Abstract
Vascular dementia (VaD), the second most prevalent type of dementia, is caused by reduced blood supply to the brain that results in cognitive impairment. Despite the efforts of numerous studies, the pathological mechanisms behind VaD remain unclear. The aim of the present study was to identify candidate genes that undergo changes in hippocampal DNA methylation owing to VaD. A genome-wide DNA methylation analysis was performed, using methylated DNA-binding domain sequencing. VaD model rats with cognitive impairment induced by bilateral common carotid artery occlusion were confirmed using the radial arm maze test. A total of 1,180 differentially methylated genes (DMGs) were identified, and functional annotation analysis revealed the DMGs to be enriched in 10 Gene Ontology biological processes. Network analysis using the STRING database indicated that seven genes were closely connected. Rats in the VaD model group demonstrated relative hypomethylation in the promoter region and increased mRNA expression of the hippocampal genes vascular endothelial growth factor (VEGFA) and kinase insert domain receptor, but only differences in VEGFA mRNA expression levels were determined to be statistically significant. In conclusion, these preliminary data from the functional annotation of hippocampal DMGs in the promoter region highlighted candidate genes for VaD that may contribute to the elucidation of its pathophysiology.
Keywords: DNA methylation, vascular dementia, cognitive impairment, vascular endothelial growth factor, kinase insert domain receptor
Introduction
Vascular dementia (VaD) is the second most prevalent form of dementia, after Alzheimer's disease (AD), and accounts for ~20% of all dementia cases (1). According to world prevalence data, the number of individuals affected by dementia is expected to increase to 70 million by 2030 (2). VaD is characterized by hypoxia, oxidative stress, and inflammation, which may lead to symptoms of mood disorders and difficulties with problem solving, memory, thinking, reasoning, and executive functions such as judgment (3–5). Moreover, the white matter, basal ganglia, and hippocampus are vulnerable to damage, and disruption of the blood-brain barrier subsequently leads to memory impairment (6–8). Although numerous studies on the pathophysiology of VaD have been conducted, the pathological mechanisms behind this disorder have yet to be fully understood (3,8–10). Studies into the genetic basis of dementia have been conducted since 1993, when it was reported that apolipoprotein E (APOE) 4 was a risk factor for AD (11).
Previous genetics studies on VaD have identified several single-nucleotide polymorphisms (SNPs), such as methylenetetrahydrofolate 677C/T (rs1801133), paraoxonase 1 L55M (rs854560), transforming growth factor-β1 +29C/T (rs1800470), and tumor necrosis factor-α −850C/T (rs1799724) (12). Although SNPs may be crucial to verifying the pathogenesis of the disease, VaD is more affected by lifestyle-associated risk factors, such as age, diabetes, hypertension, and the metabolic syndrome, compared with AD (8). Therefore, epigenetic-associated changes in the expression of a gene (that is, without alterations in the DNA sequence) may present a more suitable approach for studying VaD (13).
Epigenetic changes include DNA methylation, histone modification, chromatin remodeling, and microRNA regulation (14). DNA methylation, one of the most widely studied epigenetic changes that regulate many cellular processes (15), is mediated by DNA methyltransferases during early development and throughout the lifespan of an organism (16). Methyl group additions to the cytosine residue that precedes a guanine, the so-called cytosine-guanine (CpG) dinucleotides, have been reported to significantly modify gene expression (17,18). Previous studies have concentrated mainly on how the position of the methylation in the transcriptional unit affects its association to gene expression. The majority of these studies focused on CpG islands (CGIs), which are dense repetitions of CpG nucleotides located mainly in the gene promoter region and are known to repress long-term gene expression (13,19,20). The methyl-CpG-binding domain proteins function by binding to the promoter region of the transcription site and blocking gene expression (21).
To the best of the authors' knowledge, there are currently no studies that used the epigenetic approach of VaD. The present study may be meaningful as an initial study on the genome-wide DNA methylation differences in VaD. The aim of the present study was to investigate the differences in DNA methylation in the hippocampus, which serves an important role in VaD pathogenesis.
Materials and methods
Animal preparation
Male Wistar rats (n=4; weight, 80±10 g; age 4 weeks; Orient Bio, Seongnam, Korea) were used in this experiment. Animals were housed in a controlled environment at 25±2°C, 50–55% humidity, and 12-h light/dark cycle, with ad libitum access to food and water. The rats were randomly divided into two groups: The sham-operated animals (Sham group; n=2) and BCCAO-operated animals (VaD group; n=2). The BCCAO or sham surgery was performed when the rats were 12 weeks old, and the animals were sacrificed 6 weeks post-surgery. The experimental procedures were performed in accordance with the animal care guidelines of The National Institutes of Health (Bethesda, MD, USA), and all animal experiments were approved by The Committee for the Care and Use of Laboratory Animals at Kyung Hee University [Seoul, Korea; approval no. KHUASP(SE)-15-082].
BCCAO model establishment
The BCCAO model is an animal model of permanent VaD; it is established through the permanent ligation of the common carotid artery to induce chronic cerebral hypoperfusion, which leads to damage to the white matter, disruption of the blood-brain barrier, nerve injury of the cerebral cortex and hippocampus, oxidative stress, and inflammation (22). Rats were anesthetized with 50 mg/kg body weight Zoletil 50 (Virbac Laboratories, Carros, France); body temperature was maintained at 36.5–37.5°C using a heating pad. Following a midline incision to expose the common carotid arteries, the vessel ligations were carefully performed to avoid damage to the vagus nerve. Each carotid artery was double ligated with 3-0 silk sutures (Ailee, Seoul, Korea). Similar surgical procedures were performed on the Sham-operated animals, except for the vessel ligation step. To prevent postoperative pain and infection, the animals were maintained in a warm and clean environment during a three day recovery period.
Radial arm maze test
A behavioral study was conducted to determine whether the VaD model had been induced by the BCCAO surgery. The radial arm maze test was conceived by Olton and Samuelson in 1976 to measure spatial working memory in rats (23). The radial arm maze apparatus comprises eight arms (length, 50 cm; width, 10 cm), spaced equidistantly. The apparatus was placed 1 m above the floor, and a small water bowl was located at the end of each arm. The rats were deprived of water for 48 h and subsequently moved to the room where the radial arm maze was located and allowed acclimate for 1 h to prevent bias caused by anxiety. Rats were placed into the apparatus and were allowed to move freely for 8 min and to drink water. Three observers that were independent of the present study were recruited to count the number of errors in revisitation of the same arm each time and to calculate the data. If their spatial working memory were perfect, the rats would explore a novel arm each time to find water.
DNA extraction and methylation profiling
To explore the DMGs, genomic DNA was extracted from 10 mg hippocampal tissue using the DNeasy Blood & Tissue kit (Qiagen, Inc., Valencia, CA, USA), according to the manufacturer's protocol.
In the present study, the MBD-seq method was used, owing to its low cost, high specificity, and efficiency (24–26). Genomic DNA (1.5 µg) was sent to the Theragen Etex Bio Institute (Suwon, Korea) for the generation of recombinant methyl-binding domain protein to enrich 5-methylcytosine-modified regions of the genome for subsequent massively parallel sequencing analysis. Fragmentation of the genomic DNA was achieved with a Covaris S220 ultrasonicator (duty cycle, 10%; intensity, 5; 200 cycles per burst for 180 sec; Covaris, Woburn, MA, USA) to obtain sequences with an average length of 200 bp. Methylated fragments were captured using the MethylCap kit (Diagenode Inc., Denville, NJ, USA), according to the manufacturer's protocol. The captured DNA fragments were purified using QIAquick PCR purification columns (Qiagen, Inc.), and the purified DNA was used for library preparation. Quantification of the captured DNA was performed with the Quant-iT PicoGreen dsDNA Assay kit (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA), and 20 ng of chromatin immunoprecipitation (ChIP)-enriched, fragmented input DNA was used in the TruSeq ChIP Library Preparation kit (Illumina, Inc., San Diego, CA, USA), according to the manufacturer's protocol. The library was examined with a bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA, USA) and quantified. The denatured and amplified libraries were loaded onto Illumina NextSeq 500 using the High Output v2 kit (1×72 cycles, 2 index reads; Illumina, Inc.).
Bisulfite genomic sequencing
The aforementioned genomic DNA (500 ng) from each hippocampal sample was treated by bisulfite conversion with the EZ-96 DNA Methylation kit (Zymo Research Corp., Irvine, CA, USA), according to the manufacturer's protocol. To determine the methylation patterns in the genes, the following primers were used: Vascular endothelial growth factor A (VEGFA)-1, forward 5′-AGGTGAGGTTTGAGTTTTTTATTTA-3′, reverse 5′-CTAAACCATCAAACACCCAAAAA-3′; VEGFA-2, forward 5′-TTGTAGGGTTTTATTTTGTTATTAGG-3, reverse 5′-AAATACAAATATCCACTACACCCTC-3′; kinase insert domain receptor (KDR)-1, forward 5′-GAAGTTTTTTTAAGTGGTTTATTTTGTT-3′, reverse 5′-AAAAACTTTTCAAAATCCAAATTCA-3′; and KDR-2, 5′-GGATTTTGAAAAGTTTTTTGGGTT-3′, reverse 5′-ATATAACTTTCTTTCCATTCCTTCCA-3. Both genes were analyzed by using two primers divided into 300–400 bp for accurate analysis. PCR amplification of these regions was achieved by using a PCR KOD FX kit (Toyobo Life Science, Osaka, Japan). Amplification was carried out with the following thermocycling conditions: 95°C for 4 min; 35 cycles of 95°C for 30 sec, 55°C for 30 sec, 72°C for 30 sec and final extension at 72°C for 7 min. PCR products were purified using a QIAquick PCR purification kit (Qiagen, Inc.) and quantified using Quant-iT™ PicoGreen™ dsDNA Assay kit (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. PCR product sizes were confirmed through agarose gel electrophoresis. Polymerase chain reaction (PCR) products were subsequently processed to produce a DNA sequencing library using the TruSeq Nano DNA LT kit (Illumina Inc.), according to the manufacturer's protocol. For each library, an amplified PCR product size of approximately 450 bp was determined with a bioanalyzer using the Agilent DNA 1000 ChIP kit, and library quantification was performed on a CFX96 Real-Time PCR Detection System (Bio-Rad Laboratories Inc., Hercules, CA, USA). Serial sequencing of each library was conducted using Illumina MiSeq, and the generation of clusters of DNA libraries in flow cells and of 250 bp paired-end reads (2×250) were performed using the MiSeq 500 Cycle v2 kit (Illumina, Inc.). The raw image data were transformed into sequence data by base calling and stored in the FASTQ format.
Reverse transcription-quantitative PCR (RT-qPCR)
RT-qPCR was performed using the T100 thermal cycler and CFX-96 real-time PCR detection systems (Bio-Rad Laboratories, Inc.). RNA was extracted from hippocampal whole tissue (10 mg) using the RNeasy mini kit (Qiagen, Inc.). cDNA synthesis using the ReverTra Ace-α kit (Toyobo Life Sciences) according to the manufacturer's protocol. PCR was performed with the Thunderbird SYBR qPCR Mix (Toyobo Life Science); all cDNA samples were diluted 1:10. The PCR mixture (20 µl) contained cDNA (2 ng/µl), 1X SYBR Green qPCR mix, and 250 µM of each primer set. The following primers were used: VEGFA forward, 5′-CCTCCTCCTCCTGGGAAC-3′ and reverse, 5′-AGGGTAAGCCACTCACACA-3′; KDR forward, 5′-GCAGCCAAGTCCGAATCC-3′ and reverse, 5′-TCCCGCATCTCTTTCACTCA-3′; GAPDH forward, 5′-CTCCCATTCTTCCACCTTTGAT-3′ and reverse, 5′-CACCACCCTGTTGCTGTAG-3. Thermocycling conditions were as follows: Initial denaturation at 95°C for 1 min, followed by a single cycle of denaturation at 95°C for 10 sec, annealing at 58°C for 10 sec, and extension at 72°C for 20 sec. Data were analyzed with CFX Manager v1.5 software (Bio-Rad Laboratories, Inc.) and the relative expression of genes were quantified using the 2−ΔΔCq method (27).
Data analysis
MBD-seq reads were mapped with Bowtie v1.0.0 (www.bowtie-bio.sourceforge.net). Genome-wide and meta-gene profile analyses were performed using in-house PERL and R scripts. Highly methylated regions (HMRs) were called with MACS v1.4.2 (www.liulab.dfci.harvard.edu/MACS; P<0.01), and differentially methylated regions (DMRs) were analyzed using an in-house PERL script. DMGs were selected by in-house scripts, and were defined with at least 1 DMR between a region located 2 kbp upstream and 500 bp downstream from the transcription start site. Gene Ontology (GO) and functional annotation analysis for significant genes were performed on the DAVID Bioinformatics Resource v6.8 server (david.abcc.ncifcrf.gov) (28). To identify potential functional pathways, protein-protein interaction (PPI) network analysis was conducted using the STRING v10.0 database (www.string-db.org). Putative binding sites at the promoter regions of VEGFA and KDR were also analyzed. Transcription factor binding sites were predicted by AliBaba2.1 using the TRANSFAC 4.0 transcription factor database (www.gene-regulation.com/pub/programs/alibaba2/index.html).
Statistical analysis
Statistical analyses were performed using SPSS v22.0 (IBM Corp., Armonk, NY, USA). All data are presented as the mean ± standard error of the mean. All experiments were repeated at least three times. Statistical comparisons between groups were processed with the unpaired Student t-test. P<0.05 was considered to indicate a statistically significant difference.
Results
Confirmation of cognitive impairment induced by BCCAO
Alternation behavior in the radial arm apparatus was recorded to assess the spatial working memory performance. Rats in the VaD group had a significantly higher number of errors (12.00±2.07) in the radial arm maze test compare with rats in the Sham group (5.14±1.28; P<0.05; Fig. 1). These results confirmed that cognitive impairment had been induced by BCCAO.
Figure 1.
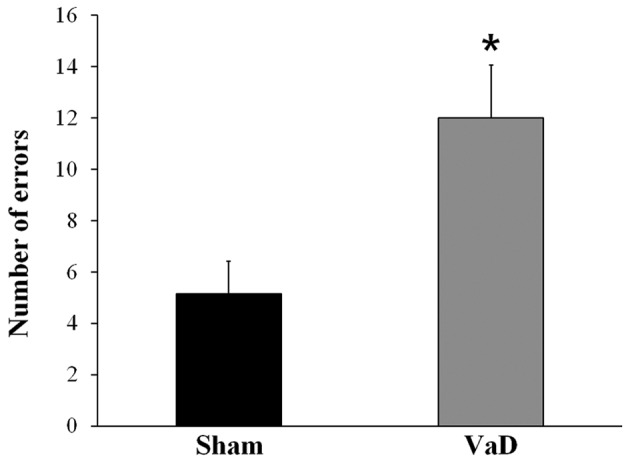
Radial arm maze test for cognitive impairment. The number of errors in the radial arm maze test were examined between sham-operated rats and VaD model rats, and were used to confirm reduced cognitive function in VaD rats. The data are presented as the mean ± standard error of the mean. *P<0.05 vs. Sham. VaD, vascular dementia.
Identification of DMRs
The aim of the present study was to uncover the DNA methylation changes associated with cognitive impairment by comparing the gene methylated regions in an animal model of VaD. To investigate the difference in DNA methylation between the Sham and VaD groups, DMRs were analyzed in the promoter regions that may affect the regulation of mRNA expression. Promoter regions were defined as being between 2 kbp upstream and 500 bp downstream from the transcription start site. DMRs were identified by HMR peaks with a fold enrichment ratio of >2.0 between Sham and VaD samples, or if a HMR peak occurred in only one sample.
Of the 112,665 reliable HMRs detected, a total of 95,238 DMRs were identified, of which 1,250 (1.3%) DMRs were specifically located in the promoter region. Of the 27,030 hypermethylated DMRs detected, 397 (1.5%) were associated with the promoter, and of the 68,208 hypomethylated DMRs detected, 853 (1.3%) were associated with the promoter.
Functional annotation of DMGs
A total of 1,180 DMGs were identified based on the detected promoter region DMRs, of which 384 (32.5%) were hypermethylated and 796 (67.5%) were hypomethylated. There were fewer hypermethylated DMGs identified in the VaD group compared with the Sham group, with fold enrichment ratios ranging from 20 to 2,388 (log2 ratios of 1.00–11.22). GO analysis of the DMG sets was performed using DAVID to verify the functions associated with the genes.
The top 10 GO biological processes for the 1,180 genes are listed in Table I. The 10 functional categories of GO biological processes: Regulation of nucleotide biosynthetic process; regulation of cyclic (c)AMP biosynthetic process; multicellular organism reproduction; regulation of lyase activity; cAMP-mediated signaling; regulation of adenylate cyclase activity; sexual reproduction; G-protein signaling, coupled to cyclic nucleotide second messenger; antigen receptor-mediated signaling pathway; and spermatogenesis. STRING identified interactions among proteins coded by 72 genes. PPI network analysis demonstrated that seven DMGs were closely connected, including cluster of differentiation 247, ζ-chain-associated protein kinase 70, protein tyrosine phosphatase receptor type C, lymphocyte-specific protein tyrosine kinase, KDR, epidermal growth factor receptor, and VEGFA (Fig. 2). Among these, VEGFA and KDR were selected as candidate genes for VaD for further validation studies, due to their high connectivity in the PPI network and association with VaD.
Table I.
Top 10 GO biological processes for 1,180 differentially methylated genes.
| GO term | Count | Fold enrichment | P-value | FDR | Genes |
|---|---|---|---|---|---|
| Regulation of nucleotide biosynthetic process | 18 | 3.37 | <0.001 | <0.01 | ACR, ADCY3, GNAZ, AVP, LHCGR, ABCA1, SSTR5, S1PR3, ADRB2, HRH3, HTR7, GIPR, GNAS, NOS3, ADM2, GUCA2B, GLP1R, GNG7 |
| Regulation of cAMP biosynthetic process | 16 | 3.34 | <0.001 | <0.01 | ACR, ADCY3, GNAZ, AVP, LHCGR, ABCA1, SSTR5, S1PR3, ADRB2, HRH3, HTR7, GIPR, GNAS, ADM2, GLP1R, GNG7 |
| Multicellular organism reproduction | 45 | 1.84 | <0.001 | <0.01 | ACR, NANOS3, RAD51C, PCDHA8, H1FNT, SPATA20, FKBP6, LHCGR, PCDHGA9, ADAD1, MAEL, HP, BCL2L1, SOHLH1, SOHLH2, KRT9, VDR, WNT4, POU5F2, NOS3, DAZL, TGM4, FAS, PIWIL4, DND1, SPATA3, B4GALNT1, PCDHGA12, EGFR, B4GALT1, AVP, PCDHGA11, MAK, TNP1, CELSR2, THEG, KDR, APRT, LEP, REC8, ESR2, TXNDC2, P2RX1, TXNDC8, VEGFA |
| Regulation of lyase activity | 15 | 3.28 | <0.001 | <0.01 | ACR, ADCY3, GNAZ, LHCGR, SSTR5, S1PR3, ADRB2, HRH3, HTR7, GIPR, GNAS, NOS3, ADM2, GLP1R, GNG7 |
| cAMP-mediated signaling | 15 | 3.13 | <0.001 | <0.01 | ADCY3, GNAZ, LHCGR, RIMS2, LEP, SSTR5, S1PR3, ADRB2, HTR7, GIPR, GNAS, ADM2, GLP2R, MC3R, GLP1R |
| Regulation of adenylate cyclase activity | 14 | 3.26 | <0.001 | <0.01 | ACR, ADCY3, GNAZ, LHCGR, SSTR5, S1PR3, ADRB2, HRH3, HTR7, GIPR, GNAS, ADM2, GLP1R, GNG7 |
| Sexual reproduction | 38 | 1.85 | <0.001 | <0.01 | ACR, NANOS3, RAD51C, PCDHA8, H1FNT, SPATA20, FKBP6, PCDHGA9, ADAD1, MAEL, HP, BCL2L1, SYCP2, SOHLH1, SOHLH2, KRT9, WNT4, POU5F2, CD4, NOS3, DAZL, FAS, PIWIL4, DND1, SPATA3, B4GALNT1, PCDHGA12, B4GALT1, PCDHGA11, MAK, TNP1, CELSR2, THEG, LEP, REC8, TXNDC2, ADAM1A, TXNDC8 |
| G-protein signaling, coupled to cyclic nucleotide second messenger | 14 | 3.18 | <0.001 | <0.01 | ADCY3, GNAZ, LHCGR, SSTR5, S1PR3, ADRB2, HRH1, HTR7, GIPR, GNAS, ADM2, GLP2R, MC3R, GLP1R |
| Antigen receptor-mediated signaling pathway | 9 | 4.67 | <0.001 | <0.01 | PTPRC, LAT2, CD19, LAX1, CD247, LCK, ZAP70, CACNB4, SKAP1 |
| Spermatogenesis | 28 | 2.05 | <0.001 | <0.01 | NANOS3, RAD51C, H1FNT, SPATA20, PCDHA8, FKBP6, ADAD1, PCDHGA9, MAEL, HP, BCL2L1, SOHLH1, SOHLH2, KRT9, POU5F2, FAS, SPATA3, PIWIL4, B4GALNT1, PCDHGA12, PCDHGA11, MAK, TNP1, CELSR2, THEG, REC8, TXNDC2, TXNDC8 |
cAMP, cyclic adenosine monophosphate; FDR, false discovery rate; GO, Gene Ontology.
Figure 2.
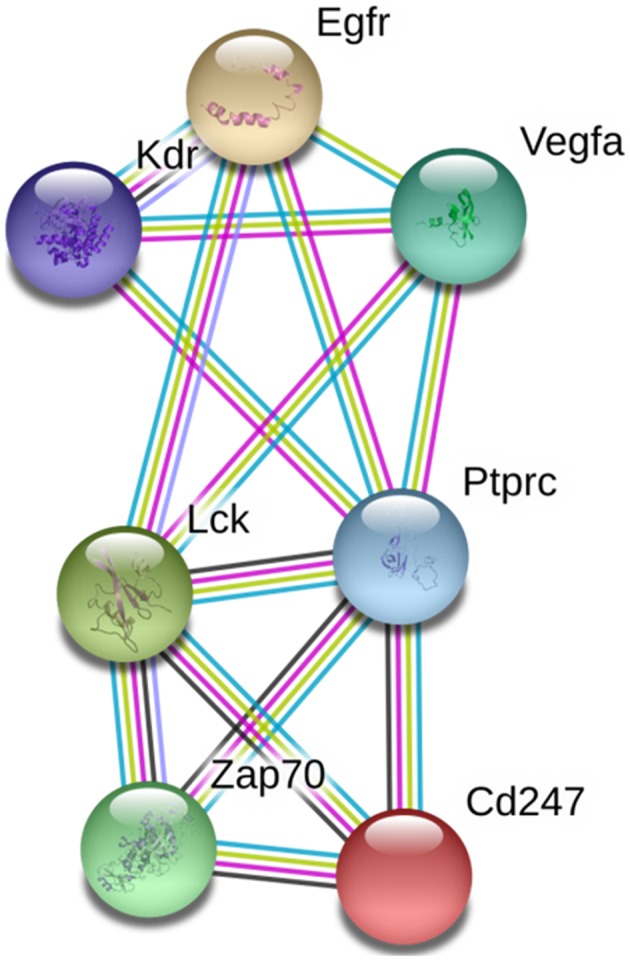
PPI network analysis. PPI network analysis was conducted using STRING, and seven genes were demonstrated to be well-connected in the VaD group. PPI, protein-protein interaction; cAMP, cyclic adenosine monophosphate; Egfr, epidermal growth factor receptor; Kdr, kinase insert domain receptor; Lck, lymphocyte-specific protein tyrosine kinase; Ptprc, protein tyrosine phosphatase receptor type C; VaD, vascular dementia; VEGFA, vascular endothelial growth factor A; Zap, ζ-chain-associated protein kinase.
Validation of VEGFA methylation and relative mRNA expression levels
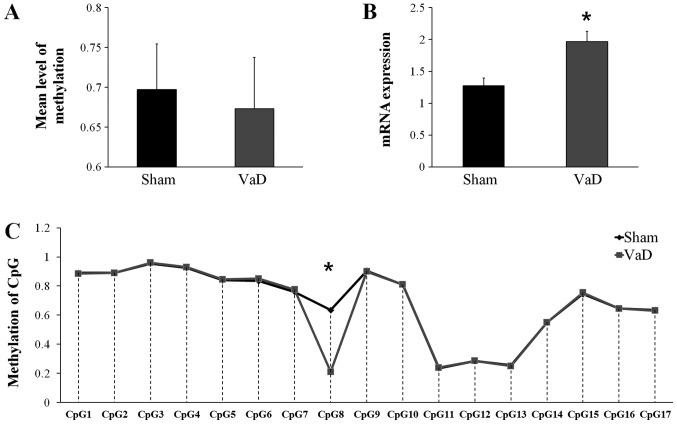
Bisulfite sequencing was used to confirm the differences in promoter region methylation of VEGFA. Although the mean CGI methylation status of VEGFA was slightly higher in the Sham group (0.6971±0.0575) compare with the VaD group (0.6729±0.0647; Fig. 3A), the difference was not statistically significant. However, VEGFA mRNA expression was significantly higher in the VaD group (1.9647±0.2804) compared with expression in the Sham group (1.2759±0.2111; P>0.05; Fig. 3B), and the methylation status in CpG8 was significantly lower in the VaD group compared with the Sham group (Fig. 3C). CpG8 is the eighth CGI in the promoter region of VEGFA, 544 bp away from the transcription start site, in the forward strand.
Figure 3.
Analysis of the VEGFA promoter region. (A) DNA methylation and (B) reverse transcription-quantitative polymerase chain reaction analysis of mRNA expression levels of VEGFA. (C) DNA methylation pattern of VEGFA promoter region detected by bisulfite modification of DNA sequencing. The data are presented as the mean ± standard error of the mean. *P<0.05 vs. Sham. CpG, cytosine-guanine dinucleotide; VaD, vascular dementia; VEGFA, vascular endothelial growth factor A.
Validation of KDR methylation and relative mRNA expression
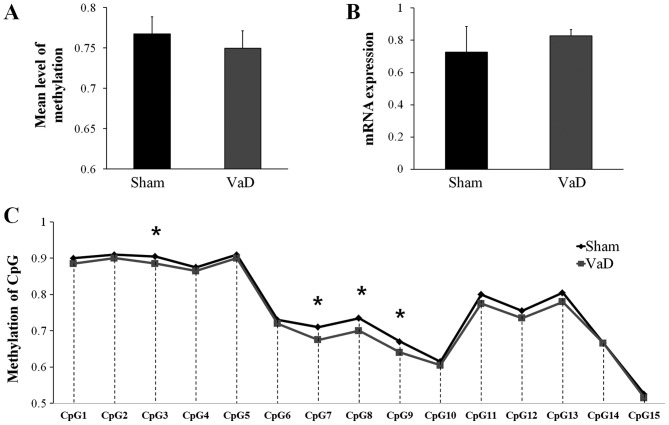
Although the mean methylation level of the CGIs in KDR promoter region was higher in the Sham group (0.7673±0.1177) than in the VaD group (0.7497±0.1185), the difference was not statistically significant (Fig. 4A). KDR mRNA expression levels in the Sham group (0.7275±0.1568) were lower compared with expression in the VaD group (0.8276±0.0383), but the difference was not statistically significant (Fig. 4B). However, significant differences were identified in the methylation status of four CGIs (CpG3, CpG7, CpG8, and CpG9) in the promoter region on the forward strand between the two groups (Fig. 4C).
Figure 4.
Analysis of the KDR promoter region. (A) DNA methylation and (B) reverse transcription-quantitative polymerase chain reaction analysis of mRNA expression levels of KDR. (C) DNA methylation pattern of KDR promoter region detected by pyrosequencing. The data are presented as the mean ± standard error of the mean. *P<0.05 vs. Sham. CpG, cytosine-guanine dinucleotide; VaD, vascular dementia.
Predicted transcription factor binding analysis
To further examine the regulatory roles of site-specific DNA methylation in the promoter region of VEGFA and KDR, predicted transcription factor analysis was performed by AliBaba2.1 programs using the TRANSFAC 4.0 transcription factor database. CpG8 in VEGFA was predicted to be a binding site for serum response factor (SRF). Putative transcription factor binding sites for CpGs of KDR were as follows: SRF in CpG3, activator protein (AP)-4 in CpG7 and CpG8, and immunoglobulin transcription factor (ITF) 2 in CpG9 (data not shown).
Discussion
The present study investigated DMGs in the hippocampus between Sham and VaD model animals, as well as their functional categories, to identify candidate genes that may be associated with cognitive function. To verify the successful establishment of the BCCAO-induced VaD model rat model, cognitive impairment was confirmed by the commonly used radial arm maze test (29,30).
Alterations of DNA methylation in the promoter region are a way to regulate gene expression (31,32). It has been suggested that specific diseases, including neurodegenerative disease, may occur by characterized hypermethylated and hypomethylated CpGs (33). Gene expression may be regulated by promoter-site CGI methylation, where hypermethylation of the CpGs suppresses gene transcription, and hypomethylation of the promoter region allows transcription factor binding and activation of gene transcription (34,35). A previous methylation analysis using 122 cortical tissues from patients with AD revealed that the Ankyrin 1 gene was hypermethylated in the superior temporal gyrus and prefrontal cortex (36). In another study using 708 autopsied brains from individuals with AD, 11 DMRs were identified, which were associated with several genes, including ankyrin-1, cadherin-23, disco-interacting protein homolog A, inactive rhomboid protein 2, 60S ribosomal protein L13 and E3 ubiquitin-protein ligase RNF34 (37). In the present study, 72 DMGs were identified and annotated to 10 categories of GO biological processes and may be associated with VaD. Through PPI network analysis, seven genes were classified as closely interacting and may be related to the VEGF signaling pathway.
VEGFA and KDR are involved in the VEGF signaling pathway (38–40). In the present study, rats in the VaD group exhibited relative hypomethylation, although this was not significant. However, VEGFA mRNA expression increased in the hippocampus, compared with rats in the Sham group. A number of putative transcription factors that bind to the hypomethylated sites were analyzed in the present study, and these may contribute to increased VEGFA expression. VEGFA is a crucial regulator of angiogenesis, skeletal growth, and ovarian angiogenesis (41). In the present study, CpG8 in the VEGFA promoter was confirmed to be hypomethylated in the VaD group and was predicted to be a binding site for the transcription factor SRF. SRF is a downstream mediator of VEGF signaling pathway and serves a crucial role for VEGF-induced angiogenesis (42,43). KDR is a VEGF receptor and is crucial for endothelial cell proliferation, survival, and migration, as well as neuroprotection or improved neurovascular coupling (41,44,45). In the present study, putative transcription factors, such as SRF, Ap-4, and ITF-2, were investigated if they were predicted bind to hypomethylated CpG in KDR, but there were few studies on the association between these transcription factors and KDR. Consistent with previous studies, upregulated VEGFA mRNA expression in the VaD model rat was confirmed; this may serve an important role in compensating for cellular damage through neurobehavioral recovery and neurovascular remodeling after hypoxic brain injury (46,47). It has also been demonstrated that increased expression of VEGFA under hypoxic conditions and increased VEGF-promoted angiogenesis and recovery of neurological damage in animal models of stroke (48,49). In the present study, hippocampal damage caused by VaD may have increased the expression of VEGFA to compensate for the impaired cognitive function through neurovascular remodeling via angiogenesis.
In summary, using methylation analysis, the present study identified DMGs in the hippocampal tissues of Sham and VaD model rats. These preliminary data may contribute to our understanding of the pathophysiology of VaD, which may lead to the development of methods for the recovery of cognitive function; however, further investigations are required.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from The National Research Foundation of Korea funded by The Ministry of Education, Science and Technology (grant no. NRF-2017R1A2B4012775).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
All authors designed the experiments. YooJK, MKS and JML were responsible for the animals, establishment of the animal model and the behavioural test. JMP and YJK performed the methylation profiling analysis, polymerase chain reaction and wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All animal experiments were approved by The Committee for the Care and Use of Laboratory Animals at Kyung Hee University [Seoul, Korea; approval no. KHUASP(SE)-15-082].
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Kalaria RN, Maestre GE, Arizaga R, Friedland RP, Galasko D, Hall K, Luchsinger JA, Ogunniyi A, Perry EK, Potocnik F, et al. Alzheimer's disease and vascular dementia in developing countries: Prevalence, management, and risk factors. Lancet Neurol. 2008;7:812–826. doi: 10.1016/S1474-4422(08)70169-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prince M, Bryce R, Albanese E, Wimo A, Ribeiro W, Ferri CP. The global prevalence of dementia: A systematic review and metaanalysis. Alzheimers Dement. 2013;9(63):e2. doi: 10.1016/j.jalz.2012.11.007. 75. [DOI] [PubMed] [Google Scholar]
- 3.Iadecola C. The pathobiology of vascular dementia. Neuron. 2013;80:844–866. doi: 10.1016/j.neuron.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gill R, Tsung A, Billiar T. Linking oxidative stress to inflammation: Toll-like receptors. Free Radic Biol Med. 2010;48:1121–1132. doi: 10.1016/j.freeradbiomed.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu H, Zhang J. Cerebral hypoperfusion and cognitive impairment: The pathogenic role of vascular oxidative stress. Int J Neurosci. 2012;122:494–499. doi: 10.3109/00207454.2012.686543. [DOI] [PubMed] [Google Scholar]
- 6.Park HR, Park M, Choi J, Park K, Chung HY, Lee J. A high-fat diet impairs neurogenesis: Involvement of lipid peroxidation and brain-derived neurotrophic factor. Neurosci Lett. 2010;482:235–239. doi: 10.1016/j.neulet.2010.07.046. [DOI] [PubMed] [Google Scholar]
- 7.Chen J, Cui X, Zacharek A, Cui Y, Roberts C, Chopp M. White matter damage and the effect of matrix metalloproteinases in type 2 diabetic mice after stroke. Stroke. 2011;42:445–452. doi: 10.1161/STROKEAHA.110.596486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Venkat P, Chopp M, Chen J. Models and mechanisms of vascular dementia. Exp Neurol. 2015;272:97–108. doi: 10.1016/j.expneurol.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khan A, Kalaria RN, Corbett A, Ballard C. Update on vascular dementia. J Geriatr Psychiatry Neurol. 2016;29:281–301. doi: 10.1177/0891988716654987. [DOI] [PubMed] [Google Scholar]
- 10.Murray ME, Meschia JF, Dickson DW, Ross OA. Genetics of vascular dementia. Minerva Psichiatr. 2010;51:9–25. [PMC free article] [PubMed] [Google Scholar]
- 11.Travis J. New piece in alzheimer's puzzle. Science. 1993;261:828–830. doi: 10.1126/science.8346434. [DOI] [PubMed] [Google Scholar]
- 12.Sun J, Tan L, Wang H, Tan M, Tan L, Li J, Xu W, Zhu X, Jiang T, Yu J. Genetics of vascular dementia: Systematic review and meta-analysis. J Alzheimers Dis. 2015;46:611–629. doi: 10.3233/JAD-143102. [DOI] [PubMed] [Google Scholar]
- 13.Jaenisch R, Bird A. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):S245–S254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 14.Fessele KL, Wright F. Primer in genetics and genomics, article 6: Basics of epigenetic control. Biol Res Nurs. 2018;20:103–110. doi: 10.1177/1099800417742967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maloney B, Lahiri DK. Epigenetics of dementia: Understanding the disease as a transformation rather than a state. Lancet Neurol. 2016;15:760–774. doi: 10.1016/S1474-4422(16)00065-X. [DOI] [PubMed] [Google Scholar]
- 16.Niculescu MD, Zeisel SH. Diet, methyl donors and DNA methylation: Interactions between dietary folate, methionine and choline. J Nutr. 2002;132(8 Suppl):2333S–2335S. doi: 10.1093/jn/132.8.2333S. [DOI] [PubMed] [Google Scholar]
- 17.Cong L, Jia J, Qin W, Ren Y, Sun Y. Genome-wide analysis of DNA methylation in an APP/PS1 mouse model of alzheimer's disease. Acta Neurol Belg. 2014;114:195–206. doi: 10.1007/s13760-013-0267-6. [DOI] [PubMed] [Google Scholar]
- 18.Sanchez-Mut JV, Gräff J. Epigenetic alterations in alzheimer's disease. Front Behav Neurosci. 2015;9:347. doi: 10.3389/fnbeh.2015.00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones PA. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 20.Borgel J, Guibert S, Li Y, Chiba H, Schübeler D, Sasaki H, Forné T, Weber M. Targets and dynamics of promoter DNA methylation during early mouse development. Nat Genet. 2010;42:1093–1100. doi: 10.1038/ng.708. [DOI] [PubMed] [Google Scholar]
- 21.Valinluck V, Tsai H, Rogstad DK, Burdzy A, Bird A, Sowers LC. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Res. 2004;32:4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farkas E, Luiten PG, Bari F. Permanent, bilateral common carotid artery occlusion in the rat: A model for chronic cerebral hypoperfusion-related neurodegenerative diseases. Brain Res Rev. 2007;54:162–180. doi: 10.1016/j.brainresrev.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 23.Olton DS, Collison C, Werz MA. Spatial memory and radial arm maze performance of rats. Learn Motiv. 1977;8:289–314. doi: 10.1016/0023-9690(77)90054-6. [DOI] [Google Scholar]
- 24.Harris RA, Wang T, Coarfa C, Nagarajan RP, Hong C, Downey SL, Johnson BE, Fouse SD, Delaney A, Zhao Y, et al. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat Biotechnol. 2010;28:1097–1105. doi: 10.1038/nbt.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lan X, Adams C, Landers M, Dudas M, Krissinger D, Marnellos G, Bonneville R, Xu M, Wang J, Huang TH, et al. High resolution detection and analysis of CpG dinucleotides methylation using MBD-seq technology. PLoS One. 2011;6:e22226. doi: 10.1371/journal.pone.0022226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Serre D, Lee BH, Ting AH. MBD-isolated genome sequencing provides a high-throughput and comprehensive survey of DNA methylation in the human genome. Nucleic Acids Res. 2010;38:391–399. doi: 10.1093/nar/gkp992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C (T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. doi: 10.1186/gb-2003-4-9-r60. [DOI] [PubMed] [Google Scholar]
- 29.Sim YJ. Treadmill exercise alleviates impairment of spatial learning ability through enhancing cell proliferation in the streptozotocin-induced alzheimer's disease rats. J Exerc Rehabil. 2014;10:81–88. doi: 10.12965/jer.140102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hussein AM, Aher YD, Kalaba P, Aher NY, Dragačević V, Radoman B, Ilic M, Leban J, Beryozkina T, Ahmed ABMA, et al. A novel heterocyclic compound improves working memory in the radial arm maze and modulates the dopamine receptor D1R in frontal cortex of the sprague-dawley rat. Behav Brain Res. 2017;332:308–315. doi: 10.1016/j.bbr.2017.06.023. [DOI] [PubMed] [Google Scholar]
- 31.Huidobro C, Fernandez AF, Fraga MF. The role of genetics in the establishment and maintenance of the epigenome. Cell Mol Life Sci. 2013;70:1543–1573. doi: 10.1007/s00018-013-1296-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goldberg AD, Allis CD, Bernstein E. Epigenetics: A landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 33.Klein H, De Jager PL. Uncovering the role of the methylome in dementia and neurodegeneration. Trends Mol Med. 2016;22:687–700. doi: 10.1016/j.molmed.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 34.Ling C, Groop L. Epigenetics: A molecular link between environmental factors and type 2 diabetes. Diabetes. 2009;58:2718–2725. doi: 10.2337/db09-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu B, Russanova VR, Gravina S, Hartley S, Mullikin JC, Ignezweski A, Graham J, Segars JH, DeCherney AH, Howard BH. DNA methylome and transcriptome sequencing in human ovarian granulosa cells links age-related changes in gene expression to gene body methylation and 3′-end GC density. Oncotarget. 2015;6:3627–3643. doi: 10.18632/oncotarget.2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lunnon K, Smith R, Hannon E, De Jager PL, Srivastava G, Volta M, Troakes C, Al-Sarraj S, Burrage J, Macdonald R, et al. Methylomic profiling implicates cortical deregulation of ANK1 in alzheimer's disease. Nat Neurosci. 2014;17:1164–1170. doi: 10.1038/nn.3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Jager PL, Srivastava G, Lunnon K, Burgess J, Schalkwyk LC, Yu L, Eaton ML, Keenan BT, Ernst J, McCabe C, et al. Alzheimer's disease: Early alterations in brain DNA methylation at ANK1, BIN1, RHBDF2 and other loci. Nat Neurosci. 2014;17:1156–1163. doi: 10.1038/nn.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma J, Sawai H, Ochi N, Matsuo Y, Xu D, Yasuda A, Takahashi H, Wakasugi T, Takeyama H. PTEN regulate angiogenesis through PI3K/akt/VEGF signaling pathway in human pancreatic cancer cells. Mol Cell Biochem. 2009;331:161–171. doi: 10.1007/s11010-009-0154-x. [DOI] [PubMed] [Google Scholar]
- 39.Waldner MJ, Neurath MF. Targeting the VEGF signaling pathway in cancer therapy. Expert Opin Ther Targets. 2012;16:5–13. doi: 10.1517/14728222.2011.641951. [DOI] [PubMed] [Google Scholar]
- 40.Lee S, Chen TT, Barber CL, Jordan MC, Murdock J, Desai S, Ferrara N, Nagy A, Roos KP, Iruela-Arispe ML. Autocrine VEGF signaling is required for vascular homeostasis. Cell. 2007;130:691–703. doi: 10.1016/j.cell.2007.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ferrara N, Gerber H, LeCouter J. The biology of VEGF and its receptors. Nat Med. 2003;9:669–676. doi: 10.1038/nm0603-669. [DOI] [PubMed] [Google Scholar]
- 42.Franco CA, Li Z. SRF in angiogenesis: Branching the vascular system. Cell Adh Migr. 2009;3:264–267. doi: 10.4161/cam.3.3.8291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chai J, Jones MK, Tarnawski AS. Serum response factor is a critical requirement for VEGF signaling in endothelial cells and VEGF-induced angiogenesis. FASEB J. 2004;18:1264–1266. doi: 10.1096/fj.03-1232fje. [DOI] [PubMed] [Google Scholar]
- 44.Ahmad S, Hewett PW, Wang P, Al-Ani B, Cudmore M, Fujisawa T, Haigh JJ, le Noble F, Wang L, Mukhopadhyay D, Ahmed A. Direct evidence for endothelial vascular endothelial growth factor receptor-1 function in nitric oxide-mediated angiogenesis. Circ Res. 2006;99:715–722. doi: 10.1161/01.RES.0000243989.46006.b9. [DOI] [PubMed] [Google Scholar]
- 45.Morin-Brureau M, Lebrun A, Rousset MC, Fagni L, Bockaert J, de Bock F, Lerner-Natoli M. Epileptiform activity induces vascular remodeling and zonula occludens 1 downregulation in organotypic hippocampal cultures: Role of VEGF signaling pathways. J Neurosci. 2011;31:10677–10688. doi: 10.1523/JNEUROSCI.5692-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dzietko M, Derugin N, Wendland M, Vexler Z, Ferriero D. Delayed VEGF treatment enhances angiogenesis and recovery after neonatal focal rodent stroke. Transl Stroke Res. 2013;4:189–200. doi: 10.1007/s12975-012-0221-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shimotake J, Derugin N, Wendland M, Vexler ZS, Ferriero DM. Vascular endothelial growth factor receptor-2 inhibition promotes cell death and limits endothelial cell proliferation in a neonatal rodent model of stroke. Stroke. 2010;41:343–349. doi: 10.1161/STROKEAHA.109.564229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Waltenberger J, Mayr U, Pentz S, Hombach V. Functional upregulation of the vascular endothelial growth factor receptor KDR by hypoxia. Circulation. 1996;94:1647–1654. doi: 10.1161/01.CIR.94.7.1647. [DOI] [PubMed] [Google Scholar]
- 49.Yang J, Liu H, Liu X. VEGF promotes angiogenesis and functional recovery in stroke rats. J Invest Surg. 2010;23:149–155. doi: 10.3109/08941930903469482. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.