Abstract
Functional redundancy, understood as the functional overlap of different genes, is a double-edge sword. At the one side, it is thought to serve as a robustness mechanism that buffers the deleterious effect of mutations hitting one of the redundant copies, thus resulting in pseudogenization. At the other side, it is considered as a source of genetic and functional innovation. In any case, genetically redundant genes are expected to show an acceleration in the rate of molecular evolution. Here, we tackle the role of functional redundancy in viral RNA genomes. To this end, we have evaluated the rates of compensatory evolution for deleterious mutations affecting an essential function, the suppression of RNA silencing plant defense, of tobacco etch potyvirus (TEV). TEV genotypes containing deleterious mutations in presence/absence of engineered functional redundancy were evolved and the pattern of fitness and pathogenicity recovery evaluated. Genetically redundant genotypes suffered less from the effect of deleterious mutations and showed relatively minor changes in fitness and pathogenicity. By contrast, nongenetically redundant genotypes had very low fitness and pathogenicity at the beginning of the evolution experiment that were fully recovered by the end. At the molecular level, the outcome depended on the combination of the actual mutations being compensated and the presence/absence of functional redundancy. Reversions to wild-type alleles were the norm in the nonredundant genotypes while redundant ones either did not fix any mutation at all or showed a higher nonsynonymous mutational load.
Keywords: compensatory evolution, experimental virus evolution, genetic redundancy, multifunctional proteins, tobacco etch virus, viral suppressors of RNA silencing
Introduction
Viruses are well known vectors of horizontal gene transfer (HGT) among hosts species (Penadés et al. 2015). In some well documented cases, viral genes have been inserted into cellular genomes, serving as sources of genetic and functional innovations (Koonin 2010; Liu et al. 2010). One of the most illustrative examples of such virus-mediated innovations is the acquisition of syncytin, an essential protein for the morphogenesis of placenta, from an endogenous retrovirus at the origin of the lineage of placental mammals (Mi et al. 2000). Gene trafficking goes in both directions, and viruses sometimes acquire genes from the genome of their host cells, or from other viruses coinfecting the same host. For example, phylogenetic studies have suggested that many Mimivirus genes have been acquired by HGT from the host (Moreira and Brochier-Armanet 2008). A second good example of HGT from host to virus is the homologue of the 70 kDa heat shock protein encoded in the genome of plant closteroviruses (Dolja et al. 2006). Although HGT is thought to be an essential mechanism for virus evolution, strong selection against increasing genome size may hinder its importance, especially for small RNA viruses (Zwart et al. 2014; Willemsen, Zwart, Higueras, et al. 2016). Accommodation of a new gene into the already compacted genome of the recipient virus would necessarily involve changes in the regulation of gene expression, retuning interactions among all viral proteins and between them and the host proteome, and optimization of codon usage of the newly acquired gene to the characteristic bias of the recipient virus. Right upon insertion, the exogenous sequence will generally have a negative impact in the fitness of the recipient virus (Dolja et al. 1993; Zwart et al. 2014). Despite any possible long-term benefit, shortsighted selection will purge these low fitness genomes from a population numerically dominated by the wild-type virus. Even in isolation, these longer genomes will have a disadvantage relative to deletion mutants that will be generated during their replication (Zwart et al. 2014; Willemsen, Zwart, Higueras, et al. 2016).
In a recent series of studies, we have explored the constraints to innovation in the prototypical genome of a plant (+)-sense ssRNA monopartite virus belonging to the picorna-like superfamily, Tobacco etch virus (TEV; genus Potyvirus, family Potyviridae). More precisely, we have explored the short-term cost of acquiring new exogenous genes and their evolutionary fate (Zwart et al. 2014; Willemsen, Zwart, Higueras, et al. 2016; Willemsen et al. 2017), the fitness burden imposed by changes in gene order (Majer et al. 2014; Willemsen, Zwart, Tromas, et al. 2016) and the effect on virus fitness of the HGT of an essential viral gene into the host genome and its constitutive expression (Tromas et al. 2014). In addition, using a segmented plant RNA virus, Alfalfa mosaic virus, we have also been exploring the fitness consequences of genome segmentation and multipartition (Sánchez-Navarro et al. 2013; Wu et al. 2017). For its direct relationship with the work reported in this article, we would mention here the cases in which we added new genes into TEV genome. We generated a number of artificial genomes carrying insertions of heterologous sequences from different origins. First, the AlkB domain from Nicotiana tabacum, which is involved in correcting alkylation damages in nucleic acids (Willemsen et al. 2017). Second, the 2 b gene from Cucumber mosaic virus (CMV), which is a suppressor of RNA silencing (RSS) (Willemsen et al. 2017). Third, the eGFP gene encoding for a version of the green fluorescent protein (Zwart et al. 2014). The first two cases would provide the virus with new (AlkB) or redundant (2b) functions, whereas the eGFP would, in principle, not provide any useful function to the virus but just increase its genome length. In all cases, engineered genomes were evolved by serial passages in N. tabacum, with at least five independent evolutionary lineages. At the end of the experimental evolution, the evolved viral populations were phenotypically (viral load, relative fitness, infectivity, and pathogenicity) and genetically (Illumina NGS study of genetic variability within each evolved lineage) characterized. In summary, eGFP and AlkB genes were always removed from the genome at a rate that depended on the demographic conditions, recombination rate, cloning site, and the exogenous gene itself (Willemsen et al. 2018). Interestingly, the 2 b RSS was pervasively retained in all evolved lineages (Willemsen et al. 2017). Furthermore, functional analyses demonstrated that the 2 b protein was able of complementing the effect of strong and weak hyposuppressor mutants negatively affecting the TEV RSS multifunctional protein HC-Pro (Willemsen et al. 2017). Therefore, the engineered TEV/2b virus encodes for evolutionarily stable functional redundancy in its genome. In this context, functional redundancy means that two or more genes are performing the same function and that inactivation of one of these genes has little or no effect on the biological phenotype. Functional redundancy may open the possibility, at least in theory, for the HC-Pro multifunctional protein to relax purifying selection on protein domains involved in the RSS function and to optimize some of its other activities as protease, cell-to-cell movement or as helper-component during aphid horizontal transmission (a trait that should be irrelevant in our experimental system, because transmission is mechanical) (for a recent review on HC-Pro functions, see Valli et al. 2018).
To experimentally explore the role of functional redundancy, provided by 2 b, on the evolutionary fate of a strong (hereafter named as FINK; see below) and a weak hyposuppressor (hereafter named as AS13; see below) mutants affecting the RSS activity of HC-Pro, we have performed an evolution experiment starting with two different TEV genetic backgrounds. The first consists on the wild-type TEV genotype into which we introduced the FINK and AS13 mutations, respectively. The second genetic background consists on the TEV/2b engineered genotype into which we introduced the same set of mutations. We sought to test the following hypotheses: 1) functional redundancy will compensate for the inefficient RSS activity of HC-Pro hyposuppressor mutants, slowing down the rate of compensatory (phenotypic and genotypic) evolution compared with the nonredundant case. 2) By buffering the effect of mutations in HC-Pro RSS activity, mutational load in this sequence may further increase.
Materials and Methods
Viral Constructs and Stocks
Mutant genotypes TEV-FINK, TEV-FINK/2b, TEV-AS13, and TEV-AS13/2b were constructed by site-directed mutagenesis as previously described (Torres-Barceló et al. 2008; Willemsen et al. 2017). TEV/2b virus variants contain the 2 b gene from CMV. FINK and AS13 mutant genotypes contain nucleotide substitutions within the central RSS region of the HC-Pro protein. The strong hyposuppressor mutant FINK consists in the G548U nucleotide substitution leading to the R183I amino acid change (positively charged hydrophilic by noncharged hydrophobic) in the highly conserved FRNK motif within the RNA-binding domain (RBD) A (Shiboleth et al. 2007; Kung et al. 2014). The weak hyposuppressor AS13 double mutant contains the A896C and A899C nucleotide substitutions leading to E299A and D300A amino acid changes (negatively charged and hydrophilic by noncharged hydrophobic) within the RBD B domain (Kasschau et al. 1997). The viability and fitness effects of these mutants, relative to the wild-type TEV, were assessed in detail in previous works (Torres-Barceló et al. 2008; Willemsen et al. 2017). Homogenized stock tissue from plants infected with these four viral genotypes was preserved at −80°C and used as stock for initiating the evolution experiments described below.
Experimental Evolution
A variable number of independent evolution lineages (as indicated in parentheses) were stablished for each of the five engineered genotypes: TEV-FINK (2), TEV-FINK/2b (5), TEV-AS13 (3), and TEV-AS13/2b (5). In all cases, 100 mg of the corresponding tissue-infected stocks were resuspended with an equal volume of phosphate buffer (50 mM KH2PO4, pH 7.0, 3% polyethylene glycol 6000) and 5 μl of this mixture used to mechanically inoculate the third true leaf of 20 4- or 5-week-old N. tabacum L. cv Xanthi NN plants dusted with Carborundum. In the case of TEV-AS13, no infected plants were obtained for one lineage and thus we used the other successful lineage (L2) to create a replicate (L3) that was maintained independently afterward. Inoculated plants were kept in a Biosafety Level-2 greenhouse chamber at 25°C under a 16-h natural sunlight (supplemented with 400 W high-pressure sodium lamps as needed to ensure a minimum light intensity of PAR 50 μmol/m2/s) and 8-h dark photoperiod. The passage duration was determined by the appearance of at least one symptomatic plant per lineage between 9- and 15-day postinfection (dpi). Those lineages in which no plant displayed symptoms 15 dpi, were left to grow for five extra days. Leaves above the inoculated leaf were collected from all symptomatic plants, stored at −80°C and used for subsequent serial passages. Then, 100 mg of plant tissues were homogenized into a fine powder with liquid N2 using a Mixer Mill MM400 (Retsch GmbH, Haan, Germany). At each passage, the third true leaf of 10 (for TEV-FINK/2b and TEV-AS13/2b) or 15–20 (for TEV-FINK and TEV-AS13) 4- or 5-week-old plants were mechanically inoculated as just described. For those lineages in which the viral load was very low, volumes of phosphate buffer were reduced in order to equilibrate the genome equivalents of all evolved viral lineages. A total of five serial passages were conducted.
Evaluation of Pathogenicity Traits
After inoculation, plants were observed every day for the presence and severity of symptoms and the number of symptomatic plants recorded. Three different pathogenicity-related traits were estimated from these data. First, the percentage of symptomatic plants at the end of the experiment (t = 20 dpi) was used as an estimate of infectivity (i). Second, infectivity time-series data were submitted to a Kaplan–Meier regression analysis of survival times and the median time to the appearance of symptoms (ST50) estimated. Third, using the infectivity time-series data, we also estimated the area under the disease progress stairs curve (AUDPS) (Simko and Piepho 2012):
| (1) |
where ik stands for the frequency of infected plants at k dpi.
Fourth, each day the severity of symptoms (S) was recorded for each individual plant according to the following semiquantitative scale: S = 0, no symptoms; S = 1, curly leaves and disperse chlorotic spots; S = 2, incipient vein etching; S = 3, extensive and strong etching and necrotic tissue; and S = 4, generalized necrosis and plant death. The average severity of symptoms, , as evaluated at each dpi (t) for the set of plants was fitted to the following logistic regression model (Campbell and Madden 1990):
| (2) |
where Smax represents the maximal symptoms scored, τhalf the half-time elapsed from inoculation to observing the maximal symptoms developed and b stands for the steepness of the symptoms development curve, that is, the inverse of the speed at which plants develop the maximal symptoms.
The six pathogenicity-related traits evaluated for each lineage, L, after each evolution passage, P, were analyzed using multivariate analysis of variance methods (MANOVA) and fitted to the following multivariate linear model:
| (3) |
where Xijk is the vector of observed trait values for lineage k of viral genotype j observed at passage i, μ the vector of grand mean values, ɛijk the vector of errors assumed to be distributed as a multivariate Gaussian. G is the factor accounting for the virus genotype (i.e., TEV-FINK, TEV-FINK/2b, TEV-AS13, and TEV-AS13/2b), L is nested within G [L(G)], and P was considered as a covariable. Interactions between the covariable and the two factors have also been incorporated into the model. The significance of each component in the model has been evaluated with the Wilk’s Λ statistic. Associations between pathogenicity traits were evaluated using a partial correlation coefficient controlling for G, rp. A factorial analysis was also performed and the six variables were reduced into a single principal component (PC1) that explained up to 79.73% of total variance. For the shake of summarizing the evolution of pathogenicity into visually appealing plots, PC1 will be presented instead of each single trait.
All statistical analyses described in this section and hereafter were performed using SPSS version 23 software (IBM, Armonk, NY).
Evaluation of Within-Host Fitness
Viral load was evaluated as a measure of within-host replicative fitness. Viral load was only determined after passages 1 and 5. Total plant RNA extractions (RNAt) were performed from 100 mg of tissue powder with the Plant RNA Isolation Mini kit (Agilent Technologies, Santa Clara, CA). Viral accumulation (TEV genomes per ng of RNAt) was quantified by RT-qPCR as previously described (Zwart et al. 2014). Briefly, reactions were performed in 20 μl volume using the GoTaq 1-Step RT-qPCR System (Promega, Fitchburg, WI) following the instructions recommended by the manufacturer in an ABI StepOne Plus Real-time PCR System (Applied Biosystems, Foster City, CA). The cycling conditions were as follow: an RT phase of 15 min at 37°C followed by 10 min at 95°C; a PCR phase consisting in 40 cycles of 10 s at 95°C, 34 s at 60°C, and 30 s at 72°C; and a final phase of 15 s at 95°C, 1 min at 60°C, and 15 s at 95°C. The following specific TEV primers designed in the CP region were used: forward 5′-TTGGTCTTGATGGCAACGTG-3′ and reverse 5′-TGTGCCGTTCAGTGTCTTCCT-3′. The StepOne Software v.2.2.2 (Applied Biosystems) was used to analyze quantitative data. All ancestral and evolved viral lineages were normalized in order to equilibrate genome equivalents in the sample viral inoculums of the first and second passages.
Viral load data were fitted to a generalized linear model (GLM) with a Gamma distribution and a logarithmic link function (based on the smallest BIC value among a set of alternative models). The model equation is now written as:
| (4) |
where Vijkl stands for the viral load for the l-th replicate plant (3) from lineage k of genotype j at passage i (in this case 1 or 5) and all other factors are as described in the previous section. The significance of each component in the model was evaluated by means of likelihood ratio tests (LRT) that asymptotically follow a χ2 probability distribution. The magnitude of effects was evaluated using the statistic (proportion of total variability in viral load attributable to each factor in the model). Conventionally, values < 0.05 are considered as small, 0.05 ≤ < 0.15 as medium and ≥ 0.15 as large effects.
All raw data are available at LabArchives under doi: 10.6070/H418352M.
Evaluation of Insert Stability by RT-PCR
Reverse transcription and amplification reactions (RT-PCR) were performed in 20 μl volume using the GoTaq 1-Step RT-qPCR System (Promega) as indicated before. Input RNAt samples consisted in 150–300 ng from the selected plants per each lineage of passages 1 and 5 and two primers flanking the 2 b gene region: forward PC96-97F 5′-TACGATATTCCAACGACTG-3′ and reverse PC92-96R 5′-GCAAACTGCTCATGTGTGG-3′. Amplification protocol was as follows: an RT phase of 15 min at 45°C plus 2 min at 95°C and a PCR phase consisting in three cycles of 15 s at 95°C and 1 min at 67°C and 37 cycles of 15 s at 95°C and 1 min at 65°C. Size and quality of PCR products were confirmed by electrophoresis on 1.5% agarose gels. TEV mutants rendered products of 651 or 298 pb according with the presence or absence of 2 b, respectively.
Sequencing
To generate the consensus sequence of each mutant and lineage from passages 1 and 5, accurate RT-PCR products were obtained. RT reactions for the HC-Pro and 2 b genes were performed using the AccuScript Hi-Fi reverse transcriptase (Agilent Technologies) and reverse primers HCPro-R2545 5′-AATTGAAGGGGAGACTATTGCCAG-3′, that flanks the TEV HC-Pro cistron, or the PC92-96R flanking the 5′ CP cistron following manufacturer’s instructions. Then, viral genomes were amplified using Phusion DNA polymerase (Thermo Scientific, Waltham, MA), 1 μl of the RT reactions and primers PC96-97F and PC92-96R (2 b gene) or HCPro-F 5′-GGATGGGATGTTGGTGGATGCTC-3′ and HCPro-R 5′-CCTTGTGTGACCATATCTCGGTTC-3′ (HC-Pro gene). PCR protocol for HC-Pro and 2 b amplicons was as follows: initial denaturation 30 s at 98°C, five cycles with denaturation step of 10 s at 98°C, annealing step of 20 s at 57°C and elongation step of 30 s at 72°C followed with 27 cycles in the same conditions except that annealing step was undergone at 65°C. Amplicon sizes, 1,473 bp for HC-Pro and 651/298 for 2 b, were confirmed by electrophoresis on 1% agarose gels. Consensus sequences of all evolved lineages at passages 1 and 5 were obtained using Sanger sequencing and an ABI 3130 XL Genetic Analyzer (Applied Biosystems) in IBMCP’s DNA Sequencing Unit (www.ibmcp.csic.es/en/services/dna-sequencing-unit; last accessed July 12, 2018). Sequences were assembled using GENEIOUS version 11 software (www.geneious.com; last accessed July 12, 2018).
Results
Evolution of Pathogenicity-Related Traits
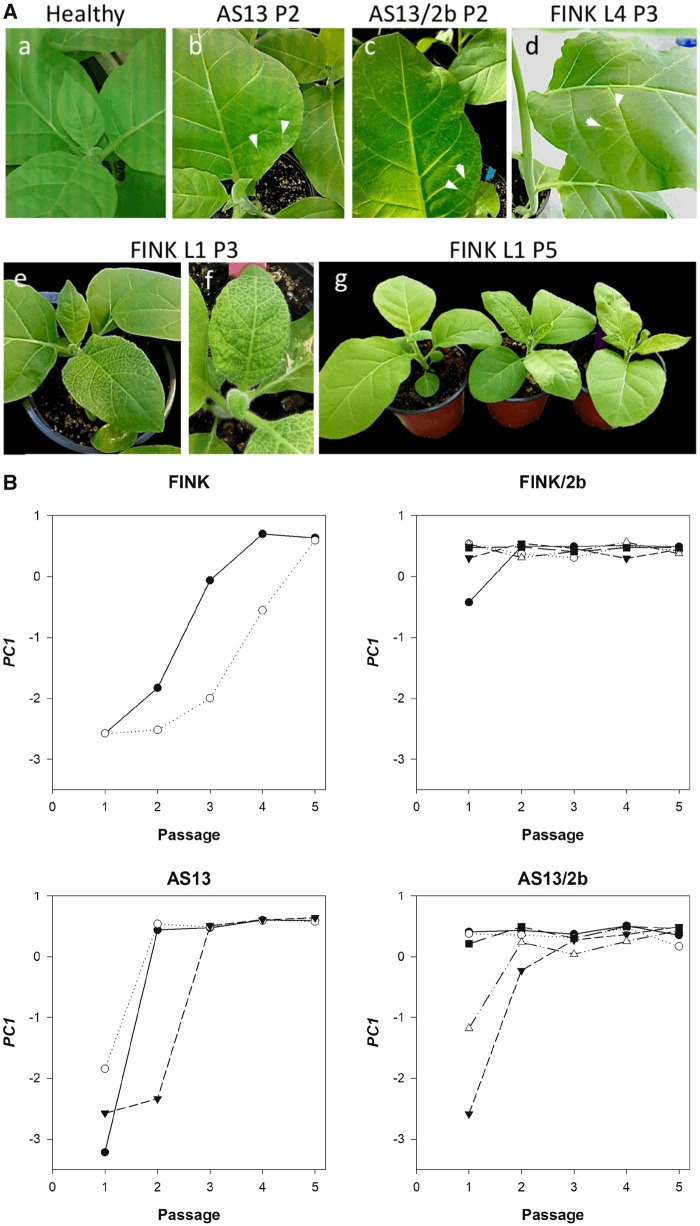
Figure 1A illustrates some representative examples of the changes in symptomatology observed along the evolution experiment. For instance, panels (b) and (c) compare the drastic difference in the type of symptoms induced by TEV-AS13, consisting in disperse chlorotic spots (would correspond to S = 1 in the symptoms scoring system described in Materials and Methods section), and TEV-AS13/2b which shows a more generalized spotted chlorotic disease (S = 2) but without the characteristic etching of wild-type TEV (see plants on the right in panel g). Panel (d) shows the symptoms generated by lineage L4 of TEV-FINK at passage 3, again consisting in disperse chlorotic spots (S = 1), while lineage L1 of TEV-FINK at the same passage had already reverted to the typical etching and vein necrosis of TEV (panels e and f; S = 3). Panel (g) shows the differences between a noninfected plant (left; also panel a) and two plants infected with lineage L1 of TEV-FINK after the last passage (S = 3). In summary, both viral genotypes expressing the 2 b RSS protein induced stronger symptoms than their corresponding counterparts. In all lineages, symptoms severity increased during the evolution experiment.
Fig. 1.
—(A) Symptoms induced by the ancestral and evolved TEV genotypes. Upper panel: from left to right, (a) healthy plant, (b) chlorotic spots observed in a plant of passage 2 (P2) infected with one of the TEV-AS13 lineage, (c) the same for a plant infected with a one of the TEV-AS13/2b lineages, and (d) a plant from the third passage (P3) infected with lineage L4 of TEV-FINK. White arrows indicate the location of some chlorotic spots. Lower panel: (e) and (f) plants of passage P3 infected with lineage L1 of TEV-FINK showing the typical etching of TEV, (g) comparison of plants from the fifth passage (P5): healthy (left) or infected with lineage L1 of TEV-FINK (right), thus displaying no symptoms or the typical etch mosaic of TEV, respectively. (B) Evolution of the first principal component (PC1) summarizing the six pathogenicity-related traits. See text for a detailed description of the contribution of the different traits into PC1. Different symbols correspond to the different independently evolved lineages.
After each passage, infectivity (i) and symptoms severity data (S) were fitted to equations (1) and (2), respectively; five different pathogenicity-related traits were inferred: i, AUDPS, Smax, τhalf, and b. In addition, time-course infectivity data were also analyzed using the Kaplan–Meier regression method and the median time to infection, ST50, was also estimated. First, we explored the dependencies and associations between these six pathogenicity-related traits using partial-correlation coefficients controlling for the viral genotype (G) factor. Table 1 shows the associations obtained. Partial correlations identify two groups of pathogenicity-related traits that respond in a highly parallel manner. In the first group, AUDPS, Smax, and i all show significant positive correlations. In the second group, ST50, τhalf, and b show significant pairwise positive correlations. However, correlations between traits belonging to different groups are all significantly negative (table 1).
Table 1.
Partial Correlations (rp), Controlling for Viral Genotype, between the Different Pathogenicity-Related Traits Measured
| i | AUDPS | ST50 | Smax | τhalf | b | |
|---|---|---|---|---|---|---|
| i | 1 | 0.841 | −0.841 | 0.847 | −0.424 | −0.645 |
| AUDPS | 1 | −0.997 | 0.989 | −0.506 | −0.834 | |
| ST50 | 1 | −0.989 | 0.502 | 0.835 | ||
| Smax | 1 | −0.483 | −0.811 | |||
| τhalf | 1 | 0.716 | ||||
| b | 1 |
Note.—In all cases, 72 d.f. and P < 0.001.
i, infectivity; AUDPS, area under the disease progress stairs curve; ST50, time to observe infections symptoms inferred from a Kaplan–Meier survival regression analysis; Smax, maximal score value for the observed symptoms; τhalf, half-time to develop the maximal symptoms; b, steepness of the symptoms development curve.
To explore the overall effect of different factors on these pathogenicity-related traits, we fitted the MANOVA model shown in equation (3) to the multivariate data. Table 2 shows the results of this analysis. The covariable and the two factors in the linear model are all significant: pathogenicity increases with passages, overall differences among viral genotypes exist and significant heterogeneity exist among lineages started from the same ancestral viral genotype (table 2). In addition, the interaction between passage (covariable) and genotype was also highly significant, indicating that the rates of pathogenicity recovery were different between viral genotypes (table 2). Finally, within each viral genotype, lineages also differed in their rates of pathogenicity evolution, although in this case the significance level is moderate (table 2; P = 0.021).
Table 2.
Multivariate Analysis of Variance (MANOVA) of the Six Pathogenicity-Related Traits
| Source of Variation | Wilk’s Λ | F | d.f. Hypothesis | d.f. Error | P |
|---|---|---|---|---|---|
| Intersection | 0.001 | 7,669.225 | 6 | 40 | <0.001 |
| Passage | 0.267 | 18.267 | 6 | 40 | <0.001 |
| Genotype | 0.115 | 7.265 | 18 | 113.622 | <0.001 |
| Lineage (Genotype) | 0.070 | 2.143 | 66 | 219.490 | <0.001 |
| Passage×Genotype | 0.230 | 4.294 | 18 | 113.622 | <0.001 |
| Passage×Lineage (Genotype) | 0.141 | 1.469 | 66 | 219.490 | 0.021 |
Note.—The model is described by equation (3).
Given the strong dependencies among the six pathogenicity-related variables, we applied a principal components analysis and found that 79.73% of the observed variability was explained by the major eigenvalue (λ1 = 4.784) of the variance–covariance matrix (the second eigenvalue was λ2 = 0.828 < 1 and negligible). Therefore, to represent the six variables into a single summary statistic, we computed the first principal component in the direction of λ1, which results in the following expression:
| (5) |
The biological meaning of this PC1 is straightforward: it gives positive weights to AUDPS, Smax, and i and negative weights to ST50, τhalf, and b, in accordance with the results of the correlation analyses shown above. The three first variables measure frequency and severity of symptoms while last three are related with the time necessary for symptoms to be developed. In other words, evolving viruses are rising in pathogenicity by increasing the positively weighted traits (i.e., increasing infectivity or the severity of symptoms) and/or by reducing the negatively weighted ones (i.e., shorter times to develop symptoms). PC1 < 0 values would correspond to viruses that produce very few infected plants and of weak symptoms that take long to be observable. By contrast, PC1 > 0 values will be characteristic of viruses that are highly infectious and quickly induce strong symptoms. Figure 1B illustrates the evolution of pathogenicity as described by PC1: a faster recovery of pathogenicity in TEV-AS13 and TEV-FINK compared with their respective 2b-carrying counterparts. However, after the first passage, a larger amount of variance exists among the lineages carrying the 2 b than among the noncarrying ones (fig. 1B). Lineage L1 of TEV-FINK/2b shows a lower PC1 than the other four lineages TEV-FINK/2b; this difference was erased after passage 2. Likewise, lineages L8 and L11 of TEV-AS13/2b also show remarkable low values of PC1 after the first passage, much lower than the other two TEV-AS13/2b lineages and in the same range of values observed for the genotypes not expressing the 2 b RSS. Thereafter passage 3, differences among the five TEV-AS13/2b lineages disappeared.
To gain further insights into the role of RSS functional redundancy during the evolution of pathogenicity, we performed pairwise post hoc Bonferroni tests using PC1 as variable in an ANOVA with the same structure than equation (3). Firstly, we compared the difference between TEV-FINK and TEV-FINK/2b (fig. 1B, panels FINK vs. FINK/2b). The mean value of PC1 for TEV-FINK was −1.018 ±0.101, while the mean value for TEV-FINK/2b was 0.411 ±0.064, being the difference highly significant (P < 0.001). Therefore, the expression of 2 b buffers the strongly deleterious effect of the mutation R182I in the highly conserved FRNK motif of the RBD A domain of HC-Pro, which leads to a weak infection phenotype and hyposuppression activity in other potyviruses (Lin et al. 2007; Shiboleth et al. 2007; Kung et al. 2014), thus relaxing selection to revert or compensate this amino acid replacement. By contrast, in absence of a backup for the RSS activity, strong selection operates upon TEV-FINK to recover its ancestral HC-Pro activity which also correlates with a stronger symptom induction and pathogenicity. Secondly, we compared the difference between TEV-AS13 and TEV-AS13/2b (fig. 1B, panels AS13 vs. AS13/2b). The mean value of PC1 for TEV-AS13 was −0.264 ±0.186, while the mean value for TEV-AS13/2b was 0.152 ±0.144, being this difference not significant (P = 0.092). This result suggests that 2 b is not compensating the effect of the deleterious E299A/D300A mutations in the RBD B domain of HC-Pro to the same extent than it does for the R182I mutation in the RBD A domain, and thus selection may operate both in TEV-AS13 and TEV-AS13/2b to improve HC-Pro activity. Thirdly, we compared the difference between TEV-FINK and TEV-AS13 (fig. 1B, panels FINK vs. AS13). In agreement with our above argumentation, these two genotypes show statistically similar profiles of pathogenicity recovery (P = 0.050). Finally, we compared the differences between TEV-FINK/2b and TEV-AS13/2b (fig. 1B, panels FINK/2b vs. AS13/2b). Supporting our above conclusion that functional redundancy provided by 2 b buffers deleterious mutational effects in the FRNK motif but not in the RBD B domain, the mean PC1 values of these two genotypes are significantly different (P = 0.009), being greater for TEV-FINK/2b. In subsequent sections, we will explore the molecular basis of this evolutionary recoveries.
Compensatory Evolution of Within-Host Replicative Fitness
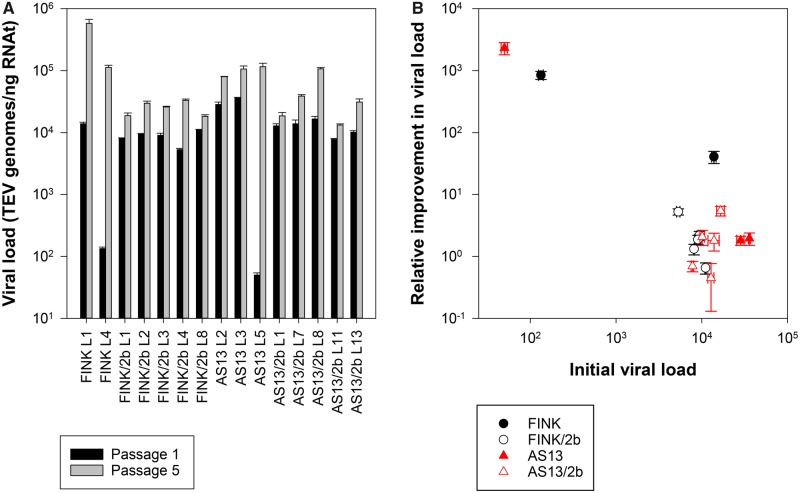
As a proxy to within-host viral fitness, we have quantified viral load by RT-qPCR (number of viral genomes per ng of RNAt extracted from 100 mg of plant tissue), as described in Materials and Methods, after the first and last evolution passages. The resulting values are shown in figure 2A. In all cases, significant improvements in viral load has been observed. Table 3 shows the results of fitting the GLM model described by equation (4) to the data. The three factors have a highly significant effect on viral load. First, viral load at passage 5 is always greater than at passage 1 and the effect is of very large magnitude ( = 0.990). The largest differences are observed for the two lineages of TEV-FINK and lineage L5 of TEV-AS13 (fig. 2A). Second, consequently, the magnitude of improvement in viral load significantly varies among viral genotypes, being on an average larger for TEV-FINK and TEV-AS13 than for the corresponding genotypes carrying the 2 b. Nonetheless, the magnitude of this effect is the smallest observed ( = 0.235) yet still can be considered as large. Third, heterogeneity can be observed between independent lineages evolved from the same ancestral genotype, being the effect also very large in magnitude. Fourth, the magnitude of these two effects actually depends on the passage, as indicated by the two significant interaction terms in table 3.
Fig. 2.
—(A) Change in viral load (a proxy of within-host replicative fitness) observed between the first and the last evolution passages. (B) Negative correlation between viral load of the nonevolved genotypes and the magnitude of improvement at the end of the evolution experiment. In both panels, error bars represent ±1 SD.
Table 3.
Generalized Linear Model Testing for the Effect of the Different Factors on Viral Load (within-host fitness)
| Source of Variation | LRTa | d.f. | P | Magnitude of the Effectb | Powerc |
|---|---|---|---|---|---|
| Intersection | 300.916 | 1 | <0.001 | 0.936 | 1 |
| Passage | 449.300 | 1 | <0.001 | 0.990 | 1 |
| Genotype | 272.237 | 3 | <0.001 | 0.235 | 0.226 |
| Lineage (Genotype) | 426.675 | 11 | <0.001 | 0.989 | 1 |
| Passage×Genotype | 491.470 | 4 | <0.001 | 0.976 | 1 |
| Passage×Lineage (Genotype) | 375.156 | 11 | <0.001 | 0.978 | 1 |
Note.—The fitted model assumes a Gamma distribution of errors and a logarithmic link function and is shown in equation (4).
Likelihood ratio test.
Evaluated as .
Conventionally, values >0.8 are considered as a standard for adequacy.
Next, we sought to evaluate whether the observed changes in fitness were directly proportional to the magnitude of the perturbation induced by the corresponding mutations in HC-Pro RSS activity, as it would be expected from reversions to the wild-type sequence, or pervasively deviate from the linear relationship, as expected for the accumulation of compensatory mutations at other sites in the HC-Pro (or, in principle, elsewhere in the genome) (Moore et al. 2000; Sanjuán et al. 2005). To test this hypothesis, we first computed the relative change in viral load as , where VL(G)(t) is the viral load estimated for lineage L of genotype G at the indicated t passage number. As illustrated in figure 2B, the lower the viral load measured at passage 1, the larger the relative gain in fitness (linear regression: R2 = 0.754, F1,13 = 39.890, P < 0.001), thus supporting the null hypothesis of fitness recoveries being proportional to the actual deleterious effect. Indeed, fitting a quadratic model to the data in figure 2B does not significantly improve the goodness of fit (partial F test: F1,12 = 3.686, P = 0.079), thus further rejecting the alternative hypothesis of second-site compensatory mutations. This conclusion of a predominance of reversions over second-site compensatory mutations will be confirmed in the sections below by determining the sequence of HC-Pro in all lineages.
Stability of the 2 b Insert
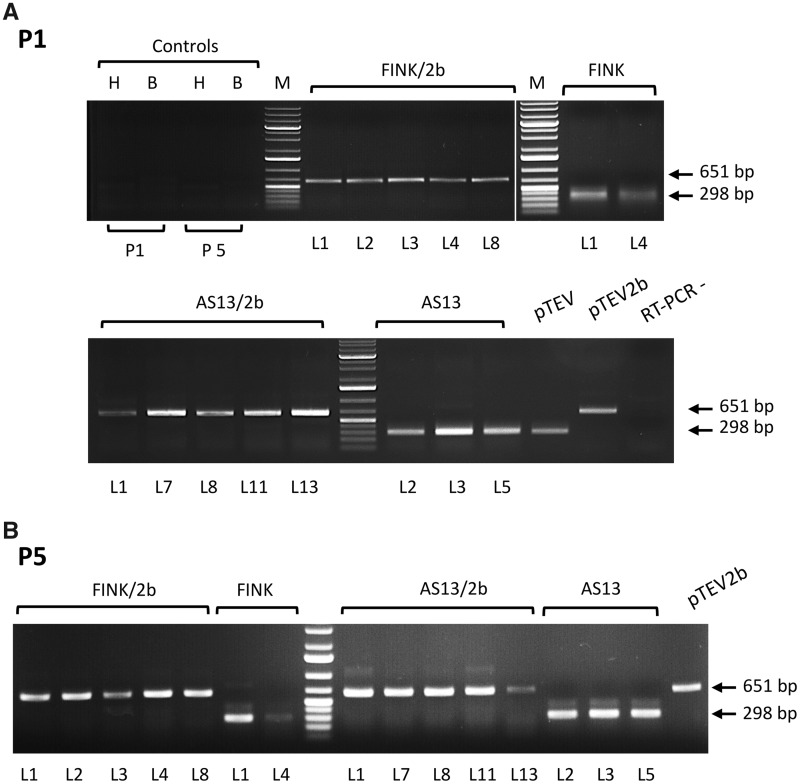
Willemsen et al. (2017) found that, among several other exogenous coding sequences inserted in the genome of TEV, 2 b was particularly stable and functionally able to compensate the severe fitness costs associated with mutations in fundamental residues of HC-Pro. Here, we have evolved engineered genotypes of TEV encoding 2 b in a situation in which its RSS activity would be required to complement the deleterious effect of mutations in the virus’ natural RSS HC-Pro. However, a possible outcome from our evolution experiment would be the evolved viruses to recover fitness by compensating or reverting mutations in HC-Pro and subsequently removing partial or totally the 2 b. To explore this possibility, and the evolutionary stability of 2 b, we have confirmed the presence of 2 b in all the corresponding evolved lineages by RT-PCR using a pair of primers specifically designed to amplify the region into which 2 b was inserted (see Materials and Methods for details). Figure 3A shows the results of this study in all lineages after passage 1 and figure 3B the same but after the last passage 5. All evolved 2b-carrying lineages have retained the insert (all have the expected amplicon size of 651 bp). Therefore, this experiment confirms the evolutionary stability of the 2 b inserted into the TEV genome and that the observed patterns of recovery of fitness and pathogenicity did not rely in removing this exogenous genetic material but in point mutations.
Fig. 3.
—Confirmation of the presence of 2b in the evolved viral lineages. Agarose gels (1%) with conventional RT-PCR products of the region encompassing the 2b inserted into the TEV genome. Expected amplicon sizes are indicated on the right panels with arrows: 298 bp for wild-type TEV and 651 bp for TEV/2b. No deletions or reorganizations were observed in any lineage or passage. (A) Amplicons of all virus lineages from the first (P1) passage. (B) Amplicons from the fifth (P5) passage. Healthy (H), buffer-inoculated (B) plant extracts and water (RT-PCR−) were used as negative RT-PCR controls. PCR products obtained from the original full-length cDNA virus clones of TEV (pTEV) and TEV/2b (pTEV2b) are included as positive controls of insertion size. RT-PCR −: negative RT-PCR control.
Very interestingly, not a single mutation, not even in a polymorphic state, has been observed in the 2 b gene (fig. 4C), thus supporting the high evolutionary stability of this exogenous gene into TEV genome.
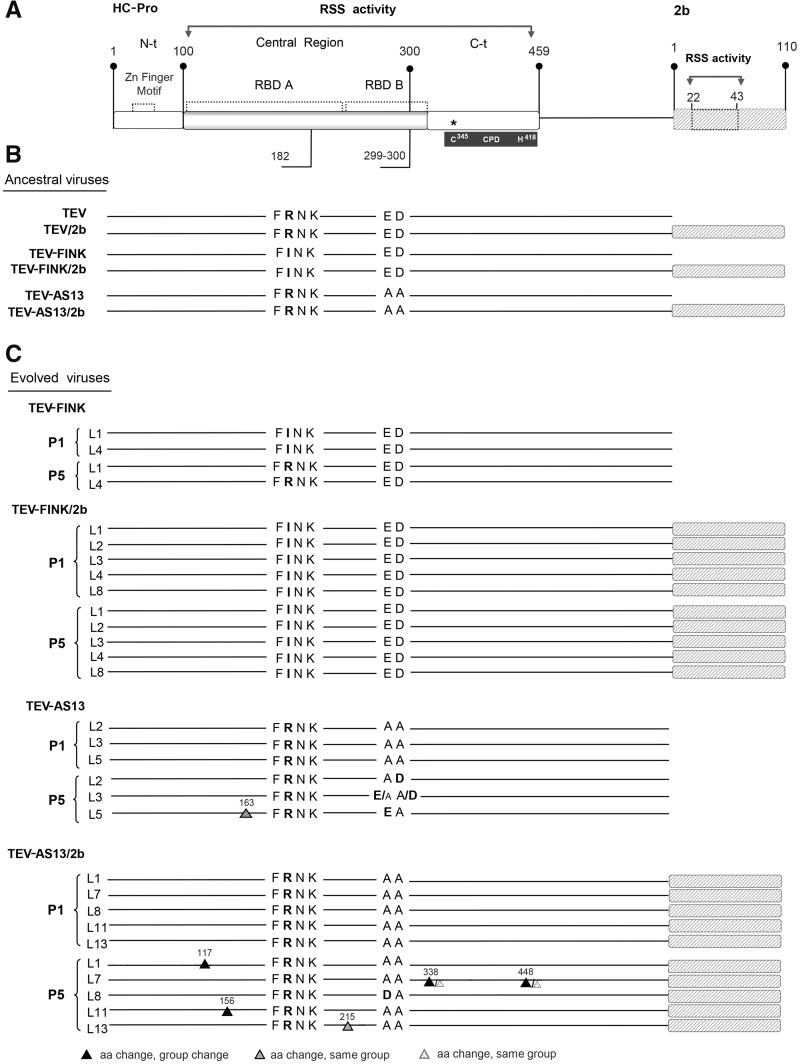
Fig. 4.
—(A) Schematic representation of TEV HC-Pro and CMV 2b viral suppressors of RNA silencing proteins as present in the engineered TEV-FINK/2b and TEV-AS13/2b genomes. The main structural and functional region/domains and relevant motifs of HC-Pro and 2b are indicated. RSS: RNA silencing-suppression region. RBD: RNA-binding domains within the RSS. CPD: cysteine proteinase domain. Position of the TEV FRNK and ED amino acid motifs indicated with a horizontal line below the corresponding RBD regions. Position of the C345 and H418 dyad amino acids of the catalytic CPD in the Ct of the central region, are indicated by an asterisk. (B) Ancestral virus genotypes used in this work with the relevant amino acids and 2b insertion marked by a gray box. (C) Same as in panel (B) but indicating the mutations found in the two genes in evolving viral lineages after one (P1) and five (P5) passages. The position and type of additional mutations found are marked with triangles. Large triangles denote amino acid replacements fixed in the consensus sequence (>50%) while small ones represent minority viral subpopulations. For additional details, see supplementary table S1, Supplementary Material online.
Molecular Changes Observed in HC-Pro
To characterize possible point mutations responsible for the observed phenotypic recoveries in presence and absence of functional redundancy, we have obtained the consensus sequences of the HC-Pro and 2b genes in all 15 evolved lineages. Supplementary table S1, Supplementary Material online, shows all observed mutations. For guiding purposes, figure 4A shows a schematic representation of the HC-Pro and 2b genes, indicating the two HC-Pro RBD involved in RSS activity and the precise location of the two motifs mutagenized on each genotype (fig. 4B). Figure 4C shows a schematic representation of the consensus sequences obtained for all 15 evolved lineages at passages 1 and 5. The two TEV-FINK lineages had reverted the strong hyposuppressor mutant R183I to the wild-type arginine (fixed change), thus restoring the FRNK motif. In sharp contrast, none of the five TEV-FINK/2b shows such a reversion after passage 5, all retaining the mutated FINK sequence motif.
Regarding the three TEV-AS13 lineages, all had partially reverted the two deleterious mutations in a complex manner. Firstly, lineage L2 only reverted mutation D300A to the wild-type aspartic acid, being the change fixed. Secondly, lineage L5 only reverted mutation E299A to the wild-type glutamic acid residue, being also a fixed substitution. In addition, L5 also incorporates a fixed I163L (noncharged hydrophobic by noncharged hydrophobic) nonsynonymous mutation in the RBD A domain. Thirdly, lineage L3 shows polymorphisms in residues 299 and 300 of the protein alternating E299A/A299E and D300A/A300D. In both cases, the most frequent mutation corresponds to the wild-type allele (glutamic or aspartic acids, respectively), thought with the consensus sequencing approach here employed we cannot distinguish between having two or four haplotypes. Again, in sharp contrast, none of the TEV-AS13/2b lineages had reverted the knockdown alanine mutations into the wild-type amino acids. However, lineage L8 has incorporated a mutation to aspartic acid into residue 299, which is not a strict reversion, thought it brings back a negative charge into this motif. All other TEV-AS13/2b lineages contain additional nonsynonymous mutations elsewhere in the HC-Pro (fig. 4C). Lineage L1 has a C117Y (noncharged of moderate hydropathy by noncharged hydrophobic) fixed substitution in the RBD A domain. Lineage L11 has a A156T (noncharged hydrophobic by noncharged hydrophilic) fixed substitution in the RBD A domain. Lineage L13 has a A215V (noncharged hydrophobic by noncharged hydrophobic) fixed substitution in the RBD B. Interestingly, lineage L7 has two polymorphic amino acid substitutions, Y338C (noncharged hydrophobic by noncharged of moderate hydropathy) that is close to the C345 active site of the cysteine proteinase domain (CPD) (Oh and Carrington 1989; Kasschau et al. 1997), and G448D (noncharged hydrophobic by negatively charged hydrophilic) in the C-terminal region outside the RBD B domain (again, our consensus sequencing approach does not allow to conclude if two or four alleles are coexisting in the viral population).
Evaluating the Topography of the Local Fitness Landscape Defined by Mutations E299A and D300A in the RBD B Domain
In the previous section, we found that lineages TEV-AS13 L2 only reverted mutation D300A to the wild-type aspartic acid and that lineage L5 only reverted mutation E299A to the wild-type glutamic acid residue, but not complete reversion to the wild-type ED motif configuration has been observed. A simple explanation for this lack of double reversion is that generating the double mutant is a low probability event since it is proportional to the square of the mutation rate. A second possibility, perhaps more tantalizing, is that the four genotypes for this motif (wild-type E299/D300, single mutants E299/A300 and A299/D300 and double mutant A299/A300) define a rugged landscape in which the single mutants correspond to valleys that cannot be traversed by selection. To explore this possibility, we have used the accumulation data shown in figure 2A to calculate the fitness of each of the four genotypes (G) as (Carrasco et. al. 2007), where t = 15 dpi is the time at which viral load, V, was estimated (see Materials and Methods section). Fitness data were then fitted to the following GLM with a Normal distribution and identity link function , where E represents the allele at the 299 locus and D the allele at the 300 locus. The second and third term in the model represent the main fitness effects of the alanine mutations at these two loci and the fourth term represents the interaction between the two loci, that is, epistasis. The intersection (χ2 = 89.055, 1 d.f., P < 0.001, = 0.986) and the two main factors (χ2 = 6.023, 1 d.f., P = 0.014, = 0.249; and χ2 = 4.292, 1 d.f., P = 0.038, = 0.185, respectively) were significant, thus supporting the deleterious effect of the E299A and D300A mutations by themselves, with mutation E299A having a stronger deleterious effect than D300A (larger value). However, the epistatic term was not significant (χ2 = 1.130, 1 d.f., P = 0.288, = 0.052), thus supporting the notion of a multiplicative smooth landscape around these four genotypes. Therefore, we conclude that the lack of observed full reversion at these two loci is not explained by a rugged fitness landscape but more likely to the lack of evolutionary time to fix the two reversions in a sequential manner.
Association between Pathogenicity and Viral Accumulation
Although this section deviates from the main discussion about the role of functional redundancy on compensatory evolution, we believe it is worth present and discussing, though in short, an interesting observation. A still controversial hypothesis brought forward to explain the evolution of parasite’s virulence assumes the existence of a positive association between virulence and within-host accumulation (Froissart et al. 2010). Increased accumulation will facilitate transmission at the cost of damaging the host. However, observations with plant viruses are highly variable regarding their support to this hypothesis. In some cases, a lack of positive association has been observed between accumulation and virulence (Moury et al. 2001; Carrasco et al. 2007). In other cases, the predicted positive correlation was there (Doumayrou et al. 2013) or found to be dependent on the particular combination of host- and virus-genotypes (Pagán et al. 2007; Agudelo-Romero et al. 2008). Contributing to confusion, in some other cases even a negative correlation was found (Rodríguez-Cerezo et al. 1991; Poulicard et al. 2010). Given that we have estimated pathogenicity, which is a proxy to virulence for TEV, and within-host replication for a set of viral genotypes, it is possible to test this hypothesis in our particular pathosystem. A significant positive partial correlation, controlling for virus genotype, gave support to the hypothesis (rp = 0.514, 27 d.f., P = 0.004).
Discussion
The evolutionary consequences of functional redundancy have been discussed at length. On the one hand, functional redundancy may be the raw material for natural selection to evolve new functions (Kondrashov et al. 2002) or to evolve moonlighting proteins with overlapping yet suboptimal activities (Lynch and Force 2000; Fares 2014; Espinosa-Cantú et al. 2015). On the other hand, by creating mutational robustness, functional redundancy may contribute to pseudogenization and gene lost, specially under strong selection for genome reduction, as it may be the case for intracellular parasites (Mendoça et al. 2011), including viruses. In this study, we have explored the role of functional redundancy for an essential viral function, the ability to suppress the RNA silencing defense of the host cells. After engineering TEV genomes containing deleterious mutations in their own RSS (a strong functional knockdown in the FRNK motif of RBD A or a weak one in the ED motif of RBD B; fig. 4B), we generated additional genomes expressing an exogenous RSS, the CMV 2 b gene. After evolving independent lineages from each one of these four genotypes, we evaluated pathogenicity- and fitness-related traits and characterized the consensus sequence for HC-Pro and 2 b in each evolved lineage. We have observed that functional redundancy affects the rate of compensatory evolution both at the phenotypic and at the genotypic level. At the phenotypic level, we have observed that, on an average, genetically redundant viruses expressing the 2 b RSS were more pathogenic and had higher fitness before starting evolution and experienced relatively smaller changes in these traits by the end of the evolution experiment (figs. 1B and 2A). By contrast, nonredundant genotypes had very low pathogenicity and fitness at the beginning of the evolution experiment and, by the last passage, had reached values similar to those of the redundant genotypes (figs. 1B and 2A).
Even though our genotyping study was limited to population consensus sequences, we made a number of interesting observations (fig. 4C). First, the patterns of genomic evolution were completely different for genetically redundant and nonredundant genomes. While the strongly deleterious mutation FINK reverted to the wild-type motif FRNK in all the nonredundant evolved genomes, not a single reversion was observed in the redundant ones. Likewise, the mutations E299A and D300A partially reverted (to glutamic acid at position 299 or to aspartic acid at position 300 and it may have fully reverted in lineage L3), but never in the redundant genomes. Interestingly, the TEV-FINK/2b lineages did not show additional mutations anywhere else in the HC-Pro cistron, while four out of five TEV-AS13/2b lineages had fixed a number of nonsynonymous mutations affecting the RBD A (L1 and L11), RBD B (L13), or the C-terminal domain (2 mutations in L7). At this stage, the observation of additional mutations fixed along HC-Pro in the TEV-AS13/2b lineages is compatible with the two hypotheses exposed above. On the one hand, according to the hypothesis of functional redundancy boosting the evolution of new functions, these extra mutations may be improving some of the additional functions performed by HC-Pro, after 2 b taking the role as RSS (which will support the hypothesis of functional redundancy allowing to generate or improve novel functions). In this way, stands out the two mutations observed in lineage L7 (fig. 4C) affecting the protease and movement domains, specially the first which renders a new cysteine residue (T338C) close to the C345 active site of the CPD (Oh and Carrington 1989; Kasschau et al. 1997). While the second, a G448D, involves a replacement in the SGLE phosphoserine motif without affecting the active serine. In addition, mutations fixed in lineages L1, L11, and L13 also affect the movement domain (fig. 4C) (Plisson et al. 2003; Valli et al. 2018). On the other hand, according to the hypothesis of functional redundancy buffering the effect of deleterious mutations and driving to pseudogenization and gene lost, the observed mutations may have a deleterious effect. However, under the pseudogenization hypothesis, we would expect to also observe fixation of synonymous mutations and, based on previous observations (reviewed in Willemsen et al. 2018), a number of in-phase deletions. Given that this is not the case, we believe our data, albeit circumstantially, are better supporting the first hypothesis. In any case, for both hypotheses, an acceleration in the rate of molecular evolution is expected in the genetically redundant lineages (Kondrashov et al. 2002), as we have observed for the TEV-AS13/2b ones. Clearly, site-directed mutagenesis experiments followed by biochemical and physiological characterization of the protease and movement activities of these mutants would help to distinguish between these two competing hypotheses.
Interestingly, we have also observed that the magnitude of the phenotypic improvement was proportional to the original fitness effect (fig. 2B): nonredundant genotypes (TEV-FINK and TEV-AS13) recovered proportionally more fitness than their corresponding genetically redundant counterparts (TEV-FINK/2b and TEV-AS13/2b, respectively). This proportionality suggests that most of the mutations involved in the pathogenicity- and fitness-recovery process were reversions rather than second-site compensatory mutations (Moore et al. 2000; Sanjuán et al. 2005). Compensatory mutations are defined as those providing a disproportionally large fitness effect in the mutant genetic background compared with their effect in the wild-type background (i.e., sign epistasis), coupled with retention of the original deleterious mutation. In this regard, our consensus sequencing results show a combination of effects: nonredundant genotypes have reverted mutations to the wild-type alleles, while redundant TEV-AS13/2b genotypes (but not TEV-FINK/2b) have fixed extra mutations that, as we just discussed in the previous paragraph, may be compensatory if they are improving the protease and/or movement activities.
An interesting question that pops up from our results is the clear difference in patterns of molecular evolution between the strong and weak hyposuppressor mutants. Lineages with the more deleterious mutation in the RBD A FRNK motif either revert to wild-type (TEV-FINK) or remained unchanged in presence of 2 b (TEV-FINK/2b). By contrast, lineages with the weaker mutation in the RBD B ED motif were more variable in the observed outcome, as just discussed in the previous two paragraphs. Why is so? FRNK has been shown to be essential for the RSS activity of HC-Pro by directly binding to the siRNAs (Shiboleth et al. 2007; Kung et al. 2014). Therefore, mutations affecting this motif are completely abolishing the RSS activity (Shiboleth et al. 2007; Kung et al. 2014). In nongenetically redundant lineages, selection for reversion is maximally strong, thus almost deterministically ensuring the fixation of the required amino acid change. In the functionally redundant lineages, by contrast, RSS activity is taken over by 2 b, relaxing the necessity of reverting isoleucine into arginine in the FRNK motif. By contrast, after discarding the possibility of evolutionary inaccessibility due to the existence of fitness valleys, a possible explanation is that mutations in the ED motif retain a residual RSS activity (Torres-Barceló et al. 2008) and may be involved into other functions without fully removing them. This being the case, the nonredundant genotypes are under weaker selection than in the case of the FRNK motif and this is why we observe partial reversions and fixation of additional mutations. This observation is in good agreement with a previous compensatory evolution experiment performed with TEV-AS13 in which all evolved lineages fixed second-site compensatory mutations, but not reversions, (Torres-Barceló et al. 2010).
Another intriguing observation, aligned with the previous work by Willemsen et al. (2017), is the high genetic and evolutionary stability of the 2 b insertion. Remarkably, in both studies, the 2 b has been always retained without even fixing a single nucleotide substitution. What fitness benefit does 2b provide to TEV? A first hypothesis would be that functional redundancy increases mutational robustness. However, as recently shown by Willemsen et al. (2018), the mutational robustness of engineered TEV genomes expressing additional genes is inversely proportional to genome length and in the case of 2b was not different than wild-type (see fig. 3 in Willemsen et al. 2018). Therefore, an increase in mutational robustness is not a likely benefit of expressing the 2b protein. A second hypothesis is that 2b facilitates TEV replication and/or cell-to-cell and systemic spread by interfering with the plant silencing suppression pathway in stages other than those affected by HC-Pro. HC-Pro sequesters sRNAs duplexes, relieving translational repression by the endonuclease AGO1, and inhibits the sRNA methylation activity of the methylase HEN1 (Ivanov et al. 2016; Valli et al. 2018). CMV 2b physically binds to AGO1 and provokes inhibition of the cleavage activity of the RISC machinery (Zhang et al. 2006) and also inhibits the production of RDR1-dependent viral siRNAs that confer systemic resistance by directing noncell autonomous antiviral silencing (Guo and Ding 2002; Díaz-Pendón et al. 2007). Therefore, both RSSs proteins have overlapping functions but also differ in some aspects of their mechanisms of action, thus supporting the hypothesis of the 2b insertion was selected under our serial passages regime because it increased virus accumulation beyond the level of HC-Pro by itself.
HC-Pro is a multifunctional protein composed by domains involved in overlapping functions (Plisson et al. 2003; Valli et al. 2018): mediator in vector transmission, RSS, cell-to-cell movement, and cysteine protease. It has been proposed that multifunctionality conflicts with optimization of every function (Guillaume and Otto 2012; Fares 2014) and therefore a tradeoff between all these functions is expected. Engineered functional redundancy relaxes these evolutionary constraints and allows multifunctional proteins to optimize the functions that are not being provided in cis, in our case the RSS. The results we are reporting here for the TEV-AS13/2b lineages may represent an early step in this division of labor and specialization molecular evolutionary pathway. Longer evolution experiments and additional characterization of the adaptive value of fixed mutations will seed light into this interesting question.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Paula Agudo for excellent technical assistance. This work was supported by Spain’s Agencia Estatal de Investigación—FEDER grant BFU2015-65037-P to S.F.E. and by a fellowship from the Dominican Republic’s Ministerio de Educación Superior, Ciencia y Tecnología to S.M.R. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the article.
Literature Cited
- Agudelo-Romero P, De la Iglesia F, Elena SF.. 2008. The pleiotropic cost of host-specialization in tobacco etch potyvirus. Infect Genet Evol. 8(6):806–814. [DOI] [PubMed] [Google Scholar]
- Campbell CL, Madden LV.. 1990. Introduction to plat disease epidemiology. New York (NY): Wiley-Interscience; p. 173–175. [Google Scholar]
- Carrasco P, De la Iglesia F, Elena SF.. 2007. Distribution of fitness and virulence effects caused by single-nucleotide substitutions in Tobacco etch virus. J Virol. 81(23):12979–12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Pendón JA, Li F, Li WX, Ding SW.. 2007. Suppression of antiviral silencing by Cucumber mosaic virus 2b protein in Arabidopsis is associated with drastically reduced accumulation of three classes of viral small interfering RNAs. Plant Cell 19(6):2053–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolja VV, Herndon KL, Pirone TP, Carrington JC.. 1993. Spontaneous mutagenesis of a plant potyvirus genome after insertion of a foreign gene. J Virol. 67(10):5968–5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolja VV, Kreuze JF, Valkonen JPT.. 2006. Comparative and functional genomics of closteroviruses. Virus Res. 117(1):38–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doumayrou J, Avellan A, Froissart R, Michalakis Y.. 2013. An experimental test of the transmission-virulence trade-off hypothesis in a plant virus. Evolution 67(2):477–486. [DOI] [PubMed] [Google Scholar]
- Espinosa-Cantú A, Ascensio A, Barona-Gómez F, DeLuna A.. 2015. Gene duplication and the evolution of moonlighting proteins. Front Genet. 6:227.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fares MA. 2014. The evolution of protein moonlighting: adaptive traps and promiscuity in the chaperonins. Biochem Soc Trans. 42(6):1709–1714. [DOI] [PubMed] [Google Scholar]
- Froissart R, Doumayrou J, Vuillaume F, Alizon S, Michalakis Y.. 2010. The virulence-transmission trade-off in vector-borne plant viruses: a review of (non-)existing studies. Philos Trans R Soc B 365(1548):1907–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillaume F, Otto SP.. 2012. Gene functional trade-offs and the evolution of pleiotropy. Genetics 192(4):1389–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo HS, Ding SW.. 2002. A viral protein inhibits the long range signaling activity of the gene silencing signal. EMBO J. 21(3):398–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov KI, et al. . 2016. Molecular insights into the function of the viral RNA silencing suppressor HC-Pro. Plant J. 85(1):30–45. [DOI] [PubMed] [Google Scholar]
- Kasschau KD, Cronin S, Carrington JC.. 1997. Genome amplification and long-distance movement functions associated with the central domain of tobacco etch potyvirus helper component-proteinase. Virology 228(2):251–262. [DOI] [PubMed] [Google Scholar]
- Kondrashov FA, Rogozin IB, Wolf YI, Koonin EV.. 2002. Selection in the evolution of gene duplications. Genome Biol. 3(2):research0008.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV. 2010. Taming of the shrewd: novel eukaryotic genes from RNA Viruses. BMC Biol. 8:2.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung YJ, et al. . 2014. Genetic analyses of the FRNK motif function of Turnip mosaic virus uncover multiple and potentially interactive pathways of cross-protection. Mol Plant Microbe Interact. 27(9):944–955. [DOI] [PubMed] [Google Scholar]
- Lin SS, Wu HW, Jan FJ, Hou RF, Yeh SD.. 2007. Modifications of the helper component-protease of Zucchini yellow mosaic virus for generation of attenuated mutants for cross protection against severe infection. Phytopathology 97(3):287–296. [DOI] [PubMed] [Google Scholar]
- Lynch M, Force A.. 2000. The probability of duplicate gene preservation by subfunctionalization. Genetics 154(1):459–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, et al. . 2010. Widespread horizontal gene transfer from double-stranded RNA viruses to eukaryotic nuclear genomes. J Virol. 84(22):11876–11887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majer E, et al. . 2014. Relocation of the NIb gene in the tobacco etch potyvirus genome. J Virol. 88(8):4586–4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S, et al. . 2000. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403(6771):785–789. [DOI] [PubMed] [Google Scholar]
- Mendoça AG, Alves RJ, Pereira-Leal JB.. 2011. Loss of genetic redundancy in reductive genome evolution. PLoS Comput Biol. 7(2):e1001082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore FBG, Rozen DE, Lenski RE.. 2000. Pervasive compensatory adaptation in Escherichia coli. Proc R Soc B 267(1442):515–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira D, Brochier-Armanet C.. 2008. Giant viruses, giant chimeras: the multiple evolutionary histories of Mimivirus genes. BMC Evol Biol. 8:12.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moury B, Cardin L, Onesto JP, Candresse T, Poupet A.. 2001. A survey of Prunus necrotic ringspot virus in rose and its variability in rose and Prunus spp. Phytopathology 91(1):84–91. [DOI] [PubMed] [Google Scholar]
- Oh CS, Carrington JC.. 1989. Identification of essential residues in potyvirus proteinase HC-Pro by site-directed mutagenesis. Virology 173(2):692–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagán I, Alonso-Blanco C, García-Arenal F.. 2007. The relationship of within-host multiplication and virulence in a plant-virus system. PLoS One 2(8):e786.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penadés JR, Chen J, Quiles-Puchalt N, Carpena N, Novick RP.. 2015. Bacteriophage-mediated spread of bacterial virulence genes. Curr Opin Microbiol. 23:171–178. [DOI] [PubMed] [Google Scholar]
- Plisson C, et al. . 2003. Structural characterization of HC-Pro, a plant virus multifunctional protein. J Biol Chem. 278(26):23753–23761. [DOI] [PubMed] [Google Scholar]
- Poulicard N, Pinel-Galzi A, Hebrard E, Farguette D.. 2010. Why Rice yellow mottle virus, a rapid evolving RNA plant virus, is not efficient at breaking rymv1-2 resistance. Mol Plant Pathol. 11(1):145.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Cerezo E, Klein PG, Shaw JP.. 1991. A determinant of disease symptom severity is located in the 3’-terminal noncoding región of the RNA of a plant virus. Proc Natl Acad Sci USA. 88(21):9863–9867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiboleth YM, et al. . 2007. The conserved FRNK box in HC-Pro, a plant suppressor of gene silencing, is required for small RNA binding and mediates symptoms development. J Virol. 81(23):13135–13148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Navarro JA, Zwart MP, Elena SF.. 2013. Effect of the number of genome segments on primary and systemic infections with a multipartite plant RNA virus. J Virol. 87(19):10805–10815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuán R, Cuevas JM, Moya A, Elena SF.. 2005. Epistasis and the adaptability of an RNA virus. Genetics 170(3):1001–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simko I, Piepho HP.. 2012. The area under the disease progress stairs: calculation, advantage, and application. Phytopathology 102(4):381–389. [DOI] [PubMed] [Google Scholar]
- Torres-Barceló C, Daròs JA, Elena SF.. 2010. Compensatory molecular evolution of HC-Pro, an RNA-silencing suppressor from a plant RNA virus. Mol Biol Evol. 27(3):543–551. [DOI] [PubMed] [Google Scholar]
- Torres-Barceló C, Martín S, Daròs JA, Elena SF.. 2008. From hypo- to hypersuppression: effect of amino acid substitutions on the RNA-silencing suppressor activity of tobacco etch potyvirus HC-Pro. Genetics 180(2):1039–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tromas N, Zwart MP, Forment J, Elena SF.. 2014. Shrinkage of genome size in a plant RNA virus upon transfer of an essential viral gene into the host genome. Genome Biol Evol. 6(3):538–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valli AA, Gallo A, Rodamilans B, López-Moya JJ, García JA.. 2018. The HC-Pro from the Potyviridae family: an enviable multitasking helper component that every virus would like to have. Mol Plant Pathol. 19(3):744–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen A, Carrasco JL, Elena SF, Zwart MP.. 2018. Going, going, gone: predicting the fate of genomic insertions in plant RNA viruses. Heredity doi: 10.1038/s41437-018-0086-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen A, Zwart MP, Ambrós S, Carrasco JL, Elena SF.. 2017. 2b or not 2b: experimental evolution of functional exogenous sequences in a plant RNA virus. Genome Biol Evol. 9(2):297–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen A, Zwart MP, Higueras P, Sardanyés J, Elena SF.. 2016. Predicting the stability of homologous gene duplication in a plant RNA virus. Genome Biol Evol. 8(9):3065–3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen A, Zwart MP, Tromas N, et al. . 2016. Multiple barriers to the evolution of alternative gene orders in a positive-strand RNA virus. Genetics 202(4):1503–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Zwart MP, Sánchez-Navarro JA, Elena SF.. 2017. Within-host evolution of segments ratio for the tripartite genome of Alfalfa mosaic virus. Sci Rep. 7(1):5004.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, et al. . 2006. Cucumber mosaic virus-encoded 2b suppressor inhibits Argonaute 1 cleavage activity to counter plant defense. Genes Dev. 20(23):3255–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwart MP, Willemsen A, Daròs JA, Elena SF.. 2014. Experimental evolution of pseudogenization and gene loss in a plant RNA virus. Mol Biol Evol. 31(1):121–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.