Abstract
BRAFV600 mutations occur in multiple nonmelanoma tumors, but no US Food and Drug Administration–approved BRAF-targeted therapies exist for these cancers. BRAF inhibitor vemurafenib was recently found to demonstrate activity across various BRAF-mutated nonmelanoma cancer types. However, most tumors ultimately become resistant to BRAF-targeted monotherapy. To identify whether co-occurring genomic alterations drive resistance to BRAF-targeted therapies, we analyzed next-generation sequencing data from 30 advanced BRAF-mutated nonmelanoma cancers treated with BRAF inhibitor monotherapy. Kaplan-Meier survival analysis and Cox proportional hazard regression analysis were performed and hazard ratios (HR) with 95% confidence intervals (CI) were calculated. All statistical tests were two-sided. We identified a strong association between co-occurring PI3K-mTOR pathway aberrations and primary resistance to BRAF-targeted therapy. PI3K-mTOR pathway aberrations were associated with a statistically significant reduction in progression-free survival (HR = 15.0, 95% CI = 3.6 to 63.0, P < .001) and overall survival (HR = 19.2, 95% CI = 3.7 to 100.0, P < .001). This suggests that co-occurring genomic alterations may predict response and resistance to BRAF inhibitor therapy and identify subgroups of BRAF-mutated nonmelanomas cancers.
The BRAF oncogene is mutated in 50% of cutaneous melanomas and up to 10% of nonmelanomas, leading to constitutive activation of mitogen-activated protein kinase (MAPK) signaling (1). While vemurafenib, cobimetinib, dabrafenib, and trametinib are US Food and Drug Administration (FDA)–approved BRAFV600-targeted therapies for metastatic melanoma with response rates higher than 50% (2), no FDA-approved therapy exists for BRAF-mutated nonmelanoma cancers. In many of these tumors, the BRAFV600 mutation is associated with an aggressive phenotype and decreased progression-free survival (PFS) and overall survival (OS). A recent “basket” study of vemurafenib in BRAFV600-mutated nonmelanoma cancers demonstrated clinical activity (3). However, many patients exhibit primary and secondary resistance. The role of concurrent genomic alterations leading to alternative survival pathway activation has yet to be clinically examined and mechanisms of resistance to BRAF-targeted therapy remain unknown in nonmelanomas harboring a BRAF mutation. To elucidate mechanisms of resistance to BRAF-targeted monotherapy, we analyzed the genomic landscape of these tumors and the association between co-occurring mutations and resistance to therapy, PFS, and OS.
We analyzed Clinical Laboratory Improvement Amendments (CLIA)-certified next-generation sequencing data from BRAFV600 nonmelanoma cancers treated with BRAF inhibitor monotherapy at UT MD Anderson Cancer Center between May 2012 and January 2016 (3). All patients provided written informed consent, and institutional review board authorization was obtained. Kaplan-Meier survival analysis and Cox proportional hazard regression analysis were performed and hazard ratios with 95% confidence intervals calculated in order to determine whether survival on BRAF-inhibitor monotherapy was associated with co-occurring mutation status. All statistical tests were two-sided, and a P value of less than .05 was considered statistically significant. P values for the hazard ratios were computed via Wald tests from the Cox proportional hazard models. The proportional hazards assumption was verified using plots and tests based on the rescaled Schoenfeld residuals. TIBCO Spotfire S + 8.2 for Windows was used to make calculations.
Of the 30 patients with BRAFV600-mutated tumors treated with BRAF inhibitor monotherapy at our institution, 11 (36.7%) had non–small cell lung cancer, five (16.7%) had colorectal cancer, four (13.3%) had cholangiocarcinoma, three (10.0%) had thyroid cancer, three (10.0%) had Erdheim Chester disease, two (6.7%) had glioblastoma, one (3.3%) patient had salivary gland carcinoma, and one (3.3%) patient had unknown primary. Three of the five colorectal cancers, including two with PI3K-mTOR pathway co-occurring alterations, were treated with BRAF inhibitor in combination with cetuximab. Three subsets of co-occurring genomic alterations were identified: 14 (46.6%) had no co-occurring alterations, five (16.6%) had PI3K-mTOR pathway alterations, and 11 (36.7%) had “other” mutations, most commonly TP53 (n = 11), SMAD4 (n = 4), LKB1 (n = 2), and IDH1 (n = 2). The tumor type breakdown, co-occurring alterations, and progression-free survival are shown in Table 1.
Table 1.
Table indicating tumor histology, all aberrations, progression-free survival (in days) and disease status at last follow-up of each BRAF-mutated nonmelanoma cancer evaluated
| Histology | Co-occurring mutations | Progression-free survival, d | Disease progression |
|---|---|---|---|
| Non–small cell lung | TP53 (E286V) | 61 | Yes |
| Non–small cell lung | – | 98 | Yes |
| Non–small cell lung | TP53 (G154V) | 57 | Yes |
| Non–small cell lung | SMAD4 (C499R) | 425 | No |
| Non–small cell lung | – | 121 | Yes |
| Non–small cell lung | SETD2 (H2514fs) | 554 | No |
| Non–small cell lung | – | 350 | No |
| Non–small cell lung | TP53 (R248W) | 216 | No |
| Non–small cell lung | AKT (D46E) | 279 | No |
| Non–small cell lung | NNF1 (3314 + 1G>A (splice)), NTRK3 (L560H), STK11 (920 + 1G>T (splice)), ITGB3 (S85N), KEAP1 (G333S), LPHN3 (N1311K), LRP1B (D2307E), UBR5 (R1427S) | 126 | Yes |
| Non–small cell lung | IDH1 (R132C), ARID2 ((splice site 92 + 1gA)) | 131 | Yes |
| Colorectal | TP53 (R175H) | 56 | Yes |
| Colorectal | – | 112 | Yes |
| Colorectal† | TP53 (R213*), MLL3 (G2568R), TLR4 (G480F), SMAD4 (P356R), PTPRT (V239F), WHSC1 (R976K) | 252 | Yes |
| Colorectal† | PTEN (R130*), TP53 (R248Q), SMAD4 (D124fs*), ATM (R3008C) | 77 | Yes |
| Colorectal† | PIK3CA (E545K), TP53 (C176Y), SMAD4 (C361C) | 72 | Yes |
| Cholangiocarcinoma | IDH1 (R132H) | 100 | Yes |
| Cholangiocarcinoma | – | 236 | Yes |
| Cholangiocarcinoma | LKB1 (F354L), MYC amplification, MYST3 amplification | 305 | No |
| Cholangiocarcinoma | PIK3R1 (splice site 1119-1 G>A) | 17 | Yes |
| Anaplastic thyroid | PIK3CA (I391M) | 56 | Yes |
| Anaplastic thyroid | TP53 (R280K) | 184 | Yes |
| Papillary thyroid | – | 86 | Yes |
| Erdheim Chester Disease | – | 66 | Yes |
| Erdheim Chester Disease | – | 799 | No |
| Erdheim Chester Disease | – | 376 | No |
| Salivary gland carcinoma | – | 84 | Yes |
| Glioblastoma | PTEN (P339fs*2) | 44 | Yes |
| Glioblastoma | CDK2NA (H66fs*54), NOTCH1 (E1567K), KRAS (G13D) | 455 | Yes |
| Unknown primary | TP53 (R110C), TP53 (P278S) | 100 | Yes |
Treated with cetuximab in addition to BRAF inhibitor. - = no co-occurring mutation present.
To investigate whether co-occurring mutations are associated with survival, we analyzed PFS and OS in each patient. Eight patients (26.7%) had ongoing response without progression on BRAF inhibitor monotherapy (range PFS = 222–805 days). Six of eight durable responders have no co-occurring alterations, two of eight have “other” mutations. Both patients with non-mTOR pathway somatic gene mutations who have yet to progress have metastatic adenocarcinoma of the lung, one with a SMAD4 C499R mutation and the other with a TP53 R248W mutation. The six patients without co-occurring mutations had non–small cell lung cancer (n = 3), Erdheim-chester disease (n = 2), and cholangiocarcinoma (n = 1). All patients with PI3K-mTOR pathway mutations progressed within 77 days (metastatic colorectal cancer [n = 2], anaplastic thyroid cancer [n = 1], glioblastoma [n = 1], and cholangiocarcinoma [n = 1]).
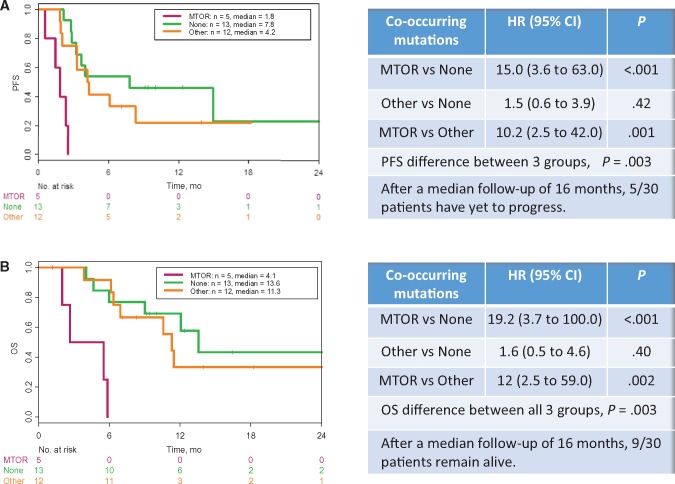
Kaplan-Meier analyses demonstrate a statistically significant reduction in PFS (hazard ratio [HR] = 15.0, 95% confidence interval [CI] = 3.6 to 63.0, P = .0002) and OS (HR = 19.2, 95% CI = 3.7 to 100.0, P < .001) in patients whose tumors harbored co-occurring PI3K-mTOR pathways mutations compared with those with “other” co-occurring mutations and no co-occurring mutations, as seen in Figure 1. Tumors with co-occurring PI3K-mTOR pathway aberrations had a median PFS of 1.8 months (95% CI = 1.4 to not reached [NR]) compared with tumors with other co-occurring mutations (4.2 months, 95% CI = 3.3 to NR) and no co-occurring mutations (7.8 months, 95% CI = 3.2 to NR). OS in patients with tumors harboring co-occurring mTOR pathway aberrations had a median OS of 4.1 months (95% CI = 2.0 to NR) compared with tumors with other co-occurring mutations (11.3 months, 95% CI = 6.9 to NR) and without co-occurring genomic alterations (13.6 months, 95% CI = 9.0 to NR).
Figure 1.
Progression-free survival and overall survival by co-occurring mutations in patients with BRAF-mutated nonmelanoma cancers treated with BRAF-inhibitor monotherapy. Kaplan-Meier survival analysis and Cox proportional hazard regression analysis for progression-free survival (A) and overall survival (B) were performed in 30 patients with BRAF-mutated nonmelanoma cancers treated with BRAF inhibitor monotherapy. Patients were stratified by tumors harboring no co-occurring mutations (None) in green, mTOR pathway mutations (MTOR) in purple, or other mutations (Other) in orange. Statistical tests were two-sided. CI = confidence interval; HR = hazard ratio; OS = overall survival; PFS = progression-free survival.
While BRAF mutations, amplifications, and fusions have been identified in a number of nonmelanoma cancers, the mutational landscape of these tumors has yet to be defined. We identified three distinct subsets of co-occurring molecular alterations in BRAF-mutated nonmelanoma cancers and found a statistically significant association between co-occurring mutations and PFS and OS. Specifically, 20% of our patients’ tumors across all histologies had mutations in PI3K-mTOR pathway genes, and this subset was associated with de novo resistance to BRAF inhibitor monotherapy and statistically significantly shorter PFS and OS (Figure 1).
In melanoma, resistance to BRAF-targeted therapy results from reactivation of the MAPK pathway, activation of parallel signaling pathways including the PI3K-mTOR pathway, or ineffective modulation of the immune system (4). Our study suggests that parallel activation of mTOR signaling may be one mechanism contributing to de novo resistance to BRAF-targeted therapy in nonmelanoma cancers as well. In preclinical colorectal cancer models, parallel activation of the PI3K-mTOR pathway has been implicated as a mechanism of resistance to BRAF inhibition (5). Clinically, resistance to BRAF inhibition may be mediated through the epidermal growth factor receptor pathway as well (6). In our cohort, despite the addition of cetuximab, survival was statistically significantly worse in tumors harboring PI3K-mTOR pathway mutations. Given the known crosstalk between the MAPK and mTOR signaling pathways (7), it is plausible that mTOR activation may also mediate acquired resistance to BRAF-targeted therapy in the patients who initially respond to BRAF inhibiton as well. This remains to be studied and also highlights the utility of re-biopsying a patient at the time of progression.
We acknowledge that the lack of serial biopsies was a limitation of our study, as was the sample size. While PI3K-mTOR pathway co-occurring alterations were identified across tumor types in our study, a larger sample size would allow for a better understanding of the role of tumor histology in response and resistance to BRAF-targeted therapy. Additionally, hot spot next-generation sequencing was utilized in our study, and whole-genome sequencing may lead to a more comprehensive pathway analysis in this subset of tumors in the future.
Co-occurring genomic alterations may help predict response and resistance to targeted therapies in BRAF-mutated cancers in a histology-independent manner. This study highlights the importance of implementing next-generation sequencing testing and enrolling BRAF-mutated cancer patients in larger genotype-matched trials when feasible (8). A phase I trial combining BRAF inhibitor vemurafenib and mTOR inhibitor everolimus is underway (NCT01596140).
Funding
This work was supported in part by The Cancer Prevention and Research Institute of Texas (RP1100584), the Sheikh Khalifa Bin Zayed Al Nahyan Institute for Personalized Cancer Therapy, 1U01 CA180964, The National Center for Advancing Translational Sciences (NCATS) grant UL1 TR000371 (Center for Clinical and Translational Sciences), The Bosarge Family Foundation, and the MD Anderson Cancer Center Support Grant (P30 CA016672).
Note
This was presented in part at the 2016 American Society of Clinical Oncology (ASCO) Annual Meeting.
The funders had no role in the design of the study; the collection, analysis, or interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication.
Vivek Subbiah receives research funding for clinical trials from Roche/Genentech, Novartis, Bayer, GSK, Nanocarrier, Vegenics, Northwest Biotherapeutics, Berghealth, Incyte, Fujifilm, Pharmamar, D3, Pfizer, Multivir, Amgen, Abbvie, and Bluprint medicines.
References
- 1. Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;4176892:949–954. [DOI] [PubMed] [Google Scholar]
- 2. Long GV, Stroyakovskiy D, Gogas H, et al. Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. N Engl J Med. 2014;37120:1877–1888. [DOI] [PubMed] [Google Scholar]
- 3. Hyman DM, Puzanov I, Subbiah V, et al. Vemurafenib in multiple nonmelanoma cancers with BRAF V600 mutations. N Engl J Med. 2015;3738:726–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Welsh SJ, Rizos H, Scolyer RA, Long GV.. Resistance to combination BRAF and MEK inhibition in metastatic melanoma: Where to next? Eur J Cancer. 2016;62:76–85. [DOI] [PubMed] [Google Scholar]
- 5. Mao M, Tian F, Mariadason JM, et al. Resistance to BRAF inhibition in BRAF-mutant colon cancer can be overcome with PI3K inhibition or demethylating agents. Clin Cancer Res. 2013;193:657–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Corcoran RB, Ebi H, Turke AB, et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discov. 2012;23:227–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mendoza MC, Er EE, Blenis J.. The Ras-ERK and PI3K-mTOR pathways: Cross-talk and compensation. Trends Biochem Sci. 2011;366:320–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Meric-Bernstam F, Brusco L, Shaw K, et al. Feasibility of large-scale genomic testing to facilitate enrollment onto genomically matched clinical trials. J Clin Oncol. 2015;3325:2753–2762. [DOI] [PMC free article] [PubMed] [Google Scholar]