Abstract
Background: We previously derived and validated a bronchial epithelial gene expression biomarker to detect lung cancer in current and former smokers. Given that bronchial and nasal epithelial gene expression are similarly altered by cigarette smoke exposure, we sought to determine if cancer-associated gene expression might also be detectable in the more readily accessible nasal epithelium.
Methods: Nasal epithelial brushings were prospectively collected from current and former smokers undergoing diagnostic evaluation for pulmonary lesions suspicious for lung cancer in the AEGIS-1 (n = 375) and AEGIS-2 (n = 130) clinical trials and gene expression profiled using microarrays. All statistical tests were two-sided.
Results: We identified 535 genes that were differentially expressed in the nasal epithelium of AEGIS-1 patients diagnosed with lung cancer vs those with benign disease after one year of follow-up (P < .001). Using bronchial gene expression data from the AEGIS-1 patients, we found statistically significant concordant cancer-associated gene expression alterations between the two airway sites (P < .001). Differentially expressed genes in the nose were enriched for genes associated with the regulation of apoptosis and immune system signaling. A nasal lung cancer classifier derived in the AEGIS-1 cohort that combined clinical factors (age, smoking status, time since quit, mass size) and nasal gene expression (30 genes) had statistically significantly higher area under the curve (0.81; 95% confidence interval [CI] = 0.74 to 0.89, P = .01) and sensitivity (0.91; 95% CI = 0.81 to 0.97, P = .03) than a clinical-factor only model in independent samples from the AEGIS-2 cohort.
Conclusions: These results support that the airway epithelial field of lung cancer–associated injury in ever smokers extends to the nose and demonstrates the potential of using nasal gene expression as a noninvasive biomarker for lung cancer detection.
The diagnostic evaluation of lung cancer among high-risk current and former smokers with lesions found on chest imaging represents a growing clinical challenge due to the implementation of lung cancer screening (1). While the National Lung Cancer Screening Trial (NLST) demonstrated a 20% relative reduction in lung cancer–related mortality through annual screening of high-risk smokers by low-dose chest CT (LDCT), approximately 25% of CT-screened subjects had a pulmonary lesion, of which over 95% were ultimately diagnosed as benign (2). While there are guidelines for the management of pulmonary nodules (3), unnecessary invasive procedures (including surgical lung biopsy) are frequently performed on patients who are ultimately diagnosed with benign disease (4,5). There is a clear and growing need to develop additional diagnostic approaches for evaluating pulmonary lesions to determine which patients should undergo CT surveillance or invasive biopsy.
Our group and others have established that airway gene expression signatures can serve as biomarkers for the presence of smoking-related lung diseases including lung cancer and COPD (6–8). Previous work has shown that chronic exposure to tobacco smoke results in an airway-wide field of injury with both reversible and irreversible alterations in bronchial airway epithelial cell gene expression upon smoking cessation (9,10). Importantly, gene expression profiles from cytologically normal bronchial epithelial cells obtained via endobronchial brushings can serve as a biomarker that distinguishes ever smokers with lung cancer from those with benign lung disease (6) independently of clinical risk factors (11). More recently, Whitney et al. reported a 23-gene bronchial genomic lung cancer classifier (12), which was validated in two prospective clinical trials (13). While the high sensitivity and negative predictive value of this classifier suggest that it can be used to assign patients to CT surveillance, it is dependent upon cells obtained during bronchoscopy. Although bronchoscopy-related complications are uncommon (14,15), bronchoscopy is not always chosen as a diagnostic modality based on the size/location of the lung lesion, pretest risk of disease, patient preference, and degree of underlying lung disease.
Given concordant response of nasal and bronchial epithelium to tobacco smoke (16) and the validated performance of the bronchial genomic classifier for lung cancer, we sought to test the hypothesis that cancer-associated expression differences might also be detectable in nasal epithelium and related to those found in bronchial epithelium. Detecting cancer-associated gene expression in nasal epithelium would suggest its potential to serve as a less invasive biospecimen for lung cancer detection and potentially expand the clinical settings in which airway gene expression could be used for this purpose.
Methods
For a complete description of the methods, please see the Supplementary Materials and Methods (available online).
Experimental Design
Patients were enrolled at 28 medical centers in the United States, Canada, and Europe as part of two prospective studies within the Airway Epithelial Gene Expression in the Diagnosis of Lung Cancer (AEGIS) clinical trials (registered as NCT01309087 and NCT00746759). All study protocols were approved by the institutional review board at each medical center, and written informed consent was obtained from all patients prior to enrollment. Inclusion and exclusion criteria have been previously described (13). All patients were current or former cigarette smokers (>100 cigarettes in their lifetime) undergoing bronchoscopy as part of their diagnostic workup for clinical suspicion of lung cancer, and all samples were collected prospectively prior to diagnosis. Patients were followed for up to one year postbronchoscopy until a final diagnosis of lung cancer or benign disease was made. Lung tumor stage was assessed using the TNM staging system (17). In this study, 554 nasal epithelium samples were selected and profiled using microarrays (see Supplementary Materials and Methods, available online). We were limited by patients with a benign diagnosis and matched them approximately 2:3 with patients diagnosed with lung cancer. Microarray data generated from bronchial epithelium samples from 299 patients together with their clinical annotations were obtained from Whitney et al. (12).
Microarray Processing
All procedures were performed as previously described (7) using Affymetrix Gene 1.0 ST microarrays. CEL files from nasal and bronchial samples passing quality control (Supplementary Materials and Methods, available online) were processed separately using the Robust Multichip Average (RMA) algorithm (18) and the standard Affymetrix Chip Definition File to estimate gene expression signal. ComBat (19) was used to correct for batch effects.
Characterization of Cancer Associated Genes in Nasal Epithelium
Cancer-associated gene expression profiles in nasal epithelium were identified using linear models (20) that corrected for smoking status, pack-years, sex, age, and RNA quality (RIN). Functional enrichment of the most differentially expressed genes (P < .001) was assessed using the Reactome and Gene Ontology (GO) databases and EnrichR (21). Similarities between cancer-associated gene expression profiles in nasal and bronchial epithelium were assessed using a preranked gene set enrichment analysis (GSEA) (22).
The expression of 11 gene clusters previously identified as being associated with cancer in the bronchial epithelium (12) was summarized in AEGIS-1 nasal samples by taking the mean of the cluster genes per sample. The association of each cluster mean with the presence or absence of cancer was computed using a Welch t test. Finally, the correlation of scores from a previously reported bronchial genomic lung cancer classifier (12) was evaluated in matched bronchial and nasal samples from AEGIS-1 (n = 157 patients). These analyses are described in detail in the Supplementary Materials and Methods (available online).
Derivation of Nasal Clinicogenomic Lung Cancer Classifier
We derived a clinical factor lung cancer classifier using a training set of AEGIS-1 patients (n = 517) using logistic regression to combine patient age, smoking status (current, former), time since quit (≤15 years, >15 years, unknown), and mass size (<3 cm, ≥3 cm, infiltrates). Similarly, we derived a clinicogenomic classifier using penalized logistic regression to combine all of the variables in the clinical factor model plus the score from a nasal lung cancer gene expression classifier. This gene expression classifier was derived in the nasal training set (n = 375) using a weighted voting algorithm. The most differentially expressed genes by moderated t test were included in this model. The total number of genes included in this classifier was optimized in cross-validation. Specifically, we selected the smallest number of genes that maximized cross-validation performance. A more detailed description is provided in the Supplementary Materials and Methods (available online).
Statistical Analysis
Differences in clinical covariates between patients with and without lung cancer were assessed using Fisher’s exact test (categorical variables) or Welch t test (continuous variables). Differential expression analyses were performed using linear modeling (limma R package [20]) or Welch t tests unless otherwise specified. For the differential expression analysis, a two-sided P value of less than .001 was considered evidence of statistically significant differential expression. Correlation coefficients were calculated using Pearson’s product-moment coefficient. Classification performance was assessed using standard measures including receiver operating characteristic (ROC) curve, the area under the curve (AUC), sensitivity, specificity, negative predictive value (NPV), and positive predictive value (PPV). Differences between ROC curve AUC values were assessed using DeLong’s test (23) for correlated ROC curves. Operating points for binary classification were chosen as the threshold that maximized sensitivity while maintaining 50% specificity in the training set. Differences in sensitivity and specificity between models were assessed using McNemar’s chi-square test for count data (24). Statistical differences in NPV between models were assessed using the generalized score statistic (25) for paired analyses or a proportions test for unpaired analyses. All confidence intervals (CIs) are reported as two-sided binomial 95% confidence intervals. All statistical tests were two-sided, and a P value of less than .05 was considered statistically significant.
Results
Study Population
Five hundred fifty-four nasal epithelium samples were selected for microarray profiling from a larger pool of RNA samples collected prospectively from patients with suspect lung cancer enrolled in the AEGIS clinical trials (13). Four hundred twenty-four of these samples were from patients enrolled in the AEGIS-1 trial, and 130 were from patients in the AEGIS-2 trial (Supplementary Figure 1, available online). Thirty-one patients from the AEGIS-1 cohort had an indeterminate cancer diagnosis at one year post–sample collection or were lost to follow-up and were removed from the study. Additionally, 18 nasal microarray samples from the AEGIS-1 data set that did not meet minimum quality standards were also removed. No samples were removed from the AEGIS-2 data set. The remaining 375 samples from the AEGIS-1 cohort were used as a training set in which all data analyses and biomarker derivation steps were performed, while the 130 samples from the AEGIS-2 cohort were used solely to validate the predictive model (Table 1). The distribution of cancer stages was slightly skewed toward later-stage cancers in the validation set (Supplementary Table 1, available online). Lung cancer patients tended to have larger nodules than patients with benign diagnoses in both the training and validation sets (P < .001 for both comparisons) (Supplementary Table 2, available online) while patient age was statistically significantly higher among cancer patients in the training set (P < .001). The gene expression data from these samples has been deposited in the NCBI Gene Expression Omnibus under accession number GSE80796.
Table 1.
Clinical and demographic characteristics of patients who contributed nasal epithelial samples
| Characteristic | AEGIS-1 training set (n = 375) | AEGIS-2 validation set (n = 130) | P |
|---|---|---|---|
| Cancer Status, No.* | .006 | ||
| Lung Cancer | 243 | 66 | |
| Benign Disease | 132 | 64 | |
| Smoking Status, No.* | .75 | ||
| Current | 140 | 46 | |
| Former | 235 | 84 | |
| Sex, No.* | .75 | ||
| Male | 237 | 80 | |
| Female | 138 | 50 | |
| Cumulative smoke exposure (SD, No.), pack-y† | 39.0 (26.9, 371) | 34.8 (30.7, 130) | .17 |
| Time since quit (SD, No.), y† | 7.6 (12.9, 309) | 9.4 (13.4, 120) | .21 |
| Age (SD), y† | 59.5 (10.4) | 61.7 (11.5) | .06 |
| Lesion size, No.*,‡ | .89 | ||
| >3 cm | 171 | 59 | |
| ≤3 cm | 142 | 54 | |
| Infiltrate | 44 | 17 | |
| Unknown | 18 | 0 | |
| Lesion location, No.*,§ | .16 | ||
| Central | 134 | 55 | |
| Peripheral | 114 | 31 | |
| Central and peripheral | 100 | 44 | |
| Unknown | 27 | 0 | |
| Lung cancer histological type, No.*,|| | .45 | ||
| Small cell | 40 | 8 | |
| Non–small cell | 200 | 58 | |
| Adenocarcinoma | 90 | 29 | |
| Squamous | 72 | 17 | |
| Large cell | 9 | 4 | |
| Not specified | 29 | 8 | |
| Unknown | 3 | 0 | |
| Diagnosis of benign condition, No.* | 105 | 34 | .13 |
| Infection | 36 | 7 | |
| Sarcoidosis | 21 | 12 | |
| Other | 48 | 15 |
P value calculated using two-sided Fisher’s Exact test.
P value calculated using two-sided Student’s t test.
P value calculated comparing >3 cm vs ≤3 cm vs infiltrates.
P value calculated comparing central vs peripheral vs central and peripheral.
P value calculated comparing non–small cell vs small cell.
Lung Cancer–Associated Gene Expression in Nasal Epithelium
We identified 535 genes that were differentially expressed in the nasal epithelium of AEGIS-1 patients diagnosed with lung cancer vs those with benign disease after one year of follow-up (P < .001) (Figure 1; Supplementary File 1, available online). Genes downregulated in patients with lung cancer were enriched for genes associated with DNA damage, regulation of apoptosis, and processes involved in immune system activation including the interferon-gamma signaling pathway and antigen presentation (Table 2). Among genes that were upregulated in lung cancer patients, we found enrichment for genes involved in endocytosis and ion transport (Table 2). A complete list of statistically significantly enriched pathways and GO categories (FDR < 0.05) is shown in Supplementary Files 2–4 (available online).
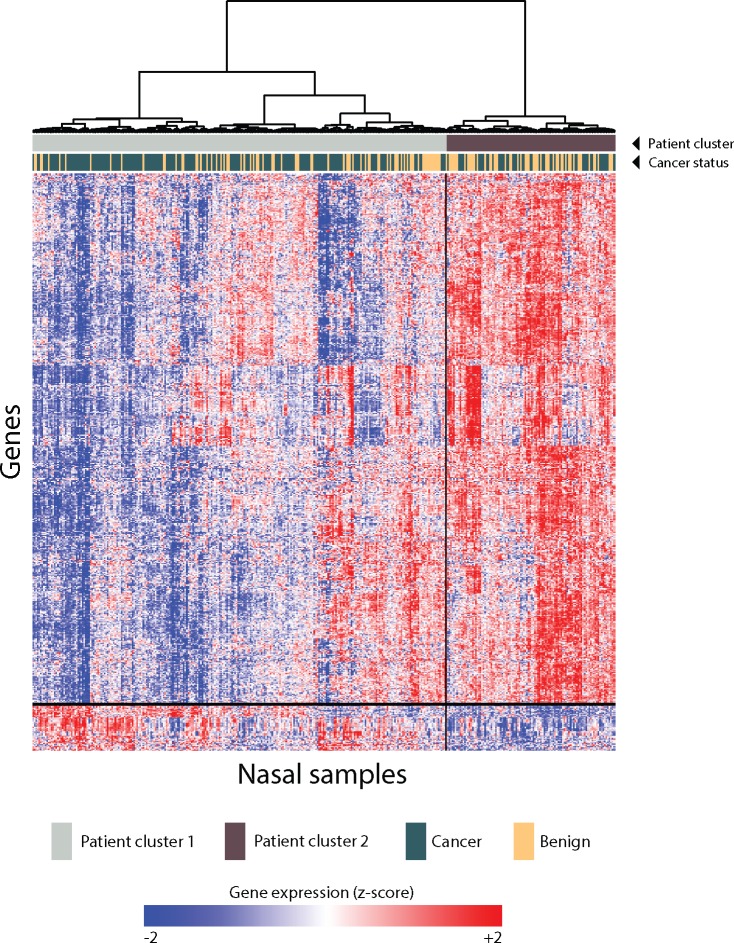
Figure 1.
Characterization of 535 cancer-associated nasal epithelial genes in the training set. Five hundred thirty-five genes were differentially expressed by cancer status in the nasal training set (P < .001) using a linear model that included cancer status, smoking status, pack-years, sex, age, and RIN as covariates. These genes were grouped into two co-expression clusters by unsupervised hierarchical clustering. Unsupervised hierarchical clustering of patients across these genes revealed two primary patient clusters.
Table 2.
Functional characterization of genes with cancer-associated expression in nasal epithelium
| Genes | False discovery rate |
|---|---|
| Downregulated genes (n = 492) | |
| DNA damage | |
| Signal transduction involved in mitotic DNA integrity checkpoint (GO:1902400) | <0.001 |
| Ubiquitin-dependent degradation of Cyclin D1 (reactome) | <0.001 |
| Regulation of apoptosis (reactome) | <0.001 |
| G1/S DNA damage checkpoints (reactome) | <0.001 |
| Immune system activation | |
| Antigen presentation and processing of exogenous antigen (GO:0019884) | <0.001 |
| Interferon-gamma signaling (reactome) | <0.001 |
| Upregulated genes (n = 43) | |
| Ion transport | |
| Response to magnesium ion (GO:0032026) | 0.01 |
| Regulation of endocytosis (GO:0030100) | 0.01 |
| Positive regulation of release of calcium ion into cytosol (GO:0010524) | 0.02 |
Similarities in Nasal and Bronchial Cancer–Associated Gene Expression
To determine if a shared pattern of cancer-related gene expression might exist between the nose and bronchus, we leveraged microarray data from 299 bronchial epithelium samples obtained from AEGIS-1 patients (12). One hundred fifty-seven of the 299 bronchial samples came from the same patients as those in our nasal training set (Supplementary Table 3 and Supplementary Figure 2, available online). Using bronchial gene expression data from the AEGIS-1 patients, we found statistically significant concordant cancer-associated gene expression alterations between the two airway sites (P < .001). GSEA (22) revealed (Figure 2A) that the genes upregulated in nasal epithelium of patients with lung cancer were among the genes most upregulated in bronchial epithelium of patients with cancer (P < .001) and that a similar relationship exists for genes downregulated in patients with cancer between the nose and bronchus (P < .001). The expression of the most concordantly differentially expressed genes is shown in Figure 2B and highlighted in Supplementary File 1 (available online).
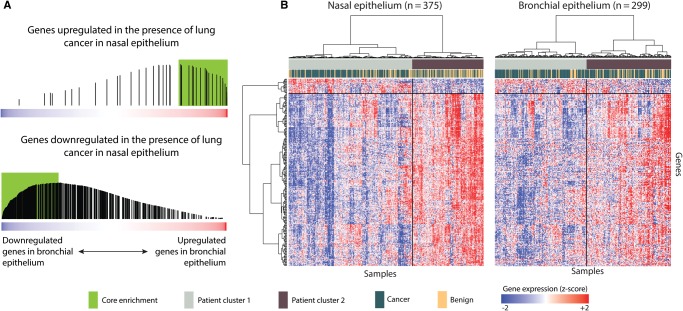
Figure 2.
Concordance between cancer-associated gene expression in bronchial and nasal epithelium. A) The 535 genes with cancer-associated expression in nasal epithelium were split into up- and downregulated gene sets, and we examined their distribution within all genes ranked from most downregulated (left) to most upregulated (right) in the bronchial epithelium of patients with cancer using gene set enrichment analysis. We found that the genes with increased expression in nasal epithelium are enriched among the genes that are most induced in the bronchial epithelium of patients with cancer (top; P < .001 by a two-sided permutation-based Kolmogorov-Smirnov-like test [22]) while the reverse was true for genes with decreased expression in nasal epithelium (bottom; P < .001 by a two-sided permutation-based Kolmogorov-Smirnov-like test [22]). Genes included in the core enrichment are shown in the green box. B) Heatmaps and hierarchical clustering of the core enrichment genes in nasal (left) and bronchial (right) samples. All statistical tests were two-sided.
To further explore the hypothesis of a shared field of nasal and bronchial lung cancer–associated injury, we examined the nasal expression of 232 genes with lung cancer–associated expression in bronchial epithelium (12). Whitney et al. grouped these genes into 11 clusters, and we found that the mean expression of eight of the 11 clusters was statistically significantly associated with lung cancer (P < .05) in our training set (Table 3), including gene clusters enriched for genes involved in cell cycle, response to retinoic acid, and the innate immune response. Based on the concordant expression of cancer-associated genes in bronchial and nasal epithelium, we computed a “bronchial” lung cancer classifier score (12) for the nasal training set samples and found that they were highly correlated with the scores computed from matched bronchial samples (n = 157, R = 0.70, P < .001) (Supplementary Figure 3, available online). Taken together, these results suggest that lung cancer–associated gene expression differences are similar in nasal and bronchial epithelium.
Table 3.
Aggregate expression of lung cancer gene clusters from bronchial epithelium in nasal epithelial samples
| Cluster | Function | No. of probesets | Direction in cancer | P* |
|---|---|---|---|---|
| 1† | Innate immune | 25 | Down | <.001 |
| 2† | Mitotic cell cycle | 47 | Down | .05 |
| 3 | Inflamation | 45 | Down | .83 |
| 4† | Resp. retinoic acid/ cell cycle | 34 | Up | .004 |
| 5 | NA | 10 | Up | .36 |
| 6 | NA | 21 | Down | .02 |
| 7† | Submucosal gland markers | 20 | Up | .01 |
| 8 | n/a | 15 | Up | .003 |
| 9† | Xenobiotic detoxification | 7 | Down | .15 |
| 10† | Cartilaginous markers | 4 | Down | .05 |
| 11 | NA | 1 | Down | .03 |
P value of two-sided t test measuring the difference in mean average expression of all genes in a cluster between cancer and benign nasal sample in the AEGIS-1 cohort.
In bronchial genomic classifier described by Whitney et al. 2015 (12).
Nasal Gene Expression as an Independent Predictor of Lung Cancer Status
To determine if nasal gene expression could serve as a predictor of lung cancer status, we selected the 30 most statistically significantly differentially expressed genes (P < .001) from among the 535 genes with cancer-associated nasal gene expression for use in a weighted-voting biomarker (Supplementary File 5, available online). The biomarker panel size of 30 genes was chosen as the smallest number of genes that achieved maximal performance in cross-validation. This biomarker had an AUC of 0.69 (n = 375, 95% CI = 0.63 to 0.75, P < .001) in cross-validation in the training set. Twenty-two of the 30 genes were also statistically significantly correlated between matched bronchial and nasal samples (mean R = 0.29, range = 0.16–0.49, P < .05).
In order to evaluate the potential for the nasal gene expression biomarker to add to clinical risk factors for lung cancer detection, we developed a clinical risk factor model and tested whether incorporating the gene-expression biomarker enhances its performance. In choosing which clinical risk factors to include, we relied on a study in which Gould et al. identified smoking status, time since quit, age, and mass size as important clinical risk factors of lung cancer for patients with solitary pulmonary nodules (26). Patient age, smoking status (current, former), time since quit (≤15 years, >15 years, unknown), and categorized mass size (<3 cm, ≥ 3 cm, infiltrates) were used to create a clinical risk factor model for lung cancer using logistic regression. The training set for this model consisted of the nasal training set used to derive the gene expression classifier as well as clinical data from an additional 142 patients from the AEGIS-1 cohort for a total training set of 517 patients for the clinical model (see Supplementary Figure 2, available online). A clinicogenomic logistic regression model that incorporated the clinical factors and the nasal gene expression classifier score was derived in the 375 training set samples with nasal gene expression.
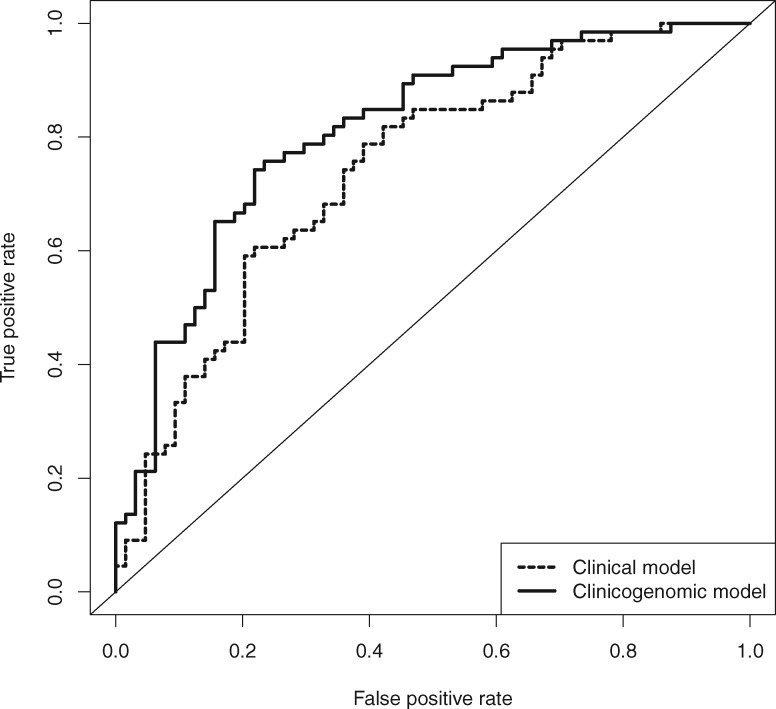
The performance of the clinical and clinicogenomic models was evaluated using an independent set of nasal samples (n = 130) from the AEGIS-2 clinical trial that were not used in the development of the classifier. The clinicogenomic model yielded an AUC of 0.81 (95% CI = 0.74 to 0.89) in the validation set, which was statistically significantly higher than the AUC of 0.74 (95% CI = 0.66 to 0.83) achieved by the clinical risk-factor model alone (P = .01) (Figure 3). Operating points for binary classification were chosen to maximize training set sensitivity with specificity of 50% or greater for both models. The addition of cancer-associated gene expression to the clinical risk factor model increased sensitivity from 0.79 (95% CI = 0.67 to 0.88) to 0.91 (95% CI = 0.81 to 0.97, P = .03) and negative predictive value from 0.73 (95% CI = 0.58 to 0.84) to 0.85 (95% CI = 0.69 to 0.94, P = .03) (Table 4). The negative likelihood ratio of the clinicogenomic classifier was consistent between training (0.18; 95% CI = 0.12 to 0.28) and validation (0.18; 95% CI = 0.08 to 0.39) sets. Additionally, in subjects with either lesion size less than 3 cm or peripheral lesions, the clinicogenomic model had a negative predictive value of 0.85 (95% CI = 0.65 to 0.96) or 0.93 (95% CI = 0.66 to 1.00), respectively (Supplementary Table 4, available online).
Figure 3.
Clinicogenomic and clinical classifier performance in the validation set. Shown are the receiver operating characteristic (ROC) curves for the clinicogenomic (solid line) and clinical (dashed line) classifiers in the independent AEGIS-2 validation set. The area under the curve (AUC) was 0.81 (95% confidence interval [CI] = 0.74 to 0.89) for the clinicogenomic classifier and 0.74 (95% CI = 0.66 to 0.83) for the clinical classifier. The difference between ROC curves was statistically significantly different (P = .01 by a two-sided Delong’s test for correlated ROC curves).
Table 4.
Classifier performance in the validation set (n = 130)
| Biomarker performance metric | Clinical model | Clinicogenomic model | P |
|---|---|---|---|
| Area under the curve (95% CI)* | 0.74 (0.66 to 0.83) | 0.81 (0.74 to 0.89) | .01 |
| Sensitivity (95% CI)† | 0.79 (0.67 to 0.88) | 0.91 (0.81 to 0.97) | .03 |
| Specificity (95% CI)† | 0.58 (0.45 to 0.70) | 0.52 (0.39 to 0.64) | .42 |
| Negative predictive value (95% CI)‡ | 0.73 (0.58 to 0.84) | 0.85 (0.69 to 0.94) | .03 |
| Positive predictive value (95% CI)‡ | 0.66 (0.54 to 0.76) | 0.66 (0.55 to 0.76) | .97 |
| Accuracy (95% CI)§ | 0.68 (0.60 to 0.76) | 0.72 (0.63 to 0.79) | .68 |
P value comparing models calculated using Delong’s two-sided test. CI = confidence interval.
P value comparing models calculated using McNemar’s two-sided chi-square test.
P value comparing models calculated using two-sided generalized score statistic.
P value comparing models calculated using two-sided Fisher Exact test.
Discussion
In this study, we have established that the lung cancer–associated airway “field of injury” detectable in bronchial airway epithelium (6,11,12) extends to the nasal epithelium. These findings strengthen the “field of injury” hypothesis in which there are gene expression alterations in normal-appearing epithelial cells throughout the entire airway of smokers with lung cancer and, intriguingly, suggest the potential for lung cancer biomarkers based on nasal epithelial gene expression.
While previous studies have validated the existence of bronchial airway gene expression alterations in patients with lung cancer and demonstrated their clinical utility in lung cancer detection (13), little is known about the physiological processes responsible for this “field of injury.” One hypothesis for the presence of lung cancer–associated alterations in nasal and bronchial gene expression is that the subset of smokers who develop lung cancer exhibit a distinct genomic response to tobacco smoke exposure throughout all airway epithelial cells, consistent with the “etiological field effect” described by Lochhead et al. for colon and other cancer types (27). This paradigm suggests that the airway gene-expression signature is a risk marker for lung cancer as opposed to a direct consequence of the presence of lung cancer based on local or systemic factors produced by the tumor or its microenvironment (ie, the “conventional field effect” defined by Lochhead et al. [27]). Consistent with the etiological field effect hypothesis, we observed a concordant downregulation of genes associated with immune system activation in patients with lung cancer in both bronchial and nasal epithelium, which might suggest that an impaired immune response sets the stage for tumorigenesis in the lung microenvironment. Alternatively, despite the distance to the tumor, these cancer-associated gene expression differences may be a direct result of factors secreted by the tumor or its microenvironment, or some other consequence of the presence of the tumor consistent with the “conventional field effect” described above.
Mechanistically, it is intriguing that a number of genes with important roles in cancer-related processes are among the differentially expressed genes. Of the genes that were downregulated in patients with lung cancer, CASP10 and CD177 were among the most correlated genes between bronchial and nasal epithelium and are associated with the induction of apoptosis and activation of the immune response, respectively. We also identified a number of genes involved in the p53 pathway that were downregulated in patients with lung cancer, including BAK1, ST14, CD82, and MUC4. BAK1 is associated with the induction of apoptosis (28,29) and has been previously shown to be downregulated in the tumors of patients with non–small cell lung cancer (NSCLC) (30). ST14 has been described as a tumor suppressor in breast cancer and its overexpression associated with the inhibition of tumor cell migration and cell invasion (31). The downregulation of CD82, which is a metastasis suppressor in prostate cancer (32), has been shown to be correlated with poor survival in patients with lung adenocarcinoma (33). MUC4, whose downregulation has been associated with increased tumor stage and poorer overall survival, has also been shown to play an oncogenic role in multiple cancers and is a tumor suppressor in NSCLC, acting as a modifier of p53 expression (34).
From a clinical perspective, we found that the addition of lung cancer–associated gene expression to established clinical risk factors improved the sensitivity and negative predictive value for detecting lung cancer; these are the key performance metrics for driving potential clinical utility in this setting (eg, allowing physicians to avoid unnecessary invasive procedures in those with benign disease). This provides the first proof of concept for the use of nasal gene expression for lung cancer detection. We elected to establish the presence of a nasal field of lung cancer–associated injury using samples from the AEGIS trial given the unique availability of matched bronchial samples, despite the fact that these patients were undergoing bronchoscopy for suspect lung cancer. The demonstration of the added value of nasal gene expression for lung cancer detection in this setting sets the stage for the development of nasal gene expression biomarkers for lung cancer in other clinical settings where bronchoscopy is not frequently used because of lesion size/location, risk of complications, or cost. In particular, it will now be of interest to develop nasal biomarkers for patients with small peripheral nodules found incidentally or via screening as our current bronchoscopy-based cohort is enriched for patients with centrally located lesions. In the clinical setting of patients with small peripheral nodules, we envision that a nasal biomarker for lung cancer with a low negative likelihood ratio (on par with the NLR we observed for the nasal biomarker in the AEGIS samples) could be used to identify nodule patients who are at low risk of malignancy and can be managed by CT surveillance.
The importance and potential impact of this study derive from several key strengths. First, the patients came from a large number of academic and community hospitals and reflect a variety of practice settings and different geographical locales; thus the diversity of alternative benign diagnoses is represented. Second, the training and validation sets came from two separate clinical trials, which minimizes the potential for the model to depend on locally confounding variables. Third, the samples were prospectively collected and cancer status was unknown at the time of collection. Fourth, we have shown that nasal gene expression identifies a source of lung cancer risk that is independent of major clinical risk factors.
There were also a number of important limitations to this study. Nasal samples used in this study were collected from patients undergoing bronchoscopy for clinical suspicion of lung cancer. As a result, our cohort was enriched for patients with larger nodules and an elevated pretest risk of lung cancer. Further, the size of our independent validation set limited our ability to assess the subgroup performance of our biomarker in patients with small and/or peripheral nodules, a clinical setting where the test may have greater clinical utility. Lastly due to our nested case/control study design, the disease prevalence in our cohort is not representative of the intended use clinical setting.
Together, the findings demonstrate the existence of a cancer-associated airway field of injury that can be measured in nasal epithelium, a biosample that can be collected noninvasively with little instrumentation or advanced training. Moreover, we find that nasal gene expression contains information about the presence of cancer that is independent of standard clinical risk factors, suggesting that nasal epithelial gene expression might aid in lung cancer detection. These findings, in particular the high NPV of the nasal clinicogenomic biomarker, suggest the potential to rule out lung cancer and set the stage for efforts to develop nasal gene expression biomarkers that might have clinical utility in avoiding unnecessary invasive procedures in settings where bronchoscopy is not used as a diagnostic procedure, including small peripheral nodules.
Funding
This research was supported by grants from the National Institutes of Health (NIH) Early Detection Research Network (U01CA152751 and U01CA214182 to AS, MEL, SD, and DE), the Department of Defense (DOD W81XWH-11‐2‐0161 to AS and ML), and the Boston University Coulter Award (0‐057‐281‐A594‐5 to AS and MEL).
This study was sponsored by Veracyte, Inc.
Notes
The study sponsor was not involved in data generation, analysis, or interpretation. The study sponsor did review a final draft of the manuscript.
The authors would like to thank Adam Gower for his implementation of the weighted voting algorithm and Yaron Gesthalter for his input and revisions to early drafts of the manuscript.
JB, AS, and MEL conceived the study. JPR performed the primary analyses. JG and CA performed preliminary analyses. GL, SZ, and YA performed the experiments. WEJ, DE, and MEL advised statistical analyses. KP and DW provided the samples. JPR, JG, CA, GL, SZ, YA, WEJ, JB, KP, DW, SD, AS, and MEL wrote the manuscript. All authors provided critical input on the manuscript and approved it for publication in its final form.
KP and DW are employees of Veracyte, Inc. AS and MEL are consultants to Veracyte, Inc. Boston University owns patents related to the subject matter of this manuscript.
Microarray CEL files and gene expression data used in this manuscript have been deposited in the NCBI Gene Expression Omnibus under accession number GSE80796.
The AEGIS Study Team: Beth Israel Deaconess Medical Center, Boston, MA: Principal Investigator: Armin Ernst and Gaetane Michaud; Co-Investigators: Adnan Majid, Sidharta Pena Gangadharan, Andres Sosa, Renelle Myers, Michael Kent, Malcolm DeCamp, Dilip Nataraj, Samaan Rafeq, David Berkowitz, Saleh Alazemi, Robert Garland; Study Coordinators: Arthur Dea, Paula Mulkern, Christina Carbone. Cleveland Clinic, Cleveland, OH: Principal Investigator: Tom Gildea; Co-Investigators: Francisco Almeida, Joseph Cicenia, Mike Machuzak; Study Coordinators: Meredith Seeley. Columbia University, New York, NY: Principal Investigator: Charles Powell; Co-Investigators: William Bulman, Joshua Sonett, Rebecca Toonkel; Study Coordinators: Kivilcim Sungur-Stasik. Georgia Clinical Research, Austell, GA: Principal Investigator: Stuart Simon; Co-Investigators: Chad Case, Alexander Gluzman, Charles Hartley, Steven Harris, Aristidis Iatridis, Jermaine Jackson, C. Coy Lassiter, Brion Lock, Kathryn McMinn, Chad Miller, Sriram Paramesh, Craig Patterson, Brett Sandifer, Samuel Szumstein, James Waldron, Christy Wilson, Paul Zolty, Stephen Strazay, Ashley Waddell; Study Coordinators: Betsy Rambo, Monica Haughton, Stacy Beasley, Penny Murray, Debra Yeager. Indiana University, Indianapolis, IN: Principal Investigator: Francis Sheski; Co-Investigators: Praveen Mathur; Study Coordinators: MaryAnn Caldwell, Annette Hempfling. Jamaica Hospital Medical Center, Jamaica, NY: Principal Investigator: Craig Thurm; Co-Investigators: Aradhana Agarwal, Akash Ferdaus; Study Coordinators: Kelly Cervellione. Louisiana State University, New Orleans, LA: Principal Investigator: Stephen Kantrow; Co-Investigators: Susan Gunn, David Welsh, Jennifer Ramsey, Jaime Palomino, Richard Tejedor; Study Coordinators: Connie Romaine. Medical University of South Carolina, Charleston, SC: Principal Investigator: Gerard Silvestri; Co-Investigators: Nicholas James Pastis, Nichole Tripician Tanner, Peter Doelken, John Terrill Huggins: Study Coordinators: Jeffrey Waltz, Katherine Taylor, Kalon Eways. National Jewish Health, Denver, CO: Principal Investigator: Ali Musani; Co-Investigators: David Hsia, Joseph Seaman, Justin Thomas: Study Coordinators: Phillip Lopez, Jami Henriksen. New York University, New York, NY: Principal Investigator: William Rom; Co-Investigators: Eric Leibert, Derrick Raptis, James Tsay, Robert Lee, Eric Bonura; Study Coordinators: Katie Schliessman, Ellen Eylers. North Florida/South Georgia Veterans Health System, Gainesville, FL: Principal Investigator: Peruvemba Sriram; Study Coordinators: Ana Thomas, Katherine Herring, Carmen Lowell. Overlake Hospital, Bellevue, WA: Principal Investigator: Amy Markezich; Co-Investigators: James Copeland, Eric Gottesman, Todd Freudenberger, William Watts: Study Coordinators: Tina Fortney. Pulmonary Associates, P.A., Phoenix, AZ: Principal Investigator: Mark Gotfried; Co-Investigators: Robert Comp, Andreas Kyprianou, James Ross, Ronald Servi; Study Coordinators: Li Yi Fu, Sherry Harker. Pulmonary and Allergy Associates, P.A., Summit, NJ: Principal Investigator: Robert Sussman; Co-Investigators: Donatella Graffino, Mark Zimmerman, Robert Restifo, Vincent Donnabella, Federico Cerrone, John Oppenheimer, Robert Capone, Jaime Cancel, Edward Dimitry, Matthew Epstein, Sue Fessler, Erwin Oei, Frederic Scoopo, Chirag Shah; Study Coordinators: Virginia Hala, Kathy Izzo, Marissa Reinton-Lim, Hazel Scherb, MaryAnn Constantino. St. Elizabeth’s Medical Center, Brighton, MA: Principal Investigator: Samaan Rafeq and Armin Ernst; Co-Investigators: Ali Ashraf, Antonio DeGorordo Arzamende, Deirdre Keogh, Ryan Chua, Ali Khodabandeh; Study Coordinators: Arthur Dea, Paula Mulkern. St. James’s Hospital, Trinity College, Dublin, Ireland: Principal Investigator: Joe Keane; Study Coordinators: Jennifer Winkles, Eliot Woodward. Temple University, Philadelphia, PA: Principal Investigator: John Travaline; Co-Investigators: Peter Bercz, Wissam Chatila, Brian Civic, Francis Cordova, Gerard Criner, Gilbert D’Alonzo, Victor Kim, Samuel Krachman, Albert Mamary, Nathaniel Marchetti, Aditi Satti, Kartik Shenoy, Irene Permet Swift, Maria-Elena Vega Sanchez, Sheila Weaver, Nicholas Panetta, Parag Desai, Fred Kueppers, Namrata Patel, Kathleeen Brennan, Alex Swift, David Ciccolella, Fred Jaffe, Jamie Lee Garfield; Study Coordinators: Carla Grabianowski, Carolina Aguiar. University of Alabama, Birmingham, AL: Principal Investigator: Mark Dransfield; Study Coordinators: Sherry Tidwell. University of British Columbia, Vancouver, BC, Canada: Principal Investigator: Stephen Lam; Co-Investigators: Annette McWilliams; Study Coordinators: Sharon Gee. University of California- Davis, Sacramento, CA: Principal Investigator: Richart Harper; Co-Investigators: Ken Yoneda, Jason Adams, Katherine Cayetano, Andrew Chan, Heba Ismail, Charles Poon, Rokhsara Rafii, Christian Sebat, Yasmeen Shaw, Matthew Sisitki, Will Tseng; Study Coordinators: Maya Juarez, Kaitlyn Kirk, Claire O’Brien. University of Missouri, Columbia, MO: Principal Investigator: Vamsi Guntur; Co-Investigators: Normand Caron, Harjyot Sohal, Casey Stahlheber, Danish Thameem, Shilpa Patel, Ousama Dabbagh, Rajiv Dhand, Rachel Kingree, Yuji Oba, Jason Goodin; Study Coordinators: Marta Fuemmeler, Angie Vick, Michel O’Donnell. University of Pennsylvania, Philadelphia, PA: Principal Investigator: Anil Vachani; Co-Investigators: Andrew Haas Colin Gillespie, Daniel Sterman; Study Coordinators: Kristina Maletteri, Karen Dengel. University of Virginia, Charlottesville, VA: Principal Investigator: George Verghese; Co-Investigators: Cynthia Brown, Elizabeth Gay, Borna Mehrad, Manojkumar Patel, Mark Robbins, C. Edward Rose, Max Weder, Kyle Enfield; Study Coordinators: Peggy Doherty. University of Wisconsin, Madison, WI: Principal Investigator: Scott Ferguson; Co-Investigators: Mark Regan, Jennifer Bierach; Study Coordinators: Michele Wolff. Vanderbilt University, Nashville, TN: Principal Investigator: Pierre Massion; Co-Investigators: Alison Miller; Study Coordinators: Gabe Garcia, Anna Ostrander, Wendy Cooper, Willie Hudson. Virginia Commonwealth University, Richmond, VA: Principal Investigator: Wes Shepherd; Co-Investigators: Hans Lee, Rajiv Malhotra, Ashutosh Sachdeva; Study Coordinators: Christine DeWilde, Anna Priday. William Jennings Bryan Dorn VAMC: Principal Investigator: Brian Smith and Andrea Mass; Study Coordinators: Justin Reynolds, Andrea Peterson, Isaac Holmes. Yale University, New Haven, CT: Principal Investigator: Gaetane Michaud; Co-Investigators: Daniel Boffa, Frank Detterbeck, Kelsey Johnson, Anthony Kim, Jonathan Puchalski, Lynn Tanoue, Kyle Bramley; Study Coordinators: Christina Carbone.
Supplementary Material
References
- 1. Wiener RS, Gould MK, Arenberg DA et al. . An official American Thoracic Society/American College of Chest Physicians policy statement: Implementation of low-dose computed tomography lung cancer screening programs in clinical practice. Am J Respir Crit Care Med. 2015;1927:881–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. National Lung Screening Trial Research Team Aberle DR, Adams AM et al. . Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med. 2011;3655:395–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rivera MP, Mehta AC, Wahidi MM. Establishing the diagnosis of lung cancer: Diagnosis and management of lung cancer, 3rd ed.: American College of Chest Physicians evidence-based clinical practice guidelines. Chest. 2013;143(5 suppl):e142S–e165S. [DOI] [PubMed] [Google Scholar]
- 4. Tanner NT, Aggarwal J, Gould MK et al. . Management of pulmonary nodules by community pulmonologists: A multicenter observational study. Chest. 2015;1486:1405–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wiener RS, Gould MK, Slatore CG, Fincke BG, Schwartz LM, Woloshin S. Resource use and guideline concordance in evaluation of pulmonary nodules for cancer: Too much and too little care. JAMA Intern Med. 2014;1746:871–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Spira A, Beane JE, Shah V et al. . Airway epithelial gene expression in the diagnostic evaluation of smokers with suspect lung cancer. Nat Med. 2007;133:361–366. [DOI] [PubMed] [Google Scholar]
- 7. Steiling K, van den Berge M, Hijazi K et al. . A dynamic bronchial airway gene expression signature of chronic obstructive pulmonary disease and lung function impairment. Am J Respir Crit Care Med. 2013;1879:933–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Blomquist T, Crawford EL, Mullins D et al. . Pattern of antioxidant and DNA repair gene expression in normal airway epithelium associated with lung cancer diagnosis. Cancer Res. 2009;6922:8629–8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Beane J, Sebastiani P, Liu G, Brody JS, Lenburg ME, Spira A. Reversible and permanent effects of tobacco smoke exposure on airway epithelial gene expression. Genome Biol. 2007;89:R201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chari R, Lonergan KM, Ng RT, MacAulay C, Lam WL, Lam S. Effect of active smoking on the human bronchial epithelium transcriptome. BMC Genomics. 2007;8:297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Beane J, Sebastiani P, Whitfield TH et al. . A prediction model for lung cancer diagnosis that integrates genomic and clinical features. Cancer Prev Res Phila Pa. 2008;11:56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Whitney D, Elashoff M, Porta-Smith K et al. . Derivation of a gene-expression classifier in a prospective study of patients undergoing bronchoscopy for suspicion of lung cancer. BMC Med Genomics. 2015;8:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Silvestri GA, Vachani A, Whitney D et al. . A bronchial genomic classifier for the diagnostic evaluation of lung cancer. N Engl J Med. 2015;3733:243–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ost DE, Ernst A, Lei X et al. . Diagnostic yield and complications of bronchoscopy for peripheral lung lesions. Results of the AQuIRE registry. Am J Respir Crit Care Med. 2015;1931:68–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tukey MH, Wiener RS. Population-based estimates of transbronchial lung biopsy utilization and complications. Respir Med. 2012;10611:1559–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang X, Sebastiani P, Liu G et al. . Similarities and differences between smoking-related gene expression in nasal and bronchial epithelium. Physiol Genomics. 2010;411:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Edge SB, Compton CC. The American Joint Committee on Cancer: The 7th edition of the AJCC Cancer Staging Manual and the future of TNM. Ann Surg Oncol. 2010;17:1471. [DOI] [PubMed] [Google Scholar]
- 18. Irizarry RA, Hobbs B, Collin F et al. . Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostat Oxf Engl. 2003;42:249–264. [DOI] [PubMed] [Google Scholar]
- 19. Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;81:118–127. [DOI] [PubMed] [Google Scholar]
- 20. Smyth GK. limma: Linear models for microarray data. In: Gentleman R, Carey VJ, Huber W, Irizarry RA, Dudoit S, eds. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Statistics for Biology and Health. New York: Springer; 2005:397–420. http://link.springer.com/chapter/10.1007/0‐387‐29362‐0_23. Accessed May 12, 2016. [Google Scholar]
- 21. Chen EY, Tan CM, Kou Y et al. . Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Subramanian A, Tamayo P, Mootha VK et al. . Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;10243:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics. 1988;443:837–845. [PubMed] [Google Scholar]
- 24. Agresti A. Categorical Data Analysis. New York: Wiley; 1990:350–354. [Google Scholar]
- 25. Leisenring W, Alonzo T, Pepe MS. Comparisons of predictive values of binary medical diagnostic tests for paired designs. Biometrics. 2000;562:345–351. [DOI] [PubMed] [Google Scholar]
- 26. Gould MK, Ananth L, Barnett PG. Veterans Affairs SNAP Cooperative Study Group. A clinical model to estimate the pretest probability of lung cancer in patients with solitary pulmonary nodules. Chest. 2007;1312:383–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lochhead P, Chan AT, Nishihara R et al. . Etiologic field effect: Reappraisal of the field effect concept in cancer predisposition and progression. Mod Pathol. 2015;281:14–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rosell R, Bivona TG, Karachaliou N. Genetics and biomarkers in personalisation of lung cancer treatment. The Lancet. 2013;3829893:720–731. [DOI] [PubMed] [Google Scholar]
- 29. Gu X, Wang J, Luo Y et al. . Down-regulation of miR-150 induces cell proliferation inhibition and apoptosis in non-small-cell lung cancer by targeting BAK1 in vitro. Tumor Biol. 2014;356:5287–5293. [DOI] [PubMed] [Google Scholar]
- 30. Singhal S, Miller D, Ramalingam S, Sun SY. Gene expression profiling of non-small cell lung cancer. Lung Cancer. 2008;603:313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang Y, Rathinam R, Walch A, Alahari SK. ST14 (suppression of tumorigenicity 14) gene is a target for miR-27b, and the inhibitory effect of ST14 on cell growth is independent of miR-27b regulation. J Biol Chem. 2009;28434:23094–23106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dong JT, Lamb PW, Rinker-Schaeffer CW et al. . KAI1, a metastasis suppressor gene for prostate cancer on human chromosome 11p11.2. Science. 1995;2685212:884–886. [DOI] [PubMed] [Google Scholar]
- 33. Adachi M, Taki T, Ieki Y, Huang CL, Higashiyama M, Miyake M. Correlation of KAI1/CD82 gene expression with good prognosis in patients with non-small cell lung cancer. Cancer Res. 1996;568:1751–1755. [PubMed] [Google Scholar]
- 34. Majhi PD, Lakshmanan I, Ponnusamy MP et al. . Pathobiological implications of MUC4 in non-small-cell lung cancer. J Thorac Oncol Off Publ Int Assoc Study Lung Cancer. 2013;84:398–407. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.