than they have SVM. For the shape features of the left hippocampus, performing PCA yielded the highest overall accuracy when utilizing LDA for classification whereas SVM yielded the same overall classification accuracy based on either the original high dimensional shape features (372 dimensions) or the features after PCA (around 12 dimensions). For the left hippocampal shape features, further dimension reduction via Student's t-tests did not help either LDA or SVM. For the right hippocampus, LDA applied to the shape features after PCA improved the overall accuracy by roughly 11% compared to the original shape features, and a further dimension reduction via Student's t-test improved the accuracy by another 13.5%. Applying SVM to the right hippocampal shape features after PCA actually decreased the accuracy by 8% compared to the original shape features, but PCA followed by Student's t-test boosted the overall classification accuracy considerably, 8% more than that obtained from using the original shape features. For the left amygdala, both LDA and SVM benefited from the PCA procedure with the improvement for LDA being more substantial than that for SVM (33% versus 8% in terms of increased overall classification accuracy). For the right amygdala, the LDA classification accuracy was improved by 19% when performing PCA whereas there was no difference in the SVM classification performance before and after PCA. Collectively, performing dimension reduction on the high dimensional shape deformation markers is much more crucial for LDA than for SVM.
3.2.2. Combined feature analysis
In our next discriminant experiment, we evaluated the discriminative power of using combinations spanning the different types of features. We first examined the combination of the two types of morphometric features, volume and shape (with shape features again being in three stages), the classification results of which are demonstrated in Table 4. Compared with the results obtained from using the two types of morphometric features separately, as shown in Table 3, we noticed that there were almost no benefits brought about by adding volume features to those of shape and sometimes the incorporation of volume served to slightly damage the shape derived results. Therefore, in our final experiment, we chose only to evaluate the combinations of shape (in the three stages) and DTI features. As shown in Table 5, for each of the four structures of interest, a combination of its shape features and its DTI features provided much better results than either did alone. The highest classification accuracy was obtained by the combination of the shape features, taken after PCA and Student's t-test for dimension reduction, and the DTI features of the right hippocampus for both LDA and SVM, with the results of utilizing SVM being even better (overall accuracy, 94.6%; sensitivity, 95.5%; specificity, 93.3%). To test our conjecture that the volume features are not needed when the more sensitive shape features are available, we returned to examine the classification results yielded by a combination of volume and DTI features as well as the complete combination of volume, shape, and DTI features. The detailed results can be found in Table A.1 and Table A.2 of the Appendix, from which it is clear that the volume features did not add discriminative power to that of the combined shape and DTI features in any of the four structures.
Table 4.
Discrimination performances (overall accuracy (Acc), sensitivity (Sen), and Specificity (Spe)) of each structure by combining the volume feature and each of the three shape features at different dimensions (original – no dimension reduction; PCA — dimension reduced via PCA; PCA + ttest — dimension reduced via PCA followed by a Student's t-test) with two classification algorithms — LDA and SVM.
| LDA | SVM | ||||||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| Original | PCA | PCA + ttest | Original | PCA | PCA + ttest | ||
| Left Hippocampus | Acc | 56.8% | 78.4% | 75.7% | 83.8% | 83.8% | 75.7% |
| Sen | 40.9% | 72.7% | 68.2% | 77.3% | 86.4% | 68.2% | |
| Spe | 80.0% | 86.7% | 86.7% | 93.3% | 80.0% | 86.7% | |
| Right Hippocampus | Acc | 59.5% | 73.0% | 86.5% | 78.4% | 73.0% | 89.2% |
| Sen | 50.0% | 59.1% | 81.8% | 72.7% | 68.2% | 86.4% | |
| Spe | 73.3% | 93.3% | 93.3% | 86.7% | 80.0% | 93.3% | |
| Left Amygdala | Acc | 51.4% | 70.3% | / | 62.2% | 70.3% | / |
| Sen | 54.5% | 63.6% | / | 59.1% | 72.7% | / | |
| Spe | 46.7% | 80.0% | / | 66.7% | 66.7% | / | |
| Right Amygdala | Acc | 29.7% | 62.2% | / | 59.5% | 48.6% | / |
| Sen | 31.8% | 50.0% | / | 59.1% | 31.8% | / | |
| Spe | 26.7% | 80.0% | / | 60.0% | 73.3% | / | |
Note: Student's t-tests were not applicable for the amygdalar shape features.
Table 5.
Discrimination performances (overall accuracy (Acc), sensitivity (Sen), and Specificity (Spe)) of each structure by combining the DTI features and each of the three shape features at different dimensions (original — no dimension reduction; PCA — dimension reduced via PCA; PCA + ttest — dimension reduced via PCA followed by a Student's t-test) with two classification algorithms — LDA and SVM.
| LDA | SVM | ||||||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| Original | PCA | PCA + ttest | Original | PCA | PCA + ttest | ||
| Left Hippocampus | Acc | 64.9% | 75.7% | 78.4% | 83.8% | 78.4% | 89.2% |
| Sen | 63.6% | 68.2% | 72.7% | 81.8% | 77.3% | 90.9% | |
| Spe | 66.7% | 86.7% | 86.7% | 86.7% | 80.0% | 86.7% | |
| Right Hippocampus | Acc | 45.9% | 89.2% | 89.2% | 81.8% | 83.8% | 94.6% |
| Sen | 36.4% | 86.4% | 81.8% | 72.7% | 86.4% | 95.5% | |
| Spe | 60.0% | 93.3% | 100.0% | 93.3% | 80.0% | 93.3% | |
| Left Amygdala | Acc | 67.6% | 73.0% | / | 64.9% | 81.1% | / |
| Sen | 68.2% | 59.1% | / | 68.2% | 72.7% | / | |
| Spe | 66.7% | 93.3% | / | 60.0% | 93.3% | / | |
| Right Amygdala | Acc | 54.1% | 67.6% | / | 64.9% | 75.7% | / |
| Sen | 54.4% | 54.4% | / | 63.6% | 72.7% | / | |
| Spe | 53.3% | 86.7% | / | 66.7% | 80.0% | / | |
Note: Student's t-tests were not applicable for the amygdalar shape features.
4. Discussion
In this study, we have investigated the abnormalities that Alzheimer's disease had induced on a population's bilateral hippocampi and amygdalas in terms of both morphometry (volume and shape) and microstructural DTI indices (FA and trace). In addition, for each structure of interest, we examined the discriminative power of each of the morphometric and microstructural biomarkers, as well as that of a variety of multi-modality combinations, when classifying between AD patients and normal aging people.
In alignment with prior studies, we found significant volumetric atrophies of the hippocampus and the amygdala in patients with AD [7–14,57]. According to our vertex-based shape analysis, we observed significant, regionally specific, surface area atrophies on both the bilateral hippocampi and the bilateral amygdalas, which is also consistent with the shape findings obtained from our previous studies [20,23] as well as other studies [15–19,21,26], although shape analyses of the amygdala are less common than those of the hippocampus.
Our vertex-based shape analyses involved testing multiple hypotheses (one per vertex of the surface) simultaneously, which in turn necessitated a multiple comparison correction procedure. Active research on the multiple-testing problem in the functional neuroimaging community has been a feature of the past decade, with the current, most popular solutions being controlling the FWER or FDR. In this study, we evaluated both of these correction procedures when performing our localized shape analysis. We found FWER-correction to be more conservative than FDR-correction; the shape group differences in a much greater number of vertices were identified as being statistically significant after FDR-correction than after FWER-correction. This finding agrees with the general understanding that FWER-correction is stricter than FDR-correction and is more likely to make pessimistic estimates. As such, FWER-correction is more appropriate when there is an overriding reason to avoid any incorrect rejections of the null hypothesis whereas FDR-correction is more appropriate when there is a good reason to tolerate an expected proportion (usually 5%) of the FDR-corrected significance values being incorrectly rejected. It is not the intent of this study to rigorously prove which correction procedure should be adopted in vertex-based statistical shape analyses, although we will say that FWER-correction seems to be more likely to provide a definite detection of the local regions that have been affected by the pathology of AD.
As suggested by the template surface subdivisions, in this dataset, the pathology of AD mainly affected the CA1 and CA2 compartments of the left hippocampus (see Fig. 3) and a portion of each of the four compartments of the right hippocampus (see Fig. 4). This agrees partially with our previous work [20] wherein the subiculum of the left hippocampus, in addition to CA1 and CA2, has also been identified as having atrophied significantly in that dataset. The main differences between these two studies lay in their total numbers of subjects involved as well as the sources of their AD patients: firstly, 23 HC and 28 AD subjects were included in this study whereas there were a total of 210 HC and 175 AD subjects in that study; secondly, participants in this study were recruited in China whereas those of the previous study were mainly recruited in the US and Europe. It is also plausible that the AD patients in these two studies were at different stages of the disease progression trajectory. With that being said, the CA1 compartment was identified as being significantly affected on the bilateral hippocampi in both studies. This subregion is recognized as the one to be affected the earliest and the most severely by the neuropathology of AD in respect of neurofibrillary tangles [58,59], neuronal loss [60,61], in vivo atrophy as measured in manual segmentations of each subregion using structural MRI [62,63], as well as localized shape deformations [20,24,64–66]. Our observed CA2 atrophy of the hippocampus has also been reported in a range of previous publications [15,20,62,65–68].
According to our amygdalar shape analysis results (Figs. 5 and 6), the localized surface area atrophy in the right amygdala is more wide spread than that in the left amygdala, with the main subregions affected being the basolateral and basomedial components. A thorough survey of the AD literature would suggest that there have been very few published amygdalar subfield morphometric analyses, with [69] being the sole example outside of previous studies by the current authors' research groups [20,23,24]. A direct comparison with the findings reported in [69] is challenging given that the subdivision criteria of the amygdalas were rather different. Compared with our previous studies, our amygdalar findings for this dataset are highly consistent.
In our DTI dataset, we found a decreased mean FA value and an increased mean trace value within each of the four structures of interest (bilateral hippocampi and amygdalas) in the AD group compared to the HC group, with all group differences having reached statistical significance except for that of the mean FA of the left amygdala. FA and trace (a linear variant of MD) are the most commonly used DTI-derived measurements. The lower FA values of the four structures in AD compared to HC may suggest a lower degree of myelination and imply that AD has induced injury and a loss of structural integrity in the corresponding structures. Meanwhile, the higher trace values may again indicate dysfunction of the four structures in AD in terms of microstructural degeneration. Similar DTI studies focusing on the hippocampus and the amygdala have been extremely rare; most DTI studies, in looking to understand AD, focused on white matter tracts such as the corpus callosum and structural connectivity between various brain regions. However, DTI statistics, such as the mean FA and mean trace values used in our study, applied to structures that have been identified as susceptible to the pathology of AD, may provide simple but powerful biomarkers for the disease, which can be all too easily overlooked.
In our structure-specific discriminant analysis of the three types of AD biomarkers (structural volume, shape deformations, and the DTI indices), we found that the hippocampus outperformed the amygdala in all situations. This superior sensitivity of the hippocampus, as an AD biomarker, justifies the much greater focus given to it relative to the amygdala in the AD research community. As demonstrated in Table 3, on average, the volumetric measurement was the least discriminative whereas shape deformations and DTI indices were case dependent, depending on the machine learning technique performing the classification. The volumetric measurement was found to provide redundant information in light of the shape deformation based features when discriminating between HC and AD; a combination of shape and volume features did not enhance and sometimes even weakened the classification performance of using shape features alone. This may be explained by shape deformations providing morphometric information which overlaps with that of volumetrics but is more detailed, and thus more sensitive. However, volume aside, a combination of the shape deformation features and the DTI indices boosted the discriminative capabilities of using either feature separately, yielding the highest accuracy of any feature combination tested. This suggests that the localized morphometrics, as represented by the shape deformation markers, and the microstructural informatics, as represented by the DTI indices, provide complementary information and their combination is of great benefit to the automated discrimination of AD and HC subjects. Overall, SVM outperformed LDA in the classifications of our experiments. However, LDA with appropriate dimension reduction strategies (such as PCA) was shown to be effective for shape deformation based classifications, at least in our setting of LDDMM. This agrees with observations from previous studies, also in the framework of LDDMM-based shape deformations [37,70,71].
In this study, the best discrimination results came from a combination of the shape deformation features and the DTI indices of the right hippocampus, with PCA followed by Student's t-test being applied to reduce the shape dimension and SVM serving as the classification technique; to be specific, only 1 subject in each group was wrongly classified (see Table 5). The right hippocampus was also found to exhibit the strongest and most significant abnormalities in all three types of features, among all four structures of interest.
The reader should note that, given the sample size in this study, a difference of 1 subject will result in a marked change; that is, 4.5% in sensitivity and 6.7% in specificity. Applying the same classification procedures to another dataset with a much larger sample size is a most important aspect of future investigation to further validate and support the discriminant conclusions drawn from this study.
The primary goal of our discriminant experiments has been to explore the potential of each of the four structures in being an efficient AD-detection biomarker through utilizing its morphometric and microstructural features. As such, emphasis is not given to identifying a single set of biomarkers (some combination of structures and features) which will yield the highest classification accuracy. With that being said, our classification results do show strength. On the same dataset, using the shape deformation features combined with the two DTI indices of a single structure (the right hippocampus) in our study yielded the same discriminative capability as that described in [40] in which the most powerful biomarker came from a combination of the FA values of 47 white matter tracts and the volumetric measurements of 95 whole-brain anatomical regions, which itself reveals the superiority in AD-versus-HC discrimination of the shape + DTI multi-modality combinations. Extracting the shape and DTI features of other anatomical structures in addition to the hippocampus and the amygdala and exploring their intelligent combination across multiple structures will be a natural extension of this work in the future.
Supplementary Material
Acknowledgments
The authors acknowledge the contribution of all participants who gave their time to be involved in this study. This study was supported by the National Natural Science Foundation of China (NSFC 81501546, 84101389, and 81171308) and the SYSU-CMU Shunde International Joint Research Institute Start-up Grant (20150306).
Abbreviations
- AD
Alzheimer's Disease
- HC
Healthy Control
- DTI
Diffusion Tensor Imaging
- CA
Cornu Ammonis
- FA
Fractional Anisotropy
- NINCDS–ADRDA
National Institute of Neurological and Communicative Disorders and Stroke and the Alzheimer's Disease and Related Disorders Association
- MTL
Medial Temporal Lobe
- MRI
Magnetic Resonance Imaging
- MCI
Mild Cognitive Impairment
- WM
White Matter
- MD
Mean Diffusivity
- LDDMM
Large Deformation Diffeomorphic Metric Mapping
- PCA
Principal Component Analysis
- LDA
Linear Discriminant Analysis
- SVM
Support Vector Machine
- TR
Repetition Time
- TE
Echo Time
- TI
Inversion Time
- FOV
Field of View
- NEX
Number of Excitations
- TIV
Total Intracranial Volume
- FWER
Familywise Error Rate
- FDR
False Discovery Rate
- LOO
Leave One Out
Footnotes
Michael I. Miller owns an equal share in Anatomyworks LLC. The terms of this arrangement have been reviewed and approved by the Johns Hopkins University, in accordance with its conflict of interest policy.
Appendix A. Supplementary data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.mri.2016.05.001.
References
- 1.McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA work group under the auspices of Department of Health and Human Services Task Force on Alzheimer's disease. Neurology. 1984;34:939–44. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 2.Dubois B, Feldman HH, Jacova C, DeKosky ST, Barberger-Gateau P, Cummings J, et al. Research criteria for the diagnosis of Alzheimer's disease: revising the NINCDS–ADRDA criteria. Lancet Neurol. 2007;6:734–46. doi: 10.1016/S1474-4422(07)70178-3. [DOI] [PubMed] [Google Scholar]
- 3.Cahill L, Babinsky R, Markowitsch HJ, McGaugh JL. The amygdala and emotional memory. Nature. 1995;377:295–6. doi: 10.1038/377295a0. [DOI] [PubMed] [Google Scholar]
- 4.Hamann SB, Ely TD, Grafton ST, Kilts CD. Amygdala activity related to enhanced memory for pleasant and aversive stimuli. Nat Neurosci. 1999;2:289–93. doi: 10.1038/6404. [DOI] [PubMed] [Google Scholar]
- 5.Phelps EA. Human emotion and memory: interactions of the amygdala and hippocampal complex. Curr Opin Neurobiol. 2004;14:198–202. doi: 10.1016/j.conb.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 6.Squire LR. Memory and the hippocampus: a synthesis from findings with rats, monkeys, and humans. Psychol Rev. 1992;99:195–231. doi: 10.1037/0033-295x.99.2.195. [DOI] [PubMed] [Google Scholar]
- 7.Du AT, Schuff N, Amend D, Laakso MP, Hsu YY, Jagust WJ, et al. Magnetic resonance imaging of the entorhinal cortex and hippocampus in mild cognitive impairment and Alzheimer's disease. J Neurol Neurosurg Psychiatry. 2001;71:441–7. doi: 10.1136/jnnp.71.4.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jack CR, Jr, Petersen RC, O'Brien PC, Tangalos EG. MR-based hippocampal volumetry in the diagnosis of Alzheimer's disease. Neurology. 1992;42:183–8. doi: 10.1212/wnl.42.1.183. [DOI] [PubMed] [Google Scholar]
- 9.Jack CR, Jr, Petersen RC, Xu YC, Waring SC, O'Brien PC, Tangalos EG, et al. Medial temporal atrophy on MRI in normal aging and very mild Alzheimer's disease. Neurology. 1997;49:786–94. doi: 10.1212/wnl.49.3.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krasuski JS, Alexander GE, Horwitz B, Daly EM, Murphy DG, Rapoport SI, et al. Volumes of medial temporal lobe structures in patients with Alzheimer's disease and mild cognitive impairment (and in healthy controls) Biol Psychiatry. 1998;43:60–8. doi: 10.1016/s0006-3223(97)00013-9. [DOI] [PubMed] [Google Scholar]
- 11.Laakso M, Soininen H, Partanen K, Helkala E, Hartikainen P, Vainio P, et al. Volumes of hippocampus, amygdala and frontal lobes in the MRI-based diagnosis of early Alzheimer's disease: correlation with memory functions. J Neural Transm Park Dis Dement Sect. 1995;9:73–86. doi: 10.1007/BF02252964. [DOI] [PubMed] [Google Scholar]
- 12.Lehericy S, Baulac M, Chiras J, Pierot L, Martin N, Pillon B, et al. Amygdalohippocampal MR volume measurements in the early stages of Alzheimer disease. AJNR Am J Neuroradiol. 1994;15:929–37. [PMC free article] [PubMed] [Google Scholar]
- 13.Bobinski M, De Leon M, Wegiel J, Desanti S, Convit A, Saint Louis L, et al. The histological validation of post mortem magnetic resonance imaging-determined hippocampal volume in Alzheimer's disease. Neuroscience. 2000;95:721–5. doi: 10.1016/s0306-4522(99)00476-5. [DOI] [PubMed] [Google Scholar]
- 14.Fox NC, Warrington EK, Freeborough PA, Hartikainen P, Kennedy AM, Stevens JM, et al. Presymptomatic hippocampal atrophy in Alzheimer's disease. A longitudinal MRI study. Brain. 1996;119(Pt 6):2001–7. doi: 10.1093/brain/119.6.2001. [DOI] [PubMed] [Google Scholar]
- 15.Apostolova LG, Dinov ID, Dutton RA, Hayashi KM, Toga AW, Cummings JL, et al. 3D comparison of hippocampal atrophy in amnestic mild cognitive impairment and Alzheimer's disease. Brain. 2006;129:2867–73. doi: 10.1093/brain/awl274. [DOI] [PubMed] [Google Scholar]
- 16.Frisoni GB, Ganzola R, Canu E, Rub U, Pizzini FB, Alessandrini F, et al. Mapping local hippocampal changes in Alzheimer's disease and normal ageing with MRI at 3 tesla. Brain. 2008;131:3266–76. doi: 10.1093/brain/awn280. [DOI] [PubMed] [Google Scholar]
- 17.Qiu A, Fennema-Notestine C, Dale AM, Miller MI. Alzheimer's disease neuroimaging initiative. Regional shape abnormalities in mild cognitive impairment and Alzheimer's disease. Neuroimage. 2009;45:656–61. doi: 10.1016/j.neuroimage.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scher A, Xu Y, Korf E, White L, Scheltens P, Toga A, et al. Hippocampal shape analysis in Alzheimer's disease: a population-based study. Neuroimage. 2007;36:8–18. doi: 10.1016/j.neuroimage.2006.12.036. [DOI] [PubMed] [Google Scholar]
- 19.Shen K, Fripp J, Mériaudeau F, Chételat G, Salvado O, Bourgeat P, et al. Detecting global and local hippocampal shape changes in Alzheimer's disease using statistical shape models. Neuroimage. 2012;59:2155–66. doi: 10.1016/j.neuroimage.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 20.Tang X, Holland D, Dale AM, Younes L, Miller MI. Shape abnormalities of subcortical and ventricular structures in mild cognitive impairment and Alzheimer's disease: detecting, quantifying, and predicting. Hum Brain Mapp. 2014;35:3701–25. doi: 10.1002/hbm.22431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson PM, Hayashi KM, de Zubicaray GI, Janke AL, Rose SE, Semple J, et al. Mapping hippocampal and ventricular change in Alzheimer disease. Neuroimage. 2004;22:1754–66. doi: 10.1016/j.neuroimage.2004.03.040. [DOI] [PubMed] [Google Scholar]
- 22.Miller MI, Younes L, Ratnanather JT, Brown T, Reigel T, Trinh H, et al. Amygdala atrophy in MCI/Alzheimer's disease in the BIOCARD cohort based on diffeomorphic morphometry. MICCAI 2012 workshop on novel biomarkers for Alzheimer's disease and related disorders. 2012 NIH Public Access. [PMC free article] [PubMed] [Google Scholar]
- 23.Miller MI, Younes L, Ratnanather JT, Brown T, Trinh H, Lee DS, et al. Amygdalar atrophy in symptomatic Alzheimer's disease based on diffeomorphometry: the BIOCARD cohort. Neurobiol Aging. 2015;36:S3–S10. doi: 10.1016/j.neurobiolaging.2014.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang X, Holland D, Dale AM, Younes L, Miller MI. The diffeomorphometry of regional shape change rates and its relevance to cognitive deterioration in mild cognitive impairment and Alzheimer's disease. Hum Brain Mapp. 2015;36:2093–117. doi: 10.1002/hbm.22758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang X, Holland D, Dale AM, Miller MI. Alzheimer's disease neuroimaging initiative. APOE affects the volume and shape of the amygdala and the hippocampus in mild cognitive impairment and Alzheimer's disease: age matters. J Alzheimers Dis. 2015;47:645–60. doi: 10.3233/JAD-150262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cavedo E, Pievani M, Boccardi M, Galluzzi S, Bocchetta M, Bonetti M, et al. Medial temporal atrophy in early and late-onset Alzheimer's disease. Neurobiol Aging. 2014;35:2004–12. doi: 10.1016/j.neurobiolaging.2014.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Medina D, deToledo-Morrell L, Urresta F, Gabrieli JDE, Moseley M, Fleischman D, et al. White matter changes in mild cognitive impairment and AD: a diffusion tensor imaging study. Neurobiol Aging. 2006;27:663–72. doi: 10.1016/j.neurobiolaging.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 28.Wang L, Goldstein FC, Veledar E, Levey AI, Lah JJ, Meltzer CC, et al. Alterations in cortical thickness and white matter integrity in mild cognitive impairment measured by whole-brain cortical thickness mapping and diffusion tensor imaging. AJNR Am J Neuroradiol. 2009;30:893–9. doi: 10.3174/ajnr.A1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bozzali M, Falini A, Franceschi M, Cercignani M, Zuffi M, Scotti G, et al. White matter damage in Alzheimer's disease assessed in vivo using diffusion tensor magnetic resonance imaging. J Neurol Neurosurg Psychiatry. 2002;72:742–6. doi: 10.1136/jnnp.72.6.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nir TM, Jahanshad N, Villalon-Reina JE, Toga AW, Jack CR, Weiner MW, et al. Effectiveness of regional DTI measures in distinguishing Alzheimer's disease, MCI, and normal aging. Neuroimage Clin. 2013;3:180–95. doi: 10.1016/j.nicl.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pierpaoli C, Jezzard P, Basser PJ, Barnett A, Di Chiro G. Diffusion tensor MR imaging of the human brain. Radiology. 1996;201:637–48. doi: 10.1148/radiology.201.3.8939209. [DOI] [PubMed] [Google Scholar]
- 32.Fellgiebel A, Yakushev I. Diffusion tensor imaging of the hippocampus in MCI and early Alzheimer's disease. J Alzheimers Dis. 2011;26(Suppl. 3):257–62. doi: 10.3233/JAD-2011-0001. [DOI] [PubMed] [Google Scholar]
- 33.Hong YJ, Yoon B, Shim YS, Cho AH, Lim SC, Ahn KJ, et al. Differences in microstructural alterations of the hippocampus in Alzheimer disease and idiopathic normal pressure hydrocephalus: a diffusion tensor imaging study. AJNR Am J Neuroradiol. 2010;31:1867–72. doi: 10.3174/ajnr.A2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller MI, Trouve A, Younes L. Geodesic shooting for computational anatomy. J Math Imaging Vis. 2006;24:209–28. doi: 10.1007/s10851-005-3624-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller MI, Younes L, Ratnanather JT, Brown T, Trinh H, Postell E, et al. The diffeomorphometry of temporal lobe structures in preclinical Alzheimer's disease. Neuroimage Clin. 2013;3:352–60. doi: 10.1016/j.nicl.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller MI, Ratnanather JT, Tward DJ, Brown T, Lee DS, Ketcha M, et al. Network neurodegeneration in Alzheimer's disease via MRI based shape diffeomorphometry and high-field atlasing. Front Bioeng Biotechnol. 2015;3:54. doi: 10.3389/fbioe.2015.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang X, Holland D, Dale AM, Younes L, Miller MI. Baseline shape diffeomorphometry patterns of subcortical and ventricular structures in predicting conversion of mild cognitive impairment to Alzheimer's disease. J Alzheimers Dis. 2015;44:599–611. doi: 10.3233/JAD-141605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Younes L, Albert M, Miller MI. BIOCARD research team. Inferring changepoint times of medial temporal lobe morphometric change in preclinical Alzheimer's disease. Neuroimage Clin. 2014;5:178–87. doi: 10.1016/j.nicl.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kloppel S, Stonnington CM, Chu C, Draganski B, Scahill RI, Rohrer JD, et al. Automatic classification of MR scans in Alzheimer's disease. Brain. 2008;131:681–9. doi: 10.1093/brain/awm319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li M, Qin Y, Gao F, Zhu W, He X. Discriminative analysis of multivariate features from structural MRI and diffusion tensor images. Magn Reson Imaging. 2014;32:1043–51. doi: 10.1016/j.mri.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 41.Magnin B, Mesrob L, Kinkingnéhun S, Pélégrini-Issac M, Colliot O, Sarazin M, et al. Support vector machine-based classification of Alzheimer's disease from whole-brain anatomical MRI. Neuroradiology. 2009;51:73–83. doi: 10.1007/s00234-008-0463-x. [DOI] [PubMed] [Google Scholar]
- 42.Morra JH, Tu Z, Apostolova LG, Green AE, Toga AW, Thompson PM. Comparison of AdaBoost and support vector machines for detecting Alzheimer's disease through automated hippocampal segmentation. IEEE Trans Med Imaging. 2010;29:30–43. doi: 10.1109/TMI.2009.2021941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Orrù G, Pettersson-Yeo W, Marquand AF, Sartori G, Mechelli A. Using support vector machine to identify imaging biomarkers of neurological and psychiatric disease: a critical review. Neurosci Biobehav Rev. 2012;36:1140–52. doi: 10.1016/j.neubiorev.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 44.Tang X, Crocetti D, Kutten K, Ceritoglu C, Albert MS, Mori S, et al. Segmentation of brain magnetic resonance images based on multi-atlas likelihood fusion: testing using data with a broad range of anatomical and photometric profiles. Front Neurosci. 2015;9:61. doi: 10.3389/fnins.2015.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang X, Oishi K, Faria AV, Hillis AE, Albert MS, Mori S, et al. Bayesian parameter estimation and segmentation in the multi-atlas random orbit model. PLoS One. 2013;8:e65591. doi: 10.1371/journal.pone.0065591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liang Z, He X, Ceritoglu C, Tang X, Li Y, Kutten KS, Oishi K, Miller MI, Mori S, Faria AV. Evaluation of cross-protocol stability of a fully automated brain multi-atlas parcellation tool. PLoS One. 2015;10:e0133533. doi: 10.1371/journal.pone.0133533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ceritoglu C, Tang X, Chow M, Hadjiabadi D, Shah D, Brown T, et al. Computational analysis of LDDMM for brain mapping. Front Neurosci. 2013;7:151. doi: 10.3389/fnins.2013.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma J, Miller MI, Younes L. A bayesian generative model for surface template estimation. Int J Biomed Imaging. 2010;2010:974957. doi: 10.1155/2010/974957. [Epub 2010 Sep 20] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vaillant M, Glaunès J. Information processing in medical imaging. Springer; 2005. Surface matching via currents. [DOI] [PubMed] [Google Scholar]
- 50.Penny WD, Friston KJ, Ashburner JT, Kiebel SJ, Nichols TE. Statistical parametric mapping: the analysis of functional brain images: the analysis of functional brain images. Academic Press; 2011. [Google Scholar]
- 51.Li Y, Shea SM, Lorenz CH, Jiang H, Chou MC, Mori S. Image corruption detection in diffusion tensor imaging for post-processing and real-time monitoring. PLoS One. 2013;8:e49764. doi: 10.1371/journal.pone.0049764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tang X, Yoshida S, Hsu J, Huisman TA, Faria AV, Oishi K, et al. Multi-contrast multi-atlas parcellation of diffusion tensor imaging of the human brain. PLoS One. 2014;9:e96985. doi: 10.1371/journal.pone.0096985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tang X, Kutten K, Ceritoglu C, Mori S, Miller MI. Simultaneous skull-stripping and lateral ventricle segmentation via fast multi-atlas likelihood fusion. SPIE medical imaging; International Society for Optics and Photonics. 2015 [Google Scholar]
- 54.Nichols T, Hayasaka S. Controlling the familywise error rate in functional neuroimaging: a comparative review. Stat Methods Med Res. 2003;12:419–46. doi: 10.1191/0962280203sm341ra. [DOI] [PubMed] [Google Scholar]
- 55.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- 56.Miller MI, Younes L, Trouvé A. Diffeomorphometry and geodesic positioning systems for human anatomy. Technology. 2014;02:36–43. doi: 10.1142/S2339547814500010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Galton CJ, Patterson K, Graham K, Lambon-Ralph MA, Williams G, Antoun N, et al. Differing patterns of temporal atrophy in Alzheimer's disease and semantic dementia. Neurology. 2001;57:216–25. doi: 10.1212/wnl.57.2.216. [DOI] [PubMed] [Google Scholar]
- 58.Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–59. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 59.Schönheit B, Zarski R, Ohm TG. Spatial and temporal relationships between plaques and tangles in Alzheimer-pathology. Neurobiol Aging. 2004;25:697–711. doi: 10.1016/j.neurobiolaging.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 60.Rössler M, Zarski R, Bohl J, Ohm TG. Stage-dependent and sector-specific neuronal loss in hippocampus during Alzheimer's disease. Acta Neuropathol. 2002;103:363–9. doi: 10.1007/s00401-001-0475-7. [DOI] [PubMed] [Google Scholar]
- 61.West MJ, Coleman PD, Flood DG, Troncoso JC. Differences in the pattern of hippocampal neuronal loss in normal ageing and Alzheimer's disease. Lancet. 1994;344:769–72. doi: 10.1016/s0140-6736(94)92338-8. [DOI] [PubMed] [Google Scholar]
- 62.Mueller SG, Schuff N, Yaffe K, Madison C, Miller B, Weiner MW. Hippocampal atrophy patterns in mild cognitive impairment and Alzheimer's disease. Hum Brain Mapp. 2010;31:1339–47. doi: 10.1002/hbm.20934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.La Joie R, Perrotin A, de La Sayette V, Egret S, Doeuvre L, Belliard S, et al. Hippocampal subfield volumetry in mild cognitive impairment, Alzheimer's disease and semantic dementia. Neuroimage Clin. 2013;3:155–62. doi: 10.1016/j.nicl.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Apostolova LG, Dutton RA, Dinov ID, Hayashi KM, Toga AW, Cummings JL, et al. Conversion of mild cognitive impairment to Alzheimer disease predicted by hippocampal atrophy maps. Arch Neurol. 2006;63:693–9. doi: 10.1001/archneur.63.5.693. [DOI] [PubMed] [Google Scholar]
- 65.Frisoni GB, Sabattoli F, Lee AD, Dutton RA, Toga AW, Thompson PM. In vivo neuropathology of the hippocampal formation in AD: a radial mapping MR-based study. Neuroimage. 2006;32:104–10. doi: 10.1016/j.neuroimage.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 66.Csernansky JG, Wang L, Swank J, Miller JP, Gado M, McKeel D, et al. Preclinical detection of Alzheimer's disease: hippocampal shape and volume predict dementia onset in the elderly. Neuroimage. 2005;25:783–92. doi: 10.1016/j.neuroimage.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 67.Bobinski M, Wegiel J, Wisniewski HM, Tarnawski M, Reisberg B, Mlodzik B, et al. Atrophy of hippocampal formation subdivisions correlates with stage and duration of Alzheimer disease. Dementia. 1995;6:205–10. doi: 10.1159/000106948. [DOI] [PubMed] [Google Scholar]
- 68.Bobinski M, Wegiel J, Tarnawski M, Bobinski M, Reisberg B, de Leon MJ, et al. Relationships between regional neuronal loss and neurofibrillary changes in the hippocampal formation and duration and severity of Alzheimer disease. J Neuropathol Exp Neurol. 1997;56:414–20. doi: 10.1097/00005072-199704000-00010. [DOI] [PubMed] [Google Scholar]
- 69.Cavedo E, Boccardi M, Ganzola R, Canu E, Beltramello A, Caltagirone C, et al. Local amygdala structural differences with 3 T MRI in patients with Alzheimer disease. Neurology. 2011;76:727–33. doi: 10.1212/WNL.0b013e31820d62d9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vaillant M, Miller MI, Younes L, Trouve A. Statistics on diffeomorphisms via tangent space representations. Neuroimage. 2004;23(Suppl. 1):S161–9. doi: 10.1016/j.neuroimage.2004.07.023. [DOI] [PubMed] [Google Scholar]
- 71.Feng J, Tang X, Tang M, Priebe C, Miller M. Machine learning in medical imaging. Springer; 2013. Metric space structures for computational anatomy; pp. 123–30. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.