Abstract
Background
Primary effusion lymphoma (PEL) is a Kaposi's sarcoma herpes virus (KSHV)–induced lymphoma that typically arises in body cavities of HIV-infected patients. PEL cells are often co-infected with Epstein-Barr virus (EBV). “PEL-like” lymphoma is a KSHV-unrelated lymphoma that arises in body cavities of HIV-negative patients. “PEL-like” lymphoma is sometimes EBV positive. The derivation of PEL/“PEL-like” cells is unclear.
Methods
Mesothelial cells were cultured from body cavity effusions of 23 patients. Cell proliferation, cytokine secretion, marker phenotypes, KSHV/EBV infection, and clonality were evaluated by standard methods. Gene expression was measured by quantitative polymerase chain reaction and immunoblotting. A mouse model of PEL (3 mice/group) was used to evaluate tumorigenicity.
Results
We found that the mesothelia derived from six effusions of HIV-infected patients with PEL or other KSHV-associated diseases contained rare KSHV+ or EBV+ mesothelial cells. After extended culture (16–17 weeks), some mesothelial cells underwent a trans-differentiation process, generating lymphoid-type CD45+/B220+, CD5+, CD27+, CD43+, CD11c+, and CD3− cells resembling “B1-cells,” most commonly found in mouse body cavities. These “B1-like” cells were short lived. However, long-term KSHV+EBV− and EBV+KSHV− clonal cell lines emerged from mesothelial cultures from two patients that were clonally distinct from the monoclonal or polyclonal B-cell populations found in the patients’ original effusions.
Conclusions
Mesothelial-to-lymphoid transformation is a newly identified invitro process that generates “B1-like” cells and is associated with the emergence of long-lived KSHV or EBV-infected cell lines in KSHV-infected patients. These results identify mesothelial cultures as a source of PEL cells and lymphoid cells in humans.
Primary effusion lymphoma (PEL) is a malignancy that predominantly arises in HIV-infected patients. PEL was first recognized as a distinct clinical entity in patients with AIDS based on its unusual presentation as a liquid malignancy generally confined to body cavities, the “indeterminate” phenotype of the tumor cells (1), and the presence of Kaposi's sarcoma herpes virus (KSHV) in the tumor cells, with or without Epstein-Barr virus (EBV) (2). KSHV+ PEL cells are of B-cell lineage as they display Ig gene rearrangements, the common leukocyte antigen CD45 (2), and the plasmablast/plasma cell marker CD138 in some cases (3) but usually lack surface and cytoplasmic Ig and the mature B-cell markers CD19 and CD20 (4). KSHV is also associated with Kaposi's sarcoma (KS), plasmablastic multicentric Castleman disease (KSHV-MCD), and a KSHV-associated inflammatory cytokine syndrome (KICS). Patients with KS, KICS, and KSHV-MCD may develop recurrent pleural effusions of unclear etiology (5).
KSHV-unrelated “PEL-like” lymphomas anatomically confined to body cavities constitute a distinct entity (6,7). These PEL-like lymphomas have a mature B-cell phenotype with CD19 and CD20 expression, tend to occur in elderly patients, and often have a favorable prognosis (6,7). A proportion of “PEL-like” lymphomas are EBV positive (7).
The body cavity site of PEL and PEL-like lymphomas suggested that this microenvironment contributes to their development and/or progression. To gain insight into this process, we focused on the mesothelium that lines these cavities (8). Here, we have examined potential mechanisms by which the mesothelium may contribute to the emergence and progression of PEL and PEL-like lymphomas.
Methods
Patients
HIV+ patients with KSHV-related diseases (17 patients) (Table 1) and patients with ovarian carcinoma (six patients) at the Clinical Center, National Cancer Institute (NCI), Bethesda, Maryland, provided biospecimens. PEL, KS, and KSHV-MCD diagnoses were confirmed pathologically. KICS diagnosis was based on a working definition (9). KSHV+ patients enrolled in aninstitutional review board (IRB)–approved protocol (NCT00006518) provided written informed consent. Samples from patients with ovarian carcinoma were IRB exempt under the Office of Human Subjects Research Protection (OHSRP) review No. 12727.
Table 1.
Selected patient data*
| Patient | Diagnosis | Age/sex | Effusion source | Effusion specimen, time from diagnosis |
HIV viral load copies/mL blood | KSHV viral load copies/106 cells |
WBC /µL effusion | RBC /µL effusion | Hg g/dL blood | CD4 cells/µL blood | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PEL | AIDS-KSHV | Blood | Effusion | |||||||||
| 81 | PEL/KS/KICS | 59/male | Pleural | Same | 8 y | Undetected | 0 | 29x106 | 1609 | 2200 | 9.3 | 187 |
| 20 | PEL | 69/male | Peritoneal | 1 mo | N/A | <20 | 6x104 | 22x107 | 2328 | 5333 | 13 | 594 |
| 18 | PEL/KS/KICS | 47/male | Pleural | 16 mo | 16 mo | <40 | 23 | 33x106 | 3278 | 79x103 | 15 | <40 |
| 11 | PEL/KS/KICS | 22/male | Pleural | 4 mo | 4 mo | 43 | 38x103 | 19x107 | 200 | 15x103 | 9 | 43 |
| 30 | PEL/KS/KICS | 33/male | Pleural | 9 mo | 26 mo | Undetected | 54 | 15x106 | 8150 | 37x103 | 13 | 96 |
| 41 | PEL/MCD/KICS | 55/male | Pleural | Same | 39 mo | <20 | 61x104 | 70x106 | 1636 | 16x103 | 11 | 363 |
| 14 | MCD/KS/DLBCL | 37/male | Pleural | N/A | 10 mo | <50 | N/D | N/D | 3800 | 37x103 | 13 | 186 |
| 19 | KS/KICS | 23/male | Peritoneal | N/A | 3 mo | <20 | 1x103 | 11x104 | 1060 | 2415 | 7.7 | 198 |
| 90 | KS/KICS | 49/male | Pleural | N/A | 30 mo | 49 | Positive | 2x106 | 89 | 46x104 | 9.3 | 224 |
| 13 | KS/KICS | 37/male | Pleural | N/A | same | 37 | 9x105 | 2x106 | N/D | N/D | 8.3 | 16 |
| 191 | KS | 48/male | Pleural | N/A | 7 y | Undetected | 0 | 2x103 | 167 | 45x104 | 9.3 | 138 |
| 188 | KS/LCBCL | 45/male | Pleural | N/A | same | 3238 | 0 | 0 | N/D | N/D | 8.9 | 46 |
| 23 | KS/MCD | 27/male | Pleural | N/A | 4 mo | 380 | 12x104 | positive | N/D | N/D | 8.9 | 24 |
| 131 | KS/MCD | 37/male | Pleural | N/A | 14 mo | Undetected | Positive | 1.5x104 | 378 | 24x103 | 11 | 37 |
| 300 | KS/MCD | 33/male | Pleural | N/A | 29 mo | Undetected | N/D | N/D | 467 | 21x103 | 11 | 61 |
| 416 | KS/KICS | 30/male | Pleural | N/A | 8 mo | Undetected | Positive | 12x104 | 156 | 40x104 | 8.8 | 34 |
| 61 | KS/MCD | 45/male | Pleural | N/A | 1 mo | <48 | 283 | 37x103 | 1050 | 13x104 | 8.2 | 330 |
DLBLC = diffuse large B cell lymphoma; Hg = hemoglobin; KICS = KSHV-associated inflammatory cytokine syndrome; KS = Kaposi's sarcoma; KSHV = Kaposi's sarcoma herpes virus; LCBCL = large cell B cell lymphoma; MCD = Multicentric Castleman Disease; N/D = not done; PEL = primary effusion lymphoma; RBC = red blood cells; WBC = white blood cells.
Cells and Proliferation
Primary cultures of mesothelial cells were established from effusions (detailed in the Supplementary Methods, available online). The PEL cell lines BC-1, BC-3, and BCBL-1 were cultured as described (10–12). Cell proliferation was measured by 3H-thymidine incorporation as described (13). Population doublings were calculated from viable cell counting as log2(Ct/Co), where Co was number cells seeded and Ct was number cells harvested; mesothelial cells when confluent status was first achieved were considered population doubling at 0.
Gene Expression and Clonality
Lentivirus for Azurite expression was generated in 293T cells (American Type Culture Collection) Manassas, VA, USA by transfection of the pLV-Azurite short hairpin RNA construct (Addgene, 36086) Cambridge, MA, USA and third-generation packaging plasmids (pMDLg/pRRE; pRSV-Rev and a VSVG envelop), as described (14). RNA was purified, and cDNA was synthesized as described (14). Quantitative polymerase chain reaction (qPCR) was performed as described in Supplementary Methods (available online) using the primers listed in Supplementary Table 1 (available online). B-cell clonality and immunoglobulin gene rearrangements were evaluated as described (15,16) and detailed in the Supplementary Methods (available online).
KSHV and EBV Sequencing
KSHV-K1 and EBV B95-8-deleted region were sequenced (17,18) and comparative sequence analysis performed as described (19) and detailed in Supplementary Methods (available online).
Immunoblotting and Immunofluorescence
Immunoblotting was performed as described in the Supplementary Methods (available online). Immunostaining is detailed in the Supplementary Methods (available online), with primary and secondary antibodies listed in Supplementary Table 2 (available online).
Mouse Model
The animal protocol (20) was approved by the Institutional Animal Care and Use Committee of the Center for Cancer Research, NCI, National Institutes of Health (NIH) (Bethesda, MD); the studies were conducted in adherence to the NIH Guide for Care and Use of Laboratory Animals. Groups of female NOD/SCID mice (three mice/group; six weeks old) were injected intraperitoneally (i.p.) with lymphoid cell lines emerging from mesothelial cultures (5x106 cells/mouse in 0.2 mL PBS) or with the PEL cell line BC-1 (10x106 cells/mouse in 0.2 mL PBS) as described (12). The mice were observed until evidence of excessive abdominal distension interfering with mouse motility or causing distress emerged, at which time the mice were killed and evaluated for evidence of ascites and intraperitoneal tumor.
Statistical Analysis
The standard deviation and standard error of the mean (SEM) were calculated by standard methods; the statistical significance of differences between two groups was calculated by two-tailed Student’s t test, where P values of less than .05 are considered statistically significant.
Results
Isolation of Mesothelial Cell Monolayers From Effusions
Pleural or peritoneal effusions from HIV-infected patients with PEL (henceforth called “PEL-effusions”; n = 6); HIV-associated KS, KSHV-MCD, and/or KICS without PEL (henceforth called “KSHV-effusions”; n = 11); and ovarian cancer (n = 6) (Table 1) generated typical cobblestone-like mesothelial monolayers (21) (Supplementary Figure 1A, available online). Cells within these monolayers were uniformly Vimentin+ (Supplementary Figure 1B, available online), consistent with a mesothelial identity (22). Similar to omentum-derived human mesothelial cells (23), effusion-derived mesothelial cells reached confluency within five to seven days during initial (1–4) passages, and this proliferative capacity declined after seven to 14 passages, particularly in mesothelial cultures from ovarian carcinoma patients, attributable perhaps to chemotherapy-induced mesothelial toxicity (Figure 1A) (23).
Figure 1.
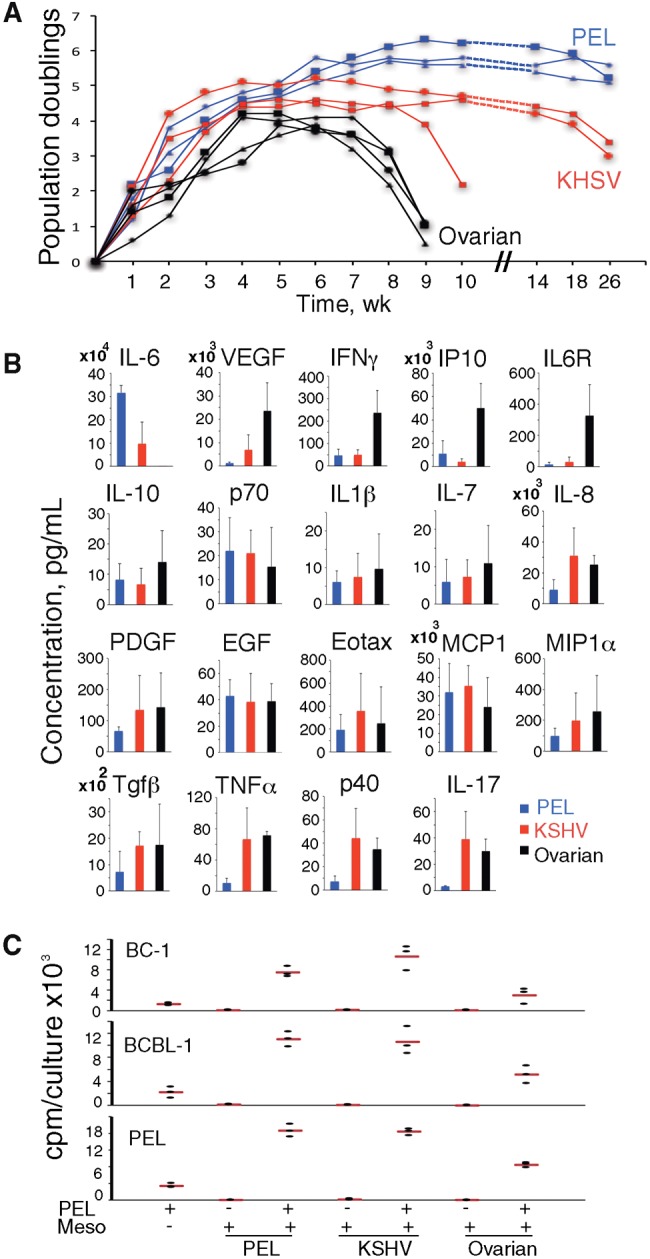
Derivation of mesothelial cells from effusions. A) Proliferative capacity of primary mesothelial cells over time in culture. Mesothelial cells from primary effusion lymphoma (PEL): PEL effusion; Kaposi's sarcoma herpes virus (KSHV): KSHV effusion; ovarian: ovarian carcinoma effusion. B) Cytokines in mesothelial conditioned medium (n = 3/group) at week 3 or 4. Bar graphs: group means (±SEM). C) Proliferation of PEL cells alone, mesothelia alone, or coculture of PEL cells over mesothelia (PEL+/Meso+). Dot plot: data from triplicate cultures from a representative experiment (three performed); the horizontal red line reflects the means. EGF = endothelial growth factor; IL = interleukin; KHSV = Kaposi's sarcoma herpes virus; PDGF = platelet-derived growth factor; PEL = primary effusion lymphoma; TNF = tumor necrosis factor; VEGF = vascular endothelial growth factor.
When cytokines were evaluated in conditioned media of mesothelial cultures (at week 3 or 4), IL-6 was unique as it was detected at statistically significantly higher levels in culture supernatants from PEL (P = .02) and KSHV (P = .03) effusions compared with cultures from ovarian cancer effusions (Figure 1B). The opposite was true for VEGF-A, IFNγ, IP-10, and IL6 receptor (R). Most of the remainder cytokines were present at similar levels in the three groups.
It was previously shown that PEL cells proliferate in response to IL-10 and IL-6 (20,24). We found that PEL cell lines BC-1, BCBL-1, and primary PEL cells proliferated better in the presence of mesothelial monolayers from PEL and KSHV effusions than in their absence (BC-1: P = .01 and P = .02, respectively; BCBL-1: P = .01 and P = .02, respectively; primary PEL: P = .005 and P = .003, respectively) (Figure 1C). This shows that mesothelial monolayers support PEL cell growth, attributable perhaps to the biologically active IL-10 and IL-6 they produce.
By immunoblotting, HIV+ patient-derived mesothelial cell lysates from passage 3 or 4 lacked the viral protein latency-associated nuclear antigen (LANA)–1, which marks KSHV infection (Supplementary Figure 1C, available online). In two cases, we could test lysates from fresh PEL cells and mesothelium from the same effusion; LANA-1 was detected in PEL but not in the mesothelium (Supplementary Figure 1C, available online).
Emergence of Lymphoid “B1-Type” Cells From Mesothelium
We monitored mesothelial cell cultures beyond the proliferative stage into “senescence” (23), characterized by enlarged/flattened cells with vacuolated nuclei and loss of monolayer integrity (Figure 2A). Initially rare, round cells with lymphoid morphology appeared within the adherent monolayer, which retained mesothelial morphology with some evidence of “senescence” (Figure 2A, week 16). By week 17, numerous lymphoid-type cells emerged, first isolated and later forming cell clusters over the monolayer (Figure 2A); this process subsided over five to 10 days. Thereafter, most lymphoid cells assumed a granular morphology and died along with the disappearance of the mesothelial monolayer (Figure 2A, week 19). Most mesothelial cultures (16/17 from HIV+ patients and 3/6 from ovarian carcinoma) displayed a similar process. The overall output of lymphoid-type cells from three KSHV-derived cultures (containing ∼3–5x105 mesothelial cells) was estimated at approximately 3 to 10.5x105 cells by daily counting the nonadherent cells.
Figure 2.
Emergence of lymphoid-type cells from mesothelial cell cultures. A) Mesothelial cell culture imaged at weeks 4, 16, 17, and 19; phase contrast microscopy. Arrows: lymphoid-type cells; asterisks: “senescent” mesothelial cells; area limited by the red dotted line is magnified in the inset. B) Sessile bodies and lymphoid-like cell in an Azurite+ mesothelial culture. C) Cytospun cells from culture supernatant of mesothelial cells at week 17. Giemsa staining showing small lymphoid-like (lymph), plasmablast/plasma cell-like (plasm), and mesothelial-like (meso) cells.
KSHV effusion-derived mesothelial cells rendered Azurite-fluorescent displayed the emergence of Azurite+ sessile bodies from the mesothelium and occasional floating Azurite+ small lymphoid-like cells (Figure 2B). Time-lapse microscopy documented emergence and movement of lymphoid-type cells from a mesothelial cell culture (Supplementary Video, available online).
Cytospun preparations of these lymphoid cells revealed lymphocytic-type and plasmablast/plasma cell-type cells; larger mesothelial-type cells were also identified (Figure 2C). The lymphocytic-type cells displayed the hematopoietic/panleukocyte CD45 marker (Supplementary Figure 2, available online) absent from mesothelial cells (25), suggesting that hematopoietic cells/leukocytes had emerged from mesothelial cultures.
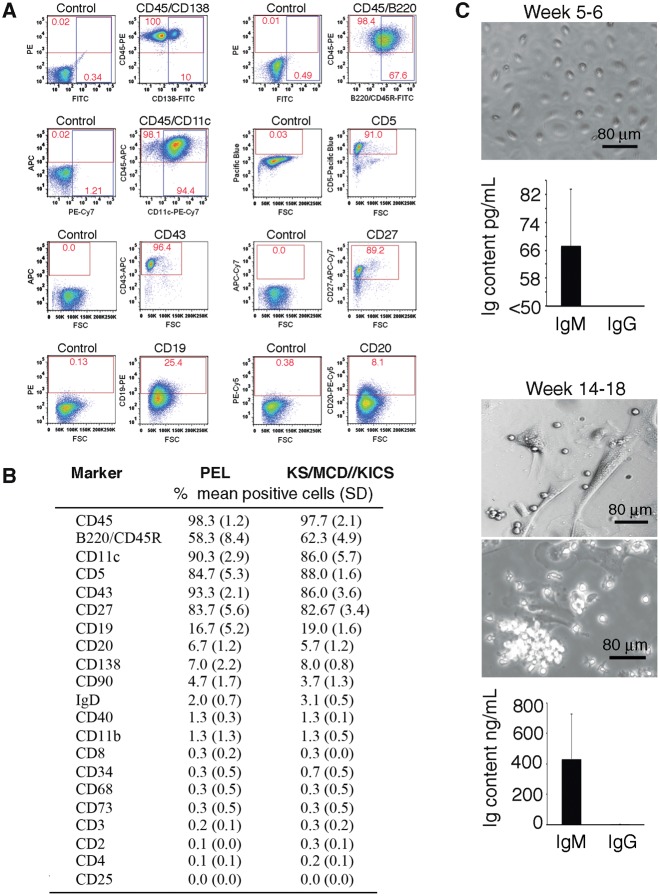
Flow cytometry (Figure 3, A and B) showed that most lymphocytic cells were CD45+, confirming their hematopoietic lineage (26). A proportion of the CD45+ cells were B220/CD45R+ (B-lymphocyte marker) and CD11c+ (dendritic cell marker also detected in some B cells [27]). A minority of CD45+ cells were CD138+ (plasma cell marker [28]). Most cells were CD5+ (mouse and human B1 cells marker, also found on T-lymphocytes [29,30]), CD27+ (human memory B-cells marker [31] also detected in human B1 cells [32] and plasmablasts [33]), and CD43+ (present on mouse B1 and human cells of various lineages, including T cells, B cell precursors, and plasma cells [29,30,34,35]). Only subsets of cells displayed the B-lineage markers CD19 and CD20. T-cell markers, including CD3; the myeloid-lineage markers CD11b and CD68; and surface IgD were generally absent.
Figure 3.
Characterization of lymphoid-like cells from mesothelial cell cultures. A and B) Nonadherent cells recovered from mesothelial cell cultures analyzed by flow cytometry. A) Representative profiles from antibody and control Ig staining; percent cells in quadrants are shown. B) Cumulative results; cultures were derived from effusions of primary effusion lymphoma (PEL) patients (n = 3) and patients with KS, KSHV-associated inflammatory cytokine syndrome (KICS), or KSHV-associated plasmablastic multicentric Castleman disease (KSHV-MCD; n = 4); percent mean positive cells (±SD) shown. C) Immunoglobulin (Ig)M and IgG content in supernatants of mesothelial cell cultures at weeks 5 to 6 and 14 to 18. Top: representative mesothelial cell monolayers at supernatant harvest. Bottom: bar graphs showing the means (n = 5; ±SD; error bars). Ig = immunoglobulin; KICS = KSHV-associated inflammatory cytokine syndrome; KS = Kaposi's sarcoma; MCD = multicentric Castleman disease; PE = primary effusion.
Mesothelial cultures initially produced little IgM (67±16.4 pg/mL; detection limit 50 pg/mL) and no IgG (week 5–6), but when lymphoid-like cells were present (week 14–18) produced more IgM (433±295 ng/mL) and IgG (6.6±2.9 ng/mL) (Figure 3C). Thus, lymphoid-type cells from mesothelial cultures resemble phenotypically B1 cells in mice (CD19+CD11b+CD5+ or CD19hiCD23− CD43+IgMhiIgDvariable) (29,36–38) and humans (CD3-CD20+CD19+CD27+CD43+CD69−CD70− with a subset CD11b+) (39–42). In the mouse, B1 cells represent the main B-cell population in the peritoneal and pleural cavities (41,43). In humans, B1 cells were reported in cord blood and adult peripheral blood (32,42). Thus, human mesothelial cell cultures can generate “B1-type” cells through a newly identified process of “mesothelial-to-lymphoid transition” (MLT).
Outgrowth of KSHV+/EBV− and KSHV−/EBV+ Cell Lines
Mesothelial cultures from patient 81 (subject with PEL, KS, and KICS) and patient 90 (subject with KS and KICS but not PEL) differed because they generated lymphoid-type cells that did not die, instead displaying sustained growth. Culture 81 produced lymphoid cells from two wells (named 81a and 81b), culture 90 from one well only. The outgrowing cells were lymphoblast-like (Figure 4A) and produced lymphoid cell-rich ascites with or without associated solid tumors when injected intraperitoneally in three of three NOD/SCID mice (20).
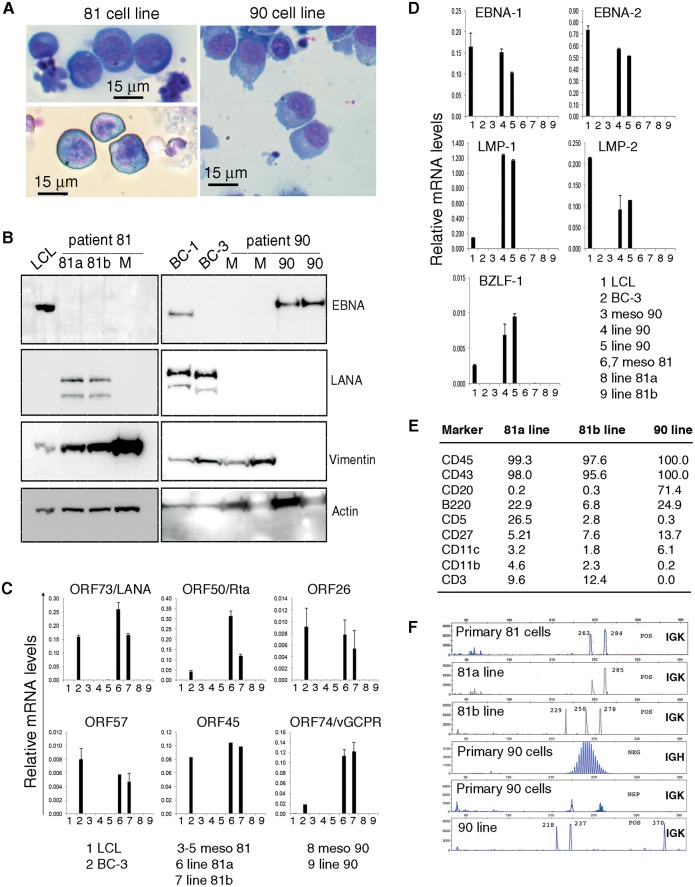
Figure 4.
Emergence of cell lines from mesothelial cultures. A) Anaplastic morphology of cytospun 81a, 81b, and 90 cells; Giemsa staining. B) Immunoblotting detection of EBV nuclear antigen (EBNA1), latency-associated nuclear antigen (LANA1), Vimentin and actin in cell lysates from patient 81 (left): 81a and 81b lines (passage 3) and mesothelial cells (M, week 4); and patient 90 (right): mesothelial cells (M, weeks 3 and 4); and 90 line (passage 1 and 4). LCL: EBV+ lymphoblastoid cell line; BC-1 (EBV+/KSHV+) and BC-3 (EBV-/KSHV+): PEL lines. Relative KSHV (C) and EBV (D) mRNA levels in mesothelial cells (Meso) from patients 81 and 90 (weeks 2–4) and cell lines 81a, 81b, and 90 (passage 1 to 3). E) Flow cytometric analysis of 81a, 81b, and 90 cell lines. Results reflect percent positive cells. F) Analysis of clonality in primary cells from patient (81 and 90) effusion and cell lines (81a, 81b, and 90) derived from mesothelial culture. EBNA = EBV nuclear antigen; EBV = Epstein-Barr virus; KSHV = Kaposi's sarcoma herpes virus; LANA = latency-associated nuclear antigen; LCL = lymphoblastoid cell line; LMP = latent membrane protein; ORF = open reading frame.
Lysates from the 81a and 81b lines were LANA1+, EBNA1−, and Vimentin+ (Figure 4B, left), whereas a lysate from early passage mesothelium (M) 81 was LANA−, EBNA1− and Vimentin+ (Figure 4B, left). Lysates from the 90 line were EBNA1+, LANA−, Vimentin− (Figure 4B, right), whereas lysates of mesothelium 90 from early passages were EBNA1−, LANA−, and Vimentin+ (Figure 4B, right).
Confirming this, a proportion of 81a and 81b cells displayed the typical speckled nuclear LANA1 staining, whereas a proportion of the 90 cells displayed nuclear EBNA2 staining (Supplementary Figure 3, available online). qPCR (Figure 4C) showed that the 81a and 81b lines express the open reading frame (ORF) 73/LANA and several other latency KSHV genes (44–46), the lytic cycle inducer ORF50/Rta (47), and the lytic transcript ORF26 (48). Line 90 expressed none of the KSHV-coded genes tested (Figure 4C), expressing instead the EBV latency genes EBNA-1, EBNA-2, LMP-1, LMP-2, and the immediate-early gene BZLF-1 (Figure 4D). The mesothelia (meso) from patients 81 and 90 did not express the KSHV- and EBV-coded genes tested (Figure 4, C and D). Culture supernatants of the 81 lines contained vIL-6 (81a: 1371 pg/mL; 81b: 721 pg/mL; BC1 culture supernatant: 911 pg/mL); culture supernatants of the 90 line did not (<250 pg/mL).
Derivation of the 81a and 81b lines from patient 81 and derivation of line 90 from patient 90 was confirmed by HLA typing. KSHV (K1 protein-encoding region) sequencing (17,19) revealed the identity of the virus from the pleural effusion of patient 81 and the 81a and 81b lines. EBV (B95-8 deletion region) sequencing (18) revealed the identity of the virus from the pleural effusion of patient 90 and the 90 line.
By flow cytometry (Figure 4E), the 81a and 81b lines displayed an “indeterminate” phenotype typical of PEL (49,50), whereas the 90 line displayed a mature B-cell phenotype. Thus, surface markers distinguish the 81a and 81b lines from the 90 line, each line sharing some of the markers of “B1-like” cells.
Analysis of clonality based on immunoglobulin (IGH and IGK loci) gene rearrangement (16,51) revealed that the mesothelial culture–derived lines are clonal (Figure 4F). The pattern of K light chain restriction of the 81b line differed from that found in the primary PEL cells from the patient’s 81 effusion, indicating that this line was not an outgrowth of the original PEL (Figure 4F). However, the K light chain restriction in the 81a line was not distinguishable from the original PEL. The clonal population detected in the EBV+ 90 line was not evident within the polyclonal primary cells from the effusion of patient 90 (Figure 4F).
Immunoglobulin (Ig) heavy chain (IGH) sequencing showed that the 90 and 81b lines display productively rearranged IGH sequences (IGHV4 and IGHV1 families, respectively) containing numerous point mutations. We did not detect a productively rearranged IGH sequence in the 81a cell line. However, 81a displays a productive rearrangement of the light chain (IGKV2) containing no mutations.
KSHV and EBV Can Infect Mesothelial Cells
Immunoblotting and qPCR did not show that the mesothelia from patients 81 and 90 were infected with KSHV or EBV during culture. However, immunofluorescence staining revealed rare KSHV (LANA-1) and EBV (LMP-2A) infection in mesothelia 81 and 90 more than four weeks prior to the emergence of the KSHV+ 81a and 81b and the EBV+ 90 cell lines (Figure 5A). The frequency of LANA+ and EBV+ cells was low in mesothelium 81 and 90 (estimated at ∼1:5000 for LANA and at ∼1:8000 for LMP2A). Only one cell had evidence of EBV and KSHV co-infection in mesothelium 81 (Figure 5B). Rare KSHV+ or EBV+ cells were also identified in mesothelia established from effusions of four distinct HIV+ patients with or without PEL (Figure 5C). No continuous cell lines grew from these four mesothelial cultures.
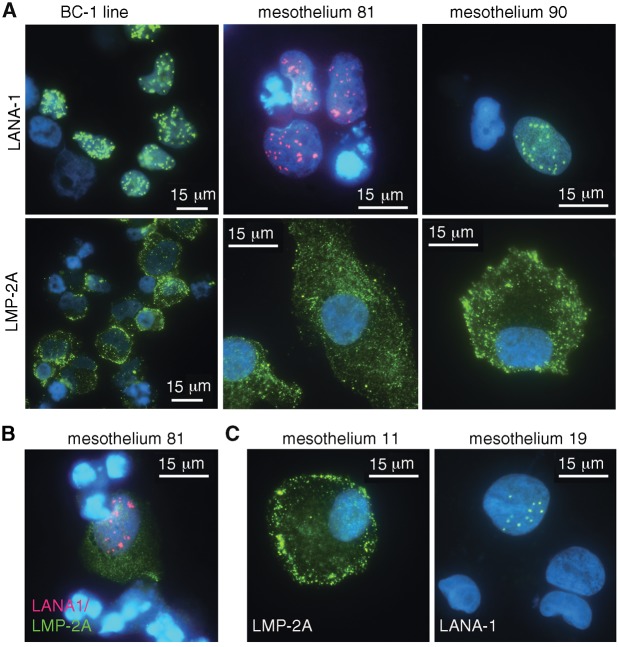
Figure 5.
Kaposi's sarcoma herpes virus (KSHV) and Epstein-Barr virus (EBV) infect primary mesothelial cells. A–C) KSHV and EBV infection in primary mesothelial cells from effusions of patients with primary effusion lymphoma (PEL; mesothelia 81 and 11) and KSHV-associated diseases but no PEL (mesothelia 90 and 19). Immunofluorescence detection of KSHV-latency-associated nuclear antigen (LANA)–1 (green or red) and EBV-latent membrane protein–2A (green); nuclear DAPI: blue. LANA = latency-associated nuclear antigen; LMP = latent membrane protein.
We tried to experimentally infect mesothelial cells with EBV and KSHV. Culture supernatant of the B95-8 marmoset cell line (contained 106 cord-blood transforming units/mL) did not infect primary mesothelial cells by EBNA2 immunostaining. Instead, rKSHV.219 virus (marks latent viral infection with GFP from the EF-1α promoter and lytic viral infection with RFP from the KSHV PAN promoter) (52) infected most primary mesothelial cells; latent infection could be established (GFP, but little/no RFP expression) (Supplementary Figure 4A, available online). Mesothelial cells infected with rKSHV were LANA-1+ (Supplementary Figure 4B, available online). Monitoring noninfected and rKSHV-infected cultures showed that rKSHV+ infection does not provide mesothelial cells with a growth advantage.
Based on these observations, we propose that occasional mesothelial cells within mesothelia that line body cavities of KSHV and/or EBV-infected individuals become naturally infected with KSHV, EBV, and infrequently both viruses (Figure 6). In the presence of activating signals (21), some mesothelial cells undergo mesothelial-to-lymphoid transition (MLT), resulting in the generation of B1-type lymphoid cells. When this process involves a KSHV and/or EBV-infected mesothelial cell, the resulting B1-type cell can be KSHV-infected, EBV-infected, or KSHV+EBV co-infected. Because these viruses can establish long-term latency in B-lineage cells and have oncogenic properties, the KSHV/EBV-infected B1-type cells acquire new capabilities, including responsiveness to mesothelium-derived cytokines and other factors, which prevents the rapid death typically observed in noninfected B1-type cells and leads to development of PEL or PEL-like tumors.
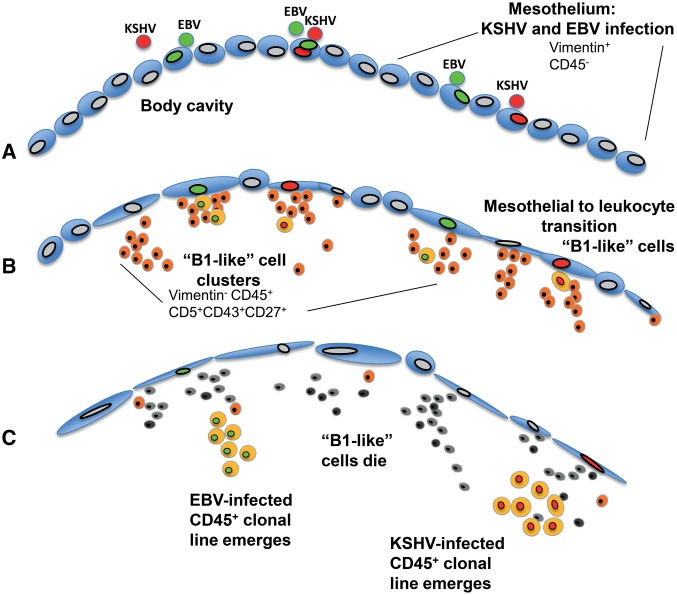
Figure 6.
A model for primary effusion lymphoma origin. A) Mesothelial cells lining body cavities become infected with endogenous Kaposi's sarcoma herpes virus (KSHV), Epstein-Barr virus (EBV), or both viruses. B) Vimentin+CD45- senescent mesothelial cells undergo mesothelial-to-lymphoid transition (MLT), giving rise to Vimentin-CD45+CD5+CD43+CD27+ “B1-like” lymphoid cells that cluster onto the mesothelium and then detach from it. C) Clonal population of CD45+, KSHV+, and EBV+ leukocytes emerge concurrent with the death of “B1-like” leukocytes. EBV = Epstein-Barr virus; KSHV = Kaposi's sarcoma herpes virus; MLT = mesothelial-to-lymphoid transition.
Discussion
The current study provides a novel and unifying insight into PEL and PEL-like lymphomagenesis. We made three observations. First, we discovered that mesothelial cell monolayers undergo a mesothelial-to-lymphoid transition, resulting in the emergence of lymphoid-type cells. This discovery extends the spectrum of mesothelial functional capabilities beyond secretion of lubricants, maintenance of surface integrity, and ability to repair (53). MLT resembles the emergence of hematopoietic cells from the endothelium of the yolk sac and the dorsal aorta identified as “endothelial-to-hematopoietic” transition (EHT) (54–56). Endothelial and mesothelial cells can undergo phenotypic and functional change through “endothelial-to-mesenchymal” (EMT) and “mesothelial-to-mesenchymal” transitions (MMT) (13,22,53,57,58). MLT, described here, may simply reflect an additional feature of mesothelial plasticity occurring invitro when mesothelial cells senesce. Mesothelial cell senescence is not an artifact of culture as it also occurs physiologically invivo and is accelerated by stress signals (23).
The second discovery we made is that the lymphocytes emerging from mesothelial cultures have a B1-like phenotype, supporting a mesothelial origin of human B1-type cells. B1 lymphocytes are the main B-cell population in murine body cavities but are rare elsewhere (29,43). Human B1-like lymphocytes have been identified in cord and peripheral blood, but to our knowledge not in body cavities (35,42,59,60). Despite their importance in immune defense (41), the origin of B1-type cells is unclear. During mouse development, B1 lymphocytes are first detected at embryonic day (E) 8.0-8.5 in the para-aortic mesoderm prior to the emergence of hematopoietic stem cells (HSCs) from the dorsal aorta (41,61), suggesting an HSC-independent origin of B1 cells (38,62,63). All mesothelia that line body cavities derive from the para-aortic splanchnopleural mesoderm. Thus, B1-type cells and mesothelia have a common developmental derivation, raising the possibility that persistence of mesodermal precursors within adult mesothelia (64) confers B1-cell differentiation potential to these mesothelia.
The third observation we made is that mesothelial cells can be infected by KSHV and EBV, a finding consistent with our previous detection of LANA1+ mesothelial-like cells in pleural effusions of three patients with pulmonary KS (65), and can yield EBV+ or KSHV+ monoclonal B-lineage lines resembling PEL or PEL-like lymphomas. The emergence of monoclonal B-lineage cell lines from mesothelial cultures suggests that PEL and PEL-like lymphoma may derive from KSHV- or EBV-infected mesothelial cells. It is noteworthy that two of the three lines so derived were clonally distinguishable from the cells originally found in the patient. It remains possible that these lines may have emerged as a result of outgrowth from rare clones of PEL- or EBV-infected cells in the original effusion. However, the evidence of MLT transition and clonal analysis suggests that a more likely explanation is that the unique clonal lines derived from the KSHV- or EBV-infected mesothelial cells. Interestingly, the “indeterminate” surface phenotype of PEL cells and cell lines, including the KSHV+ 81 lines, resembles the predominant surface phenotype of the B1-like cells. In addition, the monoclonal KSHV−/EBV+ clone resembles the KSHV− “PEL-like” lymphoma in showing a mature B-cell phenotype and EBV infection, raising the possibility that the CD20+ cell subset recovered from mesothelial cultures is a source of PEL-like lymphoma. From this perspective, body cavity–associated PEL and “PEL-like” malignancies would have a common mesothelial derivation.
There are several limitations of this study. First, the mechanisms responsible for KSHV and EBV infection of mesothelial cells lining the body cavities are not known. Second, the endogenous signals that promote MLT in mesothelial cells are not fully identified. Third, the mechanisms that underlie KSHV immortalization of mesothelium-derived lymphoid cells need to be further explored.
Funding
This work was supported by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute (NCI), National Institutes of Health, and NCI Contract No. HHSN261200800001E.
Notes
The funder had no role in the design of the study; the collection, analysis, or interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication.
GT is a co-inventor on a patent describing the measurement of vIL-6. This invention was made when GT was an employee of the US Government under 45 Code of Federal Regulations Part 7. All rights to this patent have been assigned to the US Department of Health and Human Services.
DSM, GT, HO, and MR designed and executed most of the experiments; TSU, CA, KW, KA, and RY cared for the patients and provided specimens; GT wrote the manuscript with input from DSM, TSU, and RY; TSU and RY provided critical suggestions for the project.
We thank Drs. O. Salvucci and D. Lowy for helping in various aspects of this work.
Supplementary Material
References
- 1. Knowles DM, Inghirami G, Ubriaco A, et al. Molecular genetic analysis of three AIDS-associated neoplasms of uncertain lineage demonstrates their B-cell derivation and the possible pathogenetic role of the Epstein-Barr virus. Blood. 1989;73(3):792–799. [PubMed] [Google Scholar]
- 2. Cesarman E, Chang Y, Moore PS, et al. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332(18):1186–1191. [DOI] [PubMed] [Google Scholar]
- 3. Chadburn A, Hyjek EM, Tam W, et al. Immunophenotypic analysis of the Kaposi sarcoma herpesvirus (KSHV; HHV-8)-infected B cells in HIV+ multicentric Castleman disease (MCD). Histopathology. 2008;53(5):513–524. [DOI] [PubMed] [Google Scholar]
- 4. Di Bartolo DL, Hyjek E, Keller S, et al. Role of defective Oct-2 and OCA-B expression in immunoglobulin production and Kaposi's sarcoma-associated herpesvirus lytic reactivation in primary effusion lymphoma. J Virol. 2009;83(9):4308–4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ascoli V, Calabro ML, Giannakakis K, et al. Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8-associated polyclonal body cavity effusions that mimic primary effusion lymphomas. Int J Cancer. 2006;119(7):1746–1748; author reply 1749–1750. [DOI] [PubMed] [Google Scholar]
- 6. Wu W, Youm W, Rezk SA, et al. Human herpesvirus 8-unrelated primary effusion lymphoma-like lymphoma: Report of a rare case and review of 54 cases in the literature. Am J Clin Pathol. 2013;140(2):258–273. [DOI] [PubMed] [Google Scholar]
- 7. Saini N, Hochberg EP, Linden EA, et al. HHV8-negative primary effusion lymphoma of B-cell lineage: Two cases and a comprehensive review of the literature. Case Rep Oncol Med. 2013;2013:292–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mutsaers SE. The mesothelial cell. Int J Biochem Cell Biol. 2004;36(1):9–16. [DOI] [PubMed] [Google Scholar]
- 9. Polizzotto MN, Uldrick TS, Wyvill KM, et al. Clinical features and outcomes of patients with symptomatic kaposi sarcoma herpesvirus (KSHV)-associated inflammation: Prospective characterization of KSHV inflammatory cytokine syndrome (KICS). Clin Infect Dis. 2016;62(6):730–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cesarman E, Moore PS, Rao PH, et al. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi's sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood. 1995;86(7):2708–2714. [PubMed] [Google Scholar]
- 11. Arvanitakis L, Mesri EA, Nador RG, et al. Establishment and characterization of a primary effusion (body cavity-based) lymphoma cell line (BC-3) harboring Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) in the absence of Epstein-Barr virus. Blood. 1996;88(7):2648–2654. [PubMed] [Google Scholar]
- 12. Renne R, Blackbourn D, Whitby D, et al. Limited transmission of Kaposi's sarcoma-associated herpesvirus in cultured cells. J Virol. 1998;72(6):5182–5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gasperini P, Espigol-Frigole G, McCormick PJ, et al. Kaposi sarcoma herpesvirus promotes endothelial-to-mesenchymal transition through notch-dependent signaling. Cancer Res. 2012;72(5):1157–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Salvucci O, Ohnuki H, Maric D, et al. EphrinB2 controls vessel pruning through STAT1-JNK3 signalling. Nat Commun. 2015;6:6576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Arons E, Roth L, Sapolsky J, et al. Evidence of canonical somatic hypermutation in hairy cell leukemia. Blood. 2011;117(18):4844–4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. van Dongen JJ, Langerak AW, Bruggemann M, et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: Report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2003;17(12):2257–2317. [DOI] [PubMed] [Google Scholar]
- 17. Whitby D, Marshall VA, Bagni RK, et al. Genotypic characterization of Kaposi's sarcoma-associated herpesvirus in asymptomatic infected subjects from isolated populations. J Gen Virol. 2004;85(pt 1):155–163. [DOI] [PubMed] [Google Scholar]
- 18. Parker BD, Bankier A, Satchwell S, et al. Sequence and transcription of Raji Epstein-Barr virus DNA spanning the B95-8 deletion region. Virology. 1990;179(1):339–346. [DOI] [PubMed] [Google Scholar]
- 19. Kearse M, Moir R, Wilson A, et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12):1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gasperini P, Tosato G.. Targeting the mammalian target of Rapamycin to inhibit VEGF and cytokines for the treatment of primary effusion lymphoma. Leukemia. 2009;23(10):1867–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ksiazek K, Piwocka K, Brzezinska A, et al. Early loss of proliferative potential of human peritoneal mesothelial cells in culture: The role of p16INK4a-mediated premature senescence. J Appl Physiol (1985). 2006;100(3):988–995. [DOI] [PubMed] [Google Scholar]
- 22. Foley-Comer AJ, Herrick SE, Al-Mishlab T, et al. Evidence for incorporation of free-floating mesothelial cells as a mechanism of serosal healing. J Cell Sci. 2002;115(pt 7):1383–1389. [DOI] [PubMed] [Google Scholar]
- 23. Ksiazek K, Mikula-Pietrasik J, Jorres A, et al. Oxidative stress-mediated early senescence contributes to the short replicative life span of human peritoneal mesothelial cells. Free Radic Biol Med. 2008;45(4):460–467. [DOI] [PubMed] [Google Scholar]
- 24. Sin SH, Roy D, Wang L, et al. Rapamycin is efficacious against primary effusion lymphoma (PEL) cell lines invivo by inhibiting autocrine signaling. Blood. 2007;109(5):2165–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Terada T. Immunohistochemical profile of normal mesothelium and histiocytic/methothelial hyperplasia: A case report. Int J Clin Exp Pathol. 2011;4(6):631–636. [PMC free article] [PubMed] [Google Scholar]
- 26. Wang L, Li L, Shojaei F, et al. Endothelial and hematopoietic cell fate of human embryonic stem cells originates from primitive endothelium with hemangioblastic properties. Immunity. 2004;21(1):31–41. [DOI] [PubMed] [Google Scholar]
- 27. Rubtsov AV, Rubtsova K, Kappler JW, et al. CD11c-expressing B cells are located at the T cell/B cell border in spleen and are potent APCs. J Immunol. 2015;195(1):71–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jourdan M, Caraux A, Caron G, et al. Characterization of a transitional preplasmablast population in the process of human B cell to plasma cell differentiation. J Immunol. 2011;187(8):3931–3941. [DOI] [PubMed] [Google Scholar]
- 29. Baumgarth N. The double life of a B-1 cell: Self-reactivity selects for protective effector functions. Nat Rev Immunol. 2011;11(1):34–46. [DOI] [PubMed] [Google Scholar]
- 30. Tangye SG, Ma CS, Brink R, et al. The good, the bad and the ugly—TFH cells in human health and disease. Nat Rev Immunol. 2013;13(6):412–426. [DOI] [PubMed] [Google Scholar]
- 31. Souza TA, Stollar BD, Sullivan JL, et al. Peripheral B cells latently infected with Epstein-Barr virus display molecular hallmarks of classical antigen-selected memory B cells. Proc Natl Acad Sci U S A. 2005;102(50):18093–18098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Griffin DO, Rothstein TL.. A small CD11b(+) human B1 cell subpopulation stimulates T cells and is expanded in lupus. J Exp Med. 2011;208(13):2591–2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jackson SM, Wilson PC, James JA, et al. Human B cell subsets. Adv Immunol. 2008;98:151–224. [DOI] [PubMed] [Google Scholar]
- 34. Ghosn EE, Sadate-Ngatchou P, Yang Y, et al. Distinct progenitors for B-1 and B-2 cells are present in adult mouse spleen. Proc Natl Acad Sci U S A. 2011;108(7):2879–2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Perez-Andres M, Grosserichter-Wagener C, Teodosio C, et al. The nature of circulating CD27+CD43+ B cells. J Exp Med. 2011;208(13):2565–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Choi YS, Dieter JA, Rothaeusler K, et al. B-1 cells in the bone marrow are a significant source of natural IgM. Eur J Immunol. 2012;42(1):120–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shen P, Fillatreau S.. Antibody-independent functions of B cells: A focus on cytokines. Nat Rev Immunol. 2015;15(7):441–451. [DOI] [PubMed] [Google Scholar]
- 38. Yoshimoto M. The first wave of B lymphopoiesis develops independently of stem cells in the murine embryo. Ann N Y Acad Sci. 2015;1362(1):16–22. [DOI] [PubMed] [Google Scholar]
- 39. Covens K, Verbinnen B, Geukens N, et al. Characterization of proposed human B-1 cells reveals pre-plasmablast phenotype. Blood. 2013;121(26):5176–5183. [DOI] [PubMed] [Google Scholar]
- 40. Griffin DO, Quach T, Batliwalla F, et al. Human CD11b+ B1 cells are not monocytes: A reply to “Gene profiling of CD11b+ and CD11b- B1 cell subsets reveals potential cell sorting artifacts.” J Exp Med. 2012;209(3):434–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Montecino-Rodriguez E, Dorshkind K.. B-1 B cell development in the fetus and adult. Immunity. 2012;36(1):13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bueno C, Sardina JL, Di Stefano B, et al. Reprogramming human B cells into induced pluripotent stem cells and its enhancement by C/EBPalpha. Leukemia. 2016;30(3):674–682. [DOI] [PubMed] [Google Scholar]
- 43. Hayakawa K, Hardy RR, Herzenberg LA, et al. Progenitors for Ly-1 B cells are distinct from progenitors for other B cells. J Exp Med. 1985;161(6):1554–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Majerciak V, Yamanegi K, Allemand E, et al. Kaposi's sarcoma-associated herpesvirus ORF57 functions as a viral splicing factor and promotes expression of intron-containing viral lytic genes in spliceosome-mediated RNA splicing. J Virol. 2008;82(6):2792–2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Han Z, Swaminathan S.. Kaposi's sarcoma-associated herpesvirus lytic gene ORF57 is essential for infectious virion production. J Virol. 2006;80(11):5251–5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fan X, Patera AC, Pong-Kennedy A, et al. Murine CXCR1 is a functional receptor for GCP-2/CXCL6 and interleukin-8/CXCL8. J Biol Chem. 2007;282(16):11658–11666. [DOI] [PubMed] [Google Scholar]
- 47. Lukac DM, Renne R, Kirshner JR, et al. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology. 1998;252(2):304–312. [DOI] [PubMed] [Google Scholar]
- 48. Sun R, Lin SF, Gradoville L, et al. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc Natl Acad Sci U S A. 1998;95(18):10866–10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Carbone A, Cilia AM, Gloghini A, et al. Establishment and characterization of EBV-positive and EBV-negative primary effusion lymphoma cell lines harbouring human herpesvirus type-8. Br J Haematol. 1998;102(4):1081–1089. [DOI] [PubMed] [Google Scholar]
- 50. Jenner RG, Maillard K, Cattini N, et al. Kaposi's sarcoma-associated herpesvirus-infected primary effusion lymphoma has a plasma cell gene expression profile. Proc Natl Acad Sci U S A. 2003;100(18):10399–10404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ramasamy I, Brisco M, Morley A.. Improved PCR method for detecting monoclonal immunoglobulin heavy chain rearrangement in B cell neoplasms. J Clin Pathol. 1992;45(9):770–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vieira J, O'Hearn PM.. Use of the red fluorescent protein as a marker of Kaposi's sarcoma-associated herpesvirus lytic gene expression. Virology. 2004;325(2):225–240. [DOI] [PubMed] [Google Scholar]
- 53. Mutsaers SE, Birnie K, Lansley S, et al. Mesothelial cells in tissue repair and fibrosis. Front Pharmacol. 2015;6:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Eilken HM, Nishikawa S, Schroeder T.. Continuous single-cell imaging of blood generation from haemogenic endothelium. Nature. 2009;457(7231):896–900. [DOI] [PubMed] [Google Scholar]
- 55. Lancrin C, Sroczynska P, Stephenson C, et al. The haemangioblast generates haematopoietic cells through a haemogenic endothelium stage. Nature. 2009;457(7231):892–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zovein AC, Hofmann JJ, Lynch M, et al. Fate tracing reveals the endothelial origin of hematopoietic stem cells. Cell Stem Cell. 2008;3(6):625–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sandoval P, Loureiro J, Gonzalez-Mateo G, et al. PPAR-gamma agonist rosiglitazone protects peritoneal membrane from dialysis fluid-induced damage. Lab Invest. 2010;90(10):1517–1532. [DOI] [PubMed] [Google Scholar]
- 58. Lignitto L, Mattiolo A, Negri E, et al. Crosstalk between the mesothelium and lymphomatous cells: Insight into the mechanisms involved in the progression of body cavity lymphomas. Cancer Med. 2014;3(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Descatoire M, Weill JC, Reynaud CA, et al. A human equivalent of mouse B-1 cells? J Exp Med. 2011;208(13):2563–2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Griffin DO, Holodick NE, Rothstein TL.. Human B1 cells in umbilical cord and adult peripheral blood express the novel phenotype CD20+ CD27+ CD43+ CD70. J Exp Med. 2011;208(1):67–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ito C, Yamazaki H, Yamane T.. Earliest hematopoietic progenitors at embryonic day 9 preferentially generate B-1 B cells rather than follicular B or marginal zone B cells. Biochem Biophys Res Commun. 2013;437(2):307–313. [DOI] [PubMed] [Google Scholar]
- 62. Yoshimoto M, Montecino-Rodriguez E, Ferkowicz MJ, et al. Embryonic day 9 yolk sac and intra-embryonic hemogenic endothelium independently generate a B-1 and marginal zone progenitor lacking B-2 potential. Proc Natl Acad Sci U S A. 2011;108(4):1468–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kobayashi M, Shelley WC, Seo W, et al. Functional B-1 progenitor cells are present in the hematopoietic stem cell-deficient embryo and depend on Cbfbeta for their development. Proc Natl Acad Sci U S A. 2014;111(33):12151–12156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Raftery AT. Regeneration of parietal and visceral peritoneum: An electron microscopical study. J Anat. 1973;115(pt 3):375–392. [PMC free article] [PubMed] [Google Scholar]
- 65. Bryant-Greenwood P, Sorbara L, Filie AC, et al. Infection of mesothelial cells with human herpes virus 8 in human immunodeficiency virus-infected patients with Kaposi's sarcoma, Castleman's disease, and recurrent pleural effusions. Mod Pathol. 2003;16(2):145–153. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.