Abstract
Particle size distribution, a measurable physicochemical quantity, is a critical quality attribute of drug products that needs to be controlled in drug manufacturing. The non-invasive methods of dynamic light scattering (DLS) and Diffusion Ordered SpectroscopY (DOSY) NMR can be used to measure diffusion coefficient and derive the corresponding hydrodynamic radius. However, little is known about their use and sensitivity as analytical tools for particle size measurement of formulated protein therapeutics. Here, DLS and DOSY-NMR methods are shown to be orthogonal and yield identical diffusion coefficient results for a homogenous monomeric protein standard, ribonuclease A. However, different diffusion coefficients were observed for five insulin drug products measured using the two methods. DOSY-NMR yielded an averaged diffusion coefficient among fast exchanging insulin oligomers, ranging between dimer and hexamer in size. By contrast, DLS showed several distinct species, including dimer, hexamer, dodecamer and other aggregates. The heterogeneity or polydisperse nature of insulin oligomers in formulation caused DOSY-NMR and DLS results to differ from each other. DLS measurements provided more quality attributes and higher sensitivity to larger aggregates than DOSY-NMR. Nevertheless, each method was sensitive to a different range of particle sizes and complemented each other. The application of both methods increases the assurance of complex drug quality in this similarity comparison.
Keywords: diffusion, particle size distribution, physicochemical equivalence, protein drug product, similarity
INTRODUCTION
Protein-based drug products present a challenge for quality review because of complex formulations and inherent heterogeneity in protein structures (1). Higher-resolution analytical methods are needed to ensure structural and compositional similarities between protein therapeutics containing the same drug substance. In addition to protein higher-order structure probed by NMR, circular dichroism (CD) or other approaches (2), sub-visible protein aggregation presents an immunogenicity risk to drug safety (3–5). For most solution systems, particle size measurements can be inferred from the measured diffusion coefficient of a molecule and its proportionality to hydrodynamic radius. Therefore, diffusion coefficients provide information about molecular size of protein oligomers and higher-order complexes (6). Two modern analytical methods viz. dynamic light scattering (DLS) and Diffusion Ordered SpectroscopY (DOSY) NMR are capable of non-invasively measuring molecular translational diffusion coefficients and particle size distributions (PSD) directly in drug products (7–12). In DLS, the translational diffusion of molecules is measured by virtue of decay in scattered light intensity as a function of time (13). In DOSY-NMR, the decrease in protein magnetization intensity as a function of gradient strength is measured. These intensity functions are correlation functions of molecular translational diffusion. Of note, DLS and NMR measure the ensemble averaged diffusion coefficients of all equilibrated particles. In theory, DLS weighs higher molecular weight species more heavily than smaller components because the scattered light intensity is proportional to the sixth power of molecular radii. By contrast, DOSY-NMR weighs lighter molecular species more because smaller molecules yield sharper NMR lines than do larger molecules (7,14–17). DLS and DOSY-NMR methods offer a convenient and non-invasive way to measure a particle size with reduction of convoluted data to get simpler results. Here, these methods are compared with a focus on applicability to complex protein formulations to obtain information on the protein oligomerization/ aggregation and poly-dispersity found in a given drug product solution.
Insulin forms dimer, hexamer, and higher-order structures in solution depending on the insulin concentration and the presence of Zn2+ and/or phenol (18–21). Dynamic insulin exchange among dimer, tetramer, and hexamer has been proposed previously to explain observed spectroscopic data (22,23). Formulations used for this study contain protein at sub-mM concentration and excipients such as Zn2+, m-cresol and/or phenol. These formulation conditions stabilize insulin dimer, hexamer and also higher-order structure formation (19,24,25). Hexameric or higher-order structures have also been observed for insulin analogues under these conditions (18,26–28). Therefore, insulin could be present in polydisperse form in the five formulations investigated and were designated as a model “complex protein system” for purposes of this study.
To examine if DOSY-NMR and DLS give the same result when used to measure protein diffusion coefficients, first, a test was performed on a known protein standard. Then, the same protocol was applied to five insulin drug products from different manufacturers. The results demonstrated that these two methods are sensitive to different molecular size ranges, structural forms, and, for DOSY-NMR, to exchange kinetics among oligomers. To our knowledge, these data are the first such assessment of the orthogonality of these two non-invasive particle size distribution assay methods for analyzing complex protein therapeutics.
MATERIALS AND METHODS
Protein Sample Preparation
Lyophilized powder of ribonuclease A (RNase A) (Sigma-Aldrich, St. Louis, MO) was dissolved in 75 mM PBS buffer (Quality Biological Inc., Gaithersburg, MD) to get a final protein concentration of 1 mM RNase A. The sample was filtered through 0.02-μm Antop 10 plus filter (GE Healthcare UK Limited, Buckinghamshire, UK) using a 1-mL syringe (Becton Dickinson and Company, NJ). Insulin formulations were purchased as 10-mL vials at 100 U/mL, equivalent to a concentration of 0.6 mM in monomeric form. Three different lots for each of the insulin drug products Apidra ®, Humalog ®, Humulin R ®, Novolin R ® and Novolog ® were purchased and used directly for NMR and DLS measurements without filtering. For each NMR sample, 30 μL D2O (Cambridge Isotope Laboratories, Tewksbury, MA) with 3-(trimethylsilyl)-2,2,3,3-tetradeuteropropionic acid (TMSP-d4) (Sigma-Aldrich, St. Louis, MO) was mixed with 500 μL each of insulin drug products before loading to a 5-mm NMR tube (Wilmad lab-glass, NJ). The same NMR samples were applied on polystyrene flat bottom 96-well plate (Greiner Bio-One GmBH, Frickenhausen, Germany) for DLS data acquisition.
2D DOSY-NMR
The Bruker 1H 2D DOSY pulse sequence modified with 3919 water suppression (see supplementary information for the pulse program code) was used to measure diffusion coefficients. NMR data were acquired on a Bruker 850-MHz NMR instrument equipped with a triple-gradient TXI room temperature probe capable of generating gradient field strength of 54 G/cm. The DOSY time interval and gradient pulse were set at 600 ms (Δ) and 1.2 ms (δ), respectively. A total of 64 gradient increments, linearly varied from 2 to 98%, were collected with 128 scans for each increment. The total experiment time for each sample was approximately 9 h.
NMR data processing was performed using MestReNova 11.0.3 software (Mestrelab Research S.L.). Each free induction decay (FID) series in 2D DOSY dataset was apodized using Gaussian 1-Hz broadening function, zero-filled to 128-k data points, and baseline corrected with a third-order Bernstein polynomial fit. Phase correction and baseline correction were performed on the first FID and applied to all 64 FIDs in the diffusion dimension. Bayesian DOSY processing (BDT) functionality in MestReNova was used to process the second-dimension data. A molecular species with diffusion coefficient higher than 1 × 10−7 cm2/s was not observed in the initial analysis; therefore, diffusion range limit between 1 × 10−4 and 1 × 10−7 cm2/s and a resolution factor of 98 were used for BDT processing with two repetitions. A total of 256 trace points were processed for improved resolution with a reduction factor of 1 for vertical and horizontal scales. Peak suppression functionality of MestReNova was used only for samples of drug product Apidra® to suppress strong polysorbate-20 excipient peak. Five peaks corresponding to insulin between 0 and 1.3 ppm were selected from each 2D DOSY-NMR spectrum, and the average diffusion coefficients were calculated for a drug sample. Standard deviation from five repetitions on the same lot of Novolin R® was adopted as DOSY method variation data. Reference diffusion coefficients for RNase A and insulin (pdb 3aiy) monomer, dimer and hexamer were calculated from reported crystal structures using simulation software HYDROPRO (29) and HYDRONMR (30).
Dynamic Light Scattering
DLS experiments were performed using a 96-well plate, and 110 μL of NMR sample was loaded per well. Samples were equilibrated at 25°C for 30 min. Data were acquired with acquisition time of 4 s and a total of 40 scans per well. The curve fittings were performed using regularization functionality in DYNAMICS 7.5.17 (Wyatt Technologies) with baseline limit of 0.02 and maximum sum of squares (SOS) for the correlation function fit set to 300. A default Dynals™ analysis was applied to the autocorrelation function for the calculation of diffusion coefficients (D), percent poly-dispersity (%Pd), relative scattered light intensity (%Intensity) and mass weight (%Mass) for a maximum of four species per curve. %Pd is the ratio between the width of diffusion coefficient distribution to its mean value. Normalized diffusion coefficients for DLS were calculated using equation where n is 3 or 4, DN is the normalized diffusion, Di is the diffusion coefficient of a discrete species, and Wi is the %Mass of the respective species. DLS data were obtained in duplicate for all samples, and the averaged values were reported as the results. Standard deviation from five repetitions on the same lot of Novolin R® was adopted as DLS method variation data.
RESULTS AND DISCUSSION
Method Orthogonality
To cross-check DLS and 2D DOSY-NMR for measuring protein diffusion coefficients, a standard protein RNase A was adopted as a test model. DLS and DOSY-NMR investigations were performed on identical samples. The representative 1D 1H NMR spectrum, 2D DOSY-NMR spectrum and DLS data for RNase A are shown in Fig. 1a–c, respectively. The average diffusion coefficients for RNase A obtained from DOSY-NMR and DLS fitting were 1.17 ± 0.02 and 1.19 ± 0.02 × 10−6 cm2/s, respectively. The diffusion coefficients obtained from the two methods were nearly identical within experimental error. The average %Pd obtained from DLS was 14.0 ± 0.4%, which represent a realistic metric for a mono-disperse protein solution. The poly-dispersity value of 14% was designated as the minimum requirement to assign a discrete species to be “mono-disperse” for further analysis. The diffusion coefficient of RNase A was also calculated using its reported crystal structure. The calculated translational diffusion coefficient of 1.15 × 10−6 cm2/s was obtained for RNase A monomer, consistent with experimental data. The diffusion coefficient values obtained using both methods compared very well with the literature diffusion coefficient values for RNase A (31) and demonstrate the suitability of regularization curve fitting in analyzing DLS autocorrelation curves and BDT algorithm in processing 2D DOSY-NMR spectra.
Fig. 1.
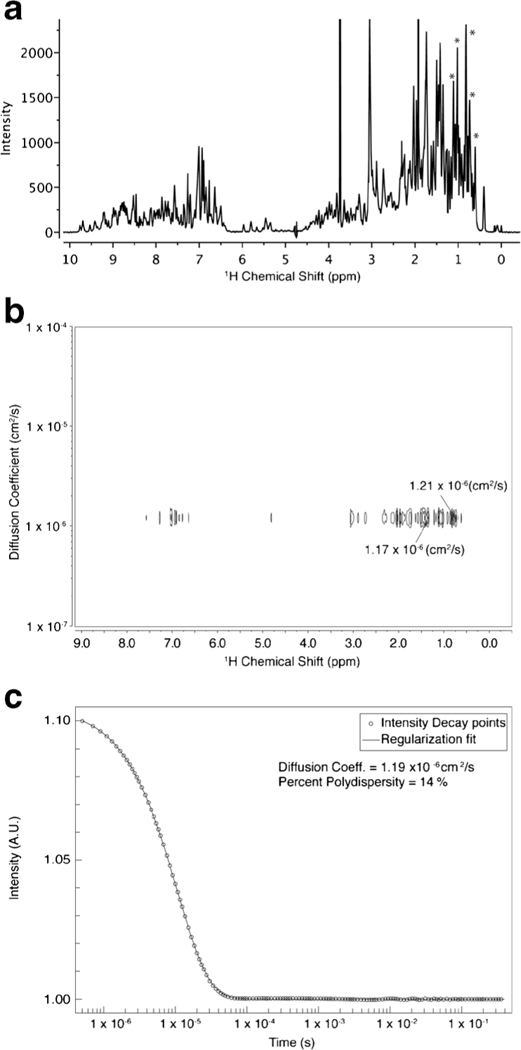
Representative data plots for two diffusion coefficient analysis methods for RNase A sample. a 1D 1H NMR spectrum. b 2D DOSY-NMR spectrum. c DLS autocorrelation curve. Strong protein methyl peaks (asterisk in a) were used to read out diffusion coefficient in a DOSY spectrum (b)
Insulin DOSY-NMR Analysis
The representative NMR spectra of 1D 1H and 2D DOSY for drug product Novolin R® are shown in Fig. 2a, b, respectively. The diffusion coefficients of insulin regular, drug substance in Novolin R®, were read directly from insulin methyl peaks. For all DOSY spectra, only one insulin species was identified. The diffusion coefficients of all the measured insulin lots ranged from 9.91 × 10−7 to 1.21 × 10−6 cm2/s (Table I), all of which were between calculated dimer and hexamer diffusion coefficient values (Fig. 3). The insulin analogue drug products (Apidra®, Novolog®, and Humalog®) showed larger diffusion coefficients, close to dimer, than that of regular insulin drug products (Novolin R® and Humulin R®). The regular insulin might be experiencing fast exchange between dimer and hexamer, which caused them to diffuse slower. NMR line broadening makes higher molecular weight species (higher than hexamer) more difficult to identify unambiguously in DOSY-NMR spectra. Thus, higher-order oligomer species could have been present but are not directly detectable by NMR due to broadened lines and low concentration.
Fig. 2.
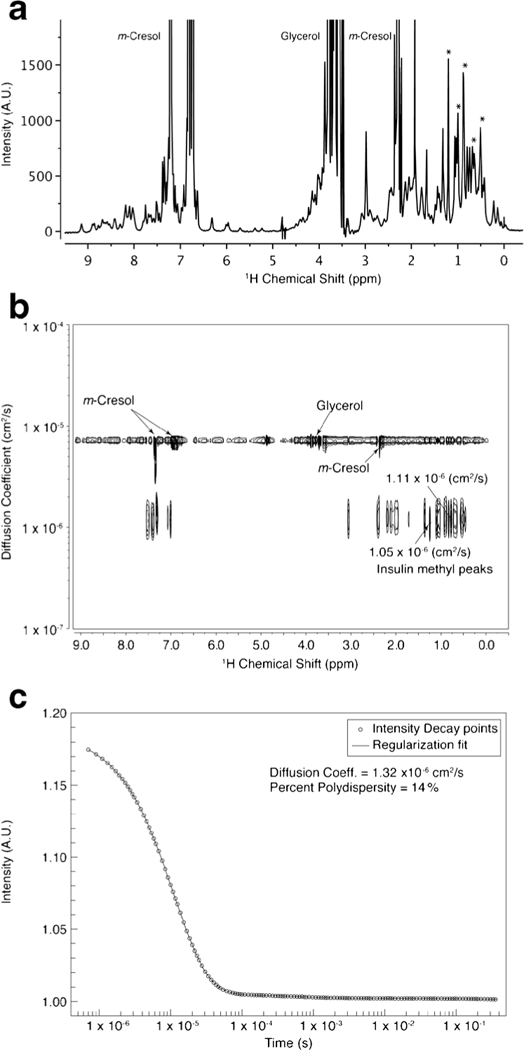
Representative data plots for diffusion coefficient analysis of insulin in drug product Novolin R®. a 1D 1H NMR spectrum of Novolin R®. b 2D DOSY-NMR spectrum. c DLS autocorrelation curve. Strong insulin methyl peaks (asterisk in a) were used to read out diffusion coefficient in a DOSY spectrum (b)
Table I.
Diffusion Coefficients for Insulin Drug Products Obtained Using DOSY-NMR
| Drug product | Drug substance | Lot no. | DOSY diffusion coefficienta (10−6 cm2/s) |
|
|---|---|---|---|---|
| Apidra® | Analogue Glulisine B3 N→K B29 K→E |
1 2 3 |
1.14 1.18 1.14 |
|
| Humalog® | Analogue Lispro B28 P→K B29 K→P |
1 2 3 |
1.08 1.17 1.06 |
|
| Humulin R® | Insulin regular | 1 | 0.99 | |
| 2 | 1.02 | |||
| 3 | 1.10 | |||
| Novolin R® | Insulin regular | 1 | 1.07 | |
| 2 | 1.07 | |||
| 3 | 1.07 | |||
| Novolog® | Analogue Aspart B28 P→D |
1 2 |
1.21 1.17 |
|
| 3 | 1.17 | |||
The averaged DOSY diffusion coefficient from all 15 lots of insulin was 1.11 × 10−6 cm2 /s and the RSD was 6%
Fig. 3.
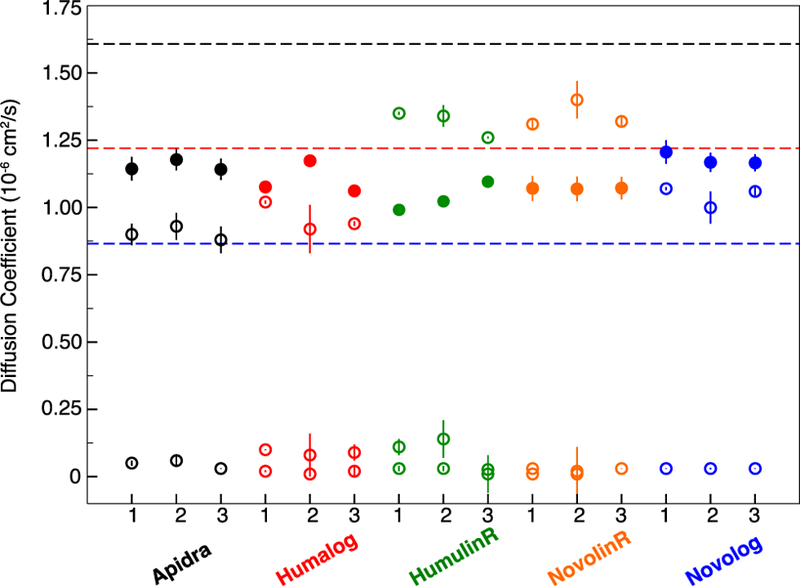
Experimental diffusion coefficients for three lots of five insulin drug products. 2D DOSY-NMR (solid circles) and DLS (open circles) results are plotted with the calculated diffusion coefficients of insulin (pdb 3aiy) monomer (1.61 × 10−6 cm2/s), dimer (1.22 × 10−6 cm2/s), and hexamer (0.87 × 10−6 cm2/s) shown as black, red, and blue dashed lines, respectively
A repetition of five DOSY measurements on one lot of Novolin R® yielded the averaged diffusion value of 1.08 × 10−6 cm2/s with relative standard deviation (RSD) of 1% (Table S1), which represented DOSY precision of diffusion coefficient measurement for insulin drug product. For each brand of insulin, three lots of drug products were subject to DOSY measurement (Table I). Among them, only Novolin R® showed the lowest inter-lot variation of 0.1%, and other inter-lot variations observed in Apidra®, Novolog®, Humulin R®, and Humalog® are above the 1% of the DOSY method precision, attributed to measurable lot-to-lot differences in these insulin products (Table S2).
Insulin DLS Analysis
A DLS signal decay curve for Novolin R® is shown in Fig. 2c. Typical DLS regularization curve fitting algorithm was applied. The calculation assumes that the correlation function is composed of several (up to four) distinct particles differing by at least one order of magnitude in diffusion coefficient. The deconvolution of a DLS curve results in weighting parameters percent intensity (%Intensity) for each species. The %Intensity is a much bigger number than %Mass, which is derived from %Intensity according to Mie’s scattering law. The %Mass normalized diffusion coefficient values were calculated as the DLS diffusion coefficient. For all lots of insulin DLS data (Tables S3 and S4), the %Intensity from insulin aggregates can be appreciable; however, aggregates of insulin were down to less than 1% in mass. Therefore, normalized diffusion coefficient values were obtained for each lot of insulin and were nearly the same as the value of the smallest species (Tables II and S4). The DLS diffusion values between 1.26 and 1.40 × 10−6 cm2/s were observed for Novolin R® and Humulin R®, both of which are regular human insulin. These values range between monomer and dimer diffusion coefficients (Fig. 3 and Table II). The DLS diffusion coefficients of the insulin analogue drug products Apidra®, Humalog®, and Novolog® were 0.91 ± 0.02, 0.96 ± 0.05, and 1.04 ± 0.04 × 10−6 cm2/s, respectively (Table III), which suggested the dominance of hexamer species for this measurement (Fig. 3) and differed from their DOSY-NMR results where the dimer species dominated. In addition, a distribution of species around the mean diffusion coefficient of each species is assumed. The %Pd values indicate the broadness and heterogeneity of the distribution. Most %Pd values for the primary species are close to the 14% value observed for RNase A, suggesting homogeneity. Some lots of Humalog ® and Humulin R® showed high %Pd of 20%, indicating higher heterogeneity in these products.
Table II.
Diffusion Coefficients for Insulin Drug Products Obtained Using DLS
| Drug product | Lot no. | Normalized diffusion coefficienta,b (10−6 cm2/s) |
% Poly-dispersityc (%Pd) | Hydrodynamic radiusd (nm) |
|---|---|---|---|---|
| Apidra® | 1 | 0.90 ± 0.04 | 9.9 ± 0.3 | 2.7 ± 0.1 |
| 2 | 0.93 ± 0.06 | 9 ± 4 | 2.6 ± 0.1 | |
| 3 | 0.88 ± 0.05 | 11 ± 1 | 2.7 ± 0.1 | |
| Humalog® | 1 | 1.02 ± 0.01 | 10.6 ± 0.3 | 2.4 ± 0.0 |
| 2 | 0.92 ± 0.09 | 30 ± 10 | 2.7 ± 0.2 | |
| 3 | 0.93 ± 0.01 | 14 ± 7 | 2.6 ± 0.0 | |
| Humulin R® | 1 | 1.35 ± 0.01 | 15 ± 1 | 1.8 ± 0.0 |
| 2 | 1.34 ± 0.04 | 11 ± 2 | 1.8 ± 0.0 | |
| 3 | 1.26 ± 0.01 | 20 ± 1 | 1.9 ± 0.0 | |
| Novolin R® | 1 | 1.31 ± 0.02 | 11 ± 1 | 1.9 ± 0.1 |
| 2 | 1.40 ± 0.07 | 13.2 ± 0.4 | 1.8 ± 0.1 | |
| 3 | 1.32 ± 0.02 | 16 ± 6 | 1.8 ± 0.0 | |
| Novolog® | 1 | 1.07 ± 0.01 | 12 ± 0.1 | 2.3 ± 0.0 |
| 2 | 1.00 ± 0.01 | 15 ± 5 | 2.4 ± 0.1 | |
| 3 | 1.06 ± 0.03 | 13 ± 3 | 2.3 ± 0.1 |
The experimental variation was from two technical repeats
The averaged DOSY diffusion coefficient from all 15 lots of insulin was 1.11 × 10−6 cm2 /s and the RSD was 17%
Percent poly-dispersity (%Pd) of the major species is shown. The %Pd value for RNAse A solution was 14.0 ± 0.4%
Hydrodynamic radius of the major specie is shown
Table III.
Comparison of Inter-Lot Diffusion Coefficient Results from DOSY-NMR and DLS
| Method | DOSY-NMR |
DLS |
|||
|---|---|---|---|---|---|
| Drug product | Inter-lot averaged diffusion coefficient (10−6 cm2/s) |
Relative standard deviation (RSD)a |
Inter-lot averaged diffusion coefficient (10−6 cm2/s) |
Relative standard deviation (RSD)a |
Inter-lot averaged %Pdb |
| Apidra® | 1.15 ± 0.02 | 2% | 0.91 ± 0.02 | 2% | 10 ± 1 |
| Humalog® | 1.10 ± 0.06 | 6% | 0.96 ± 0.05 | 5% | 20 ± 10 |
| Humulin R® | 1.04 ± 0.05 | 5% | 1.32 ± 0.05 | 4% | 15 ± 5 |
| Novolin R® | 1.07 ± 0.001 | 0.1% | 1.34 ± 0.05 | 4% | 13 ± 3 |
| Novolog® | 1.18 ± 0.02 | 2% | 1.04 ± 0.04 | 4% | 13 ± 2 |
A repeat of five DLS measurements on one lot of Novolin R® yielded a normalized diffusion coefficient value of 1.31× 10−6 cm2/s with 2% RSD, representing DLS accuracy to be 2% (Table S3). For all five brands of insulin, the inter-lot variations in diffusion coefficient are above the 2% of the DLS method precision, attributed to measurable lot-to-lot differences in these insulin products (Tables III and S4).
Comparison of the Two Methods
Different from model protein RNase A, DOSY and DLS never reported the same diffusion coefficient value on any insulin drug product. For insulin analogue products, Apidra®, Humalog® and Novolog®, the DOSY-NMR reported lower molecular weight species than DLS. For the regular insulin products, Humulin R® and Novolin R®, DOSY experiments showed smaller diffusion coefficients, between dimer and hexamer, while DLS reported dimer and higher-order oligomer species (Fig. 3). Regular insulin might experience fast exchange among monomer, dimer, hexamer and dodecamer, which could lower the averaged diffusion coefficient values measured in DOSY, but does not affect the DLS signals for all particles regardless of slow or fast exchange among them.
In addition to comparing the absolute diffusion coefficient values to assess orthogonality of DLS and DOSY-NMR measurements, the standard deviations across different lots were also compared. For each of the five brands, the inter-lot RSD from DOSY-NMR ranged between 0.1 and 6%, with Novolin R® being the most consistent solution, whereas Humalog® and Humulin R® had the largest inter-lot deviations (Table III). The inter-lot RSD values for DLS ranged between 2 and 5% with the highest variation obtained for Humalog® as well. Humalog® also has the largest %Pd of 20 ± 10% (Table III), indicating heterogeneity in its lot-to-lot comparison. The RSD for Humulin R® was slightly smaller in DLS analysis (4%) than DOSY-NMR (5%); however, the %Pd of Humulin R® was the seconded largest 15 ± 5%, meaning more heterogeneity in the primary species. Products of Apidra®, Novolog® and Novolin R® yielded less inter-lot variations and their %Pd values are also lower, less than 13% (Table III), indicating higher homogeneity in the primary species. Generally, the inter-lot variations were consistent between DLS and DOSY methods.
While the inter-lot variation is of interest for product consistency within each brand, the inter-brand variation is indicative of method sensitivity and differentiability. The range of variation in DLS diffusion coefficient values for all 15 tested lots of insulin range from 0.88 to 1.4 × 10−6 cm2/s, which was 2.5 times wider than the diffusion coefficient range from DOSY-NMR, 0.99 to 1.2 × 10−6 cm2/s (Tables I and II). In the DOSY method, the RSD of the DOSY diffusion coefficients obtained for all the tested 15 insulin lots from the five brands was only 6%. In the DLS method, the RSD from all the tested 15 lots was 17%; nearly three times more than the variation of the DOSY-NMR method. Thus, DLS differentiated insulin brands more effectively than DOSY-NMR.
CONCLUSIONS
In the present study, DOSY-NMR and DLS were demonstrated to be robust and orthogonal methods in measuring the diffusion coefficient for drug products directly and could be used to infer particle size distributions. Clinically, in general, protein aggregate size matters more than aggregate proportion in eliciting immunogenicity (32,33). Therefore, from a drug safety standpoint, a technique more sensitive to larger size aggregates (i.e., DLS) is more fit for this purpose than DOSY-NMR. For insulin drug products, trace amount of aggregates (<0.1% in mass) were detected using DLS. Within each insulin drug product, the oligomerization equilibrium and exchange kinetics are modulated by excipients and protein sequence. DLS analysis rapidly differentiated individual brands of insulin better than DOSY-NMR because of the greater sensitivity to higher molecular weight species. These higher-order oligomers were not directly observable by DOSY-NMR due to substantial line broadening. DOSY-NMR was able to evaluate lower molecular weight insulin complexes, exchange kinetics among oligomers and the diffusion properties of the individual excipients present. Importantly, the averaged diffusion coefficients observed for insulin in DOSY-NMR imply the presence of lower molecular weight complexes that are concealed from DLS.
Taken together, for protein drug quality attributes such as particle size, each result should be specified by the analytical method and the processing parameters employed. More non-invasive and orthogonal analytical methods certainly cover broader attributes of drug products, leading to tighter control of the product either within the same brand before and after manufacture change, or across the brand between the originator and a biosimilar or generic drug version. The combined results from DOSY-NMR and DLS allowed better understanding of protein oligomerization, aggregation, equilibrium and kinetics, which are affected by excipients of drug product-specific formulation. The application of orthogonal analytical methods is crucial in demonstrating part of the physicochemical equivalence between any two products even though the requirement for the sameness in drug substance identity and concentration has been met.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Darón Freedberg, Marcos Battistel, and Hugo Azurmendi for the assistance in setting up the DOSY-NMR experiments and for their helpful discussions. Support for this work comes from the US FDA CDER Critical Path funds and is gratefully acknowledged.
Footnotes
COMPLIANCE WITH ETHICAL STANDARDS
Disclaimer This article reflects the views of the author and should not be construed to represent US FDA’s views or policies.
REFERENCES
- 1.Woodcock J, Griffin J, Behrman R, Cherney B, Crescenzi T, Fraser B, et al. The FDA’s assessment of follow-on protein products: a historical perspective. Nat Rev Drug Discov 2007;6(6):437–42. [DOI] [PubMed] [Google Scholar]
- 2.Berkowitz SA, Engen JR, Mazzeo JR, Jones GB. Analytical tools for characterizing biopharmaceuticals and the implications for biosimilars. Nat Rev Drug Discov 2012;11(7):527–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahmadi M, Bryson CJ, Cloake EA, Welch K, Filipe V, Romeijn S, et al. Small amounts of sub-visible aggregates enhance the immunogenic potential of monoclonal antibody therapeutics. Pharm Res 2015;32(4):1383–94. [DOI] [PubMed] [Google Scholar]
- 4.Rosenberg AS. Effects of protein aggregates: an immunologic perspective. AAPS J 2006;8(3):E501–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hjorth CF, Norrman M, Wahlund PO, Benie AJ, Petersen BO, Jessen CM, et al. Structure, aggregation, and activity of a covalent insulin dimer formed during storage of neutral formulation of human insulin. J Pharm Sci 2016;105(4):1376–86. [DOI] [PubMed] [Google Scholar]
- 6.Philo JS. Is any measurement method optimal for all aggregate sizes and types? AAPS J 2006;8(3):E564–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gilard V, Trefi S, Balayssac S, Delsuc MA, Gostan T, Malet-Martino M, et al. Chapter 6—DOSY NMR for drug analysis A2—Holzgrabe, Ulrike. In: Wawer I, Diehl B, editors. NMR Spectroscopy in pharmaceutical analysis Amsterdam: Elsevier; 2008. p. 269–89. [Google Scholar]
- 8.Arakawa T, Philo JS, Ejima D, Tsumoto K, Arisaka F. Aggregation analysis of therapeutic proteins, part 2. Bioprocess Int 2007;5(4):36–47. [Google Scholar]
- 9.Clark TD, Bartolotti L, Hicks RP. The application of DOSY NMR and molecular dynamics simulations to explore the mechanism(s) of micelle binding of antimicrobial peptides containing unnatural amino acids. Biopolymers 2013;99(8):548–61. [DOI] [PubMed] [Google Scholar]
- 10.Li X, Shantz DF. PFG NMR investigations of tetraalkylammonium-silica mixtures. J Phys Chem C 2010;114(18):8449–58. [Google Scholar]
- 11.Li CG, Pielak GJ. Using NMR to distinguish viscosity effects from nonspecific protein binding under crowded conditions. J Am Chem Soc 2009;131(4):1368–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bocian W, Sitkowski J, Tarnowska A, Bednarek E, Kawecki R, Kozminski W, et al. Direct insight into insulin aggregation by 2D NMR complemented by PFGSE NMR. Proteins 2008;71(3):1057–65. [DOI] [PubMed] [Google Scholar]
- 13.Berne BJ, Pecora R. Dynamic light scattering: with applications to chemistry, biology, and physics New York: Dover Publications; 2000. [Google Scholar]
- 14.Panchal J, Kotarek J, Marszal E, Topp EM. Analyzing subvisible particles in protein drug products: a comparison of dynamic light scattering (DLS) and resonant mass measurement (RMM). AAPS J 2014;16(3):440–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hinton DPJ CS Diffusion ordered 2D NMR spectroscopy of phospholipid vesicles: determination of vesicle size distributions. J Phys Chem 1993;97:9064–72. [Google Scholar]
- 16.Hawe A, Hulse WL, Jiskoot W, Forbes RT. Taylor dispersion analysis compared to dynamic light scattering for the size analysis of therapeutic peptides and proteins and their aggregates. Pharm Res 2011;28(9):2302–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Demeester JDSS, Sanders N, Haustraete J. Methods for structural analysis of protein pharmaceuticals Arlington: AAPS; 2005. [Google Scholar]
- 18.Chang X, Jorgensen AM, Bardrum P, Led JJ. Solution structures of the R6 human insulin hexamer. Biochemistry 1997;36(31):9409–22. [DOI] [PubMed] [Google Scholar]
- 19.Xu Y, Yan Y, Seeman D, Sun L, Dubin PL. Multimerization and aggregation of native-state insulin: effect of zinc. Langmuir 2012;28(1):579–86. [DOI] [PubMed] [Google Scholar]
- 20.Derewenda U, Derewenda Z, Dodson EJ, Dodson GG, Reynolds CD, Smith GD, et al. Phenol stabilizes more helix in a new symmetrical zinc insulin hexamer. Nature 1989;338(6216):594–6. [DOI] [PubMed] [Google Scholar]
- 21.Teska BM, Alarcon J, Pettis RJ, Randolph TW, Carpenter JF. Effects of phenol and meta-cresol depletion on insulin analog stability at physiological temperature. J Pharm Sci 2014;103(8):2255–67. [DOI] [PubMed] [Google Scholar]
- 22.Lin MF, Larive CK. Detection of insulin aggregates with pulsedfield gradient nuclear magnetic resonance spectroscopy. Anal Biochem 1995;229(2):214–20. [DOI] [PubMed] [Google Scholar]
- 23.Hassiepen U, Federwisch M, Mulders T, Wollmer A. The lifetime of insulin hexamers. Biophys J 1999;77(3):1638–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whittingham JL, Edwards DJ, Antson AA, Clarkson JM, Dodson GG. Interactions of phenol and m-cresol in the insulin hexamer, and their effect on the association properties of B28 pro –> Asp insulin analogues. Biochemistry 1998;37(33):11516–23. [DOI] [PubMed] [Google Scholar]
- 25.Gualandi-Signorini AM, Giorgi G. Insulin formulations—a review. Eur Rev Med Pharmacol Sci 2001;5(3):73–83. [PubMed] [Google Scholar]
- 26.Smith GD, Swenson DC, Dodson EJ, Dodson GG, Reynolds CD. Structural stability in the 4-zinc human insulin hexamer. Proc Natl Acad Sci U S A 1984;81(22):7093–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ciszak E, Beals JM, Frank BH, Baker JC, Carter ND, Smith GD. Role of C-terminal B-chain residues in insulin assembly: the structure of hexameric LysB28ProB29-human insulin. Structure 1995;3(6):615–22. [DOI] [PubMed] [Google Scholar]
- 28.Palmieri LC, Favero-Retto MP, Lourenco D, Lima LM. A T3R3 hexamer of the human insulin variant B28Asp. Biophys Chem 2013;173–174:1–7. [DOI] [PubMed] [Google Scholar]
- 29.Ortega A, Amoros D. Garcia de la Torre J. Prediction of hydrodynamic and other solution properties of rigid proteins from atomic- and residue-level models. Biophys J 2011;101(4):892–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garcia de la Torre J, Huertas ML, Carrasco B. HYDRONMR: prediction of NMR relaxation of globular proteins from atomic-level structures and hydrodynamic calculations. J Magn Reson 2000;147(1):138–46. [DOI] [PubMed] [Google Scholar]
- 31.Nauman JV, Campbell PG, Lanni F, Anderson JL. Diffusion of insulin-like growth factor-I and ribonuclease through fibrin gels. Biophys J 2007;92(12):4444–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Beers MM, Bardor M. Minimizing immunogenicity of biopharmaceuticals by controlling critical quality attributes of proteins. Biotechnol J 2012;7(12):1473–84. [DOI] [PubMed] [Google Scholar]
- 33.Wang W, Singh SK, Li N, Toler MR, King KR, Nema S. Immunogenicity of protein aggregates—concerns and realities. Int J Pharm 2012;431(1–2):1–11. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


