Abstract
Spasmodic dysphonia (SD), or laryngeal dystonia, is an isolated task-specific dystonia of unknown causes and pathophysiology that selectively affects speech production. Using next-generation whole-exome sequencing in SD patients, we computed polygenic risk score from 1804 genetic markers based on a genome-wide association study in another form of similar task-specific focal dystonia, musician's dystonia. We further examined the associations between the polygenic risk score, resting-state functional connectivity abnormalities within the sensorimotor network, and SD clinical characteristics. We found that the polygenic risk of dystonia was significantly associated with decreased functional connectivity in the left premotor/primary sensorimotor and inferior parietal cortices in SD patients. Reduced connectivity of the inferior parietal cortex was correlated with the age of SD onset. The polygenic risk score contained a significant number of genetic variants lying near genes related to synaptic transmission and neural development. Our study identified a polygenic contribution to the overall genetic risk of dystonia in the cohort of SD patients. Associations between the polygenic risk and reduced functional connectivity of the sensorimotor and inferior parietal cortices likely represent an endophenotypic imaging marker of SD, while genes involved in synaptic transmission and neuron development may be linked to the molecular pathophysiology of this disorder.
Keywords: dystonia, genetic risk, imaging marker
Introduction
Spasmodic dysphonia (SD) is an isolated task-specific focal dystonia, selectively affecting speech production due to involuntary spasms in the laryngeal muscles. The family history of dystonia in up to 12% of SD patients (Blitzer et al. 1998; Kirke et al. 2015) suggests the existence of genetic risk factors, whereas the presence of structural and functional abnormalities extending beyond the basal ganglia and additionally involving the sensorimotor cortex, thalamus, and cerebellum point to neural network alterations in this disorder (Simonyan and Ludlow 2010; Battistella et al. 2015, 2016). However, exact causative pathophysiology of SD and other isolated focal dystonias remains unclear, due, in part, to low penetrance, clinical heterogeneity, and low incidence of dystonia, all of which have limited an access to the DNA samples from a large homogeneous patient cohort for the conduct of traditional genome-wide association studies (GWAS). To date, only 2 smaller-scale GWAS were performed in isolated dystonia, including patients with musician's dystonia (Lohmann et al. 2014) and cervical dystonia (Mok et al. 2014), although the findings from the latter study failed a follow-up validation (Gomez-Garre et al. 2014). A general limitation of these studies is that individual single nucleotide polymorphisms (SNPs) identified by GWAS did not capture the overall genomic or polygenic architecture of the disorder due to a very small amount of phenotypic variation explained by each SNP and a very large number of SNPs underlying the disorder risk (Purcell et al. 2009). The polygenic risk analysis, on the other hand, allows for the utilization of the predictive power of GWAS to be applied robustly in small cohorts (Purcell et al. 2009; Dudbridge 2013) in order to model the aggregate effect of alleles associated with a disorder in each individual (Dima and Breen 2015). In a number of psychiatric disorders, the polygenic risk has been further examined for its relationship with brain function and structure (Holmes et al. 2012; Sabuncu et al. 2012; Whalley et al. 2013; Lancaster et al. 2016), pointing to new avenues for personalized risk prediction and interventions (Mrazek and Lerman 2011; Tansey et al. 2012; Chatterjee et al. 2013).
In this study, we leveraged prior knowledge of available GWAS in musician's dystonia (Lohmann et al. 2014) to identify the polygenic risk of SD. We computed the polygenic risk scores in SD patients based on a GWAS in musician's dystonia (Lohmann et al. 2014) because these GWAS findings were replicated in a recent study (Nibbeling et al. 2015) and because musician's dystonia and SD share similar phenotypical features of task specificity as well as similar neural abnormalities (Granert et al. 2011; Neychev et al. 2011; Zoons et al. 2011; Ramdhani et al. 2014). In addition, we examined associations between the polygenic risk, neural network alterations, and SD clinical characteristics (i.e., disorder severity and onset) in order to identify potential imaging markers that may point to SD causative pathophysiology. We used independent component analysis (ICA) of low frequency physiological fluctuations in the resting-state blood-oxygen-level-dependent (BOLD) signal (Biswal et al. 1995) to determine brain regions whose functional connectivity within the sensorimotor network is abnormal in SD patients compared with healthy controls. Clinical characteristics of SD were derived from the review of disorder history and blinded analysis of audio-recorded voice and speech samples. Finally, we conducted a gene enrichment analysis (Huang et al. 2009) in order to determine the genetic content of the polygenic risk score that showed a significant association with the regions of abnormal functional connectivity in SD patients. Collectively, this approach allowed us to examine the relationship between the brain, genes and symptoms in SD as well as the long-envisioned genetic overlap between different forms of isolated focal dystonia (Fuchs and Ozelius 2013; Klein 2014).
Our overarching hypothesis was that the polygenic risk will be associated with brain regions of altered functional connectivity that, in turn, correlates with clinical features of SD. Specifically, because SD is a task-specific dystonia selectively affecting speech production, which is a highly learned and unique human behavior requiring complex orchestration between various sensorimotor brain regions and networks (Fuertinger et al. 2015; Simonyan and Fuertinger 2015), we hypothesized that significant relationships between the polygenic risk and brain regions of altered functional connectivity would be found within the cortical rather than subcortical regions. Based on our recent study that identified reduced functional connectivity of the sensorimotor and inferior parietal cortices to represent a potential imaging marker classifying SD from a healthy state (Battistella et al. 2016), we hypothesized that the polygenic score will also show associations with network alterations in these cortical regions.
Materials
Study Participants
A total of 57 SD patients participated in the whole-exome sequencing and resting-state functional MRI (rs-fMRI) studies (11 males/46 females, age 55.9 ± 12.0 years). The patient group included 32 subjects with the adductor form of SD (ADSD) and 25 subjects with the abductor form of SD (ABSD). Of all, 17 patients (13 ADSD and 4 ABSD) had a family history of SD and/or other isolated dystonias. None of the patients were blood relatives; only one affected member per SD family (the proband) participated in the study. In addition, 30 healthy volunteers (12 males/18 females, age 49.7 ± 9.5 years) participated in the rs-fMRI study only to contrast SD-specific functional network abnormalities. Our choice of rs-fMRI was based on prior studies showing that resting-state networks reflect the organization of task-related functional brain networks (Smith et al. 2009) and are abnormal in SD and across different forms of focal dystonia (Battistella et al. 2015, 2016). There was no significant difference in age between SD patients and healthy controls (uncorrected P = 0.07); however, these groups differed based on their gender (uncorrected P = 0.04), which was potentially due to the typical overrepresentation of females vs. males (ratio 4:1) as a clinical trait of SD (Blitzer et al. 1998; Schweinfurth et al. 2002; Ludlow et al. 2008; Kirke et al. 2015) as well as a larger number of recruited SD patients compared with healthy controls, which was necessary for the computation of the polygenic score.
All subjects were right-handed and native English speakers. All participants were Caucasian Europeans to match the cohort used in the musician's dystonia GWAS (Lohmann et al. 2014). Subjects were recruited for the study only if they had no past or present history of any neurological (other than isolated focal SD in patients), psychiatric, or laryngological problems as assessed by medical history, physical, neurological examinations, and fiberoptic nasolaryngoscopy. None of the subjects were on any medications that affected the central nervous system. None of the subjects were carriers of TOR1A/DYT1, THAP1/DYT6, TUBB4A/DYT4, or GNAL/DYT25 dystonia mutations as confirmed by our genetic screening. These stringent exclusion criteria were followed throughout the study to preserve the SD and healthy control cohorts as homogeneous and free of other CNS disorders and influences as possible, the presence of which may have confounded both imaging and genetic findings.
Diagnosis of SD was established using flexible fiberoptic nasolaryngoscopy. ADSD patients were characterized by voice breaks on the production of vowels and strained, strangled quality of voice; ABSD patients had voice breaks on the production of voiceless consonants and breathy, effortful quality of voice. Based on detailed neurological and laryngological examinations, SD patients did not have dystonia affecting any other body regions, and none had a functional (psychogenic) form of SD or other dystonias. Based on patients’ medical history, duration of SD was 15.4 ± 10.5 years with an onset of disorder at 39.8 ± 13.5 years. All patients were fully symptomatic at the time of study participation. Patients, who received botulinum toxin injections, participated in the study at least 3 months after their last injection, when fully symptomatic.
All subjects provided written informed consent prior to the study participation, which was approved by the Institutional Review Board of the Icahn School of Medicine at Mount Sinai. Some of healthy volunteers and SD patients participated in another study around the same time, which involved the identification of brain abnormalities between different SD phenotypes (Battistella et al. 2016).
Whole-Exome Sequencing and Processing
Using the Purgene procedure (Gentra Systems), genomic DNA was extracted from patients’ blood samples. Exome capture and sequencing were performed by PerkinElmer using the Agilent SureSelect V4+UTR library and an Illumina HiSeq 2000 paired-end module. The Burrows-Wheeler Aligner (BWA) (Li and Durbin 2009) was used to map reads to the reference sequence GRCh37/hg19, after which variant calling was performed using GATK v.3.3 (McKenna et al. 2010) and the HaplotypeCaller method. Both programs were run on a standard exome data processing pipeline described earlier (Linderman et al. 2014), which included read mapping, duplicate read removal, local realignment, base quality recalibration, and variant calling. The resulting Variant Call Format (VCF) files were converted to PLINK format using VCFtools. Our quality checks were based on the quality control standards set by the ENIGMA consortium (ENIGMA 2012), ensuring that the samples included in the imputation and final analysis have fewer than 5% missing genotypes. The mean depth of coverage of all 57 exomes was 72.6 with 81.8% of exomes covered at > ×20.
Polygenic Risk Scores
SD polygenic risk score, which aggregates multiple genetic variants into a single indicator of risk, was generated based on the P values and odd ratios for all markers of GWAS in musician's dystonia (Lohmann et al. 2014). Because the genetic architecture of SD is unknown, we defined a range of 8 different polygenic risk scores using 8 different threshold values, namely Pthresh = 1.0 × 10−i or 5.0 × 10−i where i = 1, 2, 3, or 4. In the first step, we included all markers available from the GWAS study (Lohmann et al. 2014), which were associated with Pthresh ranging from 1.0 × 10−4 to 0.5. Next, these variants were filtered to match the variants imputed with high confidence (r2 ≥ 0.3) from our SD exome sequences and then pruned so that no 2 variants are in significant linkage disequilibrium (LD) with each other. Based on these procedures, the polygenic score was calculated for each patient as the sum of the number of risk alleles at each of the markers, weighted by the logarithm of the odds ratios of those markers as determined from GWAS data. Consistent with an earlier study showing that a significant fraction of genetic variants may be imputed from exomes (Pasaniuc et al. 2012), we reliably imputed 3373 of the 6752 markers having uncorrected P ≤ 0.01 in the musician's dystonia GWAS (Lohmann et al. 2014). These markers were further LD-pruned to generate a list of 1804 markers contributing to the polygenic score associated with Pthresh = 0.01 (Supplementary Table). Among these 1804 markers, 120 markers were directly genotyped, and the remaining markers were imputed.
fMRI Acquisition and Analysis
All subjects were scanned on a 3 T Philips scanner equipped with an 8-channel head coil. Whole-brain rs-fMRI included acquisition of single-shot echo-planar-imaging gradient echo sequence with repetition time (TR) 2000 ms, voxel size 3 × 3 mm2, and 150 volumes. A high-resolution T1-weighted 3D gradient-echo sequence (MPRAGE) was acquired with 1 mm isotropic voxel as an anatomical reference. During the scanning, all subjects were instructed to remain still with their eyes closed, think of nothing in particular, and avoid sleeping. Head movements during scanning were minimized by cushioning the participant's head in the coil; all subjects were monitored for any movements while in the scanner.
Imaging data were analyzed as described earlier (Battistella et al. 2016). Briefly, following standard image preprocessing, 4D time series were regressed using the 6 motion parameters calculated during realignment as well as the white matter and cerebrospinal fluid mean signals, normalized to the AFNI standard Talairach-Tournoux space, and smoothed with an isotropic Gaussian kernel of 5 mm. Mean ± standard deviation root mean square motion values were 0.16 ± 0.08 in healthy controls and 0.17 ± 0.08 in SD patients, placing all subjects into the low-motion category (Oakes et al. 2005). All preprocessed time series across all subjects were concatenated and decomposed using the temporal concatenation approach implemented in the MELODIC tool of FSL software. This resulted in group-averaged spatial independent components (IC) common to both healthy and patient groups.
Based on our a priori hypothesis, we examined differences in functional connectivity between SD patients and healthy groups within the sensorimotor network only, which has been reported to be responsible for action execution and perception (Smith et al. 2009). For this, the sensorimotor IC from the group-average analysis was used to generate a subject-specific version of the spatial map and associated timeseries using dual regression (Filippini et al. 2009). First, for each subject, the group-averaged sensorimotor IC was regressed (as spatial regressors in a multiple regression) into the subject's 4D residual dataset, which resulted in a subject-specific timeseries. Next, those timeseries were regressed (as temporal regressors, again in a multiple regression) into the same 4D dataset, resulting in a subject-specific spatial map. Using AFNI software, the follow-up between-group analysis of SD patients and healthy controls was performed voxelwise using a two-sample independent t-test on the individual Z-score maps derived from dual regression; subject's age and gender were included as covariates of no interest. The significance level was set at a family-wise error (FWE)-corrected P ≤ 0.05. To ensure that our final results were not influenced by potential gender differences between the groups, we conduced an additional analysis in a subset of tightly age- and gender-matched subjects (23 SD patients vs. 23 healthy controls) using the same statistical approach but without age and gender as covariates (see Supplementary Material). This analysis confirmed the findings of our main study, suggesting a negligible, if any, effect of gender on between-group differences in functional connectivity.
We defined brain regions as abnormal based on statistically significant differences in their functional connectivity between SD patients and healthy controls. Mean Z-scores from these brain regions were extracted and used to calculate Pearson's correlation matrices at r ≥ 0.4 and within-group FWE-corrected P ≤ 0.05 for visualization of functional connectivity profiles in each group, separately.
Statistical Analysis
In SD patients, we tested a statistically significant dependence of every brain region that showed altered functional connectivity within the sensorimotor network (i.e., mean Z-score values extracted from the clusters showing significant between-group differences) on each of the polygenic scores using multiple linear regressions with 2 genetic principle components calculated from the imputed genotype data using genome-wide complex trait analysis (GCTA) (Yang et al. 2013) and 2 covariates of no interest for age and gender. The resulting P-values were corrected for multiple comparisons (conducted over the different polygenic risk scores) by permutation, in which patients’ genetic data (polygenic scores and genetic principle components) were permuted with respect to their nongenetic data (imaging abnormalities, age, and gender) (Lindquist and Mejia 2015). These permutations yielded a null distribution of datasets, in which the relationship between genetic and imaging data was randomized, yet the correlations among genetic quantities were preserved. Multiple linear regression was performed on each permuted dataset, such as for each region of imaging abnormalities, a null P value was defined as the minimum regression P over the 8 polygenic scores. For a given pair of the abnormal brain region and polygenic score, the permutation-corrected P value was calculated based on 10 800 permutations as the fraction of null P values less than the original uncorrected P value. The significance of associations between polygenic risk and the imaging abnormalities was set at a permutation-corrected P ≤ 0.05.
To examine the genetic content of the significant polygenic risk score that correlated with brain regions of abnormal functional connectivity in SD patients compared with healthy controls, we used the Gene-Enrichment and Functional Annotation Analysis tool of the Database for Annotation, Visualization and Integrated Discovery (DAVID) (version 6.7, accessed 24 June 2015) (Huang et al. 2009) to compute the annotation enrichment of the 1292 genes lying within 10 kbp of the 1804 markers of the polygenic score at a Benjamini–Hochberg false discovery rate (FDR)-corrected P ≤ 0.05.
As a final step, the brain regions of the most significant correlation with polygenic scores were examined for their relationships with SD clinical features, including the age of onset (39.8 ± 13.5 years) and severity of disorder (3.1 ± 1.6 dystonic voice breaks/sentence), using multiple regression analyses at a Benjamini–Hochberg FDR-corrected P ≤ 0.05. In each patients, SD symptom severity was assessed based on the audio recordings of production of 40 ADSD and ABSD symptom-eliciting sentences, which were blindly rated by an experienced speech-language pathologist to quantify the number of SD-characteristic voice breaks per sentence, as described earlier (Kirke et al. 2016). Patients’ age at SD onset was established based on their medical history.
Results
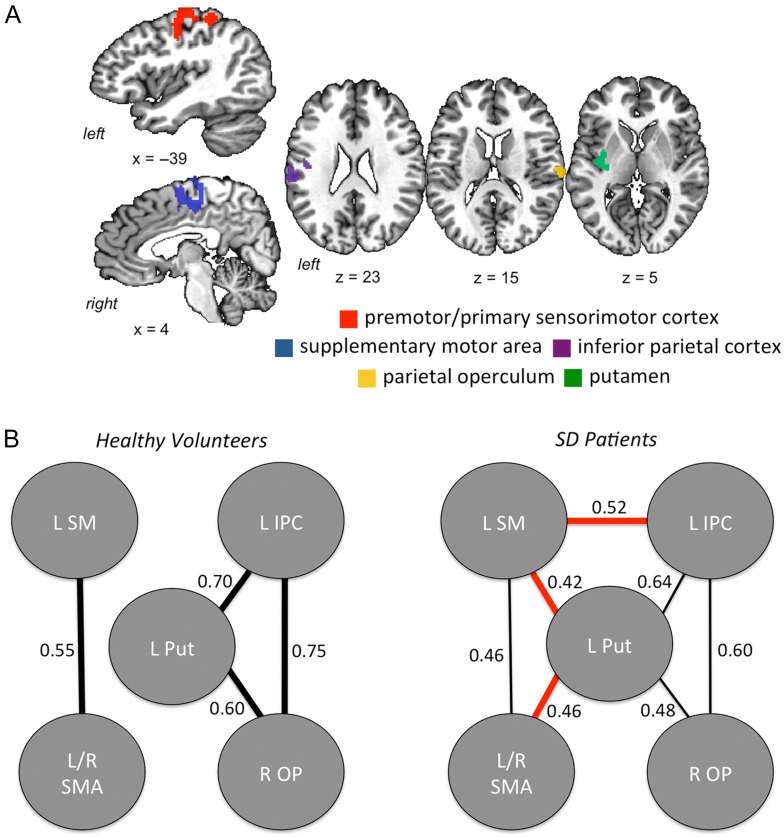
Compared with healthy controls, SD patients showed significant differences in functional connectivity within the sensorimotor network, involving left premotor/primary sensorimotor cortex, inferior parietal cortex, putamen, bilateral SMA, and right parietal operculum at FWE-corrected P ≤ 0.05 (Fig. 1A). When examining within-group connectivity profiles between these abnormal sensorimotor brain regions, we found that functional connectivity in SD patients was reduced between the left premotor/primary sensorimotor cortex and bilateral SMA as well as between the left inferior parietal cortex, putamen and right parietal operculum (Fig. 1B). Concurrently, SD patients showed enhanced functional connectivity of the left putamen with left premotor/primary sensorimotor cortex and bilateral SMA as well as between the left premotor/primary sensorimotor cortex and left inferior parietal cortex (Fig. 1B).
Figure 1.
(A) A map of significant clusters derived from between-group analysis of differences in resting-state functional connectivity within the sensorimotor network in SD patients and healthy controls at t ≥ 2.7 and FWE-corrected P ≤ 0.05. The mean Z score measures from these clusters were used in regression analysis with polygenic risk scores. (B) Visualization of functional connectivity profiles (r ≥ 0.4) within the abnormal network in SD patients and healthy controls at FWE-corrected P ≤ 0.05. In SD patients, finer lines show weaker correlations between the regions compared with healthy controls; red lines show functional connections in SD patients, which are not present in healthy controls at FWE-corrected P ≤ 0.05. LSM, left premotor primary sensorimotor cortex; LIPC, left inferior parietal cortex; L Put, left putamen; L/R SMA, left/right supplementary motor area; ROP, right operculum.
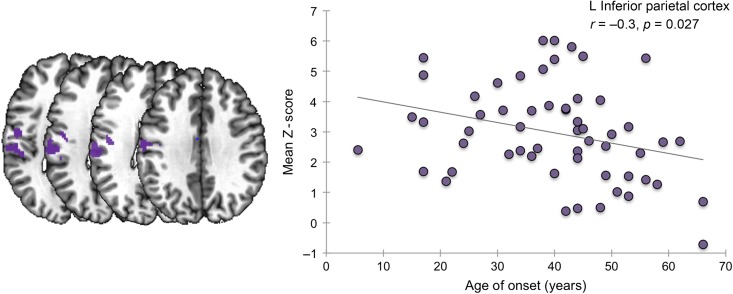
The polygenic risk scores were significantly correlated with imaging abnormalities in the left premotor/primary sensorimotor cortex (corrected P = 0.007) and left inferior parietal cortex (permutation-corrected P ≤ 0.03), with the strongest correlations found at P = 0.01 threshold of the polygenic score (Table 1). All significant correlations between network abnormalities and polygenic scores were negative (t ≤ −2.6, permutation-corrected P ≤ 0.03), indicating that connection values decreased as the polygenic scores increased (Table 1). In addition, alterations of functional connectivity in the left inferior parietal cortex, which had the strongest correlation with the polygenic score, were inversely related to the age of SD onset (r = −0.3, Benjamini–Hochberg FDR-corrected P = 0.027) (Fig. 2), indicating greater decrease of inferior parietal functional connectivity at the later age of disorder onset. No statistically significant relationships were found between functional alterations and SD severity (all Benjamini–Hochberg FDR-corrected P ≥ 0.27).
Table 1.
Regression between alterations in functional connectivity and polygenic scores
| Regions of abnormal functional connectivity (activation peak inx, y, z) | Left premotor/ sensorimotor cortex (−22, −32, 63) | Left/right supplementary motor area (2, −4, 57) | Left inferior parietal cortex (−60, −26, 23) | Right parietal operculum (62, −18, 15) | Left putamen (−30, −6, 7) | |
|---|---|---|---|---|---|---|
| Regression P-value | ||||||
| P-value threshold of polygenic score | P = 1.0 × 10−4 | 0.28 | 1.00 | 0.03* (t = −2.9) | 0.18 | 0.23 |
| P = 5.0 × 10−4 | 0.18 | 0.99 | 0.06 | 0.13 | 0.77 | |
| P = 1.0 × 10−3 | 0.05 | 1.00 | 0.05 | 0.35 | 0.72 | |
| P = 5.0 × 10−3 | 0.24 | 1.00 | 0.02* (t = −3.1) | 0.16 | 0.29 | |
| P = 1.0 × 10−2 | 0.007* (t = −2.6) | 0.83 | 0.001* (t = −3.93) | 0.52 | 0.12 | |
| P = 5.0 × 10−2 | 0.1 | 0.39 | 0.11 | 0.68 | 0.49 | |
| P = 1.0 × 10−1 | 0.14 | 0.16 | 0.16 | 0.74 | 0.89 | |
| P = 5.0 × 10−1 | 0.56 | 0.31 | 0.63 | 0.98 | 0.95 | |
Regression P-values from multiple linear regression of functional connectivity alterations versus polygenic scores, corrected using 10 800 permutations. Asterisk (*) indicates significant relationship at a permutation-corrected P ≤ 0.05. Coordinates of activation peaks are given in the standard Talairach-Tournoux space at an FWE-corrected P ≤ 0.05.
Figure 2.
A negative relationship between the mean Z score in the left inferior parietal cortex and the age at SD onset (in years). The abnormal cluster within the inferior parietal cortex is shown on the left panel.
Examination of the genetic content of the significant polygenic risk score (1292 genes lying within 10 kbp of the 1804 markers) that correlated with abnormal regions of functional connectivity between SD patients and healthy controls showed enrichment in Gene Ontology (GO) cellular components [49 genes associated with synapses (GO:0045202, Benjamini–Hochberg FDR-corrected P = ×7.910−6) and 42 genes associated with neuron projections (GO:0043005, Benjamini–Hochberg FDR-corrected P = ×3.010−4)] and biological processes (36 genes associated with synaptic transmission [GO:0007268, Benjamini–Hochberg FDR-corrected P = ×8.010−3] and 39 genes associated with neuronal development [GO:0048666, Benjamini–Hochberg FDR-corrected P = ×8.910−3]) (Table 2).
Table 2.
DAVID function annotation cluster analysis
| Category | Term | Count | P-value | Benjamini P-value |
|---|---|---|---|---|
| Annotation Cluster 2 | Enrichment Score: 4.26 | |||
| GOTERM_CC_FAT | GO:0045202 Synapse | 49 | 1.6 × 10−8 | 7.9a × 10−6 |
| GO:0030054 Cell junction | 58 | 1.0 × 10−6 | 1.7 × 10−4 | |
| GO:0044456 Synapse part | 34 | 3.4 × 10−6 | 2.8 × 10−4 | |
| GO:0045211: Postsynaptic membrane | 18 | 1.6 × 10−3 | 2.8 × 10−2 | |
| GO:0005230: Extracellular ligand-gated ion channel activity | 11 | 7.2 × 10−3 | 2.1 × 10−1 | |
| SP_PIR_KEYWORDS | Cell junction | 44 | 1.4 × 10−5 | 9.6 × 10−4 |
| Synapse | 25 | 5.6 × 10−4 | 1.9 × 10−2 | |
| Postsynaptic cell membrane | 13 | 1.6 × 10−2 | 1.8 × 10−1 | |
| Annotation Cluster 5 | Enrichment Score: 3.45 | |||
| GOTERM_CC_FAT | GO:0043005: Neuron projection | 42 | 4.4 × 10−6 | 3.0a × 10−4 |
| GO:0030425: Dendrite | 22 | 3.6 × 10−4 | 1.1 × 10−2 | |
| GO:0043025: Neuronal cell body | 17 | 2.9 × 10−2 | 2.3 × 10−1 | |
| Annotation Cluster 6 | Enrichment Score: 3.38 | |||
| GOTERM_BP_FAT | GO:0007268: Synaptic transmission | 36 | 1.8 × 10−5 | 8.0a × 10−3 |
| GO:0007267: Cell-cell signaling | 58 | 3.6 × 10−5 | 1.1 × 10−2 | |
| GO:0019226: Transmission of nerve impulse | 37 | 2.2 × 10−4 | 3.2 × 10−2 | |
| GO:0050877: Neurological system process | 74 | 2.1 × 10−1 | 9.1 × 10−1 | |
| Annotation Cluster 9 | Enrichment Score: 3.08 | |||
| GOTERM_BP_FAT | GO:0048666: Neuron development | 39 | 2.3 × 10−5 | 8.9a × 10−3 |
| GO:0030030: Cell projection organization | 40 | 6.4 × 10−5 | 1.8 × 10−2 | |
| GO:0000902: Cell morphogenesis | 39 | 6.8 × 10−5 | 1.7 × 10−2 | |
| GO:0032989: Cellular component morphogenesis | 42 | 7.4 × 10−5 | 1.8 × 10−2 | |
| GO:0030182: Neuron differentiation | 44 | 1.6 × 10−4 | 2.9 × 10−2 | |
| GO:0000904: Cell morphogenesis involved in differentiation | 28 | 4.2 × 10−4 | 5.5 × 10−2 | |
| GO:0048858: Cell projection morphogenesis | 26 | 2.1 × 10−3 | 1.5 × 10−1 | |
| GO:0048812: Neuron projection morphogenesis | 23 | 3.3 × 10−3 | 2.0 × 10−1 | |
| GO:0032990: Cell part morphogenesis | 26 | 3.8 × 10−3 | 2.1 × 10−1 | |
| GO:0031175: Neuron projection development | 25 | 7.5 × 10−3 | 2.9 × 10−1 | |
| GO:0007409: Axonogenesis | 20 | 1.0 × 10−2 | 3.3 × 10−1 | |
| GO:0007411: Axon guidance | 11 | 7.0 × 10−2 | 7.1 × 10−1 | |
From the total of 304 annotation clusters, 4 clusters showed significant enrichment in at least one of the components at a Benjamini–Hochberg FDR-corrected P ≤ 0.05.
aIndicates significant enrichment in the Gene Ontology (GO) component.
Discussion
In this first imaging genetics study of isolated focal dystonia, we demonstrated a polygenic contribution to the overall heritable risk of this disorder and defined the brain regions, genetic vulnerability of which may lead to SD development. Specifically, we identified a novel genetic relationship with abnormal functional connectivity of the left premotor/primary sensorimotor and inferior parietal cortices, with the latter showing a significant relationship with the SD age of onset. Collectively, our data provide support for polygenic influences on cortical sensorimotor alterations in SD, pointing to these brain regions as potential imaging endophenotypic markers of SD risk prediction.
The premotor and primary sensorimotor cortex, together with the basal ganglia and cerebellum, have been previously implicated in the pathophysiology of SD and other focal dystonias due to the presence of aberrant function during the resting state as well as the production of symptomatic and asymptomatic tasks (Haslinger et al. 2005; Simonyan and Ludlow 2010; Delnooz et al. 2013; Battistella et al. 2015, 2016; Lokkegaard et al. 2016). Abnormalities in these regions have been further associated with underlying alterations in the gray matter volume, cortical thickness (Simonyan and Ludlow 2012) and descending white matter pathways (Simonyan et al. 2008) in SD. Similarly, the inferior parietal cortex, which is a key region for sensorimotor processing and integration and is thought to contribute to aberrant proprioceptive input in focal dystonia (Hagura et al. 2009; Fiorio et al. 2011), has been shown to exhibit abnormal cortical thickness in SD, with the magnitude of these changes being correlated with the severity of SD symptoms (Simonyan and Ludlow 2012). In this study, we found that altered functional connectivity of the sensorimotor and inferior parietal cortices in association with the polygenic risk of SD was accompanied by a reorganization of the regional connectivity profiles in SD patients compared with healthy controls potentially to preserve the overall functional network architecture. Future computational studies modeling various interactions between these brain regions may shed light on mechanistic aspects of functional network damage in this disorder.
Despite mapping structural and functional brain alterations across different forms of focal dystonia, our understanding of whether these alterations are primary or compensatory in dystonia pathophysiology remains unclear (Neychev et al. 2011; Zoons et al. 2011; Ramdhani and Simonyan 2013; Lokkegaard et al. 2016), partly, due to difficulties in disambiguating primary from compensatory brain imaging changes (Bandettini 2009). Our current findings of significant relationships of sensorimotor and inferior parietal alterations with the polygenic risk as well as the association between reduced connectivity of the inferior parietal cortex and the age of SD onset suggest that these regions may have a primary contribution to the pathophysiology of SD. In line with this, our recent study identified that the alterations in the same brain regions have sufficient power to discriminate between the SD and healthy states (Battistella et al. 2016). Taken together, we suggest that alterations in the primary sensorimotor and inferior parietal cortices may be primary contributors to SD pathophysiology, representing potential markers for SD diagnosis and risk prediction.
Clinical and biological importance of our findings is further supported by the enrichment of genes related to synaptic transmission and neuronal development near the markers contributing to the significant polygenic score. In DYT1 and DYT11 dystonias, the causative genes were found to be highly expressed during early brain development (Breakefield et al. 2008), consistent with a view of dystonia as a neurodevelopmental disorder (Niethammer et al. 2011). The enrichment of genes related to synaptic transmission is also in line with a broad involvement of abnormal dopaminergic, GABAergic, glutamatergic, and cholinergic neurotransmission in the pathophysiology of dystonia (Breakefield et al. 2008). Altered dopaminergic and GABAergic neurotransmission has been reported in different forms of dystonia (Levy and Hallett 2002; Garibotto et al. 2011; Berman et al. 2013; Karimi and Perlmutter 2015), including SD, where striatal dopaminergic dysfunction was found both at rest and during production of symptomatic and asymptomatic tasks (Simonyan et al. 2013). Evidence for altered cholinergic transmission in dystonia comes from patients with DYT1 dystonia who respond positively to anticholinergic medications (Burke et al. 1986), whereas a mouse model with a Dyt1 deletion shows cholinergic dysregulation in the striatum (Sciamanna et al. 2012). These findings suggest that the functional enrichment of synaptic transmission and neuron development-related genes in SD patients observed in this study is unlikely due to significant LD with the genetic variants causing the disorder. Instead, the genetic variation captured by the polygenic risk score and encompassing the genes related to these biological processes may be directly relevant to the causative pathophysiology of SD.
In broader terms of dystonia pathophysiology, the polygenic risk scores generated from the SD patients’ genotypes at more than 1800 loci were derived from the results of the GWAS in musician's dystonia (Lohmann et al. 2014). As such, our findings not only provided correlates of imaging genetics of SD, but also demonstrated an overlap between the genetic etiopathophysiology of different forms of focal dystonia, which show similar clinical phenomenology of task specificity as well as similar functional brain alterations in neuroimaging studies (Pujol et al. 2000; Haslinger et al. 2010; Neychev et al. 2011; Simonyan and Ludlow 2012; Chang and Frucht 2013; Ramdhani et al. 2014).
From a methodological perspective, the use of existing smaller-sample GWAS data in musician's dystonia (Lohmann et al. 2014) may have limited the identification of a more robust genetic variation in SD. However, there are no currently available large-sample (in order of 1000+ subjects) GWAS in dystonia, partly due to the rarity of this disorder. Nevertheless, our statistically significant findings, even using data from smaller GWAS, highlight the promise of leveraging prior knowledge in order to discover genetic correlates of a rare form of focal dystonia and to identify statistically significant genotype–phenotype relationships within our cohort of SD patients. Moreover, our data provide direct evidence for the long-hypothesized genetic overlap between different forms of isolated focal dystonia. In our case, both musician's dystonia and SD are task-specific forms of focal dystonia, which makes them even more similar compared with other forms of focal dystonia (e.g., blepharospasm or cervical dystonia) (Battistella et al. 2015). Thus, building on the results from a related disorder allowed us to effectively improve the signal-to-noise ratio in a rare SD population, where straightforward association testing of individual genetic variants would likely be underpowered. In a similar fashion, we used pre-existing knowledge in the form of gene annotations to perform gene enrichment analysis, revealing a significant association of nervous system-related genes that did not emerge from single-variant analysis of the GWAS data in musician's dystonia (Lohmann et al. 2014).
In summary, our results bridged the genotype–phenotype gap in SD by showing that alterations of functional connectivity in the sensorimotor and inferior parietal cortices are associated with the polygenic risk and may represent an intermediate endophenotype and primary imaging marker of SD, while genes involved in synaptic transmission and neuron development may be linked to the molecular pathophysiology of this disorder.
Supplementary Material
Funding
We thank the Scientific Computing Department at the Icahn School of Medicine at Mount Sinai (Biomedical Research Support Shared Instrumentation Grant from the National Institutes of Health, Project # 1S10OD018522-01) and the NHLBI GO Exome Sequencing Project and its ongoing studies, which produced and provided exome variant calls for comparison: the Lung GO Sequencing Project (HL-102923), the WHI Sequencing Project (HL-102924), the Broad GO Sequencing Project (HL-102925), the Seattle GO Sequencing Project (HL-102926), and the Heart GO Sequencing Project (HL-103010). Mert R. Sabuncu was supported by NIH NIBIB K25EB013649 grant and a Bright-Focus Alzheimer's disease pilot research grant (AHAF-A2012333). This study was supported by the R01DC01180 grant to K.S. from the National Institute on Deafness and Other Communication Disorders, National Institutes of Health.
Supplementary Material
Notes
We thank Andrew Blitzer, MD, DDS, Steven J. Frucht, MD, Miodrag Velickovic, MD, and Nutan Sharma, MD, PhD, for patient referrals; Amanda Pechman, Heather Alexander, Melissa Choy, and Estee Rubien-Thomas for data acquisition, and Michael Linderman, PhD for help with the next-generation sequencing pipeline. Conflict of Interest: The authors report no conflict of interest.
References
- Bandettini PA. 2009. What's new in neuroimaging methods? Ann N Y Acad Sci. 1156:260–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battistella G, Fuertinger S, Fleysher L, Ozelius LJ, Simonyan K. 2016. Cortical sensorimotor alterations classify clinical phenotype and putative genotype of spasmodic dysphonia. Eur J Neurol. 23:1517–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battistella G, Termsarasab P, Ramdhani RA, Fuertinger S, Simonyan K. 2015. Isolated focal dystonia as a disorder of large-scale functional networks. Cereb Cortex. pii:bhv313 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman BD, Hallett M, Herscovitch P, Simonyan K. 2013. Striatal dopaminergic dysfunction at rest and during task performance in writer's cramp. Brain. 136:3645–3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal B, Yetkin FZ, Haughton VM, Hyde JS. 1995. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn Reson Med. 34:537–541. [DOI] [PubMed] [Google Scholar]
- Blitzer A, Brin MF, Stewart C. 1998. Botulinum toxin management of spasmodic dysphonia (laryngeal dystonia): a 12 year experience in more than 900 patients. Laryngoscope. 108:1435–1441. [DOI] [PubMed] [Google Scholar]
- Breakefield XO, Blood AJ, Li Y, Hallett M, Hanson PI, Standaert DG. 2008. The pathophysiological basis of dystonias. Nat Rev Neurosci. 9:222–234. [DOI] [PubMed] [Google Scholar]
- Burke RE, Fahn S, Marsden CD. 1986. Torsion dystonia: a double-blind, prospective trial of high-dosage trihexyphenidyl. Neurology. 36:160–164. [DOI] [PubMed] [Google Scholar]
- Chang FC, Frucht SJ. 2013. Motor and sensory dysfunction in musician's dystonia. Curr Neuropharmacol. 11:41–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee N, Wheeler B, Sampson J, Hartge P, Chanock SJ, Park JH. 2013. Projecting the performance of risk prediction based on polygenic analyses of genome-wide association studies. Nat Genet. 45:400–405, 405e401–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delnooz CC, Pasman JW, Beckmann CF, van de Warrenburg BP. 2013. Task-free functional MRI in cervical dystonia reveals multi-network changes that partially normalize with botulinum toxin. PLoS One. 8:e62877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dima D, Breen G. 2015. Polygenic risk scores in imaging genetics: usefulness and applications. J Psychopharmacol. 29:867–871. [DOI] [PubMed] [Google Scholar]
- Dudbridge F. 2013. Power and predictive accuracy of polygenic risk scores. PLoS Genetics. 9:e1003348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENIGMA2 Genetics support team ENIGMA2 1KGP cookbook (v3) [Online]. The Enhancing Neuroimaging Genetics through Meta-Analysis (ENIGMA) consortium. http://enigma.loni.ucla.edu/wp-content/uploads/2012/07/ENIGMA2_1KGP_cookbook_v3.doc [27 July 2012].
- Filippini N, MacIntosh BJ, Hough MG, Goodwin GM, Frisoni GB, Smith SM, Matthews PM, Beckmann CF, Mackay CE. 2009. Distinct patterns of brain activity in young carriers of the APOE-epsilon4 allele. Proc Natl Acad Sci USA. 106:7209–7214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorio M, Weise D, Onal-Hartmann C, Zeller D, Tinazzi M, Classen J. 2011. Impairment of the rubber hand illusion in focal hand dystonia. Brain. 134:1428–1437. [DOI] [PubMed] [Google Scholar]
- Fuchs T, Ozelius LJ. 2013. Genetics in dystonia: an update. Curr Neurol Neurosci Rep. 13:410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuertinger S, Horwitz B, Simonyan K. 2015. The functional connectome of speech control. PLoS Biol. 13:e1002209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garibotto V, Romito LM, Elia AE, Soliveri P, Panzacchi A, Carpinelli A, Tinazzi M, Albanese A, Perani D. 2011. In vivo evidence for GABA(A) receptor changes in the sensorimotor system in primary dystonia. Mov Disord. 26:852–857. [DOI] [PubMed] [Google Scholar]
- Gomez-Garre P, Huertas-Fernandez I, Caceres-Redondo MT, Alonso-Canovas A, Bernal-Bernal I, Blanco-Ollero A, Bonilla-Toribio M, Burguera JA, Carballo M, Carrillo F, et al. 2014. Lack of validation of variants associated with cervical dystonia risk: a GWAS replication study. Mov Disord. 29:1825–1828. [DOI] [PubMed] [Google Scholar]
- Granert O, Peller M, Jabusch HC, Altenmuller E, Siebner HR. 2011. Sensorimotor skills and focal dystonia are linked to putaminal grey-matter volume in pianists. J Neurol Neurosurg Psychiatry. 82:1225–1231. [DOI] [PubMed] [Google Scholar]
- Hagura N, Oouchida Y, Aramaki Y, Okada T, Matsumura M, Sadato N, Naito E. 2009. Visuokinesthetic perception of hand movement is mediated by cerebro-cerebellar interaction between the left cerebellum and right parietal cortex. Cereb Cortex. 19:176–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslinger B, Altenmuller E, Castrop F, Zimmer C, Dresel C. 2010. Sensorimotor overactivity as a pathophysiologic trait of embouchure dystonia. Neurology. 74:1790–1797. [DOI] [PubMed] [Google Scholar]
- Haslinger B, Erhard P, Dresel C, Castrop F, Roettinger M, Ceballos-Baumann AO. 2005. Silent event related fMRI reveals reduced sensorimotor activation in laryngeal dystonia. Neurology. 65:1562–1569. [DOI] [PubMed] [Google Scholar]
- Holmes AJ, Lee PH, Hollinshead MO, Bakst L, Roffman JL, Smoller JW, Buckner RL. 2012. Individual differences in amygdala-medial prefrontal anatomy link negative affect, impaired social functioning, and polygenic depression risk. J Neurosci. 32:18087–18100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 4:44–57. [DOI] [PubMed] [Google Scholar]
- Karimi M, Perlmutter JS. 2015. The role of dopamine and dopaminergic pathways in dystonia: insights from neuroimaging. Tremor Other Hyperkinet Mov. 5:280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirke DN, Battistella G, Kumar V, Rubien-Thomas E, Choy M, Rumbach A, Simonyan K. 2016. Neural correlates of dystonic tremor: a multimodal study of voice tremor in spasmodic dysphonia. Brain Imaging Behav. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirke DN, Frucht SJ, Simonyan K. 2015. Alcohol responsiveness in laryngeal dystonia: a survey study. J Neurol. 262:1548–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein C. 2014. Genetics in dystonia. Parkinsonism Relat Disord. 20 (Suppl 1):S137–142. [DOI] [PubMed] [Google Scholar]
- Lancaster TM, Ihssen N, Brindley LM, Tansey KE, Mantripragada K, O'Donovan MC, Owen MJ, Linden DE. 2016. Associations between polygenic risk for schizophrenia and brain function during probabilistic learning in healthy individuals. Hum Brain Mapp. 37:491–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy LM, Hallett M. 2002. Impaired brain GABA in focal dystonia. Ann Neurol. 51:93–101. [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linderman MD, Brandt T, Edelmann L, Jabado O, Kasai Y, Kornreich R, Mahajan M, Shah H, Kasarskis A, Schadt EE. 2014. Analytical validation of whole exome and whole genome sequencing for clinical applications. BMC Med Genomics. 7:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist MA, Mejia A. 2015. Zen and the art of multiple comparisons. Psychosom Med. 77:114–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann K, Schmidt A, Schillert A, Winkler S, Albanese A, Baas F, Bentivoglio AR, Borngraber F, Bruggemann N, Defazio G, et al. 2014. Genome-wide association study in musician's dystonia: a risk variant at the arylsulfatase G locus? Mov Disord. 29:921–927. [DOI] [PubMed] [Google Scholar]
- Lokkegaard A, Herz DM, Haagensen BN, Lorentzen AK, Eickhoff SB, Siebner HR. 2016. Altered sensorimotor activation patterns in idiopathic dystonia-an activation likelihood estimation meta-analysis of functional brain imaging studies. Hum Brain Mapp. 37:547–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludlow CL, Adler CH, Berke GS, Bielamowicz SA, Blitzer A, Bressman SB, Hallett M, Jinnah HA, Juergens U, Martin SB, et al. 2008. Research priorities in spasmodic dysphonia. Otolaryngol Head Neck Surg. 139:495–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok KY, Schneider SA, Trabzuni D, Stamelou M, Edwards M, Kasperaviciute D, Pickering-Brown S, Silverdale M, Hardy J, Bhatia KP. 2014. Genomewide association study in cervical dystonia demonstrates possible association with sodium leak channel. Mov Disord. 29:245–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrazek DA, Lerman C. 2011. Facilitating clinical implementation of pharmacogenomics. J Am Med Assoc. 306:304–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neychev VK, Gross RE, Lehericy S, Hess EJ, Jinnah HA. 2011. The functional neuroanatomy of dystonia. Neurobiol Dis. 42:185–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibbeling E, Schaake S, Tijssen MA, Weissbach A, Groen JL, Altenmuller E, Verbeek DS, Lohmann K. 2015. Accumulation of rare variants in the arylsulfatase G (ARSG) gene in task-specific dystonia. J Neurol. 262:1340–1343. [DOI] [PubMed] [Google Scholar]
- Niethammer M, Carbon M, Argyelan M, Eidelberg D. 2011. Hereditary dystonia as a neurodevelopmental circuit disorder: evidence from neuroimaging. Neurobiol Dis. 42:202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes TR, Johnstone T, Ores Walsh KS, Greischar LL, Alexander AL, Fox AS, Davidson RJ. 2005. Comparison of fMRI motion correction software tools. Neuroimage. 28:529–543. [DOI] [PubMed] [Google Scholar]
- Pasaniuc B, Rohland N, McLaren PJ, Garimella K, Zaitlen N, Li H, Gupta N, Neale BM, Daly MJ, Sklar P, et al. 2012. Extremely low-coverage sequencing and imputation increases power for genome-wide association studies. Nat Genet. 44:631–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol J, Roset-Llobet J, Rosines-Cubells D, Deus J, Narberhaus B, Valls-Sole J, Capdevila A, Pascual-Leone A. 2000. Brain cortical activation during guitar-induced hand dystonia studied by functional MRI. NeuroImage. 12:257–267. [DOI] [PubMed] [Google Scholar]
- Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, Sullivan PF, Sklar P, International Schizophrenia C. 2009. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 460:748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramdhani RA, Kumar V, Velickovic M, Frucht SJ, Tagliati M, Simonyan K. 2014. What's special about task in dystonia? A voxel-based morphometry and diffusion weighted imaging study. Mov Disord. 29:1141–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramdhani RA, Simonyan K. 2013. Primary dystonia: conceptualizing the disorder through a structural brain imaging lens. Tremor Other Hyperkinet Mov (N Y). 2013 Apr 18;3 pii: tre-03-152-3638-4. 10.7916/D8H70DJ7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabuncu MR, Buckner RL, Smoller JW, Lee PH, Fischl B, Sperling RA, Alzheimer's Disease, Neuroimaging I. 2012. The association between a polygenic Alzheimer score and cortical thickness in clinically normal subjects. Cereb Cortex. 22:2653–2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweinfurth JM, Billante M, Courey MS. 2002. Risk factors and demographics in patients with spasmodic dysphonia. Laryngoscope. 112:220–223. [DOI] [PubMed] [Google Scholar]
- Sciamanna G, Hollis R, Ball C, Martella G, Tassone A, Marshall A, Parsons D, Li X, Yokoi F, Zhang L, et al. 2012. Cholinergic dysregulation produced by selective inactivation of the dystonia-associated protein torsinA. Neurobiol Dis. 47:416–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonyan K, Berman BD, Herscovitch P, Hallett M. 2013. Abnormal striatal dopaminergic neurotransmission during rest and task production in spasmodic dysphonia. J Neurosci. 33:14705–14714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonyan K, Fuertinger S. 2015. Speech networks at rest and in action: interactions between functional brain networks controlling speech production. J Neurophysiol. 113:2967–2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonyan K, Ludlow CL. 2010. Abnormal activation of the primary somatosensory cortex in spasmodic dysphonia: an fMRI study. Cereb Cortex. 20:2749–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonyan K, Ludlow CL. 2012. Abnormal structure-function relationship in spasmodic dysphonia. Cereb Cortex. 22:417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonyan K, Tovar-Moll F, Ostuni J, Hallett M, Kalasinsky VF, Lewin-Smith MR, Rushing EJ, Vortmeyer AO, Ludlow CL. 2008. Focal white matter changes in spasmodic dysphonia: a combined diffusion tensor imaging and neuropathological study. Brain. 131:447–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Fox PT, Miller KL, Glahn DC, Fox PM, Mackay CE, Filippini N, Watkins KE, Toro R, Laird AR, et al. 2009. Correspondence of the brain's functional architecture during activation and rest. Proc Natl Acad Sci USA. 106:13040–13045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tansey KE, Guipponi M, Perroud N, Bondolfi G, Domenici E, Evans D, Hall SK, Hauser J, Henigsberg N, Hu X, et al. 2012. Genetic predictors of response to serotonergic and noradrenergic antidepressants in major depressive disorder: a genome-wide analysis of individual-level data and a meta-analysis. PLoS Med. 9:e1001326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalley HC, Sprooten E, Hackett S, Hall L, Blackwood DH, Glahn DC, Bastin M, Hall J, Lawrie SM, Sussmann JE, et al. 2013. Polygenic risk and white matter integrity in individuals at high risk of mood disorder. Biol Psychiatry. 74:280–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM. 2013. Genome-wide complex trait analysis (GCTA): methods, data analyses, and interpretations. Methods Mol Biol. 1019:215–236. [DOI] [PubMed] [Google Scholar]
- Zoons E, Booij J, Nederveen AJ, Dijk JM, Tijssen MA. 2011. Structural, functional and molecular imaging of the brain in primary focal dystonia—a review. Neuroimage. 56:1011–1020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.