Abstract
Background
Adenoid cystic carcinoma (ACC), an uncommon and indolent salivary gland malignancy, is characterized by varied morphologic and clinical manifestations. Molecular genetic studies of ACC identified certain structural and mutational alterations that may play a driver role in tumor development. The evolution and regional consistency of these events in ACC development progression are uncertain.
Methods
To investigate the spatial and temporal clonal landscape of ACC, whole-genome sequencing and variant analyses were performed on 34 regionally sampled primary tumors and their concurrent and metachronous metastatic deposits from eight patients.
Results
The average mutation rate per case (primary and metastasis) was 0.32 per million base pairs, and the average incidence of shared mutations in primary and metastatic specimens in each case was 21.9% (range = 0%–44.4%). The analyses revealed considerable spatial clonal differences within and between primary tumors and metastatic disease. Phylogeny formation displayed branching evolution with a main trunk and two distinct mono-splits in all cases. One of the main branches represented intratumor subclonal diversity, and the other delineated metastatic departure and progression. All metastatic tumors shared clonal linkage to their matching primary in concordance with parallel dissemination of metastasis. Synchronous metastases were genomically more similar to their primary than metachronous metastatic disease. Truncal genetic alterations included somatic mutations in the NOTCH pathway genes (NOTCH1 and SPEN) and t(6;9) associated gene fusions.
Conclusions
Our study delineated clonal and subclonal phylogeny for primary and metastatic ACC, defined early genetic drivers, and provides a conceptual framework for a rational strategy to integrate heterogeneity in clinical assessment.
The ability to predict the clinical course of individual patients with malignant solid tumors is a fundamental objective in cancer management. Achieving this goal is complicated by the diverse phenotypic features and variable clinical behaviors within and between these cancers. Furthermore, recent genomic heterogeneity studies have shown pervasive regional clonal and subclonal diversities within primary solid malignancies. Importantly, these confounding manifestations have been linked to poor outcome and therapeutic failure of cancer patients (1–3). Assessing intra- and intergenomic heterogeneity, therefore, is central to efforts to identify and validate key genetic targets for prognostication and therapeutic stratification of patients with solid malignancies.
Adenoid cystic carcinoma (ACC), in contrast to more common carcinomas, is distinguished by a spectrum of overlapping morphologic forms and indolent unpredictable clinical behaviors (4). The majority of ACC patients present with fully detectable tumors and undergo complete excision with or without postoperative radiotherapy (5). Approximately 40% of ACC patients experience progressive disease to which no effective therapy is available (6,7). Due largely to the lack of preneoplastic stage and progression models, information on key molecular events associated with early development and progression of ACC is unknown. Concerted efforts to identify novel molecular targets for effective and sustained therapy of these patients are being mounted.
Recent molecular studies of ACCs show a comparatively stable genome, with recurrent t(6:9) and t(8;9) translocations resulting in MYB-NFIB and MYBL1-NFIB gene fusions in more than 60% of tumors (8–14). The evolution and consistency of these genetic findings within and between primary tumor mass and metastatic deposits can only be achieved through structured spatial and temporal tumor tissue sampling. To accomplish this objective, we implemented a prospective regional acquisition of spatial primary tumor samples and monitored patients for recurrences. A search of our tissue database identified eight patients with regionally sampled primary tumors with concurrent and late metastatic specimens.
Using whole-genome sequencing, we studied a unique set of 42 specimens (34 tumors and eight normal) to assess the subclonal architecture and evolution of primary and metastatic ACC.
Methods
Patients and Samples
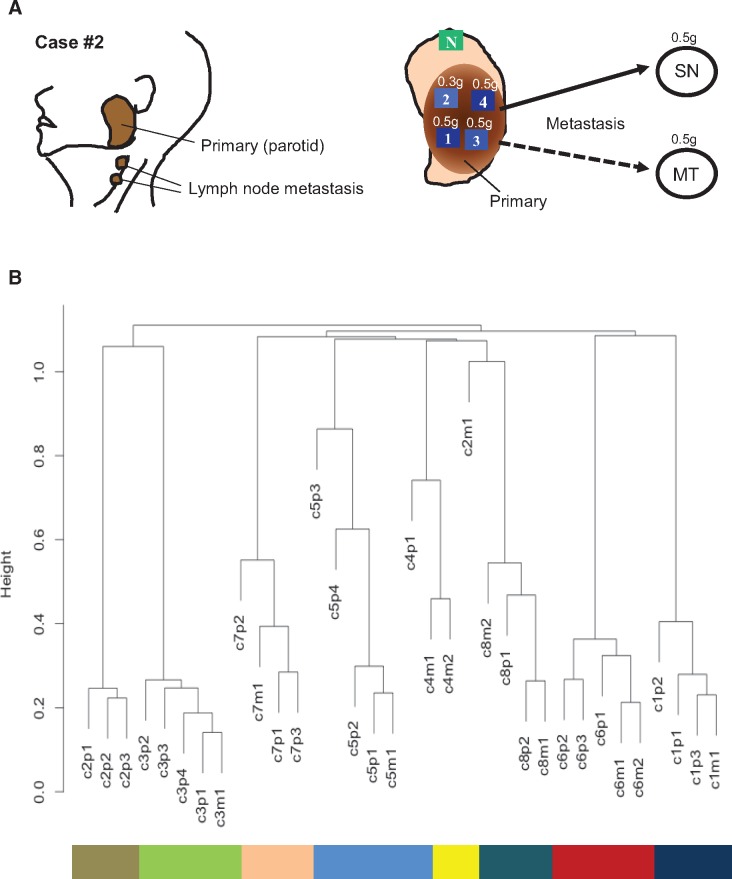
Eight adenoid cystic carcinomas from an equal number of patients with structured multiregion specimens obtained from each primary tumor and synchronous and/or metachronous metastatic deposits were retrieved from our tissue bank and formed the materials for this study (Figure 1A and Table 1). Frozen section and touch preparation was performed on each specimen, and tumor cells composed 75% to 90% of specimens used for analysis. All specimens used were obtained from patients who consented to an institutional research board protocol. Nine random samples from the eight cases were used in our previous studies (8,14).
Figure 1.
The tissue sampling and genomic clustering of adenoid cystic carcinoma (ACC) cases. A) The left panel represents one of the examples for specimens’ collection in case 2. The right panel shows our study design to characterize the intratumor heterogeneity in ACC. Multiple spatially separated specimens were obtained from resected ACC (1–4), adjacent normal tissue (N), synchronous (SN), and/or metachronous (MT) metastatic regions. Each specimen was approximately 0.3 to 0.5 g. B) Cluster dendrogram of the mutations for all 34 tissues from eight patients. The “huclust” function in R statistical programming language was utilized with complete linkage method to generate hierarchical clustering based on Euclidean distance. All samples from individual cases were clustered together except the metastasis from case 2. *Only three mutations were detected in this specimen. Of these three mutations, one (chr21 9826422 G>C) was also found in all primary specimens from case 8, leading to its displacement (see Supplementary Table 3, available online).
Table 1.
Clinico-pathologic and genome sequencing coverage information of 8 patients with adenoid cystic carcinomas in the study
| Patient | Age | Sex | ID | Tumor type | Site | Pattern | Size | Met, (y) | Coverage (>40×) |
|
|---|---|---|---|---|---|---|---|---|---|---|
| Genome, % | Coding, % | |||||||||
| 1 | 45 | F | C1p1 | Primary_1* | Pharynx | S+T | 3.5 | – | 81.6 | 88.8 |
| C1p2 | Primary_2 | |||||||||
| C1p3 | Primary_3 | |||||||||
| C1m1 | Metastasis_1* | LN | S+T | 1.7 | SN | |||||
| 2 | 42 | M | C2p1 | Primary_1 | Parotid | S+C+T | 3.8 | – | 85.6 | 92.7 |
| C2p2 | Primary_2 | |||||||||
| C2p3 | Primary_3 | |||||||||
| C2m1 | Metastasis_1 | LN | S | 0.5 | SN | |||||
| 3 | 67 | M | C3p1 | Primary_1 | Maxilla | S+T | 3.5 | – | 82.5 | 90.6 |
| C3p2 | Primary_2 | |||||||||
| C3p3 | Primary_3 | |||||||||
| C3p4 | Primary_4* | |||||||||
| C3m1 | Metastasis_1 | LN | S+T | 1 | MT (1) | |||||
| 4 | 47 | M | C4p1 | Primary_1* | Parotid | C+T | 5.5 | – | 83.6 | 91.2 |
| C4m1 | Metastasis_1 | Lung | C | 1.7 | MT (10) | |||||
| C4m2 | Metastasis_2 | |||||||||
| 5 | 43 | M | C5p1 | Primary_1* | Parotid | S+C+T | 3.5 | – | 83.7 | 90.7 |
| C5p2 | Primary_2* | |||||||||
| C5p3 | Primary_3 | |||||||||
| C5p4 | Primary_4 | |||||||||
| C5m1 | Metastasis_1 | LN | S+C | 1.4 | MT (6) | |||||
| 6 | 36 | M | C6p1 | Primary_1 | Orbital | S | 2 | – | 82.4 | 91.1 |
| C6p2 | Primary_2 | |||||||||
| C6p3 | Primary_3 | |||||||||
| C6m1 | Metastasis_1 | Scalp | S | 1.1 | MT (3) | |||||
| C6m2 | Metastasis_2 | |||||||||
| 7 | 60 | M | C7p1 | Primary_1* | FOM | C+T | 4 | – | 83.6 | 90.4 |
| C7p2 | Primary_2 | |||||||||
| C7p3 | Primary_3* | |||||||||
| C7m1 | Metastasis_1 | Lung | C | 1.3 | MT (1) | |||||
| 8 | 58 | M | C8p1 | Primary_1 | Sub-m | C+T | N/A | – | 82.2 | 90.8 |
| C8p2 | Primary_2 | |||||||||
| C8m1 | Metastasis_1 | LN | C+T | 0.7 | SN | |||||
| C8m2 | Metastasis_2* | Lung | C+T | 1.7 | MT (5) | |||||
Specimens previously used in other publications (see Supplementary Table 1, available online) (14,41). C = cribriform; FOM = floor of mouth; LN = lymph node pattern; M = metastases; MT = metachronous; S = solid; SN = synchronous; Sub-m = submandibular gland; T = tubular. (y) indicates the years after diagnosis of the primary tumor.
Whole-Genome Sequencing and Variant Detection
Details of whole-genome sequencing and variant processing have been deposited in Ingenuity Variant Analysis (see the Supplementary Methods, available online).
Phylogenetic Trees and Biological Pathway Analysis
See the Supplementary Methods (available online) for details of phylogenetic trees, dendrogram construction, candidate driver mutations, and biological pathway analyses.
MYB Expression
MYB protein expression was determined by immunohisochemistry using monoclonal anti-MYB antibody (Abcam, Cambridge, UK; ab45150, 1:25 dilution) in all cases using different primary tumor blocks. MYB full-length and truncated transcripts were assessed by reverse transcription polymerase chain reaction and 3’RACE following our previous report (9) on all five cases with t(6;9) translocation (see the Supplementary Methods, available online).
Statistical Analysis
The two-sided and two-sample unequal variance settings of the t test were utilized for evaluation of unique mutations in synchronous and metachronous metastasis. Statistical significance was defined at a P value of less than .05.
Results
Clinicopathologic Characteristic and Whole-Genome Sequencing of ACCs
Table 1 and Supplementary Figure 1 (available online) present the clinicopathologic, molecular, and genomic characteristics of patients with ACC included in the study. Whole-genome sequencing was performed on 42 samples from eight patients (23 spatially distinct primary tumor specimens, 11 synchronous and metachronous metastatic lesions, and eight matching histologically normal salivary tissue samples). In our analyses, 90.8% of the coding regions and 83.2% of the whole genome were interrogated at 40× or greater sequencing median coverage in both tumors and normal.
Identification of Somatic Mutations
Somatic mutation calls ranged from 51 175 to 84 107 variants per sample in both primary and metastatic specimens (Supplementary Figure 2, available online). Mutations of single nucleotide variants (SNVs) were commonly C/G > T/A substitutions (Supplementary Figure 3A, available online). Validation of 17 376 alterations (SNVs and Indels) was performed using Agilent targeted sequencing at 600× coverage, and mutations were called by GATK and Mutect pipeline (Supplementary Methods, available online). Further annotation-related filtering of potentially functional variants (codons, splice sites, untranslated regions (UTRs), and other noncoding regions) yielded a total of 306 somatic mutations (approximately 1.4% of targeted alterations) (Supplementary Figure 3B and Supplementary Table 3, available online).
The number of mutations per million base pairs for each sample was 0.24 (Supplementary Figure 4A, available online). A combined mutation of all samples in a given case was 0.32 per million base pairs. Our findings show that ACC as an entity falls within the low–mutation burden tumors (3,14), including differentiated thyroid carcinoma and carcinoid and small round cell tumors (Ewing’s sarcoma, neuroblastoma, medulloblastoma) (3). We estimated that one random sample of a tumor identifies on average 42.2% of the total verified mutations while three samples cover 82.9% of the mutations (Supplementary Figure 4B, available online). Moreover, interpatient comparison showed an average of less than 1.0% of the mutations to be shared by tumors of all patients.
Heterogeneity of Somatic Mutations
We then assessed intra- (primary) and intertumor (primary/metastasis) heterogeneity of specimens from all eight patients using the 306 validated mutations. The average incidence of shared mutations of combined primary and metastatic specimens of a patient was 21.9% (range = 0%–44.4%) (see Supplementary Figure 5, available online). Figure 1B presents the cluster analysis of interpatient mutational spectrum. The dendrogram shows distinctive clustering of specimens from each patient (C1–C8) except for one metastatic specimen of patient 2; this may most likely reflect an earlier metastatic seeding from primary tumor prior to subclonal split (see Figure 2, case 2).
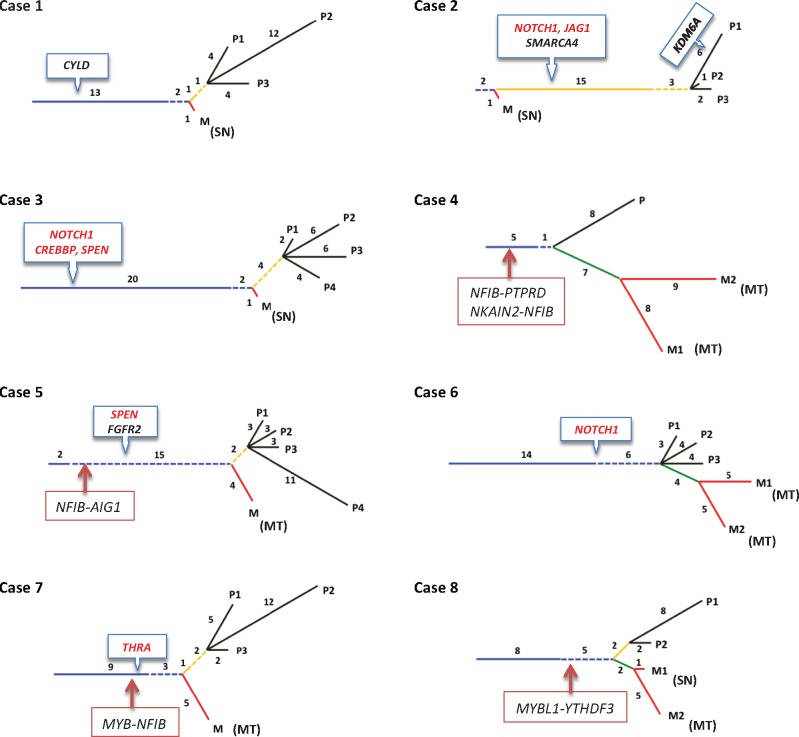
Figure 2.
The phylogenetic tree of somatic mutations for 34 tumor tissues (and eight normal tissues) from eight adenoid cystic carcinoma (ACC) cases. Ubiquitous mutations (blue): The mutations can be found in all regions of the primary tissue and metastatic tissue(s). Partial mutations (dotted blue): The mutations can be found in at least one region of the primary tissue and at least one metastatic tissue. Primary shared mutations (yellow): The mutations can be found in all regions of the primary tissues only, and (dotted yellow) more than one region of the primary tissue but not all. Metastatic shared mutations (green): The mutations can found in all metastatic tissue(s), but not in any region of primary tissues. Primary private mutations (black): The mutations can only be found in only one region of the primary tissue. Metastatic private mutations (red): The notations can only be found in only one metastatic tissue. MT = metachronous; SN = synchronous.
Phylogeny of Tumor Evolution and Progression
To delineate the clonal and subclonal evolutionary architecture of ACC, phylogeny of intraregional subclones was generated by validated mutations for each patient (Figure 2). In addition to previously delineated subgroups, we characterized an additional set of mutations to be shared by one peripheral limb of a primary and metastasis but not in all specimens, and these were labeled “partial.” All specimens from primary and metastatic lesions of a given patient showed consistent main truncal stem of variable length followed by limbal bifurcation. We considered the main trunk length to reflect the timing of subclonal progression prior to phyletic splits of primary subclonal diversity and metastasis. Interestingly, the data show evidence for regional dominance within certain primary tumors. As demonstrated in cases 1, 5, 7, and 8, only one region showed considerably higher mutation rate than other regions of the same tumor (P2 in case 1, P4 in case 5, P2 in case 7, and P1 in case 8). These findings suggest that intratumoral variations may occur in response to host loco-regional dynamics.
Interestingly, all synchronous metastases had a low number of unique mutations in comparison with metachronous metastasis within and between patients. Metachronous metastases were also associated with clear evidence for subclonal acquisition with progression. As an example, in patients 4, 6, 7, and 8 (M2 specimen), the ratios of unique mutations in metastasis in relation to corresponding primary tumors per specimen were 1.9:1, 0.57:1, 2.5:1, 0.58:1, and 1:1, respectively, while those of synchronous metastasis in patients 1, 2, 3, and 8 (M1 specimen) were statistically significantly lower (0.12:1, 0.05:1, 0.15:1 and 0.42:1; P = .04, two-sided t test), implying early seeding of the latter (Figure 2). Although ontogenetic linkage is found between metastatic deposits (green line in cases 4, 6, and 8), all metastases were traced to either the main stem and/or to one of the branches of their primary consistent with parallel dissemination.
Frequently Mutated Genes and Pathways
The 306 validated mutations by targeted sequencing were used to identify driver and important genes. This resulted in 78 unique nonsynonymous mutations in the coding regions of 73 genes (Supplementary Table 4, available online). Of these 78 mutations, 21 were nonsense, 49 were missense, and eight were splicing mutations. Fifty mutations in 47 genes were found in multiple samples (Figure 3), and 28 mutations were detected in only one sample. NOTCH1 gene mutations were identified in three different patients and comprised of a stop codon with G > T substitution (c.7400C>A, p.S2467X in patients 2 and 3) and a different frameshift mutation (c.7432_7433dupGC, p.A2478fs in patient 6). All NOTCH1 mutations were located in the proline–glutamic acid–serine–threonine (PEST) domain, as in T-ALL (15). Sequencing data also provided variant allele frequency (VAF) of gene mutations that support clonal enrichment and progression. VAF values at 96.0% and 97.2% (see Supplementary Table 5, available online) were observed for the stop mutations of NOTCH1 in patients 2 and 3. The mutations of SPEN show the VAF values at 74.5% and 81.3%. The VAFs indicate that clones with NOTCH1 and SPEN mutations occurred relatively early in tumor development. Unique metastatic mutations of the gene ACVR2A were shared in two metastatic samples from patient 4 with VAF values at 53.1% and 33.3%. Another mutation of gene POLD1 with a VAF at 54.5% existed only in one of the two metastatic samples. Although low-frequency mutations may have been missed due to the relatively low coverage, the result implies the heterogeneous development of metastatic clones during the metastasis progression.
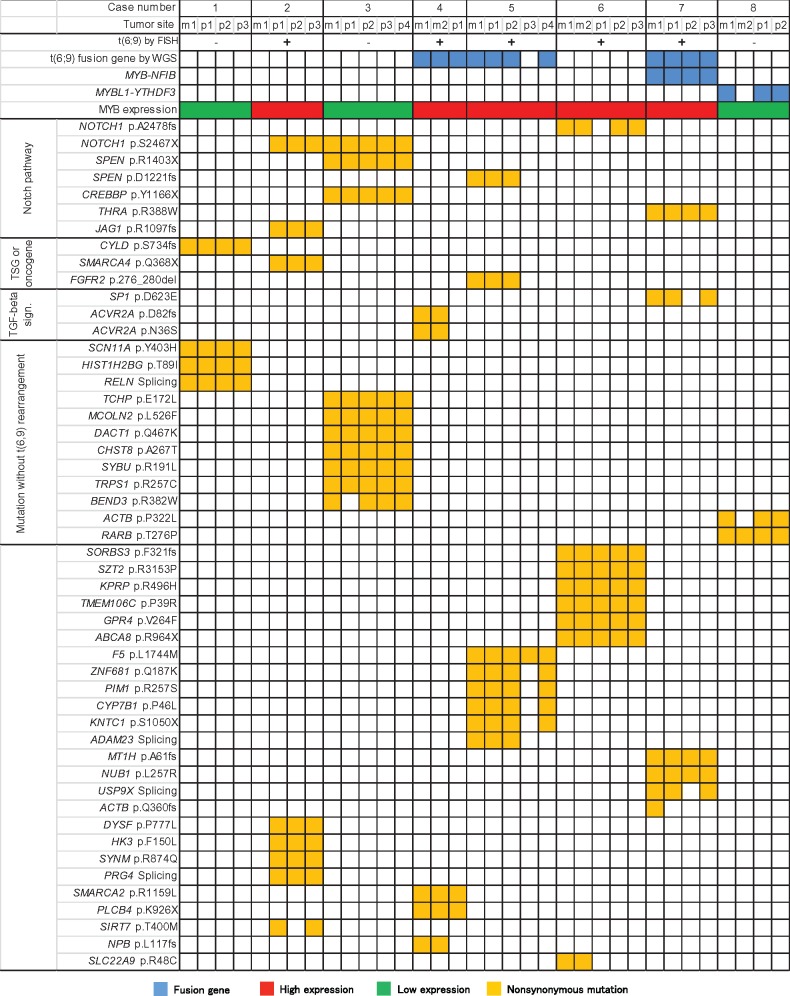
Figure 3.
Genes with somatic mutations and t(6:9) chromosome translocations from adenoid cystic carcinoma patients. *Cases 4 and 5 have unique t(6;9)-related gene fusions (see Table 1). FISH = fluorescence in situ hybridization; WGS = whole-genome sequencing. MYB-NFIB and MYBL1-YTHDF3 gene fusions were identified by both WGS and RT-PCR.
To determine potential pathways associated with these mutations, we employed the Database for Annotation, Visualization, and Integrated Discovery tool. Mutations in members of the NOTCH signaling pathway included NOTCH1 (three patients), SPEN (two patients), CREB binding protein (CREBBP; one patient), THRA (one patient), and JAG1 (one patient) in five of eight patients (Supplementary Figure 6, available online). SPEN mutations consisted of frameshift (c.3663_3664delCT, p.D1221fs) and stop (c.4207C>T, p.R1403X) mutations, and the THRA gene had a missense mutation (c.1162C>T, p.R388W). Both genes are inhibitors of the NOTCH signaling pathway (16,17). The CREBBP mutation (c.3498T>G, p.Y1166X) consisted of a stop codon, and the JAG1 gene, a member of the NOTCH pathway, had a truncating frameshift (c.3290_3303delGCTGCCCGGCTTCC, p.R1097fs) mutation. In two patients (2 and 3), mutations of multiple members of the NOTCH1 signaling pathway were found.
Structural Chromosomal Alterations
We then focused on the status of the t(6;9) chromosomal translocations and related fusion genes identified in ACC. Integrated analyses of all eight patients identified 29 chromosomal translocations (Supplementary Table 1, available online). Fluorescence in situ hybridization analyses, using MYB and NFIB probes, identified t(6;9) translocations in five patients (Table 1; Supplementary Figure 7, available online): Whole-genome sequencing identified the t(6:9) in only three of these patients (Supplementary Table 2, available online). In patient 7, who was positive for t(6;9), the MYB-NFIB fusion was identified, along with a fusion between the KIAA1244 gene (6q) and the FOCAD gene on chromosome 9p. Interestingly, the latter fusion was identified in all primary and metastatic specimens of this case. In patient 5, fusion between the NFIB and the AIG1 genes was detected in all primary and metastatic specimens. In patient 4, rearrangements and fusion of NFIB with both PTPRD (intrachromosomal) and NKAIN2 were found. Additionally, MYBL1-YTHDF3 was identified in all primary tumors and synchronous metastatic tumors in patient 8. Collectively, these findings and those reported previously (10–12) indicate a substantial complexity of the structural rearrangements associated with chromosome 6q, 8q, and 9p regions in ACC.
MYB Expression
High MYB protein expression was found in all primary tumors with t(6;9) translocation–positive cases (patients 2, 4, 5, 6, and 7) (Figure 3; Supplementary Figure 7, available online). The remaining three cases with no t(6;9) showed no MYB expression. Trancated MYB transcript was only detected in case 7 with MYB-NFIB translocation. While full-length MYB transcript was found in all four t(6;9) tumors without any MYB-NFIB formation.
Discussion
Inherent to the spatial and metastatic progression of malignant solid tumors is the emergence of regional clonal heterogeneity and functional polarization that complicate patients’ prognostication and therapeutic response. Systematic analysis of this innate phenomenon, however, has been constrained by the lack of structured sampling of tumors in a space and time. In this study, we report considerable spatial, temporal, and interpatient genomic heterogeneity in salivary ACC consistent with the physiognomic and clonal compartmentalization reported in other solid malignancies (18–21). Although the extent of intratumoral heterogeneity observed was relatively lower than those of high-grade epithelial carcinomas (8,10), a single random primary tumor sample underrepresented the subclonal diversity in a given primary ACC by at least 42.2%. We also noted a subregional clonal overgrowth within certain primary tumors, regardless of the histologic patterns, suggesting a directional response to variable local factors (vascular, nutritional, and immune response) (22). Locating these regions through imaging modalities may lead to better selection of specimens for genomic analysis. Together, these findings clearly indicate a progressive subclonal development and diversion within primary tumors requiring a sampling strategy for reliable genomic representation.
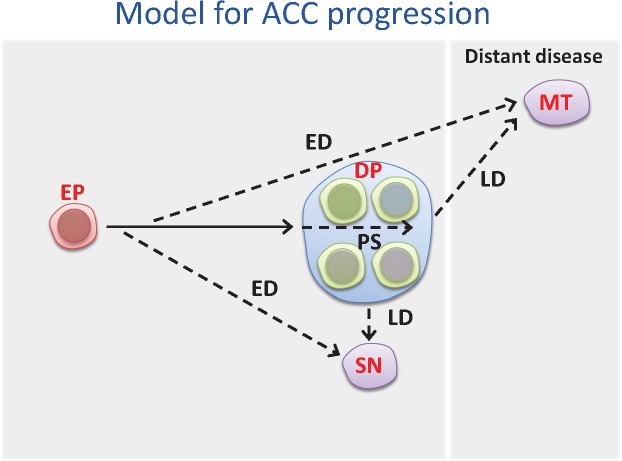
Phylogeny of the clonal data shows consistent branching evolution of primary tumor mass and metastatic dissemination of ACC. Here we propose the ACC progression model shown in Figure 4. Invariably, an early truncal formation followed by main limbal bifurcations representing intratumoral subclonal diversity and metastatic dissemination emerged. Metastatic phylogeny evolved at varying temporal stages of tumor progression for both synchronous and metachronous lesions. Interestingly, synchronous metastases appear to evolve early and to manifest close genomic resemblance to primary tumors. Paradoxically, metachronous metastasis manifested substantial subclonal differences from primary tumors (23), suggesting that dormancy and temporal adaptation to different host environments contribute to the emergence of new subclones in distant metastasis. These findings, together with the considerable intratumoral regional differences, underscore the need for a rational sampling strategy for clinical management. Moreover, we provide phylogenetic evidence that both synchronous and metachronous metastases are derived from an ancestral primary source (24) consistent with parallel dissemination. Our findings are in concordance with those of pancreatic carcinomas (24) and at variance with the metastatic cascade reported in brain tumors (25). We contend, therefore, that evidence of metastatic cascade (metastasis to metastasis) can only be supported by the lack of clonal association to primary tumors after comprehensive regional intratumoral sampling analysis (23,25,26).
Figure 4.
Schematic presentation of proposed intratumoral heterogeneity and metastatic dissemination in adenoid cystic carcinoma. The solid arrow indicates the truncal mutations during primary development. The dotted arrow with label PS indicates intra-heterogeneity in primary tumor. The dotted arrows with labels ED and LD indicate synchronous and metachronous metastasis. DP = detectable primary; ED = early dissemination; EP = early primary; LD = late dissemination; MT = metachronous met; PS = partially shared; SN = synchronous met.
An important aspect of our study is the delineation of multiple key mutations and gene rearrangements associated with ACC tumorigenesis. The finding of these truncal events is consistent with similar results by others and supports an early and/or an initiating tumorigenic role for these events in primary tumor development and metastatic progression. These alterations are comprised of somatic gene mutations in NOTCH1, CYLD, KDM6A, SMARCA4, CREBBP, SPEN, and FGFR2 (2,14). Interestingly, mutations of the NOTCH1 pathway genes were located in the trunks of four of eight primary tumors and in the branches of an additional tumor. These mutations were clustered at the hinge and the REST domains of the intracellular segment of the NOTCH1 gene, consistent with the activating mutations seen in T-ALL (27–30). Our findings, in contrast to studies of head and neck squamous and lung carcinomas (31,32), suggest that γ-secretase inhibitors may also be effective in treating a subset of ACC patients with NOTCH1 mutations (27,33,34). A a recent functional and clinical study of NOTCH1 by our group, which included some of the current tumors, highlighted the clinical relevance of this driver mutation (35). Additionally, mutation of the SPEN gene, a member of the NOTCH pathway and a known histone modification gene (15), further supports the role of the NOTCH pathway in ACC. The finding of a CYLD gene mutation in the trunk of an ACC is also of interest in view of the close similarity of ACC to basaloid dermal and salivary tumors and suggests a possible association of this gene in a subset of tumors (36,37). Similarly, FGFR2 gene mutations could be of interest if further verified as a target for therapy (37–39).
Notably, the analysis confirmed the presence of the MYB-NFIB fusions and identified novel fusion resulting from different t(6;9) translocation and additional fusions involving the NFIB gene with different gene partners. The etiologic and functional implications of these novel fusions are currently uncertain. Importantly, however, the truncal location of the t(6;9) translocation in three cases sustains an early tumorigenic role for this event in a subset of ACC. Interestingly, we only detected truncated MYB transcripts in tumors with MYB-NFIB fusion but not in t(6;9) translocation tumors with intact MYB, suggesting a differential biological function. It remains unclear, however, whether full-length and/or truncated MYB impart similar or different biological roles in ACC progression. We, however, contend that truncated MYB may play a role in a subset of ACC tumorigenesis because high intact MYB can be found in both ACC lacking MYB fusion and other salivary tumors with basaloid features (data not shown). Collectively, these findings indicate that fusions resulting from t(6;9) translocation and/or NOTCH pathway alterations constitute an early/primary event in a subset of ACC.
In this study, however, the limited size and accessibility to critical samples precluded the discussion of other relevant heterogeneity issues. As an example, the lack of small tumors for sampling did not allow for assessing the emergence of intratumoral evolution in relation to metastatic dissemination. The study also could have benefited from the availability of metastatic specimens from different organs to evaluate the effects of homing, dormancy, and organ contextual milieu on clonal evolution and pattern of dissemination. Similarly, the lack of samples from multiple concurrent metastases precluded kinetic analysis of the timing and emergence of metastatic dissemination. Additional genomic correlation of distinct regions representing variable morphologic patterns within primary and metastasis deposits would have shed some light on the underlying genomic events associated with morphologic manifestation. Higher depth of coverage would have also allowed for the assessment of the extent of rare private mutation that may have evaded detection in our analysis.
In conclusion, our data show considerable spatial, temporal, and interpatient clonal differences and evidence for parallel concurrent and metachronous metastatic disseminations. The data also support a varied timing of metastatic spread relative to primary tumor development, with progressive mutational acquisition during dormancy and adaptation at distant organ sites. The distribution of driver mutations and the evolution of dominant clones as potential therapeutic targets warrant consideration for sequential sampling of ACC patients in the context of clinical trials (22,23,32,33,40).
Funding
The study is supported in part by the National Institutes of Health (NIH) National Institute of Dental and Craniofacial Research and the NIH Office of Rare Diseases Research grant No. U01DE019765, the SGTB (Salivary Gland Biorepository) grant No. HHSN268200900039C04, the Center for Genetics and Genomics, a National Cancer Institute CA-16672 grant, and the Adenoid Cystic Carcinoma Foundation. This study further supported by the Cancer Prevention and Research Institute of Texas (R120501), the University of Texas Systems Stars Award (PS100149), the Welch Foundation Robert A. Welch Distinguished University Chair Award (G-0040), and the Kenneth D. Muller professorship (AEN).
Notes
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health. The funders had no role in study design, data collection, or analysis, decision to publish, or preparation of the manuscript.
The authors declare no competing financial interests.
The authors are grateful to Ms. Cynthia R. Mandel, Ms. Sheila Fontenot, and Ms. Ebonie Hatfield for transcribing and administrative assistance and Ms. Tamara Locke for her editorial review.
AKEN, GL, BL, YM, and PAF cowrote the manuscript. BL and PAF conducted the data analyses. XR, JianjZ, JianhZ, and YM helped with the data analysis and validation. MZ collected the samples. AKEN, BL, PAF, YM, and GL designed the study.
Supplementary Material
References
- 1. Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;36610:883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ho AS, Kannan K, Roy DM, et al. The mutational landscape of adenoid cystic carcinoma. Nat Genet. 2013;457:791–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lawrence MS, Stojanov P, Polak P, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;4997457:21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Batsakis JG, Regezi JA, Luna MA, et al. Histogenesis of salivary gland neoplasms: A postulate with prognostic implications. J Laryngol Otol. 1989;10310:939–944. [DOI] [PubMed] [Google Scholar]
- 5. Gurney TA, Eisele DW, Weinberg V, et al. Adenoid cystic carcinoma of the major salivary glands treated with surgery and radiation. Laryngoscope. 2005;1157:1278–1282. [DOI] [PubMed] [Google Scholar]
- 6. Marcinow A, Ozer E, Teknos T, et al. Clinicopathologic predictors of recurrence and overall survival in adenoid cystic carcinoma of the head and neck: A single institutional experience at a tertiary care center. Head Neck. 2014;3612:1705–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Laurie SA, Ho AL, Fury MG, et al. Systemic therapy in the management of metastatic or locally recurrent adenoid cystic carcinoma of the salivary glands: A systematic review. Lancet Oncol. 2011;128:815–824. [DOI] [PubMed] [Google Scholar]
- 8. Mitani Y, Li J, Rao PH, et al. Comprehensive analysis of the MYB-NFIB gene fusion in salivary adenoid cystic carcinoma: Incidence, variability, and clinicopathologic significance. Clin Cancer Res. 2010;1619:4722–4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mitani Y, Rao PH, Futreal PA, et al. Novel chromosomal rearrangements and break points at the t(6;9) in salivary adenoid cystic carcinoma: Association with MYB-NFIB chimeric fusion, MYB expression, and clinical outcome. Clin Cancer Res. 2011;1722:7003–7014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Persson M, Andren Y, Mark J, et al. Recurrent fusion of MYB and NFIB transcription factor genes in carcinomas of the breast and head and neck. Proc Natl Acad Sci U S A. 2009;10644:18740–18744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rao PH, Roberts D, Zhao YJ, et al. Deletion of 1p32-p36 is the most frequent genetic change and poor prognostic marker in adenoid cystic carcinoma of the salivary glands. Clin Cancer Res. 2008;1416:5181–5187. [DOI] [PubMed] [Google Scholar]
- 12. Stenman G, Sandros J, Mark J, et al. Partial 6q deletion in a human salivary gland adenocarcinoma. Cancer Genet Cytogenet. 1989;392:153–156. [DOI] [PubMed] [Google Scholar]
- 13. Zhang L, Mitani Y, Caulin C, et al. Detailed genome-wide SNP analysis of major salivary carcinomas localizes subtype-specific chromosome sites and oncogenes of potential clinical significance. Am J Pathol. 2013;1826:2048–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stephens PJ, Davies HR, Mitani Y, et al. Whole exome sequencing of adenoid cystic carcinoma. J Clin Invest. 2013;1237:2965–2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. O'Neil J, Calvo J, McKenna K, et al. Activating Notch1 mutations in mouse models of T-ALL. Blood. 2006;1072:781–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ranganathan P, Weaver KL, Capobianco AJ.. Notch signalling in solid tumours: A little bit of everything but not all the time. Nat Rev Cancer. 2011;115:338–351. [DOI] [PubMed] [Google Scholar]
- 17. Sirakov M, Boussouar A, Kress E, et al. The thyroid hormone nuclear receptor TRalpha1 controls the Notch signaling pathway and cell fate in murine intestine. Development. 2015;14216:2764–2774. [DOI] [PubMed] [Google Scholar]
- 18. Stoecklein NH, Klein CA.. Genetic disparity between primary tumours, disseminated tumour cells, and manifest metastasis. Int J Cancer. 2010;1263:589–598. [DOI] [PubMed] [Google Scholar]
- 19. Sottoriva A, Kang H, Ma Z, et al. A Big Bang model of human colorectal tumor growth. Nat Genet. 2015;473:209–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Maley CC, Galipeau PC, Finley JC, et al. Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nat Genet. 2006;384:468–473. [DOI] [PubMed] [Google Scholar]
- 21. Zhang XC, Xu C, Mitchell RM, et al. Tumor evolution and intratumor heterogeneity of an oropharyngeal squamous cell carcinoma revealed by whole-genome sequencing. Neoplasia. 2013;1512:1371–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lloyd MC, Cunningham JJ, Bui MM, et al. Darwinian dynamics of intratumoral heterogeneity: Not solely random mutations but also variable environmental selection forces. Cancer Res. 2016;7611:3136–3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hong WS, Shpak M, Townsend JP.. Inferring the origin of metastases from cancer phylogenies. Cancer Res. 2015;7519:4021–4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yachida S, Jones S, Bozic I, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;4677319:1114–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brastianos PK, Carter SL, Santagata S, et al. Genomic characterization of brain metastases reveals branched evolution and potential therapeutic targets. Cancer Discov. 2015;511:1164–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tao Y, Ruan J, Yeh SH, et al. Rapid growth of a hepatocellular carcinoma and the driving mutations revealed by cell-population genetic analysis of whole-genome data. Proc Natl Acad Sci U S A. 2011;10829:12042–12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stoeck A, Lejnine S, Truong A, et al. Discovery of biomarkers predictive of GSI response in triple-negative breast cancer and adenoid cystic carcinoma. Cancer Discov. 2014;410:1154–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wallstrom G, Anderson KS, LaBaer J.. Biomarker discovery for heterogeneous diseases. Cancer Epidemiol Biomarkers Prev. 2013;225:747–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Taylor BS, Barretina J, Maki RG, et al. Advances in sarcoma genomics and new therapeutic targets. Nat Rev Cancer. 2011;118:541–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jamal-Hanjani M, Quezada SA, Larkin J, et al. Translational implications of tumor heterogeneity. Clin Cancer Res. 2015;216:1258–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mertens F, Johansson B, Fioretos T, et al. The emerging complexity of gene fusions in cancer. Nat Rev Cancer. 2015;156:371–381. [DOI] [PubMed] [Google Scholar]
- 32. Lupinetti AD, Roberts DB, Williams MD, et al. Sinonasal adenoid cystic carcinoma: The M. D. Anderson Cancer Center experience. Cancer. 2007;11012:2726–2731. [DOI] [PubMed] [Google Scholar]
- 33. Hensley CT, Faubert B, Yuan Q, et al. Metabolic heterogeneity in human lung tumors. Cell. 2016;1644:681–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Birkbak NJ, Eklund AC, Li Q, et al. Paradoxical relationship between chromosomal instability and survival outcome in cancer. Cancer Res. 2011;7110:3447–3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ferrarotto R, Mitani Y, Diao L, et al. Activating NOTCH1 mutations define a distinct subgroup of patients with adenoid cystic carcinoma who have poor prognosis, propensity to bone and liver metastasis, and potential responsiveness to Notch1 inhibitors. J Clin Oncol. 2017;353:352–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Blake PW, Toro JR.. Update of cylindromatosis gene (CYLD) mutations in Brooke-Spiegler syndrome: Novel insights into the role of deubiquitination in cell signaling. Hum Mutat. 2009;307:1025–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fusco N, Colombo PE, Martelotto LG, et al. Resolving quandaries: Basaloid adenoid cystic carcinoma or breast cylindroma? The role of massively parallel sequencing. Histopathology. 2016;682:262–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fletcher MN, Castro MA, Wang X, et al. Master regulators of FGFR2 signalling and breast cancer risk. Nat Commun. 2013;4:2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stratton MR, Rahman N.. The emerging landscape of breast cancer susceptibility. Nat Genet. 2008;401:17–22. [DOI] [PubMed] [Google Scholar]
- 40. Greenman CD, Pleasance ED, Newman S, et al. Estimation of rearrangement phylogeny for cancer genomes. Genome Res. 2012;222:346–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mitani Y, Liu B, Rao PH, et al. Novel MYBL1 gene rearrangements with recurrent MYBL1-NFIB fusions in salivary adenoid cystic carcinomas lacking t(6;9) translocations. Clin Cancer Res. 2016;223:725–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.