Abstract
Long noncoding RNAs (lncRNAs) are associated with various diseases including cataracts. The role of lncRNA potassium voltage-gated channel subfamily Q member 1 opposite strand/antisense transcript 1 (KCNQ1OT1) on lens epithelial cell (LEC) proliferation and epithelial-mesenchymal transition (EMT) in cataracts disease remains unclear. In the present study, KCNQ1OT1 and mothers against decapentaplegic homolog (SMAD)4 expression levels were upregulated in human cataract lens posterior capsular samples and in transforming growth factor (TGF)-β2-treated SRA01/04 cells, as demonstrated by reverse transcription-quantitative polymerase chain reaction, immunohistochemical staining and western blot analyses. A further loss of function test revealed that suppression of KCNQ1OT1 inhibited the proliferation and EMT of TGF-β2-treated SRA01/04 cells. Additionally, the present study reported that increase and decrease of KCNQ1OT1 regulated SMAD4 expression, which indicated that SMAD4 may be a downstream gene of KCNQ1OT1. Finally, a constructed SMAD4 RNA interference experiment confirmed that the function of KCNQ1OT1 was to act on LEC proliferation and EMT, and this was achieved via the SMAD4 signaling pathway. The findings of the present study may provide a novel target for molecular therapy of cataracts disease.
Keywords: long noncoding RNA, potassium voltage-gated channel subfamily Q member 1 opposite strand/antisense transcript 1, mothers against decapentaplegic homolog 4, proliferation, epithelial-mesenchymal transition, cataracts
Introduction
As one of the common age-associated diseases, cataracts is a troublesome disease and is a leading cause of non-traumatic blindness worldwide. The global progression of aging has been associated with cataracts and resulting visual impairment, which has placed a burden on society. According to an epidemiological study published in 2011, ~96% of people aged >60 were reported to exhibit varying lens opacity (1). It is well known that various morphological and functional alterations of lens cells, including disordered cell cycle, DNA damage, lens epithelial cells (LECs) excessive proliferation and abnormal epithelial-mesenchymal transition (EMT) participate in the pathological formation of cataracts (2–5). Therefore, investigation of a key regulator associated with LECs' proliferation and EMT may considerably contribute to the development of molecular therapy for cataracts disease.
Long noncoding RNAs (lncRNAs) are macromolecules grouped under noncoding RNAs, with length usually >200 nucleotides. They extensively exhibit varied roles in numerous physiological and pathological processes, including cell proliferation, cell differentiation, apoptosis, tumorigenesis and EMT (6–8). LncRNA potassium voltage-gated channel subfamily Q member 1 opposite strand/antisense transcript 1 (lncKCNQ1OT1) is located on human chromosome 11p15.5 and is associated with the development of a wide array of diseases. Ren et al (9) reported that knockdown of KCNQ1OT1 suppresses the proliferation and invasion of A549 cells, as well as advanced cellular apoptosis of A549 cells. Jin et al (10) suggested that KCNQ1OT1 may promote cataractogenesis, which may be dependent upon microRNA-214 and activation of the caspase-1 pathway.
In the present study, the expression levels of KCNQ1OT1 were investigated in cataract tissue specimens and in a constructed cataract cell model induced by transforming growth factor (TGF)-β2. In addition, the function of KCNQ1OT1 in LEC proliferation and EMT was evaluated. The findings of the present study may provide insight into a novel contributor to the process of cataract formation.
Materials and methods
Patients and tissue samples collection
A total of 30 cases (17 female and 13 male; mean age 63.5 years, range 58–73 years) of fresh posterior lens capsule specimens with age-associated cataracts during phacoemulsification and paired fresh posterior lens capsule specimens without cataracts caused by ocular trauma during ophthalmectomy were collected at 4th People's Hospital of Shenyang between July 2015 and July 2017. Written informed consent was obtained from all patients for all clinical investigations conducted and the present study was approved by the Institute Research Medical Ethics Committee of 4th People's Hospital of Shenyang (Shenyang, China).
Cell culture and TGF-β2 intervention
Human LEC cell line SRA01/04 was purchased from the American Type Culture Collection (Manassas, VA, USA) and was cultured in Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (Sigma-Aldrich, Merck KGaA, Darmstadt, Germany), 100 U/ml penicillin (Baoman Biotechnology Co., Ltd., Shanghai, China), and 100 U/ml streptomycin (Baoman Biotechnology Co., Ltd.). The cell line was cultured at 37°C in a humidified atmosphere containing 5% CO2 until 80–90% confluent. For the TGF-β2 intervention, 10 ng/ml recombinant human TGF-β2 (Biolegend, Inc., San Diego, CA, USA) was applied to induce a cataract cell model according to a previous study (9).
Cell transfection
KCNQ1OT1 silencing plasmids short hairpin (sh)RNA (KCNQ1OT1 shRNA) and negative control shRNA (NC shRNA), KCNQ1OT1 overexpression plasmids (pcDNA-KCNQ1) and corresponding empty vector plasmids (pcDNA3.1), mothers against decapentaplegic homolog 4 (SMAD4) silencing plasmids (SMAD4 shRNA) and negative control shRNA (NC shRNA) were constructed by Guangzhou RiboBio Co., Ltd. (Guangzhou, China). TGF-β2-stimulated SRA01/04 cells were cultured at 60–80% confluence and were then used for a further cell transfection. Plasmids were transfected into TGF-β2-stimulated SRA01/04 cells by using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocols. Then, 48 h after transfection, the cells were used for further experimentation.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
The procedure was carried out as previously described (10). In brief, total RNAs of tissue specimens and of the cells following different interventions were extracted by TRIzol® (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocols. cDNAs were synthesis by PrimeScriptTM RT reagent kit (Takara Biotechnology Co., Ltd., Dalian, China). qPCR was performed in a total reaction volume of 50 µl with 35 cycles (denaturation at 95°C for 5 sec, annealing at 60°C for 30 sec and extension 72°C for 5 sec) using a SYBR Premix Ex Taq II kit (Takara Biotechnology Co., Ltd.) and an Applied Biosystems 7500 Fluorescent Quantitative PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The expression of KCNQ1OT1 and SMAD4 were calculated using GAPDH as an internal control by the 2−ΔΔCq method (11). The primers used were as follows: KCNQ1OT1 forward, 5′-TGCAGAAGACAGGACACTGG-3′, reverse 5′-CTTTGGTGGGAAAGGACAGA-3′; SMAD4 forward, 5′-TGGGAAGAGATCACCCTGTC-3′ and reverse 5′-CCCAACGGTAAAAGACCTCA-3′; GAPDH forward, 5′-GCACCGTCAAGGCTGAGAAC-3′, reverse 5′-TGGTGAAGACGCCAGTGGA-3′.
Western blot analysis
Total protein of tissue specimens and cells were lysed by ice-cold radio immunoprecipitation assay (RIPA) buffer (Sigma-Aldrich, Merck KGaA). Following protein concentration quantification with a Bicinchoninic Acid protein assay kit (Santa Cruz Biotechnology, Inc., Dallas, TX, USA), 40 µg protein samples were subjected to 10% SDS-PAGE and transferred onto a polyvinylidene difluoride membrane (PVDF; Amresco, LLC, Solon, OH, USA) and then blocked by 5% BSA (Cell Signaling Technology, Inc., Danvers, MA, USA) for 1 h at room temperature. Subsequently, membranes were incubated with antibodies against SMAD4 (1:5,000; cat no. ab40759; Abcam, Cambridge, MA, UK), E-cadherin (1:100; cat. no. ab76055; Abcam), fibronectin (1 µg/ml; cat. no: ab23750; Abcam) and GAPDH (1:10,000; cat. no. ab128915; Abcam) at 4°C overnight. The following day, the membranes were incubated with secondary antibodies [goat anti-rabbit IgG horseradish peroxidase (HRP), 1:2,000; cat. no. ab205718, Abcam] at room temperature for 1 h. Following washing with TBST 3 times, an ECL Western Blotting Substrate kit (cat. no. ab65623; Abcam) was used for chemiluminescence imaging with Image J software version 2X (National Institutes of Health, Bethesda, MD, USA).
Immunohistochemical (IHC) staining
The procedure was carried out as previously described (12). Briefly, the collected cataract tissues were treated orderly: 4% paraformaldehyde fixation, paraffin-embedding, section (4 µm thickness), deparaffinization, a descending series of 100, 95, 80 and 70% alcohol were applied for rehydration followed by 3% hydrogen peroxide incubation for 10 min, antigen retrieval, blocking with 10% goat serum (BioWorld Technology, Inc., St. Louis Park, MN, USA), primary antibody incubation (rabbit anti-SMAD4 antibody; 1:100; cat. no. ab40759; Abcam) at 4°C overnight, secondary antibody incubation [goat anti-rabbit IgG horseradish peroxidase (HRP), 1:2,000; cat. no. ab205718; Abcam] at 37°C for 20 min, streptavidin-horseradish peroxidase complex incubation, 3,3′-diaminobenzidine tetrahydrochloride (MedChemExpress, Monmouth Junction, NJ, USA) staining, and hematoxylin (Amresco, LLC) was used as a counterstain at room temperature for 1 min. All sections were independently assessed by two experienced pathologists and SMAD4 expression levels were evaluated by calculating the proportion of positive staining and staining intensity of tumor tissue. Sections with inconsistent results were re-examined by the original two pathologists and a senior pathologist until a consensus was attained.
5-Ethynyl-20-deoxyuridine (EdU) incorporation assay and Cell Counting Kit-8 (CCK-8) assay
The procedure was carried out as previously described (13). Briefly, SRA01/04 cells (5×103) transfected with KCNQ1OT1 or SMAD4 plasmids were seeded in a 96-well plate and cultured for 72 h. The following assays were performed according to the manufacturer's protocols of an EdU detection kit (cat. no. KGA331-500; Nanjing KeyGen Biotech Co., Ltd., Nanjing, China). The nuclei were observed under a fluorescent microscope at an excitation wavelength of 350 nm (Leica Microsystems GmbH, Wetzlar, Germany). Images were analyzed using Image-Pro Plus software version 6.0 (Media Cybernetics, Inc., Rockville, MD, USA). The quantitative data were expressed as the percentage of EdU-positive nuclei relative to total number of nuclei counted. In addition, a CCK-8 assay was performed as previously described (14). SRA01/04 cells (2×103) were cultured with DMEM supplemented with 10% FBS in a 96-well plate for 24 h and followed by transfection of ddH2O (mock group), KCNQ1OT shRNA (KCNQ1OT shRNA) and negative control shRNA (NC shRNA group) for 24 h. At days 1, 2, 3, 4 and 5 following transfection, 10 µl CCK-8 solution was added into each well and incubated at 37 °C for 4 h. The absorbance was measured at an optical density of 450 nm using a microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Statistical analysis
All experiments were repeated in triplicate and all data from three independent experiments were expressed as mean ± standard deviation. GraphPad Prism v5.0 software (GraphPad Software, Inc., La Jolla, CA, USA) was used for statistical analysis. For paired groups, the expression level of KCNQ1OT1 and SMAD4, as well as alterations in proliferative ability were analyzed by a two-tailed Student's t-test. For the comparison of more than two groups, one-way analysis of variance was conducted followed by a Student-Newman-Keuls post hoc test. Spearman correlation analysis was performed to determine the correlation between KCNQ1OT1 and SMAD4. P<0.05 was considered to indicate a statistically significant difference.
Results
Increased KCNQ1OT1 expression levels are observed in human cataract lens posterior capsular samples and in TGF-β2 treated SRA01/04 cells
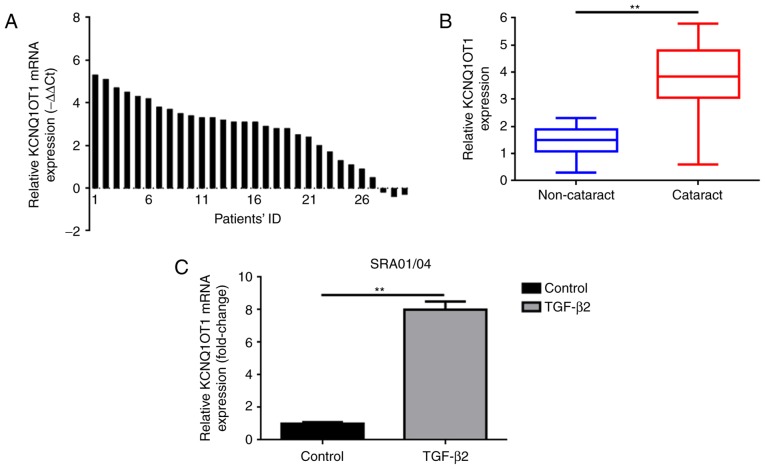
The present study investigated the expression levels of KCNQ1OT1 in 30 fresh specimens of posterior lens capsules with age-associated cataract and 30 paired posterior lens capsules specimens without age-associated cataracts via RT-qPCR analysis. As presented in Fig. 1A and B, KCNQ1OT1 was upregulated in 90% (27/30) specimens of posterior lens capsules with age-associated cataracts compared with in adjacent non-cataract specimens.
Figure 1.
Increased KCNQ1OT1 expression levels are observed in human cataract lens posterior capsular samples and in TGF-β2 induced cataract cell models. (A) KCNQ1OT1 was upregulated in 90% (27/30) tissue specimens with cataract as detected by RT-qPCR and the 2−ΔΔCq method. (B) KCNQ1OT1 was significantly upregulated in tissue specimens with cataract as detected by RT-qPCR, **P<0.01 vs. non-cataract group. (C) KCNQ1OT1 was elevated in SRA01/04 cells with the presence of TGF-β2 as observed via RT-qPCR, **P<0.01 vs. control group. All data were presented as the mean ± standard deviation from three independent experiments. ID, identification; KCNQ1OT1, potassium voltage-gated channel subfamily Q member 1 opposite strand/antisense transcript 1; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; TGF-β2, transforming growth factor-β2.
TGF-β2 was administered to SRA01/04 cells to induce a cataract cell model as previously reported (9). RT-qPCR was also applied to measure the expression of KCNQ1OT1 in the human LEC line SRA01/04 treated with or without TGF-β2. As presented in Fig.1C, significantly increased expression of KCNQ1OT1 was confirmed in SRA01/04 cells treated TGF-β2, however not in SRA01/04 cells incubated without TGF-β2.
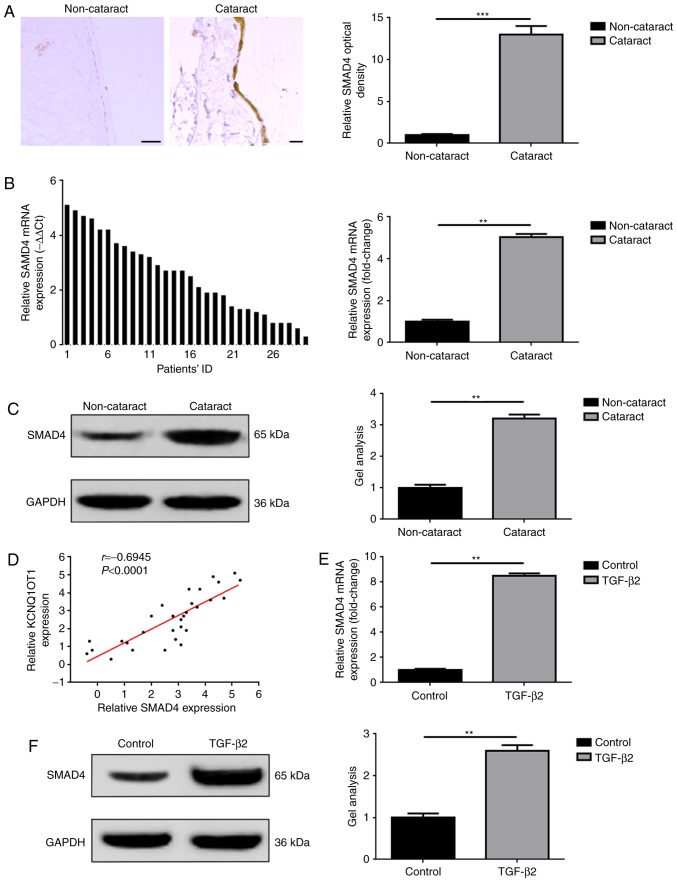
SMAD4 is overexpressed in human cataract lens posterior capsular samples and in TGF-β2 treated SRA01/04 cells
The associated effects of SMAD4, including the mediation of proliferation and EMT have been widely reported in a variety of diseases, such as cataracts (15–17). In the present study, the expression levels of SMAD4 in collected tissue specimens and in cataract cell models induced by TGF-β2 were investigated. The results were in accordance with previous studies (17). As presented in Fig. 2A-C, the overexpression of SMAD4 was detected only in tissue specimens with cataract disease, confirmed by IHC, RT-qPCR and western blot analyses. In addition, a positive correlation between KCNQ1OT1 and SMAD4 expression in cataract tissue specimens was determined by Spearman correlation analysis (Fig. 2D). Furthermore, significant upregulation of SMAD4 expression levels were detected within SRA01/04 cells treated with TGF-β2 compared with in cells not treated with TGF-β2, as determined via RT-qPCR and western blot assays (Fig. 2E and F).
Figure 2.
Elevated expression levels of SMAD4 are observed in human cataract lens posterior capsular samples and in TGF-β2 induced cataract cell models. SMAD4 was overexpressed in tissue specimens with cataract as detected by (A) immunohistochemistry, (B) RT-qPCR and (C) western blotting, **P<0.01 vs. non-cataract group. (D) A significant positive correlation between KCNQ1OT1 and SMAD4 as presented by Spearman correlation analysis. SMAD4 expression levels were elevated in SRA01/04 cells in the presence of TGF-β2 as detected by (E) RT-qPCR and (F) western blotting, **P<0.01 vs. control group. All data were presented as the mean ± standard deviation from three independent experiments. ID, identification; KCNQ1OT1, potassium voltage-gated channel subfamily Q member 1 opposite strand/antisense transcript 1; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; SMAD4, mothers against decapentaplegic homolog 4; TGF-β2, transforming growth factor-β2.
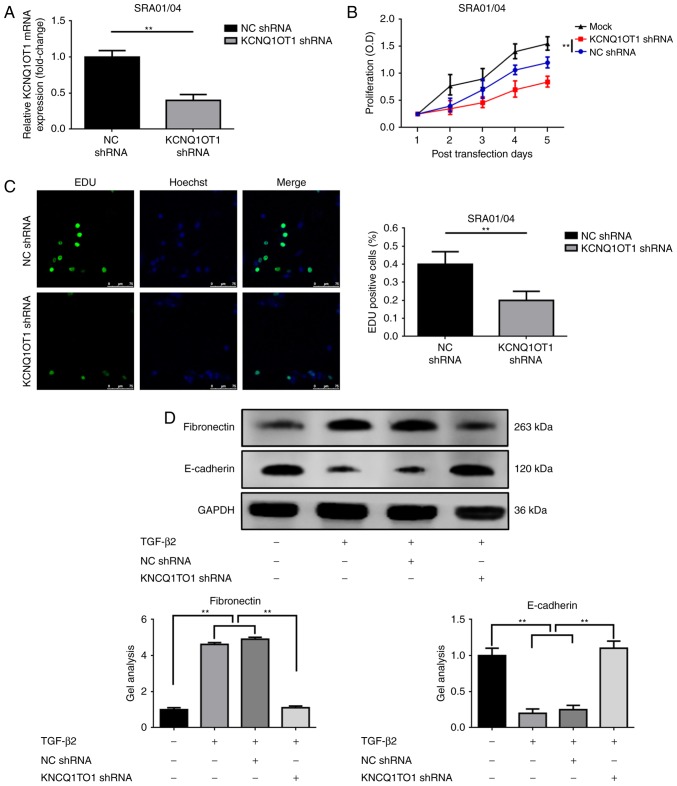
Knockdown of KCNQ1OT1 suppresses the proliferation and EMT of TGF-β2-treated SRA01/04 cells
As previously mentioned, the expression of KCNQ1OT1 in tissue and at the cellular levels were analyzed. The present study also investigated the effects of KCNQ1OT1 on LEC proliferation and EMT. Firstly, KCNQ1OT1 in TGF-β2 treated SRA01/04 cells was suppressed via transfection with KCNQ1OT1 shRNA, which was confirmed by RT-qPCR (Fig. 3A). Secondly, CCK-8 and EDU assays were performed to determine alterations in proliferative ability following the suppression of KCNQ1OT1 expression. As demonstrated in Fig. 3B and C, the downregulation of KCNQ1OT1 significantly suppressed SRA01/04 cell proliferation in the presence of TGF-β2. Lastly, the effects of KCNQ1OT1 on SRA01/04 cells EMT with TGF-β2 treatment were investigated. As presented in Fig. 3D, TGF-β2 induced the increase of fibronectin but the suppression of E-cadherin expression levels in SRA01/04 cells. This phenomenon was reversed by knockdown of KCNQ1OT1 (transfection of KCNQ1OT1 shRNA), suggesting that the suppression of KCNQ1OT1 expression inhibited EMT in TGF-β2-induced cataract cell models.
Figure 3.
Downregulation of KCNQ1OT1 suppresses the proliferation and EMT of TGF-β2-treated SRA01/04 cells. (A) KCNQ1OT1 expression was suppressed by transfection with KCNQ1OT1 shRNA as confirmed by reverse transcription-quantitative polymerase chain reaction, **P<0.01 vs. NC shRNA group. Suppression of KCNQ1OT1 expression inhibited SRA01/04 cell proliferation as determined by (B) Cell Counting Kit-8 and (C) EdU assays, **P<0.01 vs. NC shRNA group. (D) TGF-β2 induced an elevation of fibronectin but a decrease in E-cadherin expression levels (promotion of EMT), and the tendency was reversed by the suppression of KCNQ1OT1, as detected by western blotting, **P<0.01. All data were present as the mean ± standard deviation from three independent experiments. EdU, 5-Ethynyl-20-deoxyuridine; KCNQ1OT1, potassium voltage-gated channel subfamily Q member 1 opposite strand/antisense transcript 1; NC, negative control; shRNA, short hairpin RNA; TGF-β2, transforming growth factor-β2.
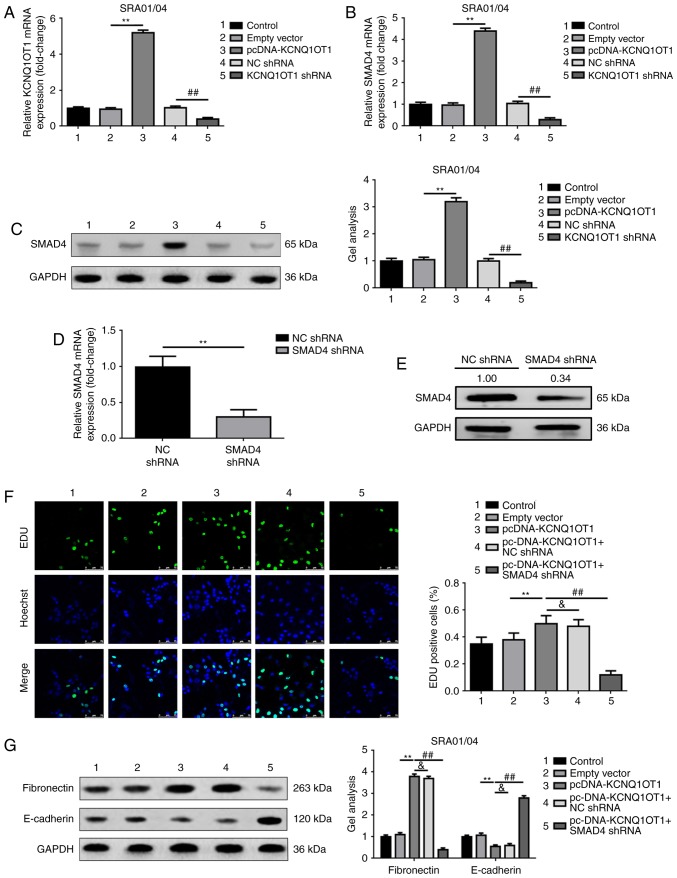
KCNQ1OT1 promotes proliferation and EMT via upregulation of SMAD4
As both KCNQ1OT1 and SMAD4 have been revealed to be involved in LEC proliferation and EMT, the association between KCNQ1OT1 and SMAD4 was investigated in the present study. The results revealed that upregulated and downregulated KCNQ1OT1 may increase or reduce the expression levels of SMAD4, respectively (Fig. 4A-C). Subsequently, RNAi was employed to determine the function of SMAD4, which may be involved in KCNQ1OT1-induced promotion of proliferation and EMT. The silencing effect of SMAD4 shRNA was firstly confirmed by RT-qPCR and western blot analyses (Fig. 4D and E). As expected, the results of the present study revealed that KCNQ1OT1-induced proliferation of SRA01/04 cells in the presence of TGF-β2 was reversed by silencing SMAD4 (co-transfection of pcDNA-KCNQ1OT1 and SMAD4 shRNA; Fig. 4F). Furthermore, the same trend was observed in the analysis of EMT, characterized by fibronectin and E-cadherin expression within SRA01/04 cells in the presence of TGF-β2 (Fig. 4G). These findings suggested that KCNQ1OT1 enhanced SRA01/04 cell proliferation and EMT via the SMAD4 signaling pathway.
Figure 4.
KCNQ1OT1 promotes proliferation and EMT via the upregulation of SMAD4. (A) Elevation and suppression of KCNQ1OT1 was confirmed by RT-qPCR, **P<0.01 vs. empty vector group, ##P<0.01 vs. NC shRNA group. Elevation and suppression of KCNQ1OT1 positively regulated SMAD4 mRNA and protein expression levels as determined by (B) RT-qPCR and (C) western blotting, respectively. **P<0.01 vs. empty vector group, ##P<0.01 vs. NC shRNA group. SMAD4 expression levels were significantly knocked down by transfection of SMAD4 shRNA, as confirmed by (D) RT-qPCR (**P<0.01 vs. NC shRNA group) and (E) western blot analyses (Relative SMAD4 protein expression in SMAD4 shRNA group was 0.34 comparing with NC shRNA group, which was 1.00). (F) Upregulation of KCNQ1OT1 (pcDNA-KCNQ1OT1 group) promoted SRA01/04 cell proliferation, this effect was reduced by silencing SMAD4 expression (pcDNA-KCNQ1OT1+SMAD4 shRNA group) as detected by EdU, **P<0.01, ##P<0.01 vs. pcDNA-KCNQ1OT1 group, &P>0.05. (G) Elevation of KCNQ1OT1 (pcDNA-KCNQ1OT1 group) promoted the progress of EMT (elevation of fibronectin, but suppression of E-cadherin expression levels), however the facilitative effect was decreased by silencing SMAD4 expression (pcDNA-KCNQ1OT1+SMAD4 shRNA group) as detected by western blotting, **P<0.01, ##P<0.01 vs. pcDNA-KCNQ1OT1 group, &P>0.05. 1, control group; 2, empty vector group; 3, pcDNA-KCNQ1OT1 group; 4, NC shRNA group; and 5, KCNQ1OT1 shRNA group. All data were presented as the mean ± standard deviation from three independent experiments. EdU, 5-Ethynyl-20-deoxyuridine; KCNQ1OT1, potassium voltage-gated channel subfamily Q member 1 opposite strand/antisense transcript 1; NC, negative control; shRNA, short hairpin RNA; SMAD4, mothers against decapentaplegic homolog 4; TGF-β2, transforming growth factor-β2.
Discussion
Cataracts are generally classified into two types of disease: Anterior subcapsular cataract (ASC) and posterior capsule opacification (PCO). ASC is a primary cataract disease characterized by star-shaped or irregular fibrotic plaques beneath the anterior capsule, resulting in notable reduction in vision due to visual axis involvement (18). PCO, known as a secondary lens opacification, is usually caused by aberrant growth of lens epithelial cells that remain in the capsular bag following cataract surgery (19). The cellular mechanism of ASC and PCO involves the proliferation, migration and EMT of LECs, leading to the transition from epithelial cells to fibroblasts, and the production of extracellular matrix proteins (collagens I, IV and fibronectin), which finally contributes to the formation of subcapsular plaques beneath the lens anterior or posterior capsule (20). At present, increasing evidence indicates that numerous factors and pathways mediate the proliferation, migration and EMT of residual LECs, and may contribute to the pathology of age-associated cataracts (21–23). Liu and Xiao (24) reported that hypoxia promotes hypoxia inducible factor-1α and facilitates EMT via the Notch homolog 1, translocation-associated (Drosophila)/snail family transcriptional repressor 1/E-cadherin pathway in SRA01/04 cells. Zhang et al (25) revealed that the silencing of mammalian target of rapamycin significantly inhibits the proliferation and migration of LEC. In the present study, the proliferation and EMT process of cataracts were investigated at the cellular level via a TGF-β2-induced cataract cell model; KCNQ1OT1 overexpression and suppression may affect the proliferation and EMT in SRA01/04 cells in the presence of TGF-β2.
LncRNA KCNQ1OT1 is located at human chromosome 11p15.5 and is generally reported as an oncogene in a variety of cancers types, including lung adenocarcinoma, hepatocellular carcinoma and breast cancer (25–27). Aberration of KCNQ1OT1 transcription is observed at a high frequency in patients with colorectal cancers (28). Gong et al (29) demonstrated that the knockdown of KCNQ1OT1 inhibits the proliferation and migration/invasive abilities of glioma cells via microRNA-370/cyclin E2 pathway. Jin et al (10) revealed that KCNQ1OT1 expression levels are elevated in human cataract lens anterior capsular samples and in SRA01/04 cell lines treated with H2O2. In the present study, the expression levels of KCNQ1OT1 were measured in the collected cataract tissue specimens. A previous report demonstrated an elevation in KCNQ1OT1 expression levels in cataract tissue specimens (30). Ectogenic TGF-β2 is widely used in cataract-associated research, particularly in LEC-associated EMT (16,31,32). In the same manner, the present study employed TGF-β2-treated SRA01/04 cells to generate a cataract cell model. The expression levels of KCNQ1OT1 were upregulated in the induced cataract cell models, as reported in the present study, which indicated that KCNQ1OT1 may function as an initiator in the formation of cataracts. Additionally, using a constructed functional experiment, the downregulation of KCNQ1OT1 inhibits SRA01/04 cell proliferation and EMT, which are the key phases in cataract formation (19).
SMAD4 belongs to the SMAD family and is a critical intracellular mediator in the TGF-β/SMAD4 signaling pathway. The effects of SMAD4 on the mediation of proliferation and EMT have been reported to be associated with cataractogenesis. Wang et al (19) suggested that microRNA-204-5p regulates EMT during human PCO by targeting SMAD4. In a SMAD4 knockout mouse study, Li et al (33) revealed that loss of SMAD4 suppresses E-cadherin expression, but upregulates N-cadherin expression, which is associated with congenital cataracts. In a cataract study, Nahomi et al (18) suggested that LEC cells treated with TGF-β2 present marked upregulation of SMAD4 and EMT-associated markers. In the present study, the expression levels of SMAD4 in cataracts and the function of KCNQ1OT1 on SMAD4 were investigated. The results of the present study revealed that SMAD4 was significantly upregulated in cataract tissue specimens and within induced cataract cell models. Furthermore, the results indicated that an increase and decrease of KCNQ1OT1 may correspondingly regulate SMAD4 expression levels. In addition, via antisense investigations, the present study proposed that SMAD4 was a downstream target of KCNQ1OT1. The effects of KCNQ1OT1 on cell proliferation and EMT were achieved via SMAD4 signaling. KCNQ1OT1 may promote cell proliferation and EMT via the regulation of SMAD4 in SRA01/04 cells.
In conclusion, the formation of age-associated cataracts is an extremely complex issue associated with several mechanisms and complex networks comprising of numerous molecules. The findings of the present study provide a novel target and insight into the pathogenesis of age-associated cataract.
Acknowledgements
The authors would like to thank Dr Xi Zhang from Institute for Cardiovascular Prevention, Ludwig-Maximilians-University for helping us prepare the manuscript.
Funding
The present study was supported by grants from National Natural Science Foundation of China (grant no. 81502333), PhD Start-up Research Foundation of Liaoning Province (grant no. 201601225).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
BC was responsible for the analysis and interpretation of data of the manuscript; JM and CL were responsible for statistical analysis; LZ was responsible for design and drafting of the manuscript.
Ethics approval and consent to participate
Written informed consent was obtained from all patients for all clinical investigations conducted and the present study was approved by the Institute Research Medical Ethics Committee of 4th People's Hospital of Shenyang (Shenyang, China).
Consent for publication
Written informed consent was obtained.
Conflict of interest
The authors declare that they have no competing interests.
References
- 1.Cruciani F, Amore F, Albanese G, Anzidei R. Investigation about causes of blindness and low vision among members of Blind and Visually Impaired Italian Union (UICI) Clin Ter. 2011;162:e35–e42. [PubMed] [Google Scholar]
- 2.Liu T, Zhang L, Wang Y, Zhang H, Li L, Bao X. Dickkopf-1 inhibits Wnt3a-induced migration and epithelial-mesenchymal transition of human lens epithelial cells. Exp Eye Res. 2017;161:43–51. doi: 10.1016/j.exer.2017.06.001. [DOI] [PubMed] [Google Scholar]
- 3.Michael R, Bron AJ. The ageing lens and cataract: A model of normal and pathological ageing. Philos Trans R Soc Lond B Biol Sc. 2011;366:1278–1292. doi: 10.1098/rstb.2010.0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zeng K, Feng QG, Lin BT, Ma DH, Liu CM. Effects of microRNA-211 on proliferation and apoptosis of lens epithelial cells by targeting SIRT1 gene in diabetic cataract mice. Biosci Rep. 2017;37 doi: 10.1042/BSR20170695. pii: BSR20170695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang L, Wang Y, Li W, Tsonis PA, Li Z, Xie L, Huang Y. MicroRNA-30a regulation of epithelial-mesenchymal transition in diabetic cataracts through targeting SNAI1. Sci Rep. 2017;7:1117. doi: 10.1038/s41598-017-01320-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu H, Zhen Q, Fan Y. LncRNA GHET1 promotes esophageal squamous cell carcinoma cells proliferation and invasion via induction of EMT. Int J Biol Markers. 2017;32:e403–e408. doi: 10.5301/ijbm.5000304. [DOI] [PubMed] [Google Scholar]
- 7.Shen Y, Dong LF, Zhou RM, Yao J, Song YC, Yang H, Jiang Q, Yan B. Role of long non-coding RNA MIAT in proliferation, apoptosis and migration of lens epithelial cells: A clinical and in vitro study. J Cell Mol Med. 2016;20:537–548. doi: 10.1111/jcmm.12755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, Zhang Y, Yang T, Zhao W, Wang N, Li P, Zeng X, Zhang W. Long non-coding RNA MALAT1 for promoting metastasis and proliferation by acting as a ceRNA of miR-144-3p in osteosarcoma cells. Oncotarget. 2017;8:59417–59434. doi: 10.18632/oncotarget.19727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ren K, Xu R, Huang J, Zhao J, Shi W. Knockdown of long non-coding RNA KCNQ1OT1 depressed chemoresistance to paclitaxel in lung adenocarcinoma. Cancer Chemother Pharmacol. 2017;80:243–250. doi: 10.1007/s00280-017-3356-z. [DOI] [PubMed] [Google Scholar]
- 10.Jin X, Jin H, Shi Y, Guo Y, Zhang H. Long Non-Coding RNA KCNQ1OT1 Promotes Cataractogenesis via miR-214 and Activation of the Caspase-1 Pathway. Cell Physiol Biochem. 2017;42:295–305. doi: 10.1159/000477330. [DOI] [PubMed] [Google Scholar]
- 11.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 12.Zhang B, Chen Y, Qiu M, Ding Z. Long noncoding RNA expression profile in HLE B-3 cells during TGF- β2-induced epithelial-mesenchymal transition. BMC Ophthalmol. 2017;17:69. doi: 10.1186/s12886-017-0461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Yang T, Zhang Z, Lu M, Zhao W, Zeng X, Zhang W. Long non-coding RNA TUG1 promotes migration and invasion by acting as a ceRNA of miR-335-5p in osteosarcoma cells. Cancer Sci. 2017;108:859–867. doi: 10.1111/cas.13201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lv J, Fan HX, Zhao XP, Lv P, Fan JY, Zhang Y, Liu M, Tang H. Long non-coding RNA Unigene56159 promotes epithelial-mesenchymal transition by acting as a ceRNA of miR-140-5p in hepatocellular carcinoma cells. Cancer Lett. 2016;382:166–175. doi: 10.1016/j.canlet.2016.08.029. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y, Yang T, Liu Y, Zhao W, Zhang Z, Lu M, Zhang W. Decrease of miR-195 promotes chondrocytes proliferation and maintenance of chondrogenic phenotype via targeting FGF-18 pathway. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18050975. pii: E975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nguyen K, Sparks J, Omoruyi FO. Investigation of the cytotoxicity, antioxidative and immune-modulatory effects of Ligusticum porteri (Osha) root extract on human peripheral blood lymphocytes. J Integr Med. 2016;14:465–472. doi: 10.1016/S2095-4964(16)60280-7. [DOI] [PubMed] [Google Scholar]
- 17.de Iongh RU, Wederell E, Lovicu FJ, McAvoy JW. Transforming growth factor-beta-induced epithelial-mesenchymal transition in the lens: A model for cataract formation. Cells Tissues Organs. 2005;179:43–55. doi: 10.1159/000084508. [DOI] [PubMed] [Google Scholar]
- 18.Nahomi RB, Pantcheva MB, Nagaraj RH. αB-crystallin is essential for the TGF-β2-mediated epithelial to mesenchymal transition of lens epithelial cells. Biochem J. 2016;473:1455–1469. doi: 10.1042/BCJ20160128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Li W, Zang X, Chen N, Liu T, Tsonis PA, Huang Y. MicroRNA-204-5p regulates epithelial-to-mesenchymal transition during human posterior capsule opacification by targeting SMAD4. Invest Ophthalmol Vis Sci. 2013;54:323–332. doi: 10.1167/iovs.12-10904. [DOI] [PubMed] [Google Scholar]
- 20.Lovicu FJ, Schulz MW, Hales AM, Vincent LN, Overbeek PA, Chamberlain CG, McAvoy JW. TGFbeta induces morphological and molecular changes similar to human anterior subcapsular cataract. Br J Ophthalmol. 2002;86:220–226. doi: 10.1136/bjo.86.2.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lovicu FJ, Shin EH, McAvoy JW. Fibrosis in the lens. Sprouty regulation of TGFβ-signaling prevents lens EMT leading to cataract. Exp Eye Res. 2016;142:92–101. doi: 10.1016/j.exer.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen X, Xiao W, Chen W, Liu X, Wu M, Bo Q, Luo Y, Ye S, Cao Y, Liu Y. MicroRNA-26a and −26b inhibit lens fibrosis and cataract by negatively regulating Jagged-1/Notch signaling pathway. Cell Death Differ. 2017;24:1431–1442. doi: 10.1038/cdd.2017.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu X, Ruan J, Ma B, Luo M. Bit1-a potential positive regulator of epithelial-mesenchymal transition in lens epithelial cells. Graefes Arch Clin Exp Ophthalmol. 2016;254:1311–1318. doi: 10.1007/s00417-016-3357-3. [DOI] [PubMed] [Google Scholar]
- 24.Liu L, Xiao W. Notch1 signaling induces epithelial-mesenchymal transition in lens epithelium cells during hypoxia. BMC ophthalmology. 2017;17:135. doi: 10.1186/s12886-017-0532-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang C, Liu J, Jin N, Zhang G, Xi Y, Liu H. SiRNA Targeting mTOR effectively prevents the proliferation and migration of human lens epithelial cells. PLoS One. 2016;11:e0167349. doi: 10.1371/journal.pone.0167349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiao W, Chen X, Li W, Ye S, Wang W, Luo L, Liu Y. Quantitative analysis of injury-induced anterior subcapsular cataract in the mouse: a model of lens epithelial cells proliferation and epithelial-mesenchymal transition. Sci Rep. 2015;5:8362. doi: 10.1038/srep08362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodriguez BA, Weng YI, Liu TM, Zuo T, Hsu PY, Lin CH, Cheng AL, Cui H, Yan PS, Huang TH. Estrogen-mediated epigenetic repression of the imprinted gene cyclin-dependent kinase inhibitor 1C in breast cancer cells. Carcinogenesis. 2011;32:812–821. doi: 10.1093/carcin/bgr017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sunamura N, Ohira T, Kataoka M, Inaoka D, Tanabe H, Nakayama Y, Oshimura M, Kugoh H. Regulation of functional KCNQ1OT1 lncRNA by β-catenin. Sci Rep. 2016;6:20690. doi: 10.1038/srep20690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gong W, Zheng J, Liu X, et al. Knockdown of Long Non-Coding RNA KCNQ1OT1 Restrained Glioma Cells' Malignancy by Activating miR-370/CCNE2 Axis. Frontiers in cellular neuroscience. 2017;11:84. doi: 10.3389/fncel.2017.00084. 28. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Wan J, Huang M, Zhao H, Wang C, Zhao X, Jiang X, Bian S, He Y, Gao Y. A novel tetranucleotide repeat polymorphism within KCNQ1OT1 confers risk for hepatocellular carcinoma. DNA Cell Biol. 2013;32:628–634. doi: 10.1089/dna.2013.2118. [DOI] [PubMed] [Google Scholar]
- 31.Chang KC, Petrash JM. Aldose reductase mediates transforming growth Factor β2 (TGF-β2)-induced migration and epithelial-to-mesenchymal transition of lens-derived epithelial cells. Invest Ophthalmol Vis Sci. 2015;56:4198–4210. doi: 10.1167/iovs.15-16557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shu DY, Wojciechowski MC, Lovicu FJ. Bone morphogenetic protein-7 suppresses TGFβ2-induced epithelial-mesenchymal transition in the lens: Implications for cataract prevention. Invest Ophthalmol Vis Sci. 2017;58:781–796. doi: 10.1167/iovs.16-20611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J, Qin Y, Zhao FK, Wu D, He XF, Liu J, Zhao JY, Zhang JS. Anterior segment dysgenesis correlation with epithelial-mesenchymal transition in Smad4 knockout mice. Int J Ophthalmol. 2016;9:943–947. doi: 10.18240/ijo.2016.07.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.