Abstract
The ability to assess toxicant exposures of 3D in vitro mammary models that recapitulate the tissue microenvironment can aid in our understanding of environmental exposure risk over time. Longitudinal studies of 3D model systems, however, are cumbersome and suffer from a lack of high-throughput toxicological assays. In this study, we establish a noninvasive and label-free optical coherence tomography (OCT)-based imaging platform for tracking exposure-response relationships in 3D human mammary epithelial organoid models. The OCT-based assay includes metrics that quantify organoid intracellular kinetic energy and cross-sectional area (CSA). We compare the results to those obtained using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) mitochondrial dye conversion assay. Both estrogen receptor (ER)-positive (MCF7) and ER-negative (MCF10DCIS.com) breast cell lines were studied, beginning one hour after exposure and continuing for several days. Six days of exposure to 17β-estradiol or the selective ER modulator 4-hydroxytamoxifen respectively increased or decreased MCF7 organoid CSA (p < .01), consistent with the role of estrogen signaling in ER-positive mammary epithelial cell proliferation. We also observed a significant decrease in the intracellular kinetic energy of MCF10DCIS.com organoids after 24 h of exposure to doxorubicin, a cytotoxic intercalating agent that causes DNA double-strand breaks (p < .01). MTT-based metabolic activity of MCF10DCIS.com organoids after 48 h of doxorubicin exposure decreased with dose in a similar manner as OCT-based energy metrics. These results demonstrate the feasibility of an OCT-based assay to quantify mammary epithelial cell toxicant response in vitro, noninvasively, longitudinally, and in the context of tissue microenvironments, providing a new high-throughput screening tool for toxicological studies.
Keywords: optical coherence tomography, mammary epithelial cell, 3D culture, toxicant response
In studies of mammary gland toxicology, there is increasing need for assays amenable to longitudinal high-throughput screening applications to capture time-dependent exposure responses, especially for environmental toxicants with long induction times. There is also a need for assays using physiologically relevant in vitro model systems. Three-dimensional in vitro organoid models that establish cell-extracellular matrix interactions recapitulate many features of in vivo tissue architecture and exhibit physiologically relevant tissue properties (Bissell, 2017; Lo et al., 2012). Importantly, as in vitro effects of exposures on 3D mammary epithelial organoids mirror environmental effects in the mammary gland (Bissell and Radisky, 2001), these models represent powerful tools for physiologically relevant studies of mammary gland toxicant responses.
The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay is commonly used in toxicant screens with cell viability as an endpoint. However, this assay has several limitations: it quantifies mitochondrial activity as a surrogate for cell viability, is unable to follow cells longitudinally, exhibits high variance, can be sensitive to culture volume and evaporation (van Tonder et al., 2015), and is challenging to implement in 3D model systems. Here we propose a novel optical coherence tomography (OCT)-based assay, which is complementary to the MTT assay and offers many advantages. OCT is a method of “optical histology,” providing depth-resolved imaging using near-infrared light scattering in a manner analogous to ultrasound imaging (Huang et al., 1991). The micrometer-scale resolution, millimeter-scale depth penetration, and high frame rate of OCT makes it uniquely suited for quantifying morphology and intracellular dynamics in 3D tissue cultures. As such, coherence imaging has garnered interest for studying 3D organoid models for which there are very few assays available, and existing assays are cumbersome (Chhetri et al., 2012; Nolte et al., 2012). Because OCT is noninvasive and does not require sample excision or killing, it enables longitudinal studies for assessing time-dependent cellular responses to drugs, toxicants, or other challenges. In combination with automated image processing methods, OCT has the potential to be high-throughput, with sufficient numbers for robust statistics. In previous studies, OCT was used to study the effects of culture time and co-culture cell seeding density on cell line-specific organoid intracellular motility (Oldenburg et al., 2015) and organoid morphology (Chhetri et al., 2012). Herein, we show the relevance of OCT-based assays for toxicology in comparison to the MTT assay, by rapidly measuring the effects of dose and exposure time simultaneously on 3D mammary tissue cultures.
There are many ways to parameterize the rich data available in time-lapse OCT imaging of 3D organoid models. Computing the OCT-based speckle fluctuation spectrum of individual organoids allows us to quantify the time scales of in-place intracellular motions (over seconds to minutes). Here we show how 2 parameters based upon this spectrum, the decay exponent (α) and fluctuation amplitude (M) defined previously (Oldenburg et al., 2015) can be effectively combined into a single “intracellular kinetic energy” metric, E = α × M. Using OCT we also track the cross-sectional area (CSA) of each organoid as a measure of cell proliferation, noting that additional morphological metrics including asphericity and lumen size have been used previously to define a premalignant phenotype (Chhetri et al., 2012). As such, the E-based kinetic energy and CSA-based cell proliferation metrics constitute a novel assay for 3D organoid model systems.
Here we apply OCT-based assays to (1) the effects of a cytotoxicant (Doxorubicin; Dox) and (2) estrogen responses in 3D mammary epithelial organoids. First, we selected the conventional chemotherapeutic Dox as a model cytotoxicant that has a well-characterized IC50 in other assays. Using this model compound we assess whether OCT-based assays are sensitive to cell killing by induced DNA double strand breaks. Second, because environmental estrogen exposure and endocrine disruptors are implicated in initiation and progression of breast cancers, particularly those of the luminal subtype (Fernandez and Russo, 2010; Trevino et al., 2015), we evaluate the responses of estrogen receptor (ER)+ MCF7 adenocarcinoma organoids to 17β-estradiol and 4-hydroxytamoxifen (4-OHT), an estrogen and selective ER modulator, respectively.
MATERIALS AND METHODS
Cell lines and reagents
Premalignant, triple-negative MCF10DCIS.com cells were purchased from Karmanos Cancer Institute (Detroit, Michigan) and were maintained in Dulbecco’s modified Eagle’s medium/F12 nutrient mix supplemented with 5% horse serum, 10 μg/ml insulin (GIBCO, Life Technologies, Carlsbad, California), 0.5 μg/ml hydrocortisone (Sigma-Aldrich, St. Louis, Missouri), 0.1 μg/ml cholera toxin (EMD Millipore, Darmstadt, Germany), 20 ng/ml Epidermal Growth Factor (Invitrogen, Life Technologies), and 50 units/ml penicillin and 50 U/ml streptomycin. MCF7 luminal adenocarcinoma cells were purchased from the American Type Culture Collection (Manassas, Virginia) and were maintained in RPMI 1640 medium (GIBCO) supplemented with 10% fetal bovine serum (Gemini Bio-Products, West Sacramento, California) and 50 U/ml penicillin and 50 units/ml streptomycin. All cells were maintained in a humidified incubator at 37 °C with 5% CO2. All compounds used for organoid culture exposures (Dox, 17β-estradiol, and 4-OHT) were purchased from Sigma-Aldrich. To study cytotoxic effects, we emphasized the MCF10DCIS.com cell line based on our previous work with this model. However, this model was not appropriate to study endocrine disruptors because it does not express ER. Therefore we used MCF7 cells for all endocrine disruptor studies.
3D culture and toxicant exposures
The 3D cultures used in this study were constructed as previously described (Chhetri et al., 2012; Oldenburg et al., 2015). Briefly, cells were seeded at a density of 30, 000 cells/cm3 into 275 μl of a biologically derived extracellular matrix comprised of a 1:1 ratio of collagen I to Matrigel (BD Biosciences, Franklin Lakes, New Jersey) (Johnson et al., 2007). In total 250 µl of the relevant culture medium was also dispensed to the apical side of each culture, and cells were refreshed every 2–3 days. For the OCT studies, MCF10DCIS.com and MCF7 cultures were grown for 10 and 14 days, respectively, to allow for formation and growth of organoids. At this time, cultures were exposed daily for up to 6 days with Dox (0, 1, or 10 µM), 4-OHT (0, 0.1, or 1 µM), or 17β-estradiol (0, 10, or 100). OCT imaging of toxicant-exposed cultures was performed prior to exposure (“0 h”), as well as at 1, 24, 48 h, and 6 days after the initial exposure. Note that media was removed from the surface of cultures immediately prior to each imaging session to reduce image artifacts, and was replaced with fresh media containing toxicant (effectively “re-dosing” cells) after imaging, and at least once per day up to 6 days. Cultures were fixed using 250 µl 4% paraformaldehyde solution overnight at 4 °C. Three cultures per condition (ie, dose) were prepared for each cell line and tracked longitudinally with OCT. OCT imaging captured an average of 6 organoids per culture, resulting in a total of n = 12–24 organoids measured at each dose and time point.
OCT imaging and extraction of metrics
The custom built, spectral-domain OCT system has been described in detail previously (Chhetri et al., 2011). Briefly, the system consists of a Ti: Sapphire laser with central wavelength of 800 nm and bandwidth of 120 nm. Images were collected using a custom spectrometer with a Dalsa Piranha line scan CCD camera, operated at 2 kHz for this study. Frames were collected at 0.876 ± 0.004 Hz in order to capture cellular dynamics (Oldenburg et al., 2015), and 300 frames were collected per time series. Representative OCT image stacks (1.8 × 1 mm) were used to generate movies at 10 × real time.
All image analyses were performed as described in our previous studies (Chhetri et al., 2012; Oldenburg et al., 2013, 2015). Briefly, manual segmentation aided via custom MATLAB scripts was employed to identify organoids within each OCT image stack for OCT fluctuation spectroscopy (Oldenburg et al., 2015). The speckle fluctuation spectra were fitted to an inverse power-law model, where α was the power exponent, and thus increasing values of α represent decreasing in-place intracellular motion at higher frequencies (Oldenburg et al., 2015). In addition, the overall fluctuation amplitude, M, was computed from a modified standard deviation normalized by pixel intensity, which provides a complementary metric of intracellular motion (Oldenburg et al., 2015). Importantly, in the current study, we propose a novel “intracellular kinetic energy” metric E = α × M that integrates the above metrics, noting that the kinetic energy of intracellular motion should be proportional to the area under the fluctuation spectral curve, and thus we might expect E as defined to scale with kinetic energy. In our definition, E is obtained using the scaled (0%–100%) values of α and M where 0% represents cell fixation and 100% represents live, untreated cells. The CSA of each organoid was manually segmented and measured from the stack-averaged OCT image using ImageJ, which provides an additional measurement of the overall size of organoids in response to exposure. The statistical method used in the study is multiple comparison t-test with Bonferroni correction.
MTT assay in 3D cultures
MCF10DCIS.com and MCF7 organoid cultures were exposed to a single dose of Dox at concentrations of 1.0 and 10.0 μM after 10 and 14 days of growth, respectively (n = 2 independent experiments). Vehicle (molecular-grade water)-treated and blank (cell-free) cultures were included as negative controls. After 48 h, an MTT dye-conversion assay was performed as follows: 37.5 μl of a 5.0 mg/ml MTT solution were added to the apical medium of each culture (Huyck et al., 2012), and cultures were incubated for 4 h at 37 °C in the dark. Cultures were then neutralized with 250 μl of DMSO and incubated overnight at 37 °C to initiate the dissolution of formazan crystals. The next day, 360 μl of 0.1 N HCl in isopropanol were added to each culture and mixed thoroughly, and the suspended cultures were transferred to individual Eppendorf tubes. Each well was washed twice with 200 μl of 0.1 N HCl in isopropanol to ensure all material was collected. Samples were then centrifuged at full speed (>20, 000 × g) for 30 s to pellet the insoluble 3D matrix and any remaining formazan precipitate, and the supernatant from each culture (containing dissolved formazan) was transferred to individual conical tubes. 200 μl of 0.1 N HCl in isopropanol were then added to each matrix/formazan pellet and mixed thoroughly to solubilize remaining formazan crystals. After centrifugation, the supernatant from each sample was again transferred to its respective conical tube. These steps (addition of 0.1 N HCl, centrifugation, and supernatant transfer) were repeated an equivalent number of times for each sample until no formazan precipitate remained in any sample. Supernatants were then centrifuged at 3000 × g for 1 min to pellet any insoluble 3D matrix components that may have been transferred to the conical tubes. Finally, supernatants were transferred into fresh conical tubes and vortexed thoroughly, and 100 μl of each sample were pipetted in triplicate to a clear 96-well plate. Absorbance (A) values were read on an ELx800 microplate reader (BioTek, Winooski, VT) at 570 and 650 nm. Blank-corrected absorbance values were calculated using Gen5 software (BioTek), and background-corrected values were obtained by subtracting blank-corrected A650 values from blank-corrected A570 values. The half maximal inhibitory concentrations (IC50) for Dox were calculated using GraphPad Prism version 7 (GraphPad Inc., La Jolla, California). Dose-response data were fitted with a variable slope sigmoidal curve using the equation , where x = the logarithm of the Dox dose and y = the response (corrected A570 value) (Sandhu et al., 2012). The top and bottom of each curve were set to 100% and 0%, respectively. All IC50 values represent 48 h of treatment with a single Dox dose.
RESULTS
Scaling of OCT Kinetic Energy Metrics for Each Cell Line
To understand the nature of the OCT-based assay, movies of live and fixed MCF10DCIS.com organoids are shown in Supplementary Figures 1A and 1B. The cross-sections of each of the organoids observed in these movies represent dozens of cells, and are speckled due to the coherent nature of OCT imaging. While it is not possible to resolve the boundaries of individual cells within each organoid, the motions are resolved to the approximate size of an individual cell (Oldenburg et al., 2015). As expected, more motion (greater kinetic energy) is observed in live relative to fixed cells, which is attributed to the ATP-driven, nanoscale movement of intracellular components. This kinetic energy is quantified by the parameters α and M for each organoid.
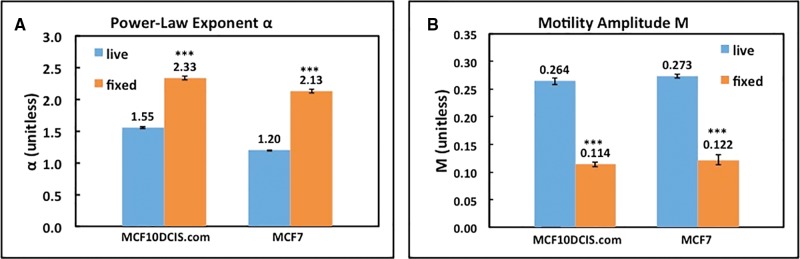
We utilized OCT to capture instantaneous measures of organoid kinetic energy and CSA. First, to establish the relevant scale for the OCT-based energy metrics, we measured α and M of mammary epithelial cell organoids before and 48 h after formalin fixation. The results, summarized in Figure 1, indicate that for both cell lines, α and M are significantly different (p < .0001) between live and fixed cells. There are also significant differences between α “live” of the 2 cell lines (p < .0001), which can be attributed to cell type-specific differences in morphology, invasive potential, and extracellular matrix interactions. Both cell lines exhibited an increase in α and a decrease in M after cell fixation, consistent with what we have reported previously (Oldenburg et al., 2015).
Figure 1.
Kinetic energy metrics α (A) and M (B) of live versus fixed MCF10DCIS.com and MCF7 cells. Error bars indicate mean ± SE. ***p < .0001 live versus fixed for each cell line.
The α and M values of live and fixed cells were used to establish a linear scale (0%–100%) corresponding to the raw values for each cell line, where the fixed values were assigned to 0%, and the live values assigned to 100%. This is a convenient way to define a typical dynamic range for each metric. This is helpful because, eg, the raw value of α increases with cell death from approximately 1.3 to 1.5 (depending on cell line) to >2.0, and scaling frees the user from unnecessarily having to remember this fact when assessing longitudinal data. However, it is important to note that 0% represents “death by fixation”, which is mechanistically different from toxicant-induced cell death. In addition, the scaled “live” values (100% representing control organoids without treatment) were determined using only the cultures presented in Figure 1, while variation in the live values in subsequent experiments may be observed depending on biologic variability in cell growth or incubation time (ie, live values of 120% are observed in data to follow).
Effect of Dox on Mammary Epithelial Cell Organoids via OCT and MTT: OCT Kinetic Energy Metrics Are Consistent with the MTT Assay
We next quantified the dose-dependent responses of MCF10DCIS.com organoids to Dox via both the OCT and MTT assays. Movies of representative OCT images (Supplementary Figs. 2A and 2B) provide a qualitative comparison of organoid activity at Day 6 between unexposed cultures and those exposed to the highest dose, where it can be observed that cells exposed to the highest dose exhibited significantly less intracellular motion. Figure 2 shows the scaled α, defined as , scaled M, defined as , where αlive = 1.55, αfixed= 2.33, Mlive= 0.264, Mfixed = 0.114 for MCF10DCIS.com, and αlive = 1.20, αfixed= 2.13, Mlive= 0.273, Mfixed = 0.122 for MCF7 (shown in Figure 1), and MTT-based metabolic activity of MCF10DCIS.com organoid cultures exposed to Dox at 48 h as a function of dose. We also computed the resulting kinetic energy E, defined as the product of scaled α and scaled M, to capture in one metric the effects of Dox on intracellular motion. According to the MTT data, MCF10DCIS.com organoids exhibit a dose-dependent loss of mitochondrial activity, a surrogate for viability, with an IC50 concentration of 3.24 × 10−6 M. Importantly, all of the OCT-based kinetic energy metrics, α, M, and E, decrease with concentration in a similar manner as the MTT values for MCF10DCIS.com organoids (Figure 2). We therefore conclude that α, M and E are good proxies for cytotoxicity in MCF10DCIS.com.
Figure 2.
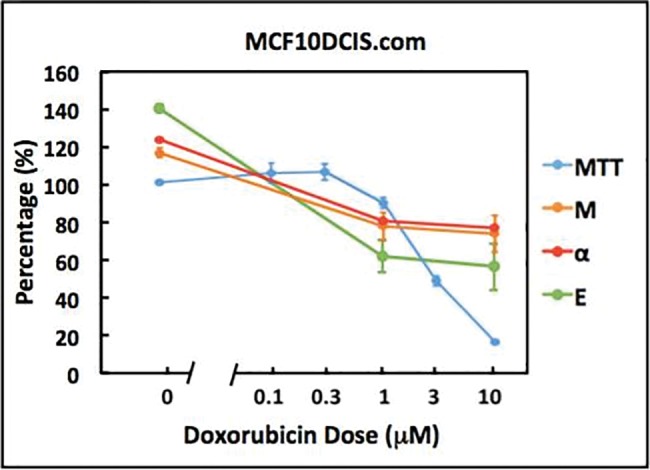
Comparison of the α, M, E, and MTT-based responses of MCF10DCIS.com cells to Dox at 48 h of exposure. Error bars represent mean ± SE.
OCT Kinetic Energy Metrics Reveal Time-Course of Cytotoxic Response
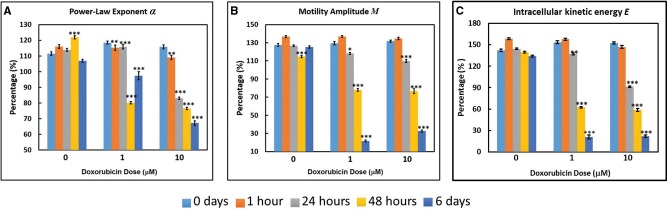
We next assessed whether the sensitivity of MCF10DCIS.com organoids to Dox-induced toxicity persists in longitudinal experiments. Figures 3A–C display the complete time- and dose-dependent responses of MCF10DCIS.com organoids to Dox exposure as measured longitudinally by OCT (The full tabular data of all measurements acquired is shown in Supplementary Table 1). Note that the scaled “live” values corresponding to 100% are defined from control organoids without treatment in Figure 1, and thus variations of live values (>100%) observed in Figure 3 arise from biologic variability in cell growth or incubation time. In pre-malignant organoids, α, M, and E decreased over exposure time for both 1 and 10 µM doses of Dox, where significant statistical differences between pre-exposure and exposed cultures were found (p < .001) at and above 1 h for α, and at and above 24 h for M and E. Although the data are presented here in time at varying doses, as an example, we also display the data of Figure 3C as dose-response in Supplementary Figure 3. The These OCT-based findings are attributed to Dox-induced cell death that suppresses intracellular motion, and demonstrate the utility of this assay in studies of chemosensitivity of mammary epithelial organoid models. The OCT-based assay also exhibits excellent stability in the control (0 µM Dox) data over the 6-day time-course, suggesting the broad applicability of this technique for measuring functional cellular changes longitudinally to avoid culture-to-culture variations. In comparison, collecting exposure time-dependent data with invasive assays such as the MTT would be subject to inter-culture variability. Importantly, because E closely tracks with α and M over multiple time points and doses, it is used as a simplified substitute for the effects of exposures on intracellular motion in the results to follow.
Figure 3.
Time- and dose-dependent responses of MCF10DCIS.com organoids to Dox as measured by the OCT-based α (A), M (B), and E (intracellular kinetic energy; C) metrics. *p < .01; **p < .001; ***p < .0001 relative to the pre-exposure time. Error bars represent mean ± SE.
OCT Kinetic Energy and Morphology Metrics Reveal Promotion and Inhibition of MCF7 Organoid Proliferation upon Estrogen and 4-OHT Exposures, Respectively
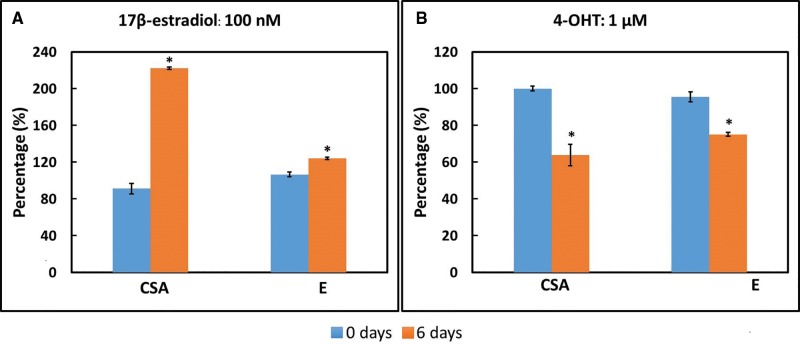
We believed the most straightforward test of the OCT platform was assessment of cell death, and having demonstrated the utility of the platform in this context, we next wondered whether we could up-modulate organoid intracellular kinetic energy by a stimulatory exposure. We therefore investigated the responses of the ER + cell line MCF7 to 17β-estradiol and 4-OHT as a function of time and dose. At the same time, organoid CSA was measured from OCT images to track changes in the size of organoids in response to exposure. This longitudinal CSA data can therefore be used as a surrogate measure of proliferation. Figure 4 shows a comparison of E and the CSA-based cell proliferation metric collected from MCF7 organoids exposed to 17β-estradiol and 4-OHT at the highest doses, comparing values obtained pre-exposure with those at the longest exposure time (6 days). (The full time- and dose-response data of MCF7 exposed to 17β-estradiol and 4-OHT are shown in Supplementary Figs. 4 and 5.) Upon exposure to 17β-estradiol, E and CSA of MCF7 organoids both increased, consistent with expected results (Figure 4A). Accordingly, E and CSA both decreased in association with 4-OHT exposure (Figure 4B). As such, E and CSA are both sensitive to the stimulatory effect of 17β-estradiol on MCF7 organoid proliferation, as well as the suppression of proliferation in 4-OHT-exposed MCF7 organoids.
Figure 4.
Comparison between CSA and E of MCF7 organoids exposed to 17β-estradiol and 4-OHT at the highest dose and longest exposure time (6 days). *p < .01 6 days versus pre-exposure (0 days).
Time-courses of the scaled CSA (defined as , where for each dose representing 100%) of MCF7 organoids exposed to 17β-estradiol and 4-OHT are shown in Figures 5A and 5B, respectively, with representative images of organoids at day 6 of exposure shown on the right-hand side. (Measured CSA data are shown in Supplementary Fig. 6.) As shown in Figure 5A, organoids exposed to the highest dose of 17β-estradiol were significantly larger in size (p < .01) at day 6 relative to the size before exposure. In experiments with 4-OHT, it is interesting to note that organoid size increased significantly (p < .01) at day 6 relative to day 0 for untreated controls (0 µM 4-OHT), which is attributed to the natural growth curve of the organoids for those cultures. In contrast, the effect of 4-OHT at the highest dose was sufficient to suppress growth, with a regression (p < .01) in organoid size at day 6 relative to pre-exposure. This suggests that 4-OHT exerts growth inhibitory effects on MCF7 cells, which is consistent with previous findings (Cormier and Jordan, 1989) and further supports the utility of noninvasive OCT imaging to longitudinal studies of toxicant exposures in physiologically relevant 3D culture systems.
Figure 5.
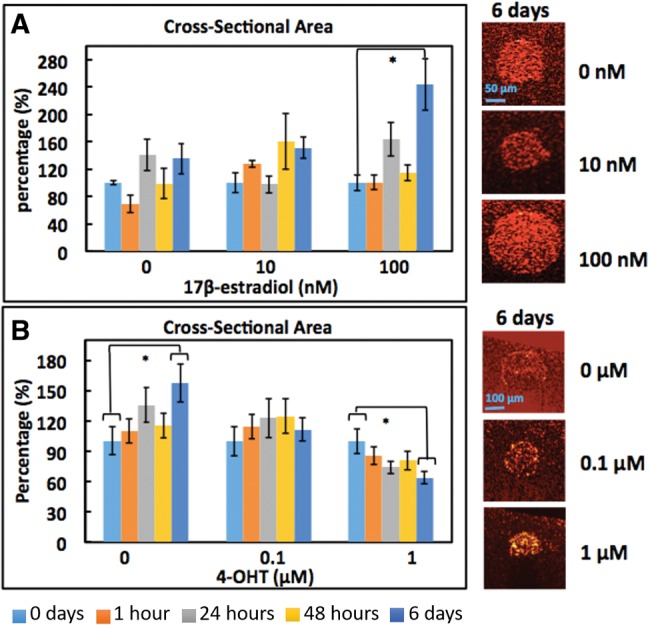
Time-course of MCF7 organoid CSA (scaled) upon exposure to 17β-estradiol (A) and 4-OHT (B). *p < .01 6 days relative to 0 days (before exposure). Error bars represent mean ± SE.
DISCUSSION
Many of the in vitro assays currently used in toxicological studies provide information about a single endpoint, including detection of cell apoptosis or cell death (eg, the TUNEL (Terminal deoxynucleotidyl transferase dUTP nick-end labeling) assay), the production of inflammatory mediators or components of the cell stress response (eg, the ELISA (enzyme-linked immunosorbent) assays), and cell viability or organelle activity (eg, the MTT assay). Moreover, inherent to many of these assays is the requirement for cell fixation, staining, and/or killing, precluding efficient longitudinal analysis. A widely used toxicological assay, the MTT assay measures cell mitochondrial activity where viable cells with active metabolism convert tetrazolium dye into colored formazan crystals. In contrast, OCT measurements are fundamentally biophysical in nature and provide metrics to image exposed cells longitudinally, tracking proliferation, kinetic energy, and morphogenesis in exposure-response studies. Interestingly, while OCT is a biophysical assay, we show in this study that it provides a functional endpoint relevant for a specific biomolecular pathway (ie, the ER modulators) and generalized cytotoxicity (ie, DNA damage). Given its ability to track this data longitudinally, in 3D, and in high-throughput, OCT provides a complement to existing toxicological assays.
Although all assays exhibit an intrinsic variability in their results, technical aspects of their implementation, especially for 3D cultures, can exacerbate variability. In applying the MTT assay to 3D cultures we noted that it was particularly difficult to collect formazan crystals trapped in the gel matrix. In comparison, longitudinal OCT avoids these technical challenges, minimizing batch variability by noninvasively imaging organoids through the closed culture lid. However, one source of variability in our experiments was the lack of controlled CO2 and temperature during the imaging scans, which typically lasted 7–10 min; this potential source of variability was controlled for by utilizing separate plates for each of 3 biological replicates in our study. In future work, the use of a commercially available microincubator can prevent this potential source of variability. Another important difference between the OCT-based assay and more traditional assays is that the spatial localization provided by OCT allows for measurement of individual organoids (in this study typically 6 per well), in comparison to a single measurement averaged over the well. By measuring multiple organoids individually, we can capture heterogeneity in cell behavior (organoid formation and toxicant responses).
In vivo studies are very important in understanding toxicant response; however, there is currently an international emphasis on replacing or reducing the use of animals in model-based research. Toward this end, 3D tissue culture models of the mammary gland provide in vivo-like context and can be employed to simulate contextual and micromechanical cues that are lacking in 2D cultures. The OCT-based platform for investigating toxicant exposure response in 3D models is able to maintain cells “in context”, and multiple time points can be collected longitudinally on a single experiment. Thus, OCT has the potential to reduce or replace the use of animals for toxicant screening. Another potential of OCT is for direct in vivo imaging of the mammary gland (Nguyen et al., 2009); while the OCT motility measurements described in this article require long imaging times that cause them to be susceptible to physiologic tissue motion, it may be possible to develop an OCT-based assay for longitudinal assessment of in vivo mammary gland response, which could drastically reduce the numbers of animals needed for toxicant screening.
In this work, we successfully revealed toxicant exposure-response relationships in 3D mammary epithelial organoid models using an OCT-based assay that quantifies aspects of kinetic energy (α, M, and E) and organoid morphology (CSA). Importantly, these OCT kinetic energy and morphology metrics can be obtained with any OCT system. Using this OCT-based assay we quantified the responses of ER- cells to a triple-negative breast cancer treatment drug Dox, revealing the time-course of the response, with statistically significant differences between pre-exposed and exposed cultures. These findings are attributed to cell death during Dox exposure that suppresses intracellular motion, and the trends in all the metrics (α, M, and E) were consistent with the MTT assay. In addition, significant changes in E and CSA of ER + organoids to estrogen and tamoxifen have also been observed. This establishes the utility of OCT as a high-throughput screening tool for assessing the effects of toxicants on mammary gland, and sets the stage for the future use of OCT in 3D longitudinal studies of environmental breast toxicants.
SUPPLEMENTARY DATA
Supplementary data are available at Toxicological Sciences online.
FUNDING
National Cancer Institute at the National Institutes of Health (R21 CA172904 to A.L.O.); National Institute of Environmental Health Sciences at the National Institutes of Health (P30 ES010126); National Science Foundation (CBET 1351473 to A.L.O.).
Supplementary Material
REFERENCES
- Bissell M. J. (2017). Goodbye flat biology - time for the 3rd and the 4th dimensions. J. Cell Sci. 130, 3–5. 10.1242/jcs.200550. 10.1242/jcs.200550 [DOI] [PubMed] [Google Scholar]
- Bissell M. J., Radisky D. (2001). Putting tumours in context. Nat. Rev. Cancer 1, 46–54. 10.1038/35094059. 10.1038/35094059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhetri R. K., Kozek K. A., Johnston-Peck A. C., Tracy J. B., Oldenburg A. L. (2011). Imaging three-dimensional rotational diffusion of plasmon resonant gold nanorods using polarization-sensitive optical coherence tomography. Phys. Rev. E Stat. Nonlin. Soft. Matter Phys. 83, 040903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhetri R. K., Phillips Z. F., Troester M. A., Oldenburg A. L., Cukierman E. (2012). Longitudinal study of mammary epithelial and fibroblast co-cultures using optical coherence tomography reveals morphological hallmarks of pre-malignancy. PLoS One 7, e49148.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormier E. M., Jordan V. C. (1989). Contrasting ability of antiestrogens to inhibit MCF-7 growth stimulated by estradiol or epidermal growth factor. Eur. J. Cancer Clin. Oncol. 25, 57–63. 10.1016/0277-5379(89)90051-5 [DOI] [PubMed] [Google Scholar]
- Fernandez S. V., Russo J. (2010). Estrogen and xenoestrogens in breast cancer. Toxicol. Pathol. 38, 110–122. 10.1177/0192623309354108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D., Swanson E., Lin C., Schuman J., Stinson W., Chang W., Hee M., Flotte T., Gregory K., Puliafito C., et al. and (1991). Optical coherence tomography. Science 254, 1178–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyck L., Ampe C., Van Troys M. (2012). The XTT cell proliferation assay applied to cell layers embedded in three-dimensional matrix. Assay Drug Dev. Technol. 10, 382–392. 10.1089/adt.2011.391. 10.1089/adt.2011.391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K. R., Leight J. L., Weaver V. M. (2007). Demystifying the effects of a three-dimensional microenvironment in tissue morphogenesis. Methods Cell Biol. 83, 547–583. 10.1016/S0091-679X(07)83023-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo A. T., Mori H., Mott J., Bissell M. J. (2012). Constructing three-dimensional models to study mammary gland branching morphogenesis and functional differentiation. J. Mammary Gland Biol. Neoplasia 17, 103–110. 10.1007/s10911-012-9251-7. 10.1007/s10911-012-9251-7 [DOI] [PubMed] [Google Scholar]
- Nguyen F. T., Zysk A. M., Chaney E. J., Kotynek J. G., Oliphant U. J., Bellafiore F. J., Rowland K. M., Johnson P. A., Boppart S. A. (2009). Intraoperative evaluation of breast tumor margins with optical coherence tomography. Cancer Res. 69, 8790–8796. 10.1158/0008-5472.CAN-08-4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte D. D., An R., Turek J., Jeong K. (2012). Tissue dynamics spectroscopy for phenotypic profiling of drug effects in three-dimensional culture. Biomed. Opt. Express 3, 2825–2841. 10.1364/BOE.3.002825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenburg A. L., Yu X., Gilliss T., Alabi O., Taylor R. M. 2nd, Troester M. A. (2015). Inverse-power-law behavior of cellular motility reveals stromal-epithelial cell interactions in 3D co-culture by OCT fluctuation spectroscopy. Optica 2, 877–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenburg A. L., Chhetri R. K., Cooper J. M., Wu W.-C., Troester M. A., Tracy J. B. (2013). Motility-, autocorrelation-, and polarization-sensitive optical coherence tomography disciminates cells and gold nanorods within 3D tissue cultures. Opt Lett. 38(15), 2923–2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu R., Rivenbark A. G., Coleman W. B. (2012). Enhancement of chemotherapeutic efficacy in hypermethylator breast cancer cells through targeted and pharmacologic inhibition of DNMT3b. Breast Cancer Res. Treat. 131, 385–399. 10.1007/s10549-011-1409-2. 10.1007/s10549-011-1409-2 [DOI] [PubMed] [Google Scholar]
- Trevino L. S., Wang Q., Walker C. L. (2015). Hypothesis: Activation of rapid signaling by environmental estrogens and epigenetic reprogramming in breast cancer. Reprod. Toxicol. 54, 136–140. 10.1016/j.reprotox.2014.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Tonder A., Joubert A. M., Cromarty A. (2015). Limitations of the 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyl-2H-tetrazolium bromide (MTT) assay when compared to three commonly used cell enumeration assays. BMC Res. Notes 8, 47. 10.1186/s13104-015-1000-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.