Abstract
Background: Clinical success of poly(ADP-ribose) polymerase inhibitors (PARPi) has been limited to repair-deficient cancers and by resistance. Oncolytic herpes simplex viruses (oHSVs) selectively kill cancer cells, irrespective of mutation, and manipulate DNA damage responses (DDR). Here, we explore potential synthetic lethal-like interactions between oHSV and PARPi.
Methods: The efficacy of combining PARPi, oHSV MG18L, and G47Δ in killing patient-derived glioblastoma stem cells (GSCs) was assessed using cell viability assays and Chou-Talalay synergy analysis. Effects on DDR pathways, apoptosis, and cell cycle after manipulation with pharmacological inhibitors and lentivirus-mediated knockdown or overexpression were examined by immunoblotting and FACS. In vivo efficacy was evaluated in two GSC-derived orthotopic xenograft models (n = 7–8 per group). All statistical tests were two-sided.
Results: GSCs are differentially sensitive to PARPi despite uniform inhibition of PARP activity. oHSV sensitized GSCs to PARPi, irrespective of their PARPi sensitivity through selective proteasomal degradation of key DDR proteins; Rad51, mediating the combination effects; and Chk1. Rad51 degradation required HSV DNA replication. This synthetic lethal-like interaction increased DNA damage, apoptosis, and cell death in vitro and in vivo. Combined treatment of mice bearing PARPi-sensitive or -resistant GSC-derived brain tumors greatly extended median survival compared to either agent alone (vs olaparib: P≤.001; vs MG18L: P = .005; median survival for sensitive of 83 [95% CI = 77 to 86], 94 [95% CI = 75 to 107], 102 [95% CI = 85 to 110], and 131 [95% CI = 108 to 170] days and for resistant of 54 [95% CI = 52 to 58], 56 [95% CI = 52 to 61], 62 [95% CI = 56 to 72], and 75 [95% CI = 64 to 90] days for mock, PARPi, oHSV, and combination, respectively).
Conclusions: The unique oHSV property to target multiple components of DDR generates cancer selective sensitivity to PARPi. This combination of oHSV with PARPi is a new anticancer strategy that overcomes the clinical barriers of PARPi resistance and DNA repair proficiency and is applicable not only to glioblastoma, an invariably lethal tumor, but also to other tumor types.
Alteration in DNA damage responses (DDRs), including DNA repair and cell cycle arrest, is a hallmark of cancer, providing targets for cancer therapy (1). Inhibitors of poly(ADP-ribose) polymerase (PARP) are an example of successfully targeting DDR for clinical efficacy in cancer with homologous recombination (HR) repair deficiencies (2). PARP is required for base excision repair (BER) and DNA single-strand break (SSB) repair (2). PARP inhibition leads to double-strand DNA breaks (DSBs). DSBs are repaired by error-free HR, or error-prone nonhomologous end joining (NHEJ) and PARP-dependent alternate NHEJ (Alt-NHEJ) (3). If HR is deficient, DSBs are repaired by NHEJ and Alt-NHEJ, which result in genomic instability. This is the basis for synthetic lethality of PARP inhibitors (PARPis) in HR-defective tumors and their development as cancer therapeutics (2,4). However, challenges for PARPi therapy remain: improving their efficacy in HR-deficient tumors, overcoming drug resistance, and expanding their use to tumors without characterized defects in HR.
Viruses, including herpes simplex virus (HSV), are actively involved in manipulating DDR (5), providing a rationale for combination with PARPi. Oncolytic HSV (oHSV), a new class of anticancer agent, is genetically engineered to selectively replicate in and kill cancer cells, amplifying and spreading within the tumor but not normal tissue (6). oHSVs have been safely administered to glioblastoma (GBM) patients (7), and oHSV talimogene laherparepvec was recently approved by the US Food and Drug Administration (FDA) for recurrent melanoma (8). GBM, a primary malignant brain tumor, has a median survival of about 15 months, which has not markedly improved (9). GBM stem cells (GSCs) are a subpopulation of highly tumorigenic cells with stem cell–like properties (10,11). GSCs maintain the genotypes/phenotypes of the patient's tumors from which they were isolated, including heterogeneous histopathology (12,13). They are important in disease progression, recurrence, and resistance to therapy, and thus they are a critical therapeutic target (10,14,15).
In this study, we investigated the therapeutic interaction between oHSV and PARPis in killing patient-derived PARPi-sensitive and -resistant GSCs and inhibiting tumor growth and how the manipulation of DDR pathways contributes to combinatorial efficacy.
Methods
Cells and Viruses
Human GSCs were isolated as previously described and cultured in EF20 medium with EGF and FGF2 (16). OHSV G47Δ (γ34.5Δ, ICP47-Us11 promoterΔ, ICP6-, LacZ+) (17), and MG18L (Us3Δ, ICP6-, LacZ+) (18) were grown and titered on Vero cells.
Cell Assays
Cell viability was measured by MTS assay after dissociated cells plated in triplicate in 96-well plates were treated at 37°C for six days. Chou-Talalay analysis was performed as described (19). For cell cycle analysis, fixed cells were stained with propidium iodide and analyzed with flow cytometry.
Immunoblotting
Cells were lysed in radioimmunoprecipitation (RIPA) buffer (Boston BioProducts) with protease and phosphatase inhibitors (Roche Diagnostics, Indianapolis, IN). After electrophoresis, proteins were transferred to PVDF membranes and incubated with primary and secondary antibodies.
In Vivo Experiments
Female athymic mice (age 7–8 weeks; National Cancer Institute, Frederick, MD) were intracerebrally implanted with GSCs as in (16) and randomly assigned to four groups. Olaparib (50 mg/kg) or vehicle was administrated intraperitoneally, and MG18L or PBS was injected intratumorally. Mice were monitored for clinical symptoms, and moribund mice were killed and the presence of tumor confirmed. All in vivo procedures were approved by the Subcommittee on Research Animal Care at Massachusetts General Hospital.
Statistical Analysis
In vitro studies (relative cell viability) were analyzed using generalized linear models, and least square means after adjustment per group are presented for the group estimates. P values were adjusted for multiple comparisons within the models using Tukey adjustment. Unpaired t test was used as indicated for two-group comparisons. Survival was analyzed by Kaplan-Meier plot, and log-rank (Mantel-Cox) test was used to compare between survival curves. Prism (GraphPad), MedCalc, and SAS software were used for analysis. P values of less than .05 were considered statistically significant. All statistical tests were two-sided.
Detailed information on these and all other methods can be found in the Supplementary Materials (available online).
Results
Sensitivity of GSCs to PARPi
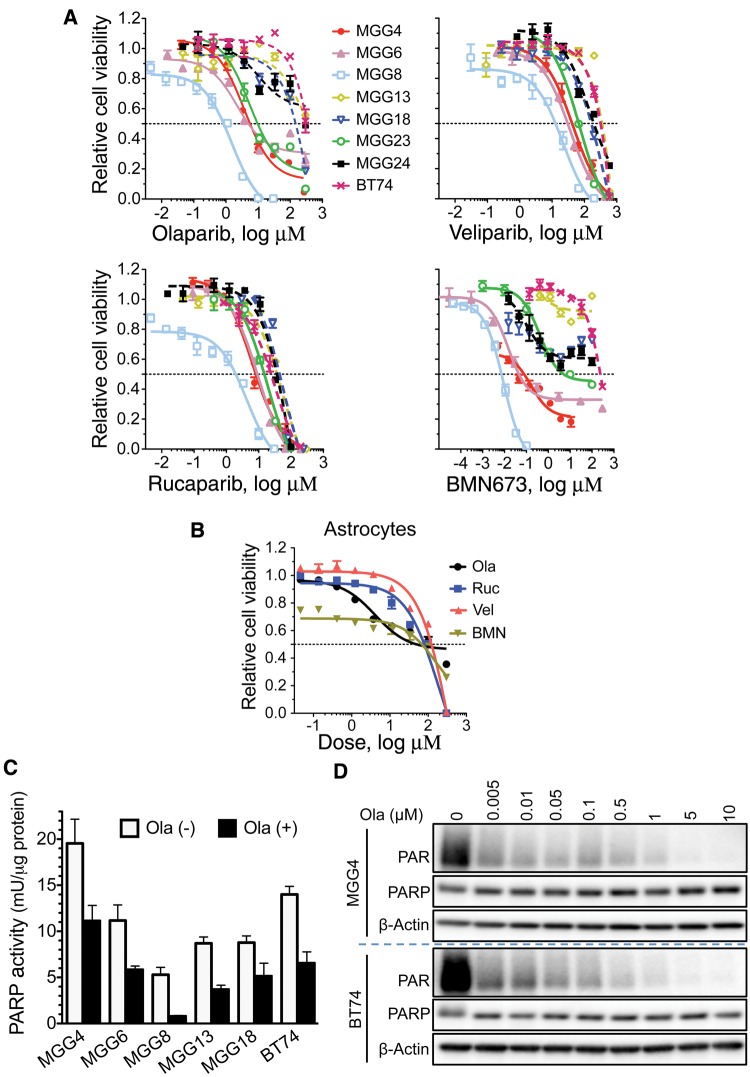
Patient-derived GSCs exhibited differential sensitivity to olaparib (Lynparza, AZD2281), an approved, potent PARP1/2 inhibitor currently in clinical trial for GBM (2). Four GSCs (MGG4, MGG6, MGG8, and MGG23) were sensitive (IC50 < 20 μM), close, or below the maximal plasma concentration (6 μg/mL or 14 μM) in patients receiving the standard 400 mg dose (20), and four (MGG13, MGG18, MGG24, and BT74) were resistant (IC50 > 100 μM) (Figure 1A;Supplementary Table 1, available online). A similar trend in sensitivity was seen with additional PARP1/2 inhibitors veliparib, rucaparib, and BMN673, which are in clinical trial, with MGG23 somewhat intermediate in sensitivity (Figure 1A;Supplementary Table 1, available online). Importantly, normal human astrocytes were relatively resistant to all four PARPis (Figure 1B;Supplementary Table 1, available online). GSC resistance was not due to PARPi inactivation as olaparib similarly inhibited PARP in GSCs, as measured by PARP enzymatic activity (Figure 1C) and PARylation (Figure 1D). For subsequent experiments, we used olaparib as a representative PARPi.
Figure 1.
Effect of poly(ADP-ribose) polymerase inhibitors (PARPis) on glioblastoma stem cell (GSC) cytotoxicity and PARP activity. A) Dose response curves for PARPis. GSCs were plated at 5000 cells/well, except MGG24 and BT74 at 8000 cells/well, and treated the next day with indicated PARPis (olaparib, veliparib, rucaparib, and BMN673) at different doses for six days, followed by MTS assay for cell viability. Nonlinear regression curves (log(inhibitor) vs response) were plotted. B) Dose response curves for PARPis on normal human astrocytes. Cells were plated at 3000 cells/well and treated as in (A). C) PARP activity, as measured by PARP Assay Kit, was inhibited in all GSCs after olaparib treatment (Ola (+), 30 μM) for 24 hours. Data are represented as mean ± SD. D) PARylated proteins (PAR), a measure of PARP activity, were detected by immunoblotting after treatment with indicated doses of olaparib for 24 hours in MGG4 and BT74. β-actin is loading control. Ola = olaparib; PARP = poly(ADP-ribose) polymerase.
Interaction of oHSV with PARPi in Killing Sensitive and Resistant GSCs in Vitro
We hypothesized that oHSV would enhance PARPi efficacy. GSCs vary in their sensitivity to killing by oHSV, either MG18L, deficient in blocking virus-induced apoptosis, or G47Δ, currently in clinical trial for recurrent glioma (7,17–19), but none were resistant and there was no association with PARPi sensitivity (Figure 2A;Supplementary Table 1, available online). We then tested whether oHSV altered PARPi sensitivity. A fixed dose of MG18L with a range of olaparib doses, or a fixed dose of olaparib with a range of MG18L doses in PARPi-sensitive (MGG4 and MGG23) and -resistant GSCs (BT74 and MGG24), shifted the combination dose response curves to lower doses in all cases compared with those for the single treatment (Figure 2, B–E, left and middle; Supplementary Figure 1A, available online). In the PARPi-sensitive GSCs, the combination of MG18L or G47Δ with olaparib was synergistic, as determined by Chou-Talalay analysis (Figure 2, B and C, right). Nontoxic doses of olaparib sensitized PARPi-resistant GSCs to MG18L and G47Δ (Figure 2, D and E, middle, right; Supplementary Figure 1B, available online), mediating a synthetic lethal-like effect. The combination effect was not due to virus replication as virus yield was not altered by olaparib at high or low multiplicity of infection (MOI) (Supplementary Figure 2, A and B, available online). Importantly, the combination did not decrease viability in normal human astrocytes (Figure 2F).
Figure 2.
The interaction of olaparib with MG18L and G47Δ in killing poly(ADP-ribose) polymerase inhibitor (PARPi)–sensitive and –resistant glioblastoma stem cells (GSCs). A) Dose response curves for MG18L (left) and G47Δ (right) in the indicated GSCs, determined as in Figure 1A. Combination of olaparib and MG18L or G47Δ in MGG4 (B), MGG23 (C), BT74 (D), MGG24 (E), and normal astrocytes (F). Left: The fixed dose of MG18L was MOI = 0.04, 0.001, 0.05, and 0.05 for (B), (C), (D), and (E), respectively, indicated with blue arrow. Middle: The fixed dose of olaparib was 1, 1, 10, and 10 µM for (B), (C), (D), and (E), respectively, indicated with brown arrow. Right: B and C) Interaction between olaparib (Ola) and MG18L or G47Δ in MGG4 and MGG23, as determined by the Chou-Talalay median effect method (19,40). Combination index < 1, = 1, and > 1 indicates synergistic, additive, and antagonistic interactions, respectively. Right: D and E) Combination of olaparib and G47Δ in BT74 (Ola = 10 μM) and MGG24 (Ola = 5 μM). Increasing virus dose is statistically significantly different from the previous dose, with or without olaparib (P < .0001). *P = .004; †P < .001; ‡P < .0001 (multiple comparisons test, Tukey). F) Combination of olaparib (10 μM, Ola (+)) and MG18L or G47Δ (0.1 MOI) in astrocytes. Cell viability was determined by MTS assay after six-day treatment and represented as mean ± SD. All statistical tests were two-sided. MOI = multiplicity of infection; Ola = olaparib; PARP = poly(ADP-ribose) polymerase.
Effect of oHSV and PARPi on DDR and Apoptosis
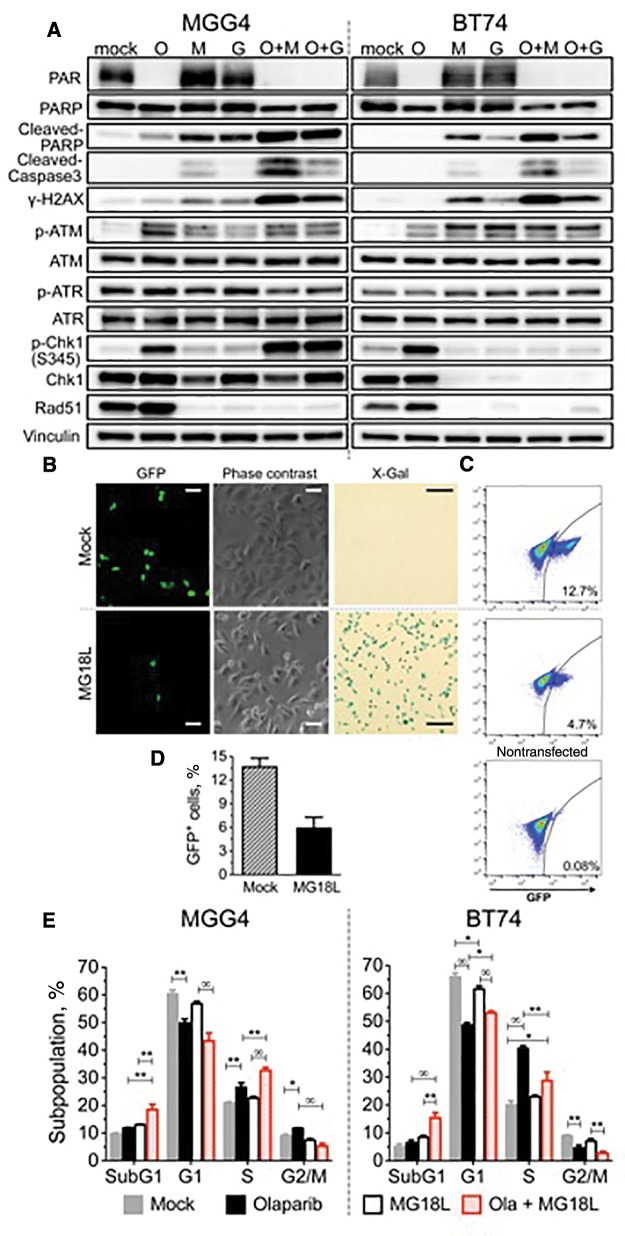
The effect of treatment on DDR pathways was examined. oHSV did not alter olaparib's inhibition of parylation (PAR) (Figure 3A;Supplementary Figure 2C, available online). We previously showed that G47Δ induces DSBs in infected GSCs (19). Both G47Δ and MG18L induced DSBs, as detected with γH2AX, in PARPi-sensitive and -resistant GSCs (Figure 3A;Supplementary Figure 2C, available online). DSBs accumulated at late times (Supplementary Figure 2D, available online), suggesting impaired DNA repair. Apoptosis, as assessed by cleaved-caspase 3 and cleaved-PARP, was induced similarly to γH2AX, with MG18L inducing more apoptosis than G47Δ (Figure 3A;Supplementary Figure 2C, available online). Both DNA damage and apoptosis were greatly and further increased after combination treatment in all GSCs (O + M, O + G) (Figure 3A;Supplementary Figure 2C, available online).
Figure 3.
Effect of oncolytic herpes simplex virus, poly(ADP-ribose) polymerase inhibitor (PARPi), and combination on DNA damage responses and cell cycle. A) PARPi-sensitive MGG4 (left) and PARPi-resistant BT74 (right) cells were treated with olaparib (O; 10 µM for MGG4 and 30 µM for BT74) or vehicle (mock) for 48 hours and then mock-infected or infected with MG18L (M) or G47Δ (G; MOI = 1) or in combination (O + M, O + G). Cells were harvested for immunoblotting at 30 hours after infection. β-actin as loading control. B) Effect of MG18L on homologous recombination. DR-GFP-transduced U2OS cells were transfected with a plasmid expressing I-SceI, and 24 hours later infected with MG18L (1MOI) or mock (PBS), followed 24 hours later by analysis of GFP-positive cells with fluorescence (left) and phase-contrast microscopy (middle). X-gal staining performed at 16 hours after MG18L infection (right), showing that almost all the cells were infected. Scale bars = 1 μm for GFP and phase contrast and 5 μm for X-gal. C) Representative fluorescence-activated cell sorting (FACS) analysis of GFP-positive cells (gated right quadrant, percent positive) from mock (top), MG18L infected (middle), and control nontransfected (lower). Total of 1X 105 cells were analyzed for each sample. D) Quantification of GFP-positive cells analyzed by FACS as in (C) from three independent experiments. Data represented as mean ± SD; P = .002 (two-sided unpaired t test). E) Cell cycle analysis of treated MGG4 (left) and BT74 (right). Cells were treated as indicated with olaparib (3 μM for MGG4 and 30 μM for BT74) and/or MG18L (MOI = 0.5) and cell cycle phases determined after 24 hours by FACS. Values are the mean of three independent experiments and represented as mean ± SD. *P < .01; **P < .001; ∞P< .0001. P values of .01 or greater are not indicated (multiple comparisons test, Tukey). In MGG4: mock vs olaparib for G2/M, P = .004. In BT74: mock vs MG18L and olaparib vs Ola+MG18L for G1, P = .005; mock vs Ola+MG18L for S, P = .002. All statistical tests were two-sided. Ola = olaparib; PARP = poly(ADP-ribose) polymerase.
ATM and ATR, DNA damage protein kinases activated by DSBs and SSBs, respectively, initiate HR repair and cell cycle checkpoints (21). ATM was activated (p-ATM) by olaparib or virus alone, but not increased with combination (Figure 3A). Activated ATR phosphorylates Chk1, a key component in DNA damage–induced cell cycle arrest and HR repair (22). P-Chk1 was strongly induced by olaparib alone in all GSCs except MGG24, and by the combination with oHSV only in MGG4 (Figure 3A;Supplementary Figure 2C, available online). oHSV infection induced Chk1 loss in MGG23, BT74, and MGG24 (Figure 3A;Supplementary Figure 2C, available online). Rad51 is a recombinase necessary for HR and a downstream target of ATM and ATR (3). Rad51 protein levels were upregulated by olaparib alone and surprisingly totally eliminated in all tested GSCs within about 30 hours of infection with MG18L or G47Δ (Figure 3A;Supplementary Figure 2, C and E, available online).
Loss of Rad51 would disable HR, so we examined whether oHSV inhibits HR in tumor cells using a I-SceI-induced DSB/HR reporter assay (23). For these studies, we used DR-GFP stably transduced U20S cells. oHSV induces Rad51 loss in U20S cells (Supplementary Figure 4B, available online). HR repair of the introduced DSBs generates functional GFP compared with non-I-SceI-transfected cells (Figure 3, B and C; mock vs nontransfected). MG18L infection of most of the cells (Figure 3B, X-gal), 24 hours after transfection, statistically significantly reduced HR repair by over 50% (Figure 3, B–D, GFP+).
Effect of oHSV and PARPi on Cell Cycle of PARPi-Sensitive and -Resistant GSCs
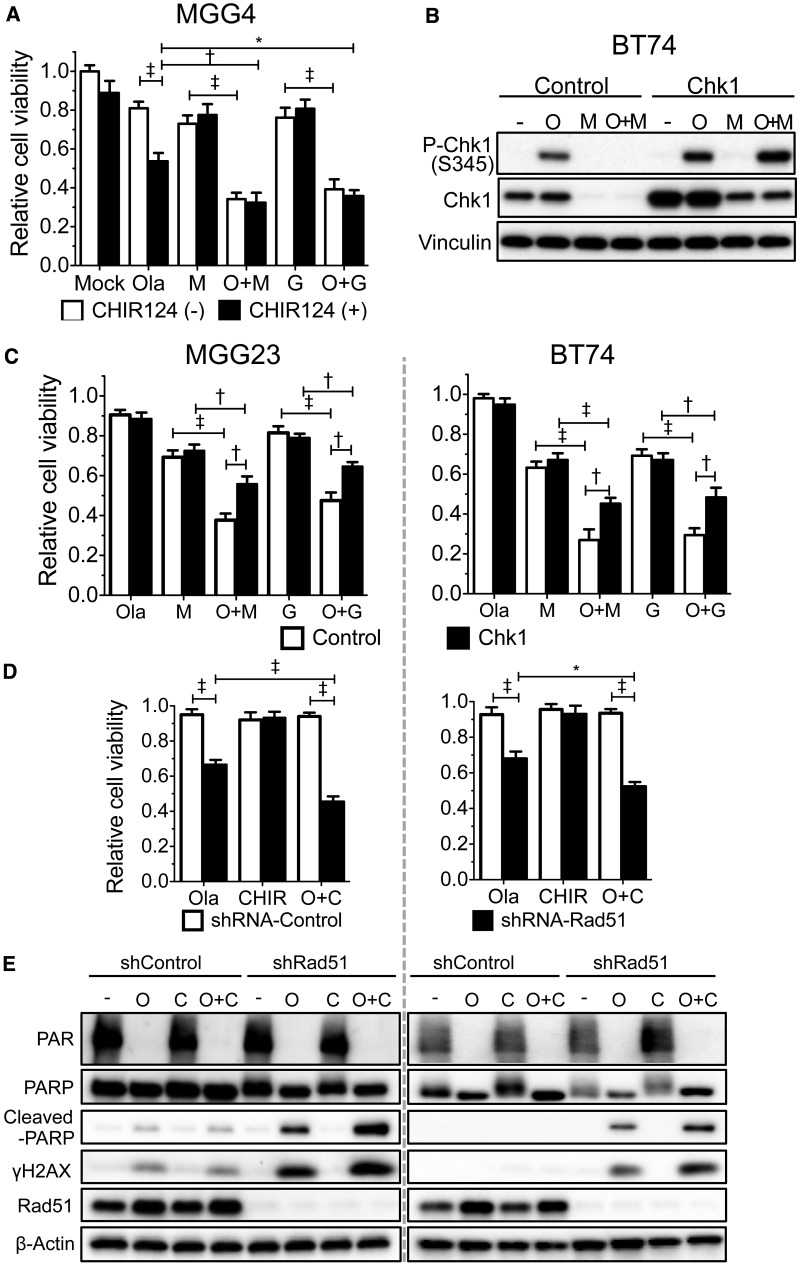
P-Chk1 phosphorylates multiple effectors for cell cycle arrest (22), so we examined treatment effects on cell cycle. Olaparib alone induced a decrease in G1 cells and some cell cycle arrest in S and G2/M phases in MGG4, while in BT74 there was a large S phase arrest and decrease in G2/M (Figure 3E;Supplementary Figure 3A, available online), which may enable time for DNA repair. MG18L did not statistically significantly alter S or G2/M phase cells (Figure 3E). However, olaparib-induced S phase arrest was further increased by MG18L in MGG4 and decreased in BT74 (Figure 3E), likely a reflection of the induction or loss of p-Chk1 (Figure 3A). In both GSCs, the combination greatly increased the sub-G1 population, indicative of dying and apoptotic cells (Figure 3E). CHIR-124, a potent and selective Chk1 kinase inhibitor (24), only abrogated cell cycle arrest when the treatment activated Chk1 (Supplementary Figure 3B, available online).
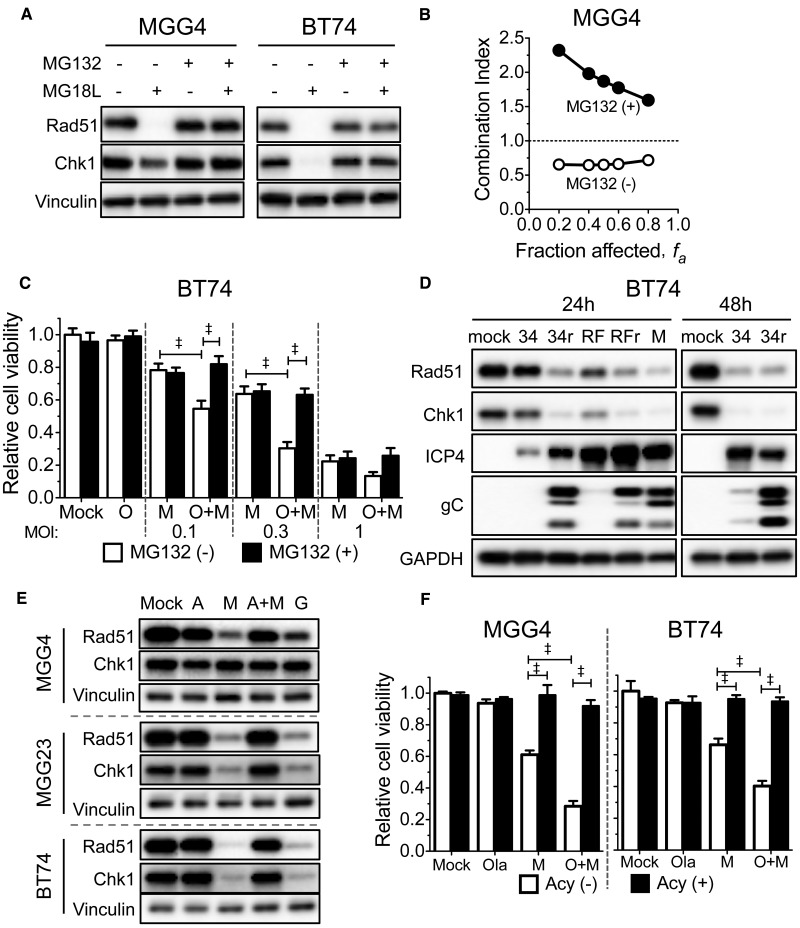
Proteasomal Degradation of Rad51 and Chk1 and oHSV Life Cycle
To determine the mechanism of protein loss, we used MG132, a reversible proteasome inhibitor (25). MG132 completely blocked MG18L-induced degradation of Rad51 and Chk1 (Figure 4A) and eliminated synergy between olaparib and MG18L in MGG4 (Figure 4B) and sensitization in BT74 (Figure 4C) at a relatively nontoxic dose that did not affect MG18L cytotoxicity (Supplementary Figure 4A, available online). HSV ICP0 is an immediate-early gene encoding a RING finger E3 ubiquitin ligase that mediates the degradation of host proteins, including DDR (26). However, ICP0 mutant viruses HSV 7134 (27) and KOS RFm (28), with delayed replication kinetics (HSV ICP4 and late gC expression), induced degradation of Rad51 and Chk1 after infection of BT74 (Figure 4D) and U20S cells (Supplementary Figure 4B, available online) like the rescued wild-type viruses (7134R and KOS RFr), demonstrating that ICP0 is not necessary for degradation. Rad51 RNA is not selectively degraded after MG18L infection (Supplementary Figure 5A, available online).
Figure 4.
The role of Rad51 and Chk1 degradation and viral DNA replication on synergy between oncolytic herpes simplex viruses (oHSV) and olaparib. A) MG132 blocks MG18L-induced degradation of Rad51 and Chk1. MGG4 and BT74 cells were treated with MG18L (MOI = 1) and/or MG132 (1 μM) as indicated for 30 hours before harvesting for immunoblot analysis and probed with antibodies to Rad51, Chk1, and vinculin as loading control. B) Requirement for proteasomal activity on synergy. MGG4 cells were treated with olaparib, MG18L, or the combination in the absence (open circles) or presence of 0.05 μM MG132 (+MG132, filled circles) for six days. Cell viability was measured by MTS assay, and the combination index determined. Combination Index < 1, = 1, and > 1 indicates synergistic, additive, and antagonistic interactions, respectively. C) BT74 cells were treated with olaparib (O; 10 μM), MG18L (M; at the doses indicated), or combination (O + M), in the absence (MG132 (-)) or presence of MG132 (0.1 μM; MG132 (+)) for six days, followed by MTS assay for cell viability, represented as mean ± SD. ‡P < .0001 (multiple comparisons tests, Tukey) between indicated pairs. D) BT74 cells were mock-infected or infected with HSV ICP0 mutants 7134 (34) and ICP0-RING finger domain mutant virus KOS RFm (RF), or rescued wild-type HSV 7134R (34r) and KOS RFr (RFr), or MG18L (M) with MOI = 10, except 7134R at 48 hours (MOI = 1). Cells were harvested for immunoblotting at 24 hours or 48 hours after infection. Membranes were probed with antibodies to Rad51, Chk1, ICP4, gC, and GAPDH as loading control. E) Acyclovir treatment blocks MG18L-induced Rad51 and Chk1 degradation. Glioblastoma stem cells (MGG4, MG23, BT74) were treated with MG18L (M, A + M; MOI = 1), G207 (G; MOI = 1), and/or acyclovir (A, A + M; 10 μM), for 30 hours before harvesting. Membranes were probed with antibodies to Rad51, Chk1, and vinculin as loading control. F) Acyclovir abrogates the combination effect of oHSV with poly(ADP-ribose) polymerase inhibitor. MGG4 (left) and BT74 (right) cells were treated with olaparib (O; 1 μM for MGG4 and 10 μM for BT74), MG18L (M; 0.05 MOI for MGG4 and 0.3 MOI for BT74), or combination (O + M) in the absence (Acy (-)) or presence of acyclovir (5 μM; A, Acy (+)) for six days, followed by MTS assay for cell viability, represented as mean ± SD. ‡P < .0001 (multiple comparisons test, Tukey) between indicated pairs. All statistical tests were two-sided.
As Rad51 and Chk1 degradation occurred coincident with gC expression (Figure 4D), which is dependent upon virus DNA replication (29), we tested whether HSV DNA replication was required. Acyclovir, a nucleoside analog inhibitor of HSV DNA replication (30), blocked virus replication (Supplementary Figure 5C, available online) and MG18L-induced degradation of Rad51 and Chk1 in GSCs (Figure 4E). It abrogated MG18L cytotoxicity, as expected, but also the combination effect with PARPi in both MGG4 and BT74 cells (Figure 4F). oHSV G207 infection of GSCs is nonpermissive (16), with a block in true late (γ2) protein synthesis (CP and SDR) (unpublished results), but no effect on virus DNA replication (Supplementary Figure 5B, available online). In contrast to acyclovir, where late proteins are also not expressed, both Rad51 and Chk1 were degraded after G207 infection, similar to MG18L (Figure 4E). This supports the necessity of HSV DNA replication for proteasomal degradation of Rad51 and/or Chk1.
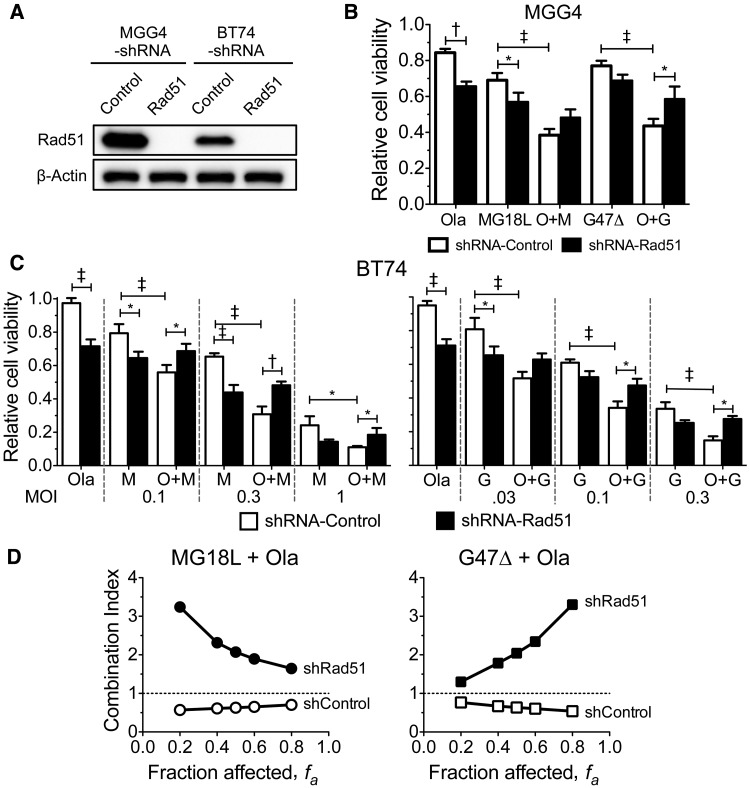
Role of Rad51 Loss in Synergy Between oHSV and PARPi
We tested whether Rad51 loss alone was necessary for synergy. Rad51 silencing, using lentivirus-mediated shRNA (Figure 5A), suppressed cell growth to some extent (Supplementary Figure 6A, available online) and sensitized both MGG4 and BT74 to olaparib or oHSV alone, but abrogated the combination effect, so that cytoxicity was similar to oHSV monotherapy (Figure 5, B and C). This was confirmed by viable cell counting (Supplementary Figure 6B, available online). The combination was now antagonistic in the Rad51-silenced MGG4 cells, while still synergistic in control shRNA-transduced cells (Figure 5D). Thus, increased sensitivity to PARPi due to Rad51 silencing compensates for the effect of oHSV-induced Rad51 loss, indicating that this mediates oHSV synergy and synthetic lethality with PARPi.
Figure 5.
Role of Rad51 knockdown on synergy between olaparib and oncolytic herpes simplex viruses. A) MGG4 or BT74 cells transduced with lentivirus expressing Rad51- or control-shRNA were grown under puromycin selection for seven days prior to immunoblot analysis. β-actin is loading control. B) MGG4 cells transduced with shRNA-control or shRNA-Rad51 were treated with olaparib (Ola; 1 μM), MG18L (0.04 MOI), G47Δ (0.04 MOI), or the combination (O + M, O + G) for six days, and cell viability was measured by MTS assay and represented as mean ± SD. *P < .01; †P < .001; ‡P < .0001 (multiple comparisons test, Tukey) between indicated pairs. MG18L shRNA-control vs shRNA-Rad51 (P = .02) and O + G shRNA-control vs shRNA-Rad51 (P = .02). C) BT74 cells transduced with shRNA-control or shRNA-Rad51 were treated with olaparib (Ola; 10 μM), MG18L (left; M), G47Δ (right; G), or combination (O + M, O + G) at the MOIs indicated for six days, and cell viability was measured by MTS assay and represented as mean ± SD. *P < .05; †P < .001; ‡P < .0001 (multiple comparisons test, Tukey) between indicated pairs. The combination of O + G statistically significantly increased viability with shRNA-Rad51 vs shRNA-control; P = .05, .01, .02 for MOIs of .03, .1, .3, respectively. The combination of O + M statistically significantly increased viability with shRNA-Rad51 vs shRNA-control; P = .02, .0005, .02 for MOIs of 0.1, 0.3, 1, respectively. M (1 MOI, shRNA-control) vs O + M (1 MOI, shRNA-control), P = .01; M (0.1 MOI, shRNA-control) vs M (0.1 MOI, shRNA-Rad51), P = .004; and G (0.03 MOI, shRNA-control) vs G (0.03 MOI, shRNA-Rad51), P = .002. D) Rad51 shRNA abrogates synergy. Interaction of olaparib and MG18L (left) or G47Δ (right) in MGG4 transduced with shRNA-control (shcontrol) or shRNA-Rad51 (shRad51). Combination index < 1, = 1, and > 1 indicates synergistic, additive, and antagonistic interactions, respectively. All statistical tests were two-sided. Ola = olaparib.
Role of Chk1 in the Combination Effect of oHSV and PARPi
Chk1 was activated in MGG4 by olaparib and in combination (Figure 3A). While CHIR-124 sensitized MGG4 to olaparib, suggesting p-Chk1 was protective, it did not alter the combination with oHSV (Figure 6A), discounting Chk1 signaling in mediating this combination effect. CHIR-124 did not sensitize BT74 to olaparib (Supplementary Figure 6C, available online). Overexpression of Chk1 in BT74, where it was degraded by oHSV, resulted in some Chk1 remaining and being activated after MG18L infection (Figure 6B;Supplementary Figure 6D, available online). This did not alter the sensitivity of MGG23 or BT74 to olaparib or oHSV alone, but partially rescued GSCs from combination cytotoxicity (Figure 6C), suggesting that oHSV-induced Chk1 degradation contributes to this.
Figure 6.
The role of Chk1 on poly(ADP-ribose) polymerase (PARP) inhibitor sensitivity of glioblastoma stem cells and in combination with oncolytic herpes simplex virus (oHSV). A) Chk1 inhibitor CHIR-124 sensitizes MGG4 to olaparib but not combination with oHSV. Olaparib (Ola; 1μM), MG18L (M; 0.04 MOI), G47Δ (G; 0.04 MOI), or the combination (O + M and O + G), with (CHIR124 (+), 0.1 μM) or without CHIR-124 (CHIR-124 (-)) for six days, and viability was measured by MTS assay. With CHIR-124, Ola was statistically significantly different from O + G (P = .003). B) Immunoblot analysis of Chk1-overexpressing BT74 (BT74-Chk1) or control-transduced (BT74-control) cells treated with mock (-), olaparib (O; 30 μM), MG18L (M; MOI = 1), or combination (O + M) and harvested after 30 hours. C) Chk1 overexpression reduces combination effect. Left: MGG23 transduced with control or Chk1 (Fig S6D) and treated with olaparib (O; 2 μM), MG18L (M; 0.002 MOI), G47Δ (G; 0.1 MOI), or combination (O + M, O + G). Right: Transduced BT74 treated with olaparib (10 μM), MG18L (0.3 MOI), G47Δ (0.1 MOI). MTS assay was performed for cell viability after six-day treatment. D) MGG23 (left) and BT74 (right), transduced with shRNA-control or shRNA-Rad51, were treated with olaparib (Ola; 2, 10 μM) or CHIR-124 (CHIR; 0.01, 1.5 μM), respectively, or combination (O + C) for six days before MTS assay. For BT74, in the presence of shRNA-Rad51, O + C was statistically significantly different from Ola (P = .001). E) Immunoblot analysis of shRNA-control or shRNA-Rad51 transduced MGG23 (left) treated with mock (-), olaparib (O; 10 μM), CHIR-124 (C; 0.1 μM), or combination (O + C) or BT74 (right) treated with olaparib (30 μM) or CHIR124 (0.2 μM). Membranes were probed with antibodies to PAR, PARP, cleaved PARP, γH2AX, Rad51, and β-actin as loading control. Cell viability is represented as mean ± SD. *P < .01; †P < .001; ‡P < .0001 (multiple comparisons test, Tukey) between indicated pairs. All statistical tests were two-sided.
We combined Chk1 inhibition with Rad51 silencing to test whether this phenocopied the effect of oHSV. The absence of Rad51 did not alter CHIR-124 sensitivity, but CHIR-124 did increase olaparib sensitivity in Rad51-silenced MGG23 and BT74 (Figure 6D). This was associated with increased apoptosis (cleaved-PARP) and DSBs (γH2AX) in Rad51-silenced GSCs treated with olaparib and additionally when combined with CHIR-124 (Figure 6E), phenocopying the effect of oHSV.
Efficacy of Combination Therapy on Survival of Mice Bearing PARPi-Sensitive and -Resistant GSC-Derived Intracerebral Tumors
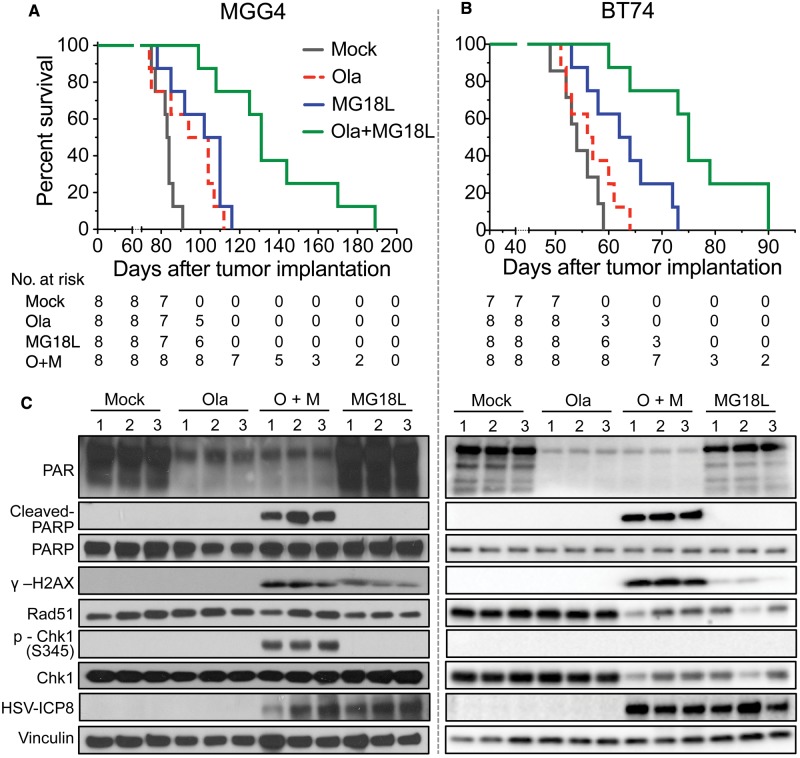
We then tested the combination therapy in vivo in PARPi-sensitive and -resistant GSC-derived brain tumors. In orthotopic MGG4 xenografts, olaparib alone statistically significantly prolonged survival (median survival = 94 days, 95% confidence interval [CI] = 75 to 107), compared with mock (median survival = 83 days, 95% CI = 77 to 86, P = .03), similar to the prolongation seen with a single intratumoral injection of MG18L (median survival = 102 days, 95% CI = 85 to 110, P = .003) compared with mock. The combination of olaparib and MG18L further extended survival (median survival = 131 days, 95% CI = 108 to 170; olaparib + MG18 vs MG18, P = .005, and vs olaparib, P = .001) by over 50% compared with mock (Figure 7A). In the BT74 model, MG18L alone, as in MGG4, statistically significantly prolonged survival (median survival for MG18L = 62 days, 95% CI = 56 to 72; mock = 54 days, 95% CI = 52 to 58, P = .009), whereas olaparib alone had no effect (median survival = 56 days, 95% CI = 52 to 61) (Figure 7B). However, the combination of olaparib and MG18L further extended survival (median survival = 75 days, 95% CI = 64 to 90; olaparib + MG18 vs MG18, P = .005, and vs olaparib, P = .0004) (Figure 7B), suggesting a synthetic lethal-like effect in vivo.
Figure 7.
Efficacy of combination therapy in glioblastoma stem cell–derived intracerebral tumors. A) Mice implanted intracerebrally with 2 x 105 MGG4 cells were treated with olaparib (Ola; 50 mg/kg) in 10% DMSO/10% 2-hydroxyl-propyl-β-cyclodextrine/PBS or vehicle, administered intraperitoneally starting on day 17 postimplantation with six cycles of five-day on and two-day off, and MG18L (1 X 106 pfu) or PBS intratumorally injected on day 19. Mock vs olaparib (P = .03) or vs MG18L (P = .003); olaparib + MG18L vs MG18L (P = .005) or vs olaparib (P = .001); log-rank test. B) Mice implanted intracerebrally with 1 x 105 BT74 cells were treated with olaparib or vehicle administered intraperitoneally daily for 26 days starting on day 9 postimplantation, and MG18L or PBS intratumorally injected on day 11. Mock vs MG18L (P = .009); olaparib + MG18L vs MG18L (P = .005) or vs olaparib (P = .0004); log-rank test. C) DNA damage responses induced by olaparib (Ola), MG18L, or combination (O + M) in vivo. Intracerebral tumors were established with MGG4 (left) and treated with olaparib (Ola) or vehicle (mock) intraperitoneally starting at day 61 after implantation for five days and/or MG18L (2 X 106 pfu) injected on day 63, and harvested on day 65. BT74 tumors (right) were treated similarly, except with olaparib starting on day 42, MG18L injected on day 44, and death on day 46. Lysates from tumor-bearing hemispheres from individual mice (1, 2, 3) were electrophoresed and probed with antibodies to PAR, poly(ADP-ribose) polymerase (PARP), cleaved PARP, γH2AX, Rad51, p-Chk1 (S345), Chk1, HSV-ICP8, and vinculin as loading control. All statistical tests were two-sided. Ola = olaparib; PARP = poly(ADP-ribose) polymerase.
DNA Damage and Apoptosis After Combination Therapy in Vivo
To gain mechanistic insight, we examined DDR in tumor lysates. PARP inhibition demonstrated olaparib blood-brain barrier transit and tumor penetration (PAR) (Figure 7C). MG18L infected all injected tumors to a similar extent, as detected by HSV early protein ICP8 expression (HSV-ICP8) (Figure 7C). DNA damage was induced by MG18L alone and greatly increased when combined with olaparib (γH2AX) (Figure 7C). Strikingly, apoptosis was only induced after combination treatment in both MGG4 and BT74 (cleaved-PARP) (Figure 7C). This is consistent with the in vitro results, showing that increased DNA damage and apoptosis likely contributed to the combination effect in vivo. Rad51 and Chk1 were clearly reduced in BT74 with MG18L alone or in combination (Figure 7C, right). In MGG4, four out of six tumors exhibited a decrease in Rad51 after MG18L treatment (Figure 7C, left), with Rad51 statistically significantly lower in MG18L treated (mean = 0.51 ± 0.07) than mock alone (mean = 0.71 ± 0.06, P = .04) or mock combined with olaparib (mean = 0.87 ± 0.10, P = .01) (Supplementary Figure 7, available online). P-Chk1 was only detected after combination treatment in MGG4 (Figure 7C, left).
Discussion
Here we report that oHSV sensitizes GSCs to PARPi killing in a synthetic lethal-like fashion in vitro and in vivo through targeting DDR. PARPi sensitivity in cancer cells is usually due to genetic alterations in DDR genes, particularly HR repair, as demonstrated with BRCA1 and BRCA2 mutated breast and ovarian cancer (2). In cancer cells without identified DNA repair deficiencies, pharmacological inhibition of DDR can induce synthetic lethality with PARPi (4). We hypothesized that oHSV would induce synthetic lethality with PARPi as it induces DNA damage and modulates DDR (19). In contrast to our previous studies with temozolomide (19), here we identify a new activity of oHSV, proteasomal degradation of Rad51, mediating synergy with PARPi. Two oHSVs with different mutations and cancer selectivity mechanisms, G47Δ and MG18L (6), similarly synergized with olaparib in PARPi-sensitive and -resistant GSCs. In contrast to oncolytic adenovirus (31), we did not find any effect of olaparib on oHSV replication.
Activation of Chk1 by ATR mediates cell cycle arrest to allow time for DNA repair and prevent cells with damaged DNA from entering mitosis (22). P-Chk1 was induced by olaparib alone in all GSCs, except MGG24, and resulted in S phase arrest. Virus infection led to loss of Chk1 protein, completely in PARPi-resistant BT74 and MGG24 and to a lesser extent in PARPi-sensitive GSCs. This was unexpected as HSV-1 was previously shown not to affect the stability of ATR pathway proteins, including Chk1 (5). Chk1 inhibition reportedly synergized with olaparib in prostate cancer cells (32), similar to MGG4. Chk1 inhibitor blocked olaparib-induced S phase arrest when Chk1 was activated but did not sensitize MGG23 or BT74 cells to olaparib. Conversely, overexpression of Chk1 counteracted oHSV-induced degradation and partially abrogated oHSV and olaparib combination cytotoxicity, coincident with Chk1 activation.
HR is a major repair pathway for DSBs, using an intact sister chromatid as a template for Rad51-catalyzed DNA strand exchange to guide error-free repair (3). Unexpectedly, oHSV infection induced a loss of Rad51 within 30 hours in all GSCs tested and inhibited HR. The most obvious viral proteins, ICP0 and Us3, were not involved. oHSV-induced Rad51 loss was abrogated by MG132 or acyclovir but not G207, which replicates its DNA but doesn't express γ2 proteins, demonstrating that this was due to proteasome-dependent degradation and required viral DNA replication. We postulate that something in the physical structure of HSV-replicating DNA and/or its location in the nucleus alters Rad51 so it is degraded. Rad51 shRNA-transduced GSCs were more sensitive to olaparib than control shRNA-transduced GSCs, in line with previous studies of cancer cell lines (33,34). Rad51 silencing also increased the cytotoxicity of oHSV in GSCs; however, the combination of oHSV and olaparib was now antagonistic. Thus, synergy between olaparib and oHSV is mediated by viral inhibition of Rad51. This would suggest that the synergistic effects with oHSV would not occur in cells with HR repair deficiencies that are already synthetic-lethal with PARPi.
In vivo, olaparib alone statistically significantly extended the survival of athymic mice bearing only PARPi-sensitive GSC-derived tumors. In contrast, MG18L alone improved overall survival in both tumor models, illustrating the broad efficacy of oHSV. Most importantly, the combination of olaparib with MG18L greatly increased survival in both PARPi-sensitive and -resistant GSC-derived tumors, indicating a synergistic effect in vivo. This is in contrast to TMZ and PARPi, where there was no combination effect in MGMT unmethylated xenografts (35). In both GSCs, combination therapy induced the greatest DNA damage and apoptosis. P-Chk1 was detected only in MGG4 after combination treatment, which might be a nonconsequential response to DNA damage or contribute to efficacy in vivo and not in vitro.
Our study is not without limitations. The large diversity between different patient GSCs, genomically and phenotypically (13,36,37), means we have likely missed the full range of complexity despite examining eight different GSCs. While we demonstrated a novel potential mechanism regulating Rad51 degradation, we have not defined how viral DNA replication might promote degradation. The treatment schedule in vivo was not optimized, such as with multiple administrations of oHSV that improve efficacy, nor was pharmacologic synergy in vivo demonstrated. The in vivo studies were performed in immunodeficient mice. It is possible that the combination effect would be altered in immunocompetent mouse models because of oHSV and/or PARPi effects on immune cells (38,39).
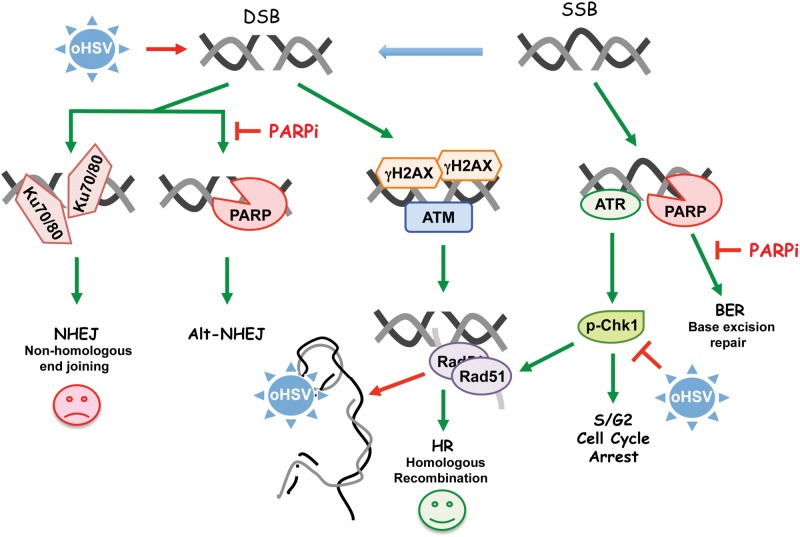
These studies describe a novel synthetic lethal-like combination therapy for targeting GSCs with PARPi and oHSV. We propose a model (Figure 8) in which PARPi treatment of GSCs blocks BER and activates DDR, including S phase arrest through Chk1 activation, while oHSV induces DSBs, inhibits HR repair through degradation of Rad51 because of DNA replication, and alters the cell cycle through Chk1 degradation, with the combination driving repair toward lethal NHEJ or other cell death pathways. Therefore, this combination strategy could be effective for GBM irrespective of molecular subtypes and DDR pathways that may be deficient in individual patients. This unique ability of oHSV to disrupt multiple interconnected components of DDR makes it a powerful therapeutic to combine with other DNA-damaging agents or inhibitors of DDR. Because of oHSV’s broad efficacy against most solid tumors, this novel combination strategy should be applicable to other cancer stem cells and tumors.
Figure 8.
Model for combination therapy of poly(ADP-ribose) polymerase inhibitors (PARPi) and inducing synthetic lethality in glioblastoma stem cells. Oncolytic herpes simplex virus (oHSV) induces DSBs (detected by γH2AX) that activate ATM to recruit repair proteins. oHSV DNA replication promotes the degradation of Rad51 and Chk1, blocking homologous recombination and disturbing cell cycle processes. DNA single strand breaks (SSBs) activate ATR, which phosphorylates Chk1. PARP binds to DNA breaks to facilitate BER and Alt-NHEJ and blocks Ku protein binding for NHEJ. Therefore, PARPis inhibit BER and facilitate SSB conversion to DSB, and inhibit Alt-NHEJ and facilitate NHEJ, which is detrimental. Red perpendicular mark indicates inhibition (degradation), green arrow indicates activation, and red arrow indicates detrimental activity. BER = base excision repair; DSB = DNA double-strand break; HR = homologous recombination; NHEJ = nonhomologous end joining; oHSV = oncolytic herpes simplex virus; PARPi = poly(ADP-ribose) polymerase inhibitor; SSB = DNA single-strand break.
Funding
This work was supported in part by grants from the National Institutes of Health (R01 CA160762 to SDR, R01NS032677 to RLM, and F31CA192453 to CP). SDR was supported in part by the Thomas A. Pappas Chair in Neurosciences.
Notes
The study sponsors had no role in the design of the study; the collection, analysis, or interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication. RLM and SDR are co-inventors on patents relating to oncolytic herpes simplex viruses, filed by Georgetown University and Massachusetts General Hospital, which have received royalties. The other authors have declared that no conflict of interest exists.
We thank other members of the Molecular Neurosurgery Laboratory for technical assistance and Melissa Humphrey for production and purification of oHSV stocks. We thank Alona Muzikansky (MGH Biostatistics Center) for statistical assistance and analysis of the data. We thank David M. Knipe (Department of Microbiology, Harvard Medical School, Boston, MA) for providing the HSV-1 ICP0 mutant and rescued viruses, and Maria Jasin (Memorial Sloan Kettering Cancer Center, New York, NY) for providing the DR-U20S cells.
Supplementary Material
References
- 1. O'Connor MJ. Targeting the DNA damage response in cancer. Mol Cell. 2015;60(4):547–560. [DOI] [PubMed] [Google Scholar]
- 2. Scott CL, Swisher EM, Kaufmann SH.. Poly (ADP-ribose) polymerase inhibitors: Recent advances and future development. J Clin Oncol. 2015;33(12):1397–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dietlein F, Thelen L, Reinhardt HC.. Cancer-specific defects in DNA repair pathways as targets for personalized therapeutic approaches. Trends Genet. 2014;30(8):326–339. [DOI] [PubMed] [Google Scholar]
- 4. Lord CJ, Tutt AN, Ashworth A.. Synthetic lethality and cancer therapy: Lessons learned from the development of PARP inhibitors. Annu Rev Med. 2015;66:455–470. [DOI] [PubMed] [Google Scholar]
- 5. Smith S, Weller SK.. HSV-I and the cellular DNA damage response. Future Virol. 2015;10(4):383–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Peters C, Rabkin SD.. Designing herpes viruses as oncolytics. Mol Ther Oncolytics. 2015;2:15010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ning J, Wakimoto H.. Oncolytic herpes simplex virus-based strategies: Toward a breakthrough in glioblastoma therapy. Front Microbiol. 2014;5:303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kohlhapp FJ, Kaufman HL.. Molecular Pathways: Mechanism of action for talimogene laherparepvec, a new oncolytic virus immunotherapy. Clin Cancer Res. 2016;22(5):1048–1054. [DOI] [PubMed] [Google Scholar]
- 9. Johnson DR, O'Neill BP.. Glioblastoma survival in the United States before and during the temozolomide era. J Neurooncol. 2012;107(2):359–364. [DOI] [PubMed] [Google Scholar]
- 10. Stopschinski BE, Beier CP, Beier D.. Glioblastoma cancer stem cells—from concept to clinical application. Cancer Lett. 2013;338(1):32–40. [DOI] [PubMed] [Google Scholar]
- 11. Suva ML, Rheinbay E, Gillespie SM, et al. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell. 2014;157(3):580–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lee J, Kotliarova S, Kotliarov Y, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9(5):391–403. [DOI] [PubMed] [Google Scholar]
- 13. Wakimoto H, Mohapatra G, Kanai R, et al. Maintenance of primary tumor phenotype and genotype in glioblastoma stem cells. Neuro Oncol. 2012;14(2):132–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tamura K, Aoyagi M, Ando N, et al. Expansion of CD133-positive glioma cells in recurrent de novo glioblastomas after radiotherapy and chemotherapy. J Neurosurg. 2013;119(5):1145–1155. [DOI] [PubMed] [Google Scholar]
- 15. Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–760. [DOI] [PubMed] [Google Scholar]
- 16. Wakimoto H, Kesari S, Farrell CJ, et al. Human glioblastoma-derived cancer stem cells: Establishment of invasive glioma models and treatment with oncolytic herpes simplex virus vectors. Cancer Res. 2009;69(8):3472–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Todo T, Martuza RL, Rabkin SD, et al. Oncolytic herpes simplex virus vector with enhanced MHC class I presentation and tumor cell killing. Proc Natl Acad Sci U S A. 2001;98(11):6396–6401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kanai R, Wakimoto H, Martuza RL, et al. A novel oncolytic herpes simplex virus that synergizes with phosphoinositide 3-kinase/Akt pathway inhibitors to target glioblastoma stem cells. Clin Cancer Res. 2011;17(11):3686–3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kanai R, Rabkin SD, Yip S, et al. Oncolytic virus-mediated manipulation of DNA damage responses: Synergy with chemotherapy in killing glioblastoma stem cells. J Natl Cancer Inst. 2012;104(1):42–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rolfo C, Swaisland H, Leunen K, et al. Effect of food on the pharmacokinetics of olaparib after oral dosing of the capsule formulation in patients with advanced solid tumors. Adv Ther. 2015;32(6):510–522. [DOI] [PubMed] [Google Scholar]
- 21. Marechal A, Zou L.. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb Perspect Biol. 2013;5(9):a012716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang Y, Hunter T.. Roles of Chk1 in cell biology and cancer therapy. Int J Cancer. 2014;134(5):1013–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pierce AJ, Johnson RD, Thompson LH, et al. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13(20):2633–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tse AN, Rendahl KG, Sheikh T, et al. CHIR-124, a novel potent inhibitor of Chk1, potentiates the cytotoxicity of topoisomerase I poisons in vitro and in vivo. Clin Cancer Res. 2007;13(2 Pt 1):591–602. [DOI] [PubMed] [Google Scholar]
- 25. Kisselev AF, Goldberg AL.. Proteasome inhibitors: From research tools to drug candidates. Chem Biol. 2001;8(8):739–758. [DOI] [PubMed] [Google Scholar]
- 26. Boutell C, Everett RD.. Regulation of alphaherpesvirus infections by the ICP0 family of proteins. J Gen Virol. 2013;94(Pt 3):465–481. [DOI] [PubMed] [Google Scholar]
- 27. Cai WZ, Schaffer PA.. Herpes simplex virus type 1 ICP0 plays a critical role in the de novo synthesis of infectious virus following transfection of viral DNA. J Virol. 1989;63(11):4579–4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Orzalli MH, DeLuca NA, Knipe DM.. Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proc Natl Acad Sci U S A. 2012;109(44):E3008–E3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Johnson PA, MacLean C, Marsden HS, et al. The product of gene US11 of herpes simplex virus type 1 is expressed as a true late gene. J Gen Virol. 1986;67(Pt 5):871–883. [DOI] [PubMed] [Google Scholar]
- 30. Furman PA, St Clair MH, Fyfe JA, et al. Inhibition of herpes simplex virus-induced DNA polymerase activity and viral DNA replication by 9-(2-hydroxyethoxymethyl)guanine and its triphosphate. J Virol. 1979;32(1):72–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Passaro C, Volpe M, Botta G, et al. PARP inhibitor olaparib increases the oncolytic activity of dl922-947 in in vitro and in vivo model of anaplastic thyroid carcinoma. Mol Oncol. 2015;9(1):78–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li L, Chang W, Yang G, et al. Targeting poly(ADP-ribose) polymerase and the c-Myb-regulated DNA damage response pathway in castration-resistant prostate cancer. Sci Signal. 2014;7(326):ra47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tahara M, Inoue T, Sato F, et al. The use of olaparib (AZD2281) potentiates SN-38 cytotoxicity in colon cancer cells by indirect inhibition of Rad51-mediated repair of DNA double-strand breaks. Mol Cancer Ther. 2014;13(5):1170–1180. [DOI] [PubMed] [Google Scholar]
- 34. Quiros S, Roos WP, Kaina B.. Rad51 and BRCA2—New molecular targets for sensitizing glioma cells to alkylating anticancer drugs. PLoS One. 2011;6(11):e27183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gupta SK,, Kizilbash SH, Carlson BL. et al. Delineation of MGMT hypermethylation as a biomarker for veliparib-mediated temozolomide-sensitizing therapy of glioblastoma. J Natl Cancer Inst. 2016;108(5):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rheinbay E, Suva ML, Gillespie SM, et al. An aberrant transcription factor network essential for Wnt signaling and stem cell maintenance in glioblastoma. Cell Rep. 2013;3(5):1567–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Piccirillo SG, Colman S, Potter NE, et al. Genetic and functional diversity of propagating cells in glioblastoma. Stem Cell Reports. 2015;4(1):7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ning J, Wakimoto H, Rabkin SD.. Immunovirotherapy for glioblastoma. Cell Cycle. 2014;13(2):175–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Huang J, Wang L, Cong Z, et al. The PARP1 inhibitor BMN 673 exhibits immunoregulatory effects in a Brca1(-/-) murine model of ovarian cancer. Biochem Biophys Res Commun. 2015;463(4):551–556. [DOI] [PubMed] [Google Scholar]
- 40. Chou TC. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev. 2006;58(3):621–681. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.