Despite recent advances in detection and management, lung cancer remains the most frequent cause of cancer-related mortality. According to a World Health Organization report, lung cancer accounted for 1.59 million deaths worldwide in the year 2012 alone (http://www.who.int/mediacentre/factsheets/fs297/en/). While current targeted therapies (such as inhibitors for EGFR and ALK) help some patients, more widely applicable biomarkers/molecular targets are urgently needed.
In this issue of the Journal, Kim et al. report that USE1, a Uba6-specific ubiquitin conjugating E2 enzyme, is frequently overexpressed (92.45%) in lung cancers compared with normal lung tissues (1). While sample size was relatively limited (106 paired cancer and adjacent nontumor lung tissues), the strikingly high frequency of USE1 overexpression in tumors suggests that USE1 may serve as a useful lung cancer biomarker and perhaps a novel molecular target.
The ubiquitin-conjugating enzyme USE1/UBE2Z is in the middle of an E1-E2-E3 ubiquitin transfer cascade and plays a major role in protein homeostasis (proteostasis). It functions downstream of and specifically with the ubiquitin-activating enzyme (E1) UBA6 to attach ubiquitin or FAT10 via ubiquitin-ligase E3 (eg, UBR1-3) to proteins to mark them for degradation by the proteasome (2,3). Embryonic ablation of the UBA6-USE1 system in mice is lethal, and its brain-specific knockout results in abnormal neuron patterning, with the afflicted mice displaying impaired social behavior (4). In this report, Kim et al. explored the clinical significance of USE1 in lung cancers.
Comparing paired cancer and adjacent nontumor lung tissues, Kim et al. found that USE1 protein levels were statistically significantly elevated in the cancer specimens. Further, this elevation was not due to increased transcription as USE1 mRNA levels were not changed, suggesting either increased translation or decreased degradation of the USE1 protein in these cancer tissues. Indeed, at least in some cases (13.2%), the elevation in USE1 protein expression seemed to be due to missense mutations in a region (D-box) of the USE1 protein that mediates interaction with the APC/C E3 ubiquitin ligase complex. These mutations in D-box greatly compromise the interaction of USE1 proteins with Cdc20 and Cdh1, the co-activator-substrate-binding (CASB) subunits of the APC/C complex, and increase the half-life of mutated proteins more than two-fold.
It is worth noting that both of the two CASB subunits of APC/C associate with, and mediate the degradation of, the USE1 proteins. APC/C activity is tightly regulated during cell cycle, and one of the mechanisms to achieve this regulation is via association and activation of the CASB subunits. Cdc20 associates in early mitosis and mediates substrate destruction in mitosis, while Cdh1 replaces Cdc20 in anaphase and subsequently catalyzes substrate degradation in late mitosis and during G1 phase (5). The expression pattern of USE1 during cell cycle (high in the G2/M phase followed by rapid degradation during metaphase) suggests a major role of APC/CCdc20 in mediating this process. It would be informative to see whether Emi1, an inhibitor of APC/C that is degraded in prometaphase (6), is involved in regulating USE1 degradation.
Does USE1 overexpression provide a survival advantage to cancer cells? Indeed, it does. The authors demonstrated that exogenous overexpression of USE1 in established lung cancer cell lines increased proliferation, migration, and invasion through membranes coated with extracellular matrix. Furthermore, in a xenograft experiment in nude mice, tumor growth of USE1-overexpressing cancer cells was found to be approximately three-fold greater than that of control cells. These results are consistent with the expression profile of USE1 in lung cancer patients. The authors went on to show that knockdown of USE1 expression by shRNA suppressed proliferation, colony formation, and invasion of cancer cells in vitro. Of note, these effects were observed both in A549 (high endogenous level of USE1) and in TC1 (low endogenous level of USE1) lung cancer cell lines, albeit with more dramatic effect in the former, emphasizing the critical role of USE1 in the aforementioned processes. These in vitro observations were confirmed in a xenograft model, where induction of shRNA against USE1 resulted in a substantial decrease in tumor growth in vivo (more than an 87% reduction compared with tumors expressing a control shRNA).
How does USE1 exert these functions? One possibility is that it mediates the degradation of a tumor suppressor. Although little is known about the clientele modified by the Uba6-USE1 pathway, the tumor suppressor RGS4 is a recognized substrate of this pathway. As a GTPase-activating protein for the Gα subunit of heterotrimeric G proteins, RGS4 suppresses tumor growth and metastasis and is expressed at lower levels in non–small cell lung cancer compared with normal lung tissues (see Figure 1) (7). It would be interesting to determine whether USE1 knockdown in lung cancer has any effect on RGS4 expression.
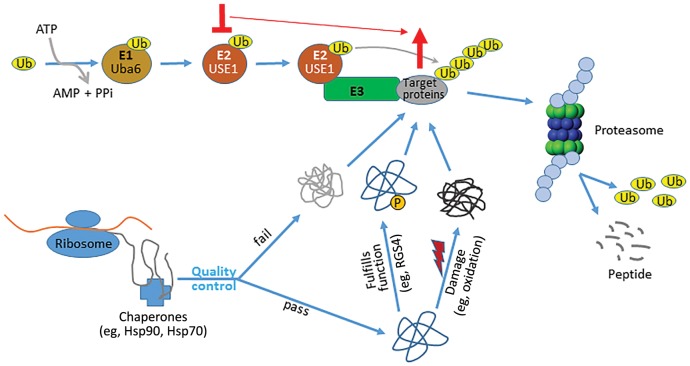
Figure 1.
USE1 is a critical ubiquitin-conjugating enzyme in lung cancer cells. Nascent proteins synthesized at ribosomes undergo folding and maturation with the assistance of chaperones such as Hsp70 and Hsp90. Correctly folded proteins are translocated to other parts of the cell and perform their physiological functions, eg, activated transmembrane GPCRs stimulate signaling by promoting the exchange of GDP with GTP on Gα, while regulator of G-protein signaling (RGS) proteins inhibit Gα signaling by promoting hydrolysis of the bound GTP. These proteins are subsequently marked, such as by phosphorylation, for degradation. Incorrectly folded, or correctly folded but damaged (such as by oxidation), proteins are also degraded to maintain proteostasis. The major protein degradation pathway, ubiquitination, starts with charging the ubiquitin-activating enzyme E1 (eg, Uba6) with a ubiquitin moiety. The ubiquitin is in turn transferred to a ubiquitin-conjugating enzyme E2 (eg, USE1) and subsequently to proteins targeted for degradation via association of the E2 with a ubiquitin-ligase E3. Ubiquitinated proteins are eventually degraded by the proteasome, and the free ubiquitin molecules are recycled. Perturbation of the ubiquitination pathway (eg, knockdown of USE1) disrupts proteostasis and results in accumulation of misfolded and/or damaged proteins, ultimately leading to cell death via apoptosis. In addition to these general effects, functional disruption of a specific component of the pathway, such as USE1, would also cause accumulation of its substrates (eg, RGS4), thus enhancing or prolonging their physiological function (eg, RGS4 as a tumor suppressor).
A second, although not mutually exclusive, possibility is that USE1 overexpression may exert its prosurvival function by modulating proteostasis in cancer cells. Proteostasis is an essential process that includes regulation of protein biogenesis, folding, trafficking, and degradation. Because of their high degree of mutation, challenging environmental conditions (eg, oxidative stress, nutrient deprivation, etc.), and unregulated cell division, cancer cells display increased dependence on the proteostasis machinery to clear damaged or hopelessly misfolded proteins (Figure 1). Indeed, it has been shown that cancer cells are more sensitive than normal cells to transient inhibition of molecular chaperones such as Hsp90 and Hsp70 (8). Further, cancer cells are sensitive to inhibition of protein degradation, and proteasome inhibitors have been shown to be clinically effective in treating patients with multiple myeloma and mantle cell lymphoma (9). Conceivably, USE1 might participate in the degradation of damaged proteins, and its inhibition, like that of the proteasome, might disrupt cancer cell proteostasis with deleterious consequences. Indeed, targeting the ubiquitin-activating enzyme UBE1 by chemical and genetic approaches has been identified as a possible anticancer strategy (10). As an important player in the alternative ubiquitination pathway, USE1 is likely to play a vital role in proteostasis in lung cancer cells, raising the possibility of its utility as a biomarker, and perhaps also as a viable molecular target in lung cancer.
Funding
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Note
The authors have no conflicts of interest to declare.
References
- 1. Kim SJ, Lee TH, Nam SH, et al. The E3 ligase APC/C promotes ubiquitylation-mediated degradation of Uba6-specific-E2 (USE1) to suppress lung tumorigenesis. J Natl Cancer Inst. 2016;1093:djw227 doi:10.1093/jnci/djw227. [Google Scholar]
- 2. Lee PC, Sowa ME, Gygi SP, et al. Alternative ubiquitin activation/conjugation cascades interact with N-end rule ubiquitin ligases to control degradation of RGS proteins. Mol Cell. 2011;433:392–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aichem A, Pelzer C, Lukasiak S, et al. USE1 is a bispecific conjugating enzyme for ubiquitin and FAT10, which FAT10ylates itself in cis. Nat Commun. 2010;1:13. [DOI] [PubMed] [Google Scholar]
- 4. Lee JY, Kwak M, Lee PC.. Impairment of social behavior and communication in mice lacking the Uba6-dependent ubiquitin activation system. Behav Brain Res. 2015;281:78–85. [DOI] [PubMed] [Google Scholar]
- 5. Zhou Z, He M, Shah AA, et al. Insights into APC/C: From cellular function to diseases and therapeutics. Cell Div. 2016;11:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hansen DV, Loktev AV, Ban KH, et al. Plk1 regulates activation of the anaphase promoting complex by phosphorylating and triggering SCFbetaTrCP-dependent destruction of the APC inhibitor Emi1. Mol Biol Cell. 2004;1512:5623–5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cheng C, Yue W, Li L, et al. Regulator of G-protein signaling 4: A novel tumor suppressor with prognostic significance in non-small cell lung cancer. Biochem Biophys Res Commun. 2016;4693:384–391. [DOI] [PubMed] [Google Scholar]
- 8. Miyata Y, Nakamoto H, Neckers L.. The therapeutic target Hsp90 and cancer hallmarks. Curr Pharm Des. 2013;193:347–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McCormack PL. Bortezomib: A review in mantle cell lymphoma in previously untreated patients unsuitable for stem-cell transplantation. BioDrugs. 2015;293:207–14. [DOI] [PubMed] [Google Scholar]
- 10. Xu W, Lukkarila JL, da Silva SR, et al. Targeting the ubiquitin E1 as a novel anti-cancer strategy. Curr Pharm Des. 2013;1918:3201–3209. [DOI] [PubMed] [Google Scholar]