Abstract
Microorganisms engage in complex interactions with other organisms and their environment. Recent studies have shown that these interactions are not limited to the exchange of electron donors. Most microorganisms are auxotrophs, thus relying on external nutrients for growth, including the exchange of amino acids and vitamins. Currently, we lack a deeper understanding of auxotrophies in microorganisms and how nutrient requirements differ between different strains and different environments. In this Opinion article, we describe how the study of auxotrophies and nutrient requirements among members of complex communities will enable new insights into community composition and assembly. Understanding this complex network over space and time is crucial for developing strategies to interrogate and shape microbial communities.
The interactions of microorganisms that coexist in nature are essential for global nutrient cycling and have a profound role in human health and disease. These interspecies interactions can have beneficial, neutral or harmful effects on members of the community. Beneficial or neutral interactions can be defined as commensalism, whereby the association between different species is beneficial to individuals of one species but has no effect on the other; mutualism, whereby microorganisms that coexist benefit from one another; or neutralism, whereby the association does not affect the microorganisms involved in the association. In addition, microorganisms can interact antagonistically in relationships that are classified as amensalism, whereby the association between different species is detrimental to individuals of one species but not to those of the other; competition, whereby all partners are disadvantaged by the presence of others; parasitism, whereby one microorganism benefits at the expense of their host organism; or predation, whereby one organism feeds on the other organism1. The sum of these interactions shapes the composition and function of the microbial community in an ecosystem. Until recently, interactions have been viewed as successional activities of various members, each providing the energy source for the next member. However, on the basis of recent studies, a new picture has emerged, which emphasizes interdependencies and convoluted networks of microorganisms that are not limited to the exchange of electron donors for growth2 but include the exchange of amino acids2–6, vitamins7–9 and other cofactors10,11 (FIG. 1). For example, microorganisms rely on other members of the microbial community to acquire signalling molecules, such as small peptides12 or siderophores13, for growth. Additionally, microorganisms depend on partners for the detoxification of inhibitory molecules (such as xenobiotics or prohibitive concentrations of metabolites)14 or the reduction of reactive oxygen species (ROS) by ROS-scavenging organisms15. Although we acknowledge the importance of these cofactors, in this article, we mainly focus on microbial interactions that involve the exchange of amino acids and vitamins.
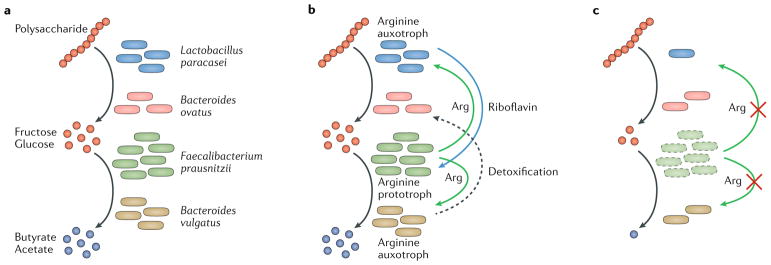
Fig. 1. Nutrient cycling in microbial communities.
a | Classic view of nutrient exchange involving successional interactions among members in a microbial community. Extracellular polysaccharides (for example, inulin, xylan or arabinogalactan) can be metabolized by human-associated bacteria, such as Bacteroides ovatus25 or Lactobacillus paracasei92, into different monosaccharides (for example, glucose or fructose). Breakdown products from saccharolytic bacteria in the gut can be converted to short-chain fatty acids (SCFAs; for example, acetate or butyrate) by primary fermenters, such as Faecalibacterium prausnitzii26 and Bacteroides vulgatus25, in a sequential manner. b | Expanding the classic view of electron donor exchange. Successional interactions among members in a microbial community (as described in part a) depend on the exchange of amino acids and vitamins. L. paracasei provides riboflavin (vitamin B2)93 for F. prausnitzii. Moreover, the arginine (Arg) prototroph F. prausnitzii supplies arginine for L. paracasei and B. vulgatus, which are both arginine auxotrophs. B. vulgatus additionally supports the growth of B. ovatus through the detoxification of inhibitory substances25. c | Perturbations (such as those that occur with antibiotic treatment or in a disease state) lead to changes in the dynamic equilibrium of microbial communities and alter the interactions between microorganisms. Reduced synthesis of arginine by F. prausnitzii limits its availability to arginine auxotrophs. In turn, growth of auxotrophs is decreased under nutrient-depleted conditions. The decrease in abundance of auxotrophs limits the availability of by-products, such as glucose and SCFAs, for other members in the community.
Prototrophic microorganisms can synthesize all the nutrients that are required for their growth from minimal medium without the addition of supplements. However, many microorganisms in nature (and in culture collections) are auxotrophic; that is, they are unable to synthesize all of the vital nutrients. Access to these nutrients is therefore essential, and the lack of these nutrients inhibits cell division and growth. Several explanations could account for the frequent occurrence of auxotrophic microorganisms. First, the energetic costs of producing certain metabolites are offset by obtaining them from the environment, from a different microorganism or from the host16,17. In metabolite-rich environments, this could select for the loss of biosynthetic genes, thus promoting auxotrophic genotypes3. Additionally, in spatially structured communities, local exchange among cooperative bacteria is increased and could increase reciprocity16,18,19. Bacteria can exhibit multiple auxotrophies at once, which results in various degrees of nutrient requirements. Furthermore, the severity of the growth defects under nutrient-limiting conditions is dependent on which step in the biosynthetic pathway is lost20.
Whereas metabolic interactions among community members (that is, substrate utilization patterns) are highly dynamic in nature, are often tightly regulated and can change over time, nutrient requirements are hardwired into the genome and therefore remain constant. This disposition defines the network in which microorganisms can thrive and results in a defined group of potential partners that allocate the necessary nutrients. In this Opinion article, we postulate that these genome-encoded requirements have profound implications for microbial interactions and thus the overall microbial network. How microbial networks are assembled and maintained and how these communities react to perturbations depend in part on the nutrient requirements of the individual members within a community. Similar to our own social network, which changes and evolves over time within certain parameters, microorganisms also have distinct preferences for partners and interactions based on their auxotrophies. We discuss how auxotrophies can shape the structure of environmental and host-associated microbial communities and how interdependencies can influence pathogenicity. The principles outlined here are universal and can be applied to many microbial communities and environments and are not restricted to interactions between the human microbiota and its host.
Auxotrophies in microbial communities
A classic view of electron donor exchange
Interspecies interactions are essential for the composition and function of the microbiome. It has been suggested that microbial communities, for example, in soil, sediments or the human gut, rely mainly on cross-feeding of electron donors for growth21. Thus, the survival of individual microorganisms depends in large part on the whole community to guarantee carbon flow and exchange of by-products. The classic view of nutrient cycling involves successional interactions of various microbial community members, each providing the energy source for the next member in a cascade-like fashion (FIG. 1a). A model example is the degradation of complex organic matter (for example, cellulosic biomass) in an anaerobic digester in which organic material is converted into short-chain fatty acids (SCFAs), carbon dioxide and methane by different microorganisms in a community22. Another example is the metabolic activity of the many members of the human gut microbiota, which aids food digestion and provides the host with essential re-evaluate the foundation of microbial network structure in ways that were previously unfeasible.
Amino acid and vitamin auxotrophies
Whereas previous work on interspecies interactions has mainly focused on cross-feeding of electron donors, recent studies have highlighted the exchange of amino acids and vitamins in microbial communities (BOX 1) and have shown that these interactions can greatly contribute to their composition5,31,32. Comparative analyses of microbial genomes indicate that more than 98% of all the microorganisms sequenced so far lack essential pathways or key genes for the synthesis of amino acids33,34. The majority of microorganisms are thus auxotrophic and require extracellular sources of amino acids, vitamins and/or cofactors for their survival. It is important to note that the metabolic costs and energy requirements for the synthesis of amino acids, vitamins and cofactors vary substantially. For example, aromatic amino acids, such as phenylalanine, tryptophan, tyrosine and histidine, are energetically more costly to synthesize than simpler amino acids such as glycine or serine35. Moreover, metabolic costs for the synthesis of amino acids, vitamins and cofactors can vary among different microorganisms owing to the use of different pathways and differences nutrients23,24. For example, a study using a gnotobiotic mouse model demonstrated that extracellular digestion of inulin increases the growth rate of Bacteroides ovatus, a prominent bacterium in the gut25. In turn, by-products from inulin catabolism can be used by primary fermenters such as Faecalibacterium prausnitzii and Bacteroides vulgatus. The sequential action of different members of the gut microbiota involving glycolytic and fermentation pathways generates the metabolic input (for example, SCFAs, lactic acid and hydrogen) for a diverse set of microorganisms, including sulfate-reducing bacteria, acetogens and methanogens26–28. The exchange of electron donors is a well-established driving force in microbial ecology, and textbook knowledge of these exchanges is often consulted when interpreting microbiome data and describing microbial networks. However, electron donor exchange alone can be insufficient to explain the dynamic interactions and apparent metabolic redundancies among microorganisms. Genome analysis, often combined with computational modelling approaches, has provided new insight into the nutrient requirements of various microorganisms that inhabit the human body. The ability to investigate multiple microorganisms simultaneously despite our lack of success in cultivating them in the laboratory29,30 has opened the door to in proteome allocation (such as amino acid composition)36 and are also dependent on the growth stage of the organism37. Therefore, the energetic burden for different microorganisms is not evenly distributed within a community.
Box 1. Experimental and computational methods to map microbial interactions.
Experimental methods
Co-culturing: co-culture experiments are providing widely applied validation for cross-feeding among microorganisms16,33,50. These experiments are constrained to a limited number of microorganisms and are most frequently performed with two members. Furthermore, co-culturing is mostly restricted to cultivated microorganisms, and the outcome is dependent on the cultivation method and medium composition, reducing its applicability to natural systems29,30.
Time course shotgun metagenomic sequencing: the composition of microbial communities can change dramatically over time. Temporal profiling at a strain-level resolution is fundamental to study longitudinal dynamics of microbial communities76. Shotgun metagenomics in combination with differential binning permits the retrieval of nearly complete genomes from complex microbial communities and enables identification of auxotrophies and nutrient requirements78.
Functional profiling using metatranscriptomics, metaproteomics and metabolomics: high-throughput functional profiles of microbial communities can be obtained by sequence-based and mass spectrometry-based omic methods76,78. Metatranscriptomics, metaproteomics and metabolomics measure mRNA, protein or metabolite levels, respectively. These approaches can be combined to determine active metabolic pathways under a certain condition and at a distinct time point and can assist in resolving interaction networks.
Computational methods
Genome comparison analysis: genome comparison analysis can be performed based on reference genomes or metagenomic data77. Genome-based prediction of auxotrophies can indicate potential cooperative or competitive interactions79. In silico analysis of metabolic pathways, confirmed by experimental data, can yield models of carbon, energy and nutrient flow in microbial communities78,80.
Metagenomic and functional profiling: meta-omics data in general entail a strong computational analysis component. Meta-omics data analysis requires the combination of multiple statistical and/or computational methods81,82 and depends on well-curated reference databases83–85.
Modelling
Genome-scale metabolic models: genome-scale metabolic models involve the creation of a metabolic network in which enzymatic reactions are linked by substrates and products (metabolites). Constraint-based modelling that uses, for example, flux balance analysis can be applied to predict metabolic phenotypes under different growth conditions86,87. Simulations using genome-scale reconstructions are frequently validated by experimental results and can be used to predict genotype–phenotype relationships and interactions among microorganisms, as well as interactions between microorganisms and their host88.
Dynamic modelling: computational and mathematical theoretical models are being used to contextualize complex omics data that were generated by high-throughput experimental techniques89. Such modelling approaches include ordinary differential equations90 and agent-based models91 and can simulate dynamic changes of regulatory networks (signalling pathways and metabolic pathways) and microbial communities. Mathematical modelling of microbial community dynamics has been reviewed previously89.
Auxotrophic bacteria rely on other members of the community, the host or the environment to provide essential nutrients. Among human-associated microbiota, several prominent bacteria, such as Bifidobacterium thermophilum, Eubacterium rectale and Staphylococcus aureus, are unable to synthesize various essential amino acids4. The source of these amino acids for these microorganisms can differ depending on the environment. For example, bacteria in the human gut can obtain amino acids from the diet, other members in the resident microbiota or the host itself for protein synthesis38. Several abundant bacteria in the gut (for example, Bacteroides spp.) and on the skin (for example, Propionibacterium spp.) are unable to synthesize essential vitamins such as cobalamin (vitamin B12) or pantothenate (vitamin B5)23,39. Some members of the Bacteroidetes phylum are missing some or all of the genes that are necessary for the synthesis of B12 (REF.9). However, these organisms possess several B12-dependent enzymes that are essential for the metabolism of sugars, amino acids and fatty acids23, which suggests a distinct need for B12. Common bacteria in the gut could potentially provide vitamin B12 to the microbial community independent of the diet23,25.
Nutrient uptake
Auxotrophies can be identified by missing or incomplete pathways in the genome, but determining if and how an organism is using the nutrients is much more challenging. For example, an essential amino acid can be acquired as a free amino acid in the form of a dipeptide, tripeptide or oligopeptide, or as part of a complex protein that originates from decaying cells40. The reason for our current inability to determine the exact source of amino acids lies in the challenge of annotating and correctly assigning transport reactions. Functional assignment of specific transporters is therefore fundamental for unravelling metabolic exchanges and delineating network structure. However, currently, the substrates of only ~25% of all bacterial membrane transporters have been assigned41. An additional challenge is predicting the substrate specificity of known transporters. Transporters can be highly promiscuous, and prediction of substrate specificity on the basis of sequence homology alone can be difficult, thus leading to generic annotations in which multiple transporters are assigned identical substrates41,42. This apparent redundancy is rarely explored when new transporters are described and can contribute to inaccurate interpretations of transport reactions and thus misidentification of the capabilities of certain bacterial members in a community. For example, a considerable proportion of bacteria in the human gut encodes transporters for corrinoids (for example, cobalamin, with vitamin B12 being the most well-known member of this group) in their genomes23. Most of these transporters are generically annotated as cobalamin transporters. However, microorganisms can use a wide range of corrinoids and corrinoid precursors and potentially show a distinct preference for certain substrates43,44. For instance, a study evaluating three functionally homologous vitamin B12 transporters (designated BtuB1, BtuB2 and BtuB3) in Bacteroides thetaiotaomicron revealed that these transporters exhibit distinct preferences for corrinoids that contain adenine or benzimidazole over B12-containing 5,6-dimethylbenzimidazole45. The wildtype strain containing all three functional transporters showed competitive advantages compared with knockout strains, which are only able to use one single corrinoid.
Network dynamics and cross-feeding
Nutrients derived from microorganisms
Abundant auxotrophies require most bacteria to obtain vital nutrients from other microorganisms or the host. These nutrients have to be available on a continuous basis to sustain growth over time. Expanding the classic view of electron donor exchange, microorganisms need interacting partners not only to provide suitable carbon and energy sources but also to supply essential nutrients. Currently, it is not known whether the organisms that provide electron donors and nutrients are identical or whether they differ from each other and/or over time. Longitudinal studies often report significant variation in the species composition of microbial communities with seemingly similar metabolic functions46,47. These changes are often explained by subtle genomic differences between microorganisms. For example, differences in transport affinity enable some organisms to uptake a substrate more effectively than its competitors. Alterations in enzyme specificity and enzyme abundance can provide additional growth advantages. However, another plausible explanation is the difference in auxotrophies between organisms with similar metabolic profiles, resulting in community dynamics that are in part dependent on nutrient availability. These interactions can promote positive, negative or neutral effects on the fitness of the community members, resulting in complex relationships. Temporal shifts in the community composition strongly affect the interaction between cooperative bacteria. For instance, the decrease in abundance of one microorganism directly affects the availability of by-products for their partner. This necessitates a rearrangement of the interaction network or forces microorganisms to reprogram their proteome during nutrient limitations48,49 (FIG. 2).
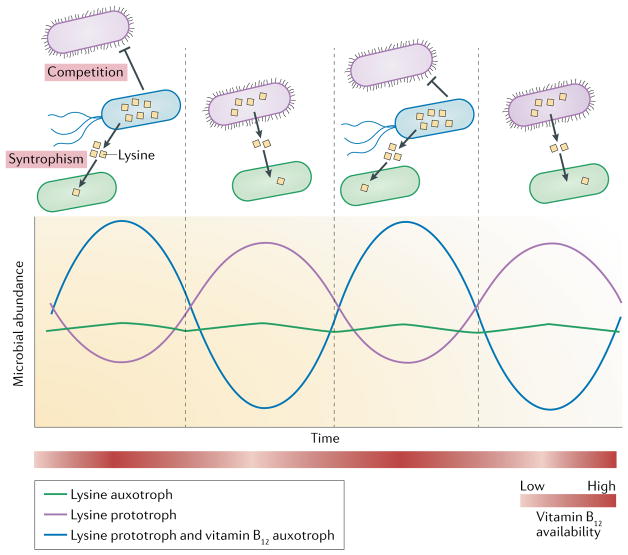
Fig. 2. Dynamic of interactions.
Oscillations in nutrient availability (red gradient) considerably affects the community composition over time. A vitamin B12 auxotroph (shown in blue) relies on other members of the community or external sources to obtain this amino acid. In turn, the vitamin B12 auxotroph provides lysine to a member in the community that is a lysine auxotroph (shown in green). When vitamin B12 becomes limited, the abundance of B12 auxotrophs decreases (blue line). As a result, lysine auxotrophs in the community interact with other members in the community (shown in purple) to obtain lysine and to maintain growth (green line). Thus, the decrease in abundance of one member (blue line) can be beneficial for competitors (purple line) and leads to changes in community composition and the interaction network.
Although evaluating the consequences of auxotrophies for complex communities can be challenging (BOX 1), insights into community assembly and dynamics have been gained from enrichment cultures5 and synthetic communities33. A recent study generated a syntrophic (that is, cross-feeding) community composed of amino-acid-auxotrophic Escherichia coli strains. Starting from the prototrophic E. coli MG1655 strain (that is, a strain that can synthesize all of its nutrients), 14 derivative auxotrophic strains were obtained, each containing a gene knockout for one essential amino acid33. None of the derivative strains were able to grow independently without supplementation of the appropriate amino acid. The authors probed all possible pairwise syntrophic interactions and observed statistically significant synergistic and cooperative growth in pairwise co-cultures. Energetically expensive and rare amino acids (that is, arginine, isoleucine, lysine, methionine, phenylalanine, threonine and tryptophan) supported improved cross-feeding in these co-cultures compared with energetically inexpensive and more common amino acids. To create a scenario closer to natural microbial communities in which auxotrophic bacteria rely on each other to survive and to form a robust steady-state community, the authors combined all 14 auxotrophic strains. After 400 generations, auxotrophic pairs that were able to establish strong cooperative interactions and thus grew the fastest (for example, by sharing arginine, lysine, methionine and threonine) dominated the consortium, showing that amino acid auxotrophies dramatically shaped the microbial community. Similar cooperation was observed in co-culture experiments involving two amino-acid-auxotrophic Saccharomyces cerevisiae strains50. The survival of both auxotrophic strains was observed for a wide range of initial strain ratios, supporting a variety of conditions permissible for cooperation. These observations are consistent with growth dynamics and varying abundances of different microorganisms in naturally occurring cooperative systems44,51.
Finally, interactions of microorganisms often rely on close proximity. Computational and in vivo studies have shown that spatial structure of microbial communities is a determining factor for cooperation and can drive community dynamics16,52,53. Co-culture experiments involving Bacillus subtilis and the endophytic fungus Serendipita indica revealed that thiamine (vitamin B1) is a key nutrient for the B1 auxotroph S. indica. However, successful growth of the co-culture is achieved only when these microorganisms are cultivated in a spatially organized environment that provides optimal conditions for cooperative interactions16,53.
Host-derived nutrients
Understanding potential interactions and contributing partners in communities in which nutrients are provided externally (for example, by the host) is challenging. For example, nutrient availability in the rhizosphere or the human gut is highly dynamic and can vary on different timescales. Physiological variation in human host physiology can be dependent on the time of the day54 (for example, sleep–wake cycles, food intake or changes in hormone levels), over different seasons55 or over the course of a lifetime56. Although multiple studies have delineated the effect of the host diet on the composition of the microbiome55,57, little work has been done to mechanistically resolve the effect of a change in diet on the microbial interaction network. Mice subjected to a Western-like diet with a high fat and low fibre content over several generations showed a progressive loss of microbial diversity, which could not be recovered after the reintroduction of a high-fibre diet58. Unravelling the social network and reintroducing beneficial taxa (that is, probiotics) to the microbiota could provide potential treatments of recurrent diseases linked to a Western diet58. Additionally, it was recently shown that certain compounds in the human diet can select for hypervirulent strains. For example, intake of trehalose can increase the virulence of a Clostridioides difficile strain59, which suggests that dietary intervention strategies (such as the use of prebiotics) could select against potential pathogens. Understanding and predicting the effects of diet on assembly, structure, composition and maintenance of the microbiota would be an important step towards rationally controlling the microbiome and gut function.
Cooperation within communities
Determining cooperation and competition on the basis of nutrient exchange within microbial communities can be a demanding task. The challenge lies not only in identifying interaction partners based on auxotrophies in complex communities but also in the fact that single microorganisms engage in multiple and promiscuous interactions depending on nutrient and energy availabilty33. Although successful examples using co-cultures have elucidated the nature of the interactions between two or more microorganisms33,50, studies involving complex microbial communities are currently rare. Given the dynamic nature and complexity of host-associated communities, most studies have been focused on microbial systems in the absence of a eukaryotic host. One such example is microbial-mediated anaerobic ammonium oxidation (anammox), which represents one of the most energy-efficient biotechnological methods for nitrogen removal from wastewater60. Metagenomic and metatranscriptomic analysis of anammox granules revealed that bacteria that belong to the phyla Chloroflexi and Chlorobi are auxotrophic for vitamins B1, B7 and B12. However, Brocadia spp. from the family Planctomycetaceae contain complete metabolic pathways for vitamins B1, B7 and B12. Increased expression of genes involved in pathways for B1, B7 and B12 synthesis in Brocadia spp. indicates that these bacteria may support the B vitamin requirements of the entire community61. A host-associated study evaluated the long-term persistence of the microbiota after faecal transplantation in patients infected with C. difficile62 and identified Bacteroides spp. as the most common donor microorganism in recipients after transplantation. Metagenomic data obtained 2 years after faecal transplantation showed that B. vulgatus and B. ovatus exhibit long-term persistence and co-occurrence in the gastrointestinal tracts of the recipients. Independent studies have shown how cooperation evolved between these two Bacteroides spp45. B. ovatus has its fitness increased by digesting fibre, for example, the polysaccharide inulin, from the diet25. In contrast to other Bacteroides spp. (for example, B. thetaiotaomicron) that use mostly extracellular breakdown products of fibre as growth substrate63, B. ovatus not only digests inulin extracellularly but also digests it intracellularly, as B. ovatus has the apparatus to take up and metabolize inulin inside the cell. However, the presence of additional saccharolytic enzymes on the cell surface confers a competitive advantage to B. ovatus. The mechanism behind cooperation between B. ovatus and B. vulgatus is currently unknown, and it has been suggested that B. ovatus supplies nutrients to B. vulgatus while B. vulgatus benefits from B. ovatus through detoxification of inhibitory substances or by the provision of an essential growth factor25.
Competition within communities
Several studies have described the robustness and plasticity of the healthy human microbiota and have linked it to high diversity54,64,65. The currently accepted theory asserts that a highly diverse ecosystem can endure perturbations and at the same time adapt to changes in the environment, resulting in a stable but flexible healthy state. It has been proposed that harbouring multiple metabolically redundant species results in high competition for resources in the ecosystem, ultimately leading to increased ecosystem stability66. Competition within microbial communities is also an important mechanism by which pathogen overgrowth can be restricted. Commensal microorganisms have established a niche in the human body by developing highly efficient approaches to uptake and metabolize available nutrients and to protect their environment against competing microorganisms. Under normal (that is, healthy) conditions, human pathogens experience colonization resistance, having to invade niches and compete for nutrients and spaces that are already occupied by adapted resident bacteria67. Acute infection, prolonged malnutrition or antibiotic usage can result in dramatic changes in the microbiome. Hence, the new state of the microbiome after a severe perturbation may not resemble the original composition68 (FIG. 3). Abrupt disruption of the dynamic equilibrium of microbial communities can considerably alter the interactions between microorganisms and between microorganisms and their host, thus creating a propitious environment for pathogens (FIG. 3). The availability of resources can affect the nature of the interaction in microbial communities. A study using amino-acid-auxotrophic yeast strains demonstrated that changes in amino acid availability can modulate the types of interaction between species. Depending on the resource availability, the interaction of these strains changed from obligatory and facultative mutualism to competition and parasitism69. Unravelling these dependencies and the resulting dynamic interactions of members in a community would be of great importance for microbiome research.
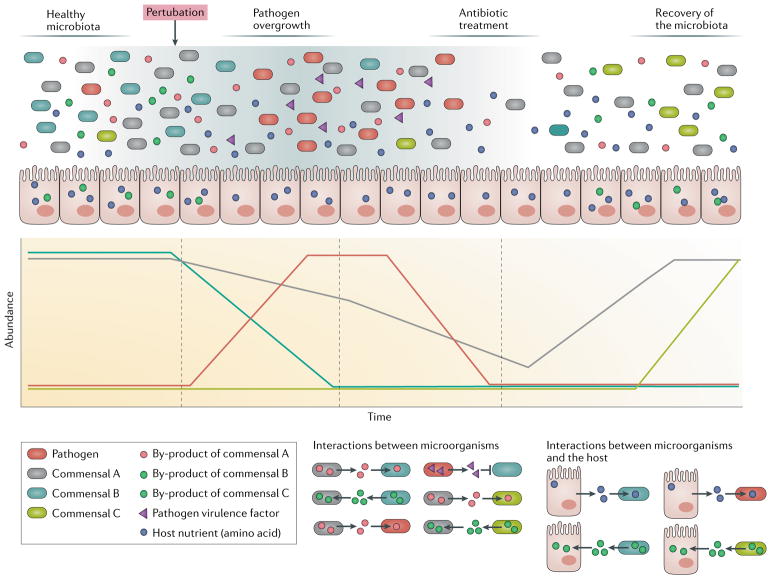
Fig. 3. Microbial community perturbation and resilience.
A healthy, human microbial community exchanges nutrients among commensal members (indicated by grey, blue and green ovals) and the host through an intricate network of interactions. Coloured lines indicate microbial abundance (total number of cells) over time. The healthy microbiota consumes available nutrients (blue dots), produces metabolites for other members (different species or strains) and for the host (red and green dots), and inhibits pathogen growth. Upon perturbation, the abundance of and composition in the gut commensal microbial community can change, creating an opportunity for pathogen overgrowth. During infection, amino-acid-auxotrophic pathogens, such as Staphylococcus aureus71, Bacillus anthracis94 or Streptococcus pyogenes4 (red ovals), compete with commensal bacteria for host amino acids (blue dots) and promote physiological changes in the host by releasing toxins (purple triangles), thus triggering inflammation. Antibiotic treatment leads to a decrease in microbial abundance that affects not only commensals but also pathogens. Over time, the abundance and diversity of the microbiota recovers. However, the composition of the new microbiota may differ to the initial state, which results in new interactions among commensals to maintain functionality and equilibrium of the microbial community67.
With the rare exception of C. difficile, which is also able to grow in vitro in the presence of carbon dioxide and hydrogen as the sole carbon and energy source, respectively70, all pathogenic bacteria known to date are heterotrophs. Several human pathogens are auxotrophic for amino acids, which confines their network4. S. aureus, a human pathogen commonly found on skin71, has been directly associated with the inflammatory skin disease atopic dermatitis. Comparison between lesional and non-lesional skin in patients with atopic dermatitis has shown that the number of S. aureus cells can surpass tenfold in patients with lesional skin72. Pan-genomic analysis for 64 S. aureus strains predicted that all strains are auxotrophic for niacin and thiamine, whereas strain-specific auxotrophies were predicted for riboflavin, guanine, leucine, methionine, cysteine and valine, among others73. These auxotrophic pathogens therefore rely either on their microbial network or on the host to provide these essential nutrients. Excessive overgrowth of specific strains of S. aureus71 suggests that nutritional resources have to be provided in sufficient quantities to support this overgrowth. The microbial network would thus have to synthesize and share adequate amounts of nutrients, rendering S. aureus dependent on commensal bacteria of the skin microbiota. Alternatively, large amounts of essential amino acids supporting S. aureus overgrowth could originate from the host. Moreover, two or more S. aureus strains could mutually support each other during infection. To our knowledge, there are no examples of commensal bacteria that overproduce nutrients during infections, which suggests that pathogens obtain nutrients from the host. Indeed, during an infection, pathogens often release a wide array of virulence factors to outcompete their commensal rivals and promote host tissue damage. For example, several gastrointestinal pathogens disrupt the epithelial barrier, thus increasing permeability to facilitate the release of nutrients74. Consequently, the abundance of nutrients minimizes the competition among commensals and pathogens and thus renders interactions based on auxotrophies insignificant. However, at what stage a pathogen switches its source of nutrition from other microorganisms to the host is currently unknown. Future studies that focus on identifying the source of critical nutrients that support pathogenic overgrowth will be crucial to increasing our understanding of disease initiation and exacerbation and could potentially lead to novel treatment strategies.
Outlook
Although auxotrophies and the role of amino acid and vitamin exchange have been identified recently in several microbial communities5,6,31,32,61, the dynamics of these complex interaction networks have only been revealed in simple co-culture experiments. However, interactions between multiple species can change over time according to nutrient availability and proteome allocation4. Currently, the identification of dynamic interactions and dependencies in natural microbial communities is challenging because members of such communities can engage in multiple and promiscuous interactions. However, defining these interactions could provide new avenues to treating diseases that have been linked to microorganisms75. Understanding multilevel interactions in a natural community requires a combination of longitudinal studies and advanced computational tools76. Auxotrophies vary substantially between strains of the same species7,73 and cannot be inferred from phylogenetic markers such as 16 S ribosomal RNA. Therefore, genome information at the strain level is required to study the effects of auxotrophies on community composition and interactions; however, longitudinal microbiome studies at a strain-level resolution have been scarce. One reason for the lack of strain-level data is the difficulty in which high quality, nearly complete genomes are retrieved77. Although this has been achieved for communities of low complexity5, it is still challenging for complex communities and thus hampers strain identification and the correct determination of auxotrophies (BOX 1). The enhancement of sequencing technology (for example, improved read length) and computational tools (for example, refined genome assembly) would greatly benefit the elucidation of interaction dynamics. Further insight into the intertwined network of interactions among microorganisms as well as microorganisms and their hosts at the strain-level could pave the way for new treatment options, such as the use of prebiotics or probiotics to not only target a single pathogen but also modulate the entire microbial network. Owing to the lack of knowledge about interactions, most current prebiotic and probiotic interventions have not been designed rationally. And although some treatment strategies have proved to be beneficial, the mechanism behind those effects is mostly unknown. Knowledge about the nutrient requirements of members of the microbiome, including pathogens and their social network, could enable rational interventions and would potentially result in new tailored treatment strategies.
Acknowledgments
Research in the authors’ laboratory was supported in part by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under award number AR071731. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Work in the authors’ laboratory was also supported by the National Science Foundation under grant number 1332344 and the US Department of Energy, Office of Science, Office of Biological & Environmental Research Awards DESC0012586, DE-SC0012658 and DE-SC0018344.
Footnotes
Author contribution
K.Z. and L.S.Z. researched data for the article, made substantial contributions to discussions of the content, wrote the article and reviewed and edited the manuscript before submission.
Competing interests
The authors declare no competing interests.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Reviewer information
Nature Reviews Microbiology thanks Felix Sommer, Jan Roelof van der Meer and the other anonymous reviewer(s) for their contribution to the peer review of this work.
References
- 1.Little AEF, Robinson CJ, Peterson SB, Raffa KF, Handelsman J. Rules of engagement: Interspecies interactions that regulate microbial communities. Annu Rev Microbiol. 2008;62:375–401. doi: 10.1146/annurev.micro.030608.101423. [DOI] [PubMed] [Google Scholar]
- 2.Mitri S, Richard Foster K. The genotypic view of social interactions in microbial communities. Annu Rev Genet. 2013;47:247–273. doi: 10.1146/annurev-genet-111212-133307. [DOI] [PubMed] [Google Scholar]
- 3.D’Souza G, et al. Less is more: Selective advantages can explain the prevalent loss of biosynthetic genes in bacteria. Evolution. 2014;68:2559–2570. doi: 10.1111/evo.12468. [DOI] [PubMed] [Google Scholar]
- 4.Yu X, Walker DH, Liu Y, Zhang L. Amino acid biosynthesis deficiency in bacteria associated with human and animal hosts. Infect Genet Evol. 2009;9:514–517. doi: 10.1016/j.meegid.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Embree M, Liu JK, Al-bassam MM, Zengler K. Networks of energetic and metabolic interactions define dynamics in microbial communities. Proc Natl Acad Sci USA. 2015;112:15450–15455. doi: 10.1073/pnas.1506034112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu YF, et al. Metabolic capability and in situ activity of microorganisms in an oil reservoir. Microbiome. 2018;6:5. doi: 10.1186/s40168-017-0392-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodionova IA, et al. Genomic distribution of B-vitamin auxotrophy and uptake transporters in environmental bacteria from the Chloroflexi phylum. Environ Microbiol Rep. 2015;7:204–210. doi: 10.1111/1758-2229.12227. [DOI] [PubMed] [Google Scholar]
- 8.Croft MT, Lawrence AD, Raux-Deery E, Warren MJ, Smith AG. Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature. 2005;438:90–93. doi: 10.1038/nature04056. [DOI] [PubMed] [Google Scholar]
- 9.Wexler AG, Goodman AL. An insider’s perspective: Bacteroides as a window into the microbiome. Nat Microbiol. 2017;2:1–11. doi: 10.1038/nmicrobiol.2017.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wyn-Jones RG. Ubiquinone deficiency in an auxotroph of Escherichia coli requiring 4-hydroxybenzoic acid. Biochem J. 1967;103:714–719. doi: 10.1042/bj1030714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gruss A, Borezée-durant E, Lechardeur D. In: Advances in microbial physiology. Poole RK, editor. Elsevier; 2012. pp. 70–111. [DOI] [PubMed] [Google Scholar]
- 12.Nichols D, et al. Short peptide induces an ‘Uncultivable’ microorganism to grow in vitro. Appl Environ Microbiol. 2008;74:4889–4897. doi: 10.1128/AEM.00393-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Onofrio AD, et al. Siderophores from neighboring organisms promote the growth of uncultured bacteria. Cell Chem Biol. 2010;17:254–264. doi: 10.1016/j.chembiol.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koppel N, Rekdal VM, Balskus EP. Chemical transformation of xenobiotics by the human gut microbiota. Microbiota. 2017;356:1–11. doi: 10.1126/science.aag2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morris JJ, Kirkegaard R, Szul MJ, Johnson ZI, Zinser ER. Facilitation of robust growth of Prochlorococcus colonies and dilute liquid cultures by ‘helper’ heterotrophic bacteria. Appl Environ Microbiol. 2008;74:4530–4534. doi: 10.1128/AEM.02479-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Germerodt S, et al. Pervasive selection for cooperative cross-feeding in bacterial communities. PLoS Comput Biol. 2016;12:1–21. doi: 10.1371/journal.pcbi.1004986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harvey E, Heys J. Quantifying the effects of the division of labor in metabolic pathways. J Theor Biol. 2014;360:222–242. doi: 10.1016/j.jtbi.2014.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verbruggen E, et al. Spatial structure and interspecific cooperation: Theory and an empirical test using the mycorrhizal mutualism. Am Nat. 2012;179:E133–E146. doi: 10.1086/665032. [DOI] [PubMed] [Google Scholar]
- 19.Kreft J. Biofilms promote altruism. Microbiology. 2004;150:2751–2760. doi: 10.1099/mic.0.26829-0. [DOI] [PubMed] [Google Scholar]
- 20.Guzmán GI, et al. Model-driven discovery of underground metabolic functions in Escherichia coli. Proc Natl Acad Sci USA. 2015;112:929–934. doi: 10.1073/pnas.1414218112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morris BEL, Henneberger R, Huber H, Moissleichinger C. Microbial syntrophy: interaction for the common good. FEMS Microbiol Rev. 2013;37:384–406. doi: 10.1111/1574-6976.12019. [DOI] [PubMed] [Google Scholar]
- 22.Lin L, Yu Z, Li Y. Sequential batch thermophilic solid-state anaerobic digestion of lignocellulosic biomass via recirculating digestate as inoculum –part II: microbial diversity and succession. Bioresour Technol. 2017;241:1027–1035. doi: 10.1016/j.biortech.2017.06.011. [DOI] [PubMed] [Google Scholar]
- 23.Degnan PH, Taga ME, Goodman AL. Vitamin B12 as a modulator of gut microbial ecology. Cell Metab. 2014;20:769–778. doi: 10.1016/j.cmet.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zilberman-schapira G, et al. The gut microbiome in human immunodeficiency virus infection. BMC Med. 2016;14:1–11. doi: 10.1186/s12916-016-0625-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rakoff-Nahoum S, Foster KR, Comstock LE. The evolution of cooperation within the gut microbiota. Nature. 2016;533:255–259. doi: 10.1038/nature17626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lopez-Siles M, Duncan SH, Garcia-Gil LJ, Martinez-Medina M. Faecalibacterium prausnitzii: from microbiology to diagnostics and prognostics. ISME J. 2017;11:841–852. doi: 10.1038/ismej.2016.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fischbach MA, Sonnenburg JL. Eating for two: How metabolism establishes interspecies interactions in the gut. Cell Host Microbe. 2011;10:336–347. doi: 10.1016/j.chom.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rey FE, et al. Metabolic niche of a prominent sulfate-reducing human gut bacterium. Proc Natl Acad Sci USA. 2013;110:13582–13587. doi: 10.1073/pnas.1312524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zengler K, et al. Cultivating the uncultured. Proc Natl Acad Sci USA. 2002;99:15681–15686. doi: 10.1073/pnas.252630999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oberhardt MA, et al. Harnessing the landscape of microbial culture media to predict new organism–media pairings. Nat Commun. 2015;6:8493. doi: 10.1038/ncomms9493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Romine MF, Rodionov DA, Maezato Y, Osterman AL, Nelson WC. Underlying mechanisms for syntrophic metabolism of essential enzyme cofactors in microbial communities. ISME. 2017;11:1434–1446. doi: 10.1038/ismej.2017.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hibberd MC, et al. The effects of micronutrient deficiencies on bacterial species from the human gut microbiota. Sci Transl Med. 2017;9:eaal4069. doi: 10.1126/scitranslmed.aal4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mee MT, Collins JJ, Church GM, Wang HH. Syntrophic exchange in synthetic microbial communities. Proc Natl Acad Sci USA. 2014;111:E2149–E2156. doi: 10.1073/pnas.1405641111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mee MT, Wang HH. Engineering ecosystems and synthetic ecologies. Mol Biosyst. 2012;8:2470–2483. doi: 10.1039/c2mb25133g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaleta C, Schäuble S, Rinas U, Schuster S. Metabolic costs of amino acid and protein production in Escherichia coli. Biotechnol J. 2013;8:1105–1114. doi: 10.1002/biot.201200267. [DOI] [PubMed] [Google Scholar]
- 36.Heizer EM, Jr, et al. Amino acid cost and codon-usage biases in 6 prokaryotic genomes: a whole-genome analysis. Mol Biol Evol. 2004;23:1670–1680. doi: 10.1093/molbev/msl029. [DOI] [PubMed] [Google Scholar]
- 37.Zuñiga C, et al. Predicting dynamic metabolic demands in the photosynthetic eukaryote Chlorella vulgaris. Plant Physiol. 2018;176:450–462. doi: 10.1104/pp.17.00605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neis EPJG, Dejong CHC, Rensen SS. The role of microbial amino acid metabolism in host metabolism. Nutrients. 2015;7:2930–2946. doi: 10.3390/nu7042930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stackebrandt E, Cummins CS, Johnson JL. In: The Prokaryotes. Falkom S, editor. Springer–Verlag; New York: 2006. pp. 400–418. [Google Scholar]
- 40.Burkovski A, Kramer R. Bacterial amino acid transport proteins: occurrence, functions, and significance for biotechnological applications. Appl Microbiol Biotechnol. 2002;58:265–274. doi: 10.1007/s00253-001-0869-4. [DOI] [PubMed] [Google Scholar]
- 41.Genee HJ, et al. Functional mining of transporters using synthetic selections. Nat Chem Biol. 2016;12:1015–1022. doi: 10.1038/nchembio.2189. [DOI] [PubMed] [Google Scholar]
- 42.Rodionov DA, et al. A novel class of modular transporters for vitamins in Prokaryotes. J Bacteriol. 2009;191:42–51. doi: 10.1128/JB.01208-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Allen RH, Stabler SP. Identification and quantitation of cobalamin and cobalamin analogues in human feces. Am J Clin Nutr. 2008;87:1324–1335. doi: 10.1093/ajcn/87.5.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seth EC, Taga ME. Nutrient cross-feeding in the microbial world. Front Microbiol. 2014;5:350. doi: 10.3389/fmicb.2014.00350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Degnan PH, Barry NA, Mok KC, Taga ME, Goodman AL. Human gut microbes use multiple transporters to distinguish vitamin B12 analogs and compete in the gut. Cell Host Microbe. 2014;15:47–57. doi: 10.1016/j.chom.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Caporaso JG, et al. Moving pictures of the human microbiome. Genome Biol. 2011;12:R50. doi: 10.1186/gb-2011-12-5-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gajer P, et al. Temporal dynamics of the human vaginal microbiota. Sci Transl Med. 2012;4:132ra52. doi: 10.1126/scitranslmed.3003605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Merchant SS, Helmann JD. Elemental economy: microbial strategies for optimizing growth in the face of nutrient limitation. Adv Microb Physiol. 2014;60:91–210. doi: 10.1016/B978-0-12-398264-3.00002-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bren A, Hart Y, Dekel E, Koster D, Alon U. The last generation of bacterial growth in limiting nutrient. BMC Syst Biol. 2013;7:27. doi: 10.1186/1752-0509-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shou W, Ram S, Vilar JMG. Synthetic cooperation in engineered yeast populations. Proc Natl Acad Sci USA. 2007;104:1877–1882. doi: 10.1073/pnas.0610575104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schink B. Synergistic interactions in the microbial world. Antonie Van Leeuwenhoek. 2002;81:257–261. doi: 10.1023/a:1020579004534. [DOI] [PubMed] [Google Scholar]
- 52.Jiang X, et al. Impact of spatial organization on a novel auxotrophic interaction among soil microbes. 2017 doi: 10.1038/s41396-018-0095-z. Preprint at bioRxiv. 195339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Harcombe WR, et al. Metabolic resource allocation in individual microbes determines ecosystem interactions and spatial dynamics. Cell. 2010;7:1104–1115. doi: 10.1016/j.celrep.2014.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thaiss CA, et al. Microbiota diurnal rhythmicity programs host transcriptome oscillations. Cell. 2016;167:1495–1510. doi: 10.1016/j.cell.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 55.Smits SA, et al. Seasonal cycling in the gut microbiome of the Hadza hunter-gatherers of Tanzania. Science. 2017;357:802–806. doi: 10.1126/science.aan4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Greenhalgh K, Meyer KM, Aagaard KM, Wilmes P. The human gut microbiome in health: establishment and resilience of microbiota over a lifetime. Environ Microbiol. 2016;18:2103–2116. doi: 10.1111/1462-2920.13318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thaiss CA, et al. Persistent microbiome alterations modulate the rate of post-dieting weight regain. Nature. 2016;540:544–551. doi: 10.1038/nature20796. [DOI] [PubMed] [Google Scholar]
- 58.Sonnenburg ED, et al. Diet-induced extinctions in the gut microbiota compound over generations. Nature. 2016;529:212–215. doi: 10.1038/nature16504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Collins J, et al. Dietary trehalose enhances virulence of epidemic Clostridium difficile. Nature. 2018;553:291–294. doi: 10.1038/nature25178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kartal B, Kuenen JG, Van Loosdrecht MCM. Sewage treatment with anammox. Science. 2010;328:702–703. doi: 10.1126/science.1185941. [DOI] [PubMed] [Google Scholar]
- 61.Lawson CE, et al. Metabolic network analysis reveals microbial community interactions in anammox granules. Nat Commun. 2017;8:15416. doi: 10.1038/ncomms15416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kumar R, et al. Identification of donor microbe species that colonize and persist long term in the recipient after fecal transplant for recurrent Clostridium difficile. Biofilms Microbiomes. 2017;3:1–12. doi: 10.1038/s41522-017-0020-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cuskin F, et al. Human gut Bacteroidetes can utilize yeast mannan through a selfish mechanism. Nature. 2016;517:165–169. doi: 10.1038/nature13995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee SM, et al. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature. 2013;501:426–429. doi: 10.1038/nature12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Riddle MS, Connor BA. The traveling microbiome. Curr Infect Dis Rep. 2016;18:29. doi: 10.1007/s11908-016-0536-7. [DOI] [PubMed] [Google Scholar]
- 66.Coyte KZ, Schluter J, Foster KR. The ecology of the microbiome: Networks, competition, and stability. Science. 2015;350:663–666. doi: 10.1126/science.aad2602. [DOI] [PubMed] [Google Scholar]
- 67.Sommer F, Anderson JM, Bharti R, Raes J, Rosenstiel P. The resilience of the intestinal microbiota influences health and disease. Nat Rev Microbiol. 2017;15:630–638. doi: 10.1038/nrmicro.2017.58. [DOI] [PubMed] [Google Scholar]
- 68.Gunderson LH. Ecological resilience - In theory and application. Annu Rev Ecol Syst. 2000;31:425–439. [Google Scholar]
- 69.Hoek TA, et al. Resource availability modulates the cooperative and competitive nature of a microbial cross-feeding mutualism. PLoS Biol. 2016;14:e1002540. doi: 10.1371/journal.pbio.1002540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Köpke M, Straub M, Dürre P. Clostridium difficile is an autotrophic bacterial pathogen. PLoS ONE. 2013;8:e62157. doi: 10.1371/journal.pone.0062157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Byrd AL, et al. Staphylococcus aureus and Staphylococcus epidermidis strain diversity underlying pediatric atopic dermatitis. Sci Transl Med. 2017;9:eaal4651. doi: 10.1126/scitranslmed.aal4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nakatsuji T, et al. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci Transl Med. 2017;9:eaah4680. doi: 10.1126/scitranslmed.aah4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bosi E, et al. Comparative genome-scale modelling of Staphylococcus aureus strains identifies strain-specific metabolic capabilities linked to pathogenicity. Proc Natl Acad Sci USA. 2016;113:E3801–E3809. doi: 10.1073/pnas.1523199113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ribet D, Cossart P. How bacterial pathogens colonize their hosts and invade deeper tissues. Microbes Infect. 2015;17:173–183. doi: 10.1016/j.micinf.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 75.Byrd BAL, Segre JA, Koch R. Adapting Koch’s postulates. Science. 2016;351:224–226. doi: 10.1126/science.aad6753. [DOI] [PubMed] [Google Scholar]
- 76.Franzosa EA, et al. Sequencing and beyond: integrating molecular ‘omics’ for microbial community profiling. Nat Rev Microbiol. 2015;13:360–372. doi: 10.1038/nrmicro3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Parks DH, et al. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat Microbiol. 2017;2:1533–1542. doi: 10.1038/s41564-017-0012-7. [DOI] [PubMed] [Google Scholar]
- 78.Zuñiga C, Zaramela L, Zengler K. Elucidation of complexity and prediction of interactions in microbial communities. Microb Biotechnol. 2017;10:1500–1522. doi: 10.1111/1751-7915.12855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Markowitz VM, et al. IMG: the integrated microbial genomes database and comparative analysis system. Nucleic Acids Res. 2012;40:115–122. doi: 10.1093/nar/gkr1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tan J, Zuniga C, Zengler K. Unraveling interactions in microbial communities — from co-cultures to microbiomes. J Microbiol. 2015;53:295–305. doi: 10.1007/s12275-015-5060-1. [DOI] [PubMed] [Google Scholar]
- 81.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mallick H, et al. Experimental design and quantitative analysis of microbial community multiomics. Genome Biol. 2017;18:228. doi: 10.1186/s13059-017-1359-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Turnbaugh PJ, et al. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Thompson LR, et al. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature. 2017;551:457. doi: 10.1038/nature24621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang M, et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat Biotechnol. 2016;34:828–837. doi: 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Durot M, Bourguignon P, Schachter V. Genome-scale models of bacterial metabolism: reconstruction and applications. FEMS Microbiol Rev. 2009;33:164–190. doi: 10.1111/j.1574-6976.2008.00146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bordbar A, Monk JM, King ZA, Palsson BO. Constraint-based models predict metabolic and associated cellular functions. Nat Rev Genet. 2014;15:107–120. doi: 10.1038/nrg3643. [DOI] [PubMed] [Google Scholar]
- 88.Shoaie S, et al. Quantifying diet-induced metabolic changes of the resource. Cell Metab. 2015;22:320–331. doi: 10.1016/j.cmet.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 89.Song HS, Cannon W, Beliaev A, Konopka A. Mathematical modeling of microbial community dynamics: A methodological review. Processes. 2014;2:711–752. [Google Scholar]
- 90.Xiao Y, et al. Mapping the ecological networks of microbial communities. Nat Commun. 2017;8:2042. doi: 10.1038/s41467-017-02090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.An G, Mi Q, Dutta-Moscato J, Vodovotz Y. Agent-based models in translational systems biology. Wiley Interdiscip Rev Syst Biol Med. 2009;1:159–171. doi: 10.1002/wsbm.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kaplan H, Hutkins RW. Metabolism of fructooligosaccharides by Lactobacillus paracasei 1195. Appl Environ Microbiol. 2003;69:2217–2222. doi: 10.1128/AEM.69.4.2217-2222.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Thakur K, Tomar SK, De S. Lactic acid bacteria as a cell factory for riboflavin production. Microb Biotechnol. 2015;9:441–451. doi: 10.1111/1751-7915.12335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Terwilliger A, et al. Bacillus anthracis overcomes an amino acid auxotrophy by cleaving host serum proteins. J Bacteriol. 2015;197:2400–2411. doi: 10.1128/JB.00073-15. [DOI] [PMC free article] [PubMed] [Google Scholar]