Abstract
The scaled recombination parameter ρ is one of the key parameters, turning up frequently in population genetic models. Accurate estimates of ρ are difficult to obtain, as recombination events do not always leave traces in the data. One of the most widely used approaches is composite likelihood. Here we show that popular implementations of composite likelihood estimators can often be uniformly improved by optimizing the trade-off between bias and variance. The amount of possible improvement depends on parameters such as the sequence length, the sample size, and the mutation rate, and can be considerable in some cases. It turns out that ABC, with composite likelihood as a summary statistic, also leads to improved estimates, but now in terms of the posterior risk. Finally, we demonstrate a practical application on real data from Drosophila.
1. Introduction
In diploid organisms, homologous chromosomes are paired during meiosis. In this process, pieces of DNA are frequently exchanged between the chromosomes, leading to a mixture of maternal and paternal genetic information. This process is called recombination. By producing new combinations of alleles and breaking up the linkage between genes, recombination increases the variation in a population, making it an important evolutionary force. Recombination rates vary between species and across the genome. Knowing the respective recombination rates is of great importance in several situations. It is e.g. necessary for understanding the process of recombination itself. At the population level, knowing the population recombination rate ρ = 4Ner, with Ne being the effective population size and r being the recombination rate per base pair (bp), is important for the analysis of population genetic data. For instance, as recombination reduces the amount of linkage disequilibrium (LD) between segregating sites (Hill and Robertson, 1986) and positive selection tends to produce areas of high LD, recombination helps to localize signals of selection in DNA sequence data, see e.g. Sabeti et al. (2002, 2006); O’Reilly et al. (2008).
However, obtaining accurate estimates of the recombination rate is challenging as not all historical recombination events leave traces in a corresponding sample of DNA sequences. Even the best estimation methods available provide estimates that exhibit a considerable amount of uncertainty. The literature suggests several different methods for estimating ρ, including the computation of lower bounds on the number of recombination events (Hudson and Kaplan, 1985; Wiuf, 2002; Myers and Griffiths, 2003), the calculation of moments or other summary statistics (Hudson, 1987; Wall, 2000; Batorsky et al., 2011), and regression based methods (Lin et al., 2013). Approaches based on maximum likelihood are used commonly as well.
Due to the high computational effort, these methods are often either approximate likelihood methods (Hey and Wakeley, 1997; Hudson, 2001; Fearnhead and Donnelly, 2002; McVean et al., 2002; Li and Stephens, 2003; Wall, 2004) or, if they are full likelihood methods, they still approximate the likelihood e.g. via importance sampling or Markov chain Monte Carlo (MCMC) algorithms (Griffiths and Marjoram, 1996; Kuhner et al., 2000; Fearnhead and Donnelly, 2001).
Hobolth and Jensen (2014) describe a method to estimate the recombination rate based on Markov approximations to the tree building process of the ancestral recombination graph (McVean and Cardin, 2005; Marjoram and Wall, 2006). Other methods for estimating recombination rates use approximate Bayesian computation (ABC), which is a Bayesian method that avoids the calculation of a likelihood function, e.g. Lopes et al. (2014); Arenas et al. (2015). For an overview on ABC see for instance Beaumont et al. (2002).
Some of the methods are implemented as software packages such as LDhat (McVean and Auton, 2007) or LDhelmet (Chan et al., 2012). These programs use the composite likelihood method of Hudson (2001) or, more precisely, a modification of the latter by McVean et al. (2002) implementing a finite-sites mutation model. A good overview on composite likelihood methods is provided by Varin et al. (2011). LDhat and LDhelmet also permit to estimate recombination rates that vary across the genome by combining composite likelihood with a Bayesian approach using a reversible jump Markov chain Monte Carlo (rjMCMC) algorithm (Green, 1995).
In this paper, we investigate whether there is room for improving composite likelihood estimators. As a measure of performance for an estimator ρ̃ of ρ, we focus on the mean squared error
The MSE provides the expected squared distance between true parameter and its estimate and may be decomposed into the sum of variance and squared bias:
| (1) |
Estimators that can be uniformly improved with respect to the MSE are called inadmissible in the statistical literature, see e.g. Berger (2013). In classical statistics, shrinkage sometimes leads to such a uniform improvement, see e.g. Gruber (1998). In a population genetic context, Futschik and Gach (2008) showed that Watterson’s estimator of the scaled mutation parameter θ is inadmissible, and provided a uniformly better estimator by shrinkage, i.e. multiplying the original estimator with a suitable constant c < 1. In subsequent sections, we show that such uniform improvements are often also possible for composite likelihood estimators of ρ.
For our practical computations, we will use the composite likelihood estimator implemented in LDhelmet, and also consider an older estimator provided by LDhat. Our focus is on stretches of DNA with constant recombination. For recombination landscapes the approach would need to be applied separately on each segment with distinct recombination rate. We do this when we apply our method to real data in section 5.
The remainder of this paper is structured as follows: The composite likelihood method of McVean et al. (2002) for estimating ρ and an alternative version implemented in LDhelmet are explained in section 2, as well as our method of improvement. In section 3, we explore the improvement of two implementations of the composite likelihood method by LDhat and LDhelmet and present simulations and results. We briefly discuss an alternative approach for improving the estimation of ρ based on ABC in section 4. An example for the application of our method to real data is shown in section 5. Section 6 concludes this paper by a discussion and an outlook.
2. Estimating the population recombination rate by composite likelihood and possible improvements
In this section we explain how composite likelihood has been implemented for estimating ρ. Further, we explain our approach to improve composite likelihood estimates.
2.1. A composite likelihood estimate of ρ
The composite likelihood method of McVean et al. (2002) extends the method of Hudson (2001) by permitting repeated mutations to occur at a site during the history of a sample. However, these (reversible) mutations are assumed to lead to no more than two alleles segregating. The estimation process is carried out in four steps: At first the population mutation rate θ per site is estimated. Hereby an approximate finite-sites version of Watterson’s estimator is used. The second step is to classify every pair of segregating sites into sets of equivalent configurations. In the next step, the likelihood of each of these sets is estimated under the value of θ from step 1 and a range of values for ρ using the importance sampling method of Fearnhead and Donnelly (2001). At last, ρ is estimated for the whole sequence by combining the likelihoods from all pairs of segregating sites. The estimated ρ is the value with the highest composite log likelihood (McVean et al., 2002).
The described method is implemented in the software packages LDhat and LDhelmet, with the latter package implementing some more accurate approximations (Chan et al., 2012). The improvement in accuracy results for instance from solving a system of recursion equations for the computation of the pairwise likelihoods instead of applying importance sampling, and the implementation of a quadra-allelic mutation model instead of a biallelic one.
In the following, we denote the estimator provided by the function pairwise in LDhat by ρ̂. Furthermore ρ̆ signifies the estimator implemented as max_lk in LDhelmet. LDhelmet can only estimate the crossing over type of recombination (see e.g. Cromie and Smith (2007)), while LDhat contains also an option to estimate the rate of gene conversion. Here, our focus is on the estimation of the rate of crossing over.
2.2. Improved estimation of ρ
In order to improve the estimators of ρ introduced in subsection 2.1 we will optimize the trade-off between bias and variance. This is related to the statistical concept of shrinkage, see Gruber (1998), and Bayesian statistics. With shrinkage, bias is introduced for the sake of reducing the variance. If the gain in variance is larger than the loss due to additional bias, this leads to an improvement in terms of the MSE. A uniform improvement over the whole parameter range, however, can be achieved only under certain circumstances. A famous example is the James-Stein estimator of the mean of a multivariate normal distribution (Stein, 1956).
Bayes estimators on the other hand are constructed to minimize the weighed (with respect to the prior distribution) integral of an error measure such as the MSE.
It will turn out that our considered estimators are biased already. In order to optimize the trade-off between bias and variance, the required correction may therefore either lead to a decrease or an increase in bias, depending on the relative magnitudes of the two sources of error.
As no explicit formulas are available for the bias and the variance of composite likelihood estimators of ρ, we model bias and variance using regression based on simulated data. As will be shown in section 3.1, the following general model captures bias and variance of both ρ̂ and ρ̆ very accurately.
| (2) |
| (3) |
We now investigate a generic rescaled estimator ρ̃* := c · ρ̃ with a positive constant c. Straightforward calculations lead to
| (4) |
and
| (5) |
Hence
| (6) |
In order to obtain an estimator that improves ρ̃, we minimize MSEρ(c · ρ̃) in c. This leads to
| (7) |
This constant cannot directly be used for improving ρ̃ as it depends on the unknown ρ. One possible strategy would be to insert ρ̃ instead of ρ in (7). This approach worked reasonably well for Watterson’s estimator of θ in Futschik and Gach (2008), but did not always lead to a uniformly improved estimator.
Alternatively, with S denoting the set of possible values of ρ (i.e. the parameter space), take c* = c(ρ*) satisfying
as modifying constant with ρ̃*. This will lead to a uniform improvement, if either supρ∈S [c(ρ)] < 1 or infρ∈S [c(ρ)] > 1. Otherwise we get c(ρ*) = 1, and the original estimator remains unchanged, i.e. ρ̃* = ρ̃.
3. Application to ρ̂ and ρ̆
In the following, we explore bias and variance of ρ̂ and ρ̆ and compare these estimators in terms of the MSE. Then we apply our method of improvement.
For the simulations concerning ρ̂ and ρ̆, we used the following simulation setup: For specified values of ρ, DNA sequence data was generated by the program msms (Ewing and Hermisson, 2010). The output of msms was transformed into fasta files via ms2dna (Haubold and Pfaffelhuber, 2013). For each of these fasta files, ρ was estimated by ρ̆ or ρ̂. Our analysis was then performed in R (R-Core-Team, 2013).
3.1. Variance, bias, and MSE of ρ̂ and ρ̆
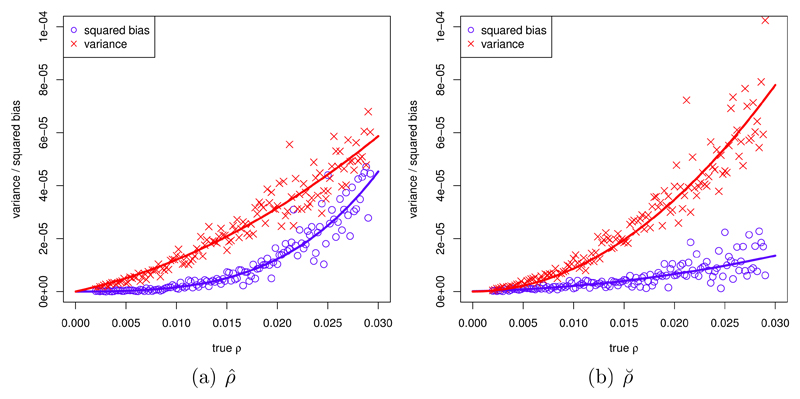
Using extensive simulation runs, we explored variance and bias of ρ̂ and ρ̆. Figure 1 provides a typical example.
Figure 1.
Squared bias and variance of ρ̂ and ρ̆ in 1/bp2; true ρ in 1/bp. Each plot symbol is based on 100 independent simulation runs (missing values that occured were removed), the curves display the resulting estimated regression relationships. Model parameters: θ=0.01/bp, n=20, l=5001 bp. Estimated regression coefficients (see equations (8), (9), (14), (15)): b1 = −7.67 · 10−2, c1 = −4.92, b2 = 8.24 · 10−4, c2 = 3.78 · 10−2; a3 = −3.31 · 10−4, b3 = −1.12 · 10−1, c4 = 8.67 · 10−2.
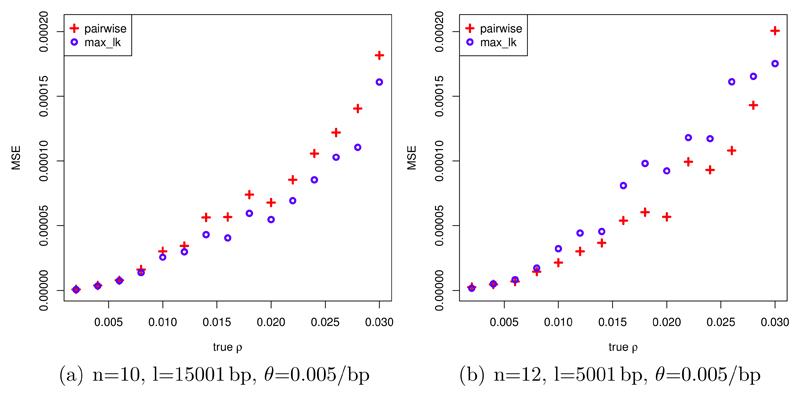
Using our simulations, figures 2 (a) and (b) show the MSE of ρ̂ and ρ̆ as functions of the true recombination rate ρ for various combinations of sample size (n), sequence length (l) and mutation rate (θ). For (a), with n=10, l=15001 bp, and θ=0.005/bp, ρ̆ performs uniformly better than ρ̂ in terms of the MSE, while under (b), where n=12, l=5001 bp, θ=0.005/bp, ρ̂ outperforms ρ̆ for almost all considered values of ρ.
Figure 2.
MSE of ρ̂ (pairwise) and ρ̆ (max_lk) for different values of ρ with different values of the parameters sample size (n), sequence length (l) and θ in (a) and (b); calculation of MSE from 50 independent simulations per value of ρ.
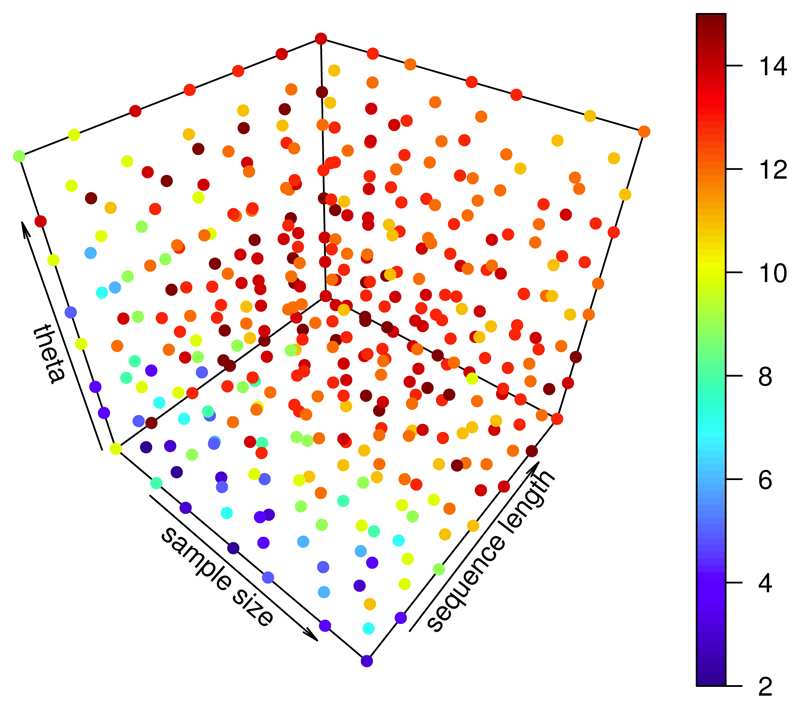
Over a large range of configurations of the parameters n, l and θ, figure 3 provides an overall picture of the relative performance of ρ̂ and ρ̆ in terms of the MSE. For each scenario we considered 15 different true ρ values. The color coded score shows for how many of these 15 values the MSE of ρ̆ is smaller than the MSE of ρ̂. Apart from the situations where the sequence length is very short and at the same time θ is small, the MSE of ρ̆ is smaller than the MSE of ρ̂ for most or sometimes all considered values of ρ. Thus, for the scenarios we consider, ρ̆ outperforms ρ̂ in the majority of cases.
Figure 3.
Each dot displays the number of cases out of 15 values of ρ ∈ [0.002, 0.03]/bp, for which the MSE of ρ̆ is smaller than that of ρ̂. Parameter ranges: θ : (0.005/bp - 0.023/bp), n: (7 - 22), l: (3001 bp - 17501 bp); MSE estimated from 47 independent simulations per value of ρ.
An estimated value θ̂ of the population mutation rate θ needs to be provided with ρ̂ and ρ̆. According to our observation, inaccuracies in θ̂ affect the estimators of ρ only slightly. Indeed, the differences in MSE(ρ̆) and MSE(ρ̂) tend to be negligible when using Watterson’s estimator, compared to the improved version proposed in Futschik and Gach (2008).
3.2. Improving ρ̆
Using regression with our simulated data, we estimated bias and variance of ρ̆. As some of the estimated coefficients did not turn out to be significantly different from zero, we dropped the corresponding terms and simplified our models (4)–(6). This led to
| (8) |
| (9) |
| (10) |
We first corrected for the constant bias by substracting the intercept a3, resulting in the estimator ρ̆2 = ρ̆ – a3. The optimal modifying constant for ρ̆2 turns then out to be
| (11) |
which is independent of ρ. The approximate computation of cm uses estimates for the regression coefficients in (8) and (9).
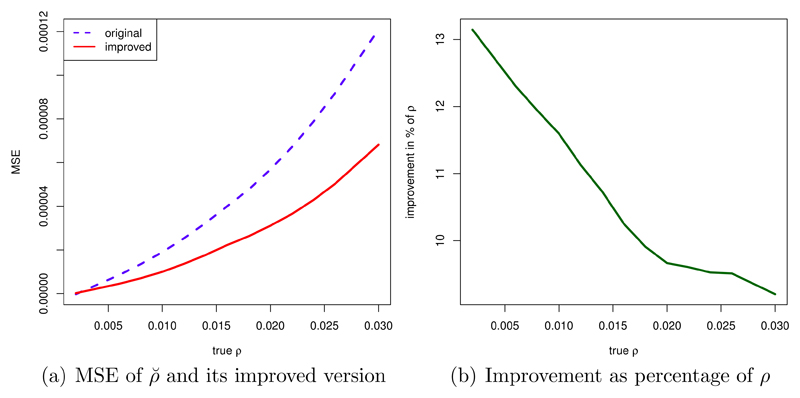
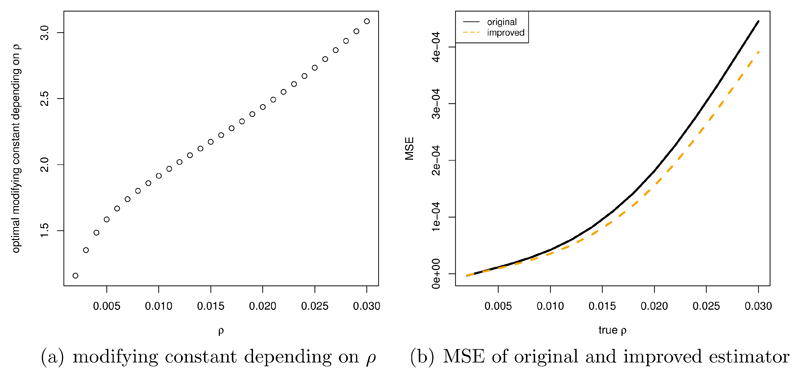
As an example, consider a model with θ=0.02/bp, n=10, l=15001 bp. Figure 4 (a) plots the MSE of the original estimator ρ̆, as well as the improved version ρ̆*. The improvement as percentage of ρ (shown in figure 4 (b)) is noticeable under this scenario.
Figure 4.
(a): MSE of ρ̆ (original and improved); (b) improvement as percentage of ρ. Parameters: θ=0.02/bp, n=10, l=15001 bp; results are based on 75 simulations per value of ρ for estimating cm, and 75 independent simulations per value of ρ to obtain the MSE.
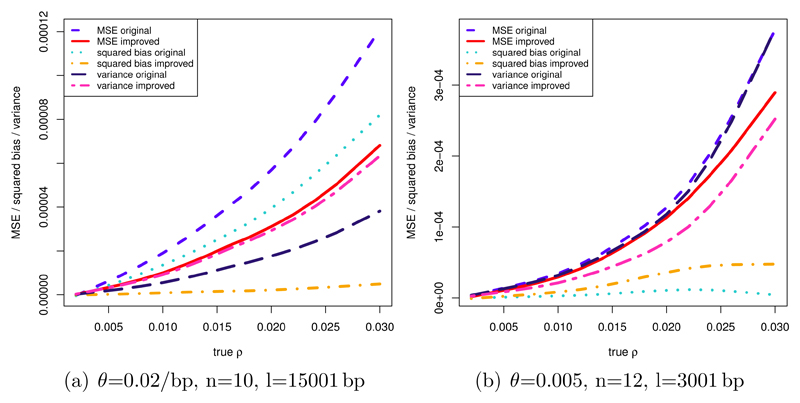
With θ=0.02/bp, n=10, l=15001 bp, the improved MSE results from a large bias reduction. The variance increases, but to a smaller extent, see figure 5 (a). Here cm = 1.289. For the parameters θ=0.005/bp, n=12, l=3001 bp cm = 0.816, and the MSE is improved due to a reduction in the variance, while the bias increases, see figure 5 (b).
Figure 5.
MSE, variance and squared bias in (1/bp)2 of original and improved estimators for different scenarios. True ρ in 1/bp. Calculation of modifying constant based on 75 simulations per value of true ρ, calculation of MSE, variance, and bias based on 75 different simulations per value of true ρ. (a) θ=0.02/bp, n=10, l=15001 bp. (b) θ=0.005/bp, n=12, l=3001 bp.
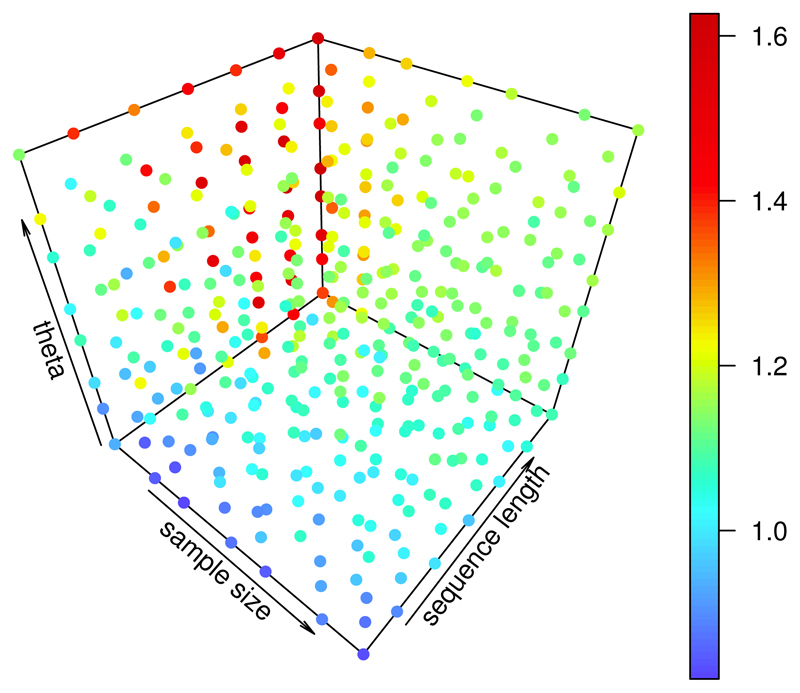
Figure 6 shows cm (color coded) depending on the model parameters. Overall, the constant increases with θ and the sequence length l, and decreases with the sample size n.
Figure 6.
Dependence of the optimal modifying constant (color coded) on the parameters θ (0.005/bp - 0.023/bp), n (7 - 22) and l (3001 bp - 17501 bp); calculation of MSE from 47 independent simulations per value of ρ.
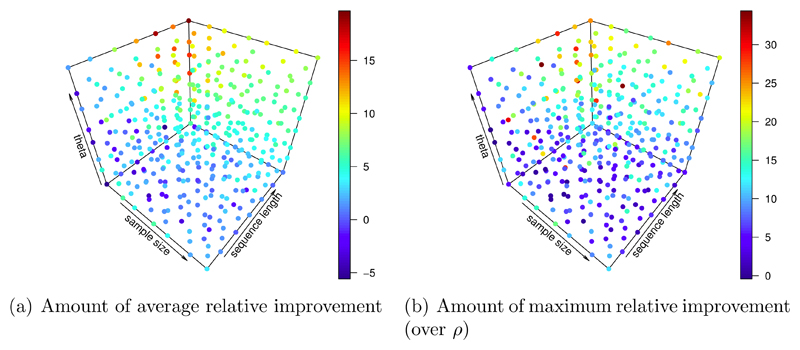
The corresponding average improvement (over all considered values of ρ) achieved relative to the true value of ρ is presented in figure 7 (a). Figure 7 (b) shows the maximum relative improvement over ρ. In some cases the achieved gains are large. We observed such cases in particular when θ and the sequence length are large and the sample size is small.
Figure 7.
Amount of relative improvement averaged over ρ (a), and maximum relative improvement (b) of ρ̆ in percent (color coded). The parameter ranges θ: (0.005/bp - 0.023/bp), n: (7 - 22), and l: (3001 bp - 17501 bp) were considered. Simulation effort: 24 simulations per value of ρ for calculating cm, 23 simulations per value of ρ for the MSE estimates.
Under some parameter combinations, the estimated shrinkage constants are nearly one, and there is not much room for uniform improvement. The original ρ̆ and ρ̆* are then nearly identical, and the noise in the estimated regression coefficients may occasionally even lead to a marginal worsening. This could be avoided by setting cm = 1, if its estimated value differs by less than ϵ from one, with ϵ denoting a bound on the simulation noise.
As it would be tedious to carry out a large amount of simulations to obtain modifying constants for each new model configuration, we fitted a regression model in order to quantify the dependence of the optimal modifying constant cm and the bias correction term a3 on the parameters sample size n, sequence length l, and mutation rate θ. By exploiting smoothness, this formula often (but not always) provides slightly more accurate estimates than those we obtained from individual simulations under single parameter combinations. This is since the smoothing reduces the random noise in the estimated coefficients. The following model provides a good fit.
| (12) |
For a3, we got
| (13) |
3.3. Improving ρ̂
As with ρ̆, there is also room for improving ρ̂. Our simulated data suggest the following formulas, describing the dependence of bias, variance, and MSE on ρ.
| (14) |
| (15) |
| (16) |
Since the nonzero coefficients differ for ρ̂ and ρ̆, the modifying constant has a different structure now:
| (17) |
Depending on ρ, cm(ρ) may take values both smaller and larger than one under some scenarios. In such situations we work with modifying constants cm(ρ̂). However, for small values of l this approach works less satisfactory.
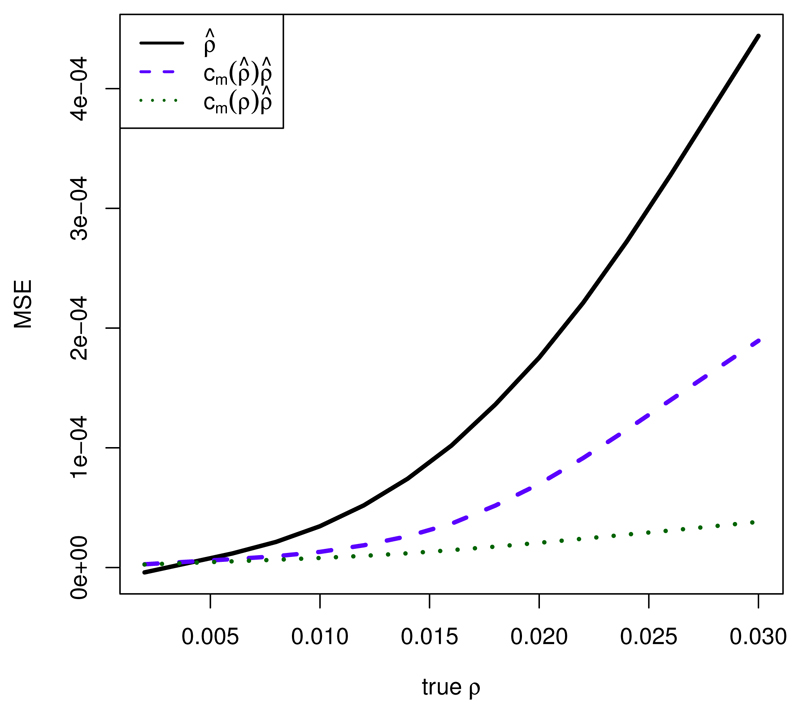
We first consider again the scenario θ = 0.02/bp, n = 10, and l = 15001 bp, using the same simulated data as with ρ̆. In Figure 8, the MSE is shown both for ρ̂ as well as for cm(ρ̂)ρ̂. Except for the smallest values of ρ, MSE(cm(ρ̂)ρ̂) < MSE(ρ̂).
Figure 8.
Dependence of MSE on ρ for ρ̂, cm(ρ̂)ρ̂, and cm(ρ)ρ̂; θ = 0.02/bp, n = 10, l = 15001 bp; 25 independent simulations per value of ρ for calculating the optimal modifying constant, 25 different independent simulations per value of ρ for calculation of the MSE.
Not unexpectedly, the errors MSE(cm(ρ)ρ̂) would be even smaller with the theoretically optimal cm(ρ). But this does not help in practice, as the true ρ will be unknown.
Under the scenario θ = 0.008/bp, n = 7, l = 15001 bp, the optimal modifying constant is monotonically increasing in ρ and always larger than one. When using the minimum of cm(ρ) over the considered range of ρ, we obtain a uniformly improved MSE. Figure 9 (a) displays cm(ρ) depending on ρ, and figure 9 (b) shows the MSE depending on ρ for the original and the improved estimator with cm = 1.158, the optimal modifying constant for ρ = 0.002/bp.
Figure 9.
θ = 0.008/bp, n = 7, l = 15001 bp; 25 simulations per value of ρ for calculation of the modifying constant, 25 simulations per value of ρ for calculation of the MSEs. (a) Optimal modifying constant depending on ρ; ρ in 1/bp. (b) MSE in (1/bp)2 of original and improved ρ̂ estimator with cm = 1.158 for ρ in 1/bp.
4. Approximate Bayesian computation
Approximate Bayesian Computation (ABC) is a method to approximate the posterior distribution of one or more parameters of interest when no closed form expression is available for the likelihood. According to Bayes’ rule it holds that
| (18) |
where ℙ(ρ|D) is the posterior probability of the parameter ρ given the data D, ℙ(D|ρ) is the likelihood, ℙ(ρ) the prior, and ℙ(D) = ∫ ℙ(D|ρ)ℙ(ρ) dρ. With ABC, a sample from an approximate posterior is simulated without directly using the likelihood. Instead, a sample is simulated under parameters randomly drawn from the prior distribution.
Parameter values that lead to simulated data close to the observed data D are taken as sample of the posterior distribution. The comparison of the simulated data sets with the observed one is carried out in terms of low dimensional but informative summary statistics. For our calculations we used the rejection algorithm of Pritchard et al. (1999), as well as the regression algorithm of Beaumont et al. (2002). Both algorithms are provided in the R-software package abc (Csillery et al., 2012). While the rejection algorithm is the most basic version of ABC, a regression correction of the accepted parameter values usually gives a better approximation to the posterior.
4.1. Our application of ABC
ABC is often used with easy to compute summary statistics for the unknown parameters. In Lopes et al. (2014) for instance, ρ (as well as θ and the non-synonymous synonymous rate ratio) is inferred from summary statistics like the number of segregating sites, moments of the heterozygosity, and several other measures. Here, we used only ρ̆ as a single but sophisticated summary statistic. In a different context, the combination of ABC with a composite likelihood approach has been investigated by Ruli et al. (2015).
Bayesian estimators are known to minimize the posterior risk, which is in our case the integrated MSE weighted with the prior. Being an approximate approach, ABC may be expected to lead to estimators that are not too far from optimizing the posterior risk.
We noticed that the performance of ABC was slightly better when we used an equidistant grid of values of ρ instead of a sample from the (uniform) prior. This effect has been observed also in the context of Quasi - Monte Carlo methods, see e.g. Caflisch (1998). In this spirit, we took parameter values uniformly on a narrow equidistant grid, and generated data under these parameter values. We then used ρ̆ on each data set to obtain simulated summary statistics. We used the same simulated data as in in section 3.1. In particular, we used 100 collections of fasta files for 141 equidistant values of ρ between 0.002/bp and 0.03/bp. As with cross-validation, each fasta file was once considered as the observed data set while the remaining fasta files were treated as a sample from the prior distribution. By iterating over all possible “real data” sets, we estimated bias and variance of the ABC posterior mean and median. Missing values were removed which led to slightly fewer than 100 simulations for some values of ρ. For our computations, we used the package abc in R.
4.2. Results for ABC
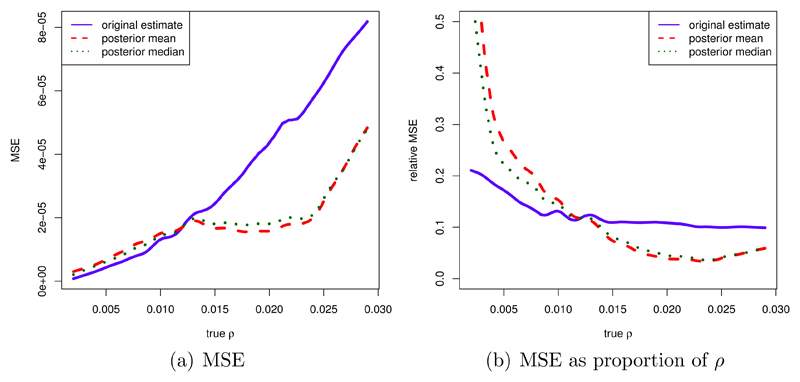
The regression algorithm outperformed the rejection algorithm (not shown here). After testing different tolerance levels, we decided on a tolerance level of 40 %, i.e. 40 % of the parameter values sampled from the prior have been accepted for the posterior. Figure 10 (a) shows the MSE depending on the true ρ for the original ρ̆ estimator as well as for the ABC based estimator. The MSE as a proportion of the true value of ρ, i.e. the MSE divided by ρ2, is displayed in figure 10 (b). With ABC, we obtain considerably improved MSE values when the true recombination rate is larger than approximately 0.015/bp, while for smaller recombination rates the MSE increases by a small amount. When measured as a proportion of ρ, this increase can be quite large, however, for small recombination rates. As ABC estimators, both posterior mean and posterior median gave quite similar results.
Figure 10.
MSE of original and improved estimates in (1/bp)2 (a) and MSE divided by ρ2 (b) for ρ in 1/bp. Calculation based on 100 simulations per value of ρ, tolerance of 40 % in ABC.
5. Example on real data
For ten haploid sequenced individuals of a Drosophila melanogaster population from Raleigh we looked at sequence data from the X chromosome. The data is available at http://pooldata.genetics.wisc.edu/dgrp_sequences.tar.bz2, http://johnpool.net/genomes.html. We considered sequentially 1000 pieces of 10Kb length and used ρ̆ to estimate for each piece a constant recombination rate. For θ we used 0.008/bp, as in Chan et al. (2012), where ρ was estimated for the same Drosophila population.
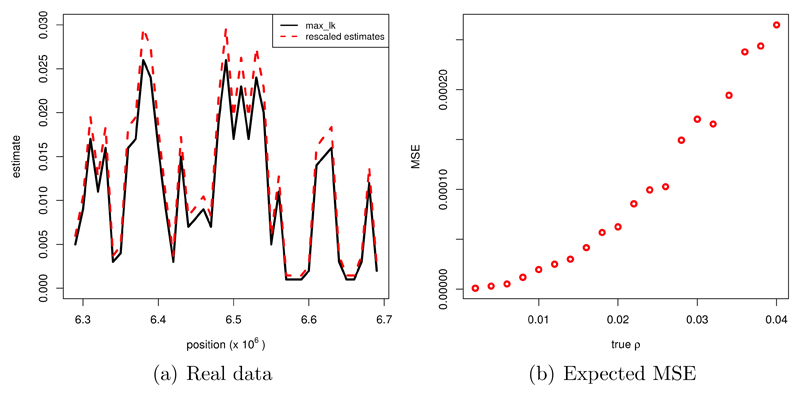
We calculated the optimal modifying constant cm and the constant term of the bias a3 for the underlying parameters according to (12) and (13) and obtained cm = 1.13, a3 = −2.83 · 10−4. We substracted a3 from each estimate and multiplied the result by cm. As cm is larger than 1, we increased the estimates by our method. In figure 11 (a) we show the original and the modified estimates for a range of values of ρ.
Figure 11.
(a) Original and rescaled estimates for a certain range of values of ρ; pieces of 10Kb length for sequence data of the X chromosome from a Drosophila melanogaster population (DGRP from Raleigh); 10 haploid individuals, θ=0.008/bp. (b) MSE in (1/bp)2 against ρ in 1/bp; n=10, l=10000 bp, θ=0.008/bp, calculation based on 100 simulated values per value of true ρ.
For understanding which accuracy can be expected, we show in figure 11 (b) the MSE plotted against the true value of ρ for θ=0.08/bp, n=10 and l=10 Kb.
The population recombination rate ρ is a parameter often needed in population genetic inference. More accurate estimates of ρ can therefore influence also the quality of estimation of other population genetic parameters, and may be beneficial for detecting signs of selection in population genetic data.
6. Discussion
We proposed an approach for improving composite likelihood estimators of ρ. In particular, we looked at versions of the composite likelihood method of Hudson (2001), as implemented in the software packages LDhat and LDhelmet (ρ̂ and ρ̆). As our simulations show, even these sophisticated widely used estimators still exhibit room for improvement with relatively little effort.
Although the rescaling factors used are not exact but estimated from simulations, our approach usually led to improved estimators, often considerably. Under some parameter configurations however, the original and the modified estimators were nearly identical. In such cases, the estimated rescaling constants was very close to one, and the estimation noise influenced whether a marginal improvement was seen or not.
In some cases the optimal rescaling factor cm for ρ̂ turned out to be both larger and smaller than one, depending on the unknown value of ρ. In such cases, we inserted ρ̂ instead of ρ in the formula for cm. Apart from very small values of ρ, this approach also led to improved estimators.
In order to apply our proposed rescaled estimator without having to carry out simulations, we present a formula for computing the modifying constant over a wide range of sample sizes, mutation rates and sequence lengths. We make such a formula also available for a sometimes helpful bias correction by an additive constant. Additionally we provide an R package on http://www.jku.at/ifas/content/e98868/employee_groups_wiss98976/employees144622/subdocs237646/content296458/ModifyMaxLkAndPairwise.zip where these formulas as well as cm(ρ̂) are implemented.
Notice that the MSE of the modified version of ρ̂ is larger than that of the rescaled ρ̆ in most cases. Averaged over the 15 different values of ρ, the rescaled ρ̆ estimator outperformed the modified ρ̂ estimator in 98.7 % of the scenarios. Thus, in general, we recommend the use of the rescaled ρ̆ estimator ρ̆*.
Additionally we presented a Bayesian approach based on ABC with ρ̆ as summary statistic. The resulting estimator showed a reduced posterior risk with respect to the MSE.
We also applied our method to real data from a Drosophila melanogaster population (DGRP from Raleigh). To fit possible local variation in ρ, we divided the sequence into smaller intervals of equal length for which we estimated ρ separately.
In future work, we plan to derive a method to identify segments of constant recombination rates. There might be not only room for improving the estimators themselves, but also for improving the partitions.
Acknowledgments
We thank Christian Schlötterer and Claus Vogl for helpful comments.
The work has been carried out at the Vienna Graduate School of Population Genetics funded by the Austrian Science Fund (FWF): DK W1225-B20.
Footnotes
Author Disclosure Statement
The authors confirm that no competing financial interests exist.
Contributor Information
Kerstin Gärtner, Email: Kerstin.Gaertner@vetmeduni.ac.at, Vienna Graduate School of Population Genetics, Institut für Populationsgenetik, Vetmeduni Vienna, 1210 Vienna, Austria, Phone: +43 1 25077 4336, Fax: +43 1 25077 4390.
Andreas Futschik, Department of Applied Statistics, Johannes Kepler University, 4040 Linz, Austria, Phone: +43 732 2468 6822, Fax: +43 732 2468 6800.
References
- Arenas M, Lopes JS, Beaumont MA, et al. Codabc: A computational framework to coestimate recombination, substitution and molecular adaptation rates by approximate bayesian computation. Molecular biology and evolution. 2015;32(4):1109–1112. doi: 10.1093/molbev/msu411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batorsky R, Kearney MF, Palmer SE, et al. Estimate of effective recombination rate and average selection coefficient for HIV in chronic infection. Proceedings of the National Academy of Sciences. 2011;108(14):5661–5666. doi: 10.1073/pnas.1102036108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont MA, Zhang W, Balding DJ. Approximate Bayesian computation in population genetics. Genetics. 2002;162(4):2025–2035. doi: 10.1093/genetics/162.4.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger JO. Statistical decision theory and Bayesian analysis. Springer Science & Business Media; 2013. [Google Scholar]
- Caflisch RE. Monte carlo and quasi-monte carlo methods. Acta numerica. 1998;7:1–49. [Google Scholar]
- Chan AH, Jenkins PA, Song YS. Genome-wide fine-scale recombination rate variation in Drosophila melanogaster. PLoS Genet. 2012;8(12):e1003090. doi: 10.1371/journal.pgen.1003090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie GA, Smith GR. Branching out: meiotic recombination and its regulation. Trends in Cell Biology. 2007;17(9):448–455. doi: 10.1016/j.tcb.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Csillery K, Francois O, Blum MGB. abc: an R package for approximate Bayesian computation (ABC): R package: abc. Methods in Ecology and Evolution. 2012;3(3):475–479. [Google Scholar]
- Ewing G, Hermisson J. MSMS: a coalescent simulation program including recombination, demographic structure and selection at a single locus. Bioinformatics. 2010;26(16):2064–2065. doi: 10.1093/bioinformatics/btq322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearnhead P, Donnelly P. Estimating recombination rates from population genetic data. Genetics. 2001;159(3):1299–1318. doi: 10.1093/genetics/159.3.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearnhead P, Donnelly P. Approximate likelihood methods for estimating local recombination rates. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2002;64(4):657–680. [Google Scholar]
- Futschik A, Gach F. On the inadmissibility of Watterson’s estimator. Theoretical Population Biology. 2008;73(2):212–221. doi: 10.1016/j.tpb.2007.11.009. [DOI] [PubMed] [Google Scholar]
- Green PJ. Reversible jump Markov chain Monte Carlo computation and Bayesian model determination. Biometrika. 1995;82(4):711–732. [Google Scholar]
- Griffiths RC, Marjoram P. Ancestral inference from samples of DNA sequences with recombination. Journal of Computational Biology. 1996;3(4):479–502. doi: 10.1089/cmb.1996.3.479. [DOI] [PubMed] [Google Scholar]
- Gruber M. Improving Efficiency by Shrinkage: The James-Stein and Ridge Regression Estimators. Vol. 156 CRC Press; 1998. [Google Scholar]
- Haubold B, Pfaffelhuber P. ms2dna, v. 1.16: Convert simulated haplotype data to DNA sequences. 2013 Available at: http://guanine.evolbio.mpg.de/bioBox/
- Hey J, Wakeley J. A coalescent estimator of the population recombination rate. Genetics. 1997;145(3):833–846. doi: 10.1093/genetics/145.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill WG, Robertson A. Linkage disequilibrium in finite populations. Theoretical and Applied Genetics. 1986;38(6):226–231. doi: 10.1007/BF01245622. [DOI] [PubMed] [Google Scholar]
- Hobolth A, Jensen JL. Markovian approximation to the finite loci coalescent with recombination along multiple sequences. Theoretical Population Biology. 2014;98:48–58. doi: 10.1016/j.tpb.2014.01.002. [DOI] [PubMed] [Google Scholar]
- Hudson RR. Estimating the recombination parameter of a finite population model without selection. Genetical Research. 1987;50(03):245–250. doi: 10.1017/s0016672300023776. [DOI] [PubMed] [Google Scholar]
- Hudson RR. Two-locus sampling distributions and their application. Genetics. 2001;159(4):1805–1817. doi: 10.1093/genetics/159.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Kaplan NL. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics. 1985;111(1):147–164. doi: 10.1093/genetics/111.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhner MK, Yamato J, Felsenstein J. Maximum likelihood estimation of recombination rates from population data. Genetics. 2000;156(3):1393–1401. doi: 10.1093/genetics/156.3.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Stephens M. Modeling linkage disequilibrium and identifying recombination hotspots using single-nucleotide polymorphism data. Genetics. 2003;165(4):2213–2233. doi: 10.1093/genetics/165.4.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K, Futschik A, Li H. A fast estimate for the population recombination rate based on regression. Genetics. 2013;194(2):473–484. doi: 10.1534/genetics.113.150201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes JS, Arenas M, Posada D, et al. Coestimation of recombination, substitution and molecular adaptation rates by approximate Bayesian computation. Heredity. 2014;112(3):255–264. doi: 10.1038/hdy.2013.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marjoram P, Wall JD. Fast ’coalescent’ simulation. BMC genetics. 2006;7(1):16. doi: 10.1186/1471-2156-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean G, Auton A. LDhat 2.1: a package for the population genetic analysis of recombination. 2007 Available at: http://www.stats.ox.ac.uk/˜mcvean/LDhat/manual.pdf.
- McVean G, Awadalla P, Fearnhead P. A coalescent-based method for detecting and estimating recombination from gene sequences. Genetics. 2002;160(3):1231–1241. doi: 10.1093/genetics/160.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean GAT, Cardin NJ. Approximating the coalescent with recombination. Philosophical Transactions of the Royal Society B: Biological Sciences. 2005;360(1459):1387–1393. doi: 10.1098/rstb.2005.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers SR, Griffiths RC. Bounds on the minimum number of recombination events in a sample history. Genetics. 2003;163(1):375–394. doi: 10.1093/genetics/163.1.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly PF, Birney E, Balding DJ. Confounding between recombination and selection, and the Ped/Pop method for detecting selection. Genome Research. 2008;18(8):1304–1313. doi: 10.1101/gr.067181.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Seielstad MT, Perez-Lezaun A, et al. Population growth of human Y chromosomes: a study of Y chromosome microsatellites. Molecular Biology and Evolution. 1999;16(12):1791–1798. doi: 10.1093/oxfordjournals.molbev.a026091. [DOI] [PubMed] [Google Scholar]
- R-Core-Team. R: A Language and Environment for Statistical Computing. 2013. [Google Scholar]
- Ruli E, Sartori N, Ventura L. Approximate Bayesian computation with composite score functions. Statistics and Computing. 2015 [Google Scholar]
- Sabeti PC, Reich DE, Higgins JM, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419(6909):832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- Sabeti PC, Schaffner SF, Fry B, et al. Positive natural selection in the human lineage. Science. 2006;312(5780):1614–1620. doi: 10.1126/science.1124309. [DOI] [PubMed] [Google Scholar]
- Stein C. Inadmissibility of the usual estimator for the mean of a multivariate normal distribution. Proceedings of the Third Berkeley symposium on mathematical statistics and probability. 1956;1:197–206. [Google Scholar]
- Varin C, Reid N, Firth D. An overview of composite likelihood methods. Statistica Sinica. 2011;21(1):5–42. [Google Scholar]
- Wall JD. A comparison of estimators of the population recombination rate. Molecular Biology and Evolution. 2000;17(1):156–163. doi: 10.1093/oxfordjournals.molbev.a026228. [DOI] [PubMed] [Google Scholar]
- Wall JD. Estimating recombination rates using three-site likelihoods. Genetics. 2004;167(3):1461–1473. doi: 10.1534/genetics.103.025742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiuf C. On the minimum number of topologies explaining a sample of DNA sequences. Theoretical Population Biology. 2002;62(4):357–363. doi: 10.1016/s0040-5809(02)00004-7. [DOI] [PubMed] [Google Scholar]