Abstract
Rationale: Lung function and chronic obstructive pulmonary disease (COPD) are heritable traits. Genome-wide association studies (GWAS) have identified numerous pulmonary function and COPD loci, primarily in cohorts of European ancestry.
Objectives: Perform a GWAS of COPD phenotypes in Hispanic/Latino populations to identify loci not previously detected in European populations.
Methods: GWAS of lung function and COPD in Hispanic/Latino participants from a population-based cohort. We performed replication studies of novel loci in independent studies.
Measurements and Main Results: Among 11,822 Hispanic/Latino participants, we identified eight novel signals; three replicated in independent populations of European Ancestry. A novel locus for FEV1 in ZSWIM7 (rs4791658; P = 4.99 × 10−9) replicated. A rare variant (minor allele frequency = 0.002) in HAL (rs145174011) was associated with FEV1/FVC (P = 9.59 × 10−9) in a region previously identified for COPD-related phenotypes; it remained significant in conditional analyses but did not replicate. Admixture mapping identified a novel region, with a variant in AGMO (rs41331850), associated with Amerindian ancestry and FEV1, which replicated. A novel locus for FEV1 identified among ever smokers (rs291231; P = 1.92 × 10−8) approached statistical significance for replication in admixed populations of African ancestry, and a novel SNP for COPD in PDZD2 (rs7709630; P = 1.56 × 10−8) regionally replicated. In addition, loci previously identified for lung function in European samples were associated in Hispanic/Latino participants in the Hispanic Community Health Study/Study of Latinos at the genome-wide significance level.
Conclusions: We identified novel signals for lung function and COPD in a Hispanic/Latino cohort. Including admixed populations when performing genetic studies may identify variants contributing to genetic etiologies of COPD.
Keywords: Hispanic/Latino, genome-wide association study, single-nucleotide polymorphisms, lung function, chronic obstructive pulmonary disease
At a Glance Commentary
Scientific Knowledge on the Subject
Lung function and chronic obstructive pulmonary disease (COPD) are heritable traits. Prior genome-wide association studies (GWAS) of lung function have identified numerous genetic variants associated with COPD risk; however, much of the individual variance in lung function remains unexplained. Furthermore, most of these studies have been limited to participants of European ancestry. Including multiethnic populations in GWAS research may identify novel variants that contribute to the etiology of lung function.
What This Study Adds to the Field
This study, the largest GWAS of lung function and COPD to exclusively include Hispanic/Latino participants, identified eight novel signals, of which three replicated in independent populations. A novel locus for FEV1 (rs4791658; ZSWIM7) replicated in a cohort of European ancestry. A locus for FEV1/FVC (rs145174011; HAL), in a previously identified region for FEV1/FVC in individuals of European ancestry and for percent emphysema in a Hispanic population, remained independent in conditional analyses but did not replicate. Admixture mapping identified a novel region associated with Amerindian ancestry and FEV1, which included a candidate variant (rs4133185) in the AGMO gene that replicated. In addition, we identified a SNP (rs7709630) for COPD, which replicated in individuals of European ancestry. Several loci previously identified in European samples were also associated with lung function traits among Hispanic/Latino participants in the Hispanic Community Health Study/Study of Latinos at the genome-wide significance level. These results emphasize the importance of including admixed populations when performing genetic studies to identify variants that may contribute to the genetic etiology of pulmonary function and COPD.
The Hispanic/Latino population is the largest and fastest-growing minority population in the United States, accounting for 17% of the U.S. population currently and an estimated 33% by 2060 (1). The Hispanic/Latino population in the United States is genetically diverse, with a mixture of European, African, and Amerindian genetic ancestries (2–4).
Chronic obstructive pulmonary disease (COPD), characterized by persistent airflow obstruction (5, 6), is the third-leading cause of death in the United States (7). The prevalence of COPD in some Hispanic groups is lower than in non-Hispanic white individuals and African Americans in the United States (8–10). In the HCHS/SOL (Hispanic Community Health Study/Study of Latinos), the largest and best-characterized cohort of Hispanics/Latinos in the United States, the prevalence of COPD varied substantially by Hispanic/Latino heritage, with Puerto Ricans and Cubans having a much higher prevalence of COPD than other Hispanic/Latino groups (10). This difference in HCHS/SOL was explained by smoking and asthma history; however, other investigators have noted that Hispanic ethnicity is inversely associated with COPD phenotypes compared with white individuals (11, 12) and have hypothesized that genetic differences may lower risk of COPD in some Hispanic groups (11, 13).
Examination of genetic risk among non-European populations may reveal novel variants that yield new pathways to treatments, such as PCSK9 (14). We are aware of only two genome-wide association studies (GWAS) of lung function or COPD-related phenotypes that analyzed Hispanics separately (13, 15). One identified a novel variant near MAN2B1 that was associated with percent emphysema on computed tomography (CT) imaging (15), and the other identified two loci that approached genome-wide statistical significance for COPD (13). This literature contrasts with that among persons of European ancestry, in whom GWAS have identified multiple loci in genes for lung function and COPD (16–29)
To improve our current understanding of the genetic architecture of lung function–related traits in Hispanics/Latinos, we performed a meta-analysis for FEV1, FEV1/FVC, airflow limitation, and COPD among six Hispanic/Latino groups in the HCHS/SOL cohort (10, 30). Findings were replicated in European, Hispanic, and African populations. Some of the results have been previously reported in abstract form (31).
Methods
Study Sample
HCHS/SOL is a community-based cohort study of 16,415 self-identified Hispanic/Latino persons aged 18 to 74 years recruited from four U.S. communities. The study design, cohort recruitment (10, 30), and baseline clinical examination (32) have been previously described. Institutional Review Boards at each field center approved study protocols, and written informed consent was obtained from all participants.
For genetic analyses, HCHS/SOL participants were classified in six genetic analysis groups: Central American, Cuban, Dominican, Mexican, Puerto Rican, and South American. The genetic ancestry structure of these groups, including principal components (PCs) plots for all individuals, has been previously reported (33). Henceforward, we will refer to the six genetic analysis groups as Hispanic/Latino ancestry groups.
Exclusion criteria were lack of valid spirometric or genetic data, missing covariates, near-exclusive Asian ancestry, and participants who were not classified in the Hispanic/Latino ancestry groups. Details are provided in the online supplement.
Phenotypic and Outcome Measures
Spirometry was conducted in accordance with American Thoracic Society/European Respiratory Society guidelines (34) using a dry-rolling sealed spirometer, as previously described (10). Participants with prebronchodilator FEV1/FVC ratios less than 70% or the lower limit of normal were selected for post-bronchodilator spirometry, the former defined by National Health and Nutrition Examination Survey reference equations (35), because those from HCHS/SOL (36) were not available during data collection.
Airflow limitation was defined as a prebronchodilator FEV1/FVC ratio less than 70% and COPD as a post-bronchodilator FEV1/FVC ratio less than 70% (5). Control subjects had normal lung function, defined as a prebronchodilator FEV1/FVC ratio greater than 70% and FVC greater than 80% predicted.
Genotyping, Quality Control, and Imputation
Consenting HCHS/SOL subjects were genotyped at Illumina on the HCHS/SOL custom 15041502 B3 array. We applied standard quality-assurance and quality-control methods (37), as previously described (33). Genome-wide imputation was performed with the full, cosmopolitan 1000 Genomes Project phase 1 reference panel (38); associations for novel loci were confirmed with 1000 Genome phase 3 imputation panel. Details are provided in the online supplement.
Statistical Analyses
See the online supplement for details.
Quantitative lung function analyses
Analyses of FEV1 and FEV1/FVC ratio used linear mixed models, stratified by Hispanic/Latino ancestry group, and adjusted for age, age squared, sex, height, height squared, study center, smoking status, pack-years, sampling weights, and the first five PCs, as fixed effects. We used random effects for genetic relatedness (kinship) and household and community (block unit) to account for environmental correlation. Results from each Hispanic/Latino ancestry group were then meta-analyzed using the MetaCor method (39). Genome-wide significance threshold was defined as P < 5 × 10−8, the Bonferroni adjustment for 1 million independent tests (40). A priori, we planned additional meta-analyses stratified by never versus ever smoking.
Analyses for airflow limitation and COPD
We analyzed airflow limitation and COPD in pooled analyses implementing the GMMAT software (41) to fit a logistic mixed model adjusting for age, sex, study center, smoking status, pack-years, sampling weights, the first five PCs, and Hispanic/Latino ancestry groups, as fixed effects and with random effects for kinship and block unit. Participants younger than 45 years were excluded.
Admixture mapping and analyses
Local ancestry estimates were previously inferred in the HCHS/SOL (42). A genome-wide admixture mapping scan was performed using a linear mixed model, as described above, testing European, African, and Amerindian ancestries at each available local ancestry interval (LAI). Statistical significance for admixture mapping was set at P < 5.68 × 10−5 on the basis of previous simulation results for HCHS/SOL (43). After discovering a genome-wide significant LAI association (at the admixture mapping level), we identified a candidate variant and performed conditional admixture mapping analysis (44).
Replication of lung function SNPs
We pursued replication of novel loci associated with FEV1 and FEV1/FVC in populations with Hispanic (13, 45–49), African-American (45, 46), and European representation (16). We performed a look-up in each individual study and then meta-analyzed across studies. For airflow limitation and COPD, we performed regional replication in publically available GWAS results of airflow obstruction in participants of European ancestry (22). For admixture mapping signals, we performed a look-up in the UK Biobank results for individuals of European ancestry (https://sites.google.com/broadinstitute.org/ukbbgwasresults).
Generalization analyses of previously reported SNPs
We looked up previously reported SNPs, their effects, SEs, and P values in HCHS/SOL (50).
Results
The mean age of the 11,822 HCHS/SOL participants with valid lung function and genetic data was 46 ± 14 years. Thirty-nine percent had ever smoked cigarettes, with median pack-years of 7.5. The COPD analysis included 363 and 5,253 individuals with and without COPD, respectively. Characteristics of the participants are shown in Table 1. Manhattan and quantile-quantile (Q-Q) plots for FEV1, FEV1/FVC, airflow limitation, and COPD are shown in Figures E1, E2, E3, and E4 in the online supplement. Genomic inflation factors ranged from 1.020 to 1.026 in quantitative lung function analyses and 0.988 to 1.011 in analyses of COPD.
Table 1.
Characteristics of Participants in Hispanic Community Health Study/Study of Latinos
| Central American | Cuban | Dominican | Mexican | Puerto Rican | South American | Combined | |
|---|---|---|---|---|---|---|---|
| No. of participants | 1,315 | 2,138 | 1,063 | 4,438 | 2,000 | 868 | 11,822 |
| Age, yr | 44 ± 13 | 49 ± 13 | 45 ± 14 | 44 ± 14 | 48 ± 14 | 46 ± 13 | 46 ± 14 |
| Male sex, % | 40 | 47 | 35 | 39 | 43 | 40 | 41 |
| Height, cm | 160 ± 9 | 164 ± 9 | 162 ± 9 | 161 ± 9 | 163 ± 9 | 161 ± 9 | 162 ± 9 |
| BMI, kg/m2 | 30 ± 6 | 29 ± 6 | 29 ± 6 | 30 ± 6 | 31 ± 7 | 29 ± 5 | 30 ± 6 |
| Smoking (never), % | 68 | 52 | 77 | 65 | 48 | 66 | 61 |
| Smoking (former), % | 19 | 19 | 13 | 19 | 20 | 20 | 19 |
| Smoking (current), % | 13 | 29 | 10 | 16 | 32 | 14 | 20 |
| Pack-years of smoking*, median (IQR) | 4.5 (1.7–14) | 18.5 (6–36) | 8.2 (3.4–18.3) | 4.0 (1.3–10.5) | 11.0 (4–25.5) | 4.9 (1.5–16) | 7.5 (2.4–21) |
| Prebronchodilator FEV1, L | 2.88 ± 0.74 | 2.80 ± 0.81 | 2.70 ± 0.75 | 2.97 ± 0.78 | 2.70 ± 0.83 | 2.95 ± 0.79 | 2.86 ± 0.80 |
| Prebronchodilator FEV1/FVC, % | 81.5 ± 5.9 | 78.7 ± 8.3 | 81.8 ± 7.2 | 80.8 ± 6.1 | 79.3 ± 7.9 | 80.1 ± 6.5 | 80.3 ± 7 |
| COPD, n (%)† | 24 (1.8) | 124 (5.8) | 24 (2.3) | 85 (1.9) | 86 (4.3) | 20 (2.3) | 363 (3.1) |
| Airflow limitation, n (%)‡ | 60 (4.6) | 260 (12.2) | 62 (5.8) | 207 (4.7) | 216 (10.8) | 49 (5.7) | 854 (7.2) |
Definition of abbreviations: BMI = body mass index; COPD = chronic obstructive pulmonary disease; IQR = interquartile range.
Data are presented as mean ± SD unless otherwise noted.
Pack-years of smoking in former or current smokers.
COPD defined as post-bronchodilator FEV1/FVC ratio less than 0.70.
Defined as prebronchodilator FEV1/FVC ratio less than 0.70.
GWAS of Lung Function across All Hispanic/Latino Ancestry Groups
Across all Hispanic/Latino ancestry groups, seven signals achieved genome-wide statistical significance, of which two were novel for lung function (Table 2). The lead novel SNP, rs4791658, in the locus associated with FEV1 (Figure 1A) was an intron variant in the gene ZSWIM7 on chromosome 17 (β = 33.4, P = 4.99 × 10−9). Its minor allele frequency (MAF) ranges from 0.39 to 0.49 across Hispanic/Latino ancestry groups (Figure 1B). SNP rs4791658 neared genome-wide significance for FEV1 among ever smokers (β = 46.8, P = 1.53 × 10−6). Effect size and P values for rs4791658 across all lung function and COPD-related traits are shown in Table E1.
Table 2.
Genome-Wide Significant Loci Associated with FEV1 and FEV1/FVC
| SNP rsID | Trait | Position Build 37 Chr: Basepair | Gene/Nearest Gene(s) (Function) | Effect Allele | EAF | Genotyped or Imputed SNP | oevar* | N | β | SE | P Value |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs76656601 | FEV1/FVC | 6: 168428548 | KIF25 (intron) | G | 0.996 | Imputed | 0.7 | 11,822 | 4.70 | 0.77 | 1.31 × 10−9 |
| rs4791658 | FEV1 | 17: 15884792 | ZSWIM7†(intron) | G | 0.541 | Imputed‡ | 0.996 | 11,822 | 33.35 | 5.70 | 4.99 × 10−9 |
| rs145174011 | FEV1/FVC | 12: 96378700 | HAL†(intron) | T | 0.997 | Imputed | 0.98 | 11,822 | 4.85 | 0.85 | 9.59 × 10−9 |
| rs28593428 | FEV1/FVC | 21: 35632170 | KCNE2 | C | 0.902 | Imputed | 0.996 | 11,821 | 0.80 | 0.14 | 1.45 × 10−8 |
| rs262113 | FEV1/FVC | 6: 142824950 | GPR126 | T | 0.812 | Imputed† | 0.999 | 11,822 | −0.57 | 0.10 | 2.83 × 10−8 |
| rs74444778 | FEV1/FVC | 7: 42553708 | LOC105375250 (intron) | G | 0.999 | Imputed | 0.914 | 11,822 | 6.17 | 1.12 | 3.61 × 10−8 |
| rs115745680 | FEV1 | 5: 154407079 | GEMIN5 KIF4B | A | 0.997 | Imputed | 0.981 | 11,822 | 313.24 | 57.32 | 4.63 × 10−8 |
Definition of abbreviations: β (FEV1) = per-allele change in FEV1 (ml); β (FEV1/FVC) = per-allele change in FEV1/FVC %; Chr = chromosome; EAF = effect allele frequency; GEMIN5 = gem nuclear organelle–associated protein 5; GPR126 = G protein–coupled receptor 126; HAL = histidine ammonia-lyase; KCNE2 = potassium channel, voltage-gated subfamily E regulatory β subunit 2; KIF4B = kinesin family member 4B; KIF25 = kinesin family member 25; N = sample size used in testing the given variant; nearest gene = the nearest genes on each side of the intergenic marker; oevar = observed/expected variance; ZSWIM7 = zinc finger, SWIM-type containing 7.
Analyses were performed using data imputed to 1000 Genome phase 1 data and stratified by Hispanic/Latino ancestry group and were adjusted for age, age squared, sex, height, height squared, smoking status, pack-years, sampling weights, first five principal components, Hispanic/Latino ancestry groups, kinship, and block unit. Within-subgroup estimates were then meta-analyzed. Previously unreported associations are in bold. Retrospectively, association analyses using genetic data imputed to 1000 Genome phase 3 imputation panel were performed in novel loci.
oevar is provided by MACH software as a measure of imputation quality.
Results for analyses using genetic data imputed to 1000 Genome phase 3 imputation panel.
Genotyped SNP in locus.
Figure 1.
Regional association plots and forest plots of genome-wide significant loci associated with FEV1 and FEV1/FVC meta-analyzed across all Hispanic/Latino ancestry groups. For each locus, we provide regional association plots with correlations between the reference SNP (the SNP with the lowest P value) and other SNPs in the region. The reference SNP is a purple diamond (genotyped SNP) or upside down triangle (imputed SNP). Other SNPs in the region are depicted as circles (genotyped SNPs) and Xs (imputed SNPs). The correlations (r2) are calculated from the group of interest and are indicated by the colors shown on the plot. For each locus, we also provide a forest plot comparing the SNP–trait association testing results across the Hispanic/Latino ancestry groups. (A) Regional association plot for the FEV1 locus (lead SNP rs4791658) on chromosome 17. (B) Forest plot for the FEV1 locus (lead SNP rs4791658) on chromosome 17. (C) Regional association plot for the FEV1/FVC locus (lead SNP rs145174011) on chromosome 12. (D) Forest plot for the FEV1/FVC locus (lead SNP rs192375903) on chromosome 12. EAfreq = effect allele frequency.
The lead novel SNP, rs145174011, in the locus associated with FEV1/FVC (Figure 1C) was an intronic variant in the gene HAL on chromosome 12 (β = 4.86, P = 9.59 × 10−9). The SNP is rare, with an MAF that ranges from 0.001 to 0.008 across Hispanic/Latino ancestry groups (Figure 1D). The minor allele does not exhibit an outlier effect (Figure E5). Sensitivity analysis using rank-normalized residuals is shown in the online supplement. This locus is near the gene CCDC38, which was previously identified for FEV1/FVC in individuals of European Ancestry (16), and the gene SNRPF, which was previously identified for percent emphysema on CT imaging in racially/ethnically diverse participants (15). In conditional analysis adjusting for rs1036429 (CCDC38) and rs7957346 (SNRPF) in HCHS/SOL, rs145174011 (HAL SNP) remained associated with FEV1/FVC (β = 4.92, P = 6.4 × 10−9), suggesting its association with FEV1/FVC is independent of previously identified variants. The findings for rs145174011 among ever and never smokers are concordant and trended toward significance (Table E1). Regional association plots and forest plots of previously reported genome-wide significant associations for FEV1 and FEV1/FVC meta-analyzed across all Hispanic/Latino ancestry groups are shown in Figure E6.
Stratification by Smoking Status
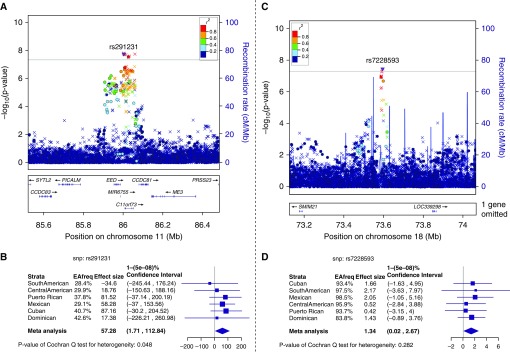
In the meta-analysis across all Hispanic/Latino ancestry groups stratified by smoking status, we identified one novel locus among ever smokers for FEV1 and one novel locus among never smokers for FEV1/FVC (Table 3). The lead variant in the locus associated with FEV1 in ever smokers was an indel variant (P = 1.66 × 10−8) on chromosome 11. The top SNP, rs291231 (β = 57.28, P = 1.92 × 10−8), in this locus has an MAF that ranges from 0.28 to 0.42 across Hispanic/Latino ancestry groups. This SNP lies between the genes EED and CCDC81 (Figures 2A and 2B). The SNP-by-pack-years interaction trended toward significance (P-interaction = 0.06), and the genotype effect became greater at higher pack-years (Figure E7).
Table 3.
Loci Associated with FEV1 and FEV1/FVC Ratio at Genome-Wide Significance in Analyses Stratified by Smoking Status
| SNP rsID | Trait | Position Build 37 Chr: Basepair | Nearest Gene(s) | Effect Allele | EAF | Genotyped or Imputed SNP | oevar* | N | β | SE | P Value |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ever smokers | |||||||||||
| Indel (no rs#) | FEV1 | 11: 86006803 | EED CCDC81 | GACA | Imputed† | 1.017 | 56.95 | 10.09 | 1.66 × 10−8 | ||
| rs291231 | FEV1 | 11: 86007090 | EED CCDC81 | G | 0.341 | Imputed† | 0.999 | 4,595 | 57.28 | 10.19 | 1.92 × 10−8 |
| rs9974878 | FEV1/FVC | 21: 35642446 | KCNE2 | G | 0.887 | Imputed† | 0.994 | 4,594 | 1.28 | 0.23 | 3.58 × 10−8 |
| Never smokers | |||||||||||
| rs116726860 | FEV1/FVC | 2: 155486042 | KCNJ3 GALNT13 | T | 0.992 | Imputed | 0.897 | 7,227 | 3.49 | 0.60 | 6.04 × 10−9 |
| rs7228593 | FEV1/FVC | 18: 73575704 | SMIM21 LOC339298 | G | 0.948 | Imputed† | 0.997 | 7,227 | 1.34 | 0.24 | 3.47 × 10−8 |
Definition of abbreviations: β (FEV1) = per-allele change in FEV1 (ml); β (FEV1/FVC) = per-allele change in FEV1/FVC %; Chr = chromosome; EAF = effect allele frequency; EED = genes embryonic ectoderm development; GALNT13 = polypeptide N-acetylgalactosaminyltransferase 13; indel = indel mutation; KCNE2 = potassium channel, voltage-gated subfamily E regulatory β subunit 2; KCNJ3 = potassium voltage-gated channel subfamily J member 3; N = sample size used in testing the given variant; nearest gene = the nearest genes on each side of the intergenic marker; oevar = observed/expected variance; SMIM21 = small integral membrane protein 21.
Analyses were stratified by Hispanic/Latino ancestry group and smoking status. Model adjusted for age, age squared, sex, height, height squared, smoking status, pack-years, sampling weights, first five principal components, Hispanic/Latino ancestry groups, kinship, and block unit. Within-subgroup estimates were then meta-analyzed. Previously unreported associations are in bold.
oevar is provided by MACH software as a measure of imputation quality.
Genotyped SNP in locus.
Figure 2.
Regional association plots and forest plots of genome-wide significant loci associated with FEV1 and FEV1/FVC stratified by smoking status and meta-analyzed across all Hispanic/Latino ancestry groups. For each locus, we provide regional association plots with correlations between the reference SNP (the SNP with the lowest P value) and other SNPs in the region. The reference SNP is a purple diamond (genotyped SNP) or upside down triangle (imputed SNP). Other SNPs in the region are depicted as circles (genotyped SNPs) and Xs (imputed SNPs). The correlations (r2) are calculated from the group of interest and are indicated by the colors shown on the plot. For each locus, we also provide a forest plot comparing the SNP–trait association testing results across the Hispanic/Latino ancestry in smoking stratified group of interest. (A) Regional association plot for the FEV1/FVC locus among never smokers (lead SNP rs7228593) on chromosome 18. (B) Forest plot for the FEV1/FVC locus among never smokers (lead SNP rs7228593) on chromosome 18. (C) Regional association plot for the FEV1 locus among ever smokers (lead SNP rs291231) on chromosome 11. (D) Forest plot for the FEV1 locus among ever smokers (lead SNP rs291231) on chromosome 11. EAfreq = effect allele frequency.
The top SNP (rs7228593) in the locus associated with FEV1/FVC in never smokers (β = 1.34, P = 3.47 × 10−8) is located between the genes SMIM21 and LOC339298 (Figure 2C) on chromosome 18. The findings in never smokers for airflow limitation (odds ratio, 0.46; P = 2.63 × 10−4) and COPD (odds ratio, 0.36; P = 0.005) were directionally concordant and trended toward significance (Table E1). Its MAF varies significantly across different Hispanic/Latino ancestry groups (Figure 2D), with the highest in participants from the Dominican ancestry group (MAF = 0.16) and lowest in Mexican ancestry group (MAF = 0.02). Forest plots and regional association plot of remaining genome-wide significant associations for FEV1 and FEV1/FVC stratified by smoking status are shown in Figure E8.
GWAS of COPD
We identified two previously unreported SNPs for COPD (Table E2 and Figure E9). SNP rs7709630 is an intron in the PDZD2 gene on chromosome 5 (P = 1.56 × 10−8, MAF = 0.11), and SNP rs2286351 is a noncoding transcript variant in CDRT15P1 on chromosome 17 (P = 1.97 × 10−8, MAF = 0.09). Lung function association analyses of these COPD SNPs are reported in Table E3.
Admixture Mapping
Manhattan plots from admixture mapping analyses for FEV1 and FEV1/FVC testing Amerindian, African, and European ancestry counts individually in LAIs are shown in Figures E10 and E11. We identified one novel significant local ancestry association region of Amerindian ancestry for FEV1 (Figure E10A) on chromosome 7 (β = −37.51, P = 2.86 × 10−6). There were no significant associations in the European versus others or African versus others analyses.
We identified a candidate variant (rs4133185) within this novel LAI region (β = −31.44, P = 7.43 × 10−7; Table E4 and Figure 3) with considerable differences in ancestry-specific allele frequencies (Figure 4). The lead SNP, rs4133185, in this candidate variant was an intron variant in the gene AGMO. The admixture mapping signal is less significant after conditional analyses adjusting for rs4133185 (Table E5 and Figures 4 and E12).
Figure 3.
Regional association plot and forest plot of candidate variant identified in local ancestry interval region associated with Amerindian ancestry and FEV1 meta-analyzed across all Hispanic/Latino ancestry groups. For each locus, we provide regional association plots with correlations between the reference SNP (the SNP with the lowest P value) and other SNPs in the region. The reference SNP is a purple diamond (genotyped SNP) or upside-down triangle (imputed SNP). Other SNPs in the region are depicted as circles (genotyped SNPs) and Xs (imputed SNPs). The correlations (r2) are calculated from the group of interest and are indicated by the colors shown on the plot. We also provide a forest plot comparing the SNP–trait association testing results across the Hispanic/Latino ancestry groups. (A) Regional association plot for the FEV1 locus (lead SNP rs4133185) on chromosome 7. (B) Forest plot for the FEV1 locus (lead SNP rs4133185) on chromosome 7. EAfreq = effect allele frequency.
Figure 4.
Amerindian admixture mapping region on chromosome 7 for FEV1. The left panel provides the admixture mapping results as two lines, with the blue line representing results from the primary analysis and the green line representing results from the conditional analysis; the association results in the same region are represented as circles. The genome-wide significant threshold for admixture mapping in the HCHS/SOL (Hispanic Community Health Study/Study of Latinos) data set is the horizontal gray dashed line. The blue and green lines and circles are given as −log10(P value) against genomic positions. The red-filled triangle corresponds to the SNP used in the conditional analysis (rs4133185). The right panel provides the ancestry-specific effect allele frequencies for the SNP used in the conditional analysis (rs4133185), as estimated by ancestry-specific allele frequency estimation applied on HCHS/SOL data set (44).
Replication
Characteristics of the participants in the replication cohorts are shown in Table E4. Of the two novel loci identified for lung function, only the FEV1 locus on chromosome 17 successfully replicated (replication P = 3.37 × 10−5; adjusted threshold for statistical significance was P < 9.2 × 10−4) but only in the SpiroMeta consortium look-up (n = 94,612) (16). The candidate variant (rs4133185), identified through admixture mapping, associated with Amerindian ancestry and FEV1 replicated in the UK Biobank (β = −8.3, P = 0.004). SNP rs291239, a genotyped SNP in the novel locus on chromosome 11 identified for FEV1 among ever smokers, approached statistical significance for replication in the meta-analysis across all cohorts with admixed populations of African ancestry (replication P = 0.007); however, the direction of effect was discordant in the individual replication cohorts (Table E7).
The novel SNP (rs7709630; PDZD2) associated with COPD regionally replicated in publically available results from the SpiroMeta-CHARGE (Cohorts for Heart and Aging Research in Genomic Epidemiology) GWAS meta-analysis of airflow obstruction among participants of European ancestry (22). The smallest P value in the replication region was for rs409839 (P = 8.9 × 10−4), and the adjusted threshold for statistical significance was P < 4.2 × 10−3. The other loci were not successfully replicated.
Generalization Analyses of Previously Identified Lung Function Loci
Using the false discovery rate–adjusted look-up threshold, several of the previously reported SNPs for FEV1, FEV1/FVC, and COPD identified in European populations were generalized to Hispanic/Latino populations (Tables E8–E10 and Figure E13). See online supplement for details.
Discussion
In this GWAS of 11,822 Hispanic/Latino HCHS/SOL participants, we identified eight novel signals, of which three replicated in independent cohorts of European ancestry. Three novel signals were associated with lung function. A novel locus in ZSWIM7 (rs4791658) associated with FEV1 replicated, and a rare variant in HAL (rs145174011) associated with FEV1/FVC did not replicate; it is in a previously identified region but remained significant in conditional analyses, suggesting an independent effect. Admixture mapping identified a novel LAI region, with a candidate variant in the AGMO gene (rs4133185), associated with Amerindian ancestry and FEV1, which replicated. In smoking-stratified analyses, a novel locus (rs291231) associated with FEV1 among 4,595 ever smokers approached statistical significance for replication in cohorts with admixed populations of African ancestry, and a novel SNP for COPD in PDZD2 (rs7709630) regionally replicated in individuals of European ancestry. In addition, we confirmed loci previously identified in European samples (KCNE2 [16] and GPR126 [26]) for FEV1/FVC in this Hispanic/Latino sample.
Several SNPs in ZSWIM7 were associated with FEV1 at genome-wide significance and replicated in independent cohorts of European ancestry. ZSWIM7 mutations were associated with colorectal adenomatous polyposis (51), and in a GWAS of SNP-by-smoking interaction, ZSWIM7 neared significance for systolic blood pressure (52). ZSWIM7 is a highly conserved eukaryotic regulator of homologous recombination and plays an important role in error-free DNA repair processes for DNA double-strand breaks (DSBs) (53–55). Unrepaired DSBs trigger cell senescence, apoptosis, and proinflammatory responses, all of which are established mechanisms in the pathogenesis of COPD (56–58). Oxidative stress and cigarette smoke cause DSBs (59, 60), and DSBs have been implicated in the pathogenesis of COPD (60). Furthermore, patients with COPD have a greater number of unrepaired DSBs, which are associated with a higher expression of markers for senescence, apoptosis, and proinflammatory phenotypes compared with asymptomatic smokers and nonsmokers (60). In our study, the effect size of ZSWIM7 for FEV1 was of greater magnitude among ever smokers, which is consistent with known effects of cigarette smoke causing DSBs. Thus, ZSWIM7, through its regulation of homologous recombination and role in error-free DNA repair processes for DSBs, is relevant to the pathogenesis of COPD.
A rare variant in HAL (rs145174011) was associated with FEV1/FVC among all participants. The minor allele was associated with decreases in FEV1/FVC of 4.9% and 6.3% (all participants and ever smokers, respectively), and with decreases in FEV1 of 100 ml and 173 ml (all participants and ever smokers, respectively). It is possible that this variant was not successfully replicated because of lack of power, given that it is a rare variant in African and Hispanic populations and is not polymorphic in European populations (61).
The HAL SNP (rs145174011) is near two previously identified loci for pulmonary traits (CCDC38 and SNRPF). CCDC38 has been associated with FEV1/FVC in individuals of European ancestry (16), and SNRPF has been associated with percent emphysema on CT imaging in racially/ethnically diverse participants in the MESA (Multi-Ethnic Study of Atherosclerosis) Lung Study (15). In the MESA Lung Study, linkage disequilibrium (LD) between SNRPF and CCDC38 accounted for much of the effect seen in white individuals and less so in African Americans and Chinese; however, the observed effect of SNRF on percent emphysema on CT imaging in Hispanics appeared to be independent from CCDC38 (15). In our study, the association of SNP rs145174011 (HAL locus) with FEV1/FVC was maintained in analyses conditioning on previously reported loci. Collectively, these data suggest there are three independent loci associated with COPD-related phenotypes in this region.
Although the HAL locus contained numerous SNPs, they were all imputed rare variants and did not replicate; therefore, further investigation, such as whole-genome sequencing of this region and functional studies, is required to refine these loci, confirm association, and understand their biological implications for COPD.
The HAL gene encodes the enzyme histidine ammonia lyase (62, 63)—the first step in histidine catabolism (64). Low levels of histidine have been associated with increased inflammation and oxidative stress (65, 66), and decreased concentrations of histidine were associated with advanced COPD and emphysema (67). Recently, whole-exome sequencing in African Americans identified three rare loss-of-function variants in HAL that were associated with increased histidine levels and were inversely related to coronary heart disease risk in African American and European American populations. Thus, a functional HAL variant may decrease histidine levels and increase susceptibility to COPD and emphysema through its effects on oxidative stress.
The region associated with Amerindian ancestry and FEV1 on chromosome 7 contained a candidate variant (rs4133185) within the AGMO gene. This SNP, rs4133185, has large differences in ancestry-specific allele frequencies (∼0.8 for Amerindian and ≤0.2 for European and African) and is associated with obstructive physiology, as measured by FEV1 (P = 7.43 × 10−7). In the conditional analysis, rs4133185 explains part, but not all, of the admixture signal. SNP rs4133185 may not be the causal variant, and the presence of multiple SNPs within the LAI region may explain the residual signal. A prior report showed an association of AGMO with FVC in a multiethnic population (68). Future whole-genome sequencing studies are needed for better fine mapping of this region.
The chromosome 11 locus (rs291231) associated with FEV1 among ever smokers nominally replicated in the meta-analysis across all cohorts with admixed populations of African ancestry. However, the direction of effect was discordant in some replication cohorts. Different directions of associations may be due to different patterns of LD with the causal variant between populations. The minor allele in rs291231 might be protective in ever smokers, as demonstrated by a per-allele increase in FEV1 of 57.28 ml.
The chromosome 18 locus associated with FEV1/FVC among never smokers contained multiple SNPs. The lead SNP (rs7228593) varies in MAF across different Hispanic/Latino ancestry groups in HCHS/SOL (0.02–0.16), is common in African populations (MAF = 0.39), and is not polymorphic in European populations (MAF = 0) (69). These variable allele frequencies and small sample size for replication may have contributed to lack of replication. Differing environmental exposures and LD may explain the observed genetic association of FEV1/FVC among never smokers in HCHS/SOL and lack of replication in independent cohorts of different ancestry backgrounds.
In the COPD analysis, SNP rs7709630, a genotyped intron variant in PDZD2, was a single SNP association with COPD. This genomic region replicated for airflow obstruction in an independent population of European ancestry (22), suggesting a true association. The CDRT15P1 locus only contained imputed SNPs and did not replicate, thus reducing the probability that this is a true association with COPD.
We hypothesize that novel loci did not replicate because of small sample size of the Hispanic/Latino replication cohorts along with differing environmental exposures, differing LD, and the complex racial admixture (2) that varies by different Hispanic/Latino ancestry groups. Furthermore, the novel locus for FEV1/FVC in HAL and the chromosome 18 locus for FEV1/FVC among never smokers have marked differences in MAF between race and ethnic populations, suggesting genetic diversity between races in these loci. The highest MAF for the lead SNPs in the HAL and in the chromosome 18 locus were observed in African populations (MAF = 0.005 and 0.38, respectively) compared with virtually no variability in European populations for these loci (MAF = 0) (61). The lack of variance of these loci in European populations limits the ability to replicate these loci, as the vast majority of GWAS of lung function have been performed in studies of European ancestry. Evidence for replication of the novel locus on chromosome 11 for FEV1 in ever smokers was stronger in the meta-analysis of cohorts with admixed populations that include African ancestry compared with the meta-analysis of all cohorts (P = 0.007 and P = 0.04, respectively), supporting the need for additional studies on admixed populations to identify possible ethnicity- or race-specific variants that may elucidate novel pathways in the pathogenesis of COPD.
The study has several potential limitations. This population-based cohort of diverse Hispanic/Latino participants is subject to population stratification. Multiple loci demonstrated varying MAF across Hispanic/Latino groups, highlighting the complex racial admixture of Hispanic/Latino populations and raising the possibility of residual population stratification. However, we adjusted our analysis for PCs, and our inflation factors ranged from 0.988 to 1.026, which indicates good control of population stratification with small inflation. The COPD analysis consisted of 363 and 5,253 participants with and without COPD (respectively), which is a relatively small sample size for a GWAS. We optimized COPD phenotype by limiting the analysis to age greater than or equal to 45 years and used post-bronchodilator spirometry measurements to define COPD.
In conclusion, we identified novel biologically plausible signals associated with clinically important pulmonary measures with evidence for replication in ZSWIM7, AGMO, and PDZD2; confirmed previous reports of association with FEV1/FVC in KCNE2 and GPR126; and established that loci previously identified in European populations are generalizable in Hispanic/Latino populations. There is a paucity of genetic studies for lung function that include underrepresented minority populations, such as Hispanic/Latino (2, 70) and African populations (14, 25, 71, 72). Our findings emphasize the importance of including admixed and multiracial populations when performing genetic studies of complex diseases and have the potential to advance our understanding of genetic risks for lung disease affecting Hispanic/Latino populations.
Acknowledgments
Acknowledgment
The authors thank the investigators, participants, and staff of the HCHS/SOL (Hispanic Community Health Study/Study of Latinos) for their contributions to this study. The HCHS/SOL investigators thank the NHLBI and the research institutions, study investigators, and field staff for their support in creating this study. They also thank the study participants, without whom this endeavor would not have been possible.
Footnotes
The baseline examination of HCHS/SOL (Hispanic Community Health Study/Study of Latinos) was performed as a collaborative study supported by NHLBI contracts N01-HC65233 (University of North Carolina), N01-HC65234 (University of Miami), N01-HC65235 (Albert Einstein College of Medicine), N01-HC65236 (Northwestern University), and N01-HC65237 (San Diego State University). The following institutes, centers, and offices contributed to the first phase of HCHS/SOL through a transfer of funds to the NHLBI: National Institute on Minority Health and Health Disparities, National Institute on Deafness and Other Communication Disorders, National Institute of Dental and Craniofacial Research (NIDCR), National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), National Institute of Neurological Disorders and Stroke, and NIH Office of Dietary Supplements. The Genetic Analysis Center at the University of Washington was supported by NHLBI and NIDCR contracts HHSN268201300005C AM03 and MOD03. Additional analysis support was provided by NIDDK grant 1R01DK101855-01 and American Heart Association grant 13GRNT16490017. Genotyping efforts were supported by the NIH Department of Health and Human Services grant HSN26220/20054C, National Center for Advancing Translational Science/Clinical Translational Science Institute grant UL1TR000124, and NIDDK Diabetes Research Center grant DK063491. This manuscript has been reviewed by the HCHS/SOL Publications Committee for scientific content and consistency of data interpretation with previous HCHS/SOL publications. T.S. is supported by NHLBI grant 1R35HL135818. A.A.D. is supported by NIH/NHLBI grant K01HL118714 and the Brigham and Women’s Hospital Minority Faculty Career Development Award. S.J.L. is supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences. R.G.B. is supported by NIH/NHLBI grants R01-HL077612 and R01-HL093081. For funding information for COPD-Costa Rica, MESA Lung, 1982 Pelotas Birth Cohort Study, and SpiroMeta, see the online supplement.
Author Contributions: Study design: K.M.B., J.C.C., and R.G.B.; data collection: S.J.L., F.P.H., L.A., I.P.H., B.L.H., R.C.K., A.M.M., D.R., S.S.R., M.S.-Q., A.M.S., M.D.T., J.C.C., and R.G.B.; data analysis: T.S., A.M., F.P.H., M.S.A., W.C., and L.V.W.; obtaining funding: R.C.K., C.C.L., S.S.R., and R.G.B.; drafting manuscript: K.M.B.; critical revision of manuscript: K.M.B., T.S., S.J.L., A.M., F.P.H., Q.Y., M.S.A., L.A., W.C., S.D.T., A.A.D., I.P.H., B.L.H., R.C.K., C.C.L., A.M.M., J.V.M., E.C.O., D.R., S.S.R., M.S.-Q., A.M.S., M.D.T., L.V.W., J.C.C., and R.G.B.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1164/rccm.201707-1493OC on February 2, 2018
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.U.S. Census Bureau. 2012 national population projections. Table 4. Projections of the population by sex, race, and Hispanic origin for the United States: 2015 to 2060. 2012 [accessed 2016 Oct 10]. Available from: https://www.census.gov/data/tables/2012/demo/popproj/2012-summary-tables.html.
- 2.Brehm JM, Celedón JC. Chronic obstructive pulmonary disease in Hispanics. Am J Respir Crit Care Med. 2008;177:473–478. doi: 10.1164/rccm.200708-1274PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hunninghake GM, Weiss ST, Celedón JC. Asthma in Hispanics. Am J Respir Crit Care Med. 2006;173:143–163. doi: 10.1164/rccm.200508-1232SO. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bryc K, Velez C, Karafet T, Moreno-Estrada A, Reynolds A, Auton A, et al. Genome-wide patterns of population structure and admixture among Hispanic/Latino populations. Proc Natl Acad Sci USA. 2010;107:8954–8961. doi: 10.1073/pnas.0914618107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vestbo J, Hurd SS, Agustí AG, Jones PW, Vogelmeier C, Anzueto A, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am J Respir Crit Care Med. 2013;187:347–365. doi: 10.1164/rccm.201204-0596PP. [DOI] [PubMed] [Google Scholar]
- 6.Vogelmeier CF, Criner GJ, Martinez FJ, Anzueto A, Barnes PJ, Bourbeau J, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive lung disease 2017 report: GOLD executive summary. Am J Respir Crit Care Med. 2017;195:557–582. doi: 10.1164/rccm.201701-0218PP. [DOI] [PubMed] [Google Scholar]
- 7.Miniño AM, Xu J, Kochanek KD. Deaths: preliminary data for 2008. Natl Vital Stat Rep. 2010;59:1–52. [PubMed] [Google Scholar]
- 8.Kosacz NM, Croxton TL, Kiley JP, Weinmann GG, Wheaton AG, Ford ES, et al. Chronic obstructive pulmonary disease among adults: United States, 2011. MMWR. 2012;61:938–943. [PubMed] [Google Scholar]
- 9.Díaz AA, Celli B, Celedón JC. Chronic obstructive pulmonary disease in Hispanics: a 9-year update. Am J Respir Crit Care Med. 2018;197:15–21. doi: 10.1164/rccm.201708-1615PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barr RG, Avilés-Santa L, Davis SM, Aldrich TK, Gonzalez F, Henderson AG, et al. Pulmonary disease and age at immigration among Hispanics: results from the Hispanic Community Health Study/Study of Latinos. Am J Respir Crit Care Med. 2015;193:386–395. doi: 10.1164/rccm.201506-1211OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruse S, Sood A, Petersen H, Liu Y, Leng S, Celedón JC, et al. New Mexican Hispanic smokers have lower odds of chronic obstructive pulmonary disease and less decline in lung function than non-Hispanic whites. Am J Respir Crit Care Med. 2011;184:1254–1260. doi: 10.1164/rccm.201103-0568OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diaz AA, Come CE, Mannino DM, Pinto-Plata V, Divo MJ, Bigelow C, et al. Obstructive lung disease in Mexican Americans and non-Hispanic whites: an analysis of diagnosis and survival in the National Health and Nutritional Examination Survey III follow-up study. Chest. 2014;145:282–289. doi: 10.1378/chest.13-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen W, Brehm JM, Manichaikul A, Cho MH, Boutaoui N, Yan Q, et al. A genome-wide association study of chronic obstructive pulmonary disease in Hispanics. Ann Am Thorac Soc. 2015;12:340–348. doi: 10.1513/AnnalsATS.201408-380OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burkart KM, Manichaikul A, Wilk JB, Ahmed FS, Burke GL, Enright P, et al. APOM and high-density lipoprotein cholesterol are associated with lung function and per cent emphysema. Eur Respir J. 2014;43:1003–1017. doi: 10.1183/09031936.00147612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manichaikul A, Hoffman EA, Smolonska J, Gao W, Cho MH, Baumhauer H, et al. Genome-wide study of percent emphysema on computed tomography in the general population: the Multi-Ethnic Study of Atherosclerosis Lung/SNP Health Association Resource Study. Am J Respir Crit Care Med. 2014;189:408–418. doi: 10.1164/rccm.201306-1061OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soler Artigas M, Loth DW, Wain LV, Gharib SA, Obeidat M, Tang W, et al. Genome-wide association and large-scale follow up identifies 16 new loci influencing lung function. Nat Genet. 2011;43:1082–1090. doi: 10.1038/ng.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soler Artigas M, Wain LV, Miller S, Kheirallah AK, Huffman JE, Ntalla I, et al. Sixteen new lung function signals identified through 1000 genomes project reference panel imputation. Nat Commun. 2015;6:8658. doi: 10.1038/ncomms9658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soler Artigas M, Wain LV, Repapi E. Obeidat ME, Sayers I, Burton PR, et al.; SpiroMeta Consortium. Effect of five genetic variants associated with lung function on the risk of chronic obstructive lung disease, and their joint effects on lung function. Am J Respir Crit Care Med. 2011;184:786–795. doi: 10.1164/rccm.201102-0192OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hancock DB, Soler Artigas M, Gharib SA, Henry A, Manichaikul A, Ramasamy A, et al. Genome-wide joint meta-analysis of SNP and SNP-by-smoking interaction identifies novel loci for pulmonary function. PLoS Genet. 2012;8:e1003098. doi: 10.1371/journal.pgen.1003098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pillai SG. A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet. 2009;5:e1000421. doi: 10.1371/journal.pgen.1000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilk JB. A genome-wide association study of pulmonary function measures in the Framingham Heart Study. PLoS Genet. 2009;5:e1000429. doi: 10.1371/journal.pgen.1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilk JB, Shrine NRG, Loehr LR, Zhao JH, Manichaikul A, Lopez LM, et al. Genome wide association studies identify CHRNA5/3 and HTR4 in the development of airflow obstruction. Am J Respir Crit Care Med. 2012;186:622–632. doi: 10.1164/rccm.201202-0366OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cho MH, Boutaoui N, Klanderman BJ, Sylvia JS, Ziniti JP, Hersh CP, et al. Variants in FAM13A are associated with chronic obstructive pulmonary disease. Nat Genet. 2010;42:200–202. doi: 10.1038/ng.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Castaldi PJ, Cho MH, Litonjua AA, Bakke P, Gulsvik A, Lomas DA, et al. COPDGene and Eclipse Investigators. The association of genome-wide significant spirometric loci with chronic obstructive pulmonary disease susceptibility. Am J Respir Cell Mol Biol. 2011;45:1147–1153. doi: 10.1165/rcmb.2011-0055OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cho MH, McDonald M-LN, Zhou X, Mattheisen M, Castaldi PJ, Hersh CP, et al. NETT Genetics, ICGN, ECLIPSE and COPDGene Investigators. Risk loci for chronic obstructive pulmonary disease: a genome-wide association study and meta-analysis. Lancet Respir Med. 2014;2:214–225. doi: 10.1016/S2213-2600(14)70002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hancock DB, Eijgelsheim M, Wilk JB, Gharib SA, Loehr LR, Marciante KD, et al. Meta-analyses of genome-wide association studies identify multiple loci associated with pulmonary function. Nat Genet. 2010;42:45–52. doi: 10.1038/ng.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Repapi E, Sayers I, Wain LV, Burton PR, Johnson T, Obeidat M, et al. Genome-wide association study identifies five loci associated with lung function. Nat Genet. 2010;42:36–44. doi: 10.1038/ng.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wain LV, Shrine N, Artigas MS, Erzurumluoglu AM, Noyvert B, Bossini-Castillo L, et al. Genome-wide association analyses for lung function and chronic obstructive pulmonary disease identify new loci and potential druggable targets. Nat Genet. 2017;49:416–425. doi: 10.1038/ng.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hobbs BD, de Jong K, Lamontagne M, Bosse Y, Shrine N, Artigas MS, et al. COPDGene Investigators; ECLIPSE Investigators. LifeLines Investigators, SPIROMICS Research Group, International COPD Genetics Network Investigators, UK BiLEVE Investigators, International COPD Genetics Consortium. Genetic loci associated with chronic obstructive pulmonary disease overlap with loci for lung function and pulmonary fibrosis. Nat Genet. 2017;49:426–432. doi: 10.1038/ng.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.LaVange LM, Kalsbeek WD, Sorlie PD, Avilés-Santa LM, Kaplan RC, Barnhart J, et al. Sample design and cohort selection in the Hispanic Community Health Study/Study of Latinos. Ann Epidemiol. 2010;20:642–649. doi: 10.1016/j.annepidem.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burkart KM, Stilp AM, Sofer T, London SJ, Davis ST, Celedon JC, et al. Genome-wide association study (GWAS) of lung function among Hispanic/Latino individuals of diverse backgrounds. The Hispanic Community Health Study/Study of Latinos [abstract] Am J Respir Crit Care Med. 2015;191:A1069. [Google Scholar]
- 32.Sorlie PD, Avilés-Santa LM, Wassertheil-Smoller S, Kaplan RC, Daviglus ML, Giachello AL, et al. Design and implementation of the Hispanic Community Health Study/Study of Latinos. Ann Epidemiol. 2010;20:629–641. doi: 10.1016/j.annepidem.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Conomos MP, Laurie CA, Stilp AM, Gogarten SM, McHugh CP, Nelson SC, et al. Genetic diversity and association studies in us hispanic/latino populations: applications in the Hispanic Community Health Study/Study of Latinos. Am J Hum Genet. 2016;98:165–184. doi: 10.1016/j.ajhg.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pellegrino R. Interpretative strategies for lung function tests. Eur Respir J. 2005;26:948–968. doi: 10.1183/09031936.05.00035205. [DOI] [PubMed] [Google Scholar]
- 35.Hankinson JL, Odencrantz JR, Fedan KB. Spirometric reference values from a sample of the general U.S. population. Am J Respir Crit Care Med. 1999;159:179–187. doi: 10.1164/ajrccm.159.1.9712108. [DOI] [PubMed] [Google Scholar]
- 36.LaVange L, Davis SM, Hankinson J, Enright P, Wilson MR, Barr RG, et al. Spirometry reference equations from HCHS/SOL (Hispanic Community Health Study/Study of Latinos) Am J Respir Crit Care Med. 2017;196:993–1003. doi: 10.1164/rccm.201610-1987OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laurie CC, Doheny KF, Mirel DB, Pugh EW, Bierut LJ, Bhangale T, et al. GENEVA Investigators. Quality control and quality assurance in genotypic data for genome-wide association studies. Genet Epidemiol. 2010;34:591–602. doi: 10.1002/gepi.20516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.1000 Genomes Project Consortium. Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sofer T, Shaffer JR, Graff M, Qi Q, Stilp AM, Gogarten SM, et al. Meta-analysis of genome-wide association studies with correlated individuals: application to the Hispanic Community Health Study/Study of Latinos (HCHS/SOL) Genet Epidemiol. 2016;40:492–501. doi: 10.1002/gepi.21981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pe’er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol. 2008;32:381–385. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- 41.Chen H, Wang C, Conomos Matthew P, Stilp Adrienne M, Li Z, Sofer T, et al. Control for population structure and relatedness for binary traits in genetic association studies via logistic mixed models. Am J Hum Genet. 2016;98:653–666. doi: 10.1016/j.ajhg.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Browning SR, Grinde K, Plantinga A, Gogarten SM, Stilp AM, Kaplan RC, et al. Local ancestry inference in a large US-based Hispanic/Latino study: Hispanic Community Health Study/Study of Latinos (HCHS/SOL) G3 (Bethesda) 2016;6:1525–1534. doi: 10.1534/g3.116.028779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sofer T, Baier LJ, Browning SR, Thornton TA, Talavera GA, Wassertheil-Smoller S, et al. Admixture mapping in the Hispanic Community Health Study/Study of Latinos reveals regions of genetic associations with blood pressure traits. PLoS One. 2017;12:e0188400. doi: 10.1371/journal.pone.0188400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang QS, Browning BL, Browning SR. ASAFE: ancestry-specific allele frequency estimation. Bioinformatics. 2016;32:2227–2229. doi: 10.1093/bioinformatics/btw220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bild DE, Bluemke DA, Burke GL, Detrano R, Diez Roux AV, Folsom AR, et al. Multi-ethnic study of atherosclerosis: objectives and design. Am J Epidemiol. 2002;156:871–881. doi: 10.1093/aje/kwf113. [DOI] [PubMed] [Google Scholar]
- 46.Kaufman JD, Adar SD, Allen RW, Barr RG, Budoff MJ, Burke GL, et al. Prospective study of particulate air pollution exposures, subclinical atherosclerosis, and clinical cardiovascular disease: the Multi-Ethnic Study of Atherosclerosis and Air Pollution (MESA Air) Am J Epidemiol. 2012;176:825–837. doi: 10.1093/aje/kws169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Victora CG, Barros FC. Cohort profile: the 1982 Pelotas (Brazil) birth cohort study. Int J Epidemiol. 2006;35:237–242. doi: 10.1093/ije/dyi290. [DOI] [PubMed] [Google Scholar]
- 48.Horta BL, Gigante DP, Gonçalves H, dos Santos Motta J, Loret de Mola C, Oliveira IO, et al. Cohort profile update: the 1982 Pelotas (Brazil) birth cohort study. Int J Epidemiol. 2015;44:441a–441e. doi: 10.1093/ije/dyv017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen W, Brehm JM, Boutaoui N, Soto-Quiros M, Avila L, Celli BR, et al. Native American ancestry, lung function, and COPD in Costa Ricans. Chest. 2014;145:704–710. doi: 10.1378/chest.13-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sofer T, Heller R, Bogomolov M, Avery CL, Graff M, North KE, et al. A powerful statistical framework for generalization testing in GWAS, with application to the HCHS/SOL. Genet Epidemiol. 2017;41:251–258. doi: 10.1002/gepi.22029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spier I, Kerick M, Drichel D, Horpaopan S, Altmüller J, Laner A, et al. Exome sequencing identifies potential novel candidate genes in patients with unexplained colorectal adenomatous polyposis. Fam Cancer. 2016;15:281–288. doi: 10.1007/s10689-016-9870-z. [DOI] [PubMed] [Google Scholar]
- 52.Sung YJ, de las Fuentes L, Schwander KL, Simino J, Rao DC. Gene–smoking interactions identify several novel blood pressure loci in the Framingham Heart Study. Am J Hypertens. 2015;28:343–354. doi: 10.1093/ajh/hpu149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martín V, Chahwan C, Gao H, Blais V, Wohlschlegel J, Yates JR, et al. Sws1 is a conserved regulator of homologous recombination in eukaryotic cells. EMBO J. 2006;25:2564–2574. doi: 10.1038/sj.emboj.7601141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Godin SK, Meslin C, Kabbinavar F, Bratton-Palmer DS, Hornack C, Mihalevic MJ, et al. Evolutionary and functional analysis of the invariant SWIM domain in the conserved Shu2/Sws1 protein family from Saccharomyces cerevisiae to Homo sapiens. Genetics. 2015;199:1023–1033. doi: 10.1534/genetics.114.173518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lou Z, Chen J. Cellular senescence and DNA repair. Exp Cell Res. 2006;312:2641–2646. doi: 10.1016/j.yexcr.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 56.Tuder RM, Petrache I. Pathogenesis of chronic obstructive pulmonary disease. J Clin Invest. 2012;122:2749–2755. doi: 10.1172/JCI60324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.MacNee W, Tuder RM. New paradigms in the pathogenesis of chronic obstructive pulmonary disease I. Proc Am Thorac Soc. 2009;6:527–531. doi: 10.1513/pats.200905-027DS. [DOI] [PubMed] [Google Scholar]
- 58.Neofytou E, Tzortzaki EG, Chatziantoniou A, Siafakas NM. DNA damage due to oxidative stress in chronic obstructive pulmonary disease (COPD) Int J Mol Sci. 2012;13:16853–16864. doi: 10.3390/ijms131216853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Caramori G, Adcock IM, Casolari P, Ito K, Jazrawi E, Tsaprouni L, et al. Unbalanced oxidant-induced DNA damage and repair in COPD: a link towards lung cancer. Thorax. 2011;66:521–527. doi: 10.1136/thx.2010.156448. [DOI] [PubMed] [Google Scholar]
- 60.Aoshiba K, Zhou F, Tsuji T, Nagai A. DNA damage as a molecular link in the pathogenesis of COPD in smokers. Eur Respir J. 2012;39:1368. doi: 10.1183/09031936.00050211. [DOI] [PubMed] [Google Scholar]
- 61. National Center for Biotechnology Information. dbSNP: Rs192375903; build 38 [accessed 2016 Nov 1]. Available from: https://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?rs=192375903.
- 62.Suchi M, Sano H, Mizuno H, Wada Y. Molecular cloning and structural characterization of the human histidase gene (HAL) Genomics. 1995;29:98–104. doi: 10.1006/geno.1995.1219. [DOI] [PubMed] [Google Scholar]
- 63.Yu B, Li AH, Muzny D, Veeraraghavan N, de Vries PS, Bis JC, et al. Association of rare loss-of-function alleles in HAL, serum histidine levels and incident coronary heart disease. Circ Cardiovasc Genet. 2015;8:351–355. doi: 10.1161/CIRCGENETICS.114.000697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Taylor RG, Lambert MA, Sexsmith E, Sadler SJ, Ray PN, Mahuran DJ, et al. Cloning and expression of rat histidase: homology to two bacterial histidases and four phenylalanine ammonia-lyases. J Biol Chem. 1990;265:18192–18199. [PubMed] [Google Scholar]
- 65.Niu Y-C, Feng R-N, Hou Y, Li K, Kang Z, Wang J, et al. Histidine and arginine are associated with inflammation and oxidative stress in obese women. Br J Nutr. 2012;108:57–61. doi: 10.1017/S0007114511005289. [DOI] [PubMed] [Google Scholar]
- 66.Watanabe M, Suliman ME, Qureshi AR, Garcia-Lopez E, Barany P, Heimburger O, et al. Consequences of low plasma histidine in chronic kidney disease patients: associations with inflammation, oxidative stress, and mortality. Am J Clin Nutr. 2008;87:1860–1866. doi: 10.1093/ajcn/87.6.1860. [DOI] [PubMed] [Google Scholar]
- 67.Ubhi BK, Cheng KK, Dong J, Janowitz T, Jodrell D, Tal-Singer R, et al. Targeted metabolomics identifies perturbations in amino acid metabolism that sub-classify patients with copd. Mol Biosyst. 2012;8:3125–3133. doi: 10.1039/c2mb25194a. [DOI] [PubMed] [Google Scholar]
- 68.Wyss AB, Sofer T, Lee MK, Terzikhan N, Nguyen JN, Lahousse L, et al. Multiethnic meta-analysis identifies new loci for pulmonary function. bioRxiv. [DOI] [PMC free article] [PubMed]
- 69. National Center for Biotechnology Information. dbSNP: Rs7228593; build 38 [accessed 2016 Nov]. Available from: https://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?rs=7228593.
- 70.Pouladi N, Bime C, Garcia JGN, Lussier YA. Complex genetics of pulmonary diseases: lessons from genome-wide association studies and next-generation sequencing. Transl Res. 2016;168:22–39. doi: 10.1016/j.trsl.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Loth DW, Artigas MS, Gharib SA, Wain LV, Franceschini N, Koch B, et al. Genome-wide association analysis identifies six new loci associated with forced vital capacity. Nat Genet. 2014;46:669–677. doi: 10.1038/ng.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wain LV, Shrine N, Miller S, Jackson VE, Ntalla I, Artigas MS, et al. Novel insights into the genetics of smoking behaviour, lung function, and chronic obstructive pulmonary disease (UK BiLEVE): a genetic association study in UK biobank Lancet Respir Med 20153769–781.[Published erratum appears in Lancet Respir Med 4:e4.] [DOI] [PMC free article] [PubMed] [Google Scholar]