Abstract
The Cell Division-Cycle-14 gene encodes a dual-specificity phosphatase necessary in yeast for exit from mitosis. Numerous disparate roles of vertebrate Cell Division-Cycle-14 (CDC14A) have been proposed largely based on studies of cultured cancer cells in vitro. The in vivo functions of vertebrate CDC14A are largely unknown. We generated and analyzed mutations of zebrafish and mouse CDC14A, developed a computational structural model of human CDC14A protein and report four novel truncating and three missense alleles of CDC14A in human families segregating progressive, moderate-to-profound deafness. In five of these families segregating pathogenic variants of CDC14A, deaf males are infertile, while deaf females are fertile. Several recessive mutations of mouse Cdc14a, including a CRISPR/Cas9-edited phosphatase-dead p.C278S substitution, result in substantial perinatal lethality, but survivors recapitulate the human phenotype of deafness and male infertility. CDC14A protein localizes to inner ear hair cell kinocilia, basal bodies and sound-transducing stereocilia. Auditory hair cells of postnatal Cdc14a mutants develop normally, but subsequently degenerate causing deafness. Kinocilia of germ-line mutants of mouse and zebrafish have normal lengths, which does not recapitulate the published cdc14aa knockdown morphant phenotype of short kinocilia. In mutant male mice, degeneration of seminiferous tubules and spermiation defects result in low sperm count, and abnormal sperm motility and morphology. These findings for the first time define a new monogenic syndrome of deafness and male infertility revealing an absolute requirement in vivo of vertebrate CDC14A phosphatase activity for hearing and male fertility.
Introduction
Cell Division-Cycle 14A gene (CDC14A, OMIM 603504) encodes an evolutionarily conserved dual-specificity phosphoprotein-phosphatase that dephosphorylates proteins at tyrosine or serine/threonine residues. The function of CDC14 in Saccharomyces cerevisiae is well understood (1), where it is an essential regulator of the FEAR-MEN mitotic pathway, cytokinesis and ribosomal deoxyribonucleic acid (DNA) condensation (2–5). Temperature-sensitive yeast cdc14-mutants die when grown at the restrictive temperature as they are unable to transit through mitosis to interphase (5). This yeast cdc14 conditional lethal phenotype can be rescued by expressing either human CDC14A or its paralogue CDC14B (6). Owing to the evolutionary conservation of amino acid sequence and retention of mitotic function when expressed in yeast (7), CDC14A was initially assumed to be necessary for progression through the cell cycle in human cell lines and in Caenorhabditis elegans (8,9). However, the phenotype of a genomic deletion of the C. elegans cdc14 gene is extra cell divisions of multiple cell lineages, ruling out an essential role in mitosis (10). In animal cells, protein phosphatase-1 (PP1) and PP2A, not CDC14A, counteract mitotic kinases (11), leaving unsettled the in vivo functions of CDC14A in higher organisms (12).
Many other disparate functions ascribed to CDC14A stem from cell-free biochemical studies and in vitro experiments using cell lines in which endogenous CDC14A expression was either partially reduced with RNAi or ectopically over-expressed. Based on these studies, which often employed human cancer cells, the reported functions for CDC14A include DNA double strand break repair (13), chromosome segregation (14,15), microtubule integrity (16), modulating tumor suppressor protein p53 (17) and regulating the centrosome cycle and the actin cytoskeleton (9,18–20). Recently, two recessive variants of CDC14A were associated with human deafness (21), and zebrafish morpholino-oligonucleotide knockdown of cdc14aa expression indicated a role in kinocilia length regulation (21,22).
Human deafness is a clinically and genetically heterogeneous disorder. There are at least 400 different syndromes in which hearing loss (HL) is co-inherited with extra-auditory features. Additional features that accompany human deafness are in many instances identified only after the underlying genes have been identified (23), animal models studied and additional clinical assessments obtained. In many large families segregating deafness, the underlying pathogenic variants are unknown but the phenotypes have been genetically mapped to chromosomal intervals. This has been the case for autosomal recessive deafness at the DFNB32 locus that was mapped to chromosome 1p13.3–22.1 (OMIM 608653) (24). Here we report eight families segregating different homozygous recessive mutant alleles of CDC14A associated with human deafness DFNB32. In the present study, some deaf males homozygous for variants of CDC14A are also infertile. Like deafness, male infertility is a common, genetically heterogeneous disorder (25) and could have been an incidental finding. After engineering a variety of different recessive mutations of mouse Cdc14a, we consistently observed that females were deaf and fertile, while deaf males were infertile. Using CRISPR/Cas9 editing, we introduced a single amino acid substitution that ablates phosphatase activity resulting in profound deafness and male, but not female, infertility in mouse. These data support the conclusion that phosphatase activity of mammalian CDC14A in vivo is not essential for cell division, but is necessary for hearing and male fertility.
Results
DFNB32-linked families segregating deafness
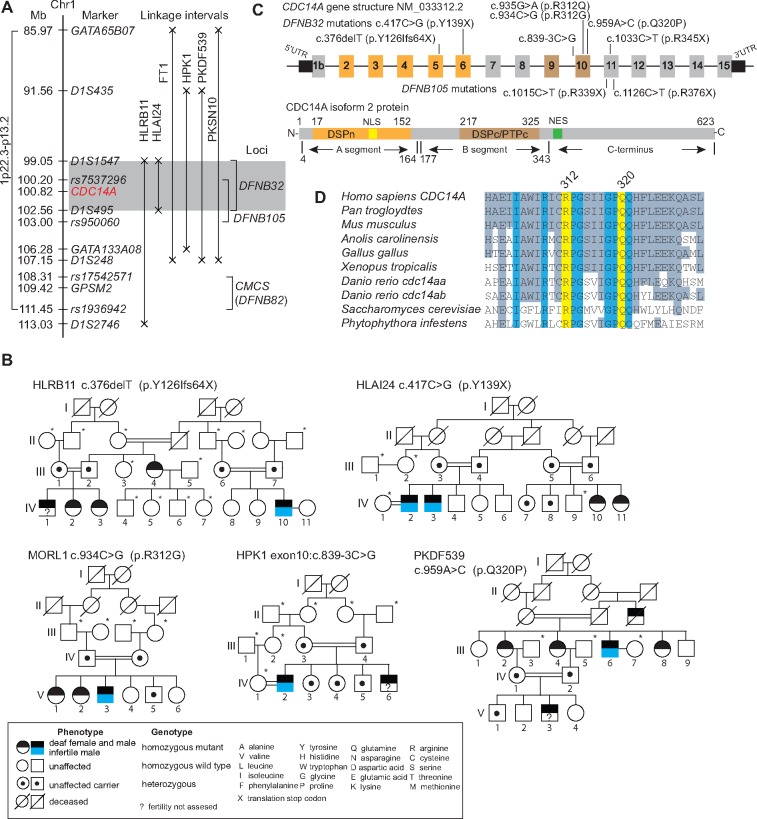
We initially sought to identify the molecular genetic explanation for DFNB32-linked deafness segregating in five Pakistani families (HLRB11, HLAI24, HPK1, PKDF539, PKSN10) and one Tunisian family (FT1), which was investigated in the original report of DFNB32-deafness (24). Each of the six families has a different homozygous haplotype of the markers closely linked to DFNB32 suggesting six different mutant alleles (Supplementary Material, Fig. S2A). The linkage interval in the HLAI24 family is comprised of a haplotype defined by meiotic recombinations at flanking short tandem repeat (STR) markers D1S1547 and D1S495, which refined the linkage interval to 3.51 Mb (Fig. 1A). The meiotic breakpoints in families HLAI24, FT1, HPK1, PKDF539 and PKSN10 exclude GPSM2 (Fig. 1A). Mutations of GPSM2 are associated with Chudley–McCullough syndrome (CMCS, OMIM 604213), characterized by sensorineural deafness and hypoplasia of corpus callosum (26,27). However, the linkage interval on chromosome 1p for deafness segregating in family HLRB11 did not exclude GPSM2 (Fig. 1A). Sanger and/or whole exome sequencing (WES) of affected members of all six DFNB32-linked families revealed only common variants of GPSM2 predicted to be non-pathogenic. These data indicate that mutant alleles of at least one of the other 26 genes or non-coding RNAs in the DFNB32 interval (hg19, chr1: 99,048,258-102,561,767; Supplementary Material, Tables S1 and S2) are likely to be the cause of DFNB32-deafness.
Figure 1.
Deafness locus DFNB32 and recessive mutations of CDC14A. (A) Recessive deafness linked to markers for the DFNB32 locus shown in gray for a 3.5-Mb interval of chromosome 1p21.2. The recently reported redundant DFNB105-linkage interval (21) entirely overlaps with the DFNB32 interval defined previously by family FT1 (24). (B) Five consanguineous families segregating different homozygous variants of CDC14A. Some deaf males in these families are infertile (blue, lower half of male symbol). Asterisks indicate that gDNA are unavailable. See Supplementary Material, Figure S1 for pedigrees of families FT1, PKSN10 and MORL2. (C) Human CDC14A gene and protein schematics showing the exons in which the nine pathogenic variants occur. The longest CDC14A isoform has 623 residues encoding a dual-specificity phosphatase domain (DSPn), core dual-specificity phosphatase domain (DSPc/PTPc, residues 177–32) and a Nuclear Export Signal (residues 343–363). Also see Supplementary Material, Figure S1. (D) p.R312Q, p.R312G and p.Q320P substitutions alter conserved residues of CDC14A.
Pathogenic variants of CDC14A are associated with DFNB32 deafness
WES of affected members of two DFNB32 genetically linked families (HLRB11, HLAI24) and WES of two additional Iranian families (MORL1 and MORL2) revealed different homozygous recessive mutations in CDC14A, which encodes Cell Division-Cycle 14A protein (Fig. 1B;Supplementary Material, Fig. S1A, B and Table S2). The largest transcript of CDC14A encodes 623 residues (Fig. 1C and Supplementary Material, Fig. S2B). CDC14A also has two annotated transcription start sites (Exons 1a and 1b, NM_001319211 and NM_001319212.1), four different 3′ untranslated regions (UTRs), and an alternatively spliced Exon 8. Sanger sequencing of CDC14A from genomic DNA of affected individuals from the other four DFNB32-linked families revealed four additional homozygous mutant alleles of CDC14A (Fig. 1A;Supplementary Material, Fig. S1A, B and Table S1). Each of the eight variants of CDC14A (c.376delT, c.417C>G, c.839–3C>G c.934C>G, c.935G>A, c.959A>C, c.1033C>T and c.1126C>T, Fig. 1B;Supplementary Material, Fig. S1A and B) co-segregate with deafness and are absent from at least 150 ethnically matched (Pakistani, Tunisian or Iranian) normal control individuals (Supplementary Material, Table S2). Variants c.376delT, c.417C>G, c.935G>A, c.959A>C and c.1126C>T have allele frequencies of 0.00002438, 0.0000108, 0.0000123, 0.00000407 and 0.0000044, respectively, while the other three variants have not yet been reported in the gnomAD database (Supplementary Material, Table S2). In summary, we have identified eight variants in CDC14A: one frameshift mutation (p.Y126Ifs64X), three translational stop-gain mutations (p.Y139X, p.R345X and p.R376X) (21; Fig. 1B;Supplementary Material, Fig. S1B) and three missense mutations of two evolutionarily conserved residues (p.R312Q, p.R312G and p.Q320P, Fig. 1B and D;Supplementary Material, Fig. S1B). The eighth novel variant of CDC14A, segregating in family HPK1, alters an acceptor splice site of Exon 10 (c.839-3C>G; Fig. 1C;Supplementary Material, Fig. S1C). Analyses of leukocyte mRNA from affected and unaffected (carrier and homozygous wild-type) HPK1 family members (Fig. 1B) revealed two aberrant mRNA transcripts owing to the c.839-3C>G variant, one that skips Exon 10 and another that uses a cryptic Exon 10 acceptor splice site, adding two additional nucleotides (c.839-2 and c.839-1) to the mature mRNA. Both aberrant splicing events shift the native reading frame to introduce predicted premature termination codons (p.K279fs16X and p.K279fs10X; Supplementary Material, Fig. S1C) that could target the defective mRNA for degradation by the nonsense-mediated decay (NMD) pathway (28). If synthesized, the two truncated CDC14A proteins (p.K279fs16X and p.K279fs10X) would lack essential residues located in the dual-specificity phosphatase domain (DSPc/PTPc; Fig. 1C) that are necessary for phosphatase activity. Although we did not detect wild-type transcripts in lymphocyte cDNA from affected individuals in the HPK1 family, we cannot exclude the possible presence of wild-type CDC14A transcripts owing to specialized splicing factors (29) that circumvent the mutant acceptor splice site in cells, where CDC14A has an essential role.
Computational structure model of CDC14A protein
The missense mutations p.R312Q, p.R312G and p.Q320P (Fig. 1B;Supplementary Material, Fig. S1B) are substitutions of two highly conserved residues (Fig. 1D) that are predicted to be damaging by in silico tools MutationTaster, PROVEAN and SIFT (Supplementary Material, Table S3). To gain further insight into CDC14A dysfunction resulting from p.R312Q, p.R312G and p.Q320P, we developed a molecular structural model of the human CDC14A catalytic domain at atomic resolution, which consists of two subdomains, DSPn (residues 17–152) and DSPc/PTPc (residues 217–325; Fig. 1C). DSPn lacks critical residues essential for phosphatase catalysis but participates in phosphoprotein substrate recognition (30). The sequence encoding DSPc/PTPc contains the catalytic site where phosphotyrosine, phosphoserine and phosphothreonine residues are dephosphorylated (30). In the absence of a molecular protein structure of CDC14A at the atomic level, the human CDC14B (NP_003662) X-ray crystal structure (30), which comprises domains DSPn and DSPc/PTPc, was used as a template to model the structure of human CDC14A. The amino acid sequence alignment of these two paralogues has good correspondence in the secondary structural elements and an amino acid sequence identity of 59.76%, indicating likely comparable folds in the aligned regions (Supplementary Material, Fig. S3A). However, unlike Cdc14a mutant mice described below, we observed that homozygous Cdc14b null mutant mice have normal hearing and viability, and males and females are fertile (Supplementary Material, Fig. S4).
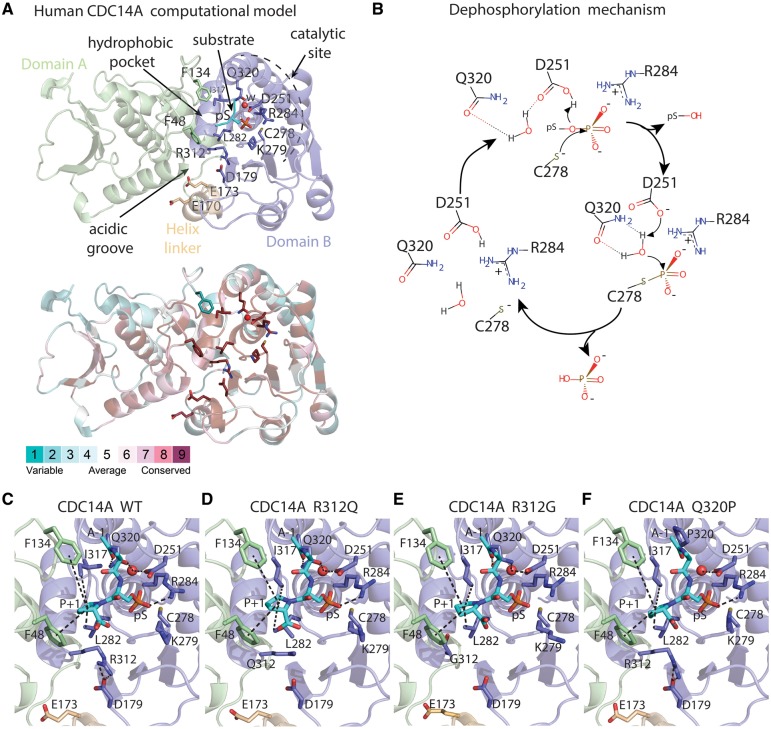
Our model of CDC14A has two segments separated by a helix of eight residues (Fig. 2A). Segments A and B have 30% sequence identity and share some common structural features (Supplementary Material, Fig. S3B and C). The phosphoprotein substrate-binding site of CDC14A can be subdivided into a hydrophobic pocket, an acidic groove and a catalytic site. The upper part of the acidic groove (residues 170–179; Fig. 2A and C) is stabilized by the salt bridge formed between the acidic aspartate-179 (D179) and basic arginine-312 (R312), the latter being substituted with a non-basic residue in the p.R312Q and p.R312G substitutions. The D179 residue is also evolutionarily conserved in CDC14A orthologues (Supplementary Material, Fig. S5) and paralogues and the equivalent residues D215 and R320 of CDC14B also form a salt bridge (30). The other acidic residues in this groove are proposed to interact with the positively charged residues in the substrate (30) (Fig. 2A). The main components involved in the catalytic reaction are glutamine-320 (Q320), which is substituted in the p.Q320P mutation, as well as aspartate-251 (D251), arginine-284 (R284) and cysteine-278 (C278) in addition to a critical water molecule (Fig. 2A). The first dephosphorylation step involves a nucleophilic attack of C278 to the phosphate group resulting in deprotonation of D251 and release of the substrate, leading to the formation of a covalent bond between the phosphate group and C278 (Fig. 2B). Next, the protonation of D251 activates the water molecule, which performs a nucleophilic attack to hydrolyze the phosphate group, essential for dephosphorylation. The interactions with residues D251 and Q320, part of the catalytic machinery, ensure polarization, optimal position and orientation of this water molecule, which is crucial for the nucleophilic attack to occur. The hydrolysis of the covalent bond between the cysteine and the phosphate induces the phosphate’s elimination from the central binding site, which is then ready to bind the next phosphorylated substrate to start the cycle again. During the cycle, the negative charge of the phosphate group is stabilized by both R284 and the positive dipole of helix 4B (Fig. 2A and B). The residues taking part in catalysis and binding of substrate are conserved among CDC14 paralogues and orthologues.
Figure 2.
Molecular model of human CDC14A wild-type (WT) and three missense variants constructed using as template the x-ray structure of human CDC14B, a paralogue of CDC14A. (A) Domains A (green) and B (light-purple), helix linker (yellow) and a phosphorylated peptide (substrate, cyan). The phosphorylated serine (pS) is proximal to a water molecule (w, red sphere) responsible for phosphate hydrolysis. Lower panel show evolutionary conservation of the residues. (B) Reaction mechanism for CDC14-mediated dephosphorylation of phosphoprotein substrates with its phosphate group oriented on top of the positive dipole of helix 4B, where the core enzyme machinery is located. (C–F) Catalytic sites in the structural models of the human CDC14A WT and substitutions p.R312Q, p.R312G and p.Q320P. The substrate is shown as cyan sticks where alanine and proline are indicated as A-1 and P + 1. Interactions involving the catalytic site residues are indicated by dashes.
In our structural model, the replacement of arginine-312 by glutamine (p.R312Q) would impair the electrostatic stabilization of the upper part of the acidic groove and may cause misfolding of the region involved in substrate recognition (Fig. 2D). Consistent with this prediction, in COS-7 cells pEGFP-CDC14A-R312Q forms aggregates that were not observed when wild-type pEGFP-CDC14A was over-expressed in COS-7 cells (Supplementary Material, Fig. S6J and K). The substitution of arginine-312 by glycine (p.R312G) segregating in family MORL1 could destabilize the upper part of the acidic groove, as introduction of glycine (small residue lacking a side chain) would increase the flexibility of this segment (Fig. 2E). This could cause protein misfolding in the region involved in substrate recognition (Fig. 2D). The substitution of glutamine-320 by a proline (p.Q320P) is predicted to prevent the polarization and favorable orientation necessary for the activation of the water molecule in the catalytic site (Fig. 2F). As a result, the p.Q320P variant of CDC14A would not hydrolyze the covalent bond between C278 and the phosphate, resulting in a catalytically inactive CDC14A, where the substrate remains bound. Although CDC14A with p.Q320P substitution is catalytically inactive, the cellular localization of EGFP-CDC14A-Q320P protein is not affected (Supplementary Material, Fig. S6C, G and L). In family PKDF539 segregating the p.Q320P substitution of CDC14A, HL is the most severe among the eight families (Fig. 3A).
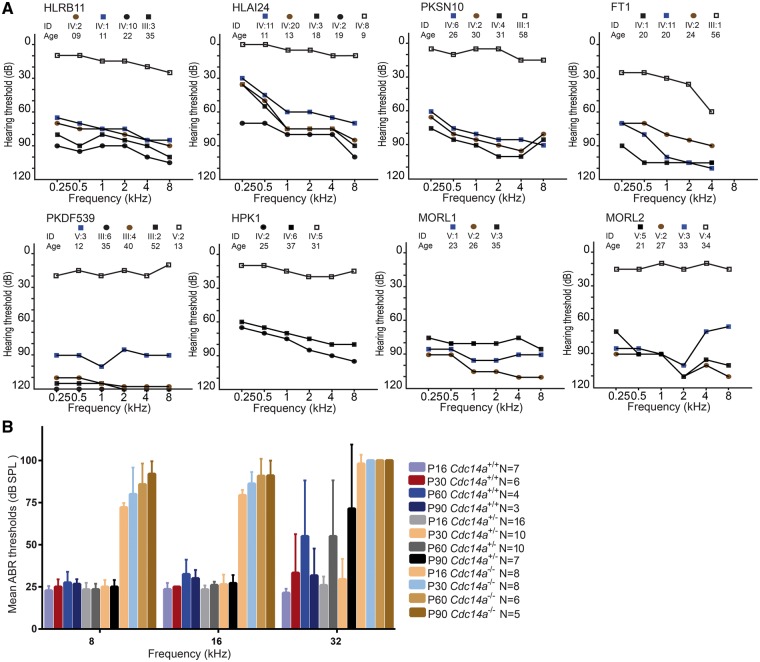
Figure 3.
Mutations of CDC14A cause progressive moderate-to-profound hearing loss in human and mouse. (A) Pure tone air conduction audiograms of right ears of affected and normal hearing heterozygous individuals of eight DFNB32 families recorded under ambient noise conditions at 0.25, 0.5, 1, 2, 4 and 8 kHz input. (B) In mouse, average auditory brainstem response (ABR) thresholds were recorded for 8, 16 and 32 kHz input at ages P16, P30, P60 and P90. The four different mutant alleles of mouse Cdc14a have indistinguishable ABR thresholds and were combined here (see Supplementary Material, Fig. S8B). ABR thresholds of right ears of wild-type (C57B6/N; Cdc14a+/+) mice, heterozygotes (Cdc14a+/tm1a, Cdc14a+/tm1b, Cdc14a+/tm1d collectively designated here Cdc14a+/−), homozygotes and compound heterozygotes (Cdc14atm1a/tm1a, Cdc14atm1b/tm1b, Cdc14atm1a/tm1b and Cdc14atm1d/tm1d,collectively designated Cdc14a−/−). Error bars are standard deviations. N = number of animals tested.
Our computational modeling predicts that a truncated CDC14A protein owing to p.R345X or p.R376X mutation would retain phosphatase activity, assuming that its mRNA escapes degradation via a nonsense-mediated mRNA decay mechanism (NMD). Human CDC14A that only includes residues from 16 to 360 can rescue cdc14-deficient yeast mutants and retains phosphatase activity (6,20). Nevertheless, the sequence of the C-terminus of CDC14A (residues 343–623), mostly absent owing to either nonsense mutation, is necessary for normal hearing. The unique amino acid sequence of the CDC14A C-terminus may have a role in intracellular localization. Enhanced green fluorescent protein (EGFP) tagged-CDC14A truncated at arginine-345 is mis-compartmentalized in transfected COS-7 cells and in the organ of Corti (Supplementary Material, Fig. S6D, I and M). In vivo, a reduced level rather than complete absence of phosphatase activity associated with the truncated forms of CDC14A may explain the less severe HL and fertility of deaf males of families PKSN10 and MORL2 (Fig. 3A and Supplementary Material, Fig. S1A).
LacZ and EGFP-CDC14A expression are consistent with endogenous CDC14A localization
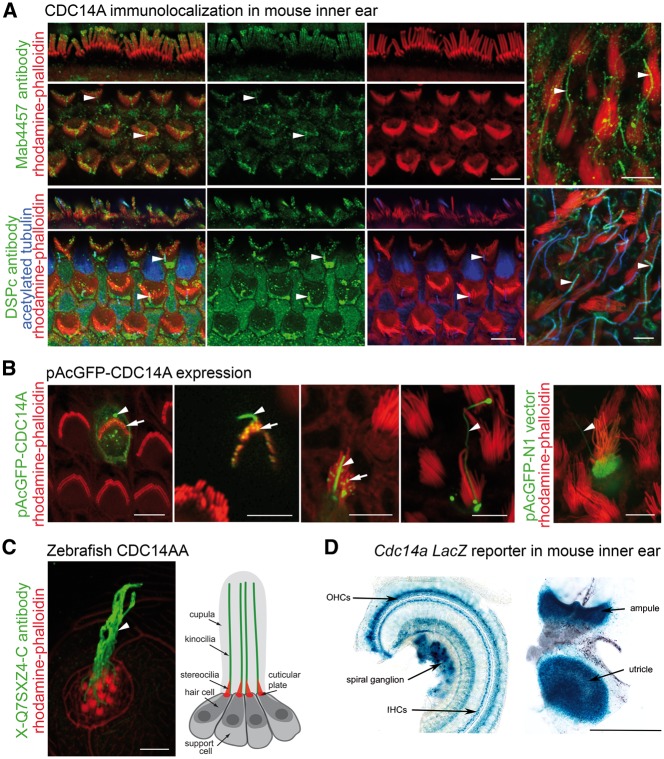
To understand the function of CDC14A in hearing, we examined the expression and localization of CDC14A in the inner ear using a custom antibody (DSP-Ab) against an epitope adjacent to the mouse DSPc/PTPc core phosphatase domain as well as four commercial polyclonal antibodies (GTX52005, NBP1-84573, 34-8100 and Mab4457) raised against epitopes of different regions of CDC14A (Supplementary Material, Fig. S7A and Table S4). Immunoreactivity of several antisera raised against CDC14A was associated with the upper one-third of stereocilia and tubulin-rich structures in hair cells including kinocilia, basal bodies (Fig. 4A) and the pericuticular ring. Some antisera also showed immunoreactivity in the nuclei of different cell types (Supplementary Material, Fig. S7B). In zebrafish neuromast cells, endogenous Cdc14aa (X-Q7SXZ4-Ab) was also present in kinocilia of hair cells (Fig. 4C). In mouse, the endogenous CDC14A localizations were reproduced by biolistically transfecting EGFP-CDC14A cDNA (longest Cdc14a transcript, NM_001080818) into inner ear sensory epithelia explants. EGFP-CDC14A protein was observed along the length of kinocilia, in basal bodies and as puncta concentrated towards the tips of stereocilia (Fig. 4B;Supplementary Material, Fig. S6A, E and F). In transfected COS-7 cells and supporting cells of the organ of Corti, EGFP-CDC14A was associated with filamentous tubulin in the cytoplasm (Supplementary Material, Fig. S6J). These observations corroborate the association of endogenous CDC14A with hair cell basal bodies, kinocilia, pericuticular necklace and with pillar cells that have extensive microtubule networks involved in transmitting mechanical sound stimuli from the basilar membrane to the reticular lamina.
Figure 4.
Expression of CDC14A in the mouse inner ear and zebrafish lateral line. (A) Endogenous CDC14A (green) is localized to the upper third of IHC stereocilia (red), near the tips of OHC stereocilia (red) and along the length of kinocilia (arrowheads) in mouse auditory (three left columns) and vestibular (right column) hair cells at P4. (B) Biolistically transfected pAcGFP-CDC14A mimics endogenous localization of CDC14A in hair cells, accumulating in kinocilia (arrowheads) and near tips of stereocilia (arrows). Expression of pAcGFP-N1 vector control fills the hair cell body, stereocilia and kinocilium. (C) Zebrafish CDC14AA is also localized along kinocilia (arrowhead and depicted in adjacent drawing) of neuromast hair cells. (D) In the organ of Corti (left) and vestibular sensory epithelium of the utricle and ampule (right), X-gal staining for β-galactosidase activity reporting CDC14A shows expression in spiral ganglion, lamina spiralis ossea, supporting cells, and hair cells. Scale bars, 5 μm in (A–C) and 500 μm in (D).
As proxy for the cell type-specific expression of endogenous Cdc14a, we examined β-galactosidase activity in mice heterozygous for a LacZ reporter inserted downstream of Exon 2 of mouse Cdc14a (Cdc14atm1a/+and Cdc14atm1b/+; Fig. 4D). Normal hearing heterozygotes show prominent β-galactosidase activity in the organ of Corti and the vestibular sensory epithelia consistent with endogenous CDC14A localization that persists at least until P60. LacZ signal is also detected in cells of the lamina spiralis ossea and in spiral ganglion (Fig. 4D). These observations suggest that the deafness of homozygous mutant Cdc14a mice, and humans with CDC14A mutations, may be owing to malfunctioning of hair cells, supporting cells and/or spiral ganglion neurons.
CDC14A genotype–phenotype relationship
In humans, different CDC14A alleles were associated with HL ranging from moderate to profound (Fig. 3A). In families HLRB11, HLAI24 and PKDF539, affected individuals in their first decade of life or early teens have significant HL but with lower thresholds than individuals in the second decade of life and older (Fig. 3A). Although we only have longitudinal audiometric data for some individuals, many affected individuals reported progressive loss of hearing with age, an observation corroborated by their parents. Moreover, in everyday life, affected individuals from the families HPK1 and PKSN10 use hearing aids for oral communication. In contrast, in family PKDF539 homozygotes for the substitution of proline-320 for glutamine (p.Q320P), predicted to abolish the phosphatase activity of CDC14A, are profoundly deaf rather than the less severe phenotype of individuals homozygous for truncating CDC14A variants, which perhaps allows for a low level of phosphatase activity (Fig. 3A). As described above, C-terminal truncations (p.R345X and p.R376X) are predicted to retain phosphatase activity while isoforms with a downstream translation start site might explain the less severe phenotype of the more N-terminal truncating mutations (p.Y126Ifx64X and p.Y139X). No vestibular problems were reported by affected individuals and no obvious balance problems were detected using Romberg and tandem gait tests.
Certain variants of human CDC14A are associated with male infertility
Inherited defects of cilia can affect multiple organ systems (31). Since we detected endogenous and over-expressed CDC14A protein in kinocilia and primary cilia, we extended the clinical history and physical evaluation in the families segregating mutations of CDC14A to include the possibility of comorbidity with other disorders. Deaf males in families HLRB11, HLAI24, HPK1, MORL1 and PKDF539 who were married stated that for many years they have been unable to father children (Fig. 1B). Semen analyses of 38-year-old (y/o) affected male IV: 2 (family HPK1) showed a low sperm count, and 75% of his sperm present were immotile and 60% had an abnormal morphology (Supplementary Material, Table S5). Individual IV: 10 (38 y/o, HLRB11) had no detectable sperm in his semen (Supplementary Material, Table S5), while two deaf brothers (HLAI24, IV: 3 18 y/o and IV: 2 19 y/o) had normal sperm counts, but 70 and 100% of their spermatozoa were immotile, respectively. Deaf male V:3 family (MORL1) has a normal sperm count but 67% were immotile. Other males in our study declined semen assessment. Nevertheless, not all deaf males with homozygous mutations of CDC14A are infertile. Deaf males IV:4, IV:6 (family PKSN10; Supplementary Material, Fig. S1A) and V:2 (MORL2; Supplementary Material, Fig. S1A) are biological fathers. Deaf females (families HLRB11, III:4 PKDF539, III:2 and III:4; PKSN10, IV:2) have multiple children (Fig. 1B and Supplementary Material, Fig. S1A).
We considered two explanations for comorbidity of deafness and male infertility in four of eight DFNB32-linked families segregating mutations of CDC14A (Fig. 1B and Supplementary Material, Fig. S1A). Since both disorders are common and genetically heterogeneous (25,32), the male infertility could be a coincidental finding. Examination of WES data of deaf infertile males IV-10 (HLRB11) and IV-2 (HLAI24) did not reveal pathogenic coding variants that might explain male infertility. Therefore, we explored the possibility that a CDC14A deficiency is associated with male infertility by examining the fertility of Cdc14a mutant mice.
Spectrum of mouse Cdc14a mutations associated with perinatal lethality, deafness and male infertility
Initially, we produced three different mutant alleles of mouse Cdc14a designated Cdc14atm1a (knockout first allele/exon trap), Cdc14atm1b (LacZ tagged and Cdc14a Exon 3 deleted producing a frameshift) and Cdc14atm1d (Exon 3 deleted; Supplementary Material, Fig. S8A). Homozygosity for any of the three alleles can result in perinatal lethality, indicating that CDC14A is also important for viability. From a cross between two heterozygous parents, 25% of pups are expected to be homozygous mutant. However, only 2.8% (12 of 436 pups) homozygous Cdc14a mutants survived to weaning, but thereafter these mice appear to have a normal life span. Unexpectedly, compound heterozygotes (Cdc14atm1a/tm1b) survive in greater numbers when compared with the either homozygote. We obtained 17 live Cdc14atm1a/tm1b pups from a total of 147 new born mice (12%), but still fewer than the expected 25% (χ2 of 10.99; P = 0.005). Hearing in the rare homozygotes (Cdc14atm1a/tm1a Cdc14atm1b/tm1b, Cdc14atm1d/tm1d) and compound heterozygotes (Cdc14atm1a/tm1b) were compared with that of heterozygotes and wild-type littermates using auditory brainstem response (ABR). At postnatal day 16 (Cdc14atm1a/tm1a, Cdc14atm1b/tm1b, Cdc14atm1d/tm1d and Cdc14atm1a/tm1b) the rare homozygotes and compound heterozygotes had only residual hearing at low frequencies (8 and 16 kHz). HL progressed to profound deafness at all frequencies by P90 (Fig. 3B and Supplementary Material, Fig. S8B). Therefore, Cdc14atm1a/tm1a Cdc14atm1b/tm1b, Cdc14atm1d/tm1d and Cdc14atm1a/tm1b mice show progressive loss of hearing, similar to the phenotype segregating in families HLRB11, HLAI24, HPK1 and PKSN10 (Fig. 3A). Absent distortion-product otoacoustic emissions (DPOAEs) at all ages tested (P16, 30, 60 and 90; Supplementary Material, Fig. S8C) indicated a loss of outer hair cell (OHC) function for both rare homozygotes and compound heterozygotes as early as P16. However, CDC14A seems unimportant for stria vascularis function as Cdc14a mutant mice have endocochlear potentials (EPs) that are in the normal range (Supplementary Material, Fig. S8D).
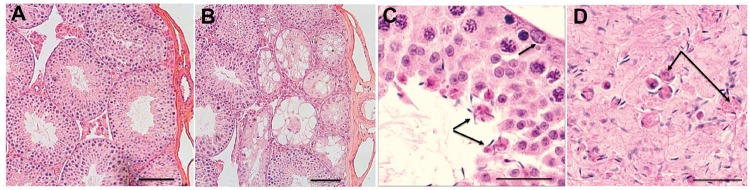
Regarding male infertility, over a 2- to 5-month period we repeatedly mated six of the rare surviving deaf male mice (Cdc14atm1b/tm1b n = 3, Cdc14atm1d/tm1d n = 2 and Cdc14atm1a/tm1b n = 1) with several demonstrably fertile wild-type B6 females. These males did not give rise to progeny, while Cdc14a homozygous mutant deaf females had many litters of healthy pups. We examined these infertile male mice for histological abnormalities of the testes. The Cdc14a mutant mice displayed varying stages of degeneration of the seminiferous tubules, primarily subcapsular and near the rete testis. The degeneration was characterized by loss of spermatogenic cells, vacuolization of sustentacular cells and partial collapse of the lumina. In addition, there were retained spermatid heads at the basement membranes and/or aggregates of spermatids around residual bodies compared with control animals (Fig. 5A–D). A significant decrease was noted in the epididymal sperm count in mutant animals compared with control littermates (Supplementary Material, Table S6). Mutant mice also had an increased number of abnormal sperm and excessive fragmentation (data not shown). We conclude that in mouse, in addition to deafness, mutant alleles of Cdc14a result in male infertility, suggesting that there is also an association of some biallelic mutant genotypes of human CDC14A with deafness and male infertility. We have named this monogenic syndrome Hearing Impairment and Infertile Male Syndrome. HIIMS is distinguished from Deafness-Infertility Syndrome (DIS OMIM 611102), which is owing to a chromosome 15q deletion of STRC and CATSPER2, genes associated with deafness and male infertility, respectively (33,34).
Figure 5.
Histopathology of mouse testes. (A) Cross section of 3-month-old wild-type mouse testes showing normal morphology of the seminiferous tubules in rete testes. (B) Cross-section of 3-month-old homozygous Cdc14atm1d/tm1d testes show degeneration of seminiferous tubules characterized by vacuolization of the epithelium. (C) Spermatids attached to residual bodies (arrows) in the seminiferous tubule of homozygous Cdc14atm1b/tm1b. (D) Epididymis of a homozygous Cdc14atm1b/tm1b with increased cell debris and sperm attached to globules of residual bodies (arrows). Defects shown in (C) and (D) are absent from the wild-type littermates (data not shown). Scale bars, 100 μm in (A, B) and 20 μm in (C, D).
Phosphatase-null cdc14aa mutant zebrafish have normal hearing, fertility and kinocilia
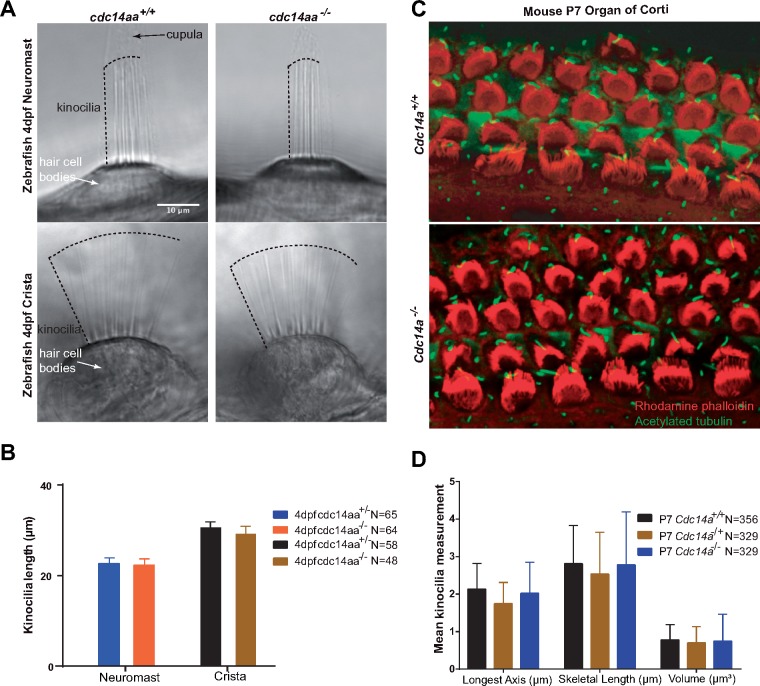
The zebrafish genome has two orthologues of human CDC14A that are designated cdc14aa and cdc14ab. Morpholino-knockdowns of cdc14aa expression were reported to cause variable shortening of inner ear kinocilia by 22.5% (21) and 14% (22). To determine if the morpholino-induced short kinocilia phenotype can be replicated with a germline mutation (35), we engineered using CRISPR/Cas9 a frame-shifting (G284fs4X; Supplementary Material, Fig. S9A–C) 7bp deletion (c.del 854_860; NM_201149) in the phosphatase domain of zebrafish cdc14aa. Its paralogue, cdc14ab, remained wild-type as confirmed by genomic DNA sequence analysis (data not shown). Based on our 3D model, cdc14aa p.G284fs4X is predicted to ablate phosphatase activity. However, for homozygous cdc14aa p.G284fs4X mutant fish, their startle response (Supplementary Material, Fig. S9D) and the variability and mean kinocilia lengths of cristae and neuromasts were indistinguishable from wild-type (Fig. 6A, B and Supplementary Material, Fig. S9E and F). Also, there were no bilateral symmetry defects as reported for a cdc14aa morpholino knockdown (22). Staining with FM1–43, a permeant styryl dye of open hair cell transduction channels (36), indicated that mechanotransduction is intact in these mutants (data not shown) and adult male fish homozygous for p.G284fs4X are also fertile.
Figure 6.
Kinocilia of hair cells in mouse organ of Corti and zebrafish neuromast and posterior crista from homozygous mutants are indistinguishable in length from wild-type. (A) Zebrafish 4 days post-fertilization (4 dpf) showing representative images of kinocilia for live-anesthetized heterozygous and homozygous mutant cdc14aa. (B) A total of 112 kinocilia (64 neuromast and 48 crista) from homozygous mutants and 123 (65 and 58) kinocilia from heterozygotes were measured. Two neuromasts and one crista examined per fish and 5 homozygous mutants and 5 heterozygotes zebrafish larvae were analyzed. Statistical analyses of mean kinocilia lengths of heterozygotes and homozygous mutant larvae show no significant difference. See Supplementary Material, Figure S9E and F for individual data points and statistical analyses. Error bars are standard deviations. (C) Representative images of P7 mouse organ of Corti from a wild-type and Cdc14atm1b/tm1b homozygous mutant mouse stained with rhodamine phalloidin (red) and acetylated tubulin (green). Lengths of kinocilia from two ears (Cdc14atm1d/tm1d) were measured using Volocity software to correct for parallax. (D) Kinocilia lengths (m) of wild-type and Cdc14atm1d/tm1d are not significantly different. Error bars are standard deviations. N is the number of hair cells examined.
Pathomorphology of auditory sensory epithelia of Cdc14a deaf mutant mice
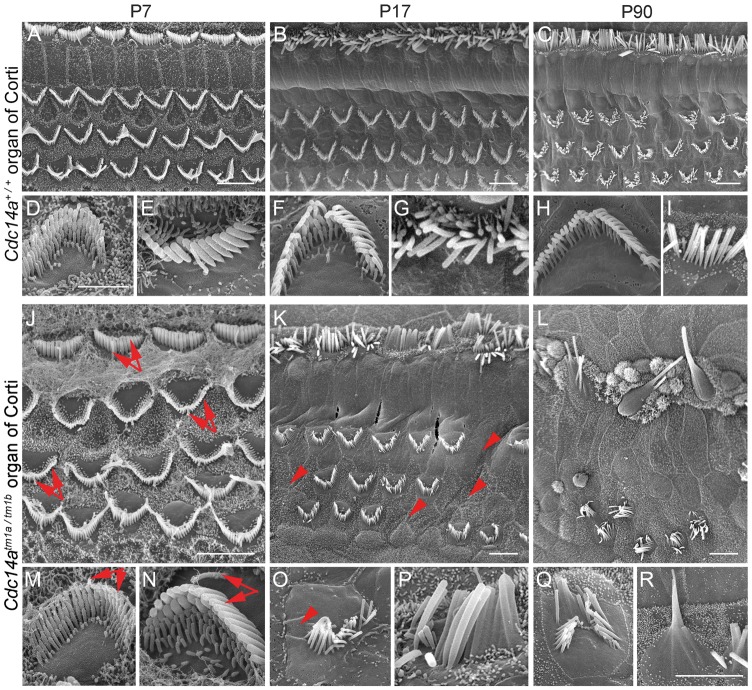
P7 inner ears of homozygous and compound heterozygous mutant mice (Cdc14atm1b/tm1b, Cdc14atm1d/tm1d and Cdc14atm1a/tm1b) were stained for acetylated tubulin and rhodamine-phalloidin to visualize the cytoskeleton. For the P7 organ of Corti from mutant mice, the morphology of hair cell stereocilia and kinocilia was indistinguishable from wild-type using either light microscopy (Fig. 6C and D) or scanning electron microscopy (SEM) (Fig. 7). By P17, 5% of mutant-derived inner and OHCs in the apical turn had degenerated and some stereocilia were fused to one another (Fig. 7). At this age, Cdc14a mutant mice have severe to profound HL and DPOAEs were absent indicating that OHCs are not functional. In the apical turn of a mutant P90 cochlea, 50% of inner and OHCs were missing when compared with 0.5% for wild-type littermates (Fig. 7). Pronounced hair cell loss in Cdc14a mutant mice at P90 was observed in the apical and basal turns of the organ of Corti with inner hair cell (IHC) degeneration preceding the loss of OHCs (Fig. 7). Progressive degeneration of hair cells paralleled the partial conservation of hearing at younger ages (P16-P30). Approximately 50% of hair cells were still present at P90 although all mutant mice were profoundly deaf suggesting that there may be an additional malfunction in the auditory system contributing to the HL of Cdc14a mutant mice.
Figure 7.
SEM images of inner (IHC) and outer hair cells (OHC) from the apical turn of the organ of Corti from wild-type and mutant Cdc14a mice showing stereocilia bundle morphology. (A–I) Three rows of OHC and one row of IHC stereocilia from the wild-type at three ages. (J–R) Comparable images from the compound heterozygous Cdc14atm1a/tm1b mouse. Higher magnification views of OHC (D, F, H and M, O, Q) and IHC (E, G, I and N, P, R) stereocilia bundles. IHCs and OHCs of Cdc14atm1a/tm1b mutants at P7 (J, M, N) appear to have normal kinocilia and stereocilia bundle morphology (arrows). At P17, for Cdc14atm1a/tm1b, some stereocilia of IHCs and OHCs fuse and undergo degeneration (K, O, P). Some OHC stereocilia and OHCs themselves are missing (K, O, arrowheads). Extensive degeneration of IHC and OHC stereocilia is observed at P90 (L, Q, R). Many OHC stereocilia bundles are missing (L), and most IHC stereocilia are degenerating (L, R). Scale bars for (A–C) and (J–L) are 5 m. A 2 m scale bar in D applies to D-I and a 2m scale bar in R applies to (M–R).
CDC14A phosphatase activity is necessary for hearing
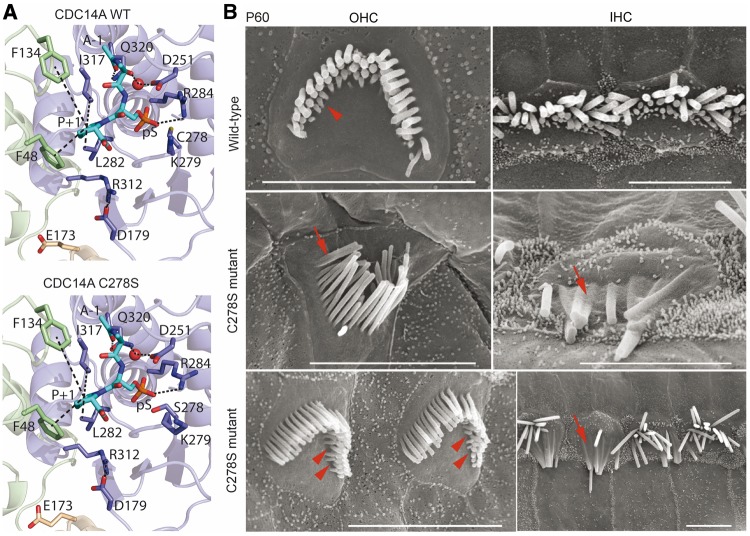
The locations of the three amino acid substitution variants of human CDC14A (p.R312Q, p.R312G and p.Q320P; Fig. 1D) segregating in families FT1, MORL1 and PKDF539, respectively, and the predicted loss of enzyme activity from our computational model (Fig. 2D–F), suggest that normal hearing requires the phosphatase activity of CDC14A. To test this prediction, we used CRISPR/Cas9 in mouse to edit codon 278, replacing cysteine with serine (c.832T>A, p.C278S; Fig. 8A). This specific substitution was chosen because biochemical studies of purified CDC14A protein demonstrated that serine at residue-278 ablates phosphatase catalytic activity while maintaining overall protein tertiary structure (20). The sulfur atom of cysteine-278 located at the center of the catalytic site is responsible for the nucleophilic attack on the phosphate (Figs 2 and 8A). The sulfur atom forms a covalent bond with the phosphate group, which is not possible if residue-278 is serine although similar enough not to impair protein folding (20,30). Consistent with the previous study (37), we observed that EGFP-C278S-CDC14A localization in inner ear hair cells is indistinguishable from that of wild-type CDC14A (data not shown).
Figure 8.
Structural model of CRISPR/Cas9 edited p.C278S variant of mouse CDC14A protein and SEM images of hair cells. (A) Catalytic site close-up view of structural models of mouse CDC14A wild-type and mutant p.C278S. The substrate (alanine-phosphorylated-serine-proline peptide) is in the binding site shown as cyan sticks where the alanine and the proline are indicated as A-1 and P + 1. The interactions involving the catalytic site residues are indicated by dashes. (B) SEM micrographs of P60 organ of Corti IHC and OHC stereocilia bundles of wild-type (upper row) and a homozygous p.C278S mutant (middle and lower rows). Wild-type OHC stereocilia bundle (upper left) shows three rows of stereocilia. In the shortest third row, most stereocilia are present (arrowhead). Homozygous p.C278S stereocilia are degenerating (middle and lower rows), fusing (arrows), and several third row OHC stereocilia are missing (arrowheads). Scale bars, 5 μm.
Our CRISPR/Cas9 oocyte injections produced 14 founder mice (F0) of which nine founders had the targeted p.C278S allele (Supplementary Material, Fig. S10). All of the ten founder mice including two homozygous C278S-CDC14A founder mice were severely-to-profoundly deaf. By SEM, inner ears of P60 deaf F0 homozygous C278S-CDC14A mice show fusion of stereocilia and degeneration of hair cells indistinguishable from other mutant alleles of Cdc14a (Fig. 8B). Homozygous F0 C278S-CDC14A male mice were infertile and showed degeneration of seminiferous tubules (data not shown) and had low sperm counts similar to other mutant alleles of Cdc14a (Fig. 5A–D). Germ line transmission of C278S-CDC14A was readily achieved. However, crosses between heterozygotes yielded only a few homozygous p.C278S mice (5 survivors/153 progeny) indicating that CDC14A phosphatase activity is also necessary to obtain an expected Mendelian ratio of the homozygous p.C278S genotype. Collectively, these data indicate that the phosphatase catalytic activity of mouse CDC14A is essential for male fertility and hearing and promotes perinatal viability.
Discussion
Dynamic reversible phosphorylation regulates the activity of numerous essential proteins (38,39). However, the in vivo functions requiring human CDC14A dual-specificity phosphatase have been enigmatic (12). Plausible indicators of the functions of mammalian CDC14A, and its paralogue CDC14B, began with observations that either of these two human genes can substitute for an otherwise lethal deficiency of endogenous cdc14 in budding and fission yeast (6). Unexpectedly, our data from human families segregating eight mutant alleles of CDC14A and Cdc14a mouse models reveal that CDC14A is required both for normal hearing and male fertility. At least in mouse, CDC14A is also advantageous for perinatal survival. The bias in ascertainment of human families segregating deafness likely precluded identifying recessive variants of human CDC14A associated with reduced viability, if they exist at all. Although the wild-type CDC14B paralogue may compensate in vivo for a loss of some CDC14A functions, and thus are shrouded, CDC14B does not substitute for CDC14A functions required for viability, fertility and hearing in mouse or loss of hearing and male infertility owing to pathogenic variants of human CDC14A.
Moderate-to-severe non-syndromic HL segregating in a Tunisian family genetically defined a chromosome 1p22.1–p13.3 locus designated DFNB32 (24). Subsequently, the DFNB82 deafness locus was genetically mapped and partially overlapped with the DFNB32 interval. This overlapping chromosomal interval has five annotated genes, none of which had mutations associated either with DFNB82 or DFNB32 phenotypes (40). Additional clinical data revealed that DFNB82-affected individuals have Chudley–McCullough syndrome and were segregating recessive pathogenic variants of GPSM2 (27). The GPSM2 gene was excluded from the chromosome 1p22.1–p13.3 linkage interval in the original DFNB32 family, consistent with the absence of pathogenic variants of GPSM2 in affected members (40). Yet, GPSM2 is mistakenly annotated as the gene underlying DFNB32 deafness (http://hereditaryhearingloss.org; December 30, 2017, date last accessed) and the DFNB105 locus is superfluous (21) as it is located entirely within the meiotic breakpoint-boundaries of the DFNB32 locus (Fig. 1A) defined by family FT1 (24).
Different mutations of CDC14A are associated with variation in the degree of HL ranging from moderate to profound. Our structural modeling predicted that the Q320 residue is essential for phosphatase catalysis, which may explain why a missense mutation (p.Q320P) in the phosphatase catalytic site was associated with severe-to-profound HL and male infertility in family PKDF539. In comparison, the C-terminal truncating mutations p.R345X (this study) and p.R376X (21) are associated with moderate-to-profound HL and males in the pedigrees are depicted as fertile. Truncating mutations of CDC14A in Exon 11 probably avoid NMD since a wild-type short isoform of CDC14A encoding 383 residues ends in Exon 11 (NM_003672) compared with the longest CDC14A isoform that has 15 exons (NM_0033312.2). Our computation model predicts that only residues 3–343 are important for phosphatase activity. In support of this prediction, truncated human CDC14A protein that includes only residues 16–365 can rescue mitosis of an otherwise lethal S. cerevisiae cdc14 mutant and is able to dephosphorylate cdk-1 in vitro (6,20). However, the human deafness phenotype of p.R345X and p.R376X points to an additional function of the CDC14A C-terminal sequence that is not conserved among its two orthologues and is perhaps involved in specifying localization to particular intracellular compartments.
CDC14 is associated with the centrosomes of many animals including C. elegans, Xenopus laevis, Gallus gallus and Homo sapiens (12). During interphase, the mother centrosome becomes the basal body of cilia, flagella and kinocilia. CDC14 of P. infestans, a plant pathogen, is also localized at the base of zoospore flagella (41). Similarly, in zebrafish, cdc14aa is localized to the base of cilia (22) and along the length of inner ear hair cell and neuromast kinocilia. Mouse CDC14A was reported to localize to kinocilia (21), while human CDC14A is associated with centrosomes and the basal bodies of cilia (9). Using several CDC14A custom and commercial antibodies we confirmed previously reported localizations of CDC14A in kinocilia and basal bodies and reveal additional localizations of CDC14A in hair cell stereocilia, as well as in nuclei and cytoplasm of different cells. When CDC14A-EGFP was over-expressed in the sensory epithelium of the mouse organ of Corti and the vestibular sensory epithelium, the EGFP signal was localized along the length of hair cell kinocilia, in their basal bodies, in stereocilia as well as in primary cilia, nuclei and cytoplasm of non-sensory cells. These data reinforce the finding of endogenous immunolocalization of CDC14A.
Abnormally short kinocilia leading to bilateral symmetry defects were reported in two independent studies that used antisense morpholinos to knock down expression of zebrafish cdc14aa (21,22). However, our mouse Cdc14a and zebrafish cdc14aa germline mutants do not have an obvious kinocilia dysmorphology. The absence of an obvious kinocilia phenotype in homozygous p.G284fs4X zebrafish that are null for phosphatase activity may be owing to activation of a genetic compensatory network by the germline mutation of cdc14aa as suggested for other genes (42), but not by morpholino-knockdown of cdc14aa expression. Alternatively, there may be functional redundancy between cdc14aa and cdc14ab, an explanation to be examined by engineering cdc14ab mutant fish and examining the phenotype of single and cdc14aa; cdc14ab double homozygous mutants. Discrepancies between morphant and germline zebrafish mutants may also be attributed to p53-dependent apoptosis, binding of the morpholino to superfluous targets or toxicity-associated developmental delays (35). Fifty percent (10 of 20) of different morphant phenotypes were not reproduced with germline mutations of the same target gene (35). We surmise from the phenotype of our germline cdc14aa mutant zebrafish and Cdc14a mutant mice that either the reported shortening of kinocilia by 22.5% is a spurious effect of morpholinos or there is compensation for the loss of CDC14A in germline mutants but not in morphants (42). We also observed that kinocilia within a single wild-type zebrafish neuromast/crista or in neighboring cells in the mouse cochlear can differ in length by 1–2 μm, similar to the variance in length for wild-type cilia in C. elegans (43). The biological significance of small length differences of sensory cilia is largely unknown (44).
SEM analyses also showed a normal appearance for inner ear auditory hair cells at P7 and P9 of Cdc14a homozygous mutants in mouse, and by P16 only a few hair cells had degenerated. These data indicate that CDC14A phosphatase is not necessary for the development of the wild-type architecture of hair bundles including transiently present kinocilia. Rather, after the onset of hearing at P17, CDC14A is required for conservation of hair cells; unless there is a developmental deficiency that manifests perhaps with sound stimulation. Our observations leave open the question as to whether CDC14A-associated deafness in humans and mice is the result of malfunctioning kinocilia, stereocilia or primary defects elsewhere that subsequently trigger degeneration of hair cells.
Infertility of some deaf males is associated with specific variants of CDC14A and defines a new monogenic syndrome (designated HIIMS) owing to insufficient phosphatase activity, while other variants with residual activity are associated with non-syndromic deafness DFNB32. There is precedent for allele-specific variants causing either a syndromic or non-syndromic HL. For example, different mutant alleles of TBC1D24 (OMIM 613577) are associated with either non-syndromic deafness, epilepsy, epilepsy and deafness or DOORS syndrome (OMIM 220500).
The phosphoprotein substrates of CDC14A critical for hearing and male fertility have not yet been identified, but the search is delimited by the subcellular localizations of CDC14A. For example, CDC14A is reported to form a complex with RN-tre, a Rab5 GTPase-activator and a protein partner of epidermal growth factor receptor-pathway-substrate 8 (EPS8), an actin filament remodeling protein (37). Mutant alleles of human EPS8 result in deafness (OMIM 615974) (45) and Eps8 mutant mice have dysfunctional diminutive stereocilia (46). During development, microvilli on the apical surface of hair cells in Eps8 mutant mice fail to elongate stereocilia. If dephosphorylation of EPS8 by wild-type CDC14A is necessary for stereocilia development, then Cdc14a mutant hair cells would have short stereocilia. Yet, mice homozygous for mutant alleles of Cdc14a initially develop wild-type-length stereocilia that begin to degenerate after P16 (Fig. 7). Regulated dephosphorylation of EPS8 by CDC14A appears unnecessary for the development of stereocilia bundle architecture, but may be necessary to maintain wild-type hair cell function. We also can’t rule out the possibility that the same phosphoprotein substrate of CDC14A is required both for maintenance of stereocilia and integrity of spermatozoa.
Key questions remain as to the reasons for reduced perinatal viability of homozygous Cdc14a mutant mice and the identity of substrates of CDC14A phosphatase required for hearing, male fertility and viability. Our findings do show that mutations of mouse Cdc14a, but not Cdc14b, cause profound deafness and male infertility and compromise survival during development. In human, deafness and male infertility for some recessive variants of human CDC14A define a new allele-specific syndrome (HIIMS) affecting hearing function in both males and females and male reproduction. This molecular genetic study has provided new insight into the multifaceted in vivo functions and clinical variability of mutant alleles of human CDC14A, mouse Cdc14a and zebrafish cdc14aa.
Materials and Methods
Statement of ethics and clinical assessments
This study was approved by the Combined Neuroscience Institutional Review Board at the National Institutes of Health (NIH), USA (protocol OH-93-N-016 to T.B.F.) In addition, approval was obtained from institutional review boards at the School of Biological Sciences (# 00005281 to S.N.) and the Centre of Excellence in Molecular Biology (FWA00001758 to S.R.), University of the Punjab, Pakistan, The Comité Consultaif d’étique à Sfax, Tunisia (to S.M.), and at the University of Iowa (#199502083 to R.J.H.S). Ascertainment was initiated after obtaining written informed consent. Zebrafish and mouse protocols were approved by the NIH Animal Use Committee, #1362-13 to K.K. and #1263-15 to T.B.F, respectively.
Probands were identified through schools for hearing impaired children in Pakistan and Iran. Pure tone thresholds were measured at 0.25, 0.5, 1, 2, 4 and 8 kHz in a quiet location for members of families HLRB11, HLAI24, HPK1, PKSN10, PKDF539 or in a sound-proof booth for members of families FT1, MORL1, MORL2. HL was classified based on averaging thresholds (0.5, 1, 2 and 4 kHz) as moderate (41–70 dB HL), severe (71–95 dB HL) and profound (>95 dB HL). Subjects were asked about the age of onset and progression of HL. Vestibular function was evaluated by tandem-gait and Romberg tests and no obvious balance problems were detected. Blood chemistries including liver, kidneys and testes function were obtained from two affected individuals per family. Semen analyses were obtained from six consenting deaf males.
Genetic and WES analyses
Families PKDF539 and FT1 were initially screened by a genome-wide linkage scan using 360 STR markers, while other families were screened by targeted STR genotyping and sequencing of known deafness genes (47). WES was performed on three affected individuals from families HLAI24 (IV:2, IV:10, IV:11), HLRB11 (IV:1, IV:2, IV:3) and two individuals (one affected and one unaffected) from families MORL1 (V:1 and V:4) and MORL2 (V:4 and V:5). Nextera Rapid Capture Exome (32 920 080-bp of exonic sequence, Illumina) was used to prepare WES libraries for members of families HLRB11 and HLAI24 and sequenced on an Illumina HiSeq 1000. Agilent SureSelect All Exon V5 bait kit was used for families MORL1 and MORL2 and libraries sequenced on an Illumina HiSeq 4000. Ninety-four percent of captured bases had greater than 10×-coverage (Supplementary Material, Table S7). The smallest DFNB32 linkage interval contains 57 991 coding nucleotides, 95.6% had >10× coverage (Supplementary Material, Table S7) while the remaining 4.4% was screened by Sanger sequencing. For families PKSN10, HPK1, PKDF539 and FT1, exons and intronic boundaries of CDC14A were also assessed for variants by Sanger sequencing.
Splicing analyses of Exon 10 c.829-3C>G variant
RNA was extracted from 1 ml of blood of two affected and one normal carrier from family HPK1 using Trizol (ThermoFisher). cDNA libraries were synthesized using Revert Aidtm Premium (Fermentas). Oligo-dT and CDC14A gene-specific primers (P1, P2 and P3) were used to prepare cDNAs. cDNA across Exon 10 was amplified by primer-pairs in Exons 9 and 11 (P4 and P5; Supplementary Material, Table S8) to study the effect of c.829-3C>G variant on Exon 10 splicing. cDNA was cloned into TOPO-TA vectors (ThermoFisher) and 16 clones were sequenced for each polymerase chain reaction (PCR)-product.
cDNA expression constructs, immunolocalization, antibody validation and LacZ staining
Full length mouse Cdc14a cDNA (NM_001080818) was PCR amplified from P7 mouse cochlear oligo(dT)-primed cDNAs using primers P5 and P6 (Supplementary Material, Table S8). PCR products were cloned into a pCR4-TOPO TA vector (Invitrogen), confirmed by Sanger sequencing and then cloned into to expression vectors pAcGFP-N1 and pGFP-C2 via In-Fusion cloning (Clontech). CDC14A with an N-terminal GFP (pGFP-C2) or a C-terminal GFP (pAcGFP-N1) are designated GFP-CDC14A and CDC14A-GFP, respectively, and were sequence verified. Three equivalent deafness-associated human mutations p.R312Q, p.R320Q and p.R345X were introduced by GeneArt Site-Directed Mutagenesis (Invitrogen) into mouse Cdc14a cDNA cloned in pGFP-C2 (Clontech).
Anti-human CDC14A antibodies were custom made (polyclonal antibody DSPc-Ab) or obtained commercially from Novus Biological (NBP1-84573), Zymed (34-8100) and Genetex (GTX52005). Catalog numbers are used to refer to each antiserum. All antibodies were evaluated and validated by co-immunolocalization assays with CDC14A-GFP transfected COS-7 cells. COS-7 cells were grown to 60–70% confluency on coverslips in Dulbecco’s modified Eagle medium supplemented with 10% FBS at 37°C and 10% CO2. Then using Fugene HD reagent (Promega), cells were transfected with cDNA expression vectors encoding an N- or a C-terminal GFP-tagged mouse CDC14A. The next day, cells were washed, fixed with 4% paraformaldehyde in 1× phosphate buffered saline (PBS), permeabilized with 0.5% Triton-X100, blocked with 2% bovine serum albumin (BSA) and 5% goat serum in 1× PBS and incubated for 2 h with an anti-CDC14A primary antibody. Cells were washed in 1× PBS and stained with Alexa Fluor 568 anti-rabbit or anti-mouse IgG (Molecular Probes) secondary antibodies followed by three washes in 1× PBS. Samples were mounted with ProLong Gold Antifade with DAPI (Molecular Probes) and imaged using a LSM780 and a 63× 1.4NA objective (Carl Zeiss). Antisera against human CDC14A were selected for immunostaining of mouse tissue only when epitopes were conserved. Antibodies 34-8100 and NBP1-84573 showed specific immunoreactivity to full length mouse CDC14A-GFP (data not shown). MAB4457 (mouse monoclonal anti-human CDC14A antibody) showed immunoreactivity only to a shorter N-terminal peptide of residues 1–106. Four commercial CDC14A antibodies did not show cross-reactivity to mouse CDC14B-GFP that was transfected into COS-7 cells (data not shown) while a positive control, commercial CDC14B antisera (NBP1-84572, Novus Biological) detected CDC14B-GFP (data not shown). Immunolocalization of zebrafish Cdc14aa was studied using polyclonal antibody X-Q7SXZ4-Ab, raised against C-terminal epitopes of zebrafish Cdc14aa (Supplementary Material, Fig. S7A and Table S4). The locations of the epitopes are shown for mouse CDC14A (Supplementary Material, Fig. S7A). Immunolocalization conducted at ages P0 to P12, P30 and P60 as described (48). Briefly, otic capsules were fixed with 4% paraformaldehyde in 1× PBS, permeabilized with 0.5% Triton-X 100, blocked with 2% BSA and 5% goat serum in 1× PBS, and incubated with anti-CDC14A primary antibodies. In some experiments, acetylated tubulin antibody was added to the primary antibody solution. After washes in 1× PBS, samples were stained with Alexa Fluor 488, 568 and 647 anti-rabbit or anti-mouse IgG (Molecular Probes) secondary antibodies followed by three washes in 1× PBS. Samples were mounted in ProLong Gold Antifade Reagent with or without DAPI (Molecular Probes) and imaged as described above.
LacZ staining was performed using Cdc14atm1a and Cdc14atm1b heterozygotes at P6, P12 and P60. Briefly, otic capsules were dissected in 1× PBS solution and incubated in fixation buffer (1× PBS with 1% formaldehyde, 0.2% glutaraldehyde, 0.02% NP-40) for 10 min at room temperature. The preparations were washed two times (30 min each) in 1× PBS solution with 0.02% NP-40 and 2 mM MgCl2 followed by incubation in a LacZ staining solution (1× PBS, 1 mg/ml X-gal; Sigma Aldrich), 5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6, 2 mM MgCl2) for 2–24 h at 37°C. After staining, otic capsules were washed once in 1× PBS containing 0.02% NP-40 and 2 mM MgCl2. Otic capsule from P60 mice were first decalcified for 3 days at RT in 2% paraformaldehyde containing 0.25 M EDTA. The softened bony capsule was peeled away and the sensory epithelia microdissected from inner ears, mounted on glass slides using Immu-Mount mounting media (ThermoFisher) and imaged on an Eclipse90i (Nikon).
COS-7 transfections and co-localization with cytoskeletal components
GFP-tagged full length wild-type and mutant mouse Cdc14a (NM_001080818) cDNA constructs were transfected into COS-7 cells grown on coverslips in 6-well dishes (Costar). All plasmids were transfected at a final concentration of 0.02 μg/μl/well. Mouse acetylated tubulin, mouse α- and γ-tubulin primary antibodies (T7451, T9026 and T6557, Sigma Aldrich) and β-tubulin antibody (ab11309-200, abcam) were used to highlight microtubular network while rhodamine-phalloidin (Life Technologies) stained F-actin.
Computational modelling methods of CDC14A
Human CDC14B structure (PDB identifier: 1hoe) (49) was identified as a template for structural modeling of human CDC14A (NP_201569) and mouse CDC14A (NP_001074287) using HHpred (50), which used as input the Hidden Markov-model (HMM) human CDC14A (NP_201569) profile obtained with HHblits (51) after three iterations against the UniProt20 database (February 2016). The sequence coverage, correspondence between secondary structural elements and the 64% sequence identity with conserved domains confirmed human CDC14B as a suitable template among those obtained with HHpred. The sequence alignment between the query sequence human CDC14A (NP_201569) and the template generated by HHpred covered residues 4–343 in CDC14A and 42–379 in the template CDC14B. Similar alignments were obtained with MSAProbs (52) and MUSCLE (53). Human (NP_201569) and mouse CDC14A (NP_001074287) sequences were aligned using MSAProbs (52). The homology models containing domains A and B (residues 4–343) of human CDC14A (NP_201569) (UniProt Q9UNH5–2) and mutants p.R312Q, p.R312G, p.Q320P and mouse CDC14A isoform 1 and p.C278S were obtained after 2000 iterations of Modeller (54). The selected model had the lowest MolPDF score, and subsequently evaluated using ProQ2 (55) and PROCHECK (56). The ProQ2 scores for the final models were all ∼0.76 compared with 0.969 for the template and only one residue (Lysine-197), was found to be in a disallowed region according to PROCHECK. The crystallographic water molecules as well as the substrate (A-pS-P) present in the template were also included in each model with no additional distance restraints. The structural superimposition of domains A and B for the human CDC14A (NP_201569) model was performed using TM-align (57), and a similar alignment was obtained with Pymol (Molecular Graphics System v1.8). The root-mean-square deviation of atomic positions (RMSD) obtained between domains was of 3.1 Å. Similar results were obtained when the structures of domains A and B in the template structure (CDC14B) were compared.
Cdc14a and Cdc14b mutant mouse models
Four different mutant alleles of mouse Cdc14a were developed from ES cells obtained from KOMP. B6N ES cells [HEPD0623_4_G09; Cdc14a knockout first allele, Cdc14atm1a(EUCOMM)Hmgu (herein abbreviated Cdc14atm1a)] were obtained from Eucomm (Fig. 3; Supplementary Material, Fig. S1). These ES cells have a 6-kb insertion in Intron 2 that includes two FRT sites flanking a cassette that includes an Engrailed Exon 2 splice acceptor site (EN2SA), a -galactosidase reporter (LacZ) and a neomycin selection cassette (NeoR; Supplementary Material, Fig. S5A). Three loxP sites flank both the NeoR and Exon 3. Transcripts from the Cdc14atm1a allele splice from Exon 2 to the En2SA donor site of the exon trap in Intron 2. Blastocyst injections of the Cdc14atm1a(EUCOMM)Hmgu ES cells (HEPD0623_4_G09) were made at University of Michigan Transgenic Animal Model Core generating Cdc14atm1a(EUCOMM)Hmgu chimeric mice.
Germline transmission of the Cdc14atm1a allele was achieved when tm1a chimeras were mated to wild-type albino B6N females. Cdc14aTm1a heterozygotes (Supplementary Material, Fig. S8A) were crossed to a wild-type [B6N Tg(ACTB-cre)2Mrt, https://www.jax.org/strain/019099)] to obtain body-wide LacZ expression while deleting Exon 3 (i.e. Cdc14atm1b(EUCOMM)Hmgu, abbreviated Cdc14atm1b). Additionally, heterozygous Cdc14aTm1a mice were mated to ACTB: FLPe B6N (https://www.jax.org/strain/019100) to obtain the conditional allele Cdc14atm1c(EUCOMM)Hmgu (abbreviated Cdc14atm1c) in which Exon 3 is floxed. A Cdc14atm1c mouse was crossed to B6N Tg(ACTB-cre)2Mrt to obtain Cdc14atm1d(EUCOMM)Hmgu, which has a body-wide deletion of Exon 3 (Cdc14atm1d allele) and LacZ has been deleted (Supplementary Material, Fig. S8A). Heterozygous Cdc14atm1a, Cdc14atm1b and Cdc14atm1d mice were subsequently crossed to obtain homozygous mutants for each allele. Mice were genotyped at P1–P21 using PCR primers P6–P9 (Supplementary Material, Fig. S8A and Table S8). The Cdc14b mutant mouse [B6.129P2(Cg)-Cdc14btm1.2Pzg/J] has a frame-shifting deletion (58) of Exon 2 and was obtained from JAX and maintained on a B6N background.
Auditory testing
ABRs and DPOAE were obtained using Tucker-Davis Technologies (TDT; Alachua) hardware (RZ6 Processor) and software (BioSigRZ v5.1) as described (48). ABRs were conducted at P16, P30, P60 and P90.
EP measurements
Mice were anesthetized with tribromoethanol at a dose of 0.35 mg/g body weight. EP measurements were made in the basal turn of the cochlea by a round-window approach through the basilar membrane of the first turn (59). Glass microelectrodes preparation was previously described (60,61). Anoxia was induced by intramuscular injection of succinylcholine chloride (0.1 µg/g) after establishment of deep anesthesia followed by additional injection of tribromoethanol. Data were recorded digitally (Digidata 1440A and AxoScope 10; Axon Instruments) and analyzed using Clampfit10.
Quantification of kinocilia length of Cdc14a mutant mouse
Kinocilia in the P7 organ of Corti were immunostained with primary kinocilin (KNCN) antibody (NBP2-48844, Novus Biological) and Alexa Fluor 488, anti-rabbit IgG secondary antibodies. Kinocilin localizes along the length of kinocilia at P7 (62). Full lengths of kinocilia in consecutive sections of apical turns were imaged by performing Z-stacks using a LSM780 and a 63 × 1.4 NA objective (Zeiss). Kinocilia skeletal lengths, longest axis and volume of 300–400 cells were measured using Volocity software (Perkin Elmer).
Helios gene gun transfections of inner ear sensory epithelia
Mouse P2–P3 organ of Corti and vestibular sensory epithelial explants were cultured and transfected with both N- and C-terminal EGFP-tagged full-length mouse CDC14A in separate experiments using a Helios gene gun as described (63). Gene gun-mediated transfections were also done using mouse Cdc14a cDNA expression constructs bearing human-equivalent R312Q, Q320P or R345X mutations and an N-terminal EGFP. After 24 h of transfection, cultures were fixed overnight in 4% paraformaldehyde at 4°C and stained with acetylated tubulin antibody (blue) and rhodamine-phalloidin. Samples were mounted with ProLong Gold Reagent with or without DAPI and imaged using LSM780 Zeiss confocal microscope equipped with 63× 1.4 NA objective (Zeiss, Inc.).
Testes histopathology, sperm count and motility
Five mutants (Cdc14atm1b/tm1b n = 2, Cdc14atm1a/tm1b n = 1, Cdc14atm1d/tm1d n = 1 and Cdc14aC278S/C278S n = 1), five wild-type littermates and two heterozygotes (Cdc14atm1d/+, Cdc14atm1b/+) were included for testes histology and sperm analyses. Tissues required for microscopic evaluation (right testis and epididymis) were weighed and fixed in modified Davidson’s solution. Tissues were trimmed, processed routinely, embedded in paraffin and stained with hematoxylin and eosin for light microscopic evaluation of spermatogenesis (Charles River Laboratories). Microscopic evaluation was conducted by two board-certified veterinary pathologists. Pathologists were blinded to the genotype and reported that each mutant had histological abnormal findings when compared with the wild-type and heterozygous littermates. Sperm from the cauda epididymis were used for motility analyses (Charles River Laboratories). A validated Hamilton Thorne IVOS instrument was utilized for sperm count and motility assessment. The left epididymis was immediately placed in a petri dish containing 2 ml of 1% BSA dissolved in Dulbecco’s PBS warmed to 36–41°C and sperm allowed to disperse. Approximately 9 μl was loaded into the pre-warmed stage of the sperm analyzer. The analyzer was programmed to select fields until a minimum count of 200 sperm or a maximum of 20 fields were achieved. The percent of motile sperm was determined for each animal. For the total sperm count, the left epididymis was minced in 2 ml of media and two sperm smear slides were prepared and dried. The minced epididymides were then diluted with 8 ml of deionized water and homogenized. Each sample was briefly vortexed and a 100 μl sample of each homogenate was transferred to a reaction vial containing a dye which stains the head of the sperm. A 9-μl aliquot of the stained sperm was placed into a 20-μm deep Cell-Vu glass slide and loaded into the analyzer. Twenty fields per nimal were analyzed and the number of epididymal sperm determined. The counts reported were adjusted for caudal epididymal weight. For morphological examination, two eosin stained slides were prepared for each animal from the caudal epididymis a minimum of 200 sperm cells per animal were examined by light microscopy for morphological development.
Scanning electron microscopy
Inner ears from P7, P9, P17, P60 and P90 mutant and wild-type mouse were dissected from temporal bones, fixed for 2 h at room temperature in 2.5% glutaraldehyde in 0.1 M cacodylate buffer (EMS) augmented with 2 mM CaCl2, dehydrated in a graded series of ethanol (EtOH), processed for SEM as described (48) and imaged using a Hitachi S-4800 SEM. Harvested cauda epididymis was incubated for 5 min at 37°C in pre-warmed (37°C) 1% BSA buffer to allow sperm to disperse in the medium. Epididymis plasma was fixed with cold 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer for 2 h at 4°C. Fixative was removed by centrifugation at 1600 rpm for 5 min, and the sperm pellet was washed in cacodylate buffer and post-fixed in 1% osmium tetroxide, pH 7.2–7.4, for 1 h. Then sperm were transferred to poly-l-lysine coated glass cover slips, dehydrated in ascending graded ethanol (50, 70, 95 and 100%), critical-point dried using CO2 (CPD 030, Bal-Tec) and sputter-coated with 4 nm of platinum (Q150T-ES, Quorum Technologies).
Zebrafish husbandry and CRISPR/Cas9 editing
Adult Danio rerio were housed using standard methods on a 14-h light, 10-h dark cycle. Larvae were examined at 3–7 days post fertilization (dpf) unless stated otherwise. Zebrafish larvae were raised in E3 embryo media (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2 and 0.33 mM MgSO4) at 30°C. A CRISPR/Cas9 target within Exon 10 of cdc14aa (NM_201149) was selected using CRISPR tools (http://crispr.mit.edu/). A 58-mer oligo and 80-mer (Supplementary Material, Table S8) oligos were annealed to form an 118 nt guide DNA by PCR extension at 72°C for 10 min (Supplementary Material, Table S8) (64) and purified (Agencourt AMPure XP-beads, Beckman-Coulter). Guide RNA was synthesized (HiScribe T7 High Yield RNA, NEB), purified by isopropanol/sodium acetate precipitation and quantified (Agilent small RNA quantification kit). Cas9 RNA was transcribed from pT3TS-nCas9n plasmid using mMESSAGE-mMACHINE T3 (Life Technologies) and quantified (NanoDrop 2000c, ThermoFisher). Next, 2–5 nl of 50 ng/μl gRNA, 50 ng/μl Cas9 RNA and 0.05% phenol red (Sigma-Aldrich) were injected into zebrafish zygotes using a Pneumatic PicoPump (WPI). Eight to sixteen 5-day-old embryos were selected to test efficiency of injected gRNA and to identify mutations of cdc14aa. Each larva was placed in a tube with lysis buffer, DNA was extracted and a 500 bp genomic region surrounding the CRISPR targeted sequence was amplified and cloned into a TOPO vector and four clones per larva sequenced (Supplementary Material, Fig. S9). Remaining injected embryos were grown to adulthood to identify cdc14aa mutants. Each founder was outcrossed to wild-type TAB5 fish and the F1 generation was genotyped at 3 months of age. Heterozygotes for the same deletion or insertion were crossed to obtain homozygous mutants.
Phenotyping homozygous zebrafish mutants
To visualize mature hair cells and assay mechanotransduction, 5 days post fertilization (dpf) larvae were immersed in a 3-μM solution of FM1–43 (Thermo) in E3 (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2 and 0.33 mM MgSO4) for 30 s. Larvae were immediately washed three times in E3 and mounted on their sides in 2% low-melt agarose in E3 media containing 0.03% MESAB (Sigma). Anesthetized larvae were imaged by confocal microscopy. Kinocilia length measurements in phenotypically wild-type heterozygous siblings and cdc14aa mutants were made on live zebrafish larvae at 5 dpf. To measure neuromast kinocilia length, each larva was individually anesthetized with 0.03% MESAB in E3 embryo media. Larvae were mounted dorsally with tungsten pins onto a Sylgard matrix (Dow Corning). To add contrast for neuromast kinocilia measurements, 0.10 µm #16688 polybead-microspheres (Polysciences) were added to E3 media at 5% (v/v). To measure kinocilia length in crista, larvae were mounted on their sides in 1% low-melt agarose (E3 media plus 0.03% MESAB). Images of kinocilia were acquired on a Nikon C2 (60× water/1.0 NA) confocal microscope. A 488-nm solid state laser was used to excite and a transmitted PMT detector was used to collect z-stacks of neuromasts and cristae, with images taken every 0.5 µm. Kinocilia lengths were examined from one medial crista and two primary posterior lateral line neuromasts (L2 and L3) per larva. Images were analyzed using ImageJ. Two tailed t-test and Mann–Whitney test were used to compare populations.
Startle response
The auditory function of zebrafish was evaluated using a DanioVision chamber and EthoVision XT software (Noldus). Heterozygous male zebrafish were crossed with heterozygous females and embryos collected. Ninety-six 6 dpf larvae were transferred to a 96-well plate (1× 1 cm wells) and placed into the DanioVision chamber and allowed to equilibrate to 28°C. Zebrafish movement was recorded for 10 s before and after initiation of the tapping function. Recordings were made for a series of tapping stimuli (Levels 1–8) with a resting interval of 10 min. The startle response was measured from activity plots.
CRISPR/Cas9 genome editing of the endogenous mouse Cdc14a gene
The serine to cysteine substitution at codon-278 was created by CRISPR-mediated genome editing directly in fertilized eggs of C57BL/J (Jackson Laboratory) as described (65). Three target sequences for guide RNA (gRNA) near to the target codon were initially selected using the ranking tool CRISPR Design (http://crispr.mit.edu). Selected gRNAs were synthesized by T7 in vitro transcription using a MegaScript T7 kit (Ambion) as described (65,66). The gRNAs were then compared for their activity to introduce a double strand break at the target site in mouse embryonic fibroblasts with a Surveyor assay kit (data not shown) (Integrated DNA Technology). gRNA gCD-1 (Supplementary Material, Table S8) was chosen. A single stranded mutagenic DNA oligonucleotide (126-mer) carrying the c.832T>A transversion (NM_001080818) was designed based upon the position and orientation of the guide RNA gCD-1 (65) (Supplementary Material, Table S8) used to create the p.C278S substitution. To generate the c.832T>A point mutation in the genome of fertilized eggs, C57BL6/J donor eggs were microinjected with a mixture of SpCas9 protein (50 ng/µl, PNA Bio), and gRNA gCD-1 (50 ng/µl) and the mutagenic oligo (50 ng/µl) as described (66). F0 founders carrying the mutation (c.832T>A; C278S) were evaluated by PCR with forward (P12; Supplementary Material, Table S8) and reverse primers (P13, Supplementary Material, Table S8) and the mutation (c.832T>A; p.C278S) was confirmed in F0 founders by sequencing (Supplementary Material, Fig. S10). The c.832T>A (p.C278S) mutant allele was assigned the symbol Cdc14aem1Tbf (MGI: 5810960).
Supplementary Material
Supplementary Material is available at HMG online.
Authors’ Contributions
Conceptualization, A.I., T.B.F., I.A.B., S.N., K.S.K.; Manuscript first draft, A.I., I.A.B., K.K., S.N., T.B.F.; Ascertained families and obtained clinical data, A.I., R.B., A.M., A.M.W., I.B., S.M., S.N., S.R., K.K., H.N.; Supervised work in Tunisia, S.M.; Supervised and designed study of Iranian families, H.A. and R.J.H.S.; Structural modeling, C.F-F; Sequencing/genotyping, WES, A.I., I.B., R.B., S.N., R.J.M., M.L., T.C., A.M., H.A., H.Y., U.S., A.U.R., A.B.; Zebrafish genetics/CRISPR/Cas9 mutagenesis, A.I., A.J.B., K.S.K., T.B.F.; CRISPR/Cas9 mutagenesis of mouse Cdc14a, L.D., A.I., Mouse histology, M.S.; SEM, confocal microscopy, gene gun transfections, I.A.B., A.I.; Mouse ABR, E.A.W. and T.S.F.; EP measurements, R.O., M.H.
Supplementary Material
Acknowledgements
We thank the study participants and Muhammad Akhtar, Zeeshan Ahmed, Carmen Brewer, Steve Boyden, Patrick Diers, Dennis Drayna, Meghan Drummond, Elizabeth Driver, Joe Duda, Linda Kooistra, James McGehee, Memoona Ramzan, Keri Richards, Thomas Saunders, Jamie Sexton, Muhammad Usman, Philine Wangemann, Edward Wilcox and Barbara Zwiesler.
Conflict of Interest statement. None declared.
Funding
The work was supported in part by HEC, Islamabad to S.R., Ministry of Higher Education and Scientific Research Tunisia to S.M., NIH Intramural Research Programs NEI ZIC-EY000458-09 to L.D.), NIMH MH002946 for C.F.-F., NIDCD/NIH DC003544, DC002842 and DC012049 to R.J.H.S., NIDCD DC000080 to T.S.F., NIDCD/NIH Intramural Research Program DC000088-03 to M.H., NIDCD/NIH Intramural Research Program DC000086 to R.J.M, NIDCD DC000085-01 to K.S.K., NIH R01TW007608 and HEC 3288 to S.N. and funds from the NIDCD/NIH Intramural Research Program DC000039 to T.B.F. This work utilized computational resources of the NIH HPC Biowulf cluster.
References
- 1. Wood J.S., Hartwell L.H. (1982) A dependent pathway of gene functions leading to chromosome segregation in Saccharomyces cerevisiae. J. Cell. Biol., 94, 718–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Higuchi T., Uhlmann F. (2005) Stabilization of microtubule dynamics at anaphase onset promotes chromosome segregation. Nature, 433, 171–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stegmeier F., Visintin R., Amon A. (2002) Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell, 108, 207–220. [DOI] [PubMed] [Google Scholar]
- 4. Sullivan M., Higuchi T., Katis V.L., Uhlmann F. (2004) Cdc14 phosphatase induces rDNA condensation and resolves cohesin-independent cohesion during budding yeast anaphase. Cell, 117, 471–482. [DOI] [PubMed] [Google Scholar]
- 5. Visintin R., Craig K., Hwang E.S., Prinz S., Tyers M., Amon A. (1998) The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol. Cell, 2, 709–718. [DOI] [PubMed] [Google Scholar]
- 6. Li L., Ernsting B.R., Wishart M.J., Lohse D.L., Dixon J.E. (1997) A family of putative tumor suppressors is structurally and functionally conserved in humans and yeast. J. Biol. Chem., 272, 29403–29406. [DOI] [PubMed] [Google Scholar]
- 7. Vazquez-Novelle M.D., Esteban V., Bueno A., Sacristan M.P. (2005) Functional homology among human and fission yeast Cdc14 phosphatases. J. Biol. Chem., 280, 29144–29150. [DOI] [PubMed] [Google Scholar]
- 8. Gruneberg U., Glotzer M., Gartner A., Nigg E.A. (2002) The CeCDC-14 phosphatase is required for cytokinesis in the Caenorhabditis elegans embryo. J. Cell. Biol., 158, 901–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mailand N., Lukas C., Kaiser B.K., Jackson P.K., Bartek J., Lukas J. (2002) Deregulated human Cdc14A phosphatase disrupts centrosome separation and chromosome segregation. Nat. Cell Biol., 4, 317–322. [DOI] [PubMed] [Google Scholar]
- 10. Saito R.M., Perreault A., Peach B., Satterlee J.S., van den Heuvel S. (2004) The CDC-14 phosphatase controls developmental cell-cycle arrest in C. elegans. Nat. Cell Biol., 6, 777–783. [DOI] [PubMed] [Google Scholar]
- 11. Wurzenberger C., Gerlich D.W. (2011) Phosphatases: providing safe passage through mitotic exit. Nat. Rev. Mol. Cell Biol., 12, 469–482. [DOI] [PubMed] [Google Scholar]
- 12. Mocciaro A., Schiebel E. (2010) Cdc14: a highly conserved family of phosphatases with non-conserved functions? J. Cell Sci., 123, 2867–2876. [DOI] [PubMed] [Google Scholar]
- 13. Lin H., Ha K., Lu G., Fang X., Cheng R., Zuo Q., Zhang P. (2015) Cdc14A and Cdc14B redundantly regulate DNA double-strand break repair. Mol. Cell Biol., 35, 3657–3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schindler K., Schultz R.M. (2009) The CDC14A phosphatase regulates oocyte maturation in mouse. Cell Cycle, 8, 1090–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yuan K., Hu H., Guo Z., Fu G., Shaw A.P., Hu R., Yao X. (2007) Phospho-regulation of HsCdc14A By Polo-like kinase 1 is essential for mitotic progression. J. Biol. Chem., 282, 27414–27423. [DOI] [PubMed] [Google Scholar]
- 16. St-Denis N., Gupta G.D., Lin Z.Y., Gonzalez-Badillo B., Veri A.O., Knight J.D., Rajendran D., Couzens A.L., Currie K.W., Tkach J.M.. et al. (2016) Phenotypic and interaction profiling of the human phosphatases identifies diverse mitotic regulators. Cell Rep., 17, 2488–2501. [DOI] [PubMed] [Google Scholar]
- 17. Li L., Ljungman M., Dixon J.E. (2000) The human Cdc14 phosphatases interact with and dephosphorylate the tumor suppressor protein p53. J. Biol. Chem., 275, 2410–2414. [DOI] [PubMed] [Google Scholar]
- 18. Chen N.P., Uddin B., Hardt R., Ding W., Panic M., Lucibello I., Kammerer P., Ruppert T., Schiebel E. (2017) Human phosphatase CDC14A regulates actin organization through dephosphorylation of epithelial protein lost in neoplasm. Proc. Natl. Acad. Sci. U.S.A, 114, 5201–5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen N.P., Uddin B., Voit R., Schiebel E. (2016) Human phosphatase CDC14A is recruited to the cell leading edge to regulate cell migration and adhesion. Proc. Natl. Acad. Sci. U.S.A., 113, 990–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kaiser B.K., Zimmerman Z.A., Charbonneau H., Jackson P.K. (2002) Disruption of centrosome structure, chromosome segregation, and cytokinesis by misexpression of human Cdc14A phosphatase. Mol. Biol. Cell, 13, 2289–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Delmaghani S., Aghaie A., Bouyacoub Y., El Hachmi H., Bonnet C., Riahi Z., Chardenoux S., Perfettini I., Hardelin J.P., Houmeida A.. et al. (2016) Mutations in CDC14A, encoding a protein phosphatase involved in hair cell ciliogenesis, cause autosomal-recessive severe to profound deafness. Am. J. Hum. Genet., 98, 1266–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Clement A., Solnica-Krezel L., Gould K.L. (2012) Functional redundancy between Cdc14 phosphatases in zebrafish ciliogenesis. Dev. Dyn., 241, 1911–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Friedman T.B., Riazuddin S. (2014) Nonsyndromic deafness: it Ain't necessarily so In Popper A., Fay R. (eds), Perspectives on Auditory Research, Vol. 50 Springer, New York, pp. 149–161. [Google Scholar]
- 24. Masmoudi S., Tlili A., Majava M., Ghorbel A.M., Chardenoux S., Lemainque A., Zina Z.B., Moala J., Mannikko M., Weil D.. et al. (2003) Mapping of a new autosomal recessive nonsyndromic hearing loss locus (DFNB32) to chromosome 1p13.3-22.1. Eur. J. Hum. Genet., 11, 185–188. [DOI] [PubMed] [Google Scholar]
- 25. Neto F.T., Bach P.V., Najari B.B., Li P.S., Goldstein M. (2016) Spermatogenesis in humans and its affecting factors. Semin. Cell. Dev. Biol., 59, 10–26. [DOI] [PubMed] [Google Scholar]
- 26. Chudley A.E., McCullough C., McCullough D.W. (1997) Bilateral sensorineural deafness and hydrocephalus due to foramen of Monro obstruction in sibs: a newly described autosomal recessive disorder. Am. J. Med. Genet., 68, 350–356. [DOI] [PubMed] [Google Scholar]
- 27. Doherty D., Chudley A.E., Coghlan G., Ishak G.E., Innes A.M., Lemire E.G., Rogers R.C., Mhanni A.A., Phelps I.G., Jones S.J.. et al. (2012) GPSM2 mutations cause the brain malformations and hearing loss in Chudley–McCullough syndrome. Am. J. Hum. Genet., 90, 1088–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. He F., Jacobson A. (2015) Nonsense-mediated mRNA decay: degradation of defective transcripts is only part of the story. Annu. Rev. Genet., 49, 339–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nakano Y., Jahan I., Bonde G., Sun X., Hildebrand M.S., Engelhardt J.F., Smith R.J.H., Cornell R.A., Fritzsch B., Bánfi B., Barsh G.S. (2012) A mutation in the Srrm4 gene causes alternative splicing defects and deafness in the Bronx Waltzer mouse. PLoS Genet., 8, e1002966.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gray C.H., Good V.M., Tonks N.K., Barford D. (2003) The structure of the cell cycle protein Cdc14 reveals a proline-directed protein phosphatase. EMBO J., 22, 3524–3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lee J.E., Gleeson J.G. (2011) Cilia in the nervous system: linking cilia function and neurodevelopmental disorders. Curr. Opin. Neurol., 24, 98–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Griffith A.J., Friedman T.B. (2016). Hereditary hearing loss In Wackym P.A., Snow J.B. (eds), Ballenger's Otorhinolaryngology Head and Neck Surgery, 18th edn.People’s Medical Publishing House, USA, pp. 329–345. [Google Scholar]
- 33. Avidan N., Tamary H., Dgany O., Cattan D., Pariente A., Thulliez M., Borot N., Moati L., Barthelme A., Shalmon L.. et al. (2003) CATSPER2, a human autosomal nonsyndromic male infertility gene. Eur. J. Hum. Genet., 11, 497–502. [DOI] [PubMed] [Google Scholar]
- 34. Vona B., Hofrichter M.A., Neuner C., Schroder J., Gehrig A., Hennermann J.B., Kraus F., Shehata-Dieler W., Klopocki E., Nanda I.. et al. (2015) DFNB16 is a frequent cause of congenital hearing impairment: implementation of STRC mutation analysis in routine diagnostics. Clin. Genet., 87, 49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kok F.O., Shin M., Ni C.W., Gupta A., Grosse A.S., van Impel A., Kirchmaier B.C., Peterson-Maduro J., Kourkoulis G., Male I.. et al. (2015) Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Dev. Cell, 32, 97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Meyers J.R., MacDonald R.B., Duggan A., Lenzi D., Standaert D.G., Corwin J.T., Corey D.P. (2003) Lighting up the senses: FM1-43 loading of sensory cells through nonselective ion channels. J. Neurosci., 23, 4054–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lanzetti L., Margaria V., Melander F., Virgili L., Lee M.H., Bartek J., Jensen S. (2007) Regulation of the Rab5 GTPase-activating protein RN-tre by the dual specificity phosphatase Cdc14A in human cells. J. Biol. Chem., 282, 15258–15270. [DOI] [PubMed] [Google Scholar]
- 38. Barford D., Das A.K., Egloff M.P. (1998) The structure and mechanism of protein phosphatases: insights into catalysis and regulation. Annu. Rev. Biophys. Biomol. Struct., 27, 133–164. [DOI] [PubMed] [Google Scholar]
- 39. Fahs S., Lujan P., Kohn M. (2016) Approaches to study phosphatases. ACS Chem. Biol., 11, 2944–2961. [DOI] [PubMed] [Google Scholar]
- 40. Walsh T., Shahin H., Elkan-Miller T., Lee M.K., Thornton A.M., Roeb W., Abu Rayyan A., Loulus S., Avraham K.B., King M.C.. et al. (2010) Whole exome sequencing and homozygosity mapping identify mutation in the cell polarity protein GPSM2 as the cause of nonsyndromic hearing loss DFNB82. Am. J. Hum. Genet., 87, 90–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ah-Fong A.M.V., Judelson H.S., Yu J.-H. (2011) New role for Cdc14 phosphatase: localization to basal bodies in the oomycete phytophthora and its evolutionary coinheritance with eukaryotic flagella. PLoS One, 6, e16725.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rossi A., Kontarakis Z., Gerri C., Nolte H., Holper S., Kruger M., Stainier D.Y. (2015) Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature, 524, 230–233. [DOI] [PubMed] [Google Scholar]
- 43. Cornils A., Maurya A.K., Tereshko L., Kennedy J., Brear A.G., Prahlad V., Blacque O.E., Sengupta P. (2016) Structural and functional recovery of sensory cilia in C. elegans IFT mutants upon aging. PLoS Genet., 12, e1006325.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ishikawa H., Marshall W.F. (2011) Ciliogenesis: building the cell's antenna. Nat. Rev. Mol. Cell Biol., 12, 222–234. [DOI] [PubMed] [Google Scholar]
- 45. Tavazzani E., Spaiardi P., Zampini V., Contini D., Manca M., Russo G., Prigioni I., Marcotti W., Masetto S. (2016) Distinct roles of Eps8 in the maturation of cochlear and vestibular hair cells. Neuroscience, 328, 80–91. [DOI] [PubMed] [Google Scholar]
- 46. Manor U., Disanza A., Grati M., Andrade L., Lin H., Di Fiore P.P., Scita G., Kachar B. (2011) Regulation of stereocilia length by myosin XVa and whirlin depends on the actin-regulatory protein Eps8. Curr. Biol., 21, 167–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Naz S., Imtiaz A., Mujtaba G., Maqsood A., Bashir R., Bukhari I., Khan M.R., Ramzan M., Fatima A., Rehman A.U.. et al. (2017) Genetic causes of moderate to severe hearing loss point to modifiers. Clin. Genet., 91, 589–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Morozko E.L., Nishio A., Ingham N.J., Chandra R., Fitzgerald T., Martelletti E., Borck G., Wilson E., Riordan G.P., Wangemann P.. et al. (2015) ILDR1 null mice, a model of human deafness DFNB42, show structural aberrations of tricellular tight junctions and degeneration of auditory hair cells. Hum. Mol. Genet., 24, 609–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pflugrath J.W., Wiegand G., Huber R., Vertesy L. (1986) Crystal structure determination, refinement and the molecular model of the alpha-amylase inhibitor Hoe-467A. J. Mol. Biol., 189, 383–386. [DOI] [PubMed] [Google Scholar]
- 50. Soding J., Biegert A., Lupas A.N. (2005) The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res., 33, W244–W248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Remmert M., Biegert A., Hauser A., Soding J. (2011) HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat. Methods, 9, 173–175. [DOI] [PubMed] [Google Scholar]
- 52. Liu Y., Schmidt B., Maskell D.L. (2010) MSAProbs: multiple sequence alignment based on pair hidden Markov models and partition function posterior probabilities. Bioinformatics, 26, 1958–1964. [DOI] [PubMed] [Google Scholar]
- 53. Edgar R.C. (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics, 5, 113.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sali A., Blundell T.L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol., 234, 779–815. [DOI] [PubMed] [Google Scholar]
- 55. Wallner B., Elofsson A. (2003) Can correct protein models be identified? Protein Sci., 12, 1073–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. (1993) Procheck: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- 57. Zhang Y., Skolnick J. (2005) TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res., 33, 2302–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wei Z., Peddibhotla S., Lin H., Fang X., Li M., Rosen J.M., Zhang P. (2011) Early-onset aging and defective DNA damage response in Cdc14b-deficient mice. Mol. Cell Biol., 31, 1470–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wangemann P., Nakaya K., Wu T., Maganti R.J., Itza E.M., Sanneman J.D., Harbidge D.G., Billings S., Marcus D.C. (2007) Loss of cochlear HCO3- secretion causes deafness via endolymphatic acidification and inhibition of Ca2+ reabsorption in a Pendred syndrome mouse model. Am. J. Physiol. Renal Physiol, 292, F1345–F1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Noben-Trauth K., Latoche J.R., Neely H.R., Bennett B., Schrijver I. (2010) Phenotype and genetics of progressive sensorineural hearing loss (Snhl1) in the LXS set of recombinant inbred strains of mice. PLoS One, 5, e11459.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Choi B.Y., Kim H.-M., Ito T., Lee K.-Y., Li X., Monahan K., Wen Y., Wilson E., Kurima K., Saunders T.L.. et al. (2011) Mouse model of enlarged vestibular aqueducts defines temporal requirement of Slc26a4 expression for hearing acquisition. J. Clin. Invest., 121, 4516–4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Leibovici M., Verpy E., Goodyear R.J., Zwaenepoel I., Blanchard S., Laine S., Richardson G.P., Petit C. (2005) Initial characterization of kinocilin, a protein of the hair cell kinocilium. Hear. Res., 203, 144–153. [DOI] [PubMed] [Google Scholar]
- 63. Belyantseva I.A. (2016) Helios® gene gun-mediated transfection of the inner ear sensory epithelium: recent updates. Methods Mol. Biol., 1427, 3–26. [DOI] [PubMed] [Google Scholar]
- 64. Varshney G.K., Pei W., LaFave M.C., Idol J., Xu L., Gallardo V., Carrington B., Bishop K., Jones M., Li M.. et al. (2015) High-throughput gene targeting and phenotyping in zebrafish using CRISPR/Cas9. Genome Res., 25, 1030–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Richardson C.D., Ray G.J., DeWitt M.A., Curie G.L., Corn J.E. (2016) Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat. Biotechnol., 34, 339–344. [DOI] [PubMed] [Google Scholar]
- 66. Wang H., Yang H., Shivalila C.S., Dawlaty M.M., Cheng A.W., Zhang F., Jaenisch R. (2013) One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell, 153, 910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.